The microscopic alga Picocystis sp. strain ML is responsible for recurrent algal blooms in Mono Lake, CA. This organism was characterized by only very little molecular data, despite its prominence as a primary producer in saline environments.
ABSTRACT
The microscopic alga Picocystis sp. strain ML is responsible for recurrent algal blooms in Mono Lake, CA. This organism was characterized by only very little molecular data, despite its prominence as a primary producer in saline environments. Here, we report the draft genome sequence for Picocystis sp. strain ML based on long-read sequencing.
ANNOUNCEMENT
Mono Lake, a hypersaline soda lake in eastern California, sustains a population of the oxygenic alga Picocystis sp. strain ML. Previous studies have described this strain, a close relative to Picocystis salinarum (1), as a major primary producer in the lake (2). Members of the genus Picocystis have also been cultivated from East Africa (3, 4), Inner Mongolia, China (5), and, more recently, Peru (6). Despite its role as a primary producer and its global distribution, nothing is known of its genomic potential. A recent study characterized a bloom using metagenomic and transcriptomic approaches and suggested that Picocystis sp. strain ML produced photosynthetic transcripts, potentially producing oxygen, at low-light depths (7). Here, we report the draft genome sequence of Picocystis sp. strain ML, which was previously estimated to be 23 Mbp (8).
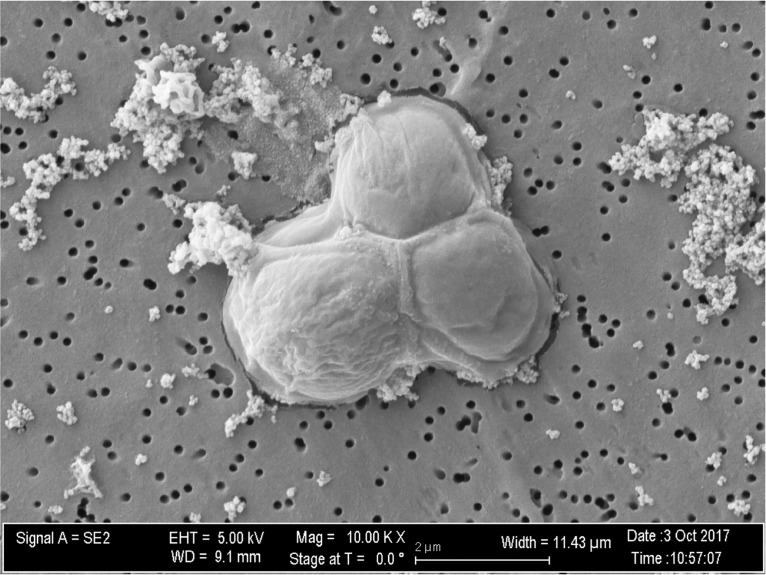
A sample collected from 20-m depth in Mono Lake (7) was inoculated into L1 liquid medium (product number MKL150L; National Center for Marine Algae). Upon visualization of growth, a sample was spread onto L1 agar (1.0% [wt/vol]) for isolation, and a single colony was used to inoculate L1 medium for DNA extraction. Axenic status was determined by a lack of growth in marine purity broth (9) and using scanning electron microscopy (SEM) (Fig. 1). Volumes of 250 µl were passed through 0.1-µm polycarbonate filters, and the retained cells were fixed (0.75% ruthenium red, 50% glutaraldehyde, 1 M HEPES) and sputter coated (5 nm AuPd) with a Hummer V1 sputtering system (Anatech USA). Samples were viewed for axenic status on a Zeiss NEON field emission gun-SEM (FEG-SEM) dual-beam high-resolution system with an energy selective backscatter (EsB) detector (Zeiss). High-molecular-weight DNA was extracted via a modified cetyltrimethylammonium bromide (CTAB) extraction, purified with Sera-Mag SpeedBeads (GE) via the AMPure XP protocol (Agencourt), and quantified using a Qubit fluorometer (final concentration, 166 µg/ml). A genomic library was prepared using the PacBio SMRTbell template prep kit 1.0-SPv3 (Pacific Biosciences). The final library was size selected at 10 kb (Blue Pippin; Sage Science) and sequenced on a PacBio Sequel (Pacific Biosciences) using 4 single-molecule real-time (SMRT) cells via 2.0 chemistry, with a 10-h movie. To remove any contaminating bacterial sequences, reads were filtered with custom scripts for those that taxonomically matched the phylum Chlorophyta based on Kaiju version 1.6.2 classification (10). After quality control, a total of 251,086 reads were assembled using Canu version 1.6, generating 318 contigs, with a coverage of 40.77×, a total assembly length of 29.6 Mbp, a GC content of 53.6%, and an N50 value of 154 kbp (11).
FIG 1.
Scanning electron microscopy (SEM) image of Picocystis sp. strain ML on a polycarbonate filter. Scale bar = 2 µm.
A total of 14 small subunit (SSU) rRNA regions were found using RNAmmer version 1.2, closely matching (99.7% ± 0.67%) that of P. salinarum L7 (GenBank accession number AF153313) or matching (99%) the chloroplast of P. salinarum CCMP:1897 (GenBank accession number KJ746599) based on BLAST alignment (12, 13). RepeatMasker version 4.0.7 was used to mask repetitive elements (0.11%) with RMBlast version 2.6.0 (14) before gene prediction using AUGUSTUS trained to Chlamydomonas reinhardtii, the most closely related green algal model (15). A total of 5,613 coding regions were detected, of which 40.4% were characterized using BLASTKoala version 2.1 (16). The final assembly represents the first publicly available draft genome sequence of Picocystis sp. strain ML.
