Abstract
The formation of the potent neurotoxic methylmercury (MeHg) is a microbially mediated process that has raised much concern because MeHg poses threats to wildlife and human health. Since boreal forest soils can be a source of MeHg in aquatic networks, it is crucial to understand the biogeochemical processes involved in the formation of this pollutant. High-throughput sequencing of 16S rRNA and the mercury methyltransferase, hgcA, combined with geochemical characterisation of soils, were used to determine the microbial populations contributing to MeHg formation in forest soils across Sweden. The hgcA sequences obtained were distributed among diverse clades, including Proteobacteria, Firmicutes, and Methanomicrobia, with Deltaproteobacteria, particularly Geobacteraceae, dominating the libraries across all soils examined. Our results also suggest that MeHg formation is also linked to the composition of non-mercury methylating bacterial communities, likely providing growth substrate (e.g. acetate) for the hgcA-carrying microorganisms responsible for the actual methylation process. While previous research focused on mercury methylating microbial communities of wetlands, this study provides some first insights into the diversity of mercury methylating microorganisms in boreal forest soils.
Introduction
Mercury (Hg) is a potent toxin that might cause severe negative effects on wildlife and human health1. The toxicity of Hg is of such concern that 128 countries have signed the Minamata Convention, a global treaty that entered into force in August 2017 with the explicit objective to reduce Hg emissions and protect human health and the environment. High Hg emissions in the past have led to high present-day Hg levels in different parts of the atmosphere, oceans and terrestrial ecosystems2,3. Fish consumption is the main pathway for human Hg exposure4. High Hg levels in freshwater fish, not seldom at concentrations unsafe for human consumption, have in recent decades raised much concern in many boreal regions5–7. In these freshwater boreal catchments, the link between amount of atmospheric Hg deposited and the Hg accumulated in food-webs is complex8. Attention has therefore been directed to understand processes within the catchment soils that may contribute to the formation and mobilisation of readily bioavailable methylmercury (MeHg) in runoff water. Local biogeochemistry in catchment soils may for example influence the magnitude and timing of the response in fish Hg concentrations following decreased deposition loadings. One example of this is seen in a 40-year monitoring dataset from Canada showing flat or increasing trends of Hg in freshwater fish up to 2012 although Hg deposition has decreased during recent decades7.
Because Hg has a strong affinity for reduced sulphur or thiol (RSH) functional groups of soil organic matter (OM)9,10, the increased atmospheric deposition of Hg during the industrialisation period has resulted in high Hg concentrations in organic-rich soils11. As a consequence, the OM-rich soils, characteristic of the boreal biome, have retained Hg deposition from both natural and anthropogenic emissions, and now represent an important global Hg stock9,12. For example, recent atmospheric deposition has increased the stock of Hg in the organic-rich upper layer of Swedish forest soils by a factor of three to four11,13. This is of special concern because soil OM has been identified as an important vector of Hg and methylmercury (MeHg) transport from catchments to surface waters in boreal areas14,15. Indeed, the mobilisation of inorganic Hg (Hg(II)) and, the more harmful, MeHg from soils by means of OM-mediated transport has been linked to MeHg accumulation in lake sediments within catchments15,16 and in fish17. The role of forest soils is thus evident from the increase in MeHg export from forests to aquatic ecosystems16,18,19, and the subsequent bioaccumulation in downstream fish17,20. Since forest soils are an important site for MeHg formation21, it is crucial to understand the processes and the organisms involved in MeHg formation in boreal soils.
The methylation of Hg(II) to MeHg is biologically mediated22 and takes place under oxygen deficient conditions such as those of flooded soils, sediments15, anoxic water columns23 and suspended particles of aquatic systems24,25. Typical forest environments with high MeHg formation are wetlands26, organic-rich riparian soils27, and soils that have been recently become water logged soils after forest harvest21. Specific strains of sulphate-reducing bacteria28,29, iron reducing bacteria (FeRB)30,31, methanogens32 and Firmicutes33 have the capability to methylate Hg(II). However, a number of factors controlling microbial activity and/or the geochemical speciation of inorganic Hg(II) will govern MeHg formation in the environment21,34–36. For example, increases in temperature might lead to increases in biological activity and subsequently also higher Hg(II) methylation rates37. Redox potential also seems to be a key factor as suboxic and mildly reducing conditions seem to promote high Hg(II) methylation rates, whereas anoxic and strongly reducing conditions might lead to elevated sulphide concentrations that eventually prevent Hg(II) from being available for methylation37. Sulphur plays a major role in influencing Hg(II) methylation by directly affecting the activity of some methylating bacteria (e.g. sulphate reducing bacteria, SRB) and/or control the availability of Hg(II) for methylation10. Specific organic matter (OM) compounds can promote Hg(II) methylation by enhancing bacterial activity15, but also by defining Hg(II) speciation38 and Hg(II) availability39,40. OM can also facilitate Hg(II) methylation by inhibiting mercury sulphide (HgS(s)) precipitation or enhance HgS(s) dissolution thereby providing available Hg(II) for methylating microorganisms41. High OM concentrations might also decrease Hg methylation by formation of high mass molecular mass complexes that hamper Hg(II) availability39. Recently it has been concluded that the availability of Hg(II) depends heavily on the S(−II) concentration in porewater and the RSH(aq)/RSH(ads) molar ratio of dissolved OM38. Besides all the geochemical factors that might directly or indirectly affect Hg(II) availability and methylation, a recent study suggest that the composition of the combined bacterial community may also influence the structure of Hg(II) methylating communities42. Together these studies, mainly performed in aquatic systems, highlight the importance of geochemical conditions for determining the availability of Hg(II) and the activity and composition of the microbial communities involved, directly or indirectly, in MeHg formation.
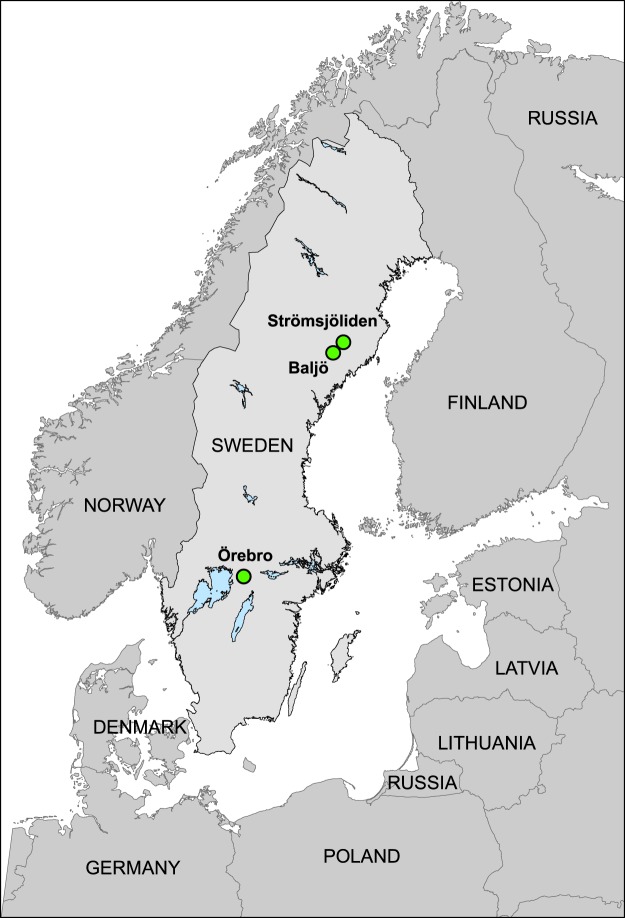
The identification of two functional genes, hgcA and hgcB, which play essential roles in Hg(II) methylation22, provided the means to more directly characterise the complexity of microbial communities involved in the formation of MeHg in natural ecosystems. This approach has been applied to marshes, sediments and swamps in several geographic regions42–46; rice paddies in China47, and water conservation areas of the northern Everglades, USA48. However, very little work to date has been conducted to reveal the distribution of microbial groups responsible for Hg(II) methylation in forest soils within the vast boreal biome. To the best of our knowledge, no studies have directly described the composition and the spatial variation in Hg(II) methylating microbial communities in such forests. Therefore, the primary goal of this paper was to describe Hg(II) methylating microbial communities in various boreal forest soils and identify characteristics important for shaping these communities. High-throughput next generation sequencing of amplified 16S rRNA and hgcA genes combined with molecular barcoding and detailed soil geochemical characterisations were performed to study the Hg(II) methylating microbial communities in 200 soil samples from three different boreal forest regions (Fig. 1) in order to shed light on the biogeography of microorganisms responsible for MeHg formation in the boreal landscape.
