Abstract
Sporadic Alzheimer’s disease (AD) is the most common cause of dementia. However, representative experimental models of AD have remained difficult to produce because of the disease’s uncertain origin. The Polycomb group protein BMI1 regulates chromatin compaction and gene silencing. BMI1 expression is abundant in adult brain neurons but down-regulated in AD brains. We show here that mice lacking one allele of Bmi1 (Bmi1+/−) develop normally but present with age cognitive deficits and neurodegeneration sharing similarities with AD. Bmi1+/− mice also transgenic for the amyloid beta precursor protein died prematurely and present aggravated disease. Loss of heterochromatin and DNA damage response (DDR) at repetitive DNA sequences were predominant in Bmi1+/− mouse neurons and inhibition of the DDR mitigated the amyloid and Tau phenotype. Heterochromatin anomalies and DDR at repetitive DNA sequences were also found in AD brains. Aging Bmi1+/− mice may thus represent an interesting model to identify and study novel pathogenic mechanisms related to AD.
Introduction
Alzheimer’s disease of the late-onset sporadic form (AD) is the most common dementia1. At the genetic level, carriers of the apolipoprotein E4 (APOE4) allele have a higher risk to develop the disease with age2. Despite this, the etiology of AD remains largely elusive, with old age representing the greatest risk factor. Familial AD (FAD) is an early-onset, autosomal dominant disorder, that is associated with mutations in amyloid beta precursor protein (APP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2), and representing less than 5% of all Alzheimer’s disease cases3. Short-term memory deficits and behavioral changes are the earliest known manifestations of AD and FAD. While brain atrophy is generally restricted to the limbic system and hippocampus at the disease’s onset, neurodegeneration eventually spreads to all cortical areas, resulting in loss of up to 30% of the brain mass. At the histo-pathological level, the disease is characterized by the accumulation of amyloid plaques and intra-neuronal phospho-Tau (p-Tau) tangles. Early loss of synapses and, later, of neurons, is also a characteristic of the disease3. While numerous transgenic mouse models of FAD have been developed that could recapitulate several hallmarks of the disease, experimental mouse models of AD are rare because the disease’s origin remains uncertain4–6.
Nucleosomes are the basic building unit of chromatin and are constituted of a 147 base pairs of double-stranded DNA wrapped around a histone octamer7. Post-translational modification of histones can modify chromatin compaction and stability. BMI1 (B-cell specific Moloney murine leukemia virus integration site 1) is a core component of the Polycomb Repressive Complex 1 (PRC1). The PRC1 can ubiquitylate histone H2A at lysine 119 (H2Aub) through the E3-ubiquitin ligase activity of Ring1a/b, its catalytic unit8–11. The main classical function of BMI1 is to maintain chromatin compaction and gene silencing at developmental and senescence-associated loci. The Ink4a locus, which encodes for the p16Ink4a and p19Arf tumor suppressor proteins, is one of the main targets of BMI1 repressive activity, and cells deficient for Bmi1 undergo rapid senescence12,13. Bmi1-null mice present axial skeleton defects, reduced post-natal growth and lifespan, as well as cerebellar degeneration14. The BMI1 protein is also recruited at DNA break sites where it can promote DNA damage response (DDR) and repair15,16. Bmi1-deficiency in mice is furthermore associated with accumulation of DNA damage and reactive oxygen species (ROS), both of which can be improved through co-deletion of Chk2 or p5317,18. More recently, BMI1 was found to be abundant at heterochromatin regions containing repetitive DNA sequences (namely the constitutive heterochromatin). BMI1 knockdown resulted in reduced histone H3 trimethylation at lysine 9 (H3K9me3) and in transcriptional activation of tandem repeats in mammalian cells19.
We recently reported that the expression of BMI1 is reduced in AD brains and in AD neurons produced from the differentiation of patient-specific induced pluripotent stem cells. Notably, acute BMI1 inactivation in mature human neurons could recapitulate most AD-related neuronal pathologies20. Herein, we show that mice lacking one Bmi1 allele (Bmi1+/−) develop normally, but display with age a pathology sharing some similarities with AD. Crosses between Bmi1+/− mice and transgenic mice carrying a mutant human APP gene revealed genetic cooperation in disease onset and severity. Furthermore, loss of heterochromatin and DDR at repetitive DNA sequences in neurons was characteristic of Bmi1+/− mice and AD brains. Experimental evidences in mice furthermore suggested a significant contribution of the DDR to the observed neuronal pathology. Loss of one Bmi1 allele in mice thus predisposes to age-related neurodegeneration sharing some similarities with AD.
Methods
Human samples
Paraffin-embedded human brains were obtained from the department of pathology of Maisonneuve-Rosemont Hospital. Frozen post-mortem human cortices are a gift from the Douglas Hospital Brain Bank. The human samples were used accordingly to the Maisonneuve-Rosemont Hospital Ethic Committee (Approval ID #2012-481, 11065). All samples were obtained after informed consent of the patients. Samples were confirmed by histo-pathological analysis as non-demented controls, FAD or AD cases. The list of human samples is described in Supplemental Table 1.
Animals
Mice (male and female) were used in compliance with- and with the approval of the Animal Care Committee of the Maisonneuve-Rosemont Hospital Research Center (Approval ID #2009-40; #2009-42; # 2011-23). The FAD mouse model (Tg-Thy-1APPSwDutIowa in C57BL/6 J background) was purchased from The Jackson laboratory21. Chk2+/− mice were obtained from N. Motoyama. p53+/− and Ink4a+/− mice were obtained from The Jackson Laboratory17,22. Bmi1+/− mice were a generous gift from M. van Lohuizen (Netherlands)14. All mice were maintained in the C57BL/6 J genetic background. Bmi1+/− mice were crossed with either Chk2+/− or p53+/− mice, to obtain Bmi1+/−/Chk2+/− or Bmi1+/−/p53+/− mice. Bmi1+/−/Chk2+/− or Bmi1+/−/p53+/− mice were then intercrossed to obtain Bmi1−/−/Chk2−/− or Bmi1−/−/p53−/− mice. From several breeding, we obtained the expected Mendelian ratio for Bmi1−/−/Chk2−/− mice (12.5%), but not for Bmi1−/−/p53−/− mice (1 animal out of 200 offspring or 0.5%), suggesting that the Bmi1/p53 double mutant is lethal23. A similar strategy was used to generate Bmi1−/−/Ink4a−/− mouse embryos in order to perform the neuronal cultures. Wild type (Bmi1+/+) and Bmi1−/− animals used for comparison were littermates of the double mutants.
Murine neuronal cultures
After dissociation of E18.5 cortices, cells were plated at 1.5 × 105 cells/well on poly-L-lysine-coated 8-well cultures slides (BD Biosciences). Cells were maintained in normal medium composed of Neurobasal-A Medium (Invitrogen), Glutamax-I (Gibco), gentamycin (50 µg/ml; Gibco), B27 supplement (Gibco), nerve growth factor (50 ng/ml; Invitrogen) and BDNF (0,5 ng/ml; Invitrogen). For the assays using inhibitors, we added 2 μM of DMSO, 2 μM of ATM/ATR inhibitor (CGK733; Millipore) or 10 μM of ATM inhibitor (KU55933; KuDOS Pharmaceuticals) to the cell culture medium for 16 hours. NAC was added daily at 5 mM into the culture medium for 7 days18.
Amyloid fractions and Immunoprecipitation
Cortices were minced and homogenized in a 10 ml TBS 1 × (50 mM Tris-Cl, pH 7.5. 150 mM NaCl)/protease inhibitor cocktail solution (Roche Diagnostics), using a tissue grinder. The homogenates were sonicated and ultra-centrifuged at 175,000 g for 1 h at 4 °C. The supernatants were collected as the soluble cellular fraction24. The dense pellet was re-homogenized in a 10 ml TBS 1×/1% Triton X-100/protease inhibitor cocktail solution, followed by sonication and centrifugation at 100,000 g for 1 h at 4 °C. The supernatants were recuperated as the insoluble cellular fraction24. For immunoprecipitation, protein samples and cellular fractions were incubated overnight with the primary antibody followed by incubation with the protein A/G beads (Millipore) for 2 hours the second day. Beads with bound immunocomplexes were washed with RIPA buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.1%Tween 20; protease inhibitors Complete) and bound proteins were subsequently heat eluted with 1x Laemmli buffer.
Western blot
Cortices were minced and homogenized in 5 ml of complete mini protease inhibitor cocktail solution (100 mM Tris-HCl, pH7.5; 150 mM NaCl; 0.1% Tween 20; 2% SDS; protease inhibitors Complete; Roche Diagnostics) using a tissue grinder. The homogenates were sonicated and quantified using the Bradford reagent. For Western Blot, proteins were resolved by SDS-PAGE electrophoresis and transferred to a nitrocellulose blotting membrane (Bio-Rad). Membranes were incubated with the primary antibodies, treated with the corresponding horseradish peroxidase-conjugated secondary antibodies (Sigma), and developed using the Immobilon Western (Millipore).
Immunohistochemistry
Tissues were immersed in Formalin or 4% paraformaldehyde/3% sucrose in 0.1 M phosphate buffer, pH 7.4 for 16 h at room temperature on a rocking platform. Samples were washed three times in PBS and embedded in paraffin according to standard protocols. 8 µm thick sections were mounted on Super-Frost glass slides (Fisher Scientific) and processed for immunohistochemistry staining. Paraffin-embedded slices were analyzed by using the Vectastain® ABC kit (Vector) according to the manufacturer instructions. Peroxidase substrates used are the Vector® VIP (Violet) (Vector), and DAB (brown) (Sigma)18. Observations were made using the Zeiss imager Z2 microscope and images were captured with a digital camera. To obtain comparable results, we selected pyramidal neurons from the frontal cortex to quantify the number of chromocenter/neuron. Pyramidal neuron identity was determined by the expression of NeuN, the cell morphology (using phase contrast imaging), the large nuclear diameter (when compared to astrocytes), and the presence of chromocenters and of a large nucleolus.
