Abstract
Cysteine persulfide and polysulfide are produced in cells and exist in abundance in both low MW and protein fractions. However, the mechanism of regulation of the formation of cellular cysteine polysulfides and the physiological functions of cysteine persulfides/polysulfides produced in cells are not fully understood. We recently demonstrated that cysteinyl‐tRNA synthetase (CARS) is a novel cysteine persulfide synthase. CARS is involved in protein polysulfidation that is coupled with translation. In particular, mitochondria function in biogenesis and bioenergetics is also supported and up‐regulated by cysteine persulfide derived from mitochondrial CARS (also known as CARS2). Here, we provide an overview of recent advances in reactive persulfide research and our understanding of the mechanisms underlying the formation and the physiological roles of reactive persufides, with a primary focus on the formation of cysteine persulfide by CARS and the most fundamental mitochondrial bioenergetics mediated by persulfides, that is, sulfur respiration.
Linked Articles
This article is part of a themed section on Chemical Biology of Reactive Sulfur Species. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v176.4/issuetoc
Abbreviations
- 8‐nitro‐cGMP
8‐nitroguanosine 3,5‐cyclic monophosphate
- CARS
cysteinyl‐tRNA synthetase
- CBS
cystathionine β‐synthase
- CPERS
CysSSH synthase
- CSE
cystathionine γ‐lyase
- CysSSH
cysteine persulfide
- Cys‐tRNA
cysteinyl‐tRNA
- Drp1
dynamin‐related protein 1
- EcCARS
Escherichia coli CARS
- ETC
electron transport chain
- GSSH
glutathione persulfide
- HPE‐IAM
β‐(4‐hydroxyphenyl)ethyl iodoacetamide
- Kcat
catalytic rate constant
- KO
knockout
- NRF2
nuclear factor‐erythroid‐2‐related factor 2
- PLP
pyridoxal phosphate
- PUNCH‐P
puromycin‐associated nascent chain proteomics
- PUNCH‐PsP
PUNCH‐P modified for polysulfides proteomics
- RNS
reactive nitrogen oxide species
- WT
wild type
Introduction
Reactive persulfide species including cysteine persulfide (CysSSH) are found in abundant quantities in both prokaryotic and eukaryotic cells (Toohey, 2011; Fukuto et al., 2012; Ida et al., 2014; Shimizu, et al., 2017; Peng, et al., 2017; Akaike et al., 2017). Various forms of persulfide/polysulfide species exist in cells as cysteine‐containing low MW compounds such as glutathione persulfide (GSSH) and CysSSHs in proteins. Persulfide/polysulfide species have a unique redox property that differs from that of simple thiols because of the additional sulfur atoms. CysSSH can act as a strong nucleophile and an antioxidant and may play an important role in regulating oxidative stress and redox signalling in cells (de Beus et al., 2004: Mustafa et al., 2009; Vandiver et al., 2013; Ida et al., 2014; Gao et al., 2015; Yang et al., 2015; Kasamatsu et al., 2016). ROS are produced during inflammation and infection and induce oxidative stress. ROS and reactive nitrogen oxide species (RNS), which are produced by the reaction of ROS and NO, cause various chemical modifications such as oxidation and nitration of biomolecules, which results in cell damage (Beckman et al., 1990; van der Vliet and Cross, 2000; Halliwell, 2007). Some electrophiles that depend on ROS and RNS for formation, however, are endogenous signalling molecules and mediate redox signalling (Forman et al., 2004; Sies, 2014). These electrophilic signalling molecules include nitro derivatives and oxidized derivatives of fatty acids and 8‐nitroguanosine 3′,5′‐cyclic monophosphate (8‐nitro‐cGMP) (Sawa et al., 2007; Uchida and Shibata, 2008; Rudolph and Freeman, 2009; Schopfer et al., 2011). We have studied the signalling mechanisms and physiological roles of 8‐nitro‐cGMP and found that the signalling by this compound is regulated by CysSSH in cells. CysSSH effectively reacted with 8‐nitro‐cGMP to form 8‐SH‐cGMP and thereby nullified the electrophilic signalling activity of 8‐nitro‐cGMP (Nishida et al., 2012; Ida et al., 2014; Ihara et al., 2017). We developed a quantitative analytical system for the persulfide/polysulfide species of cysteine and related compounds that involved LC‐MS‐MS and found that different amounts of various persulfide/polysulfide species were produced in mammalian cells (Ida et al., 2014). We identified two transsulfuration enzymes, cystathionine β‐synthase (CBS) and cystathionine γ‐lyase (CSE), that participate in endogenous CysSSH formation (Nishida et al., 2012; Ida et al., 2014). CBS and CSE can catalyse CysSSH production via C‐S cleavage of cystine. It was also found that appreciable amounts of CysSSH exist in different cellular proteins. We recently demonstrated, for the first time, that cysteinyl‐tRNA synthetase (CARS) can act as a novel CysSSH‐producing enzyme (Akaike et al., 2017). Biochemical analyses revealed that CARS can produce CysSSH from a substrate cysteine. CARS can also mediate direct incorporation of CysSSH into proteins during translation, which results in the formation of protein persulfides and polysulfides. Of great importance is our discovery that CysSSH‐producing activity is critically involved in the regulation of mitochondrial function – a new physiological role of persulfide/polysulfide (Figure 1). This article provides an overview of recent advances in reactive persulfide research and discusses our understanding of the formation mechanism and physiological roles of reactive persufides, with a primary focus on unique mechanisms of CARS‐dependent CysSSH production and the fundamental versatile physiological functions of reactive persufides in various organisms, for example, sulfur respiration in mitochondria.
Figure 1.
