Granulocyte colony-stimulating factor receptor (G-CSFR), encoded by the CSF3R gene, represents a major regulator of neutrophil production and function in mammals, with inactivating extracellular mutations identified in a cohort of neutropenia patients unresponsive to G-CSF treatment. This study sought to elucidate the role of the zebrafish G-CSFR by generating mutants harboring these inactivating extracellular mutations using genome editing.
KEYWORDS: blood diseases, cytokine receptors, developmental hematopoiesis, neutropenia, zebrafish
ABSTRACT
Granulocyte colony-stimulating factor receptor (G-CSFR), encoded by the CSF3R gene, represents a major regulator of neutrophil production and function in mammals, with inactivating extracellular mutations identified in a cohort of neutropenia patients unresponsive to G-CSF treatment. This study sought to elucidate the role of the zebrafish G-CSFR by generating mutants harboring these inactivating extracellular mutations using genome editing. Zebrafish csf3r mutants possessed significantly decreased numbers of neutrophils from embryonic to adult stages, which were also functionally compromised, did not respond to G-CSF, and displayed enhanced susceptibility to bacterial infection. The study has identified an important role for the zebrafish G-CSFR in maintaining the number and functionality of neutrophils throughout the life span and created a bona fide zebrafish model of nonresponsive neutropenia.
INTRODUCTION
Granulocyte colony-stimulating factor (G-CSF), also known as colony-stimulating factor 3 (CSF3), acts via its cognate cell surface receptor, G-CSFR, or CSF3R, to play a key role in both developmental and “emergency” granulopoiesis, augmenting the proliferation, differentiation, and survival of myeloid progenitors and their neutrophilic granulocyte progeny (1). In patients with severe congenital neutropenia (SCN), levels of circulating neutrophils are extremely low, defined as <0.5 × 109/liter, typically <10% of average counts, and these patients suffer from ongoing episodes of opportunistic bacterial infections that are potentially life threatening (2–4). G-CSF is utilized in the clinic to successfully treat the majority of neutropenia cases, but a group of patients remain unresponsive to the therapy (5). Within this patient cohort, “crippling” mutations within the extracellular domain of G-CSFR have been identified (1).
The zebrafish is an established model for the investigation of hematopoiesis, immunity, and related disorders (6, 7). Like mammals, zebrafish undergo distinct waves of hematopoiesis (8), which are regulated by conserved cytokine receptor signaling (9–17). Zebrafish possess a single G-CSFR, encoded by the csf3r gene orthologue, which has been shown to have a conserved role in developmental myelopoiesis (11). Zebrafish are also highly amenable to genetic manipulation, including genome editing (18). A recent publication described the use of this approach to generate a csf3r mutant that exhibited a persistent basal deficit in neutrophils (19).
Mutant zebrafish lines that harbored G-CSFR variants based on those observed in neutropenia patients unresponsive to G-CSF were generated by genome editing. These csf3r mutant fish possessed severely reduced numbers of neutrophils during primitive, definitive, and emergency hematopoiesis, which continued into adulthood. Importantly, the mutant fish displayed functional deficits, including increased susceptibility to bacterial infection and lack of responsiveness to exogenous G-CSF, with decreased survival of adult neutrophils. Collectively, this work identifies a key and ongoing role for zebrafish G-CSFR in maintaining levels of functional neutrophils and establishes a bona fide model of neutropenia unresponsive to G-CSF.
RESULTS
Generation of a csf3r mutant zebrafish based on neutropenia-associated mutations.
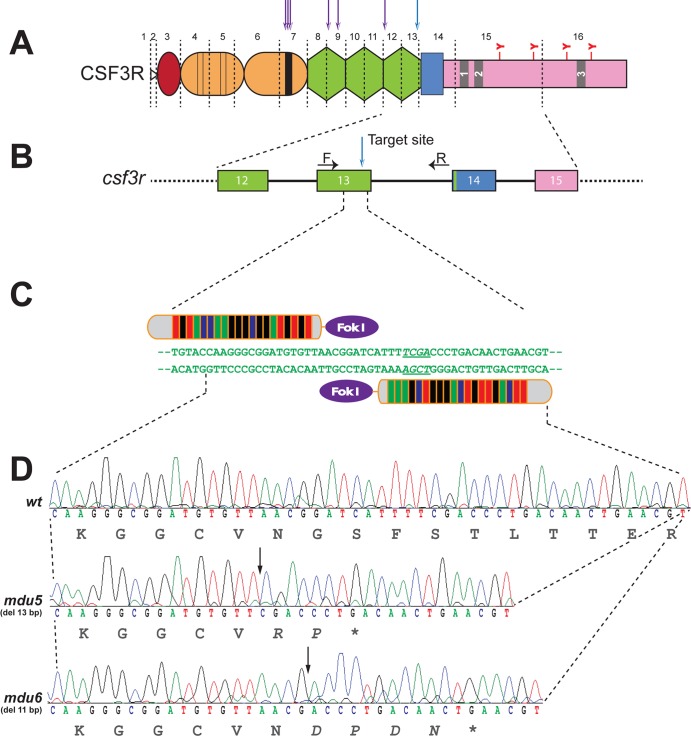
A variety of truncating mutations within the extracellular region of the human G-CSFR that are associated with SCN and chronic idiopathic neutropenia (CIN) have been described (Fig. 1A). To generate similar variants of zebrafish G-CSFR, transcription activator-like effector nuclease (TALEN)-mediated genome editing was employed to target csf3r exon 13, which encodes the most membrane-proximal fibronectin type III-like domain of the extracellular region (Fig. 1B and C). TALEN-injected embryos were raised to adulthood and crossed with wild-type fish, and F1 progeny were screened for mutations by restriction fragment length polymorphism (RFLP) with TaqI that were confirmed by sequencing. This identified two mutant alleles, csf3rmdu5 and csf3rmdu6, representing 13-bp and 11-bp deletion mutations, respectively (Fig. 1D). Each caused a frameshift that introduced a premature stop codon, leading to truncated G-CSFR proteins lacking all transmembrane and intracellular sequences. Fish carrying these alleles were outcrossed twice, and heterozygous carriers were then incrossed to generate homozygote mutants for further analysis.