Senescence-associated β-galactosidase assay
Senescence-associated (SA) β-galactosidase staining was detected histochemically at pH 6 as described previously25. Briefly, tissue slices were fixed in PBS containing 1% formaldehyde, 0.4% glutaraldehyde, and 0.02% Igepal. After three washes, slices were exposed to the X-gal solution (1 mg/ml X-gal; 5 mM K3Fe(CN)6; 5 mM K4Fe(CN)6; 1 mM MgCl2, in PBS; pH 6.0). Slides were mounted and observations made under a microscope.
Antibodies
Rabbit anti-p53 (Santa Cruz Biotechnology), rabbit anti-p-JNK (Invitrogen), mouse anti-synaptophysin (Sigma), mouse anti-amyloid clone DE2B4 (Abcam), rabbit anti-amyloid clone FCA3542 (Calbiochem), mouse anti-amyloid MOAB2 (Novus; NBP2-13075)), pan anti-Tau k9JA (DAKO); mouse anti-p-Tau clone AT-8 (Thermo scientific), mouse anti-p-Tau clone PHF1 (a gift from Dr. Davies, Albert Einstein College of Medicine), mouse anti-NeuN (Chemicon), rabbit anti-cleaved caspase-3 (Cell Signaling), rabbit anti-GFAP (Dako; Z0334), Iba1 (Wako; 019-19741), mouse anti-Bmi1 clone F6 (Millipore), mouse anti-Bmi1 (Abcam), mouse anti-p-ATM (Novus), rabbit anti-p-ATR (Santa Cruz Biotechnology), rabbit anti-H3K9me3 (Abcam), mouse anti-H2Aub clone E6C5 (Millipore), mouse anti-HP1 (Millipore), mouse anti-β-actin (Sigma), mouse anti-tubulin (Sigma), mouse anti-H3 (Upstate), and rabbit anti-mouse IgG (Upstate).
RT and real-time PCR
All primers were designed to flank individual exons and tested by PCR in RT+ and RT− control extracts. Total RNA was isolated using TRIzol reagent (Invitrogen). The RNA was treated with DNaseI for 20 min prior to reverse transcription (RT) and PCR for satellite repeats, which are intron-less. Reverse transcription was performed using 1 µg of total RNA and the MML-V reverse transcriptase (Invitrogen). Real-time PCR was performed using the Platinum SYBRGreen SuperMix (Invitrogen) and a Real-Time PCR apparatus (BioRad). Primers sets used were as follow: p16Ink4a (F) 5_-CAACGCCCCGAACTCTTTC-3_; (R) 5_-GCAGAAGAGCTGCTACGTGAAC-3_, p19Arf (F) 5_-GGCTAGAGAGGATCTTGAGAAGAGG-3_; (R) 5_-GCCCATCATCATCACCTGGTCCAGG-3_. Primers sets used for Bmi1, IAP, Maj Sat, Min Sat, Line, Sine and IAP were as described19.
Chromatin Immunoprecipitaion (ChIP) assay
ChIP was performed using the ChIP Assay kit (Upstate). Briefly, 50 mg of cortical tissue was frozen for 1 hour and then homogenized at RT according to the manufacturer’s protocol. The tissue was sonicated on ice for 10 sec at 30% amplitude to shear the chromatin (Branson Digital Sonifier 450, Crystal Electronics, On. Canada). Sonicated materials were immunoprecipitated using designated antibodies. Fragments were then amplified by real-time PCR in triplicates. Human primers sets used were as follow: MCBOX (F) 5′-AGGGAATGTCTTCCCATAAAAACT-3′; (R) 5′-GTCTACCTTTTATTTGAATTCCCG-3′; SATIII (F) 5′-AATCAACCCGAGTGCAATCNGAATGG-3′; (R) 5′-TCCATTCCATTCCTGTACTCGG-3′; SATa (F) 5′-AAGGTCAATGGCAGAAAAGAA-3′; (R) 5′-CAACGAAGGCCACAAGATGTC-3′; ACTIN (F) 5′-CCTCAATCTCGCTCTCGCTC-3′; (R) 5′-CTCTAAGGCTGCTCAATGTCA-3′; ß-GLOBIN (F) 5′-GGCTGTCATCACTTAGACCTC-3′; (R) 5′-GGTTGCTAGTGAACACAGTTG-3′;5′. Mouse primers sets used were as follow: MajSAT 5′- GGCGAGAAAACTGAAAATCACG-3′, 5′-CTTGCCATATTCCACGTCCT-3′; MinSAT 5′- TTGGAAACGGGATTTGTAGA-3′, 5′-CGGTTTCCAACATATGTGTTTT-3′; LINE 5′-TGGCTTGTGCTGTAAGATCG-3′, 5′-TCTGTTGGTGGTCTTTTTGTC-3′; SINE1 5′-GAGCACACCCATGCACATAC-3′, 5′-AAAGGCATGCACCTCTACCACC-3′; Actin 5′-TCGATATCCACGTGACATCCA-3′; 5′-GCAGCATTTTTTTACCCCCTC-3′; HoxA7 5′-GTGGGCAAAGAGTGGATTTC-3′; 5′-CCCCGACAACCTCATACCTA-3′; β-major 5′-CAGTGAGTGGCACAGCATCC-3′; 5′-CAGTCAGGTGCACCATGATGT-3′. ChIP-qPCR data was analyzed according to the Percent Input method19. First, the raw Ct of the diluted 1% input fraction is adjusted by subtracting 6.64 cycles (i.e. log2 of the dilution factor 100). Subsequently, the percent input of each IP fraction is calculated according to this equation: 100*2(Adjusted Input Ct-Ct(IP).
Behavioral tests
Behavioral assays were performed at the Douglas Hospital Institution, Montreal, Canada under the supervision of Dr. Rocheford. Male mice were first accustomed to the test environment prior to testing. Groups of 15-month old male WT (n = 8) and Bmi1+/− (n = 8) mice were tested successively and were habituated, trained, and evaluated at the same time of day for each experiment. Behavioral tests included Morris water maze test and Barnes maze assay. In the Morris water maze assay, a hidden escape platform is immersed just below water level with visual cues (color) around the pool in plain eyesight of the mouse. Upon release, the mouse navigates around the pool seeking an exit while various parameters are recorded, including the time spent in each quadrant of the pool and the time taken to reach the platform (latency). Barnes maze assay includes a circular platform containing 20 circular holes around its circumference and visual cues (colored shapes) surrounding the surface in plain eyesight of the mouse. The platform is intensely lit by an overhead lamp, and underneath one of the holes there is an exit box. Different parameters are analyzed comprising latency to escape and number of errors.
LTP assay
For this study, we used 15-month old male WT and Bmi1+/− mice. Under deep anesthesia, brains were removed and coronal brain slices (350 µm thickness) containing the hippocampus were cut in hyperosmotic, ice-cold and carbogenated (bubbled by 95%O2/5%CO2 to maintain the pH at 7.4) slice cutting solution (in mM: 252 sucrose, 2.5 KCl, 4 MgCl2, 0.1 CaCl2, 1.25 KH2PO4, 26 NaHCO3 and 10 glucose) using a Vibratome. Freshly cut slices were placed in an incubating chamber with carbogenated artificial cerebrospinal fluid (ACSF (~310 mOsmol/L) consists of (mM): 125 NaCl, 2.5 KCl, 1 MgCl2, 2 CaCl2, 1.25 NaH2PO4, 26 NaHCO3 and 25 glucose). Slices were recovered at 32 °C for one hour and subsequently maintained at room temperature. Carbogenated ACSF containing bicuculline methbromide (5 µM) to block GABAA receptor-mediated inhibitory synaptic current was used to perfuse slices in all recordings. Postsynaptic responses, which were evoked by stimulating the Schaffer collateral-commissural pathway via constant current pulses (0.08 ms) delivered through a tungsten bipolar electrode (FHC), were recorded from the hippocampal CA1 region, amplified by a Multiclamp 700B (Axon) and stored in a PC for offline analysis using Clampfit (Axon). All recordings were performed at room temperature. Field excitatory postsynaptic potential (fEPSP) was evoked at 0.05 Hz and detected by an ACSF-filled glass electrode that was placed in the stratum radiatum of the hippocampal CA1 region. Long-term potentiation (LTP) of fEPSP was induced by a high-frequency tetanus (100 Hz, 100 pulses). Student’s t-test was used to compare changes in fEPSP slope at 60 minutes after tetanus between WT and Bmi1+/− groups. In order to determine if fEPSPs are potentiated by tetanus within each mouse group, paired Student’s t-test was used to compare fEPSP slope recorded before (baseline) and at 60 minutes after tetanus.
Statistical analysis
The Student’s t-test for unpaired samples was used to measure statistical differences. An analysis of variance (ANOVA) followed by the Dunnett test was used for multiple comparisons with one control group. In each figures, the criterion for significance (P value) was mentioned.
