Staphylococcus aureus nitric oxide synthase (saNOS) is a major contributor to virulence, stress resistance, and physiology, yet the specific mechanism(s) by which saNOS intersects with other known regulatory circuits is largely unknown. The SrrAB two-component system, which modulates gene expression in response to the reduced state of respiratory menaquinones, is a positive regulator of nos expression.
KEYWORDS: SrrAB, Staphylococcus aureus, bacterial nitric oxide synthase, cell respiration, metabolomics, sepsis, two-component system
ABSTRACT
Staphylococcus aureus nitric oxide synthase (saNOS) is a major contributor to virulence, stress resistance, and physiology, yet the specific mechanism(s) by which saNOS intersects with other known regulatory circuits is largely unknown. The SrrAB two-component system, which modulates gene expression in response to the reduced state of respiratory menaquinones, is a positive regulator of nos expression. Several SrrAB-regulated genes were also previously shown to be induced in an aerobically respiring nos mutant, suggesting a potential interplay between saNOS and SrrAB. Therefore, a combination of genetic, molecular, and physiological approaches was employed to characterize a nos srrAB mutant, which had significant reductions in the maximum specific growth rate and oxygen consumption when cultured under conditions promoting aerobic respiration. The nos srrAB mutant secreted elevated lactate levels, correlating with the increased transcription of lactate dehydrogenases. Expression of nitrate and nitrite reductase genes was also significantly enhanced in the nos srrAB double mutant, and its aerobic growth defect could be partially rescued with supplementation with nitrate, nitrite, or ammonia. Furthermore, elevated ornithine and citrulline levels and highly upregulated expression of arginine deiminase genes were observed in the double mutant. These data suggest that a dual deficiency in saNOS and SrrAB limits S. aureus to fermentative metabolism, with a reliance on nitrate assimilation and the urea cycle to help fuel energy production. The nos, srrAB, and nos srrAB mutants showed comparable defects in endothelial intracellular survival, whereas the srrAB and nos srrAB mutants were highly attenuated during murine sepsis, suggesting that SrrAB-mediated metabolic versatility is dominant in vivo.
INTRODUCTION
Staphylococcus aureus is a highly adaptable mammalian pathogen which can infect nearly every tissue and organ system. This is due in part to its coordinated production of a wide spectrum of cell surface adhesins, secreted toxins, and tissue-degrading enzymes, as well as its impressive metabolic versatility. As a facultative anaerobe, the metabolism of S. aureus is highly fluid, retaining the ability to fluctuate between aerobic and anaerobic respiration and/or mixed acid fermentation (1, 2). In fact, S. aureus metabolism is predicted to be one of the most complex in terms of estimated metabolite numbers (1), and genome-scale reconstruction of its metabolic networks has predicted approximately 1,250 potential metabolic reactions and 1,400 metabolites (3). S. aureus can utilize several major metabolic pathways, including complete glycolytic (Embden-Meyerhof-Parnas), pentose phosphate, and tricarboxylic acid (TCA) pathways. This bacterium also has major metabolic pathways for fermentation as well as a complex branched respiratory chain with a variety of components.
S. aureus nitric oxide synthase (saNOS) has been established as an important modulator of stress resistance (4–7), virulence (4, 6), and, most recently, aerobic respiration (8, 9) and nitrate-based respiration (10). A second major regulator of S. aureus physiology (11–15), nitrosative stress resistance (16–18), and virulence (15, 19–21) is the staphylococcal respiratory regulator (SrrAB) two-component system, which is hypothesized to sense and respond to the reduced state of the respiratory menaquinone pool (17). SrrAB was identified by transposon sequencing to be an important contributor to osteomyelitis infection, which was shown to be a highly anoxic in vivo environment (22). In the same study, transcriptome analysis revealed that nos expression was significantly downregulated in a srrA mutant during low-oxygen growth (22). Furthermore, RNA microarray analysis of S. aureus wild-type and srrAB mutants exposed to nitrosative stress showed that SrrAB upregulates anaerobic metabolism (narG, pflB, and nrdG), nitrosative stress (scdA, hmp), and cytochrome biosynthesis (qox, cta) genes (17), all of which were also shown in our recent transcriptome sequencing (RNA-seq) study to be upregulated in a nos mutant growing under aerobic respiratory conditions (9). These results, combined with the previous observation that inactivation of both nos and srrAB conferred a reduced aerobic growth phenotype (8), are highly suggestive of a previously unrecognized interplay between saNOS and SrrAB in the regulation of S. aureus physiology and virulence. Therefore, in the current study, we characterized the interrelationship between the genes for these two proteins using a combination of genetic, molecular, and physiological approaches.
RESULTS
Effects of srrAB and nos mutation on gene expression.
Congruent with RNA microarray analysis of a clonal complex 8 (CC8)/USA300 (community-acquired methicillin-resistant S. aureus [MRSA]) srrA mutant (22), we found that low-oxygen expression of nos was downregulated 4-fold in a srrAB mutant of UAMS-1 (see Fig. S1 in the supplemental material), a methicillin-susceptible S. aureus (MSSA) strain belonging to the USA200/CC30 clade (23). RNA-seq analysis of a UAMS-1 nos mutant grown to late exponential phase under conditions promoting aerobic respiration showed that it had several SrrAB-dependent genes (17, 22) upregulated ≥2-fold, including those encoding nitrosative stress genes (hmp, scdA), respiratory cytochromes (cta, qox), and anaerobic metabolism (ldh2, pfl) (9). These results suggest that SrrAB may facilitate an adaptive response to the nos mutant’s altered respiratory phenotype (9). To ascertain if SrrAB is involved in mediating expression of some of these genes, real-time PCR was performed on RNA isolated from mid-exponential-phase aerobic cultures of the wild type and isogenic nos mutant, srrAB mutant, and nos srrAB double mutant (Table 1). This analysis revealed that mutation of srrAB in the nos mutant reversed the induction of hmp and scdA observed in the nos single mutant but did not affect expression of qox. Expression of ldh2 in the nos and srrAB single mutants was slightly elevated compared to that in the wild-type strain (1.4- and 2-fold, respectively), whereas expression was additively upregulated in the double mutant (4.7-fold) (Table 1).
TABLE 1.
Real-time PCR analysis of SrrAB-regulated and metabolomics-associated genes
| Function/annotation | Gene identifiera | Fold change in expression inb
: |
||||
|---|---|---|---|---|---|---|
| nos mutant/pMK4 | srrAB mutan/pMK4 | nos srrAB mutan/pMK4 | nos srrAB mutant complemented with nos | nos srrAB mutant complemented with srrAB | ||
| Flavohemoprotein | SAR0233 (hmp) | 12.78 ± 3.47 | 1.89 ± 1.19 | 1.11 ± 0.10 | 0.53 ± 0.07 | 2.33 ± 1.10 |
| Iron-sulfur cluster repair protein | SAR0256 (scdA) | 5.27 ± 0.63 | 2.34 ± 1.31 | 1.47 ± 0.07 | 0.78 ± 0.13 | 2.11 ± 0.61 |
| Pyruvate formate lyase-activating enzyme | SAR0218 (pflB) | ND | ND | ND | ND | ND |
| Quinol oxidase polypeptide III cytochrome | SAR1032 (qoxC) | 2.14 ± 0.32 | 1.71 ± 0.77 | 2.18 ± 0.13 | 1.32 ± 0.15 | 2.54 ± 0.49 |
| Lactate dehydrogenase 2 | SAR2680 (ldh2) | 1.38 ± 0.25 | 1.96 ± 0.69 | 4.71 ± 0.24 | 1.14 ± 0.14 | 2.15 ± 0.14 |
| Lactate dehydrogenase 1 | SAR0234 (ldh1) | 0.19 ± 0.03 | 7.47 ± 2.17 | 205.86 ± 37.96 | 2.69 ± 0.15 | 2.94 ± 1.12 |
| Nitrate reductase, alpha subunit | SAR2486 (narG) | 1.54 ± 0.19 | 6.79 ± 1.27 | 82.49 ± 5.36 | 7.75 ± 0.84 | 10.59 ± 3.99 |
| Nitrite reductase | SAR2489 (nirB) | 0.68 ± 0.11 | 3.91 ± 0.91 | 53.80 ± 2.58 | 5.06 ± 2.97 | 2.86 ± 0.87 |
| Formate/nitrite transporter | SAR2493 (narT) | 0.41 ± 0.09 | 0.97 ± 0.14 | 10.14 ± 1.42 | 1.12 ± 0.09 | 0.74 ± 0.06 |
| Arginine deiminase | SAR2714 (arcA) | 0.61 ± 0.09 | 1.22 ± 0.33 | 1,733.57 ± 336.18 | 1.80 ± 0.13 | 1.90 ± 0.30 |
| Ornithine carbamoyltransferase | SAR2713 (arcB) | 0.54 ± 0.08 | 0.95 ± 0.20 | 1,195.31 ± 215.91 | 1.64 ± 0.12 | 1.59 ± 0.19 |
| Glutamate N-acetyltransferase/amino acid N-acetyltransferase | SAR0184 (argJ) | 0.35 ± 0.07 | 0.50 ± 0.05 | 0.46 ± 0.02 | 0.95 ± 0.06 | 0.62 ± 0.08 |
| Argininosuccinate synthase | SAR0923 (argG) | 0.35 ± 0.06 | 0.41 ± 0.05 | 0.84 ± 0.06 | 0.86 ± 0.07 | 0.64 ± 0.03 |
| Ornithine carbamoyltransferase | SAR1142 (otc) | 0.72 ± 0.08 | 0.84 ± 0.13 | 1.16 ± 0.09 | 1.40 ± 0.07 | 1.43 ± 0.12 |
| Arginase | SAR2255 (rocF) | 0.69 ± 0.10 | 0.93 ± 0.12 | 0.84 ± 0.10 | 1.84 ± 0.11 | 1.36 ± 0.07 |
Gene identifiers refer to old locus tags of the MRSA 252 sequenced genome (GenBank accession number NC_002952.2).
Fold change in expression relative to that in the wild type at mid-exponential phase of aerobic respiratory growth. ND, not detectable.
As previously observed (9), expression of ldh1 was repressed in the nos mutant under this growth condition, and repression appeared to be dependent on SrrAB, as expression of ldh1 was upregulated 7.5-fold in the srrAB mutant and 206-fold in the nos srrAB double mutant (Table 1). Expression of nitrate reductase (nar) genes was also previously upregulated in the nos mutant during aerobic late-exponential-phase growth (9), and the nar operon and formate/nitrite transporter gene nirT are regulated by SrrAB (17, 22). Therefore, expression of genes involved in nitrate respiration/assimilation and nitrite transport (narG, nirB, nirT) was assessed in the nos mutant, srrAB mutant, and double mutant during mid-exponential-phase aerobic growth (Table 1). At this earlier time point, these genes were not significantly upregulated in the nos mutant. However, narG and nirB were modestly upregulated in the srrAB mutant (6.8- and 3.9-fold, respectively), suggesting that SrrAB has a repressive effect on expression of these genes during aerobic growth (Table 1). Furthermore, expression of narG, nirB, and nirT was highly upregulated (82-, 54-, and 10-fold, respectively) in the nos srrAB double mutant. Collectively, these results suggest that a subset of genes with altered expression profiles in the nos mutant during aerobic growth is regulated (either directly or indirectly) by SrrAB. Interestingly, our previous RNA-seq analysis also revealed that srrAB expression itself was downregulated ∼1.6-fold in the nos mutant (9), further reinforcing the idea that there may be regulatory cross talk between saNOS and SrrAB.