FIG 1.
Generation of csf3r mutants based on neutropenia-associated alleles. (A) G-CSFR and its perturbation in neutropenia. The schematic representation of the G-CSFR shows the N-terminal leader sequence (open triangle), followed by the extracellular region comprising the immunoglobulin domain (red), four conserved cysteines (thin lines), and a W-S-X-W-S motif (thick line) within the cytokine receptor homology domain (orange); three fibronectin type III-like domains (green); and a transmembrane domain (blue); as well as boxes 1 to 3 (numbered gray rectangles) within the cytoplasmic region (pink), which includes four tyrosine (Y) residues. The exon boundaries of the zebrafish G-CSFR are shown by dashed vertical lines, and the relative positions of human G-CSFR extracellular truncations associated with SCN or CIN are indicated above the diagram by arrows. (B and C) Genome targeting of zebrafish csf3r. (B) Schematic representation of the intron/exon structure of part of the csf3r gene. Exons are represented as numbered boxes colored as in panel A, with the introns represented by solid lines and spanning primers indicated by arrows. (C) Targeting of exon 13 with a TALEN pair. The relevant nucleotide sequence is shown, with its targeting by left and right TALENs indicated and the TaqI restriction site italicized and underlined. (D) csf3r mutant alleles generated. Shown are sequences of homozygous wild-type (wt) and mutant (mdu5 and mdu6) csf3r alleles, with the respective translations shown below. The csf3rmdu5 allele represents a 13-bp deletion and the csf3rmdu6 allele an 11-bp deletion, both of which cause a frameshift resulting in a small number of residues translated from an alternative reading frame, followed by a stop codon that truncates at the C terminus of the third fibronectin type III-like domain.
csf3r mutants possess reduced numbers of neutrophils throughout life.
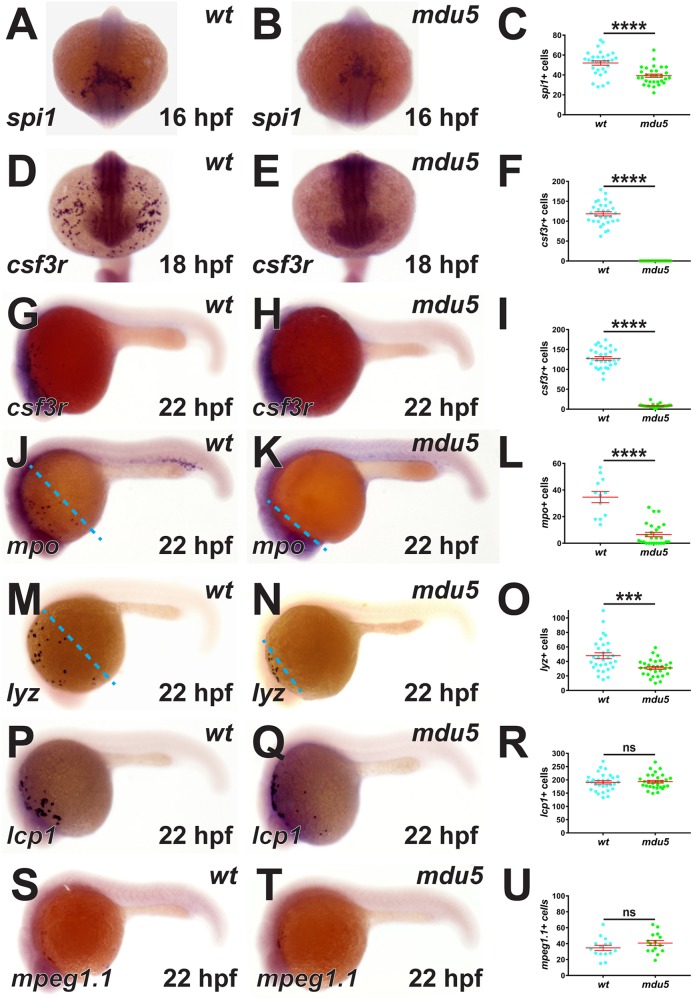
The effects of these csf3r mutations on primitive hematopoiesis were assessed first. Compared with wild-type embryos, homozygous csf3rmdu5/mdu5 mutant embryos showed a modest reduction in the myeloid progenitor marker spi1 (20) (Fig. 2A to C). Notably, an almost complete loss of csf3r expression was observed at all time points (Fig. 2D to I). Analysis with markers for mature myeloid cells demonstrated a substantial decrease in mpo+ granulocyte cells (21) (Fig. 2J to L) and a smaller, but still significant, reduction in the lyz+ leukocyte cell population (22) (Fig. 2M to O). In contrast, lcp1+ leukocytes (23) and mpeg1.1+ macrophages (24) were not affected (Fig. 2P to U). The mpo+ and lyz+ myeloid cell populations also showed a migration defect (Fig. 2J, K, M, and N), as previously noted in csf3r knockdown embryos (11). Similar results were obtained with homozygous csf3rmdu6/mdu6 mutants, and no differences were noted between heterozygote mutants and the wild types (data not shown).
FIG 2.
Effect of G-CSFR truncation on primitive myeloid cells. Homozygous wild-type (wt) and csf3rmdu5/mdu5 (mdu5) embryos were subjected to WISH with spi1 at 16 h postfertilization (hpf) (A and B); csf3r at 18 hpf (D and E) and 22 hpf (G and H); and mpo (J and K), lyz (M and N), lcp1 (P and Q), and mpeg1.1 (S and T) at 22 hpf. Individual embryos were assessed for the number of spi1+ (C), csf3r+ (F and I), mpo+ (L), lyz+ (O), lcp1+ (R), and mpeg1.1+ (U) cells, with the means and standard errors of the mean (SEM) shown in red and statistically significant differences indicated (****, P < 0.0001; ***, P < 0.001; ns, not significant). The dashed lines demarcate the extents of migration of mpo+ and lyz+ cell populations.