Results
Bmi1+/− mice develop accelerated aging features and neurological symptoms
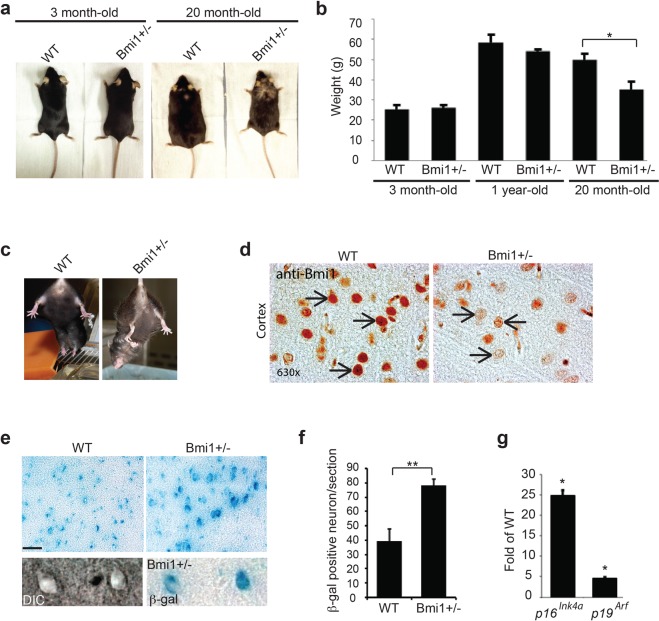
We previously reported that Bmi1+/− mice have reduced median and maximal lifespan along with apparition of lens cataracts at higher frequencies than WT littermates, suggesting that Bmi1 hemi-deficiency may accelerate the aging process18. Alopecia and weight loss are common characteristics of premature aging in mice26–30. We observed that between the ages of 15–24 months (defined here as old mice), Bmi1+/− mice exhibited alopecia and reduced body size (~25%) when compared with age-match WT littermates (Fig. 1a,b). These pathological features were not present in 3-month and 1-year old Bmi1+/− mice (Fig. 1a,b). We performed the paw-clasping test to verify if old Bmi1+/− mice presented pathological reflexes. Adult WT mice picked by the tail and slowly inclined towards a horizontal surface spread all four limbs in anticipation of contact. However, instead of limb extension, old Bmi1+/− mice displayed an irregular clasping behavior characterized by the dragging of the fore paws towards the trunk and asymmetry in limb position (Fig. 1c). This phenotype is generally associated with a neurological syndrome31.
Figure 1.
Bmi1+/− mice present accelerated aging features and neurological symptoms. (a) Photographs of 3 month-old and 20 month-old WT and Bmi1+/− mice. Note that aged Bmi1+/− mice are smaller and exhibit hair loss (alopecia) compared to WT littermates (n = 5). (b) Quantification representing the mean weight at 3 months (n = 6), 1-yr (n = 6) and 20 months (n = 6) of WT and Bmi1+/− mice. (c) Photographs showing the paw clasping reflex of aged WT (n = 5) and Bmi1+/− mice (n = 5). Note that the Bmi1+/− mouse pulls its fore limbs into its body rather than spreading them away from its trunk, and present an important asymmetry in the hind limbs’ position. (d) IHC analysis on mouse cortical sections showing robust Bmi1 expression in the nucleus of WT neurons (arrows) and weak expression in Bmi1+/− neurons (arrows). (e) Analysis of SA β-galactosidase activity in the cortices of old WT and Bmi1+/− mice. Scale bar: 20μm. (f) Quantification of the number of SA β-galactosidase positive neurons/cortical section. (h) Quantitative PCR analysis of p16Ink4a and p19Arf expression in the cortices of old WT (n = 3) and Bmi1+/− (n = 3) mice.
The Bmi1 gene operates as a central regulator of apoptosis and senescence through repression of the p19Arf/p53 and p16Ink4a/pRb tumor suppressor axes. Senescence is usually excluded in post-mitotic cells such as neurons, yet increased SA β-galactosidase activity and expression of the senescence-associated genes p19Arf and p16Ink4 have been noted in cortical neurons of aged p73+/− mice and of Bmi1−/− pups18,25,32. Furthermore, expression of p16Ink4 is considered as a biomarker of aging33. We first confirmed neuronal expression of Bmi1 in the cortex of old WT mice and its severe reduction in old Bmi1+/− mice (Fig. 1d). Next, we compared these old WT and Bmi1+/− mice for SA β-galactosidase activity and p16Ink4/p19Arf expression. We observed robust SA β-galactosidase staining in pyramidal neurons and a 2-fold increase in the total number of SA β-galactosidase-positive neurons/section in layers 2 and 3 of the frontal cortex of Bmi1+/− mice (Fig. 1e,f). This was accompanied with significant increased in p16Ink4 and p19Arf expression (Fig. 1g). These findings suggested that in addition to affect lifespan, Bmi1 hemi-deficiency resulted in the apparition of an age-dependent neurological syndrome associated with neuronal senescence.
Bmi1+/− mice show altered spatial memory and reduced long-term potentiation
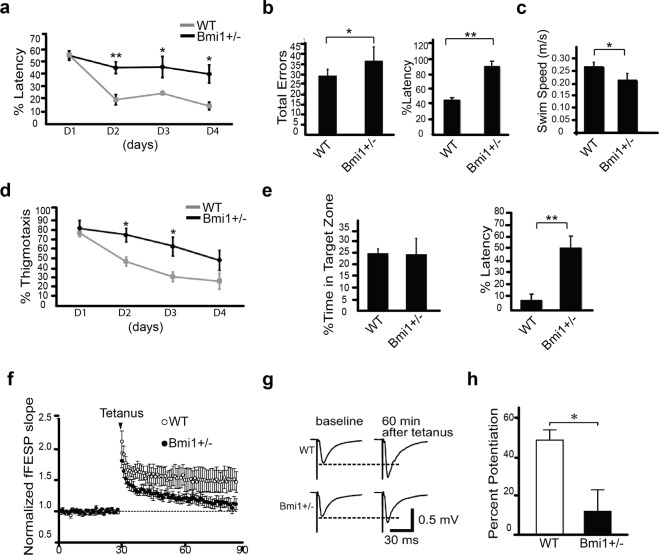
To test whether the neurological symptoms were associated with perturbed cognitive functions, we examined apparently healthy 15 month-old WT and Bmi1+/− mice for spatial learning and memory aptitude using the Morris water and Barnes maze tests. For the Morris water navigation task, mice were trained for 4 days to locate the platform (training) and then evaluated on their capacity to remember the platform location (probe test). Bmi1+/− mice performed poorly in this assay during both the training period and probe test, as demonstrated by the increased time (latency) required to reach the platform (Fig. 2a,b). Bmi1+/− mice also exhibited an anxious phenotype as demonstrated by an increased thigmotactic behavior while performing the training (Fig. 2d)34. The swimming speed of aged Bmi1+/− mice was also reduced (0.25 m/s for WT and 0.20 m/s for Bmi1+/−) (Fig. 2c). In contrast, the swimming speed of 1 year-old WT and Bmi1+/− mice was nearly identical (not shown), thus consistent with the apparition of a progressive, age-related reduction in locomotors strength. The Barnes maze test is based on mice aversion to open enlightened spaces, which ultimately prompts them to find secure shelter. Bmi1+/− mice were more prone to visiting non-target holes while searching for the hole with the escape box and the latency of escape was also increased (Fig. 2e). Although these results suggested impaired spatial memory formation, the reduction in swimming speed and the possibility of visual dysfunction23 led us to perform electrophysiological analyses to further test this possibility.
Figure 2.
Bmi1+/− mice exhibit memory and LPT impairment. (a–e) 15-month old male WT (n = 8) and Bmi1+/− (n = 8) mice were tested for behavioral changes associated with spatial memory formation deficit. (a–c) Results of Morris water maze probe test, showing time spent in the target quadrant searching for the hidden platform, mean time to reach the target platform (latency) and mean swim speed. (d) Charts representing mean thigmotaxis of Morris water maze 4-day trials. (e) Histograms representative of Barnes maze test results including mean total errors and mean latency to reach the target probe. (f–h) LTP assays on hippocampal slices of 15-month old male WT and Bmi1+/− mice. (f) Scatter plots revealed changes in the slope of field excitatory postsynaptic potential (fEPSP) recorded from the hippocampal CA1 region of WT and Bmi1+/− mice. LTP was induced by tetanus (100 Hz, 100 stimulation). Dotted line represents the baseline. (g) Representative traces of fEPSP recorded before (baseline) and 60 min after tetanus. Dotted lines represent baselines. (h) Histogram of fEPSP slope at 60 min after LTP induction. Normalized fEPSP slopes, recorded between 55–60 min after tetanus, were averaged for calculating percent potentiation. Values are mean ± SEM. *P < 0.05; **P < 0.01; Student’s t-test.
LTP is a cellular surrogate for spatial memory formation35. Using the same animals as in the behavioral tests, LTP of the Schaffer collateral pathway was induced in the CA1 region of the hippocampus. We found that a commonly used protocol for inducing LTP (100 Hz, 100 pulses) enhanced fEPSP slope in WT mice at 60 minutes after tetanus (Percent potentiation: 48.9 ± 5.2%, data from 6 slices and 3 mice). Compared with fEPSP recorded before tetanus (baseline), WT slices displayed significant potentiation (P = 0.036; paired Student’s t-test) (Fig. 2f–h). Slices from Bmi1+/− mice exhibited weaker potentiation than WT (11.7 ± 11.2%, data from 6 slices, 4 mice; transgenic vs. WT: P = 0.045, Student’s t-test) and failed to show significant potentiation when compared to baseline at 60 minutes after tetanus (P = 0.357; paired Student’s t-test) (Fig. 2f–h). Our findings demonstrated that LTP formation in Bmi1 hemi-deficient mice was significantly impaired, thus strengthening the hypothesis of impaired spatial memory formation.