Mutation of nos and srrAB impacts S. aureus aerobic growth.
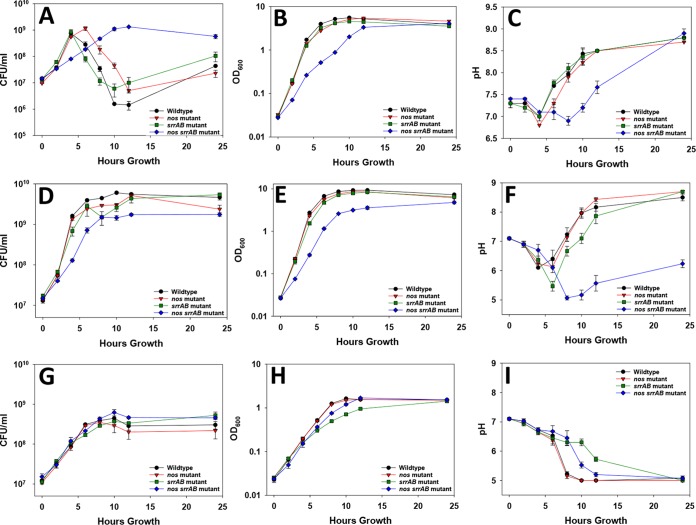
To pursue the hypothesis that SrrAB and saNOS are important for optimal aerobic growth, a series of growth curves was conducted on UAMS-1 (wild type) and its isogenic nos mutant, srrAB mutant, and nos srrAB double mutant strains (Fig. 1). In aerobic tryptic soy broth (TSB) without glucose (TSB−G) culture conditions, the wild type and srrAB mutant had comparable growth patterns, as measured by determination of the number of CFU (rapid growth from 0 to 4 h, followed by a steady decline in cell viability), whereas the nos mutant had a shifted but similar pattern (rapid growth from 0 to 6 h) (Fig. 1A). Congruent with our previous studies in TSB−G (9), the nos mutant displayed a consistent but subtle reduction in the optical density (OD) compared to that for the wild type, and interestingly, this was phenocopied by the srrAB mutant (Fig. 1B). As previously observed in a USA300 background (8), our nos srrAB double mutant of UAMS-1 also had a dramatically impaired growth phenotype which was most pronounced in aerobic TSB−G (Fig. 1A and B). This decrease in growth was also observed in aerobic TSB cultures grown in the presence of glucose (TSB+G) (Fig. 1D and E). Under both aerobic growth conditions, the nos srrAB mutant had lower pH values than the wild-type and single mutant strains (Fig. 1C and F). The impaired growth phenotype of the double mutant was partially rescued by expressing either nos or srrAB from a plasmid (Fig. S2). Low-oxygen TSB+G culture conditions (promoting fermentative growth) minimized the growth differences between the wild type, the nos mutant, and the nos srrAB mutant, and consistent with the findings of previous studies (21, 24), the srrAB mutant was more impaired (Fig. 1G and H), further supporting the role that SrrAB plays in regulating low-oxygen growth (21, 24). The pH profiles during low-oxygen growth provide further evidence of the differences in growth, as the pH drops at a slower pace during the growth of the srrAB mutant than during the growth of the other strains (Fig. 1I). Collectively, these results suggest that although both saNOS and SrrAB contribute to optimal aerobic growth, the effect of SrrAB loss is more pronounced under low-oxygen growth conditions.
FIG 1.
Growth curves of the wild type, nos mutant, srrAB mutant, and nos srrAB mutant for aerobic growth in TSB−G, aerobic growth in TSB+G, and low-oxygen TSB+G cultures. (A to C) Aerobic TSB−G cultures were inoculated to an OD600 of 0.025 in TSB−G medium, and the bacteria were grown at 37°C with aeration (250 rpm, 1:12.5 volume-to-flask ratio). (D to F) Aerobic TSB+G cultures were grown as described above for TSB−G cultures. (G to I) Low-oxygen cultures were inoculated to an OD600 of 0.025 and grown at 37°C in a static incubator (1:1.6 volume-to-flask ratio). Growth over a 24-h period was monitored (at 0, 2, 4, 6, 8, 10, 12, and 24 h) by determination of the number of CFU per milliliter (CFU/ml) by serial dilution plating (A, D, and G), OD600 measurement (B, E, and H), and pH measurement (C, F, and I). Data points represent the average from 3 independent experiments; error bars indicate the standard error of the mean (SEM).
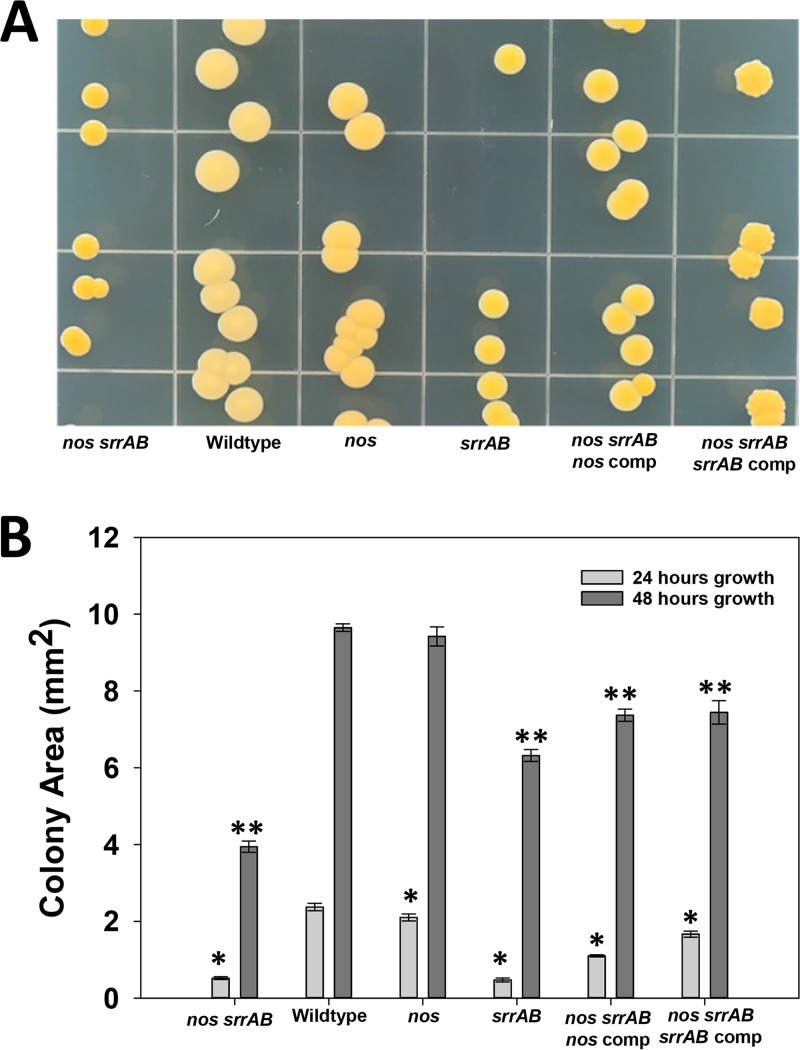
The colony morphology was assessed on aerobic tryptic soy agar (TSA) plates for the wild type and the isogenic mutants (Fig. 2). At 48 h of growth, the nos srrAB double mutant had small hyperpigmented yellow colonies compared to the nos mutant and wild type, a phenotype that was partially complemented by either nos or srrAB expressed from a plasmid (Fig. 2A). The srrAB mutant also displayed hyperpigmentation at this time point, but the colony size was larger than that of the nos srrAB double mutant. As previously reported (4), the nos mutant also had increased carotenoid pigmentation compared to the wild type, but not as much as the srrAB and double mutant strains (Fig. 2A). Colony areas (in square millimeters) were quantified at both 24 and 48 h of growth (Fig. 2B), and at 24 h, both srrAB and nos srrAB mutant colonies were similar in size; however, at 48 h, the double mutant colonies were smaller than the srrAB mutant colonies.
FIG 2.
Colony morphology of wild-type and isogenic mutant strains. (A) Cultures were grown for ∼16 h with the respective antibiotic for selection and serially diluted for plating on TSA medium with no antibiotic. The plates were incubated for 48 h at 37°C. (B) Colony areas were measured from colonies inoculated as described above on TSA plates, and measurements were taken at 24 h and 48 h of growth at 37°C using a Zeiss Semi 305 microscope. Data measurements represent those from 4 to 7 independent measurements, and error bars indicate the SEM. *, statistically significant difference (P < 0.05, Holm-Sidak test) relative to the wild type at 24 h; **, statistically significant difference (P < 0.05, Holm-Sidak test) relative to the wild type at 48 h. comp, complemented strain.
Dual mutation of nos and srrAB restricts S. aureus to fermentative metabolism during aerobic growth.
The reduced aerobic growth and pH curves of the nos srrAB mutant (Fig. 1A to F), combined with its small colony size (Fig. 2) and high expression levels of ldh1, ldh2, and other anaerobic genes (Table 1), suggested that this mutant may be restricted to fermentative metabolism. Therefore, extracellular acetate and lactate levels were quantified in all strains grown in TSB−G (Fig. 3). Peak acetate concentrations in the wild type and the nos and srrAB mutants corresponded to their transitions into stationary phase at between 4 and 6 h, and the peak acetate concentration for the nos srrAB mutant, although delayed (12 h), also correlated with stationary phase (Fig. 3A). The increase and persistence of acetate in both the nos and srrAB mutants suggested that these strains may be more reliant on substrate-level phosphorylation for energy than the wild type (Fig. 3A). In agreement with the observed overexpression of ldh1 and ldh2, the nos srrAB mutant accumulated significant levels of l-lactate by 8 h (20 mM) which disappeared by 24 h of growth (Fig. 3B). By comparison, l-lactate was detected at very low concentrations and only during early-exponential-phase growth for the wild type and the nos and srrAB mutants (Fig. 3B).
FIG 3.