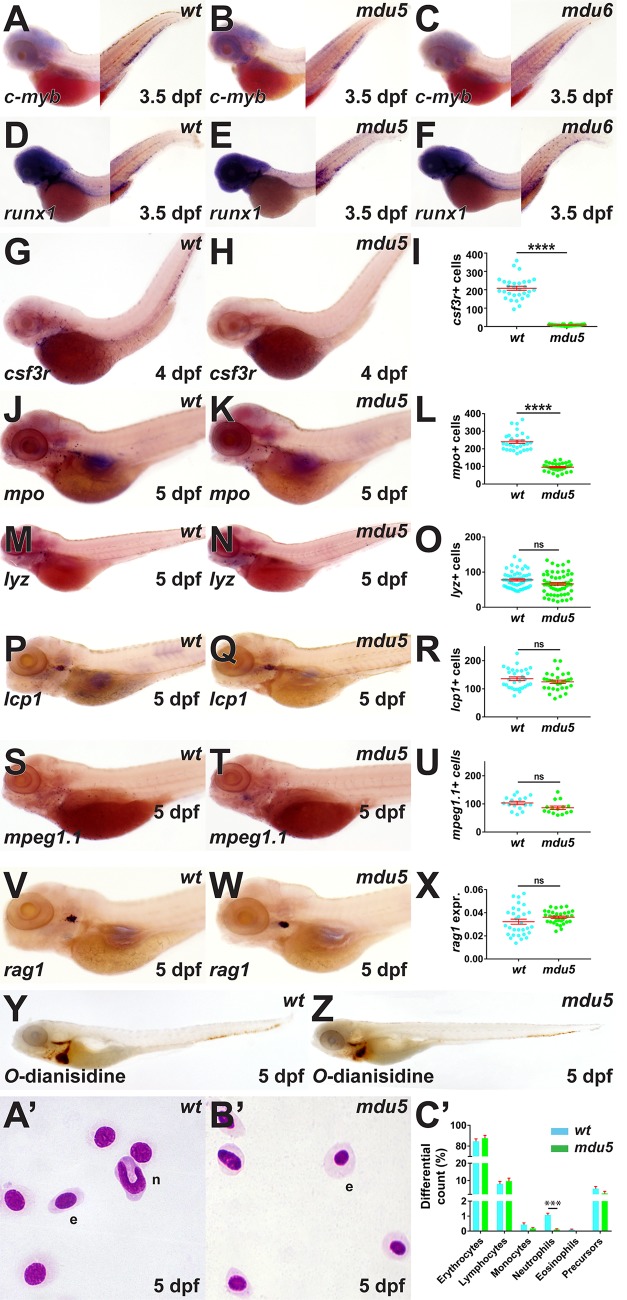
With regard to definitive hematopoiesis, no difference was observed in expression of the HSC markers c-myb (25) (Fig. 3A to C) and runx1 (26) (Fig. 3D to F) at 3.5 days postfertilization (dpf) between the wild type and the csf3rmdu5/mdu5 or csf3rmdu6/mdu6 mutant. However, csf3rmdu5/mdu5 embryos again showed negligible csf3r expression (Fig. 3G to I) and significantly decreased numbers of mpo+ cells (Fig. 3J to L) at later time points, with the number of mpo+ cells in heterozygotes again similar to those in the wild type (data not shown). In contrast, there was no difference between wild-type and csf3rmdu5/mdu5 mutants in the numbers of lyz+ (Fig. 3M to O), lcp1+ (Fig. 3P to R), and mpeg1.1+ (Fig. 3S to U) cells or the area of expression of the lymphoid cell marker rag1 (27) (Fig. 3V to X) or in the extent of staining of hemoglobinized red blood cells with O-dianisidine (21) (Fig. 3Y and Z). The csf3rmdu6/mdu6 mutant produced similar results for mpo, lyz, and rag1 (data not shown). Analysis of blood smears at 5 dpf confirmed a significant reduction in the number of circulating neutrophils in csf3rmdu5/mdu5 mutants (Fig. 3A′ to C′).
FIG 3.
Effect of G-CSFR truncation on definitive hematopoiesis. Homozygous wild-type (wt), csf3rmdu5/mdu5 (mdu5), and csf3rmdu6/mdu6 (mdu6) embryos were subjected to WISH with c-myb (A to C) and runx1 (D to F) at 3.5 dpf; csf3r at 4 dpf (G and H); and mpo (J and K), lyz (M and N), lcp1 (P and Q), mpeg1.1 (S and T), and rag1 (V and W) or whole-embryo staining with O-dianisidine (X and Y) at 5 dpf or were subjected to blood analysis at 5 dpf (A′ and B′). e, erythrocyte; n, neutrophil. Individual embryos wetre assessed for the numbers of csf3r+ (I), mpo+ (L), lyz+ (O), lcp1+ (R), and mpeg1.1+ (U) cells. The area of staining for rag1 is expressed as a ratio to eye size averaged for individual embryos (X) or blood differential counts (C′), with means and SEM shown in red and statistically significant differences indicated (****, P < 0.0001; ***, P < 0.001; ns, not significant).