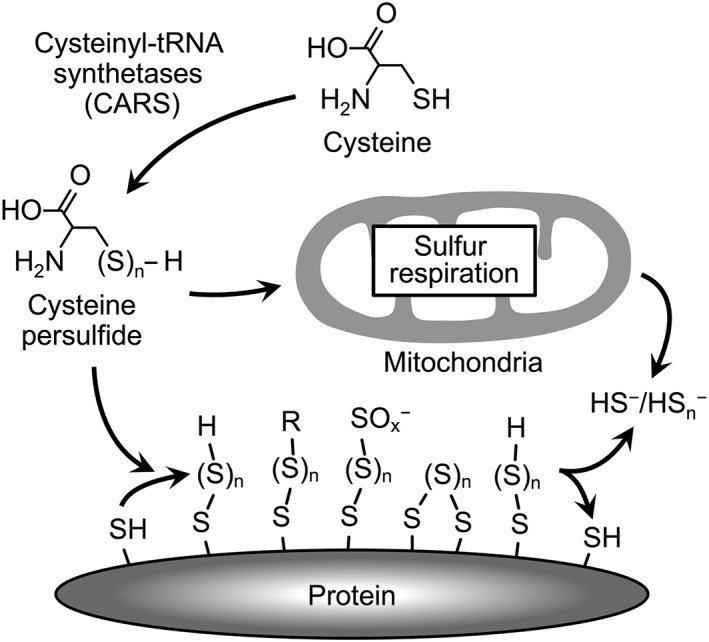
CysSSH formation, protein polysulfidation and regulation of mitochondrial function mediated by the novel CysSSH‐producing enzyme CARS. CARS produces CysSSH from cysteine. CysSSH‐producing activity of CARS is critically involved in translation‐coupled protein polysulfidation. Protein polysulfidation may be involved in the regulation of protein functions, such as expression of enzyme activity and maintenance of protein structure. CysSSH produced by mitochondrial CARS2 mediates the sulfur‐mediated bioenergetics in mitochondria (sulfur respiration). The molecular mechanisms underlying sulfur respiration are shown in Figure 5.
Unique reactive properties of cysteine persulfide and protein polysulfides
CysSSH has unique reactive properties that distinguish it from parental cysteine. Persulfide/polysulfide species of cysteine possess both nucleophilic and electrophilic properties (Fletcher and Robson, 1963; Parker and Kharasch, 1959; Abdolrasulnia and Wood, 1980). On the basis of potential nucleophilicity of polysulfides as well as their nucleophilicity of thiol residues, we can detect various polysulfide species present in cells and differentiate them from parental thiols. We recently developed a convenient method to detect proteins bearing persulfide/polysulfide moieties that is based on the biotin–polyethylene glycol‐conjugated maleimide (biotin‐PEG‐MAL) labelling gel shift assay (PMSA) (Jung et al., 2016). With this PMSA, we demonstrated such proteins in Escherichia coli and mammalian cells. To establish a selective and quantitative detection method for low MW persulfides/polysulfides and protein persulfides/polysulfides, we developed a detection method that utilizes LC‐electrospray ionization (ESI)‐MS‐MS and utilized β‐(4‐hydroxyphenyl)ethyl iodoacetamide (HPE‐IAM) as a trapping agent (Figure 2A) (Numakura et al., 2017; Akaike et al., 2017). HPE‐IAM has a mild electrophilicity and forms stable adducts with persulfides/polysulfides so that we can avoid artificial degradation of polysulfide structures during the labelling process. By using this selective and quantitative LC‐ESI‐MS‐MS analysis, we found that various proteins, such as alcohol dehydrogenase 5 (ADH5) and GAPDH, are highly polysulfidated in cells. For example, LC‐ESI‐MS‐MS analysis, after pronase digestion of HPE‐IAM‐labelled ADH5, demonstrated that 70% of cysteine residues in ADH5 were endogenously polysulfidated (Figure 2B). Additional LC‐quadrupole‐time‐of‐flight‐MS analyses identified sites of polysulfide formation and the sulfur chain length in each protein. Recent studies suggested that protein polysulfidation may be involved in the regulation and maintenance of the activity of some enzymes (Jarosz et al., 2015; Jung et al., 2016; Millikin et al., 2016). Further investigation using analytical systems we developed for persulfide/polysulfide stated above will contribute to clarify the role of protein polysulfidation in redox metabolism signalling (Akaike et al., 2017).
Figure 2.
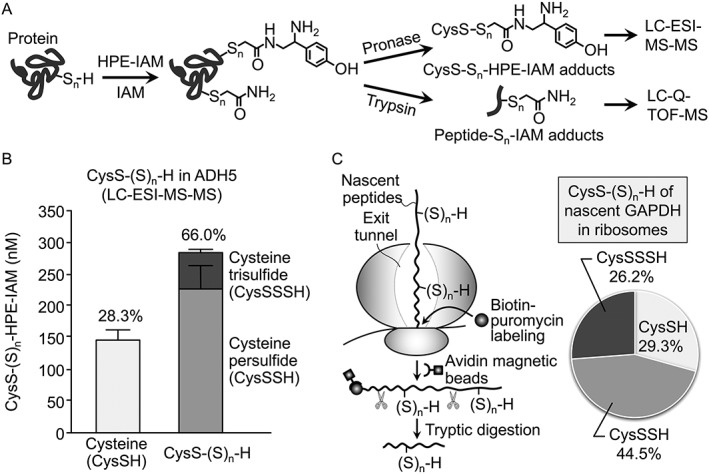
(A) Schematic illustration of specific and quantitative analytical systems for protein polysulfidation that used HPE‐IAM labelling and MS. Polysulfidated cysteine residues in proteins were modified and stabilized with HPE‐IAM or IAM. Amounts of cysteine polysulfides were quantified by LC‐ESI‐MS‐MS analysis of proteinase‐digested protein samples. The sites of polysulfidated cysteine residue in proteins were determined with LC‐quadrupole‐time‐of‐flight‐MS using trypsin digests of various proteins. (B) Quantitative identification of cysteine polysulfide levels in recombinant ADH5 protein by using HPE‐IAM labelling and LC‐ESI‐MS‐MS. In ADH5 protein, the level of polysulfidated cysteine (right column) was much higher than that of non‐polysulfidated cysteine (left column), which suggested extensive polysulfidation in ADH5 protein. (C) Schematic illustration of a new method for identification of protein polysulfidation in the nascent peptide PUNCH‐PsP (left), and the level of cysteine polysulfides incorporated into the GAPDH nascent peptide as determined by PUNCH‐PsP (right). In PUNCH‐PsP, newly synthesized peptides were labelled with biotin‐puromycin and collected with avidin‐magnetic beads. Levels of cysteine polysulfides were determined, using the analytical systems shown in (A).