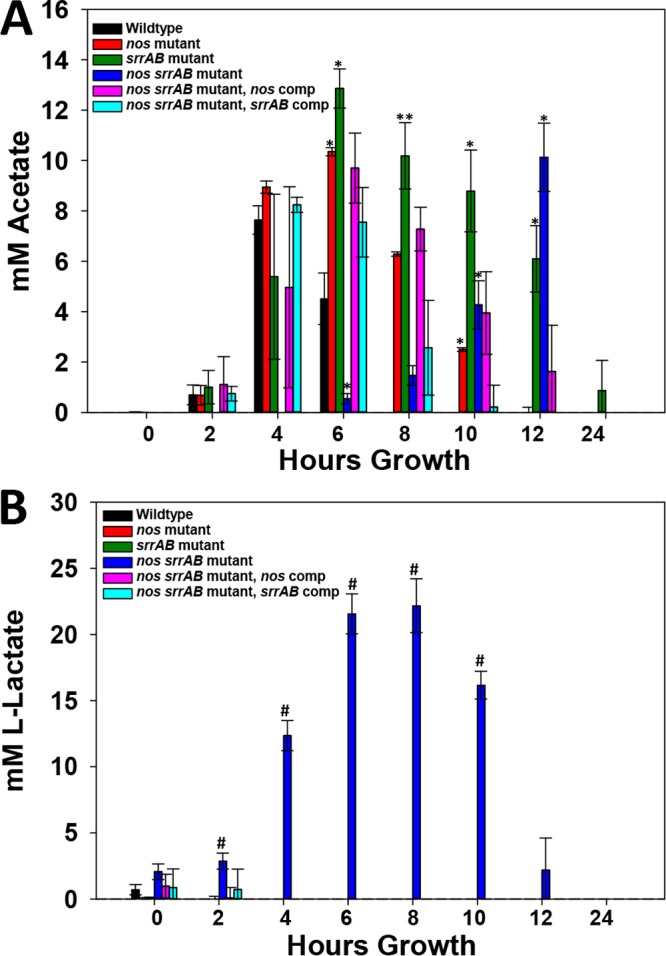
Acetate and lactate extracellular concentrations. (A) Acetate measurements. (B) l-Lactate measurements. Culture supernatants were collected at time of inoculation (0) and 2, 4, 6, 8, 10, 12, and 24 h of growth from aerobic TSB−G cultures, and acetate and lactate concentrations were measured by enzymatic analysis as described in Materials and Methods. All measurements were performed in 96-well cell culture plates using a Cytation 3 imaging reader (BioTek). Data represent the average for the following number of biological replicates: 4 for the wild type and the nos srrAB mutant, 3 for the nos mutant and srrAB mutant, and 2 for both nos srrAB-complemented strains. Each bar represents the mean concentration; error bars indicate the SEM. *, statistically significant difference (P < 0.05 Holm-Sidak test) relative to the wild type; **, statistically significant difference (P < 0.05, Dunn’s test) relative to the wild type; #, statistically significant difference (P < 0.01, two-tailed t test) relative to the wild type.
The patterns of acetate and lactate production/consumption for the nos srrAB double mutant described above, combined with its slow aerobic growth, are highly suggestive of fermentative metabolism (amino acids → acetyl coenzyme A [acetyl-CoA] → substrate-level phosphorylation to generate ATP, acetate, and lactate to maintain cellular redox balance). To determine whether or not the nos srrAB mutant can respire on oxygen, the oxygen consumption rates for all strains were determined at mid-exponential-phase growth in aerobic TSB−G cultures (9, 10) using a Clark-type electrode (Fig. 4). As previously described (9), the nos mutant had a small but not statistically significant increase in O2 consumption relative to the wild type. The srrAB mutant had a 34% reduction in relative O2 consumption relative to the wild type, whereas the nos srrAB mutant had an 83% reduction relative to the wild type (Fig. 4). The significantly impaired ability of the double mutant to respire on O2 was partially rescued when either the nos or srrAB gene was supplied in trans (Fig. 4).
FIG 4.
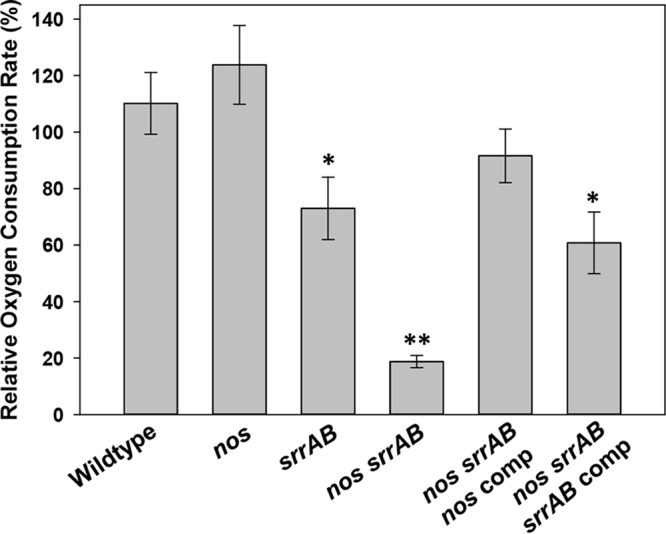
Effect of nos and srrAB mutations on oxygen consumption. Oxygen consumption was determined for the wild type, nos mutant, srrAB mutant, and complemented strains grown for 3 h and the nos srrAB mutant grown for 6 h in TSB−G. Cultures were harvested and resuspended in PBS amended with 6 mM glucose, and oxygen consumption was measured with a Clark-type electrode, normalized to the number of CFU per milliliter, and expressed as a percentage of the wild-type oxygen consumption rate. Data represent the average from 5 independent experiments; error bars indicate the SEM. *, statistically significant difference (P < 0.05, one-tailed t test) relative to the wild type; **, statistically significant difference (P < 0.01, Mann-Whitney rank-sum test) relative to the wild type.
Aerobic respiration in the nos srrAB mutant was further assessed by determining its susceptibility to gentamicin, an aminoglycoside that requires the electron transport chain for energy-dependent uptake into the cell (25). When 10× MIC of gentamicin was added to growing cultures, the wild type and the srrAB mutant were both susceptible to gentamicin killing, and in fact, the srrAB mutant died significantly faster at 1 and 2 h posttreatment (Fig. 5A). However, the nos and nos srrAB mutants were both tolerant to gentamicin killing, a phenotype rescued by complementation with a nos-expressing plasmid in both mutants but not by srrAB complementation in the double mutant (Fig. 5B). The complete tolerance of both the nos single mutant and the nos srrAB double mutant to gentamicin was unexpected, as the nos single mutant was still able to respire on O2 (Fig. 4A). These results suggest that gentamicin tolerance is directly related to saNOS function and that one or more previously observed nos mutation-altered respiratory phenotypes (9) (i.e., elevated 5-cyano-2,3-ditolyltetrazolium chloride [CTC] staining, elevated membrane potential, increased reactive oxygen species [ROS] levels) are responsible. Furthermore, complete tolerance of the nos srrAB mutant to gentamicin killing was not simply a function of its slower growth. Although this strain was overall more tolerant to killing by 20× MIC of vancomycin and 20× MIC of ciprofloxacin than the other strains, slow killing was still observed (Fig. S3).
FIG 5.
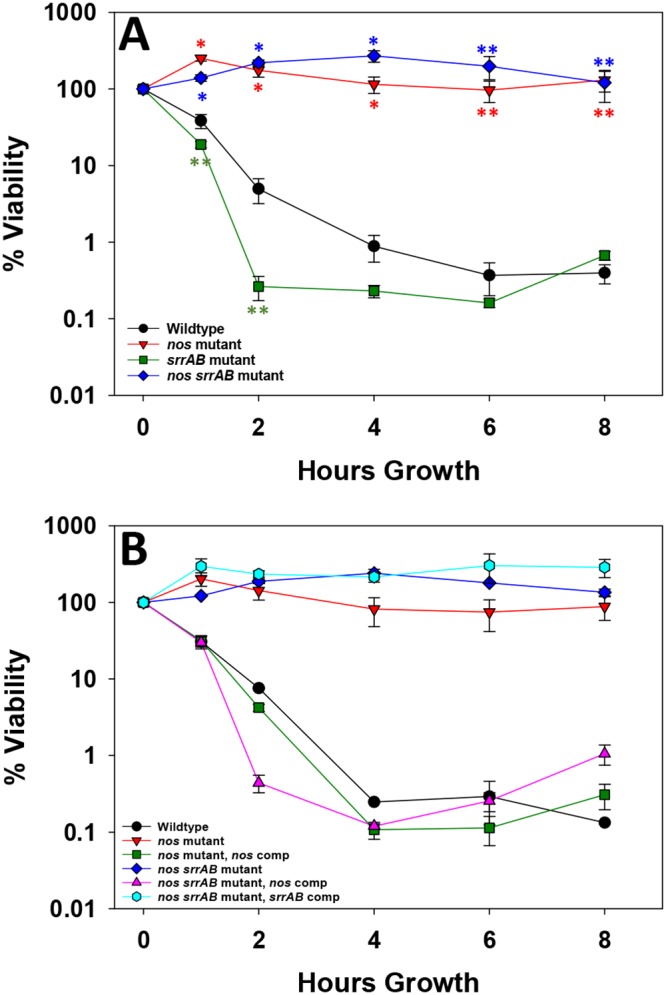
Contributions of nos and srrAB to gentamicin tolerance. Early-exponential-phase aerobic TSB−G cultures of the wild type and isogenic nos, srrAB, and nos srrAB mutants (A) as well as the nos- and nos srrAB-complemented strains (B) were treated with a final concentration of 5 µg/ml gentamicin. The numbers of CFU per milliliter were monitored at time zero (just prior to antibiotic treatment) and 1, 2, 4, 6, and 8 h posttreatment, as described in Materials and Methods. All data represent the average from 3 (A) or 2 (B) independent experiments; error bars indicate the SEM. *, P < 0.01 (one-tailed t test) compared to the wild type; **, P < 0.05 (one-tailed t test) compared to the wild type.
Nitrate assimilation can partially restore nos srrAB mutant aerobic growth.
Given that the nos srrAB mutant had significantly elevated nitrate reductase, nitrite reductase, and nitrite transporter transcript levels during aerobic growth (Table 1), both nitrate and ammonia concentrations were determined from the TSB−G culture supernatants of the wild type and the single and double mutants (Fig. S4). Relatively low levels of each metabolite were detected throughout the growth phases for all strains (range, 30 to 100 µM for nitrate and 2 to 4 mM for ammonia), and no significant differences between the wild type and the mutant strains were detected at each time point for nitrate. The nos srrAB mutant displayed reduced ammonia levels at 8 h of growth, but this was likely due to its delayed growth, as ammonia levels were comparable to those of the other strains at 24 h (Fig. S4). Next, we addressed the question of whether the nos srrAB mutant could sustain improved aerobic respiratory growth (TSB−G) if supplemented with nitrate, nitrite, or ammonia (Fig. 6 and Fig. S5). The maximal specific growth rates were determined from cultures amended with nitrate, nitrite, and ammonia. The unamended nos and srrAB single mutant and double mutant culture growth rates were reduced by 15%, 25%, and 72%, respectively (Fig. 6). The addition of nitrate, nitrite, or ammonia did not have a significant impact on the growth of the nos and srrAB mutants compared to that of the wild type (Fig. 6 and Fig. S5C and D); however, the growth of the nos srrAB mutant was significantly improved by addition of all three metabolites (Fig. 6 and Fig. S5B). These results suggest that increased nitrate assimilation partially rescues the growth of the nos srrAB mutant during aerobic respiratory growth.
FIG 6.