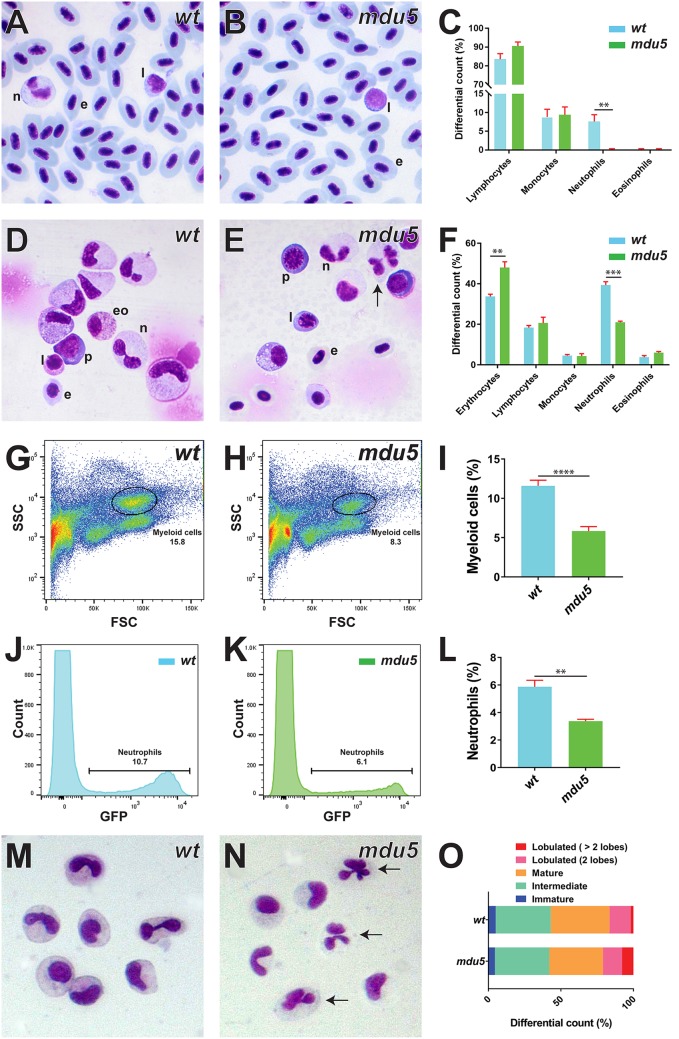
Analysis of adult homozygous csf3rmdu5/mdu5 fish confirmed that the number of blood neutrophils remained markedly reduced relative to their wild-type siblings (Fig. 4A to C). A significantly decreased neutrophil population was also observed in the kidneys of csf3rmdu5/mdu5 fish, but they were well differentiated, with hypersegmented neutrophils observed, while erythroid numbers were slightly elevated (Fig. 4D to F). Fluorescence-activated cell sorter (FACS) analysis of kidney cells from fish harboring the csf3rmdu5/mdu5 genotype on a transgenic mpo promoter-driven GFP [Tg(mpo::GFP)] background (28) identified a decrease in both the total myeloid (Fig. 4G to I) and mpo+ (Fig. 4J to L) populations. Histochemical staining of sorted myeloid cells demonstrated an increase in hypersegmented neutrophils in csf3rmdu5/mdu5 fish (Fig. 4M to O).
FIG 4.
Effect of G-CSFR truncation on adult myeloid cells. (A to F and M to O). Histological analysis of adult blood and kidney cells. Imaging (A and B, D and E, and M and N) and quantitation of indicated cell populations (C, F, and O) of Giemsa-stained blood (A to C), kidney (D to F), and sorted kidney myeloid (M to O) cells from wild-type (wt) and csf3rmdu5/mdu5 (mdu5) adult fish, with hypersegmented neutrophils indicated by arrows. e, erythrocyte; eo, eosinophil; l, lymphocyte; n, neutrophil; p, precursor. Means and SEM are shown, and statistically significant differences are indicated (***, P < 0.001; **, P < 0.01). (G to L) FACS analysis of myeloid cells. Shown are analyses of kidney myeloid (G and H) and GFP+ neutrophil (J and K) populations of wild-type (wt) and csf3rmdu5/mdu5 (mdu5) adult fish and their respective quantitations (I and L), with means and SEM shown and statistically significant differences indicated (****, P < 0.0001; **, P < 0.01).
csf3r mutants exhibit functional deficits.
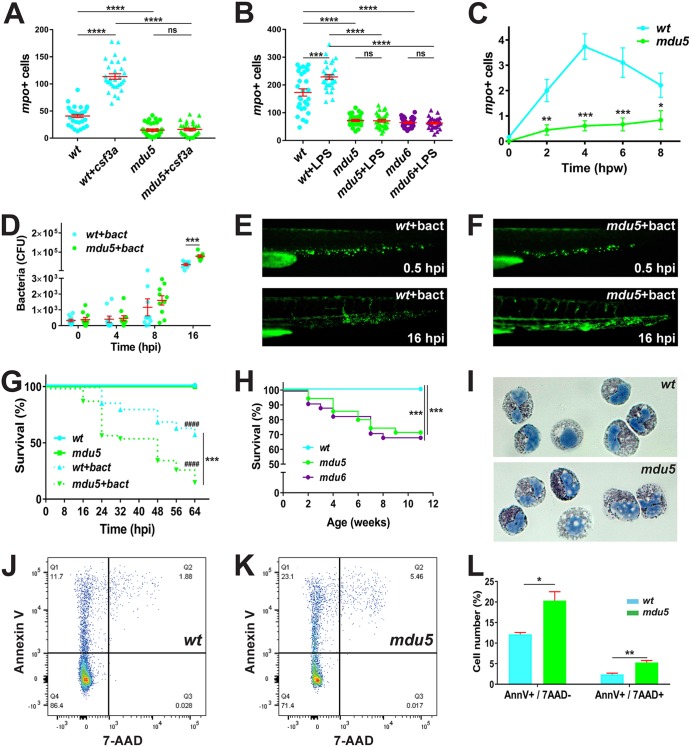
A range of functional responses were also analyzed in the csf3r mutants. Responsiveness to G-CSF was examined by injection of csf3a mRNA, which elicited a substantial increase in mpo+ cells in wild-type embryos that was not observed in csf3rmdu5/mdu5 mutant embryos (Fig. 5A). Next, lipopolysaccharide (LPS) injection was used as a model of emergency granulopoiesis (11), which also resulted in an increase in mpo+ cells in wild-type embryos but not in csf3rmdu5/mdu5 or csf3rmdu6/mdu6 mutant embryos (Fig. 5B). In response to wounding, csf3rmdu5/mdu5 mutant embryos also displayed reduced migration of mpo+ cells compared to wild-type embryos (Fig. 5C). Embryos were also subjected to infection with fluorescent Escherichia coli. Bacterial numbers increased more rapidly in csf3rmdu5/mdu5 mutants than in wild-type embryos, which reached statistical significance at 16 h postinfection (Fig. 5D), and this increase was also evident by fluorescence microscopy (Fig. 5E and F). This was reflected in significantly greater mortality of bacterially infected csf3rmdu5/mdu5 mutants than of wild-type embryos (Fig. 5G). Consistent with this, close monitoring of csf3rmdu5/mdu5 and csf3rmdu6/mdu6 mutants revealed reduced survival of juveniles under normal husbandry conditions compared to the wild type (Fig. 5H). Finally, analysis of the myeloid cell population in adult kidney marrow identified normal myeloperoxidase staining but increased numbers of vacuoles (Fig. 5I), along with significantly higher levels of apoptosis (Fig. 5J to L), in the csf3rmdu5/mdu5 mutants.