Protein polysulfidation mediated by CARS
As mentioned above, our PMSA and LC‐MS‐MS analyses clearly showed that different types of proteins are highly polysulfidated endogenously (Ida et al., 2014; Ono et al., 2014; Doka et al., 2016; Jung et al., 2016; Akaike et al., 2017). To understand how protein polysulfidation occurs in cells, we studied whether CARS can catalyse direct incorporation of CysSSH/Cys‐(S)n‐H into tRNA. CARS is an enzyme that produces cysteinyl‐tRNA (Cys‐tRNA) via cysteine and aminoacyl‐tRNA (Carter et al., 1993; Woese et al., 2000; Guo et al., 2010). Our recent work revealed for the first time that CARS from E. coli (EcCARS) produces CysSSH‐tRNA by using CysSSH as a substrate. CysSSH‐tRNA can be a substrate for protein synthesis in ribosomes as CysS‐tRNA and can lead to translation‐coupled protein polysulfidation of newly synthesized proteins. This translation‐coupled co‐translational protein polysulfidation is clearly shown by puromycin‐associated nascent chain proteomics (PUNCH‐P), as recently reported (Aviner et al., 2014), with PUNCH‐P being modified for polysulfide proteomics (PUNCH‐PsP) (Figure 2C) (Akaike et al., 2017). This PUNCH‐PsP approach allowed us to identify intact forms of CysS‐(S)n‐H residues in nascent peptides of GAPDH present only within E. coli ribosomes. In fact, the PUNCH‐PsP we conducted showed significant polysuflidation at the Cys247 residue of the nascent peptides of GAPDH protein immediately after the synthesis in the ribosomes in E. coli cells. All native forms of cysteine, CysSSH and CysSSH residues were also efficiently recovered from native whole GAPDH protein, and polysulfidation affected more than 60% of the Cys247 residues of the mature protein. Thus, the extensive and prevalent cysteine polysulfidation is now found to co‐translationally occur in ribosomes and continues to be physiologically maintained in mature proteins in cells.
In addition, we confirmed a novel function of EcCARS as a CysSSH synthase (CPERS) by showing its catalytic activity in generating CysS‐(S)n‐H, for example, CysSSH and CysSSSH, from the substrate cysteine. The CPERS activity of EcCARS depends partly on pyridoxal phosphate (PLP) but not on ATP and tRNA, the latter two being required for Cys‐tRNA biosynthesis by EcCARS. We also performed a stable isotope (34S) tracer experiment combined with LC‐MS‐MS analysis in an attempt to understand the mechanism of cysteine polysulfidation catalysed by CARS. Our analyses clearly showed cleavage of a sulfur atom from one cysteine and its transfer to another cysteine to form CysSSH (Figure 3).
Figure 3.
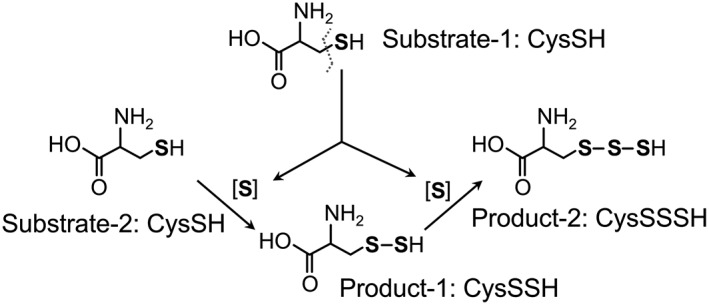
Diagram of the reaction mechanisms in the formation of CysSSH and polysulfides, as catalysed by CARSs. During the formation of cysteine persulfides, sulfur atoms are cleaved from donor cysteine (Substrate‐1) and transferred to acceptor cysteine (Substrate‐2) to form the cysteine persulfide (Product‐1). Subsequent transfer of sulfur to Product‐1 forms cysteine trisulfide (Product‐2). CysSH, cysteine; CysSSSH, cyeteine hydrotrisulfide.
CARS as a CysSSH‐producing enzyme
CARS is involved in the translation process as a canonical function by catalysing Cys‐tRNA synthesis. Our rigorous investigation of the mechanism of protein polysulfidation demonstrated that CARSs also have CPERS activity and are critically involved in protein polysulfidation (Akaike et al., 2017). Some aminoacyl‐tRNA synthetases reportedly function in physiological processes beyond translation, known as moonlighting functions of aminoacyl‐tRNA synthetases (Wakasugi and Schimmel, 1999; Guo et al., 2010). Therefore, CPERS activity in CARS is regarded as a relatively new function of aminoacyltransferase beyond translation. In fact, CARS efficiently produces CysSSH by using cysteine as a substrate, in a manner that is independent of the aminoacyltransferase reaction. Figure 3 shows the reaction mechanism of transsulfidation catalysed by CARSs that our recent study clarified. CARS has a very low K M and a high catalytic rate constant K cat, which indicates that EcCARS is quite efficient in producing CysSSH (Ono et al., 2014; Yadav et al., 2016). The K cat/K M value of CARS is almost equal to the value of EcCARS. However, CSE and CBS utilize only cystine (not cysteine) as a substrate, which is quite distinct from CARSs, which use cysteine (not cystine) for CysSSH production (Figure 4) (Ida et al., 2014). Intracellular cystine concentrations are usually at low micromolar or submicromolar levels, which are much lower than the K M value of CSE (more than 200 μM). Thus, CSE may play a minor role in directly producing CysSSH under physiologically relevant conditions. Also, the cystine/CSE reaction may not outcompete reactions with other major enzymes that metabolize cystine and GSH, which can strongly interact with and consume cysteine under physiological baseline conditions in cells and in various organs. In fact, the intracellular cysteine concentration is reportedly 100–1000 μM (Ida et al., 2014), which is apparently much higher than the K M of CARS. Also, intracellular levels of cysteine are regulated by the uptake of cystine from the extracellular space, which is mediated by xCT, the cystine‐glutamate transporter. Expression of this xCT transporter is regulated by the transcriptional factor nuclear factor‐erythroid‐2‐related factor 2 (NRF2) (Sasaki et al., 2002). Thus, NRF2 may regulate CysSSH production by CARS through the regulation of intracellular cysteine levels, which is mediated by the xCT transporter. Therefore, it is quite reasonable to suggest that CARS may be a major enzyme responsible for endogenous generation of CysS‐(S)n‐H (Figure 4).
Figure 4.