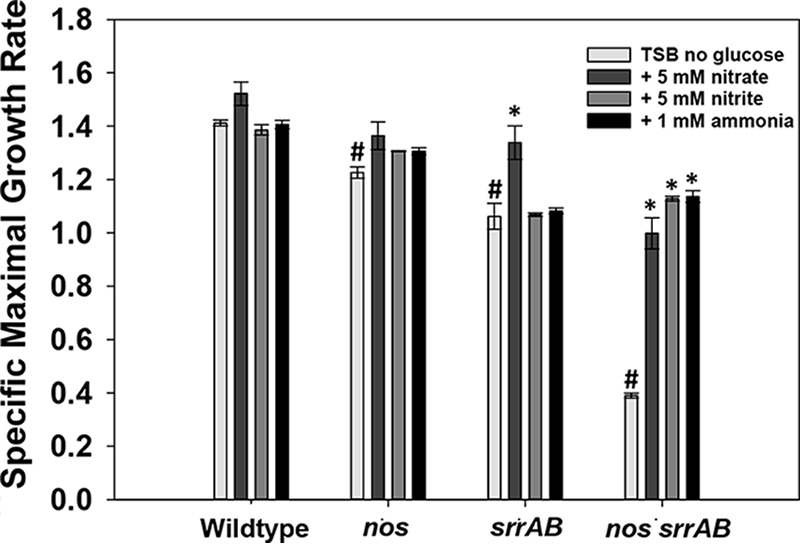
Maximum specific growth rate. The wild-type, nos mutant, srrAB mutant, nos srrAB mutant, and nos srrAB-complemented strains were inoculated to a final OD600 of 0.025 in TSB−G and were left untreated or supplemented with 5 mM nitrate, 1 mM nitrite, or 1 mM ammonia, and 200 µl was dispensed into quadruplicate wells of a Bioscreen C plate. Cultures were incubated in a Bioscreen C plate reader with maximum shaking at 37°C, and growth was monitored by measuring the OD600 every 15 min for 24 h. The maximum specific growth (µmax) rates (per hour) were determined by linear regression analysis of the slope (R2 >0.99). Data represent the average of 4 technical replicates for 3 biological samples; error bars indicate the SEM. *, P < 0.05 (Holm-Sidak test) relative to each respective untreated strain; #, P < 0.05 (Holm-Sidak test) relative to the untreated wild type.
Metabolomics profiling of nos and srrAB single mutant and nos srrAB double mutant aerobic cultures.
To further probe the metabolic changes associated with the nos srrAB mutant during mid-exponential-phase growth, targeted metabolomics analysis was performed on the wild type, the nos, srrAB, and nos srrAB mutants, and the complemented strains (Tables 2 and 3 and File S1). This analysis revealed that all three mutant strains had decreased intracellular lactate levels relative to the wild type (Table 2) and verified that the nos srrAB mutant had significantly elevated levels of secreted lactate (Table 3), which are also observed in Fig. 3B. The increased amount of excreted lactate in the nos srrAB mutant is consistent with the increased expression of ldh1 and ldh2 in this strain (Table 1). Alterations in TCA cycle intermediates relative to that in the wild type were also observed in the nos srrAB double mutant, including increased intracellular succinate and pyruvate levels and decreased extracellular α-ketoglutarate levels (Tables 2 and 3). Amino acid changes of note in the nos srrAB mutant included unique urea cycle amino acid changes (increased intracellular ornithine and citrulline levels relative to those in the wild type), significantly decreased intracellular glutamine and glutamate levels relative to those in the wild type (this pattern was also observed in the srrAB single mutant), and significantly increased levels of intracellular aromatic amino acids (tyrosine, tryptophan, phenylalanine) relative to those in the wild type and the nos and srrAB single mutants (Tables 2 and 3 and File S1). Measurement of gene expression surrounding the citrulline and ornithine nodes of the urea cycle revealed that arginine deiminase (arcA; arginine → citrulline) and ornithine carbamoyltransferase (arcB; carbamoyl-P → citrulline) were upregulated >1,000-fold, whereas the levels of otc (which is redundant with arcB), bifunctional ornithine acetyltransferase/N-acetylglutamate synthase (argJ; N-acetyl-ornithine → ornithine), argininosuccinate synthase (argG; citrulline → arginosuccinate), and arginase (rocF; arginine → urea) were not changed relative to those in the wild type (Table 1). The NAD+/NADH ratios for both the srrAB and nos srrAB mutants were also significantly higher than those for the wild type and nos mutant. Collectively, these metabolite changes confirm that the nos srrAB mutant relies on lactate fermentation, presumably to offset redox imbalance, and also acquires additional energy via arginine deamination.
TABLE 2.
Intracellular metabolite summary
| Cellular metabolite | % increase/decreasec
|
||
|---|---|---|---|
| nos mutant vs wild type | srrAB mutant vs wild type | nos srrAB mutant vs wild type | |
| Organic acids | |||
| Lactate | 69.89 | −84.46 | −66.96 |
| Citrate | 218.88 | — | — |
| Succinate | 40.38 | 119.49 | 199.61 |
| Pyruvate | — | 69.29 | 34.00 |
| Amino acids | |||
| Arginine | — | 47.08 | — |
| Asparagine | — | −34.68 | −55.58 |
| Citrulline | BLOQ | BLOQ | 260.8b |
| Glutamine | −45.35 | −47.82 | −69.83a |
| Glutamate | — | −58.36 | −65.58 |
| Ornithine | −66.12 | −62.72 | 126.74 |
| Leucine | — | 71.11 | 43.93 |
| Isoleucine | — | 57.01 | — |
| Valine | −25.43 | 51.12 | −99.95 |
| Phenylalanine | — | 47.42 | 91.48 |
| Tryptophan | — | 149.56 | 303.53 |
| Tyrosine | — | 67.33 | 181.64 |
| Methionine | — | −29.61 | — |
| Acyl-CoAs | |||
| Acetyl-CoA | — | −71.85 | — |
| Malonyl-CoA | −72.86 | −71.86 | — |
| Adenine nucleotides | |||
| NADH | — | −92.72a | −86.58 |
| NAD+ | −16.75 | 17.93 | 51.95 |
| NAD+/NADH | — | 1,603.3a | 1,069.03 |
| ATP | — | 76.77 | 85.31 |
| ADP | — | 45.63 | — |
| AMP | — | — | −70.16 |
The calculation was based on the data for 2 samples, as the value for the third sample was below limit of quantification.
The data are reported as the number of nanomoles per milligram of protein since the percent increase/decrease could not be calculated.
The data are for changes that were statistically significant (P < 0.05). —, the change was not statistically significant. BLOQ, below the limit of quantification.
TABLE 3.
Extracellular metabolite summary
| Extracellular metabolite | % increase/decreasea
|
||
|---|---|---|---|
| nos mutant vs wild type | srrAB mutant vs wild type | nos srrAB mutant vs wild type | |
| Organic acids | |||
| Lactate | — | 751.97 | 1,823.59 |
| Citrate | 4.63 | — | — |
| α-Ketoglutarate | −65.64 | −80.67 | −87.68 |
| Fumarate | −34.49 | −21.06 | −57.22 |
| Malate | 21.12 | 56.92 | 19.98 |
| Pyruvate | 388.02 | — | — |
| Succinate | — | −10.27 | — |
| Amino acids | |||
| Proline | — | 50.16 | 75.86 |
| Ornithine | 45.53 | 57.36 | 140.44 |
| Citrulline | — | — | 145.39 |
| Glycine | — | 24.33 | 23.05 |
| Glutamine | — | 60.37 | 86.99 |
| Glutamate | — | — | 31.50 |
| Threonine | — | 26.78 | 19.5 |
| Alanine | 27.22 | — | 24.46 |
| Asparagine | 25.34 | 42.93 | 39.24 |
| Aspartate | — | 26.35 | 20.02 |
The data are for changes that were statistically significant (P < 0.05). —, the change was not statistically significant.
The contributions of nos and srrAB are not synergistic with virulence and intracellular invasion.
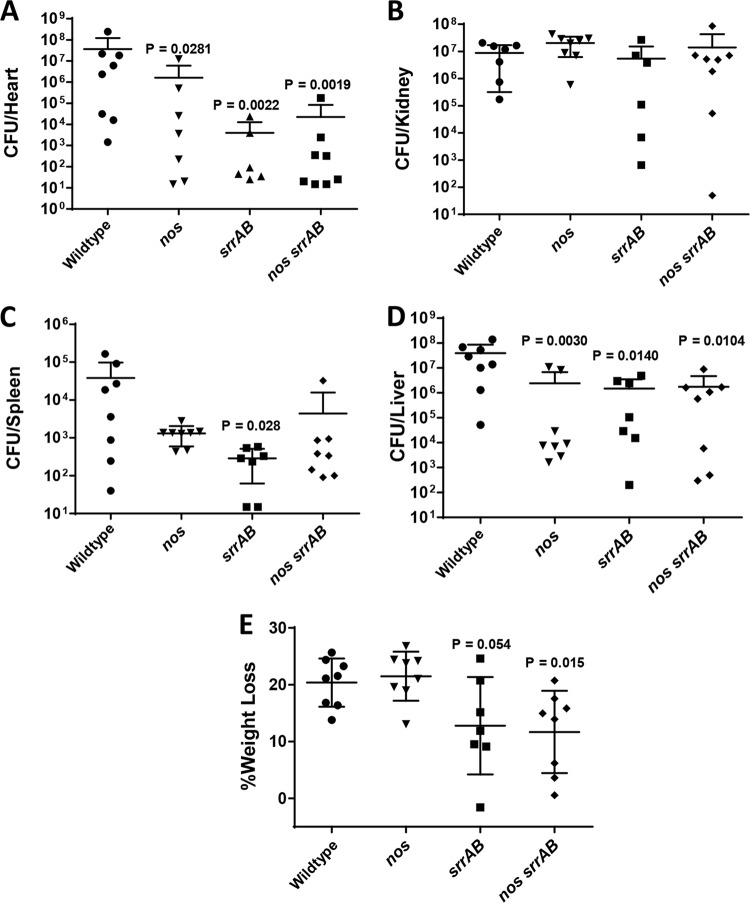
Several of the phenotypes of the nos srrAB double mutant are reminiscent of those of S. aureus small-colony variants (SCVs; slow fermentative growth, increased tolerance to antibiotics, smaller colony size on agar plates), which typically have an increased ability to persist inside nonphagocytic eukaryotic cells (26–28). We therefore assessed the wild type and isogenic mutants for their ability to invade human endothelial cells (Fig. 7). All three mutants were significantly compromised in their intracellular invasion ability relative to the wild type, but the invasion frequency was not measurably different between the single and double mutant strains in this model (P > 0.05, Kruskal-Wallis one-way analysis of variance on ranks). The virulence of the wild type and the nos, srrAB, and nos srrAB mutants was also compared in a 3-day murine sepsis model of infection (Fig. 8), which revealed differential organ colonization among the mutants and the wild type. The nos single mutant had a decrease in the number of CFU in the heart (1.5-log reduction, P = 0.0281; Fig. 8A), spleen (1.5-log reduction, not statistically significant; Fig. 8C), and liver (1.2-log reduction, P = 0.003; Fig. 8D) relative to that of the wild type, but the nos mutant and the wild type colonized the kidneys at comparable levels (Fig. 8B). Both the srrAB and nos srrAB mutants had the same pattern of liver (statistically significant for both), spleen (statistically significant for the srrAB mutant), and kidney colonization as the nos mutant, but the magnitude of the decreased numbers of CFU was much greater for these two mutant strains in the heart (3-log reduction, statistically significant for both) (Fig. 8A to D). The virulence of these strains was also assessed by monitoring the weight loss of the infected animals (Fig. 8E). At day 3 postinfection, mice infected with the nos single mutant displayed weight loss comparable to that achieved with wild-type infection (∼20%), whereas the srrAB mutant- and nos srrAB mutant-infected mice underwent reduced weight loss (∼12%) which was significantly different between the nos srrAB mutant-infected mice (P = 0.015) and the wild-type-infected mice (Fig. 8E). Similar in vivo results with respect to organ colonization and animal weight loss were obtained in a separate 4-day infection comparing the wild type and the nos srrAB mutant (Fig. S6).