Old Bmi1+/− mice present a pathology showing some features of AD
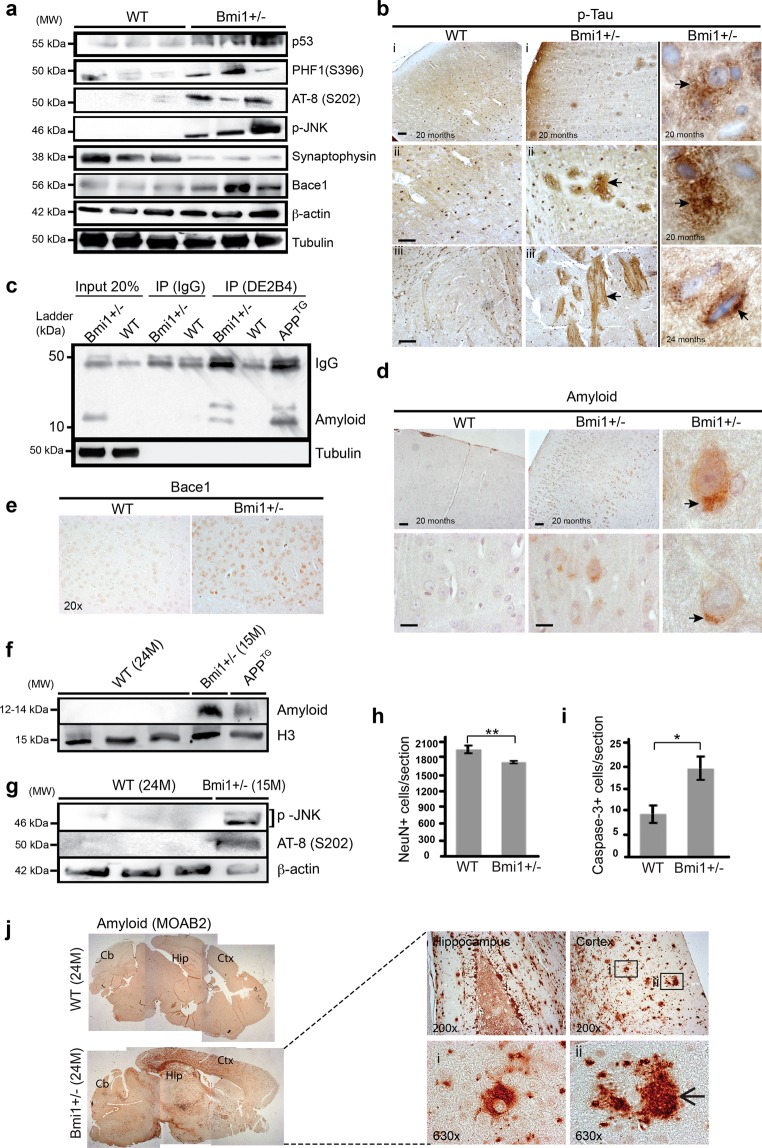
We previously showed that most neuronal anomalies in Bmi1−/− mice were mediated by p53 activation18. Using immuno-blot, we observed increased p53 levels in the cortex of 15–20 month-old Bmi1+/− mice when compared to WT littermates (Fig. 3a). Hallmarks of AD and related dementias are accumulation of p-Tau, extra-cellular amyloid plaques, reduced synaptophysin immunoreactivity and neuronal loss. By immunoblot using antibodies that recognize p-Tau at Ser202 (AT-8) or Ser396 and Ser404 (PHF1), we observed the presence of p-Tau accumulation in Bmi1+/− cortices (Fig. 3a). In cultured human neurons, BMI1 knockdown results in an increase in total Tau levels20. Similarly, we observed that total Tau levels were increased in the cortex of 22–24 month-old Bmi1+/− mice (Fig. S1a,b). Remarkably, these anomalies correlated with reduced synaptophysin levels (indicative of synaptic atrophy) and increased p-JNK immunoreactivity (Fig. 3a)36,37. By immuno-histochemistry (IHC), we observed p-Tau immunoreactivity in the neuron’s perikarya of 20 month-old WT and Bmi1+/− mice. However, large p-Tau deposits in the neuronal cell body and strong immunoreactivity in track fibers of the cortical white matter were only observed in Bmi1+/− mice (Fig. 3b). The presence of p-Tau tangles was not observed.
Figure 3.
Old Bmi1+/− mice present cortical neurodegeneration. (a) Western blot analysis on cortical extracts from 15–20 month-old WT and Bmi1+/− mice. (b) IHC for p-Tau (PHF1) on cortical sections of old WT (n = 4) and Bmi1+/− (n = 4) mice. (i) Low magnification. Scale bar: 20μm. Higher magnifications in (ii) revealed p-Tau deposits on ghost-like neurons and in (iii) p-Tau immunoreactivity in track fibers of the cortical white matter of Bmi1+/− mice. The 3 insets show images of the most severe cases observed of p-Tau accumulation in Bmi1+/− mouse neurons (arrows). Scale bar: 8μm. (c) Immuno-precipitation (IP) of the β-amyloid peptide (DE2B4 antibody) using the soluble cellular fraction of cortices from old WT and Bmi1+/− mice, and from a 6-month old APP transgenic mouse (positive control). The blot was revealed using the FCA3542 antibody. N = 2 independent experiments. (d) IHC on brain sections of old WT (n = 4) and Bmi1+/− (n = 4) mice for the expression of β-amyloid (DE2B4 antibody). Top images scale bar is 20μm; Lower images scale bar is 8μm. Arrows in the insets indicate amyloid accumulation in the neuronal cell body. (e) IHC staining for Bace1 on brain sections of old WT (n = 2) and Bmi1+/− (n = 2) mice. (f,g) Immuno-blot analysis of cortical extracts from 24 month-old WT and 15 month-old Bmi1+/− mice showing accumulation of pathological proteins in Bmi1+/− mice but not in old WT mice. N = 2 independent experiments. (h) Quantification of the number of NeuN + cortical neurons in the frontal cortex of 15–20 month-old WT (n = 5) and Bmi1+/− (n = 5) using IHC. (i) Quantification of the number of neurons positive for activated caspase-3 in the frontal cortex of 15–20 month-old WT (n = 5) and Bmi1+/− (n = 5) mice. (j) Composite images of IHC staining on whole brain sections from 24-month old WT and Bmi1+/− mice showing amyloid-positive deposits in the cortex (Ctx) and hippocampus (Hip) but not in the cerebellum (Cb) of Bmi1+/− mice. (Right panel) High-resolution images showing extra-cellular amyloid plaque-like structures (arrow) in the cortex and hippocampus of Bmi1+/− mice. Values are mean ± SEM. *P < 0.05; **P < 0.01; Student’s t-test.
We next performed immunoblot on mouse cortices using an antibody (FCA3542) that can recognize the mouse and human amyloid peptide. Notably, we could detect the presence of a ~12–14 kDa band only in the Bmi1+/− brain samples (Fig. 3c-Input 20%). This band was also revealed using FCA3542 after immunoprecipitation (IP) with another anti-amyloid antibody (DE2B4), but not after IP with IgG (Fig. 3c-IP). Antibody specificity was also confirmed using cortical extracts from human APPSwe/Dutch/Iowa (APP) transgenic mice (Fig. 3c-IP). On Bmi1+/− brain sections at 20 months of age, we observed that amyloid was preferentially found in the neuronal soma (Fig. 3d). Amyloid plaques were not observed. Cleavage of APP by the β-secretase Bace1 generates the ~12–14 kDa C99 fragment (C99). Notably, we observed increased Bace1 levels in cortical neurons of Bmi1+/− mice (Fig. 3a,e). To distinguish between accelerated aging and neurodegeneration, we compared the brains of 24 month-old WT mice with that of 15 month-old Bmi1+/− mice for the expression of the ~12–14 kDa fragment. Notably, the amyloid-related band was not detectable in the cortex of old WT mice (Fig. 3f). Likewise, p-JNK and p-Tau (AT-8) were not observed in the 24 month-old WT mice (Fig. 3g).
To evaluate the possibility of neuronal loss, we calculated the total number of cortical neurons per section in the frontal cortex using a pan-neuronal marker (NeuN). When compared to WT littermates, 15–20 month-old Bmi1+/− mice displayed a ~20% reduction in NeuN-positive neurons (Fig. 3h). To examine if the lower neuronal density observed in Bmi1+/− mice correlated with increased apoptosis, we quantified the number of neurons positive for activated caspase-3 in the frontal cortex. This revealed a ~2 fold increase in the frequency of apoptotic neurons in old Bmi1+/− mice (Fig. 3i). Reduced neuronal density and increased apoptosis were also observed in the hippocampus of 15–17 month-old Bmi1+/− mice (Fig. S2a,b), which was consistent with the behavioral and LTP results. We next analyzed the brains from rare surviving 22–25 month-old Bmi1+/− mice using antibodies against amyloid (n = 7). A single 24 month-old Bmi1+/− animal presented amyloid deposits that were present in all cortical areas, with other brain regions being generally less affected (Fig. 3j). Interestingly, the pattern of amyloid immuno-reactivity was slightly distinct when using another amyloid antibody (Fig. S3a,b). Intra-neuronal accumulation of amyloid and reduced synaptophysin immuno-reactivity was also observed in the hippocampus of another Bmi1+/− mouse (Fig. S3c,d). This revealed that old Bmi1+/− mice could spontaneously develop a disorder sharing some similarities with AD.