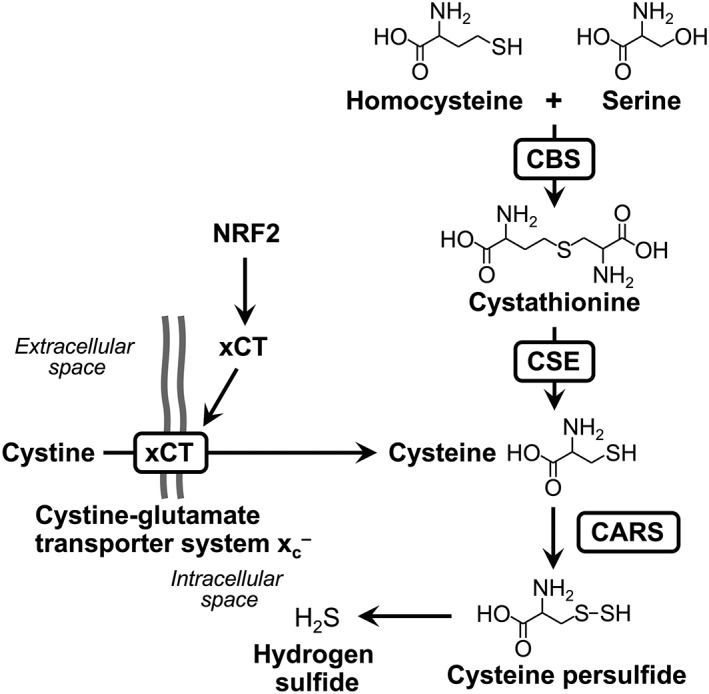
Diagram of the mechanisms involved in the cellular formation of CysSSH and H2S and regulation of this formation by CARS, CBS and CSE. CBS and CSE are involved in cysteine biogenesis, though the canonical trans‐sulfuration pathway. Cystine transport by cystine‐glutamate transporter system, mediated by xCT, also contributes to the regulation of intracellular cysteine levels. Cysteine biosynthesis and cystine uptake affect cysteine persulfide production by CARS via modulation of cellular cysteine levels.
CysSSH production by EcCARS depended on the addition of PLP. Proteomic analyses suggested that several preferential sites exist for PLP binding in EcCARS, including lysine residues at the 73KIIK76 and 266KMSK269 motifs (Akaike et al., 2017). Sequence data obtained previously suggested that several lysine residues, at the KIIK and KMSK motifs, are highly conserved in EcCARS and other homologues in different organisms, including mammals (Akaike et al., 2017). Two well‐conserved cysteine residues are bound to Zn2+ at the active centre for tRNA aminoacylation (Zhang et al., 2003). To determine the role of PLP bound to EcCARS, we generated several lysine mutants of this enzyme, with which we measured CysS‐(S)n‐H formation and translation. We found greatly reduced CysSSH and CysSSSH synthesis in various lysine‐to‐alanine mutants, compared with the wild type (WT), at K73A, K76A, K266A, K269A, and double mutants K73/76A and K266/269A, of EcCARS. All these mutant enzymes had intact protein synthesis potential as determined in the PUREfrex cell‐free protein synthesis assay (Akaike et al., 2017). The amounts of PLP bound to EcCARS were quantified by using LC‐ESI‐MS‐MS with 2,4‐dinitrophenylhydrazine. This 2,4‐dinitrophenylhydrazine‐labelling LC‐MS‐MS analysis revealed that the amounts of PLP bound to the enzymes decreased in lysine mutants compared with the WT and, more important, that PLP levels correlated well with persulfide‐producing activities. Cysteine‐to‐aspartate mutants such as C28D (and C28S) and the double mutant C28/209D, however, still had high persulfide production, similar to WT persulfide production, although these mutants had greatly reduced protein synthesis and translational activity (Akaike et al., 2017).
The three‐dimensional EcCARS structure constructed by computer modelling supported the finding that PLP binds to specific lysine residues at the 73KIIK76 and 266KMSK269 motifs of EcCARS. The computer‐based prediction suggested two possible PLP‐binding sites at K73 and K269 of the EcCARS KIIK and KMSK motifs. In addition, this computer modelling indicated that PLP‐bound motifs have a vicinal location within 10–20 Å but different from that of the ATP‐binding HIGH motif and the Zn2+‐binding active site of EcCARS for Cys‐tRNA biosynthesis. A comparable change in PLP binding capacity and stability seems to exist, caused by mutation of any of the four lysine residues, because each lysine mutation at the KIIK and KMSK motifs affected EcCARS synthesis of all CysS‐(S)n‐H to a great degree. One conceivable reason for the comparable effect is that PLP may need more than one lysine residue, rather than just one lysine binding, to realize the stable binding and full catalytic activity of CARS and thus act as CPERS during formation of CysS‐(S)n‐H. That is, PLP may need to be stabilized by binding to multiple lysine residues to achieve stable binding and demonstrate potent catalytic activity. Otherwise, reactive persulfides derived from CARS, because of their nucleophilic nature, will quickly degrade an aldehyde group of PLP, which may block the catalytic activity of PLP even properly bound to these particular lysine residues of CARS. The computational structural analysis showing close locations (within 20 Å) of these lysine residues in the KIIK and KMSK motifs supports this interpretation. These data together indicate that EcCARS has a moonlighting function as a CPERS, which is independent of its original catalytic function as an aminoacyl‐tRNA synthetase (Akaike et al., 2017).
CPERS activity of mammalian CARSs
CPERS activity of CARSs can be observed not only in bacterial CARS (EcCARS) but also in mammalian CARSs (Akaike et al., 2017). Two different mammalian CARSs have been found, one located in the cytosol, CARS1, and the other, CARS2, in the mitochondria (Hallmann et al., 2014; Coughlin et al., 2015). We now know that both CARSs (mouse CARS1 and human CARS2) show strong CysS‐(S)n‐H‐producing activities that are dependent on PLP. In fact, the CPERS activity of CARS2 correlates well with the cellular PLP levels and that CARS2 activity can be modulated by exposing the enzyme to different concentrations of PLP (Akaike et al., 2017). We utilized CARS1‐deficient and CARS2‐deficient HEK293T cells generated via the CRISPR/Cas9 system to determine how much cellular CysS‐(S)n‐H originated from CARS1 and CARS2. Although CARS1‐knockout (KO) cells are not yet available, CARS2 KO cells have been successfully established. Activity of one of the clones, with a 30 bp deletion and a 8 bp insertion downstream of the translation‐initiating codon in the CARS2 first exon, was analysed by using LC‐MS‐MS methods and the results showed that CysS‐(S)n‐H and GSSH levels were significantly decreased in CARS2 KO cells, implying that CARS2 was a major producer of persulfide/polysulfide species. Although low but detectable levels of CARS2 were still seen in CARS2 KO cells, the cells were also treated with siRNA against CARS2, and this treatment caused decreases of 67 and 42% in CysSSH and GSSH levels respectively. After knockdown of CARS1 in CARS2 KO cells, CysSSH decreased only slightly, thus indicating a predominant role of CARS2 as a CPERS.