FIG 7.
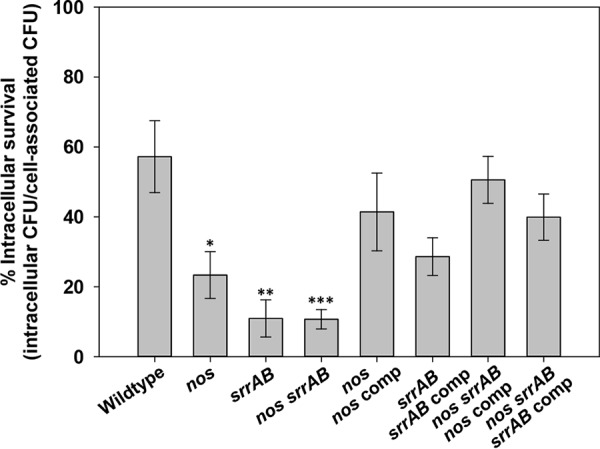
Endothelial cell invasion by the S. aureus wild-type, isogenic mutant, and complemented strains. Sixteen-hour aerobic TSB+G cultures of either wild-type (n = 7 independent experiments), isogenic nos mutant (n = 6), srrAB mutant (n = 5), nos srrAB mutant (n = 6), or complemented (n = 3) strains were used to infect confluent endothelial cells (EA.hy926 cells) for 1 h (average infection dosage range, 3 × 108 to 5 × 108 CFU ml−1) at 37°C in 5% CO2. Intracellular percent survival was determined as described in Materials and Methods and represents the ratio of intracellular to cell-associated S. aureus (intracellular plus adherent). *, P = 0.01 (t test) relative to the wild type; **, P = 0.003 (t test) relative to the wild type; ***, P = 0.001 (Mann-Whitney rank-sum test) relative to the wild type. Error bars indicate the SEM.
FIG 8.
In vivo virulence of wild type and isogenic nos and srrAB mutants. Wild-type (5.8 × 107 CFU) and isogenic nos (5.2 × 107 CFU), srrAB (3.4 × 107 CFU), and nos srrAB (3.6 × 107 CFU) mutant cultures were each retro-orbitally injected into 8-week-old C57BL/6 female mice (n = 8 mice per group). Mice were euthanized at 3 days postinfection, and organs were processed for determination of the number of CFU per organ as described in Materials and Methods. Graphs depict the calculated bacterial burdens for heart (A), kidney (B), spleen (C), and liver (D). The lowest dilution plated for heart and spleen was 10−1. (E) The percent weight loss at 3 days postinfection of each animal group is also reported. For panels A to E, the data are graphed as scatter plots with the mean (solid line) and standard deviation (error bars). Two-tailed P values are reported for all statistically significantly differences in comparisons to the wild type using a Mann-Whitney test.
DISCUSSION
Complex interconnected regulatory pathways are involved in S. aureus metabolic adaptation, which contributes to its versatility as a pathogen, and new insights into this interplay may lead to alternative treatment strategies. The results of this study, combined with those described in previous publications (8, 9), strongly suggest an important role for both saNOS and SrrAB in the modulation of S. aureus physiology. Mutation of S. aureus nos impacts both aerobic (8, 9) and anaerobic (10) nitrate-based respiration, and srrAB mutants are compromised in the induction of low-oxygen/anaerobic metabolic genes, likely contributing to the low-oxygen growth defect typically observed in srrAB mutants (15, 17, 19, 21, 22). Factors that impair electron flow during respiration, such as nitrosative stress, mutation of the quinol oxidase respiratory cytochrome, and low-O2 growth, have previously been reported to be signals to which SrrAB responds (17, 18, 22). Our previously published study of an S. aureus nos mutant revealed alterations in membrane potential and respiratory dehydrogenase activity and increased endogenous ROS levels, either of which could affect the quinone pool reduction during respiration (9). These results, combined with the nos mutant having elevated expression of SrrAB-regulated anaerobic genes (9), suggest that SrrAB coordinates gene expression in response to altered respiratory chain function, to allow optimal aerobic growth in the absence of saNOS. This theory was further evaluated in the present study by monitoring aerobic gene expression and the growth properties of a nos srrAB double mutant. Surprisingly, the results of these experiments suggest that saNOS and SrrAB act independently in the regulation of certain aerobic respiratory phenotypes but also display interplay in the control of other aspects of aerobic respiration, as discussed in more detail below.
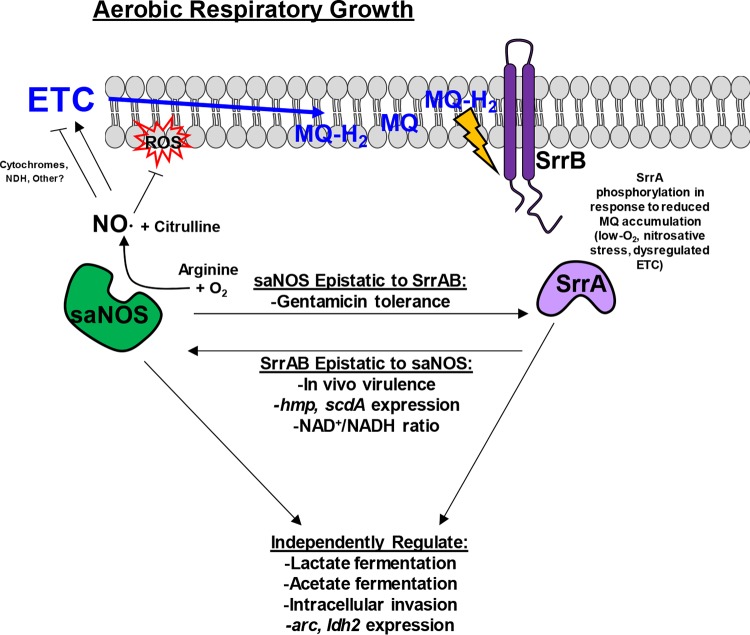
A proposed model for the regulation of aerobic respiration by saNOS and SrrAB, based on the phenotypes observed in the nos, srrAB, and nos srrAB mutants in this and other published studies, is summarized in Fig. 9. During aerobic respiratory growth, SrrB (sensor kinase) monitors the reduced state of the membrane menaquinone pool (17). Independently of SrrAB, saNOS generates nitirc oxide (NO), which acts on an as-yet-unidentified member(s) of the respiratory chain to act as a “clutch,” slowing respiration to an optimal level that limits endogenous ROS accumulation. One likely target of saNOS-derived NO is respiratory cytochromes, which are known to bind NO and slow respiration (29–33). This proposed clutch function of saNOS is supported by previous analysis of a nos mutant, which displayed increased endogenous ROS levels, increased respiratory dehydrogenase activity, an increased membrane potential, and slightly elevated oxygen consumption (9). The fact that SrrAB upregulates expression of nos during low-oxygen growth (see Fig. S1 in the supplemental material) also supports a clutch function for saNOS and demonstrates interplay between saNOS and SrrAB. The absence of saNOS likely renders aerobic respiration suboptimal, resulting in an accumulation of reduced menaquinones, which is sensed by SrrB. Cellular countermeasures that minimize the negative effects of aerobic respiration are then likely induced directly via phosphorylated SrrA and/or indirectly through other metabolic regulators, allowing aerobic growth to proceed relatively normally.
FIG 9.
Proposed model of saNOS and SrrAB interplay and regulation of aerobic respiration in S. aureus. MQ, menaquinone; ETC, electron transport chain; NDH, NADH dehydrogenase.
The absence of SrrAB appears to have a more pronounced effect on S. aureus aerobic respiratory growth, as oxygen consumption is decreased by 34% relative to that by the wild type (Fig. 4), and the NAD+/NADH ratio is highly increased (Table 2). Although these results suggest that SrrAB has a more global role than saNOS in remodeling S. aureus metabolism in response to aerobic respiratory stress, the fact that the srrAB mutant phenocopies the nos mutant with respect to aerobic growth properties (Fig. 1A to C) suggests some redundancy and independent regulation by saNOS and SrrAB. One example of this is the effect of nos and srrAB mutations on acetate excretion (Fig. 3A). Energy conservation is essential during impaired cellular respiration, in which case substrate-level phosphorylation during acetate generation drives ATP production. During aerobic growth, acetate was produced among all the strains, with the rapid increase in the accumulation of acetate corresponding to the transition into death phase (Fig. 1). Interestingly, acetate levels were sustained into stationary phase in both the nos and srrAB mutants compared to the wild type, suggesting their greater need for ATP production via substrate-level phosphorylation to potentially offset impaired cellular respiration (Fig. 3A). The srrAB mutant has altered aerobic respiration, as evident by decreased oxygen consumption, whereas in the nos mutant, respiration may negatively impact the cell due to increased endogenous ROS (9). In each case, the increased levels of acetate may represent an adaptation to suboptimal respiration. Acetate production in S. aureus also has the additional advantage of providing a stored energy source catabolized through the TCA cycle (34). Mutation of both nos and srrAB did not have an additive effect on acetate excretion; rather, acetate accumulated gradually over time as a function of growth (Fig. 3A). These results reinforce the hypothesis that saNOS and SrrAB have independent roles in affecting aerobic acetate fermentation.