Figure 1.
The location of the three field sites used in this study. Örebro (O) in the south of Sweden includes three catchments and Balsjö (B) and Strömsjöliden (S) in the north of Sweden includes three and two catchments, respectively.
Results
Bacterial community composition in boreal forest soils
We collected 200 boreal forest soil samples distributed across eight catchments in Sweden in 2012 (Tables S1 and S2). A total of 3 321 197 high quality 16S rRNA sequences remained after quality control and chimera removal (7–72 911 reads per sample). The sample with only 7 reads was removed, and we then rarefied the rest of the data to the remaining sample with the fewest reads (1692 reads). The final rarefied sequence dataset (329 940 reads) clustered into 33 158 operational taxonomic units (OTUs) using a similarity threshold of 97%. In the rarefied dataset, 35 taxa at phyla level, 69 taxa at class level, 119 taxa at order level, and 187 taxa at family level were detected from all the soil samples across three regions. The overall coverage of the forest bacterial community is reflected in the combined richness detected for random subsets of analysed samples. The logarithmic shape indicated that most of the considerable OTU richness occurring in the forest soils was accounted for in the combined dataset (Fig. S1). Among the dominant phyla across all regions (>5% relative abundance), Acidobacteria was the most abundant, followed by Proteobacteria, Planctomycetes, Bacteroidetes, Parcubacteria and Verrucomicrobia (Table 1). Combined, these phyla accounted for 77.5% of the total sequences (Table 1). Most of the previously identified clades known to contain Hg(II) methylators33,49 were detected in the present study, including Deltaproteobacteria (3.31% of the total reads), Chloroflexi (2.60% of the total reads), Firmicutes (0.77% of the total reads) and Euryarchaeota (0.66% of the total reads) (Table 1). Microbial community composition based on 16S rRNA sequences in the 34 studied MeHg hotspots (%MeHg >1%) showed a similar pattern in terms of the dominant phyla (>5% relative abundance), with Acidobacteria and Proteobacteria being the most abundant ones. However, Bacteroidetes and Chloroflexi contributed much more to the total communities at these hotspots compared to the combined dataset across all 200 samples (Table 1).
Table 1.
Comparison of the relative abundances (%) of the most abundant taxa (>2.5% of reads at phylum level) in all the samples (n = 200) with the MeHg hotspots (n = 34) based on 16S rRNA sequences.
| Most abundant taxa | Mean ± SD | Maximum | Minimum | |||
|---|---|---|---|---|---|---|
| All samples | Hotspots | All samples | Hotspots | All samples | Hotspots | |
| Acidobacteria | 36.11 ± 10.53 | 25.57 ± 8.77 | 73.64 | 49.29 | 8.10 | 9.40 |
| Proteobacteria | 13.99 ± 4.03 | 16.56 ± 2.96 | 28.13 | 27.60 | 2.90 | 8.87 |
| Alphaproteobacteria | 6.83 ± 3.01 | 7.13 ± 2.81 | 16.43 | 13.95 | 1.77 | 2.66 |
| Deltaproteobacteria | 3.31 ± 1.69 | 3.56 ± 1.38 | 13.36 | 7.15 | 0.71 | 1.30 |
| Gammaproteobacteria | 2.06 ± 1.33 | 1.48 ± 0.76 | 7.15 | 3.66 | 0.24 | 0.35 |
| Betaproteobacteria | 1.78 ± 2.13 | 4.14 ± 2.47 | 11.11 | 10.46 | 0.00 | 0.65 |
| Epsilonproteobacteria | 0.01 ± 0.03 | 0.03 ± 0.06 | 0.30 | 0.24 | 0.00 | 0.00 |
| Planctomycetes | 8.18 ± 4.21 | 5.82 ± 2.77 | 24.82 | 11.64 | 1.36 | 1.95 |
| Bacteroidetes | 6.61 ± 5.24 | 11.38 ± 7.92 | 51.60 | 51.60 | 0.41 | 1.60 |
| Parcubacteria | 6.35 ± 4.19 | 9.01 ± 5.14 | 26.36 | 24.47 | 0.06 | 2.13 |
| Verrucomicrobia | 6.28 ± 2.78 | 5.30 ± 2.31 | 14.89 | 10.64 | 0.65 | 0.65 |
| Thaumarchaeota | 3.96 ± 2.77 | 2.53 ± 2.44 | 18.44 | 14.83 | 0.00 | 0.00 |
| Actinobacteria | 3.11 ± 2.38 | 2.94 ± 1.62 | 19.86 | 6.03 | 0.47 | 0.89 |
| Chlamydiae | 2.83 ± 2.56 | 1.31 ± 1.08 | 22.87 | 3.71 | 0.24 | 0.30 |
| Chloroflexi | 2.60 ± 3.18 | 7.16 ± 5.18 | 17.79 | 15.19 | 0.00 | 0.12 |
| Others | 9.97 ± 0.89 | 12.41 ± 1.66 | 17.14 | 8.98 | 0.00 | 0.00 |
Relative abundances of classes under phylum Proteobacteria are listed with indent (SD: Standard deviation).
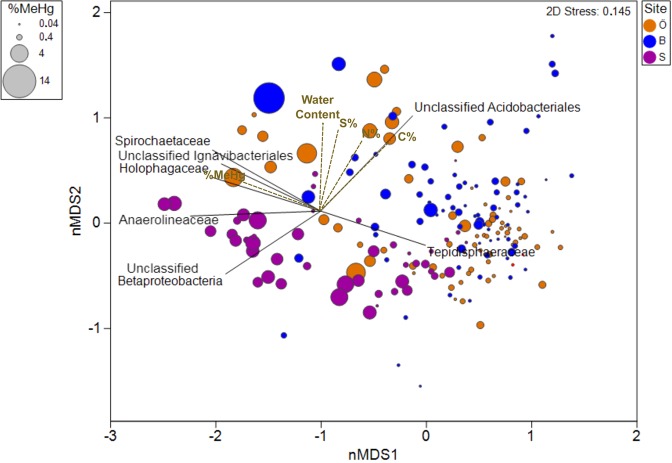
A non-metric multidimensional scaling (nMDS) plot based on 16S sequences was used to visualise the composition of the bacterial community among samples. Unclassified Acidobacteriales, Unclassified Ignavibacteriales, Spirochaetaceae, Holophagaceae, Anaerolineaceae, Betaproteobacteria and Tepisiphaeraceae were important contributing families for shaping the differences in bacterial community composition among samples (Fig. 2). Geochemical factors that were correlated (correlation coefficients > 0.5) with the bacterial composition were projected on top with longer vectors implying stronger correlations (Fig. 2). %MeHg, reflected by bubble sizes, presented a strong coupling to the bacterial community composition, which was further confirmed by %MeHg presenting a long vector among all the geochemical factors (Fig. 2). Water content, C%, S% and N% were all found to be the factors that affected the composition of soil bacterial community (Fig. 2), indicating that a supply of organic matter and nutrients in the moist soil shapes the bacterial community. This is in agreement with previous research that pointed out the contribution of nutrients and organic matter to bacterial activities and Hg(II) methylation15,37. Also, S was well correlated with both C and N (Table S3), suggesting that most of the measured sulphur in the sampled soils has likely an organic origin. This has been found as a common feature in boreal soils27,36,50.
Figure 2.
Non-metric multidimensional scaling (nMDS) of microbial community composition of all samples (family level based on 16S rRNA) overlaid with families (black line) and geochemical factors (dotted brown line) moderately correlated with biotic ordination (correlation coefficients > 0.5) (%MeHg: MeHg/THg). Relative dissimilarities (or distances) among the samples were computed according to the resemblance matrix calculated on fourth rooted family reads. The different sites Örebro (O); Balsjö (B) and Strömsjöliden (S) are color-coded.
Unclassified Fibrobacterales, Methanosaetaceae, unclassified Ignavibacteriales, Spirochaetaceae, Holophagaceae and Anaerolineaceae exhibited the highest correlations with %MeHg (Table 2). Syntrophobacteraceae, Methanosarcinaceae, Methanoregulaceae, Desulfobulbaceae, Syntrophaceae, Desulfobacteraceae and Dehalococcoidaceae, were also found relevant to the bacterial community composition in high-%MeHg sites (Table 2).
Table 2.