FIG 5.
Functional consequences of G-CSFR truncation. (A) Response to exogenous G-CSF. Wild-type (wt) and csf3rmdu5/mdu5 (mdu5) embryos at the 1- to 8-cell stage were either left uninjected or injected with csf3a mRNA (+csf3a) and analyzed at 48 h with mpo, with the numbers of mpo+ cells in individual embryos shown. Means and SEM are shown in red, and statistically significant differences are indicated. (B) Response to LPS injection. Wild-type (wt), csf3rmdu5/mdu5 (mdu5), and csf3rmdu6/mdu6 (mdu6) embryos at 48 hpf were either left uninjected or injected with LPS (+LPS) and analyzed 8 h later with mpo as described for panel A. (C) Response to wounding. Quantitation of myeloid cell recruitment at the indicated hours postwounding (hpw) in wild-type (wt) and csf3rmdu5/mdu5 (mdu5) embryos, showing means and SEM using mpo and analyzed as described for panel A. (D to G) Response to bacterial infection. Wild-type (wt) and csf3rmdu5/mdu5 (mdu5) embryos at 72 hpf were injected with PBS or fluorescent E. coli (+bact). The bacterial load was enumerated by plate counting (D), with means and SEM shown in red; visualized by fluorescence microscopy (E and F) at the indicated hours postinfection (hpi); and displayed as a Kaplan-Meier plot (G) (wt versus mdu5, ***, P < 0.001; uninfected versus infected, ####, P < 0.0001). (H) Relative juvenile survival. Wild-type (wt), csf3rmdu5/mdu5 (mdu5), and csf3rmdu6/mdu6 (mdu6) juveniles were placed in tanks on the standard recirculating aquarium system, and survival was monitored and displayed as a Kaplan-Meier plot. (I to L) Analysis of adult kidney myeloid cells. Shown are staining of sorted myeloid cells for myeloperoxidase (I) and analysis of myeloid cells for apoptosis using annexin V/7-AAD staining (J and K), with quantitation of early (AnnV+/7-AAD−) and late (AnnV+/7-AAD+) cells (L). ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
DISCUSSION
G-CSFR, encoded by the CSF3R gene, plays a major role in the production and function of neutrophilic granulocytes in mammals. CSF3R mutations leading to defective forms of G-CSFR are associated with neutropenia in humans (4), while both Csf3−/− (29) and Csf3r−/− (30, 31) mice display basal neutropenia with reduced survival of neutrophils. Zebrafish possess a conserved G-CSFR (11), which was targeted using genome editing to elucidate its role in granulopoiesis.
Zebrafish csf3r mutants showed decreased numbers of primitive myeloid cells and mature neutrophils during embryogenesis, similar to previous work in which zebrafish csf3r was ablated using morpholinos (11). We further showed that the csf3r mutants displayed reduced neutrophil numbers into adulthood, which could not be examined using transient morpholino-mediated knockdown. The decreased neutrophil numbers were in agreement with recently described csf3r knockouts containing introduced early stop codons in the N-terminal immunoglobulin (Ig) domain of the G-CSFR, which showed a selective neutrophil deficiency from embryogenesis into adulthood (19). Importantly, the zebrafish csf3r mutants we generated possessed a number of relevant functional deficits that were not reported with the Ig domain-based csf3r mutants. Specifically, like the neutropenia patients harboring equivalent mutations, our zebrafish csf3r mutants were unresponsive to G-CSF, as well as being neutropenic, and were additionally resistant to LPS-induced granulopoiesis. They were also more susceptible to bacterial infection, exhibiting increased bacterial loads and decreased survival, while the overall survival of the csf3r mutants was also compromised. Their neutrophils additionally showed reduced migration to sites of wounding in embryos and reduced survival in adults, although the neutrophils present exhibited robust differentiation in terms of nuclear segmentation and myeloperoxidase activity. These phenotypes were comparable to several observed in Csf3−/− (29) and Csf3r−/− (30, 31) mutant mice, further highlighting the strong conservation of this key aspect of leukocyte biology.
Neutropenia occurs as sporadic and autosomal dominant forms, as well as being a characteristic of a range of other disorders (4). Two major classes of CSF3R mutations have been identified in neutropenia patients, with quite different disease etiologies (2, 32). One class represents acquired mutations that cause truncation of the C terminus of the intracellular region of the receptor, resulting in sustained signaling in response to G-CSF. These mutations are associated with a strong predisposition to myelodysplastic syndromes (MDS)/acute myeloid leukemia (AML), but their contribution to SCN is likely only minor (33). The other class of mutations are mutations within the extracellular region of the receptor resulting in signaling-defective forms of the receptor (32), which have been reported in patients with SCN and CIN who are unresponsive to normal G-CSF therapy. The majority of such mutations result in large deletions that serve to delete the transmembrane and intracellular regions, along with variable amounts of the extracellular region (34–38).