Adding back WT CARS2 in CARS2 KO cells significantly increased persulfide levels. The CARS2 C78/257D mutant restored persulfide production in CARS2 KO cells, but the K124/127A and K317/320A mutants (mutants of KIIK and KMSK motifs, respectively) did not have this effect. CARS2 KO cells had greatly reduced Cys‐tRNA synthetase activity and adding back the C78/257D mutant did not increase the Cys‐tRNA synthetase activity of the KO cells, as shown by expression of mitochondrial cytochrome c oxidase subunit 1 (MTCO1, encoded by mitochondrial DNA), although full CPERS activity was restored. These data confirm that CARS2 acts as a CPERS in mammals and that this activity is distinct from CARS activity.
We also investigated the contributions of CSE and CBS to endogenous persulfide production. Silencing CSE and CBS significantly decreased intracellular cysteine levels, which in turn reduced persulfide production. In CARS2 KO cells, CSE and CBS knockdown also led to lower cysteine levels but not to lower persulfide production. These findings show that the metabolic pathways mediated by CSE/CBS in each cell line provide cysteine, regardless of CARS2 expression. In other words, CBS and CSE are involved in the cysteine biosynthesis rather than CysSSH production under physiological conditions. Furthermore, almost two‐thirds of CysSSH appears to be supplied by CARS2 in HEK293T cells, as shown by the corresponding decrease in CysSSH levels. The remaining CysSSH in CARS2 KO cells is not obtained via CSE/CBS expressed in HEK293T cells, because no additional reduction in CysSSH occurred even after CSE/CBS knockdown in CARS2 KO cells. CSE and CBS therefore most likely do not participate directly in persulfide production but instead may support cysteine biosynthesis and the supply of cysteine to CARS, at least under physiological conditions (Figure 4).
Cars2‐deficient mice revealing CPERS activity of CARSs in vivo
We investigated the CPERS actions of CARS2 in vivo by generating Cars2‐deficient mice via CRISPR/Cas9 technology (Akaike et al., 2017). We designed a guide RNA against exon 1 of Cars2. We also developed a mutant mouse line with a mutant Cars2 allele (line 1) that had a 200 bp deletion with a translation‐initiating codon in exon 1. Although the Cars2 −/− deletion was embryonically lethal, the heterozygous Cars2 +/− strain of mice are usually born without any abnormal macroscopic appearances or growth profiles, found during a 6 month observation period after birth. These mice expressed only half the mitochondrial CARS2 protein and a markedly decreased CysSSH production, relative to the WT mice. Because no substantial change in mitochondrial DNA‐encoded MTCO1 was noted, we believe that Cars2 +/− mice had intact Cys‐tRNA synthetase activity. Sulfide metabolites in the liver of Cars2 +/− mice and their WT littermates were quantified via LC‐MS‐MS analysis and HPE‐IAM, as described earlier. Compared with their WT littermates, these Cars2 +/− mice exhibited a marked difference in persulfide production. In major organs of Cars2 +/− mice, compared with those of WT mice, endogenous levels of CysSSH and its derivatives – GSSH, sulfite, thiosulfate and hydropolysulfides – were decreased by almost 50%. We also developed another strain of Cars2 +/− mice (line 2) with an alternative guide RNA that targeted Cars2 exon 3 to eliminate the possibility of off‐target effects by the guide RNA that we used to produce line 1 Cars2 +/− mice. Line 2 Cars2 +/− mice had phenotypes, almost identical to those of line 1 mice.
The finding that Cars2 +/− mice demonstrated an approximate 50% reduction in CysSSH should be emphasized, inasmuch as it suggests that Cars2 contributes almost all of the CysSSH production in mouse tissues under physiological conditions. This suggestion is confirmed by the fact that Cars2 modification did not change the expression of other sulfide‐metabolizing enzymes, such as CSE, CBS and 3‐mercaptopyruvate sulfur transferase. Our relatively long‐term observation for less than 6 months with Cars2 +/− mice did not show any difference in their macroscopic appearance, compared with the WT mice.
To investigate whether the mitochondrial CARS isoform CARS2 can generate CysSSH and provide it to the cell, we isolated mitochondria from mouse liver and determined the release of de novo‐synthesized CysSSH from these organelles. Mitochondria did indeed release a large amount of CysSSH, which lends support to the idea that CysSSH that is generated in mitochondria is discharged into the cytoplasm and sustains protein polysulfidation. CysSSH generated from whole‐cell proteins was decreased in Cars2 +/− mice, but cysteine levels were affected. In particular, production of 20–30% of CysSSH in all cellular proteins (polysulfidation) depended on CARS2 expression, not only in an in vivo study using Cars2 KO mice but also in an in vitro experiment with cell cultures, as shown by HPE‐IAM labelling LC‐MS‐MS analysis with isolation of whole cell and tissues proteins. These findings suggest that CysSSH generated from CARS2 contributes significantly to polysulfidation, which appears to be mediated by post‐translational and co‐translational processes, the former being controlled by the thioredoxin–thioredoxin reductase system, as recently reported (Doka et al., 2016). On the basis of these data, we expect that CysSSH produced in mitochondria is released into the cytoplasm and provides sulfur to proteins for polysulfidation. This evidence was the first demonstration that positively confirmed CARS2 as the major CPERS in mammals.
Regulation of mitochondrial functions by reactive persulfide species
Importantly, our recent study also demonstrated novel physiological roles of CARS as a CPERS during regulation of mitochondrial functions (Figure 5) (Akaike et al., 2017). CARS2 KO cells showed greatly modified mitochondrial morphological features (i.e. shrunken or fragmented appearance), which improved considerably when CARS2 was added back, as shown by MitoTracker Red fluorescence staining, transmission electron microscopy and immunofluorescence staining for translocase of outer mitochondrial membrane 20 and CARS2. Unlike other lysine mutants tested, WT CARS2 and the C78/258D mutant exhibited a much improved mitochondrial morphology. Related to these results, deletion of the CARS2‐activated dynamin‐related protein 1 (Drp1), a primary mitochondrial fission mediator (Akhtar et al., 2016), and adding back WT CARS2 and the C78/257D mutant, but not the K317/320A mutant, significantly attenuated Drp1 GTPase activity, thus generating CysSSH without CARS activity. Under normal culture conditions, Drp1 in HEK293T cells was extensively polysulfidated, as demonstrated by our new biotin‐PEG‐MAL capture method. Both CARS2 KO and CARS1/2 double‐knockdown cells had markedly suppressed Drp1 polysulfidation. Drp1 is probably activated by chemical depolysulfidation or by a post‐translational process maintained physiologically by the thioredoxin–thioredoxin reductase system, for example. We therefore established that Drp1 is an important signal effector molecule that is regulated in a reversible manner via a unique process involving polysulfidation and depolysulfidation.
Figure 5.