Moderate (0.5 ≤ R < 0.7) to weak (0.3 ≤ R < 0.5) Pearson correlations between families and %MeHg in all samples based on 16 S rRNA.
| Families | Correlations with %MeHg |
|---|---|
| Unclassified Fibrobacterales | 0.56 |
| Methanothrix | 0.54 |
| Unclassified Ignavibacteriales | 0.52 |
| Spirochaetaceae | 0.52 |
| Holophagaceae | 0.50 |
| Anaerolineaceae | 0.41 |
| Lentimicrobiaceae | 0.40 |
| Syntrophobacteraceae | 0.39 |
| Unclassified Phycisphaerales | 0.37 |
| Methanosarcinaceae | 0.37 |
| Methanoregulaceae | 0.35 |
| Desulfobulbaceae | 0.35 |
| Porphyromonadaceae | 0.35 |
| Rhodobiaceae | 0.33 |
| Unclassified Clostridiales | 0.32 |
| Gemmatimonadaceae | 0.30 |
| Syntrophaceae | 0.30 |
| Unclassified Omnitrophica | 0.30 |
| Nitrosomonadaceae | 0.30 |
| Desulfobacteraceae | 0.30 |
| Dehalococcoidaceae | 0.30 |
| Unclassified Obscuribacterales | −0.30 |
| Unclassified Solibacterales | −0.33 |
| Tepidisphaeraceae | −0.38 |
Families potentially involved in Hg methylation were marked in bold.
Distribution of Hg(II) methylators
Among the screened 200 soils samples, we selected those with high MeHg concentrations and %MeHg (>1%), and defined them as “MeHg hotspots” (see “MeHg hotspots” soils geochemistry descriptors in Table S4, n = 34). In 34 of these MeHg hotspots, the relative abundance of microbial families carrying representatives known to methylate Hg(II) was assessed based on hgcA sequences33,49. A total of 1 257 577 hgcA sequences remained after quality control and chimera removal (11 404–55 461 reads per sample). The hgcA dataset was rarefied to the remaining sample with the fewest reads (11 404 reads). The rarefied sequence dataset accounted a total of 387 736 reads that clustered into 573 operational taxonomic units (OTUs) using a similarity threshold of 97%. As for the 16 rRNA, the logarithmic shape indicated that most of the considerable species richness of Hg(II) methylators occurring in the forest soils was accounted for in the combined dataset (Fig. S1). Representative sequences from 22 families were found in the 34 analysed MeHg hotspots. Of all the hgcA sequences, 3.13% were not taxonomically assigned (unclassified), 0.28% were unclassified Euryarchaeota, and 7.28% could not be assigned beyond the rank of Bacteria (Unclassified Bacteria).
The majority of the sequences annotated to the level of family clustered with Deltaproteobacteria, making up 85.4% of all the hgcA reads (Table 3). The remaining classified hgcA sequences were distributed across diverse families affiliated to Firmicutes and Methanomicrobia. Unclassified Deltaproteobacteria represented up to 56% of the reads and among the identified families, Geobacteraceae were the most abundant, contributing up to 40% in Strömsjöliden. Ruminococcaceae (3.21% of all hgcA reads) occurred as another important family in the hotspots in Örebro; while methanogens and syntrophic lineages were less abundant in the hotspots based on hgcA sequences (Table 3).
Table 3.
Relative abundance of families involved in Hg(II) methylation based on hgcA sequences in 34 hotspots.
| Families | Örebro | Balsjö | Strömsjöliden |
|---|---|---|---|
| % of hgcA reads | % of hgcA reads | % of hgcA reads | |
| Unclassified Deltaproteobacteria | 43.24 ± 37.11 | 44.85 ± 30.09 | 55.69 ± 18.23 |
| Geobacteraceae | 26.79 ± 31.09 | 24.62 ± 22.22 | 39.40 ± 18.96 |
| Unclassified Bacteria | 10.72 ± 17.45 | 25.58 ± 33.67 | 1.43 ± 1.02 |
| Ruminococcaceae | 9.12 ± 18.23 | 1.52 ± 2.30 | 0.15 ± 0.04 |
| Unclassified | 6.62 ± 8.65 | 2.37 ± 3.86 | 1.27 ± 2.98 |
| Unclassified Euryarchaeota | 0.84 ± 2.22 | 0.02 ± 0.02 | 0.01 ± 0.02 |
| Desulfovibrionaceae | 0.83 ± 1.28 | 0.16 ± 0.03 | 0.02 ± 0.04 |
| Unclassified Methanomicrobiales | 0.49 ± 1.21 | 0.06 ± 0.09 | 0.03 ± 0.12 |
| Syntrophaceae | 0.35 ± 0.45 | 0.05 ± 0.00 | 0.00 ± 0.00 |
| Methanomassiliicoccaceae | 0.31 ± 0.53 | 0.02 ± 0.00 | 0.13 ± 0.05 |
| Methanoregulaceae | 0.20 ± 0.03 | 0.06 ± 0.03 | 0.00 ± 0.01 |
| Syntrophomonadaceae | 0.17 ± 0.13 | 0.02 ± 0.03 | 0.13 ± 0.04 |
| Unclassified Desulfovibrionales | 0.14 ± 0.15 | 0.02 ± 0.05 | 0.03 ± 0.04 |
| Unclassified Clostridiales | 0.06 ± 0.22 | 0.51 ± 0.19 | 0.08 ± 0.07 |
| Unclassified Firmicutes | 0.06 ± 0.02 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Unclassified Desulfuromonadales | 0.03 ± 0.00 | 0.10 ± 0.03 | 0.39 ± 0.32 |
| Desulfobulbaceae | 0.02 ± 0.02 | 0.01 ± 0.00 | 0.00 ± 0.01 |
| Desulfuromonadaceae | 0.01 ± 0.01 | 0.00 ± 0.04 | 0.00 ± 0.04 |
| Syntrophorhabdaceae | 0.01 ± 0.00 | 0.00 ± 0.02 | 0.00 ± 0.06 |
| Unclassified Deferrisoma | 0.01 ± 0.00 | 0.00 ± 0.00 | 1.18 ± 0.98 |
| Desulfarculaceae | 0.00 ± 0.02 | 0.00 ± 0.00 | 0.00 ± 0.02 |
| Pelobacteraceae | 0.00 ± 0.01 | 0.01 ± 0.01 | 0.07 ± 0.03 |
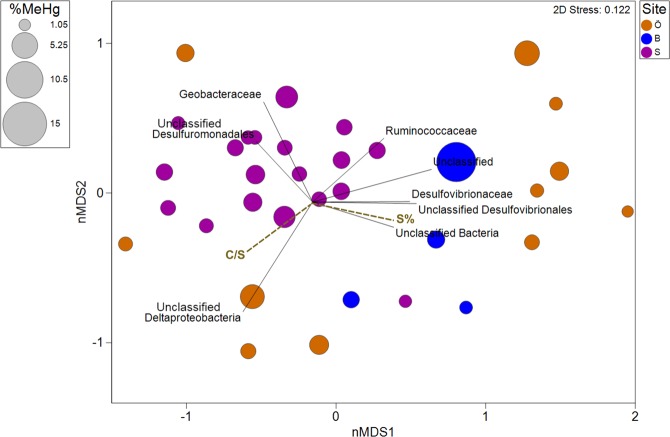
Unclassified Desulfuromonadales, Geobacteraceae, Ruminococcaceae, unclassified Desulfovibrionales, Desulfovibrionaceae, and unclassified Deltaproteobacteria seemed to contribute to differences in the composition of Hg(II) methylators in the studied soils (Fig. 3a). Among the measured geochemical parameters, the S% and the C/S seemed to have an impact on shaping the community composition of Hg(II) methylators (Fig. 3b). Moreover, Methanoregulaceae, Desulfovibrionaceae, Desulfuromonadaceae, Desulfarculaceae and Methanomassiliicoccaceae correlated positively with S% and negatively with C/S (Table S5). In the studied MeHg hotspots, S was strongly correlated with both C and N (Table S6), suggesting most of the measured sulphur in the hotspots is also likely presented in organic forms.
Figure 3.
Non-metric multidimensional scaling (nMDS) of potential Hg methylators (family level based on hgcA) in 34 hotspots overlaid with geochemical factors that were moderately correlated with the biotic ordination positions (correlation coefficients > 0.5). The different sites Örebro (O); Balsjö (B) and Strömsjöliden (S) are color-coded.