On closer examination, these inactivating, unresponsive neutropenia-associated alleles appear to form two distinct subgroups. One subgroup, representing a variety of CSF3R mutations that cluster in a region encoding the cytokine receptor homology (CRH) domain, affect a single allele but exert a dominant-negative effect on the wild-type receptor and are associated with peripheral neutropenia, as well as a myeloid maturation arrest in the bone marrow (34, 36, 39). In two of these cases, the patients responded to high doses of G-CSF, in concert with either dexamethasone (40) or stem cell factor (34). The other subgroup, representing biallelic CSF3R mutations typically leading to truncations after the CRH, is associated with neutropenia but without maturation arrest in the bone marrow (35, 37, 38), with one of the patients responding to alternative treatment with granulocyte-macrophage colony-stimulating factor (GM-CSF) (38).
The zebrafish csf3r mutant generated in this study represented an allele from the latter subgroup. Notably, these mutants were neutropenic, but without a maturation defect in the kidney marrow, and unresponsive to G-CSF, with heterozygote carriers indistinguishable from wild-type fish. Collectively, these data provide strong support for the notion that the zebrafish csf3r mutants we created faithfully recapitulate the equivalent human mutants. They provide formal proof that such mutations cause disease and suggest that this subgroup of extracellular mutations likely represent null alleles for the gene. This research has also produced an animal model to explore alternative approaches to restore neutrophil numbers in such cases.
MATERIALS AND METHODS
Fish husbandry and genetic manipulations.
Zebrafish were maintained with standard husbandry practices, following national guidelines for their care and use. Wild-type embryos at the 1-cell stage were injected with 100 pg/nl mRNA encoding TALENs (41) that targeted exon 13 of the csf3r gene and raised to adulthood. They were outcrossed with wild-type fish, and carriers of relevant mutant alleles were identified from fin clips obtained under anesthesia with benzocaine. Following an additional round of outcrossing, heterozygote mutant carriers were incrossed to generate homozygous mutant fish. One allele was also crossed onto the Tg(mpo::GFP) (28) background. All studies involving animals were approved by the Deakin University Animal Ethics Committee.
Genomic-DNA analysis.
Genomic DNA from adult fin clips and whole embryos was isolated with QuickExtract following the manufacturer’s instructions. It was subjected to PCR with csf3r-specific primers and analyzed by RFLP with TaqI (with primers 5ʹ-GATAATGACTACGGAGACCAGCG and 5ʹ-GGAAACACCGATACACACTCATAC) or subjected to high-resolution melt analysis (42) (with primers 5ʹ-CAACCACACACTAAATATCATGCCC and 5ʹ-GTGACCTTCTTGCTGCGAGGCGAC) using Precision Melt Suremix and analysis software (Bio-Rad) to identify potential mutants, which were confirmed by Sanger sequencing at the Australian Genome Research Facility.
WISH and hemoglobin staining.
Anesthetized embryos were dechorionated and fixed in 4% (wt/vol) paraformaldehyde at 4°C before whole-mount in situ hybridization (WISH) with antisense digoxigenin (DIG)-labeled gene-specific probes, as described previously (43, 44), or subjected to staining of hemoglobin with O-dianisidine (21). Quantitation was achieved by enumeration of individual cells or by measuring the area of staining relative to eye diameter using CellSens Dimension 1.6 software (Olympus) in a blind fashion on images taken on a dissecting microscope. Data from approximately 30 embryos were analyzed for significance with Student’s t test, using Welch’s correction where necessary, with data tested for normality.
In vivo analyses.
Embryos at the 1- to 8-cell stage were injected with in vitro-transcribed gcsf.a mRNA, as described previously (11). To simulate emergency hematopoiesis, embryos at 48 h postfertilization (hpf) were injected with 5 µg/ml LPS and analyzed 8 h later, as reported previously (11). Wound-healing assays were performed by transecting the tips of the caudal tail fins of embryos at 3 dpf with a scalpel after anesthesia with 0.1 mg/ml benzocaine, as described previously (22). Infection studies involved injection of embryos at 72 hpf with 2 to 5 nl of ∼9 × 1010 CFU E. coli expressing green fluorescent protein (GFP) (no. 25922GFP; ATCC) into the venous return using a published protocol (45), with bacteria quantified by plate counting and visualized and imaged using an Olympus MVX10 fluorescence microscope and DP72 camera. Survival of infected embryos and juvenile fish was monitored by regular visual inspection and displayed as a Kaplan-Meier curve, with statistical significance determined using a log-rank (Mantel-Cox) test.
In vivo and ex vivo analyses.
Adult zebrafish kidney cells were prepared in ice-cold phosphate-buffered saline (PBS) supplemented with 1 mM EDTA and 2% (vol/vol) fetal calf serum and passaged through a 40-μm sieve. The cells were analyzed using a FACSCanto II, with myeloid cells identified in a side scatter/forward scatter (SSC/FSC) plot and neutrophils by GFP fluorescence for fish on the Tg(mpo::GFP) background. Apoptosis was assessed using an annexin V-phycoerythrin (PE)/7-aminoactinomycin D (7-AAD) apoptosis detection kit (BioLegend). Myeloid cells were sorted on a FACSAria II (BD Biosciences) for further analysis. Cytospin preparations were prepared from embryonic and adult blood, adult kidney, and sorted kidney myeloid cells and stained with Giemsa stain or a myeloperoxidase kit (Sigma), and differential counts were performed. Data were analyzed for significance with Student's t test.
ACKNOWLEDGMENTS
We recognize the support of an International Research Scholarship (F.B.) and a Faculty of Health Postdoctoral Fellowship (P.R.) from Deakin University. The funder had no role in study design, data collection and interpretation, or manuscript preparation.
We thank the Deakin University Animal House staff for superb aquarium management and Siying Ye for assistance with FACS analysis.