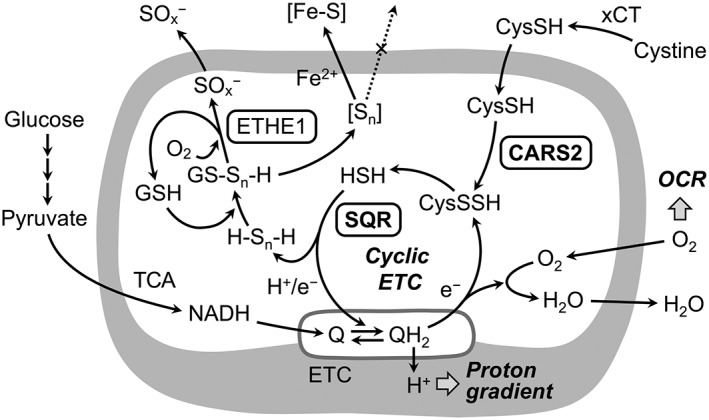
Regulation of the mitochondrial ETC mediated by CARS2 and CysSSH. CysSSH produced by CARS2 in mitochondria is reductively metabolized to CysSH and HS−, which may be further oxidized by the sulfide:quinone oxidoreductase (SQR), in a manner linked to ETC in mitochondria (cyclic ETC). The CysS‐(S)n‐H‐dependent HS− metabolism may be coupled with formation of iron–sulfur clusters (Fe–S), that is controlled by the mitochondrial ETC. Ethylmalonic encephalopathy protein 1 reportedly catalyses oxidative degradation of glutathione polysulfide (GS‐Sn‐H), whose formation is catalysed by SQR and related to cysteine persulfide production by CARS and cyclic ETC. Q/QH2, ubiquinone/ubiquinol; TCA, tricarboxylic acid.
Evidence has clearly shown the contribution of CARS2 to the biogenesis and functioning of mitochondria. Specifically, mitochondrial DNA that was normalized by nuclear DNA decreased in CARS2 KO cells but was restored by adding back WT CARS2 and the C78/257D mutant but not the lysine mutants, which implied that CARS2‐derived persulfide improved mitochondrial biogenesis. The membrane potential of mitochondria decreased in CARS2 KO cells, but it was restored or increased after adding back or overexpressing the WT and the C78/257D mutant but not after using lysine mutants. We utilized extracellular flux analysis to determine the oxygen consumption rate (OCR) in HEK293T CARS2 KO cells. This OCR in CARS2 KO cells was approximately 50% of that in WT cells, a result that agrees with the incomplete elimination of CARS2 protein and thereby reduced MTCO1 expression in CARS2 KO cells. Introducing WT CARS2 and the C78/257D mutant, but not the lysine mutant, restored the decreased OCR in CARS2 KO cells. These observations led to the novel concept that CARS2‐derived CysSSHs play an important role in the mitochondrial electron transport chain (ETC), which postulates a completely new and fundamental role of persulfides in maintaining mitochondrial bioenergetics.
During our efforts to clarify how CARS2‐derived CysSSH supports mitochondrial bioenergetics, we noted a profile of human CARS2 products in a cell‐free enzyme reaction that was quite different compared with cellular CARS2 metabolism in HEK293T cells in culture. CARS2 synthesized primarily CysSSH/CysSSSH in cell‐free solutions but, in HEK293T cells, there was preferential formation of the hydrosulfide ion HS− (from H2S) together with thiosulfate, rather than formation of CysSSH . We therefore hypothesized that the mitochondrial compartment serves as a unique metabolic environment for additional metabolism of de novo CysSSH synthesized by CARS2, perhaps being coupled with the mitochondrial ETC.
An investigation of the metabolic profile of CysSSH and its derivatives in HEK293T cells led to identification of a close relationship between CARS2‐dependent CysSSH production and ETC function. We used two methods to inhibit the cellular ETC and thereby to induce loss of mitochondrial DNA. In one, the specific inhibitor of complex III antimycin A was used and in the other, the ETC disrupter ethidium bromide was used. Both ETC treatments resulted in significantly increased CysSSH production and simultaneously reduced HS− production, as determined by HPE‐IAM labelling LC‐MS‐MS analysis. These opposite, stoichiometric relationships between CysSSH and H2S formation led to the belief that conversion of CysSSH to H2S depends on ETC activity and is mediated by the ETC in the cells. It is conceivable, therefore, that CysSSH generated from CARS2 in mitochondria is reduced by accepting an electron from the ETC, thus generating H2S.
These findings also provide strong support for the involvement of the CARS2‐CysSSH pathway in mitochondrial function because CARS2‐dependent CysSSH production is integrated into and tightly linked to the mitochondrial ETC, which is itself implicated in energy metabolism. Studies have found that low concentrations (nM range) of H2S maintained ETC function that may have been mediated by the sulfide:quinone reductase and other enzymes that oxidize sulfides to thiosulfate (Grieshaber and Völkel, 1998; Griesbeck et al., 2002; Goubern et al., 2007; Ono et al., 2014; Szabo et al., 2014; Hine et al., 2015). However, how H2S was supplied endogenously in mitochondria remained unclear. Our various studies suggest that CSE, CBS and 3‐mercapopyruvate sulfur transferase are not primary H2S sources in mitochondria in different mammalian cell lines and in vivo in mice (Nishida et al., 2012; Morikawa et al., 2012; Ono, et al., 2014; Shirozu et al., 2014; Nakano et al., 2015; Yadav et al., 2016). In fact, our recent study was the first to confirm that HS− (or H2S) was formed indirectly from CARS2 via CysSSH generation in the mitochondrial environment (Akaike et al., 2017). In addition, another study found that CysSSH may be partly responsible for endogenous formation of iron–sulfur clusters (Takahashi et al., 2017). Because iron–sulfur clusters are synthesized and utilized in complexes I–III of the ETC in mitochondria (Stehling and Lill, 2013) and are actively transported outside the mitochondria, CysSSH‐dependent metabolism of HS− may be tied to production of iron–sulfur centres of the mitochondrial ETC and cytosolic formation and maintenance of different iron–sulfur complex systems. Therefore, CARS2 serves as a major CPERS, which aids mitochondrial biogenesis and bioenergetics.