Other phenotypes of the nos, srrAB, and nos srrAB mutants suggestive of independent regulation include intracellular invasion of endothelial cells (Fig. 7), expression of the ldh2 and arginine deiminase genes (Table 1), and lactate excretion (Fig. 3B). The nos srrAB mutant had significantly impaired aerobic respiratory growth (Fig. 1) and oxygen consumption (Fig. 4) and significantly increased excreted lactate levels (Fig. 3B) relative to the wild type. Due to the compromised oxygen consumption in the nos srrAB mutant, it is likely that these cells maintain a redox balance through lactate fermentation, which corresponds to the significant induction of ldh1 and ldh2 expression observed in the double mutant (Table 1). The arginine deiminase pathway most likely provides additional ATP via substrate-level phosphorylation (35) in the nos srrAB mutant, with significant increases in both arcA and arcB expression (>1,733- and >1,195-fold, respectively; Table 1) relative to that in the wild type. Furthermore, examination of products of the arginine deaminase pathway revealed that the nos srrAB mutant had increased amounts of intracellular and extracellular ornithine, 126% and 140%, respectively (Tables 2 and 3), and an increased amount of intracellular citrulline was detected only in the nos srrAB mutant (Tables 2 and 3). Expression of genes involved in nitrate metabolism (genes for nitrate reductase, nitrite reductase, nitrite transport) was also highly upregulated in the nos srrAB mutant (Table 1), even though nitrate concentrations (100 µM) could not support aerobic respiration and there was little variation in the nitrate concentration among the strains (Fig. S4). The addition of each of nitrate, nitrite, and ammonia was able to partially rescue the growth of the nos srrAB mutant (Fig. 6 and Fig. S5), suggesting more of an assimilatory role, although we cannot exclude the possibility of nitrate-based respiration in this mutant based on these data alone. The partial rescue of the nos srrAB mutant with ammonia strongly suggests nitrogen assimilation into glutamate and glutamine (both metabolites with elevated concentrations; Tables 2 and 3), which could be used to synthesize amino acids, purines, pyrimidines, amino sugars, and NAD+ (36).
In addition to the independent regulation elicited by saNOS and SrrAB in response to suboptimal aerobic respiration, several potential epistatic relationships between these two systems were also observed (Fig. 9). Given that both the nos and nos srrAB mutants displayed complete tolerance to gentamicin killing, while the srrAB mutant was sensitive to this antibiotic, like the wild type (Fig. 5), it is likely that saNOS is epistatic to SrrAB with respect to this phenotype. This nos-dominant phenotype could be complemented only by a nos-expressing plasmid in both the nos and nos srrAB mutants. Gentamicin, an aminoglycoside antibiotic, requires active transport into the cell through an energy-dependent mechanism (37). The lack of gentamicin uptake and accumulation in cells deficient in or not capable of respiration is well documented (38–40). Studies have shown that S. aureus small-colony variant (SCV) resistance to gentamicin uptake was due to the inability to generate and maintain energized membranes from respiration, directly linking gentamicin uptake to electron transport and the generation of a proton motive force (PMF) (37). Intriguingly, the nos mutant was still able to respire on oxygen at rates comparable to those for the wild type (Fig. 4), suggesting that some other aspect of the respiratory chain that is altered in the nos mutant is responsible for its gentamicin-tolerant phenotype. One possibility is that the menaquinone redox state is altered when saNOS is absent, due to increased endogenous ROS production (9). This would be consistent with the findings of a study that implicated dysregulation of the quinone cycle in resistance to gentamicin in facultative bacteria (40), as well as a study that showed increased resistance to gentamicin in a menaquinone-deficient Bacillus subtilis mutant (25).
Several phenotypes suggestive of SrrAB epistasis to saNOS were also observed, such as expression of scdA and hmp (Table 1), significantly increased NAD+/NADH ratios (Table 2), and in vivo virulence (Fig. 8). In all cases, the phenotypes of the srrAB and nos srrAB mutants were comparable. The importance of saNOS and SrrAB to S. aureus virulence has been demonstrated in several infection models. A nos mutant was previously shown to have reduced virulence (4) and reduced abscess size and CFU burdens (6) in murine sepsis and abscess models of infection, respectively. The results of both of those studies are consistent with our current results, in which infection with the nos mutant resulted in reduced CFU burdens of the heart, spleen, and liver (Fig. 8) and no difference in weight loss for mice infected with the nos mutant from that for mice infected with the wild type (Fig. 8E). Strains with a mutation of srrAB have shown reduced virulence in anoxic sites, as in murine orthopedic implant-associated infections (41) and in intraosseous infections (22). S. aureus virulence in a murine septic arthritic model was highly influenced by SrrAB in both the presence and the absence of host nitrosative stress (18). Our data are congruent with those from previous studies, in that the srrAB mutant showed reduced virulence in most organs assessed for CFU burden, and mice infected with this mutant had less weight loss than mice infected with the wild type (Fig. 8). Although the nos srrAB mutant has altered growth properties in vitro, it did not translate to an additive effect on in vivo virulence: the double mutant phenocopied the srrAB single mutant with respect to organ CFU burdens and mouse weight loss (Fig. 8), suggesting that the influence of SrrAB on virulence is dominant in this model. Interestingly, the CFU burden in the kidneys of mice infected with strains with a mutation of nos and/or srrAB was not impacted relative to that in the kidneys of mice infected with the wild type in this 3-day model of infection (Fig. 8B). This pattern of differential organ colonization/CFU burden is reminiscent of the effects of a cytochrome mutation on S. aureus sepsis (42), in which inactivation of either qoxB or cydB resulted in significantly reduced colonization of the liver or heart, respectively, but kidney bacterial burdens were unaffected.
In summary, our current findings reinforce the importance of metabolic flexibility to the effectiveness of S. aureus as a pathogen and provide insight into the complex interconnectedness of SrrAB and saNOS, which together play an important role in mediating the switch between aerobic and anaerobic metabolism, as demonstrated by restriction of the nos srrAB mutant to fermentative metabolism and a small-colony variant-like phenotype. These data highlight a small but vital portion of S. aureus metabolic regulatory networks that also includes Rex (43), NreABC (44, 45), and ArcR (46). The potential roles of Rex, ArcR, and NreABC as additional players in regulating the fermentative phenotype of the nos srrAB mutant are the subject of ongoing studies, as well as additional experimental validation of the proposed epistatic relationships between saNOS and SrrAB.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All Staphylococcus aureus strains used in this study are listed in Table S1 in the supplemental material. Unless otherwise indicated, all experiments were performed with the strains UAMS-1 (wild type), KR1010 (UAMS-1 nos::erm mutant), KB6004 (UAMS-1 ΔsrrAB mutant), and KR1014 (nos::erm ΔsrrAB mutant), each of which harbored plasmid pMK4 (empty vector), an nos complementation plasmid, or an srrAB complementation plasmid. For each experiment, the strains were freshly streaked from −80°C glycerol stocks and grown at 37°C on tryptic soy agar (TSA) with the following selective antibiotics, as appropriate, at the indicated concentrations: erythromycin (Erm) at 2 µg/ml, tetracycline (Tet) at 5 µg/ml, and chloramphenicol (Cm) at 5 µg/ml. Individual colonies were selected from TSA plates and then grown for approximately 15 h in tryptic soy broth containing 14 mM glucose (TSB+G; with antibiotic as appropriate) with shaking at 250 rpm and at 37°C. For aerobic and low-oxygen growth experiments, cultures were inoculated from overnight cultures to a final OD at 600 nm (OD600) of 0.025. For aerobic growth curves, cultures were grown at 37°C in 40 ml of either TSB+G or TSB with no glucose (TSB−G) in 500-ml Erlenmeyer flasks (1:12.5 volume/flask ratio) and shaking at 250 rpm. For low-oxygen growth curves, cultures were grown at 37°C in TSB+G in 150 ml of medium in 250-ml Erlenmeyer flasks (1:1.7 volume/flask ratio) without shaking. For all growth curve analyses, 1-ml samples were taken every 2 h for the first 12 h and at the final time point (24 h of growth). Each sample was centrifuged (13,000 × g, 4°C for 5 min), and the cell-free supernatants were stored for acetate, lactate, nitrate, nitrite, and ammonia measurements. Growth was monitored by measuring the number of CFU per milliliter by serial dilution plating (47) and the optical density (OD600) with a Genesys 10 Bio spectrophotometer (Thermo Fisher). The supernatant pH was measured with pH indicator strips (Millipore) at each time point.
Generation of UAMS-1 nos srrAB mutant.
For generation of the nos srrAB double mutant, the temperature-sensitive allele replacement vector pTR27 (nos::erm) (4) was phage transduced from S. aureus RN4220 into KB6004 (ΔsrrAB) (48). Once its presence in the target strain was confirmed, a temperature-sensitive allele replacement event was initiated by growth at 43°C (the nonpermissive temperature for plasmid replication) on TSA plus 10 mg/ml Erm to promote chromosomal integration via homologous recombination at the nos locus. A second recombination event was induced by growing a single isolated colony in TSB (no antibiotic) for 5 days at 30°C with subculturing every 24 h. Screening for the nos insertion and loss of the vector was completed by picking and patching colonies onto TSA plus 2 µg/ml Erm and TSA plus 10 µg/ml Cm. PCR was used to confirm the presence of the nos and srrAB mutant alleles in strain KR1014. KR1014 was complemented by moving either pMKnos (the nos complement plasmid) or pIHW58 (the srrAB complement plasmid) into this strain by phage transduction. Plasmid pMK4 was also moved into both KR1014 and KB6004 by phage transduction to create empty vector strains.
RNA isolation and qPCR.
To measure mid-exponential-phase (3 h of growth for the wild type, nos mutant, srrAB mutant, and complemented nos srrAB mutant strains; 6 h of growth for the nos srrAB mutant) gene expression, cultures were inoculated to a final OD600 of 0.025 in TSB−G. At mid-exponential-growth phase, 6 ml of each culture (40 ml for the nos srrAB mutant) was centrifuged (3,901 × g, 4°C for 10 min) and cell pellets were resuspended in RNAlater. To compare the expression levels of nos between the wild type and the srrAB mutant, both strains were grown under low-oxygen conditions in TSB+G as described above and harvested by centrifugation after 6 h of growth. All pellets were stored at −80°C until further processing. RNA was extracted using a FastPrep system with Lysing Matrix B as previously described (49). RNA was further treated with Turbo DNase (Ambion Turbo DNA-free kit) to remove any contaminating DNA, and the RNA concentration and purity were quantified on a BioTek Take3 plate. For real-time PCR, cDNA was synthesized from 0.75 µg of purified RNA using an iScript reverse transcriptase kit (Bio-Rad). Expression of genes of interest was measured by real-time using iQ SYBR green super mix (Bio-Rad) and a CFX Connect real-time system (Bio-Rad). Relative fold expression normalized to that of the reference housekeeping gene sigA was calculated using the Livak method (2−ΔΔCT), as described previously (4, 50). The primers used for quantitative PCR (qPCR) are listed in Table S2. For each gene transcript, qPCR was performed on 3 biological samples with 3 technical replicates per sample.
Quantification of extracellular acetate, lactate, nitrate, and ammonia.