Phylogenetic analysis of hgcA genes
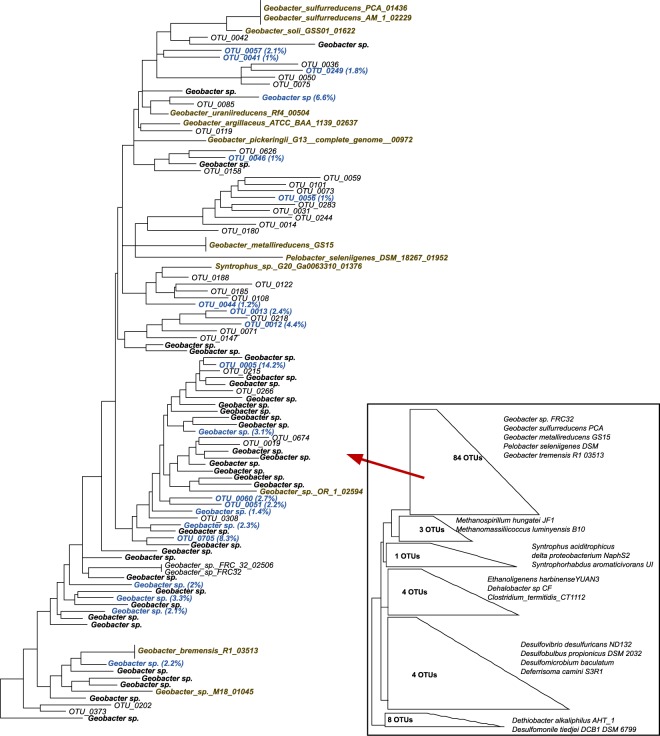
All the Proteobacteria families belonged to Deltaproteobacteria, a class with which most currently confirmed Hg(II)-methylating bacteria are affiliated51,52. When combined, the 20 most abundant OTUs accounted for 72% of the total reads. Noteworthy, phylogenetic analysis revealed that the most abundant Hg(II)-methylating OTUs (“OTU_0005”, “OTU_0705”, “OTU_0008”, and “OTU_0012”) in the studied forest soils were either taxonomically assigned as Geobacter sp. or phylogenetically related to Geobacter species (Fig. 4). Among the 20 most abundant OTUs, 17 were taxonomically annotated as Deltaproteobacteria. Among these 17 OTUs, 9 were taxonomically annotated as Geobacter and 8 were phylogenetically related to Geobacter species (Fig. 4). Summing the 9 OTUs taxonomically annotated as Geobacter with the 8 OTUs phylogenetically related to Geobacter species, the resulting 17 OTUs accounted for 62% of the total hgcA reads (Fig. 4). While the 5th most abundant OTU was taxonomically denoted as Firmicutes (Ethanoligenens), the 6th and 7th most abundant OTUs could not be annotated beyond the bacterial domain.
Figure 4.
Phylogenetic relationships of Deltaproteobacterial hgcA sequences in the studied forest soils. The 20 most abundant Deltaproteobacteria are in blue. The OTUs taxonomically assigned as Geobacter are indicated in the plot “Geobacter sp.”. OTUs non-taxonomically assigned are presented as “OTU”. Reference genomes are marked in brown. The tree was generated using RAxML (version 8.2.4) with the PROTGAMMLG model and the autoMR to choose the number of necessary bootstraps (750). Please see details of the collapsed tree in the Fig. S2.
Discussion
Community composition of Hg(II) methylators in boreal forest soils
Among the diverse microbial communities seen in the soil samples (Table 1), most of the previously identified Hg(II) methylating groups, e.g., Deltaproteobacteria, Chloroflexi, Firmicutes and Euryarchaeota could be detected (Table 3). Deltaproteobacteria have been considered a predominant Hg(II) methylating class in anaerobic soils44,47,48. In the present study, Deltaproteobacteria were also the predominant Hg(II) methylators at the hotspots with Geobacteraceae as the most represented family. When considering the OTUs that were taxonomically annotated, this family alone contributed over 30% of all hgcA reads. However, if we also account for the OTUs that were phylogenetically related to Geobacteraceae, this family might have contributed to more than 60% of the hgcA reads. The importance of Geobacteraceae could be seen at all the sampled sites and particularly in Strömsjöliden (Table 3). Iron reducing bacteria (FeRB) have previously been shown to be important for Hg(II) methylation in some environments30,31,46,51, and most Geobacter tested so far are particularly efficient at MeHg formation in the laboratory31. This suggests that the ability to methylate Hg(II), a typical feature among the Geobacteraceae, is present in the studied soils and widely distributed in terrestrial and aquatic ecosystems. However, while previous studies have quantified the contribution of SRB (i.e molybdate inhibitor) and methanogens (i.e bromoethanesulfonate inhibitor) to MeHg formation30,36,42,53,54, the lack of a specific inhibitor for iron-reduction have impeded the quantification of the contribution of FeRB to MeHg formation in environmental samples. The discovery of the hgcA pushed the state of the art and made possible to identify Hg(II) methylators in environment22,33. Our results combined with previous findings in wetlands and paddy soils44,47,48 highlight the importance of Geobacteraceae as Hg(II) methylators in boreal forest soils and potentially in a wide range of environments.
While SRB are considered to be the principal Hg(II) methylators in aquatic systems55–59, not much information is available on Hg(II) methylators in soils. However, identified SRB in the hotspots only accounted for a minor portion of Hg(II) methylators (Table 3). However, it is nevertheless plausible that at least some of the hgcA sequences annotated as unclassified Deltaproteobacteria (Table 3) could be unknown Hg(II) methylating SRB or even Hg(II) methylating sulphate-reducing syntrophs, capable of syntrophic fermentation of simple organic acids in the absence of sulphate as the terminal electron acceptor60,61. Therefore, we cannot discard the possibility that also SRB contribute significantly to Hg(II) methylation in the studied systems. A previous study based on selective inhibitors and rate measurements indeed suggested SRB played an important role in MeHg formation in boreal forest soils36. Additionally it has been demonstrated that even when SRB belong to the ‘rare biosphere’ of peatlands, they contribute significantly to respiration processes62.
Ruminococcaceae belongs to another newly confirmed representative of Hg(II) methylators, the Firmicutes33. Firmicutes contributed to Hg(II) methylating microbial communities at the water conservation areas of the Florida Everglades48 but were not detected in boreal wetlands44. In the present study, Ruminococcaceae were prominent contributors to the hgcA pool in hotspots from Örebro and in all soils from Strömsjöliden (Table 3). They could thus play a role in shaping the composition of Hg(II) methylating community as further indicated by the negative correlation though weak between Ruminococcaceae and C/S, a primary geochemical factor shaping Hg(II) methylating communities in the hotspots (Table S5 and Fig. 3b). Not much research has been devoted to the possible relationship between organic S and Hg(II) methylating Ruminococcaceae. Considering the abundance of this group in forest soils, further efforts are needed to shed light on the metabolic or physiological pathways of Hg(II) methylating Ruminococcaceae.
Methanogens were early on suspected to be responsible for Hg(II) methylation63, but not until recently were they verified as a significant source of Hg(II) methylators in various environments32,44. In the hotspots in the studied soils, they were also detected, though they were not very abundant in the Hg(II) methylating microbial community. Chloroflexi has recently been identified as potential Hg(II) methylators in the water conservation areas, paddy soils and wetlands44,48,64. The hgcA data did not confirm any significant role of this group in MeHg production in boreal forest soils (Table 3), even though 16S rRNA data revealed non-Hg(II) methylating Chloroflexi (e.g. the class Anaerolineae) in soils from all three regions (Table 1).
Previous studies have mainly explored flooded environments such as paddy soils47, boreal wetlands44 and the water areas of the Florida Everglades48. Hence our study provided important new information on the composition and diversity of Hg(II) methylating microbial communities in non flooded boreal forest soils and the boreal landscape, and in doing so identified Geobacteraceae as significant Hg(II) methylators in the terrestrial biome. The diversity of Hg(II) methylators described in this study need to be interpreted cautiously, as bias is inherent in methods employing PCR amplification of any variable target gene. The hgcA gene was only recently discovered and the optimization of the appropriate methods and, in particular the design of primers for the hgcA amplification, is still ongoing65. Additionally, DNA based methods only reveal the presence of organisms, while alternative approaches based on transcription data, proteomes or rate measurements are needed for verifying their activity. Our data nevertheless provide new insights about Hg(II) methylating microbial communities in boreal forest soils and can as such guide and serve as a resource for future research efforts in this field.