As described above, the CysSSH metabolizing pathway is critically involved in mitochondrial bioenergetics in mammalian cells (Figure 5). This suggests bioenergetics mediated by sulfur, instead of being mediated by oxygen, which is a catabolic process known as sulfur respiration in bacteria. Some photosynthetic bacteria derive their energy from sulfur compounds such as H2S as an electron donor (Shimizu et al., 2017). Also, sulfur‐oxidizing bacteria (e.g. Acidithiobacillus thiooxidans) obtain energy by oxidising H2S, using sulfide:quinone oxidoreductase (Yin et al., 2014). In Staphylococcus aureus, reactive persulfides have been reported not only in cysteine but also in CoA, as CoA‐SSH (Peng et al., 2017). In this context, our finding is the first demonstration of sulfur respiration in mammalian cells, as shown in Figure 5 (Akaike et al., 2017). The discovery of sulfur respiration in mammals may have major implications not only to fundamental biology, but also to disease pathogenesis related to energy metabolism in cancer and stem cell biology.
In conclusion, this overview of recent advances in reactive persulfide research describes our development of a new analytical approach to CysSSH quantification and our discovery of a new CysSSH‐producing enzyme, CARS. We have also detailed the novel role of CysSSH produced by CARS2 in the regulation of mitochondrial functions, including sulfur respiration. Co‐translational protein‐bound CysSSH formation may have an important role in protecting proteins from oxidative stress and in redox signal regulation. Continued elucidation of the physiological functions of reactive persulfides formed from new persulfide‐producing enzymes that we discovered will promote the development of new therapeutic agents for various diseases involving oxidative stress.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http//www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017a,b).
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank J.B. Gandy for her excellent editing of the manuscript. This work was supported in part by Grants‐in‐Aid for Scientific Research and Grants‐in‐Aid for Scientific Research on Innovative Areas (Research in a Proposed Research Area), from the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Fujii, S. , Sawa, T. , Motohashi, H. , and Akaike, T. (2019) Persulfide synthases that are functionally coupled with translation mediate sulfur respiration in mammalian cells. British Journal of Pharmacology, 176: 607–615. 10.1111/bph.14356.
References
- Abdolrasulnia R, Wood JL (1980). Persulfide properties of thiocystine and related trisulfides. Bioog Chem 9: 253–260. [Google Scholar]
- Akaike T, Ida T, Wei F‐Y, Nishida M, Kumagai Y, Alam MM et al (2017). Cysteinyl‐tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat Commun 8: 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar MW, Sanz‐Blasco S, Dolatabadi N, Parker J, Chon K, Lee MS et al (2016). Elevated glucose and oligomeric β‐amyloid disrupt synapses via a common pathway of aberrant protein S‐nitrosylation. Nat Commun 7: 10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017a). The Concise Guide to PHARMACOLOGY 2017/2018: Enzymes. Br J Pharmacol 174: S272–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD et al (2017b). The Concise Guide to PHARMACOLOGY 2017/2018: Transporters. Br J Pharmacol 174: S360–S446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviner R, Geiger T, Elroy‐Sein O (2014). Genome‐wide identification and quantification of protein synthesis in cultured cells and whole tissues by puromycin‐associated nascent chain proteomics (PUNCH‐P). Nat Protoc 9: 751–760. [DOI] [PubMed] [Google Scholar]
- Beckman JS, Beckman TW, Chen J, Marshal PA, Freeman BA (1990). Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci U S A 87: 1620–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beus MD, Chung J, Colón W (2004). Modification of cysteine 111 in Cu/Zn superoxide dismutase results in altered spectroscopic and biophysical properties. Protein Sci 13: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW Jr (1993). Cognition, mechanism, and evolutionary relationships in aminoacyl‐tRNA synthetases. Annu Rev Biochem 62: 715–748. [DOI] [PubMed] [Google Scholar]
- Coughlin CR II, Scharer GH, Friederich MW, Yu HC, Geiger EA, Creadon‐Swindell G et al (2015). Mutations in the mitochondrial cysteinyl‐tRNA synthase gene, CARS2, lead to a severe epileptic encephalopathy and complex movement disorder. J Med Genet 52: 532–540. [DOI] [PubMed] [Google Scholar]
- van der Vliet A, Cross CE (2000). Oxidants, nitrosants, and the lung. Am J Med 109: 398–421. [DOI] [PubMed] [Google Scholar]
- Doka E, Pader I, Biro A, Johansson K, Cheng Q, Ballago K et al (2016). A novel persulfide detection method reveals protein persulfide‐ and polysulfide‐reducing functions of thioredoxin and glutathione systems. Sci Adv 2: e1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Robson A (1963). The occurrence of bis‐(2‐amino‐2‐carboxyethyl) trisulfide in hydrolysates of wool and other protein. Biochem J 87: 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman HJ, Fukuto JM, Torres M (2004). Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol 287: C246–C256. [DOI] [PubMed] [Google Scholar]
- Fukuto JM, Carrington SJ, Tantillo DJ, Harrion JG, Ignarro LJ, Freeman BA et al (2012). Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxide, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem Res Toxicol 25: 769–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M et al (2015). Quantitative H2S‐mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife 4: e10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubern M, Andriamihaja M, Nübel T, Blachier F, Bouillaud R (2007). Sulfide, the first inorganic substrate for human cells. FASEB J 21: 1699–1706. [DOI] [PubMed] [Google Scholar]
- Griesbeck C, Schütz M, Schödl T, Bathe S, Nausch L, Mederer N et al (2002). Mechanism of sulfide‐quinone reductase investigated using site‐directed mutagenesis and sulfur analysis. Biochemistry 41: 11,552–11,565. [DOI] [PubMed] [Google Scholar]
- Grieshaber MK, Völkel S (1998). Animal adaptations for tolerance and exploitation of poisonous sulfide. Annu Rev Physiol 60: 33–53. [DOI] [PubMed] [Google Scholar]
- Guo M, Yang XL, Schimmel P (2010). New functions of aminoacyl‐tRNA synthetases beyond translation. Nat Rev Mol Cell Biol 11: 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B (2007). Biochemistry of oxidative stress. Biochem Soc Trans 35: 1147–1150. [DOI] [PubMed] [Google Scholar]