Bmi1 hemi-deficiency cooperates with APP-mediated neurodegeneration
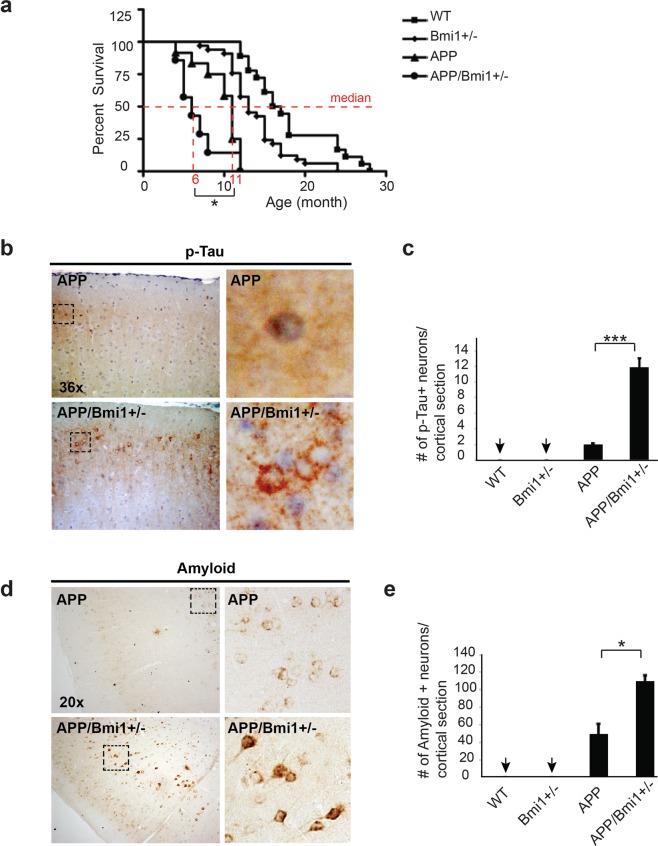
Genetic cooperation between two mutations can be indicative of protein-protein interactions and/or convergent molecular pathways. To test if Bmi1 hemi-deficiency could cooperate or not with FAD-associated mutations, we crossed Bmi1+/− mice with human APP transgenic mice. From this, we obtained 4 genotypes in F1: WT, Bmi1+/−, APP and APP/Bmi1+/− mice. Kaplan Mayer analysis revealed that although both Bmi1+/− and APP mice had reduced maximal and median lifespan compared to WT littermates, the median lifespan of the APP/Bmi1+/− animals was further reduced by ~45% when compared to APP mice (Fig. 4a). The difference was significant despite the limitations of using a small data set. When analyzed by IHC at 6 months of age for p-Tau, we observed almost no signal in WT and Bmi1+/− mice, and weak immunoreactivity in APP mice. Notably, while neuronal accumulation of p-Tau was rare in APP mice and absent in Bmi1+/− mice, it was relatively robust and abundant in APP/Bmi1+/− mice (Fig. 4b,c). Similarly, although intra-neuronal amyloid immunoreactivity was weak to absent in Bmi1+/− mice at this young age, it was easily detectable in APP mice (Fig. 4d). Notably, the number of immunoreactive neurons and the signal strength for amyloid were greatly enhanced in APP/Bmi1+/− mice (Fig. 4d,e). Other parameters of disease severity were also enhanced in APP/Bmi1+/− mice, including reactive gliosis, microglia activation, synaptic atrophy and apoptosis (Fig. S4). Taken together, these results suggested genetic cooperation between Bmi1 hemi-deficiency and mutant APP in neurodegeneration.
Figure 4.
APP/Bmi1+/− mice show genetic cooperation in disease onset and severity. (a) Kaplan-Meyer representation of the survival curves of WT (n = 11), Bmi1+/− (n = 13), APP (n = 6) and APP/Bmi1+/− (n = 6) mice. (b) IHC staining for p-Tau (PHF1) on cortical sections of 6 month-old WT (n = 3), Bmi1+/− (n = 3), APP (n = 3),and APP/Bmi1+/− (n = 3) mice. Scale bar: 20μm. Scale bar in the inset: 8μm. (c) Quantification of p-Tau-positive neurons shown in (b). (d) IHC for amyloid (DE2B4) on cortical sections of 6-month old WT (n = 3), Bmi1+/− (n = 3), APP (n = 3), and APP/Bmi1+/− (n = 3) mice. Scale bar: 20μm. (e) Quantification of amyloid-positive cells shown in (d). Values are mean ± SEM. *P < 0.05; (**) < 0.01; (***) < 0.001; Student’s t-test.
Loss of heterochromatin and DDR in cortical neurons of Bmi1+/− mice
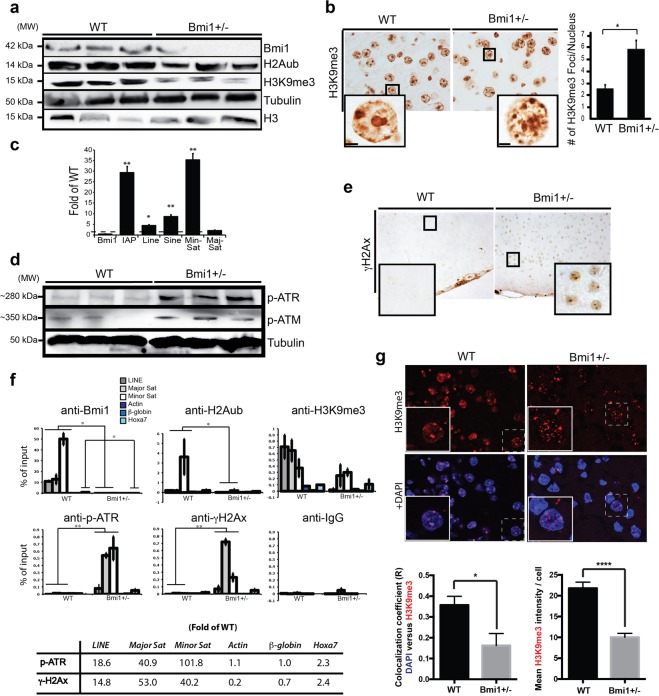
BMI1 is required for heterochromatin maintenance in somatic cells19. We thus investigated if the AD-like phenotype of Bmi1+/− mice was also associated with heterochromatin alterations. By Western blot analysis on whole cortices, we observed reduced levels for histones H2Aub and H3K9me3 in 15 month-old Bmi1+/− mice (Fig. 5a). To test if this correlated with an abnormal chromatin structure, we performed IHC. We observed that in contrast to WT mice, where 2-3 large chromocenters/neuron were present, chromocenters of Bmi1+/− neurons were smaller and more numerous, suggesting heterochromatin de-nucleation (Fig. 5b). Consistently, this was associated with transcriptional de-repression of satellite repeats, which are normally silenced by heterochromatin (Fig. 5c)38–40. One prediction of heterochromatin de-nucleation is activation of a DDR. The Ataxia Telengectasia Mutated (ATM) and Ataxia Telengectasia and RAD-3 Related (ATR) kinases work at the apex of the DDR41–43. Using immunoblot, we observed accumulation of phospho-ATM at Serine 1981 (p-ATM) and phospho-ATR at Serine 428 (p-ATR) in the cortices of Bmi1+/− mice (Fig. 5d). The histone mark γH2AX is present at sites of DNA damage44. We observed nuclear accumulation of γH2AX in cortical neurons only in Bmi1+/− mice, suggesting that the DDR occurs primarily in neurons (Fig. 5e).
Figure 5.
Loss of heterochromatin and DDR in cortical neurons of Bmi1+/− mice. (a) Western blot analysis of cortical extracts from old WT (n = 6) and Bmi1+/− (n = 6) mice. (b) IHC on cortical sections from old WT (n = 3) and Bmi1+/− (n = 3) mice with quantification of the number of H3K9me3 foci per nucleus (6 sections/sample were counted). Scale bar in the inset: 3μm. (c) Real-time RT-PCR analysis for the expression of satellite repeats in cortices from WT (n = 3) and Bmi1+/− (n = 3) mice. (d) Western blot analysis of cortical extracts from old WT (n = 3) and Bmi1+/− (n = 3) mice. (e) IHC for γH2AX on cortical sections of old WT and Bmi1+/− mice. (f) ChIP analyses were performed on frontal cortex homogenates from 15 month-old WT (n = 3) and Bmi1+/− (n = 3) mice. Quantitative PCR was performed in triplicate for each DNA sequence. All data are presented as fold of input. Bmi1 and H2Aub reduction (p < 0.05), and p-ATR and γH2AX accumulation (p < 0.01) in Bmi1+/− mice are significant (Two-way ANOVA analysis). (bottom panel) Fold differences for γH2AX and p-ATR accumulation in Bmi1+/− mice were measured relative to WT levels for each DNA sequences. (g) Cortical slices from 45-day old mice. Areas showed are located in layers 2-3 of the frontal cortex. In WT samples, large pyramidal neurons were strongly labeled by the H3K9me3 antibody and H3K9me3 co-localized with DAPI. In Bmi1+/− samples, large pyramidal neurons were weakly labeled by the H3K9me3 antibody and the H3K9me3 signal was fragmented. Bottom panels: Quantification of the images showed in (f). Values are mean ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001; Student’s t-test.
To localize DNA damage on the genome, we performed chromatin immuno-precipitation (ChIP) experiments on whole cortices. As expected, Bmi1, H3K9me3 and H2Aub were enriched at genomic repeats (Minor and Major satellite repeats and LINE elements) and at the Hoxa7 locus in WT mice and were depleted in Bmi1+/− mice (Fig. 5f)19. In contrast, while p-ATR and γH2AX were nearly absent in WT mice, they were preferentially and highly enriched at genomic repeats in Bmi1+/− mice (Fig. 5f).