REFERENCES
- 1.Liongue C, Wright C, Russell AP, Ward AC. 2009. Granulocyte colony-stimulating factor receptor: stimulating granulopoiesis and much more. Int J Biochem Cell Biol 41:2372–2375. doi: 10.1016/j.biocel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 2.Germeshausen M, Ballmaier M, Welte K. 2007. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: results of a long-term survey. Blood 109:93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 3.Berliner N. 2008. Lessons from congenital neutropenia: 50 years of progress in understanding myelopoiesis. Blood 111:5427–5432. doi: 10.1182/blood-2007-10-077396. [DOI] [PubMed] [Google Scholar]
- 4.Ward AC, Dale DC. 2009. Genetic and molecular approaches to the diagnosis of severe congenital neutropenia. Curr Opin Hematol 16:9–13. doi: 10.1097/MOH.0b013e32831952de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dale DC, Bolyard AA, Schwinzer BG, Pracht G, Bonilla MA, Boxer LA, Freedman MH, Donadieu J, Kannourakis G, Alter BP, Cham BP, Winkelstein J, Kinsey SE, Zeidler C, Welte K. 2006. The severe congenital neutropenia international registry: 10-year follow-up report. Support Cancer Ther 3:220–231. doi: 10.3816/SCT.2006.n.020. [DOI] [PubMed] [Google Scholar]
- 6.Traver D, Herbomel P, Patton EE, Murphey RD, Yoder JA, Litman GW, Catic A, Amemiya CT, Zon LI, Trede NS. 2003. The zebrafish as a model organism to study development of the immune system. Adv Immunol 81:253–330. [PubMed] [Google Scholar]
- 7.Yoder JA, Nielsen ME, Amemiya CT, Litman GW. 2002. Zebrafish as an immunological model system. Microbes Infect 4:1469–1478. doi: 10.1016/S1286-4579(02)00029-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen AT, Zon LI. 2009. Zebrafish blood stem cells. J Cell Biochem 108:35–42. doi: 10.1002/jcb.22251. [DOI] [PubMed] [Google Scholar]
- 9.Liongue C, Ward AC. 2007. Evolution of class I cytokine receptors. BMC Evol Biol 7:120. doi: 10.1186/1471-2148-7-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paffett-Lugassy N, Hsia N, Fraenkel PG, Paw B, Leshinsky I, Barut B, Bahary N, Caro J, Handin R, Zon LI. 2007. Functional conservation of erythropoietin signaling in zebrafish. Blood 110:2718–2726. doi: 10.1182/blood-2006-04-016535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liongue C, Hall CJ, O'Connell BA, Crosier P, Ward AC. 2009. Zebrafish granulocyte colony-stimulating factor receptor signalling promotes myelopoiesis and myeloid cell migration. Blood 113:2535–2546. doi: 10.1182/blood-2008-07-171967. [DOI] [PubMed] [Google Scholar]
- 12.Ma AC, Ward AC, Liang R, Leung AY. 2007. The role of jak2a in zebrafish hematopoiesis. Blood 110:1824–1830. doi: 10.1182/blood-2007-03-078287. [DOI] [PubMed] [Google Scholar]
- 13.Aggad D, Stein C, Sieger D, Mazel M, Boudinot P, Herbomel P, Levraud JP, Lutfalla G, Leptin M. 2010. In vivo analysis of Ifn-g1 and Ifn-g2 signaling in zebrafish. J Immunol 185:6774–6782. doi: 10.4049/jimmunol.1000549. [DOI] [PubMed] [Google Scholar]
- 14.Iwanami N, Mateos F, Hess I, Riffel N, Soza-Ried C, Schorpp M, Boehm T. 2011. Genetic evidence for an evolutionarily conserved role of IL-7 signaling in T cell development of zebrafish. J Immunol 186:7060–7066. doi: 10.4049/jimmunol.1003907. [DOI] [PubMed] [Google Scholar]
- 15.Zhu LY, Pan PP, Fang W, Shao JZ, Xiang LX. 2012. Essential role of IL-4 and IL-4Ralpha interaction in adaptive immunity of zebrafish: insight into the origin of Th2-like regulatory mechanism in ancient vertebrates. J Immunol 188:5571–5584. doi: 10.4049/jimmunol.1102259. [DOI] [PubMed] [Google Scholar]
- 16.Liongue C, O'Sullivan LA, Trengove MC, Ward AC. 2012. Evolution of JAK-STAT pathway components: mechanisms and role in immune system development. PLoS One 7:e32777. doi: 10.1371/journal.pone.0032777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sertori R, Liongue C, Basheer F, Lewis KL, Rasighaemi P, de Coninck D, Traver D, Ward AC. 2016. Conserved IL-2Rgc signaling mediates lymphopoiesis in zebrafish. J Immonol 196:135–143. doi: 10.4049/jimmunol.1403060. [DOI] [PubMed] [Google Scholar]
- 18.Sertori R, Trengove MC, Basheer F, Ward AC, Liongue C. 2016. Genome editing in zebrafish: a practical overview. Brief Funct Genomics 15:322–330. doi: 10.1093/bfgp/elv051. [DOI] [PubMed] [Google Scholar]
- 19.Pazhakh V, Clark S, Keightley MC, Lieschke GJ. 2017. A GCSFR/CSF3R zebrafish mutant models the persistent basal neutrophil deficiency of severe congenital neutropenia. Sci Rep 7:44455. doi: 10.1038/srep44455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward AC, McPhee DO, Condron MM, Varma S, Cody SH, Onnebo SMN, Paw BH, Zon LI, Lieschke GJ. 2003. The zebrafish spi1 promoter drives myeloid-specific expression in stable transgenic fish. Blood 102:3238–3240. doi: 10.1182/blood-2003-03-0966. [DOI] [PubMed] [Google Scholar]
- 21.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. 2001. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98:3087–3096. doi: 10.1182/blood.V98.10.3087. [DOI] [PubMed] [Google Scholar]
- 22.Hall C, Flores MV, Storm T, Crosier K, Crosier P. 2007. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol 7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbomel P, Thisse B, Thisse C. 2001. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol 238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 24.Zakrzewska A, Cui C, Stockhammer OW, Benard EL, Spaink HP, Meijer AH. 2010. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood 116:e1–e11. doi: 10.1182/blood-2010-01-262873. [DOI] [PubMed] [Google Scholar]