To measure extracellular metabolites from spent media, supernatants from TSB−G aerobic growth experiments were thawed on ice. Acetate, lactate, nitrate, and ammonia were measured by enzymatic analysis with R-Biopharm kits. Measurements were performed in 96-well cell culture plates (Costar 3596), and quantities were measured in a Cytation 3 imaging reader (BioTek). Each supernatant had 3 technical replicates and 3 biological replicates per strain per time point. To account for potential background in the media, the amount of each metabolite in sterile TSB−G was measured and subtracted from each measurement. Acetate concentrations were measured using an acetic acid kit (R-Biopharm), which couples acetate and CoA ligation by acetyl-CoA synthetase to citrate synthase and l-malate dehydrogenase to measure the NAD+ reduction. The reaction volume for acetate measurements was adjusted to 283 µl. l-Lactate concentrations were determined using a d-lactic acid/l-lactic acid kit (R-Biopharm), where lactate dehydrogenase activity is measured by the reduction of NAD+. The reaction volume for l-lactate was adjusted to 224 µl. Nitrate concentrations were determined with a nitrate kit (R-Biopharm), which uses nitrate reductase to oxidize NADPH, which can be measured spectrophotometrically at 340 nm. All procedures were conducted according to the methods outlined in the kit, with adjustment of the reaction volume to 305 µl for each assay to fit a 96-well format. Ammonia concentrations were determined using an ammonia kit (R-Biopharm), which converts ammonia and 2-oxoglutarate to l-glutamate, oxidizing NADH via glutamate dehydrogenase. NADH oxidation was measured at 340 nm spectrophotometrically.
Calculation of specific maximal growth rate.
For growth supplementation with nitrate, nitrite, and ammonia, 25 ml of TSB−G was inoculated to an OD600 of 0.025 from fresh overnight cultures and supplemented as indicated with either 5 mM sodium nitrate, 1 mM sodium nitrite, or 1 mM ammonium chloride prior to incubation. Cultures were dispensed in sterile 100-well honeycomb Bioscreen C plates in 200-µl aliquots (1:2 volume/well ratio) with four technical replicates for each culture. Cultures were incubated with maximum shaking at 37°C, and growth was monitored by measuring the OD600 every 15 min for 24 h on the Bioscreen C growth analysis system (Growth Curves USA). Maximum specific growth rates (per hour) were determined by linear regression analysis of the slope over a range of from 50 to 200 min, where the R2 value was >0.99 (51). All growth rates were determined on three independent biological replicates for each condition.
Oxygen consumption.
Oxygen consumption was measured using a 4-channel free radical analyzer (TBR-4100; World Precision Instruments) and a Clark-type electrode (ISO-Oxy-2; World Precision Instruments) as described previously (9, 17). O2 consumption was measured from aerobic TSB−G inoculated to a final OD600 of 0.025 as described above. Cultures were grown to mid-exponential phase: 3 h for the wild-type, nos mutant, srrAB mutant, and complemented strains and 6 h for the nos srrAB mutant. Cultures were harvested by centrifugation (3,901 × g, 22°C for 5 min), and spent medium was discarded. Pellets were resuspended in prewarmed phosphate-buffered saline (PBS), and then 500 µl of each resuspension was added to 12.5 ml of PBS in a glass vial, the mixture was stirred at 350 rpm and 37°C, and O2 consumption was measured after the addition of 10 µl of 100 mM glucose.
Antibiotic kill curves.
Fourteen-hour overnight cultures of each strain were diluted to a final OD600 of 0.025 in 40 ml TSB−G and grown at 37°C and 250 rpm at a 1:12.5 volume/flask ratio. Cultures were grown for 2 h and sampled for serial dilution and plating for determination of the number of CFU (at the time zero time point) prior to addition of either gentamicin (final concentration, 5 µg/ml; 10× MIC), vancomycin (final concentration, 40 µg/ml; 20× MIC), or ciprofloxacin (final concentration, 30 µg/ml; 20× MIC). Cultures were grown for 8 more hours at 37°C and 250 rpm, with samples taken at 1, 2, 4, 6, and 8 h posttreatment for serial dilution and plating for determination of the number of CFU. The number of CFU per milliliter at each time point posttreatment was expressed as a percentage (percent viability) of the number of CFU per milliliter at time zero for each culture.
Cell collection and metabolite sample preparation for metabolite analysis.
For targeted metabolomics, cultures were grown aerobically in 40 ml of TSB−G as described above and harvested at mid-exponential-growth phase (3 h for the wild type, nos mutant, srrAB mutant, and nos srrAB-complemented strains and 6 h for the nos srrAB mutant). Cells were harvested by centrifugation (3,901 × g, 4°C for 10 min) in prechilled 50-ml Falcon tubes. Culture supernatants were removed and immediately placed on ice, and 1-ml aliquots were dispensed into prechilled 1.7-ml microcentrifuge tubes and flash frozen in liquid nitrogen. One milliliter of uninoculated medium (n = 3) was also dispensed into prechilled 1.7-ml microcentrifuge tubes and flash frozen in liquid nitrogen. Cell pellets were resuspended in 2 ml of ice-cold PBS and centrifuged (3,901 × g, 4°C for 3 min). The supernatants were removed, and the pellets were immediately resuspended in 2 ml of PBS. Samples were each dispensed in five 400-µl aliquots into prechilled 1.7-ml microcentrifuge tubes and centrifuged (3,901 × g, 4°C for 3 min). After supernatant removal, the cell pellets were flash frozen in liquid nitrogen. All samples were stored at −80°C prior to analysis. All harvesting steps were performed on ice or at 4°C and were kept cold on ice. One aliquoted cell pellet per condition was used to quantify the cellular protein concentration using the Pierce bicinchoninic acid assay, and the other 4 cell pellet aliquots were shipped on dry ice, along with the collected supernatants and sterile TSB−G controls, to the Southeast Center for Integrated Metabolomics (Targeted Metabolomics Core, Sanford Burnham Prebys Medical Discovery Institute). Extraction, derivatization, and liquid chromatography-tandem mass spectrometry quantitation of the metabolites from the cell homogenates and extracellular medium samples were performed as described in our previous publication (9).
Intracellular survival assay.
Cells of the endothelial cell line EA.hy926 (ATCC) were propagated in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in 5% CO2. Intracellular invasion assays were performed as described previously (52). In brief, confluent endothelial cells grown in T75 flasks were washed with Dulbecco’s phosphate-buffered saline (DPBS) and released by 10 min of incubation in 3 ml trypsin-EDTA (0.25%). The entire volume of cells was harvested by centrifugation at 200 × g for 10 min, and the cells were resuspended in 22 ml of sterile DMEM. The cells were then dispensed in 500-µl aliquots in a 24-well plate and incubated to confluence for 48 h. S. aureus strains were grown for 16 h in TSB+G supplemented with the appropriate antibiotics at 37°C and 250 rpm. S. aureus cells from overnight cultures were harvested by centrifugation and washed a total of 3 times in DMEM (without antibiotic). After the final wash, S. aureus cells for each strain were resuspended in 1 ml DMEM at a final OD600 of 1.0. In parallel, endothelial cells were washed once with 500 µl DPBS, and then 490 µl of DMEM (without antibiotic) was added to each well. Finally, to begin infection, 10 µl of each S. aureus-DMEM suspension was added to duplicate wells containing endothelial cells. Infected cells were incubated for 1 h at 37°C in 5% CO2 and 0 rpm, and then one set of wells was washed with PBS (to remove nonadherent extracellular bacterial cells) and eukaryotic cells were completely lysed with 500 μl 0.5% Triton X-100 in PBS. The total numbers of cell-associated (adherent plus intracellular) CFU were determined via serial dilution and plating on TSA. To determine the intracellular number of CFU, DMEM was removed from the duplicate well of each infection and replaced with DMEM supplemented with 20 µg/ml of lysostaphin. The wells were incubated for 1 h as before to ensure the complete killing of extracellular S. aureus and then washed and lysed with Triton X-100 as described above. Intracellular bacterial counts were determined via serial dilution and plating on TSA, and intracellular invasion was calculated as the percentage of the intracellular bacteria relative to the number of cell-associated bacteria.
In vivo murine sepsis model.
A murine systemic infection model was performed using 8-week-old C57BL/6 female mice (The Jackson Laboratory). The wild type and isogenic nos, srrAB, and nos srrAB mutant (each harboring the pMK4 plasmid) aerobic TSB cultures were grown to mid-exponential phase and frozen at −80°C until use. Inocula were prepared by resuspending the cell pellets in PBS to acquire a target bacterial concentration of 5 × 108 CFU/ml. All mice (n = 8 per group) were then infected via retro-orbital injection with 100 µl of each wild-type or nos, srrAB, or nos srrAB mutant strain (the exact inoculum numbers for each strain are reported in the figure legend for each experiment). Mouse weights were recorded at 48-h intervals, and the behavior of the animals was monitored for signs of distress (i.e., hunched back, scruffy fur, slow movements). At 3 days postinfection, the mice were humanely euthanized and the spleen, heart, liver, and kidneys were harvested. Kidney pairs, hearts, and spleens were homogenized in Lysing Matrix H tubes (MP Biomedicals, Santa Ana, CA) containing Hanks balanced salt solution with 0.2% human serum albumin and 10 mM HEPES using a FastPrep-24 5G instrument (MP Biomedicals) according to the manufacturer's recommended settings for mouse kidneys, hearts, and spleens, respectively. The livers were cut in half, and each half was placed into a Lysing Matrix I tube (MP Biomedicals) and homogenized using the manufacturer's recommended settings for mouse liver. After homogenization, the tubes with liver tissue from each mouse were combined. Subsequent homogenates were serially diluted in PBS containing 0.1% Triton X-100 for the first dilution and solely PBS for subsequent dilutions. Diluted samples were plated on TSA plates. A second infection model was performed as described above using only wild-type and nos srrAB mutant strains (Fig. S6), and the infection proceeded for 4 days prior to humane euthanizing of the mice. These studies were conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (53). The Institutional Animal Care and Use Committee (IACUC) of The University of Kansas Medical Center approved this protocol.
Statistical analysis.
Unless otherwise indicated, all in vitro assays were conducted in at least 3 independent experiments. Statistical analysis for all in vitro data was completed with SigmaPlot software (version 13, build 13.0.0.83; Systat Software Inc., San Jose, CA). Data were tested for normality and equal variance prior to choosing the appropriate parametric or nonparametric test. For in vivo experiments, Mann-Whitney testing (two-tailed P value) was used to compare the data for each mutant to those for the wild type, using GraphPad Prism (version 7.04) software for Windows (GraphPad Software, La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Christopher Petucci and Nidhi Kapoor (Metabolomics Core, Sanford Burnham Prebys Medical Discovery Institute, and the Southeast Center for Integrated Metabolomics) for performing the targeted metabolomics analysis.
This work was supported in part by a Southeast Center for Integrated Metabolomics (SECIM) Pilot and Feasibility Award to K.C.R. (funded by SECIM NIH grant U24-DK097209), by NIH grant AI118999 to K.C.R., and, in part, by KUMC start-up funds and NIH grant AI121073 to J.L.B.
K.L.J., A.B.M., and K.C.R. conceived of and designed this study; K.L.J., A.B.M., J.N.B., S.S.O., M.J.R., M.A.M., J.L.B., and K.C.R. performed data acquisition, analysis, and interpretation; and K.L.J., A.B.M., J.N.B., S.S.O., M.J.R., J.L.B., and K.C.R. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00570-18.