- Hallmann K, Zsurka G, Moskau‐Hartmann S, Kirschner J, Korinthenberg R, Ruppert AK et al (2014). A homozygous splice‐site mutation in CARS2 is associated with progressive myoclonic epilepsy. Neurology 83: 2183–2187. [DOI] [PubMed] [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine C, Harputlugil E, Zhang Y, Ruckenstuhl C, Lee BC, Brace L et al (2015). Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 160: 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y et al (2014). Reactive cysteine persulfides and S‐polythionation regulate oxidative stress and redox signaling. Proc Natl Acad Sci U S A 111: 7606–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara H, Kasamatsu S, Kitamura A, Nishimura A, Tsutsuki H, Ida T et al (2017). Exposure to electrophiles impairs reactive persulfide‐dependent redox signaling in neuronal cells. Chem Res Toxicol 30: 1653–1684. [DOI] [PubMed] [Google Scholar]
- Jarosz AP, Wei W, Gauld JW, Auld J, Özcan F, Aslan M et al (2015). The chemical biology of protein hydropersulfides: studies of a possible protective function of biological hydropersulfide generation. Free Radic Biol Med 89: 512–521.26453916 [Google Scholar]
- Jung M, Kasamatsu S, Matsunaga T, Akashi S, Ono K, Nishimura A et al (2016). Protein polysulfidation‐dependent persulfide dioxygenase activity of ethylmalonic encephalopathy protein 1. Biochem Biophys Res Commun 480: 180–186. [DOI] [PubMed] [Google Scholar]
- Kasamatsu S, Nishimura A, Morita M, Matsunaga T, Abdul Hamid H, Akaike T (2016). Redox signaling regulated by cysteine persulfide and protein polysulfidation. Molecules 21: E1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikin R, Bianco CL, White C, Saund SS, Henriquez S, Sosa V et al (2016). The chemical biology of protein hydropersulfides: studies of a possible protective function of biological hydropersulfide generation. Free Radic Biol Med 97: 136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa T, Kajimura M, Nakamura T, Hishiki T, Nakanishi T, Yukutake T et al (2012). Hypoxic regulation of the cerebral microcirculation is mediated by a carbon monoxide‐sensitive hydrogen sulfide pathway. Proc Natl Acad Sci U S A 109: 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK et al (2009). H2S signals through protein S‐sulfhydration. Sci Signal 2: ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Ishii I, Shinmura K, Tamaki K, Hishiki T, Akahoshi N et al (2015). Hyperhomocysteinemia abrogates fasting‐induced cardioprotection against ischemia/reperfusion by limiting bioavailability of hydrogen sulfide anions. J Mol Med (Berl) 93: 879–889. [DOI] [PubMed] [Google Scholar]
- Nishida M, Sawa T, Kitajima N, Ono K, Inoue H, Ihara H et al (2012). Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat Chem Biol 8: 714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numakura T, Sugiura H, Akaike T, Ida T, Fujii S, Koarai A et al (2017). Production of reactive persulfide species in chronic obstructive pulmonary disease. Thorax 72: 1074–1083. [DOI] [PubMed] [Google Scholar]
- Ono K, Akaike T, Sawa T, Kumagai T, Wink DA, Tantillo DJ et al (2014). Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic Biol Med 77: 82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker AJ, Kharasch N (1959). The scission of the sulfur‐sulfur bond. Chem Rev 59: 583–628. [Google Scholar]
- Peng H, Shen J, Edomonds KA, Luebke JL, Hickey AK, Palmer LD et al (2017). Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species in Staphlococcus aureus. mSphere 2: e00082‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph TK, Freeman BA (2009). Transduction of redox signaling by electrophile‐protein reactions. Sci Signal 2: re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Sao H, Kuriyama‐Matsumura K, Sato K, Maebara K, Wang H et al (2002). Electrophile response element‐mediated induction of the cystine/glutamate exchange transporter gene expression. J Biol Chem 277: 44765–44771. [DOI] [PubMed] [Google Scholar]
- Sawa T, Zaki MH, Okamoto T, Akuta T, Tokuomi Y, Kim‐Mitsuyama S et al (2007). Protein S‐guanylation by the biological signal 8‐nitroguanosine 3′,5′‐cyclic monophosphate. Nat Chem Biol 3: 727–735. [DOI] [PubMed] [Google Scholar]
- Schopfer FJ, Cipollina C, Freeman BA (2011). Formation and signaling actions of electrophilic lipids. Chem Rev 111: 5997–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Shen J, Fang M, Zhang Y, Hori K, Trinidad JC et al (2017). Sulfide‐responsive transcriptional repressor SqrR functions as a master regulator of sulfide‐dependent photosynthesis. Proc Natl Acad Sci U S A 114: 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirozu K, Tokuda K, Marutani E, Lefer D, Wang R, Ichinose F (2014). Cystathionine γ‐lyase deficiency protects mice from galactosamine/lipopolysaccharide‐induced acute liver failure. Anitioxid Rexod Signal 20: 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H (2014). Role of metabolic H2O2 generation: redox signaling and oxidative stress. J Biol Chem 289: 8753–8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling O, Lill R (2013). The role of mitochondria in cellular iron‐sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harb Prespect Biol 5: a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Ransy C, Modis K, Andriamihaja M, Murghes B, Coletta C et al (2014). Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br J Pharmacol 171: 2099–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Wei FY, Watanabe S, Hirayama M, Ohuchi Y, Fujimura A et al (2017). Reactive sulfur species regulate tRNA methylthiolation and contribute to insulin secretion. Nucleic Acids Res 45: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toohey JI (2011). Sulfur signaling: is the agent sulfide or sulfane? Anal Biochem 413: 1–7. [DOI] [PubMed] [Google Scholar]
- Uchida K, Shibata T (2008). 15‐Deoxy‐delta(12,14)‐prostaglandin J2: an electrophilic trigger of cellular responses. Chem Res Toxicol 21: 138–144. [DOI] [PubMed] [Google Scholar]
- Vandiver MS, Paul BD, Xu R, Karuppagounder S, Rao F, Snowman AM et al (2013). Sulfhydration mediates neuroprotective actions of parkin. Nat Commun 4: 1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K, Schimmel P (1999). Two distinct cytokines released from a human aminoacyl‐tRNA synthetase. Science 284: 147–151. [DOI] [PubMed] [Google Scholar]
- Woese CR, Olsen GJ, Ibba M, Söll D (2000). Aminoacyl‐tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev 64: 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PK, Martinov M, Vitvitsky V, Seravalli J, Wedmann R, Filipovic MR et al (2016). Biosynthesis and reactivity of cysteine persulfides in signaling. J Am Chem Soc 138: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Qu C, Zhou Y, Konkel JE, Shi S, Liu Y et al (2015). Hydrogen sulfide promotes Tet1‐ and Tet2‐mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity 43: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Zhang X, Li X, He Z, Liang Y, Guo X et al (2014). Whole‐genome sequencing reveals novel insights into sulfur oxidation in the extremophile Acidithiobacillus thiooxidants . BMC Microbiol 14: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CM, Christian T, Newberry KJ, Perona JJ, Hoou YM (2003). Zinc‐mediated amino acid discrimination in cysteinyl‐tRNA synthetase. J Mol Biol 327: 911–917. [DOI] [PubMed] [Google Scholar]