- 25.Bertrand JY, Kim AD, Teng S, Traver D. 2008. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development 135:1853–1862. doi: 10.1242/dev.015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Vitas MR, Baas AM, Crosier PS, Crosier KE. 2002. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development 129:2015–2030. [DOI] [PubMed] [Google Scholar]
- 27.Willett CE, Zapata A, Hopkins N, Steiner LA. 1997. Expression of zebrafish rag genes during early development identifies the thymus. Dev Biol 182:331–341. doi: 10.1006/dbio.1996.8446. [DOI] [PubMed] [Google Scholar]
- 28.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. 2006. A transgenic zebrafish model of neutrophilic inflammation. Blood 108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 29.Lieschke GJ, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, Fowler KJ, Basu S, Zhan YF, Dunn AR. 1994. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor deficiency, and impaired neutrophil mobilization. Blood 84:1737–1746. [PubMed] [Google Scholar]
- 30.Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. 1996. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity 5:491–501. doi: 10.1016/S1074-7613(00)80504-X. [DOI] [PubMed] [Google Scholar]
- 31.Hermans MH, van de Geijn GJ, Antonissen C, Gits J, van Leeuwen D, Ward AC, Touw IP. 2003. Signaling mechanisms coupled to tyrosines in the granulocyte colony-stimulating factor receptor orchestrate G-CSF-induced expansion of myeloid progenitor cells. Blood 101:2584–2590. doi: 10.1182/blood-2002-07-2062. [DOI] [PubMed] [Google Scholar]
- 32.Ward AC. 2007. The role of the granulocyte colony-stimulating factor receptor (G-CSF-R) in disease. Front Biosci 12:608–618. doi: 10.2741/2086. [DOI] [PubMed] [Google Scholar]
- 33.Liongue C, Ward AC. 2014. Granulocyte colony-stimulating factor receptor mutations in myeloid malignancy. Front Oncol 4:93. doi: 10.3389/fonc.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha S, Zhu QS, Romero G, Corey SJ. 2003. Deletional mutation of the external domain of the human granulocyte colony-stimulating factor receptor in a patient with severe chronic neutropenia refractory to granulocyte colony-stimulating factor. J Pediatr Hematol Oncol 25:791–796. doi: 10.1097/00043426-200310000-00010. [DOI] [PubMed] [Google Scholar]
- 35.Papadaki HA, Kosteas T, Gemetzi C, Damianaki A, Anagnou NP, Eliopoulos GD. 2004. Acute myeloid/NK precursor cell leukemia with trisomy 4 and a novel point mutation in the extracellular domain of the G-CSF receptor in a patient with chronic idiopathic neutropenia. Ann Hematol 83:345–348. doi: 10.1007/s00277-004-0862-y. [DOI] [PubMed] [Google Scholar]
- 36.Druhan LJ, Ai J, Massullo P, Kindwall-Keller T, Ranalli MA, Avalos BR. 2005. Novel mechanism of G-CSF refractoriness in patients with severe congenital neutropenia. Blood 105:584–591. doi: 10.1182/blood-2004-07-2613. [DOI] [PubMed] [Google Scholar]
- 37.Triot A, Järvinen PM, Arostegui JI, Murugan D, Kohistani N, Dapena Díaz JL, Racek T, Puchałka J, Gertz EM, Schäffer AA, Kotlarz D, Pfeifer D, Díaz de Heredia Rubio C, Ozdemir MA, Patiroglu T, Karakukcu M, Sánchez de Toledo Codina J, Yagüe J, Touw IP, Unal E, Klein C. 2014. Inherited biallelic CSF3R mutations in severe congenital neutropenia. Blood 123:3811–3817. doi: 10.1182/blood-2013-11-535419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klimiankou M, Klimenkova O, Uenalan M, Zeidler A, Mellor-Heineke S, Kandabarau S, Skokowa J, Zeidler C, Welte K. 2015. GM-CSF stimulates granulopoiesis in a congenital neutropenia patient with loss-of-function biallelic heterozygous CSF3R mutations. Blood 126:1865–1867. doi: 10.1182/blood-2015-07-661264. [DOI] [PubMed] [Google Scholar]
- 39.Ward AC, van Aesch YM, Gits J, Schelen AM, de Koning JP, van Leeuwen D, Freedman MH, Touw IP. 1999. Novel point mutation in the extracellular domain of the granulocyte colony-stimulating factor (G-CSF) receptor in a case of severe congenital neutropenia hyporesponsive to G-CSF treatment. J Exp Med 190:497–507. doi: 10.1084/jem.190.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dror Y, Ward AC, Touw IP, Freedman MH. 2000. Combined corticosteroid/granulocyte colony-stimulating factor (G-CSF) therapy in the treatment of severe congenital neutropenia unresponsive to G-CSF: activated glucocorticoid receptors synergize with G-CSF signals. Exp Hematol 28:1381–1389. doi: 10.1016/S0301-472X(00)00544-0. [DOI] [PubMed] [Google Scholar]
- 41.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garritano S, Gemignani F, Voegele C, Nguyen-Dumont T, Le Calvez-Kelm F, De Silva D, Lesueur F, Landi S, Tavtigian SV. 2009. Determining the effectiveness of high resolution melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet 10:5. doi: 10.1186/1471-2156-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulte-Merker S, Ho RK, Herrmann BG, Nüsslein-Volhard C. 1992. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 116:1021–1032. [DOI] [PubMed] [Google Scholar]
- 44.Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 45.Fehr A, Eshwar AK, Neuhauss SC, Ruetten M, Lehner A, Vaughan L. 2015. Evaluation of zebrafish as a model to study the pathogenesis of the opportunistic pathogen Cronobacter turicensis. Emerg Microbes Infect 4:e29. doi: 10.1038/emi.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]