Interplay between bacterial communities and Hg(II) methylators
%MeHg has previously been used as a proxy for methylation efficiency66,67, and high %MeHg has also in a few cases been shown to correlate positively with the abundance of Hg(II) methylators21,68. In the current study, sites with high %MeHg featured bacterial communities different from those observed at sites with low % MeHg (Fig. 2). Although, families known to contain Hg(II) methylators (Syntrophobacteraceae, Methanosarcinaceae, Methanoregulaceae, Desulfobulbaceae, Syntrophaceae, Desulfobacteraceae and Dehalococcoidaceae; 25) were found at sites with high %MeHg, there were also positive correlations between %MeHg and families that are not known to host Hg(II) methylators, such us unclassified Fibrobacterales, Methanothrix (formerly Methanosaeta), unclassified Ignavibacteriales, Spirochaetaceae, Holophagaceae and Anaerolineaceae (Table 2). This suggests that not only the Hg(II) methylators themselves, but also the supporting and interacting bacterial communities residing in the soil environment may influence MeHg formation across the studied regions. Anaerolineaceae, Spirochaetaceae and Holophagaceae are for example known to generate acetate by fermentation processes69. Fibrobacterales, have recently been suggested to have an important role in cellulose hydrolysis in anaerobic environments, including soils70. The Ignavibacteria class was recently described (Iino et al., 2010) and the physiology and metabolic capacities of this group is still poorly known, even if a distinctive feature of this group is the ability to grow on cellulose and its derivatives with the utilization of Fe(III) oxide as electron acceptor71. It may well be that these families, which correlated well with %MeHg (Table 2) and seem to be involved in the degradation of long chain OM compounds72,73, promoted MeHg production by providing appropriate substrates (e.g. acetate) for the Hg(II) methylators. Hg(II) methylators and non-Hg(II) methylating members of Desulfobulbaceae, known to oxidise organic substrates incompletely to acetate74, might also have provided the necessary substrate to Hg(II) methylators (Table 2). Based on our results, we propose an important role of also the non-Hg(II) methylating bacterial heterotrophs in sustaining the activity of the Hg(II) methylating microorganisms and thereby influencing MeHg formation in boreal forest soils. Moreover, the correlation between Methanothrix and %MeHg deserves special attention. It has been shown that Methanothrix can establish syntrophic cooperation with Anaerolineaceae72 or Geobacteraceae75 in methanogenic degradation of long chain carbon compounds (alkanes). As our results show that Geobacteraceae are major contributors to the Hg(II) methylating microbial community (Table 3), the high correlation found between Methanothrix and %MeHg could be the result of the interaction between the non-Hg(II) methylating Methanothrix and the Hg(II) methylating Geobacteraceae. In brief, we provide novel system-level information on putative trophic interactions between non-Hg(II) methylating and the Hg(II) methylating taxa. We further suggest that more in depth studies with metagenome-level sequencing and metabolic pathway reconstruction will be a logical next step to gain a more complete understanding of how Hg(II) methylating bacterial and archaeal species interact in soils.
Conclusions
A newly developed strategy that combine high-throughput hgcA amplicon sequencing with molecular barcoding revealed diverse clades of Hg(II) methylators in forest soils. This study confirms a predominant role of Deltaproteobacteria, and in particular Geobacteraceae, as key Hg(II) methylators in boreal forest soils. Firmicutes, and in particular Ruminococcaceae, were also abundant members of the Hg(II) methylating microbial community. Besides the identified Hg(II) methylators, we suggest that the non-Hg(II)-methylating bacterial community (e.g. Anaerolineaceae, Holophagaceae and Spirochaetaceae) might have contributed to the net MeHg formation (%MeHg) by processing OM and thereby providing low molecular mass OM compounds as a substrate to Hg(II) methylators (e.g acetate). By revealing linkages between Hg(II) methylators and non- Hg(II) methylators, our results call for further community-level work on the metabolic interactions in soil microbial communities to understand Hg(II) methylation. Such studies would need to go beyond the Hg(II) methylating microbial populations. Our findings provide a better understanding of Hg(II) methylating microbial communities in forest soils and the boreal landscape.
Materials and Methods
Site description
Soil samples were collected from 200 sites in October 2012 and were distributed across eight catchments in three boreal forest regions in Sweden (Table S1 and S2). Within each of the catchments, 25 samples were collected. The most southern region Örebro (59°10′16.39″N 14°34′3.01″E) includes three catchments and the sampled soils are dominantly Podzol with Histosols76 in the lower parts of the catchments along the streams. The organic matter (O) horizons were most often thicker than 20 cm. More detailed information is given in Eklöf et al.18. Two northern regions, Balsjö (64°1′37″N 18°55′43″E) and Strömsjöliden (64°6′48˝N 19°7′36″E), are located 600–700 km north of Örebro and around 14 km apart from each other. Balsjö includes three catchments dominated by orthic Podzol, with Histosols along the streams. The O horizons were most often thicker than 10–20 cm in the lower parts and less than 10 cm higher up in the catchments. More details are given in Löfgren et al. (2009). Strömsjöliden includes two catchments and the soils are dominated by fine-grained moraine. The organic layers are most often less than a few centimetres deep. The samples with high soil MeHg concentrations and %MeHg > 1% were defined as “MeHg hotspots” (n = 34), see a summary of the soil characteristics of “MeHg hotspots” in Table S4.
The daily mean air temperatures during the 9 sampling days in September in 2012 varied between 7 and 12 °C in Örebro catchments and 4 and 11 °C in Balsjö and Strömsjöliden catchments. There were no major rain events during the sampling period and the temperature and precipitation was normal for the time of the year.
Soil sampling
Soil samples were collected with a soil coring tube (Ø = 23 mm). In each catchment, around half of the samples (n = 12) were collected systematically along the topographic fall line of the hill slope, at set distances from the stream draining the area. These samples were collected from the upper 6 cm of the O horizons or the whole O horizons if these were less than 6 cm deep. The locations of the remaining sampling sites (n = 13) were chosen by actively looking for potential hot spots for MeHg formation, such as wet patches, driving tracks and stump holes. These targeted samples were also collected from various depths, e.g. depths where groundwater levels were most frequently fluctuating were of special interest for potential Hg(II) methylation.
Single-use plastic gloves were used and soil samples for chemical analyses were collected in plastic bags or acid washed Falcon tubes and stored on ice in a cooler during transport to the laboratory (within 8 hours). Soil samples for molecular analyses were collected following adequate aseptic sampling protocols. All sampling equipment was sterilized by washing in 70% ethanol in between samples. Samples were collected in sterilized plastic tubes and frozen in liquid nitrogen directly in the field, and then stored at −80 °C until further processing and analyses.
Chemical analyses
Soil samples were analysed for total Hg (THg), MeHg, water content, and mass percentage of carbon (C), nitrogen (N) and sulphur (S). Samples were freeze-dried and ground by hand in a mortar prior to analyses for THg, C%, N% and S%. Wet and dry weights were measured to estimate the water content. Total Hg was measured using a Perkin Elmer SMS100 total Hg analyser in accordance with US EPA method 7473. The method includes a thermal decomposition step, followed by amalgamation and atomic absorption spectrophotometric detection (working range 0.05–600 ng). Reproducibility and accuracy of measurements were checked by analyses of replicate samples and reference standards. Analyses of MeHg were done by using GC-ICPMS77 on fresh samples immediately after thawing. C, N and S were analysed on dry soils packed tightly in tin capsules (Elemental Microanalysis, 6.4 mm) and subsequently measured by high temperature catalytic oxidation with a COTECH ECS 4010 elemental analyser calibrated with sulfanilamide standard (C 41.84%, N 16.27%, H 4.68%, O 18.58%, S 18.62%). Analytical precision was <±0.3% for C, ±1.5% for N and ±3.5% for S.
Microbiological analyses
16S rRNA gene
Microbial DNA was extracted from soil samples using the Power soil DNA isolation Kit (MoBio Laboratories Inc, CA, USA) and the quality of the extracted DNA was assessed by gel electrophoresis (1% agarose). Bacterial 16 S rRNA genes were amplified in two steps polymerase chain reaction (PCR) according to the protocol in Sinclair et al. (2015). Briefly, non-barcoded primers Bakt_341F and Bakt_805R (Table S7) were used for the 1st PCR step of 20 cycles. The resulting PCR products were diluted 100 times before being used as template in a 2nd PCR step of 10 cycles with similar primers carrying sample-specific 7-base DNA barcodes. All PCRs were conducted in 20 μL volume using 1.0 U Q5 high fidelity DNA polymerase (NEB, UK), 0.25 μM primers, 0.2 mM dNTP mix, and 0.4 μg bovine serum albumin. The thermal program consisted of an initial 95 °C denaturation step for 5 min, a cycling program of 95 °C for 40 seconds, 53 °C for 40 seconds, 72 °C for 60 seconds and a final elongation step at 72 °C for 7 minutes. Amplicons from the 2nd PCR were purified using the Qiagen gel purification kit (Qiagen, Germany) and quantified using a fluorescence-based DNA quantitation kit (PicoGreen, Invitrogen). The final amplicons after two PCR steps were pooled in equal proportions to obtain a similar number of sequencing reads per sample. Amplicon sequencing was carried out following the protocol described in Sinclair et al. (2015) using the MiSeq instrument. Illumina sequencing was performed by the SNP/SEQ SciLifeLab facility hosted by Uppsala University using 300 bp chemistry. Chimera identification and OTU (Operational Taxonomic Unit) clustering by denoising was done using UNOISE (from USEARCH version 9, refs69,70). SINTAX (from USEARCH version 9, ref.78) with the SILVA reference database (release 128) was used as a base to taxonomically annotate OTUs. The sequence data has been deposited to the EBI Archive under accession number PRJEB20882.