To test if heterochromatin anomalies were present before disease onset, we performed immuno-blot, IHC and confocal microscopy on cortices from 2-3 month-old WT and Bmi1+/− mice. This revealed reduced H3K9me3 levels and fragmented H3K9me3 immunostaining in Bmi1+/− pyramidal neurons when compared to WT (Figs 5g and S5a,b). Reduced H3K9me3 levels were also present in cultured embryonic day 18.5 neurons, as measured using ChIP analyses (Fig. S5c). Despite the presence of chromatin anomalies at the earliest step of neurogenesis, we failed to detect a DDR or the expression of AD-related markers in 3 month-old Bmi1+/− mice using immuno-blot (Fig. S5d). Mild accumulation of p53 was however noticed in Bmi1+/− mice (Fig. S5d). This suggested that in Bmi1+/− mice, loss of heterochromatin is a very early event in the disease process.
Amyloid and Tau pathologies can be mitigate by inhibition of the DDR
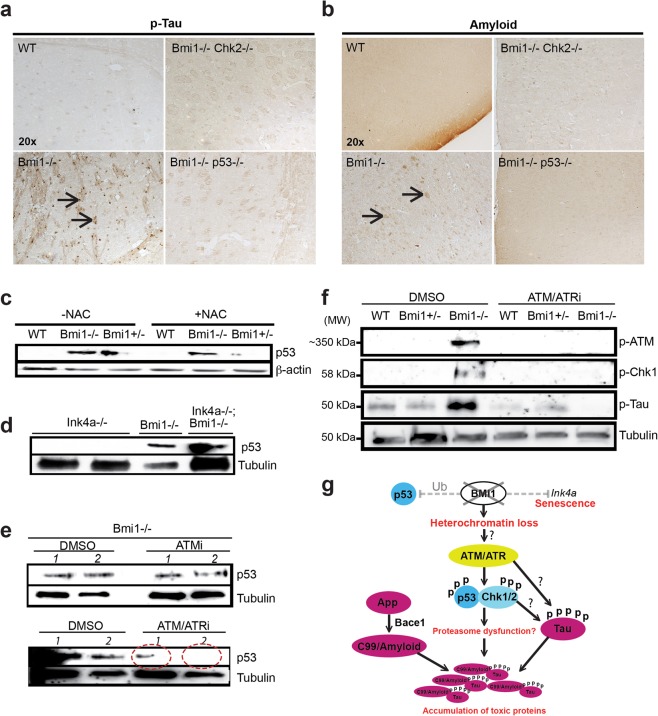
Bmi1-null (Bmi1−/−) mice are smaller than normal and die around post-natal day 3012. Yet, we could find cortical neurons immuno-reactive for p-Tau and amyloid in 25 day-old Bmi1−/− mice (Fig. 6a,b). To gain insight into the disease mechanism, we generated cohorts of 25 day-old Bmi1−/− mice carrying null mutations in p53 (Bmi1−/−/p53−/−) or Chk2 (Bmi1−/−/Chk2−/−). Chk2 encodes a kinase activated by ATM, and both ATM and Chk2 can stimulate p53 activity45,46. Notably, we found that p-Tau and amyloid immuno-reactivity were both mitigated in Bmi1−/−/p53−/−and in Bmi1−/−/Chk2−/− mice (Fig. 6a,b).
Figure 6.
Blocking the DDR mitigate the amyloid and Tau phenotype in Bmi1-null neurons. (a,b) IHC performed on cortical sections from 25 day-old mice. (a) Arrows indicate p-Tau-positive neurons in Bmi1−/− mice. (b) Arrows indicate amyloid-positive neurons in Bmi1−/− mice. These anomalies were largely prevented in the double-mutant mice. (c–f) Immunoblot were performed on embryonic day 18.5 mouse cortical neurons cultured for 7 days in vitro. (c–d) Addition of NAC or co-deletion of Ink4a could not prevent p53 accumulation in Bmi1-deficient neurons. Note the presence of p53 also in Bmi1+/− neurons. NAC was added daily to the cultures for a period of 7 days. (e,f) DMSO, ATMi or ATM/ATRi were added to the cultures 16 hours prior to protein extraction. (e) ATM/ATRi largely prevented p53 accumulation in Bmi1−/− neurons. Numbers 1 and 2 in (e) indicate two independent biological replicates. Note residual p53 expression in ATM/ATRi-treated neurons (circles with dash lines). (f) Addition of ATM/ATRi could prevent p-ATM, p-Chk1 and p-Tau accumulation in Bmi1−/− neurons. (g) Hypothetical model to explain the impact of Bmi1 inactivation in mouse neurons. Following loss of Bmi1, up-regulation of the Ink4a locus induces neuronal senescence. On the other hand, genomic instability at the heterochromatin induces a chronic DDR. The p53 protein is then stabilized by ATR/ATM-mediated kinase activity. The accumulation of p53 may be also exacerbated by the loss of Bmi1/Ring1a-mediated ubiquitin ligase and antioxidant activities. The activated ATM/Chk2, ATR/Chk1 and p53 pathways then promote Tau phosphorylation and accumulation of the C99 fragment. Elevated Bace1 levels also contribute to over-production of the C99 fragment. Accumulation of these toxic proteins contributes to neurodegeneration.
To understand the basis of p53 activation, we cultured WT, Bmi1−/− and Bmi1+/− embryonic day 18.5 cortical neurons to test if free radical-mediated DNA damage was responsible for p53 accumulation18,47,48. Using N-Acetyl Cysteine (NAC), a free radical scavenger, we found that reducing free radicals could slightly improve p53 levels in Bmi1−/− and Bmi1+/− neurons (Fig. 6c). The p19Arf protein can stabilize p53, and p19Arf is up regulated in Bmi1−/− neurons18,49. However, co-deletion of Bmi1 and Ink4a (encoding for p16Ink4a and p19Arf) could not prevent p53 accumulation (Fig. 6d). In response to DNA damage, the ATM and ATR kinases can target p53 for phosphorylation45,50,51. We found that inhibition of ATM alone could not prevent p53 accumulation, but that inhibition of ATM and ATR largely mitigated p53 accumulation in Bmi1−/− neurons (Fig. 6e). Residual p53 levels were however observed. We also observed the presence of p-ATM and phospho-Chk1 (a downstream target of ATR) in Bmi1−/− neurons and found that ATM/ATRi was sufficient to prevent their accumulation (Fig. 6f). Remarkably, p-Tau accumulation in Bmi1−/− neurons could be also prevented after exposure to ATM/ATRi (Fig. 6f). These results suggested a theoretical model where AD-related neuronal pathologies observed in Bmi1-deficient mice are triggered in part by a ATM/ATR-driven DDR working upstream of p53, but where Bmi1/Ring1 ubiquitin ligase activity and Bmi1-mediated ROS repression are also likely required to prevent p53 accumulation (Fig. 6g)52.
Loss of heterochromatin and DDR in AD brains
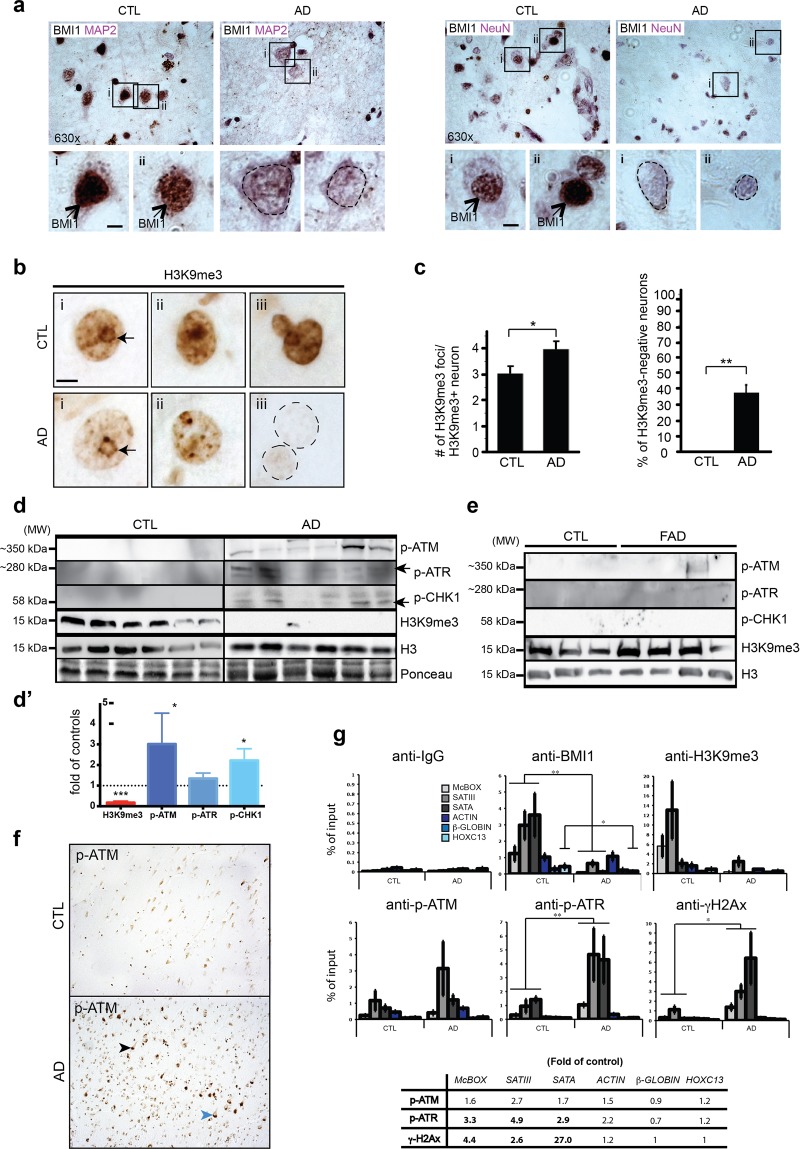
We next investigated the possible relevance of our findings in the context of AD. We first confirmed the robust expression of BMI1 in cortical neurons from aged controls and BMI1 depletion in neurons from AD patients (Fig. 7a). To test for possible heterochromatin anomalies, we performed IHC on frontal cortex sections53. In control samples, all neurons were labeled with H3K9me3 and presented ~3 heterochromatic foci (or chromocenters)/neuron (Fig. 7b,c). In AD samples, ~38% of all pyramidal neurons showed highly reduced or absent H3K9me3 nuclear staining, and the neuron’s chromocenters were smaller and more numerous (Fig. 7b,c). To test the possibility of genomic instability, we analyzed the expression of DDR proteins. By Western blot analysis, we observed accumulation of p-ATM, p-ATR and p-CHK1 in AD brains, but not in age-match controls (Fig. 7d,d’). The presence of a DDR in AD brains also correlated with reduced H3K9me3 levels (Fig. 7d). In contrast, accumulation of DDR proteins was not observed in FAD brains, with the exception of one sample presenting p-ATM accumulation (Fig. 7e). Likewise, H3K9me3 levels were unaffected in FAD brains (Fig. 7e). Using IHC, we also confirmed that DDR in AD brains occurred primarily in neurons (Fig. 7f).