REFERENCES
- 1.Liebeke M, Lalk M. 2014. Staphylococcus aureus metabolic response to changing environmental conditions—a metabolomics perspective. Int J Med Microbiol 304:222–229. doi: 10.1016/j.ijmm.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 2.Somerville GA, Proctor RA. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol Mol Biol Rev 73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee DS, Burd H, Liu J, Almaas E, Wiest O, Barabasi AL, Oltvai ZN, Kapatral V. 2009. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J Bacteriol 191:4015–4024. doi: 10.1128/JB.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sapp AM, Mogen AB, Almand EA, Rivera FE, Shaw LN, Richardson AR, Rice KC. 2014. Contribution of the nos-pdt operon to virulence phenotypes in methicillin-sensitive Staphylococcus aureus. PLoS One 9:e108868. doi: 10.1371/journal.pone.0108868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaish M, Singh VK. 2013. Antioxidant functions of nitric oxide synthase in a methicillin sensitive Staphylococcus aureus. Int J Microbiol 2013:1. doi: 10.1155/2013/312146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Sorge NM, Beasley FC, Gusarov I, Gonzalez DJ, von Kockritz-Blickwede M, Anik S, Borkowski AW, Dorrestein PC, Nudler E, Nizet V. 2013. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J Biol Chem 288:6417–6426. doi: 10.1074/jbc.M112.448738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surdel MC, Dutter BF, Sulikowski GA, Skaar EP. 2016. Bacterial nitric oxide synthase is required for the Staphylococcus aureus response to heme stress. ACS Infect Dis 2:572–578. doi: 10.1021/acsinfecdis.6b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhari SS, Kim M, Lei S, Razvi F, Alqarzaee AA, Hutfless EH, Powers R, Zimmerman MC, Fey PD, Thomas VC. 2017. Nitrite derived from endogenous bacterial nitric oxide synthase activity promotes aerobic respiration. mBio 8:e00887-17. doi: 10.1128/mBio.00887-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mogen AB, Carroll RK, James KL, Lima G, Silva D, Culver JA, Petucci C, Shaw LN, Rice KC. 2017. Staphylococcus aureus nitric oxide synthase (saNOS) modulates aerobic respiratory metabolism and cell physiology. Mol Microbiol 105:139–157. doi: 10.1111/mmi.13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinkel TL, Ramos-Montanez S, Pando JM, Tadeo DV, Strom EN, Libby SJ, Fang FC. 2016. An essential role for bacterial nitric oxide synthase in Staphylococcus aureus electron transfer and colonization. Nat Microbiol 2:16224. doi: 10.1038/nmicrobiol.2016.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao S, Huseby DL, Brandis G, Hughes D. 2017. Alternative evolutionary pathways for drug-resistant small colony variant mutants in Staphylococcus aureus. mBio 8:e00358-17. doi: 10.1128/mBio.00358-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mashruwala AA, Boyd JM. 2017. The Staphylococcus aureus SrrAB regulatory system modulates hydrogen peroxide resistance factors, which imparts protection to aconitase during aerobic growth. PLoS One 12:e0170283. doi: 10.1371/journal.pone.0170283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mashruwala AA, Guchte AV, Boyd JM. 2017. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife 6:23845. doi: 10.7554/eLife.23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Windham IH, Chaudhari SS, Bose JL, Thomas VC, Bayles KW. 2016. SrrAB modulates Staphylococcus aureus cell death through regulation of cidABC transcription. J Bacteriol 198:1114–1122. doi: 10.1128/JB.00954-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pragman AA, Ji Y, Schlievert PM. 2007. Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry 46:314–321. doi: 10.1021/bi0603266. [DOI] [PubMed] [Google Scholar]
- 16.Grosser MR, Weiss A, Shaw LN, Richardson AR. 2016. Regulatory requirements for Staphylococcus aureus nitric oxide resistance. J Bacteriol 198:2043–2055. doi: 10.1128/JB.00229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinkel TL, Roux CM, Dunman PM, Fang FC. 2013. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 4:e00696-13. doi: 10.1128/mBio.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson AR, Dunman PM, Fang FC. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol 61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- 19.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol 186:2430–2438. 10.1128/JB.186.8.2430-2438.2004. doi: 10.1128/JB.186.8.2430-2438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, Memmi G, Wolz C, Schlievert PM, Cheung A, Doring G. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol 65:1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x. [DOI] [PubMed] [Google Scholar]
- 21.Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol 183:1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR, Hinger SA, Aysanoa EE, Blanchard C, Dunman PM, Wasserman GA, Chen J, Shopsin B, Gilmore MS, Skaar EP, Cassat JE. 2015. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog 11:e1005341. doi: 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillaspy AF, Lee CY, Sau S, Cheung AL, Smeltzer MS. 1998. Factors affecting the collagen binding capacity of Staphylococcus aureus. Infect Immun 66:3170–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Throup JP, Zappacosta F, Lunsford RD, Annan RS, Carr SA, Lonsdale JT, Bryant AP, McDevitt D, Rosenberg M, Burnham MK. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392–10401. doi: 10.1021/bi0102959. [DOI] [PubMed] [Google Scholar]
- 25.Bryan LE, Van Den Elzen HM. 1977. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob Agents Chemother 12:163–177. doi: 10.1128/AAC.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proctor RA, Kriegeskorte A, Kahl BC, Becker K, Loffler B, Peters G. 2014. Staphylococcus aureus small colony variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front Cell Infect Microbiol 4:99. doi: 10.3389/fcimb.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaudaux P, Francois P, Bisognano C, Kelley WL, Lew DP, Schrenzel J, Proctor RA, McNamara PJ, Peters G, Von Eiff C. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect Immun 70:5428–5437. doi: 10.1128/IAI.70.10.5428-5437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Eiff C, Heilmann C, Proctor RA, Woltz C, Peters G, Götz F. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J Bacteriol 179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borisov VB, Forte E, Konstantinov AA, Poole RK, Sarti P, Giuffre A. 2004. Interaction of the bacterial terminal oxidase cytochrome bd with nitric oxide. FEBS Lett 576:201–204. doi: 10.1016/j.febslet.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Borisov VB, Forte E, Sarti P, Brunori M, Konstantinov AA, Giuffre A. 2006. Nitric oxide reacts with the ferryl-oxo catalytic intermediate of the CuB-lacking cytochrome bd terminal oxidase. FEBS Lett 580:4823–4826. doi: 10.1016/j.febslet.2006.07.072. [DOI] [PubMed] [Google Scholar]
- 31.Brown GC. 1995. Nitric oxide regulates mitochondrial respiration and cell functions by inhibiting cytochrome oxidase. FEBS Lett 369:136–139. doi: 10.1016/0014-5793(95)00763-Y. [DOI] [PubMed] [Google Scholar]
- 32.Brunori M, Giuffre A, Forte E, Mastronicola D, Barone MC, Sarti P. 2004. Control of cytochrome c oxidase activity by nitric oxide. Biochim Biophys Acta 1655:365–371. doi: 10.1016/j.bbabio.2003.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Giuffre A, Borisov VB, Mastronicola D, Sarti P, Forte E. 2012. Cytochrome bd oxidase and nitric oxide: from reaction mechanisms to bacterial physiology. FEBS Lett 586:622–629. doi: 10.1016/j.febslet.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 34.Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun 70:6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunin R, Glansdorff N, Pierard A, Stalon V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White D. 2007. The physiology and biochemistry of prokaryotes, 3rd ed. Oxford University Press, Inc, New York, NY. [Google Scholar]
- 37.Miller MH, Edberg SC, Mandel LJ, Behar CF, Steigbigel NH. 1980. Gentamicin uptake in wild-type and aminoglycoside-resistant small-colony mutants of Staphylococcus aureus. Antimicrob Agents Chemother 18:722–729. doi: 10.1128/AAC.18.5.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlessinger D. 1988. Failure of aminoglycoside antibiotics to kill anaerobic, low-pH, and resistant cultures. Clin Microbiol Rev 1:54–59. doi: 10.1128/CMR.1.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baverud V, Gunnarsson A, Karlsson M, Franklin A. 2004. Antimicrobial susceptibility of equine and environmental isolates of Clostridium difficile. Microb Drug Resist 10:57–63. doi: 10.1089/107662904323047817. [DOI] [PubMed] [Google Scholar]
- 40.Bryan LE, Kwan S. 1981. Aminoglycoside-resistant mutants of Pseudomonas aeruginosa deficient in cytochrome d, nitrite reductase, and aerobic transport. Antimicrob Agents Chemother 19:958–964. doi: 10.1128/AAC.19.6.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mashruwala AA, Gries CM, Scherr TD, Kielian T, Boyd JM. 2017. SaeRS is responsive to cellular respiratory status and regulates fermentative biofilm formation in Staphylococcus aureus. Infect Immun 85:e00157-17. doi: 10.1128/IAI.00157-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, Skaar EP. 2013. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio 4:e00241-13. doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pagels M, Fuchs S, Pane-Farre J, Kohler C, Menschner L, Hecker M, McNamarra PJ, Bauer MC, von Wachenfeldt C, Liebeke M, Lalk M, Sander G, von Eiff C, Proctor RA, Engelmann S. 2010. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol Microbiol 76:1142–1161. doi: 10.1111/j.1365-2958.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nilkens S, Koch-Singenstreu M, Niemann V, Gotz F, Stehle T, Unden G. 2014. Nitrate/oxygen co-sensing by an NreA/NreB sensor complex of Staphylococcus carnosus. Mol Microbiol 91:381–393. doi: 10.1111/mmi.12464. [DOI] [PubMed] [Google Scholar]
- 45.Schlag S, Fuchs S, Nerz C, Gaupp R, Engelmann S, Liebeke M, Lalk M, Hecker M, Gotz F. 2008. Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J Bacteriol 190:7847–7858. doi: 10.1128/JB.00905-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makhlin J, Kofman T, Borovok I, Kohler C, Engelmann S, Cohen G, Aharonowitz Y. 2007. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J Bacteriol 189:5976–5986. doi: 10.1128/JB.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jett BD, Hatter KL, Huycke MM, Gilmore MS. 1997. Simplified agar plate method for quantifying viable bacteria. Biotechniques 23:648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 48.Lewis AM, Matzdorf SS, Endres JL, Windham IH, Bayles KW, Rice KC. 2015. Examination of the Staphylococcus aureus nitric oxide reductase (saNOR) reveals its contribution to modulating intracellular NO levels and cellular respiration. Mol Microbiol 96:651–669. doi: 10.1111/mmi.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patton TG, Rice KC, Foster MK, Bayles KW. 2005. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol Microbiol 56:1664–1674. doi: 10.1111/j.1365-2958.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 50.Lewis AM, Rice KC. 2016. Quantitative real-time PCR (qPCR) workflow for analyzing Staphylococcus aureus gene expression. Methods Mol Biol 1373:143–154. doi: 10.1007/7651_2014_193. [DOI] [PubMed] [Google Scholar]
- 51.Perni S, Andrew PW, Shama G. 2005. Estimating the maximum growth rate from microbial growth curves: definition is everything. Food Microbiol 22:491–495. doi: 10.1016/j.fm.2004.11.014. [DOI] [Google Scholar]
- 52.Edwards AM, Massey RC. 2011. Invasion of human cells by a bacterial pathogen. J Vis Exp. doi: 10.3791/2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.