HgcA gene
Among the 50 samples selected based on having %MeHg >1%, 34 resulted in positive PCR amplification of the hgcA gene. The protein-coding gene hgcA which plays an essential role in Hg methylation was amplified with previously published hgcA primers (hgcA_ 261 F and hgcA_912R) (Table S7, 34) modified for parallelized high-throughput Illumina sequencing. HPLC-purified primers carrying Illumina adaptors at the 5′ end (hgcA_261F_Adaptor and hgcA_912R_Adaptor, Table S7) were here used for the 1st stage PCR. In the 2nd stage PCR, standard Illumina handles and barcode primers (Table S7) were used to enable pooling of all the samples for parallelized Illumina sequencing. HgcA was first amplified in 50 μL volume with 1x Phusion GC Buffer, 0.2 mM dNTP mix, 5% DMSO, 0.1 μM of each adaptor-linked primer, 7 μg/μL BSA, 4 μL extracted DNA template, and 1.0 U Phusion high fidelity DNA polymerase (NEB, UK) for an initial denaturation of 2 min at 98 °C followed by 35 cycles (10 s at 96 °C, 30 s 56.5 °C and 45 s at 72 °C), and a final extension at 72 °C for 7 min. Following this initial step, a 2nd PCR was conducted to add sample-specific molecular barcodes. Reactions were carried out in 20 μL volumes using 1x Q5 reaction buffer, 0.2 mM dNTP mix, 0.1 μM barcode primers, purified 1st PCR products and 1.0 U Q5 high fidelity DNA polymerase (NEB, UK) for an initial denaturation of 30 s at 98 °C followed by 18 cycles (10 s at 98 °C, 30 s 66 °C and 30 s at 72 °C), and a final extension at 72 °C for 2 min. The quality and size of the hgcA amplicons were assessed by gel electrophoresis and GelRed visualization on a 1% agarose gel (Invitrogen, USA) prior to purification by Agencourt AMPure XP (Beckman Coulter, USA) after both PCR steps. Quantifications of purified amplicons from the 2nd stag PCR were performed using the PicoGreen kit (Invitrogen).
Amplicons were sequenced using the same method as for the 16S rRNA gene. Forward read sequences were only used in data analysis due to long PCR product. Low quality sequences were filtered and trimmed using SICKLE79 and adapter were removed by using CUTADAPT80. Subsequent processing of reads were performed by USEARCH and clustered at 60% identity cutoff using cd-hit-est81. HMMER82 search was used for taxonomical annotation with manually curated database of Proteobacteria and sequences of Podar et al. (2015) ref.49. More details can be found in Bravo et al. (2018) ref.46.
Phylogenetic analysis
A phylogenetic analysis was performed for hgcA sequences representative for the OTUs observed for the 34 hotspots and existing hgcA entries in our curated database. The sequences were adequately curated and taxonomy homogenized using taxtastic (https://github.com/fhcrc/taxtastic) and the R-package taxize83. The obtained protein sequences were aligned with MUSCLE84 (version 3.8.1551). The alignment was trimmed to the size of the amplicon, and a tree was generated using RAxML85 (version 8.2.4) - with the PROTGAMMLG model and autoMR to choose the number of necessary bootstrap resamplings (n = 750). This tree and the corresponding alignment were used to generate a reference package for PPLACER86. The guppy tool of PPLACER was then used to classify the sequences with a likelihood threshold of 0.8.
Statistical analysis
Family-level microbial community composition in the different samples were compared using non-metric multidimensional scaling (nMDS) based on Bray-Curtis similarities and using the software PRIMER 787. Information on the common set of samples from community composition based on Bray-Curtis similarities and that from geochemical variables based on Euclidean distance was presented in one single ordination. A combined nMDS plot with bubble and vector plots of geochemical factors projected on the same ordination of community composition was constructed to reveal the relationships between community compositions and potentially explanatory geochemical variables87,88. Pearson’s correlation coefficient (R) was assessed to reveal linear relationships between variables using a significance level of alpha < 0.05.
Supplementary information
Acknowledgements
This project was carried out within the Swedish-Sino SMaREF (2013-6978) funded by the Swedish Research Council. This study was also supported by the Swedish Energy Agency (grant number 36155-1) and the Swedish Research Council (Grants 2011-7192 and 2012-3892). Sequencing was carried out at the SciLifeLab SNP/SEQ facility hosted by Uppsala University and we also acknowledge the Uppsala Multidisciplinary Centre for Advanced Computational Science (UPPMAX) for access to storage and computational resources.
Author Contributions
A.G.B., St.B., J.X., K.B., K.E. conceived the study. A.G.B. and K.E. conducted the sampling campaigns. J.X. and O.O.A. performed the laboratory analyses with guidance from A.G.B., St.B. and K.E. M.B. performed the bioinformatics analyses. J.X. and A.G.B. conducted all statistical analyses and built the figures with comments and suggestions from the rest of the co-authors. J.X., A.G.B. and St.B. wrote the manuscript with significant assistance and comments from the rest of the co-authors (i.e. M.B., K.E., O.O.A., J.K.S., K.B., U.S., E.B.).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jingying Xu, Email: jingying.xu@ebc.uu.se.
Andrea G. Bravo, Email: jandriugarcia@gmail.com
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37383-z.
References
- 1.UNEP. Global Mercury Assessment 2013: Sources, Emissions, Releases, and Environmental Transport. UNEP 42, doi:DTI/1636/GE (2013).
- 2.Amos HM, Jacob DJ, Streets DG, Sunderland EM. Legacy impacts of all-time anthropogenic emissions on the global mercury cycle. Global Biogeochem. Cycles. 2013;27:410–421. doi: 10.1002/gbc.20040. [DOI] [Google Scholar]
- 3.Lamborg C, et al. Mercury in the Anthropocene Ocean. Oceanography. 2014;27:76–87. doi: 10.5670/oceanog.2014.11. [DOI] [Google Scholar]
- 4.Karagas MR, et al. Evidence on the human health effects of low-level methylmercury exposure. Environ. Health Perspect. 2012;120:799–806. doi: 10.1289/ehp.1104494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munthe, J., Wängberg, I., Rognerud, S. & Fjeld, E. Mercury in Nordic ecosystems. (2007).
- 6.Åkerblom S, Nilsson M, Yu J, Ranneby B, Johansson K. Temporal change estimation of mercury concentrations in northern pike (Esox lucius L.) in Swedish lakes. Chemosphere. 2012;86:439–445. doi: 10.1016/j.chemosphere.2011.09.037. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi N, Tang RWK, Bhavsar SP, Arhonditsis GB. Fish mercury levels appear to be increasing lately: A report from 40 years of monitoring in the province of Ontario, Canada. Environ. Sci. Technol. 2014;48:5404–5414. doi: 10.1021/es403651x. [DOI] [PubMed] [Google Scholar]
- 8.Munthe J, et al. Recovery of mercury-contaminated fisheries. Ambio. 2007;36:33–34. doi: 10.1579/0044-7447(2007)36[33:ROMF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 9.Gu B, et al. Mercury reduction and complexation by natural organic matter in anoxic environments. Proc. Natl. Acad. Sci. 2011;108:1479–1483. doi: 10.1073/pnas.1008747108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drott A, Lambertsson L, Björn E, Skyllberg U. Importance of dissolved neutral mercury sulfides for methyl mercury production in contaminated sediments. Environ. Sci. Technol. 2007;41:2270–2276. doi: 10.1021/es061724z. [DOI] [PubMed] [Google Scholar]
- 11.Johansson K, Tyler G. Impact of atmospheric long range transport of lead, mercury and cadmium on the Swedish forest environment. Water, Air Soil Pollut. Focus. 2001;1:279–297. doi: 10.1023/A:1017528826641. [DOI] [Google Scholar]
- 12.Xu J, et al. Sources and remediation techniques for mercury contaminated soil. Environ. Int. 2014;74:42–53. doi: 10.1016/j.envint.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Shanley, J. B. & Bishop, K. In Mercury in the environment: pattern and process (ed. Banks, M. S.) 119–142, at http://pubs.er.usgs.gov/publication/70157507, (University of California Press, 2012).