Figure 7.
Loss of heterochromatin and DDR in AD brains. (a) IHC staining for BMI1 (brown staining-black arrows) and MAP2 or NeuN (violet staining) on paraffin sections from the frontal cortex of age-match control (n = 2) and AD (n = 2) patients. Weak nuclear staining for BMI1 (dash lines) was observed in AD neurons. Scale bar: 3μm. (b) IHC staining for H3K9me3 on frontal cortex sections of control (n = 5) and AD (n = 5) brains: (i) reduced peri-nucleolar heterochromatin in AD (arrows), (ii) chromocenters de-condensation in AD, and (iii) loss of H3K9me3 immunoreactivity in AD neurons (dash lines). Scale bar: 5μm. (c) Quantification of data in (b) showing the number of H3K9me3 foci/H3K9me3-positive neuron. H3K9me3-negative neurons were excluded from the analysis. Quantification of the number of H3K9me3-negative neurons, which were not found in aged controls. 6 full-fields at 630 magnification/sample were counted. Values are mean ± SEM. *P < 0.05; (**) < 0.01; Student’s t-test. (d) Immunoblot using extracts from the frontal cortex of age-match control and AD patients. (d’) Quantification of the results showed in (d). (e) Immunoblot using extracts from the hippocampus of age-match control and FAD patients. (f) IHC staining for p-ATM on paraffin sections from the frontal cortex of age-match control (n = 2) and AD (n = 2) patients. p-ATM is predominant in AD neurons and accumulates in both cytosolic (blue arrowhead) and nuclear (black arrowhead) compartments. (g) ChIP experiments were performed on frontal cortex extracts of age-match control (n = 6) and AD brains (n = 6). All data are represented as fold of input. (A) Note BMI1 and H3K9me3 enrichment at McBOX, SATIII and SATA in control samples. BMI1 was also found at HOXC13. BMI1 and H3K9me3 were depleted at all loci in AD samples (bottom). Fold differences between control and AD samples for p-ATM, p-ATR and γH2AX accumulation. Values are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; Two way-ANOVA test was performed for multiple gene analysis and Student’s t-test for single gene analysis.
To establish where the DDR was located on the genome, we performed ChIP experiments using frontal cortex samples. When compared to controls, we found significant enrichment for p-ATR and γH2AX at genomic repeats (SATIII, SATA and McBOX) in AD brains (Fig. 7g). In contrast, there was no enrichment at HOXC13, β-GLOBIN or ACTIN (Fig. 7g). Although there was a general trend for p-ATM enrichment at genomic repeats in AD samples, it was not significant. BMI1 and H3K9me3 were also enriched at HOXC13 and at genomic repeats in control brains and were depleted in AD brains (Fig. 7g). Taken together, these findings revealed remarkable similarities between the neuronal genomic instability phenotype of Bmi1+/− mice and AD patients.
Discussion
We have found that Bmi1+/− mice develop with age a neurological and progeroid syndrome characterized by alopecia, weight loss, abnormal paw clasping reflex and neuronal senescence. Memory deficit as measured by their performance in the Morris water maze and Barnes test correlated with impairment in LTP formation as well as with synaptic atrophy and neuronal loss. Pathological brain anomalies such as p-Tau and amyloid accumulation were also found in old Bmi1+/− mice. Remarkably, when compared to Bmi1+/− or APP mice alone, Bmi1+/−/APP mice showed significantly shorter lifespan and increased disease severity. Genetic and cell culture studies further revealed that inhibition of the DDR and/or p53 could mitigate the accumulation of p-Tau and amyloid in neurons from Bmi1-null mice, suggesting that the neuronal response to genomic instability plays a role in the disease process. At last, loss of heterochromatin in neurons and accumulation of DNA damage at genomic repeats were found in Bmi1+/− mice and AD brains.
One possible interpretation of our data is that Bmi1+/− mice age faster and thus present age-related neurological anomalies earlier than WT mice. This possibility is supported by our observation that some pathological marks, including p-Tau (using PHF1, which recognizes an epitope around Ser396 and Ser404) and SA β-galactosidase activity, although much less abundant, were also detected in the brains of old WT mice. However, other pathological marks, such as p-Tau (using AT-8, which recognizes Ser202), p-JNK and amyloid (C99) accumulation were not detected even in 24 month-old WT mice, suggesting that Bmi1 hemi-deficiency represents a pathological brain aging process that is distinct from end-stage physiological brain aging.
Using the Bmi1+/− and Bmi1−/− mouse models, we observed activation of DDR proteins and of p53 in neurons. This is consistent with previous work showing that Bmi1 is important to maintain genomic stability15–18,54. Interestingly, over-expression of Chk1 or Chk2 was shown to exacerbate Tau toxicity in a Drosophila model of neurodegeneration by specific phosphorylation of human Tau at Ser26255. Likewise, JNK activation may promote Tau phosphorylation32. Thus, several kinases potentially targeting Tau are activated in Bmi1+/− mice. It however remains to be investigated by which mechanisms Bace1 level is increased and why the amyloid metabolism is affected in Bmi1+/− mice. Our results showed that amyloid accumulation was mitigated after p53 deletion and we recently reported that p53 activation in human neurons might perturb the clearance of misfolded proteins by interfering with proteasome function20. Thus, a plausible mechanism to explain our results is that activated p53 also impairs proteasome function in mouse neurons. Further work is however needed to validate this.
The heterochromatin island hypothesis of aging stipulates that epigenome instability at the constitutive heterochromatin is a driving force of cellular aging56,57. This hypothesis may be particularly relevant in post-mitotic neurons, where cell division associated with telomere attrition is irrelevant to the cellular aging process. Notably, inactivation of chromatin remodeling proteins, such as those of the NURD complex, results in DDR and in cellular senescence that are preceded by heterochromatin alterations58. Furthermore, modifications of constitutive heterochromatin in response to an acute stress can result in epigenome instability and perturbation of gene expression program, which are hallmarks of cellular aging59. In mice, deletion of the SUV39h1/h2 methyltransferases leads to reduced viability, loss of H3K9me3 and genomic instability60. In Drosophila, perturbation of H3K9me2 levels through inactivation of Su(var)3-9 results in genomic instability and constitutive DDR at the heterochromatin, in both somatic and germ-line cells61. We showed here that heterochromatin alterations in Bmi1+/− mice are present at the time of neurogenesis and thus before activation of DDR proteins and neurodegeneration. Notably, this correlated with preferential accumulation of DNA damage (as marked by γH2Ax) at repetitive DNA sequences. Similarly, we found that DNA damage accumulation was predominant at genomic repeats in AD brains. Exactly why loss of heterochromatin compaction strongly correlates with DNA damage accumulation at genomic repeats is unknown. It is possible that repetitive DNA sequences are intrinsically unstable upon loss of heterochromatin compaction and thus prone to de novo DNA damage formation. It is also possible that the efficiency of DNA repair is reduced upon BMI1 deficiency or that recognition of the H3K9me3 mark is important to activate the process of DNA repair at these specific regions15,16,62. It remains however to be tested whether heterochromatin anomalies observed in Bmi1+/− mice are sufficient to induce the DDR and neurodegeneration.
Interstitial amyloid plaques and Tau tangles represent the classical pathological hallmarks of AD. While synaptic atrophy, neuronal loss and behavioral anomalies are present in 15-month old Bmi1+/− mice, the animals do not develop Tau tangles or amyloid plaques (with the exception of one animal). Rather, old Bmi1+/− mice present intra-neuronal accumulation of amyloid and show moderate levels of p-Tau deposits in the neuronal cell body and the axon. The absence of amyloid plaques formation in Bmi1+/− mice is not surprising since the mouse amyloid peptide is known to be less prone to aggregation than the human one63. Also, transgenic mice over-expressing wild-type human APP rarely develop amyloid plaques, in contrast with those over-expressing APP with FAD-associated mutations64. Notably, massive neurodegeneration was reported in APP/PSEN1 transgenic mice (APP(SL)PS1KI mice), which only develop intra-neuronal accumulation of amyloid65. Thus, intra-neuronal accumulation of oligomeric amyloid species, including the C99 fragment, may be sufficient to induce neurodegeneration66,67. Nevertheless, the absence of amyloid plaques and Tau tangles as well as the late onset neuronal disease should be taken into account before using Bmi1+/− mice as a model of AD.