During the recent bloom, no genes were observed for ammonium oxidation from ammonia-oxidizing bacteria (AOB), unlike in previous years (17), and it was speculated that Picocystis sp. strain ML was assimilating these compounds (7). This genome revealed the presence of solute carrier family (commonly referred to as SLC) ammonium transporters, illustrating the genetic potential for Picocystis sp. strain ML to take in ammonium ions, perhaps explaining the lack of ammonium oxidation transcripts in the water column and the lack of AOB in such a monoculture environment. Overall, this genome sheds light on a primary producer’s genetic potential in a unique aquatic ecosystem.
Data availability.
This whole-genome shotgun project has been deposited in DDBJ/ENA/GenBank under the accession number QYZS00000000. The version described in this paper is version QYZS01000000. Raw sequence reads have been deposited in the SRA database under BioProject number PRJNA490491. Custom scripts and software settings are available on GitHub at https://github.com/emilyjunkins/PicoML/tree/v1.0 and https://doi.org/10.5281/zenodo.2366252.
ACKNOWLEDGMENTS
We thank all participants from the 2016 International Geobiology Course and the Agouron Institute for course funding. Duke University provided sequencing capabilities via the GCB Sequencing and Genomic Technologies Shared Resource. James Floyd and the University of Oklahoma Samuel Roberts Noble Microscopy Laboratory provided imaging resources. The University of Oklahoma Supercomputing Center for Education and Research (OSCER) provided server support and archival storage prior to analysis. A California State Parks permit to the U.S. Geological Survey and Geobiology 2016 allowed us to conduct sampling on and around Mono Lake.
The mention of brand name products does not constitute an endorsement by the USGS.
REFERENCES
- 1.Lewin RA, Krienitz L, Goericke R, Takeda H, Hepperle D. 2000. Picocystis salinarum gen. et sp. nov. (Chlorophyta)–a new picoplanktonic green alga. Phycologia 39:560–565. doi: 10.2216/i0031-8884-39-6-560.1. [DOI] [Google Scholar]
- 2.Roesler CS, Culbertson CW, Etheridge SM, Goericke R, Kiene RP, Miller LG, Oremland RS. 2002. Distribution, production, and ecophysiology of Picocystis strain ML in Mono Lake, California. Limnol Oceanogr 47:440–452. doi: 10.4319/lo.2002.47.2.0440. [DOI] [Google Scholar]
- 3.Krienitz L, Bock C, Kotut K, Luo W. 2012. Picocystis salinarum (Chlorophyta) in saline lakes and hot springs of East Africa. Phycologia 51:22–32. doi: 10.2216/11-28.1. [DOI] [Google Scholar]
- 4.Schagerl M, Burian A, Gruber-Dorninger M, Oduor SO, Kaggwa MN. 2015. Algal communities of Kenyan soda lakes with a special focus on Arthrospira fusiformis. Fottea 15:245–257. doi: 10.5507/fot.2015.012. [DOI] [Google Scholar]
- 5.Fanjing K, Qinxian J, Jia E, Mianping Z. 2009. Characterization of a eukaryotic picoplankton alga, strain DGN-Z1, isolated from a soda lake in Inner Mongolia, China. Nat Resour Environ Issues 15:185–189. [Google Scholar]
- 6.Tarazona Delgado R, Terreros HM, Astocondor MM, Huatuco EM. 2017. Picocystis salinarum (Prasinophyceae, Chlorophyta) en las Salinas de Chilca, Lima, primer registro para el Perú. Arnaldoa 24:557–566. doi: 10.22497/arnaldoa.242.24208. [DOI] [Google Scholar]
- 7.Stamps BW, Nunn HS, Petryshyn VA, Oremland RS, Miller LG, Rosen MR, Bauer KW, Thompson KJ, Tookmanian EM, Waldeck AR, Loyd SJ, Johnson HA, Stevenson BS, Berelson WM, Corsetti FA, Spear JR. 2018. The metabolic capability and phylogenetic diversity of Mono Lake during a bloom of the eukaryotic phototroph Picocystis strain ML. Appl Environ Microbiol 84:e01171-18. doi: 10.1128/AEM.01171-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.dos Santos AL, Pollina T, Gourvil P, Corre E, Marie D, Garrido JL, Rodriguez F, Noel MH, Vaulot D, Eikrem W. 2017. Chloropicophyceae, a new class of picophytoplanktonic prasinophytes. Sci Rep 7:14019. doi: 10.1038/s41598-017-12412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterbury JB, Watson SW, Valois FW, Franks DG. 1986. Biological and ecological characterization of the marine unicellular cyanobacteria Synechococcus. Can Bull Fish Aquat Sci 214:71–120. [Google Scholar]
- 10.Menzel P, Ng KL, Krogh A. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Smit AFA, Hubley R, Green P. 2010. RepeatMasker Open-4.0. http://repeatmasker.org.
- 15.Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. AUGUSTUS: a Web server for gene finding in eukaryotes. Nucleic Acids Res 32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Ward BB, Martino DP, Diaz MC, Joye SB. 2000. Analysis of ammonia-oxidizing bacteria from hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl Environ Microbiol 66:2873–2881. doi: 10.1128/AEM.66.7.2873-2881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome shotgun project has been deposited in DDBJ/ENA/GenBank under the accession number QYZS00000000. The version described in this paper is version QYZS01000000. Raw sequence reads have been deposited in the SRA database under BioProject number PRJNA490491. Custom scripts and software settings are available on GitHub at https://github.com/emilyjunkins/PicoML/tree/v1.0 and https://doi.org/10.5281/zenodo.2366252.