- 14.Grigal DF. Inputs and outputs of mercury from terrestrial watersheds: a review. Environ. Rev. 2002;10:1–39. doi: 10.1139/a01-013. [DOI] [Google Scholar]
- 15.Bravo AG, et al. Molecular composition of organic matter controls methylmercury formation in boreal lakes. Nat. Commun. 2017;8:14255. doi: 10.1038/ncomms14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isidorova A, et al. The effect of lake browning and respiration mode on the burial and fate of carbon and mercury in the sediment of two boreal lakes. J. Geophys. Res. Biogeosciences. 2016;121:233–245. doi: 10.1002/2015JG003086. [DOI] [Google Scholar]
- 17.Hongve D, Haaland S, Riise G, Blakar I, Norton S. Decline of acid rain enhances mercury concentrations in fish. Environ. Sci. Technol. 2012;46:2490–1. doi: 10.1021/es3002629. [DOI] [PubMed] [Google Scholar]
- 18.Eklöf K, Meili M, Åkerblom S, von Brömssen C, Bishop K. Impact of stump harvest on run-off concentrations of total mercury and methylmercury. For. Ecol. Manage. 2013;290:83–94. doi: 10.1016/j.foreco.2012.05.039. [DOI] [Google Scholar]
- 19.Bishop K, et al. The effects of forestry on Hg bioaccumulation in nemoral/boreal waters and recommendations for good silvicultural practice. Ambio. 2009;38:378–380. doi: 10.1579/0044-7447-38.7.373. [DOI] [PubMed] [Google Scholar]
- 20.Wu P, et al. Does forest harvest increase the mercury concentrations in fish? Evidence from Swedish lakes. Sci. Total Environ. 2018;622–623:1353–1362. doi: 10.1016/j.scitotenv.2017.12.075. [DOI] [PubMed] [Google Scholar]
- 21.Eklöf K, et al. Formation of mercury methylation hotspots as a consequence of forestry operations. Sci. Total Environ. 2018;613–614:1069–1078. doi: 10.1016/j.scitotenv.2017.09.151. [DOI] [PubMed] [Google Scholar]
- 22.Parks JM, et al. The genetic basis for bacterial mercury methylation. Science (80-.). 2013;339:1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 23.Eckley CS, Hintelmann H. Determination of mercury methylation potentials in the water column of lakes across Canada. Sci. Total Environ. 2006;368:111–125. doi: 10.1016/j.scitotenv.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 24.Monperrus M, Tessier E, Amouroux D, Leynaert A. Mercury methylation, demethylation and reduction rates in coastal and marine surface waters of the Mediterranean Sea. Mar. Chem. 2007;107:49–63. doi: 10.1016/j.marchem.2007.01.018. [DOI] [Google Scholar]
- 25.Gascón Díez E, et al. Role of Settling Particles on Mercury Methylation in the Oxic Water Column of Freshwater Systems. Environ. Sci. Technol. 2016;50:11672–11679. doi: 10.1021/acs.est.6b03260. [DOI] [PubMed] [Google Scholar]
- 26.Tjerngren I, Karlsson T, Björn E, Skyllberg U. Potential Hg methylation and MeHg demethylation rates related to the nutrient status of different boreal wetlands. Biogeochemistry. 2012;108:335–350. doi: 10.1007/s10533-011-9603-1. [DOI] [Google Scholar]
- 27.Skyllberg, U. L. F., Qian, J. I. N., Frech, W. & Xia, K. Distribution of mercury, methyl mercury and organic sulphur species in soil, soil solution and stream of a boreal forest catchment. 1, 53–76 (2002).
- 28.Compeau GC, Bartha R. Sulfate-reducing Bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl. Environ. Microbiol. 1985;50:498–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King JK, Kostka JE, Frischer ME, Saunders FM, Jahnke RA. A quantitative relationship that demonstrates mercury methylation rates in marine sediments are based on the community composition and activity of sulfate-reducing bacteria. Environ. Sci. Technol. 2001;35:2491–2496. doi: 10.1021/es001813q. [DOI] [PubMed] [Google Scholar]
- 30.Fleming EJ, Mack EE, Green PG, Nelson DC. Mercury methylation from unexpected sources: molybdate-inhibited freshwater sediments and an iron-reducing bacterium. Appl. Environ. Microbiol. 2006;72:457–464. doi: 10.1128/AEM.72.1.457-464.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerin EJ, et al. Mercury methylation by dissimilatory iron-reducing bacteria. Appl. Environ. Microbiol. 2006;72:7919–7921. doi: 10.1128/AEM.01602-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamelin S, Amyot M, Barkay T, Wang Y, Planas D. Methanogens: principal methylators of mercury in lake periphyton. Environ. Sci. Technol. 2011;45:7693–7700. doi: 10.1021/es2010072. [DOI] [PubMed] [Google Scholar]
- 33.Gilmour CC, et al. Mercury methylation by novel microorganisms from new environments. Environ. Sci. Technol. 2013;47:11810–11820. doi: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 34.Skyllberg U, Qian J, Frech W, Xia K, Bleam WF. Distribution of mercury, methyl mercury and organic sulphur species in soil, soil solution and stream of a boreal forest catchment. Biogeochemistry. 2003;64:53–76. doi: 10.1023/A:1024904502633. [DOI] [Google Scholar]
- 35.Liem-Nguyen, V., Jonsson, S., Skyllberg, U., Nilsson, M. B. & Andersson, A. Effects of Nutrient Loading and Mercury Chemical Speciation on the Formation and Degradation of Methylmercury in Estuarine Sediment. Environ. Sci. Technol. 6983–6990, 10.1021/acs.est.6b01567 (2016). [DOI] [PubMed]
- 36.Kronberg RM, Jiskra M, Wiederhold JG, Björn E, Skyllberg U. Methyl mercury formation in hillslope soils of boreal forests: The role of forest harvest and anaerobic microbes. Environ. Sci. Technol. 2016;50:9177–9186. doi: 10.1021/acs.est.6b00762. [DOI] [PubMed] [Google Scholar]
- 37.Bigham GN, Murray KJ, Masue-Slowey Y, Henry EA. Biogeochemical controls on methylmercury in soils and sediments: Implications for site management. Integr. Environ. Assess. Manag. 2017;13:249–263. doi: 10.1002/ieam.1822. [DOI] [PubMed] [Google Scholar]
- 38.Liem-Nguyen V, Skyllberg U, Björn E. Thermodynamic Modeling of the Solubility and Chemical Speciation of Mercury and Methylmercury Driven by Organic Thiols and Micromolar Sulfide Concentrations in Boreal Wetland Soils. Environ. Sci. Technol. 2017;51:3678–3686. doi: 10.1021/acs.est.6b04622. [DOI] [PubMed] [Google Scholar]
- 39.Chiasson-Gould SA, Blais JM, Poulain AJ. Dissolved organic matter kinetically controls mercury bioavailability to bacteria. Environ. Sci. Technol. 2014;48:3153–3161. doi: 10.1021/es4038484. [DOI] [PubMed] [Google Scholar]
- 40.Schaefer JK, Morel FMM. High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nat. Geosci. 2009;2:123–126. doi: 10.1038/ngeo412. [DOI] [Google Scholar]
- 41.Graham AM, Aiken GR, Gilmour CC. Effect of dissolved organic matter source and character on microbial Hg methylation in Hg-S-DOM solutions. Environ. Sci. Technol. 2013;47:5746–5754. doi: 10.1021/es400414a. [DOI] [PubMed] [Google Scholar]
- 42.Bravo, A. G. et al. Methanogens and iron-reducing bacteria: the overlooked members of mercury methylating microbial communities in boreal lakes. Appl. Environ. Microbiol. 84 (2018). [DOI] [PMC free article] [PubMed]
- 43.Gilmour, C. C. et al. Impacts of Activated Carbon Amendment on Hg Methylation, Demethylation and Microbial Activity in Marsh Soils. In AGU Fall Meeting (2015).