In conclusion, we demonstrated that loss of one Bmi1 allele in mice results in age-related neurodegeneration sharing some similarities with AD. Bmi1+/− mice may thus represent an interesting animal model to identify new pathogenic mechanisms related to AD.
Supplementary information
Acknowledgements
We are grateful to D. Cécyre and J. Prud’homme for the human brain samples, J. Rochford for help with the behavioral analysis, and to Drs F. Rodier, R. Kothary, G. Ferbeyre and D. Picketts for critical reading of the manuscript. This project was supported by grants from the Canadian Institutes of Health Research (374972) and Natural Science and Engineering Research Council of Canada (2017-05504). J.E.H. and R.H. held a fellowship from the University of Montreal Molecular Biology Program. G.B. was supported by a fellowship from the Fonds de Recherche en Santé du Québec.
Author Contributions
J.E.H., R.H., W.C., M.A., N.T., P.N. and Y.C.T. performed the experiments. G.B., J.E.H. and T.P.W. wrote the manuscript.
Data Availability Statement
The authors declare that they will make available all data presented in this manuscript.
Competing Interests
G.B. is a co-founder of StemAxonTM and member of its scientific advisory board. The corporation was however not involved in this study.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37444-3.
References
- 1.Savva GM, et al. Age, neuropathology, and dementia. N. Engl. J. Med. 2009;360:2302–2309. doi: 10.1056/NEJMoa0806142. [DOI] [PubMed] [Google Scholar]
- 2.Kanekiyo T, et al. ApoE and Abeta in Alzheimer’s disease: accidental encounters or partners? Neuron. 2014;81:740–754. doi: 10.1016/j.neuron.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blennow K, et al. Alzheimer’s disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 4.Chapman PF, et al. Impaired synaptic plasticity and learning in aged amyloid precursor protein transgenic mice. Nat. Neurosci. 1999;2:271–276. doi: 10.1038/6374. [DOI] [PubMed] [Google Scholar]
- 5.Galvan V, et al. Reversal of Alzheimer’s-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc. Natl. Acad. Sci. USA. 2006;103:7130–7135. doi: 10.1073/pnas.0509695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomiyama T, et al. A mouse model of amyloid beta oligomers: their contribution to synaptic alteration, abnormal tau phosphorylation, glial activation, and neuronal loss in vivo. J. Neurosci. 2010;30:4845–4856. doi: 10.1523/JNEUROSCI.5825-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fodor BD, et al. Mammalian Su(var) genes in chromatin control. Annu. Rev. Cell Dev. Biol. 2010;26:471–501. doi: 10.1146/annurev.cellbio.042308.113225. [DOI] [PubMed] [Google Scholar]
- 8.Buchwald G, et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, et al. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J. Biol. Chem. 2006;281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 10.Satijn DP, et al. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell. Biol. 1997;17:4105–4113. doi: 10.1128/MCB.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs JJ, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 13.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Lugt NM, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 15.Facchino S, et al. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 2010;30:10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ismail IH, et al. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatoo W, et al. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J. Neurosci. 2009;29:529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdouh, M. et al. The Polycomb Repressive Complex 1 Protein BMI1 is Required for Constitutive Heterochromatin Formation and Silencing in Mammalian Somatic Cells. J. Biol. Chem. (2015). [DOI] [PMC free article] [PubMed]
- 20.Flamier A, et al. Modeling late-onset sporadic Alzheimer’s disease through BMI1 deficiency. Cell Reports. 2018;23:2653–2666. doi: 10.1016/j.celrep.2018.04.097. [DOI] [PubMed] [Google Scholar]
- 21.Bornemann KD, Staufenbiel M. Transgenic mouse models of Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2000;908:260–266. doi: 10.1111/j.1749-6632.2000.tb06653.x. [DOI] [PubMed] [Google Scholar]
- 22.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 23.Barabino A, et al. Loss of Bmi1 causes anomalies in retinal development and degeneration of cone photoreceptors. Development. 2016;143:1571–1584. doi: 10.1242/dev.125351. [DOI] [PubMed] [Google Scholar]
- 24.Kawarabayashi T, et al. Age-dependent changes in brain, CSF, and plasma amyloid (beta) protein in the Tg2576 transgenic mouse model of Alzheimer’s disease. J. Neurosci. 2001;21:372–381. doi: 10.1523/JNEUROSCI.21-02-00372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker DJ, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondratov RV, et al. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 30.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 31.Lalonde R, Strazielle C. Motor performance and regional brain metabolism of spontaneous murine mutations with cerebellar atrophy. Behav. Brain Res. 2001;125:103–108. doi: 10.1016/S0166-4328(01)00276-5. [DOI] [PubMed] [Google Scholar]
- 32.Wetzel MK, et al. p73 regulates neurodegeneration and phospho-tau accumulation during aging and Alzheimer’s disease. Neuron. 2008;59:708–721. doi: 10.1016/j.neuron.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon P, et al. Thigmotaxis as an index of anxiety in mice. Influence of dopaminergic transmissions. Behav. Brain Res. 1994;61:59–64. doi: 10.1016/0166-4328(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 35.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 36.Sclip A, et al. c-Jun N-terminal kinase regulates soluble Abeta oligomers and cognitive impairment in AD mouse model. J. Biol. Chem. 2011;286:43871–43880. doi: 10.1074/jbc.M111.297515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, et al. Pathological Axonal Death through a MAPK Cascade that Triggers a Local Energy Deficit. Cell. 2015;160:161–176. doi: 10.1016/j.cell.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M, et al. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 2004;32:5742–5749. doi: 10.1093/nar/gkh912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/S0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Q, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 43.Smith J, et al. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 44.Rogakou EP, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 45.Keramaris E, et al. Ataxia telangiectasia-mutated protein can regulate p53 and neuronal death independent of Chk2 in response to DNA damage. J. Biol. Chem. 2003;278:37782–37789. doi: 10.1074/jbc.M304049200. [DOI] [PubMed] [Google Scholar]
- 46.Chaturvedi P, et al. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–4054. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- 47.Brady CA, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang XP, et al. Two-phase dynamics of p53 in the DNA damage response. Proc. Natl. Acad. Sci. USA. 2011;108:8990–8995. doi: 10.1073/pnas.1100600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mellert H, et al. The ARF/oncogene pathway activates p53 acetylation within the DNA binding domain. Cell Cycle. 2007;6:1304–1306. doi: 10.4161/cc.6.11.4343. [DOI] [PubMed] [Google Scholar]
- 50.Lakin ND, et al. The ataxia-telangiectasia related protein ATR mediates DNA-dependent phosphorylation of p53. Oncogene. 1999;18:3989–3995. doi: 10.1038/sj.onc.1202973. [DOI] [PubMed] [Google Scholar]
- 51.Tibbetts RS, et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calao M, et al. Direct effects of Bmi1 on p53 protein stability inactivates oncoprotein stress responses in embryonal cancer precursor cells at tumor initiation. Oncogene. 2013;32:3616–3626. doi: 10.1038/onc.2012.368. [DOI] [PubMed] [Google Scholar]
- 53.Frost B, et al. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 2014;17:357–366. doi: 10.1038/nn.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chagraoui J, et al. An anticlastogenic function for the Polycomb Group gene Bmi1. Proc. Natl. Acad. Sci. USA. 2011;108:5284–5289. doi: 10.1073/pnas.1014263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iijima-Ando K, et al. A DNA damage-activated checkpoint kinase phosphorylates tau and enhances tau-induced neurodegeneration. Hum. Mol. Genet. 2010;19:1930–1938. doi: 10.1093/hmg/ddq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imai S, Kitano H. Heterochromatin islands and their dynamic reorganization: a hypothesis for three distinctive features of cellular aging. Exp. Gerontol. 1998;33:555–570. doi: 10.1016/S0531-5565(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 57.Villeponteau B. The heterochromatin loss model of aging. Exp. Gerontol. 1997;32:383–394. doi: 10.1016/S0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- 58.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 61.Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Y, et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Strooper B, et al. Production of intracellular amyloid-containing fragments in hippocampal neurons expressing human amyloid precursor protein and protection against amyloidogenesis by subtle amino acid substitutions in the rodent sequence. EMBO J. 1995;14:4932–4938. doi: 10.1002/j.1460-2075.1995.tb00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sasaguri H, et al. APP mouse models for Alzheimer’s disease preclinical studies. EMBO J. 2017;36:2473–2487. doi: 10.15252/embj.201797397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casas C, et al. Massive CA1/2 neuronal loss with intraneuronal and N-terminal truncated Abeta42 accumulation in a novel Alzheimer transgenic model. Am. J. Pathol. 2004;165:1289–1300. doi: 10.1016/S0002-9440(10)63388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LaFerla FM, et al. Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- 67.Lauritzen I, et al. The beta-secretase-derived C-terminal fragment of betaAPP, C99, but not Abeta, is a key contributor to early intraneuronal lesions in triple-transgenic mouse hippocampus. J. Neurosci. 2012;32:16243–16255a. doi: 10.1523/JNEUROSCI.2775-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that they will make available all data presented in this manuscript.