- 44.Schaefer, J. K., Kronberg, R.-M., Morel, F. M. M. & Skyllberg, U. Detection of a key Hg methylation gene, hgcA, in wetland soils. Environ. Microbiol. Rep. n/a–n/a, 10.1111/1758-2229.12136 (2014). [DOI] [PubMed]
- 45.Bravo, A. G. et al. Persistent Hg contamination and occurrence of Hg-methylating transcript (hgcA) downstream of a chlor-alkali plant in the Olt River (Romania). Environ. Sci. Pollut. Res. 1–13 10.1007/s11356-015-5906-4 (2016). [DOI] [PubMed]
- 46.Bravo AG, et al. Geobacteraceae are important members of mercury-methylating microbial communities of sediments impacted by wastewater releases. ISME J. 2018;12:802–812. doi: 10.1038/s41396-017-0007-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y-R, Yu R-Q, Zheng Y-M, He J-Z. Analysis of the microbial community structure by monitoring an Hg methylation gene (hgcA) in paddy soils along an Hg gradient. Appl. Environ. Microbiol. 2014;80:2874–2879. doi: 10.1128/AEM.04225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bae HS, Dierberg FE, Ogram A. Syntrophs dominate sequences associated with the mercury methylation-related gene hgcA in the water conservation areas of the Florida Everglades. Appl. Environ. Microbiol. 2014;80:6517–6526. doi: 10.1128/AEM.01666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Podar M, et al. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci. Adv. 2015;1:e1500675–e1500675. doi: 10.1126/sciadv.1500675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tjerngren I, Meili M, Björn E, Skyllberg U. Eight boreal wetlands as sources and sinks for methyl mercury in relation to soil acidity, C/N ratio, and small-scale flooding. Environ. Sci. Technol. 2012;46:8052–8060. doi: 10.1021/es300845x. [DOI] [PubMed] [Google Scholar]
- 51.Yu RQ, et al. Contribution of coexisting sulfate and iron reducing bacteria to methylmercury production in freshwater river sediments. Environ. Sci. Technol. 2012;46:2684–2691. doi: 10.1021/es2033718. [DOI] [PubMed] [Google Scholar]
- 52.Ranchou-Peyruse M, et al. Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: implications for environmental studies. Geomicrobiol. J. 2009;26:1–8. doi: 10.1080/01490450802599227. [DOI] [Google Scholar]
- 53.Pak K, Bartha R. Mercury methylation and demethylation in anoxic lake sediments and by strictly anaerobic bacteria. Appl. Environ. Microbiol. 1998;64:1013–1017. doi: 10.1128/aem.64.3.1013-1017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bravo, A. G. et al. High methylmercury production under ferruginous conditions in sediments impacted by sewage treatment plant discharges. Water Res. 80 (2015). [DOI] [PubMed]
- 55.Ullrich SM, Tanton TW, Abdrashitova SA, Svetlana A. Mercury in the aquatic environment: a review of factors affecting methylation. Crit. Rev. Environ. Sci. Technol. 2001;31:241–293. doi: 10.1080/20016491089226. [DOI] [Google Scholar]
- 56.Achá D, Hintelmann H, Pabón CA. Sulfate-reducing bacteria and mercury methylation in the water column of the Lake 658 of the Experimental Lake Area. Geomicrobiol. J. 2012;29:667–674. doi: 10.1080/01490451.2011.606289. [DOI] [Google Scholar]
- 57.King JK, Kostka JE, Frischer ME, Saunders FM. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl. Environ. Microbiol. 2000;66:2430–2437. doi: 10.1128/AEM.66.6.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu RQ, et al. Mercury methylation in Sphagnum moss mats and its association with sulfate-reducing bacteria in an acidic Adirondack forest lake wetland. FEMS Microbiol. Ecol. 2010;74:655–668. doi: 10.1111/j.1574-6941.2010.00978.x. [DOI] [PubMed] [Google Scholar]
- 59.Gilmour CC, Henry EA, Mitchell R. Sulfate stimulation of mercury methylation in freshwater sediments. Environ. Sci. Technol. 1992;26:2281–2287. doi: 10.1021/es00035a029. [DOI] [Google Scholar]
- 60.Plugge CM, Zhang W, Scholten JCM, Stams AJM. Metabolic flexibility of sulfate-reducing bacteria. Front. Microbiol. 2011;2:1–8. doi: 10.3389/fmicb.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McInerney MJ, et al. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 2008;1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- 62.Pester M, Bittner N, Deevong P, Wagner M, Loy A. A ‘rare biosphere’ microorganism contributes to sulfate reduction in a peatland. ISME J. 2010;4:1–12. doi: 10.1038/ismej.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood JM. Biological cycles for elements in the environment. Naturwissenschaften. 1975;62:357–364. doi: 10.1007/BF00625342. [DOI] [Google Scholar]
- 64.Liu YR, Wang JJ, Zheng YM, Zhang LM, He JZ. Patterns of Bacterial Diversity Along a Long-Term Mercury-Contaminated Gradient in the Paddy Soils. Microb. Ecol. 2014;68:575–583. doi: 10.1007/s00248-014-0430-5. [DOI] [PubMed] [Google Scholar]
- 65.Christensen GA, et al. Development and validation of broad-range aualitative and clade-specific quantitative molecular probes for assessing mercury methylation in the environment. Appl. Environ. Microbiol. 2016;82:6068–6078. doi: 10.1128/AEM.01271-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skyllberg U, et al. Net methylmercury production as a basis for improved risk assessment of mercury-contaminated sediments. Ambio. 2007;36:437–442. doi: 10.1579/0044-7447(2007)36[437:NMPAAB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 67.Drott A, Lambertsson L, Björn E, Skyllberg U. Do potential methylation rates reflect accumulated methyl mercury in contaminated sediments? Environ. Sci. Technol. 2008;42:153–158. doi: 10.1021/es0715851. [DOI] [PubMed] [Google Scholar]
- 68.Remy S, Prudent P, Probst JL. Mercury speciation in soils of the industrialised Thur River catchment (Alsace, France) Appl. Geochemistry. 2006;21:1855–1867. doi: 10.1016/j.apgeochem.2006.08.003. [DOI] [Google Scholar]
- 69.Hunger S, Gößner AS, Drake HL. Anaerobic trophic interactions of contrasting methane-emitting mire soils: Processes versus taxa. FEMS Microbiol. Ecol. 2015;91:1–14. doi: 10.1093/femsec/fiv045. [DOI] [PubMed] [Google Scholar]
- 70.Ransom-Jones E, Jones DL, McCarthy AJ, McDonald JE. The Fibrobacteres: An Important Phylum of Cellulose-Degrading Bacteria. Microb. Ecol. 2012;63:267–281. doi: 10.1007/s00248-011-9998-1. [DOI] [PubMed] [Google Scholar]
- 71.Podosokorskaya OA, et al. Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ. Microbiol. 2013;15:1759–1771. doi: 10.1111/1462-2920.12067. [DOI] [PubMed] [Google Scholar]
- 72.Liang, B. et al. Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Express5 (2015). [DOI] [PMC free article] [PubMed]
- 73.Juottonen H, et al. Distinct Anaerobic Bacterial Consumers of Cellobiose-Derived Carbon in Boreal Fens with Different CO2/CH4 Production Ratios. Appl. Environ. Microbiol. 2017;83:e02533–16. doi: 10.1128/AEM.02533-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuever, J. In The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria 9783642390, 75–86 (2014).
- 75.Holmes, D. E. et al. Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in methanogenic rice paddy soils. Appl. Environ. Microbiol. 2 (2017). [DOI] [PMC free article] [PubMed]
- 76.WRB. World Reference Base for Soil Resources 2014, update 2015. International soil classification system for naming soils and creating legends for soil maps. World Soil Resources Reports No. 106. World Soil Resources Reports No. 106, 10.1017/S0014479706394902 (2014).
- 77.Lambertsson L, Lundberg E, Nilsson M, Frech W. Applications of enriched stable isotope tracers in combination with isotope dilution GC-ICP-MS to study mercury species transformation in sea sediments during in situ. J. Anal. At. Spectrom. 2001;16:1296–1301. doi: 10.1039/b106878b. [DOI] [Google Scholar]
- 78.Edgar, R. SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv 074161, 10.1101/074161 (2016).
- 79.Joshi, N. A. & Fass, J. N. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. (2011).
- 80.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 81.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chamberlain SA, Szöcs E. taxize: taxonomic search and retrieval in R. F1000Research. 2013;2:191. doi: 10.12688/f1000research.2-191.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsen, F. A., Kodner, R. B. & Armbrust, E. V. pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 538, 10.1186/1471-2105-11-538 (2010). [DOI] [PMC free article] [PubMed]
- 87.Clarke, K. R. & Gorley, R. N. PRIMER v7: User Manual/Tutorial. Prim. Plymouth UK 192 p, 10.1111/j.1442-9993.1993.tb00438.x (2015)
- 88.Löfgren S, Ring E, von Brömssen C, Sørensen R, Högbom L. Short-term effects of clear-cutting on the water chemistry of two boreal streams in northern Sweden: a paired catchment study. Ambio. 2009;38:347–356. doi: 10.1579/0044-7447-38.7.347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.