Abstract
Research on gene expression (GE) provides insights into the physiology of a cell or group of cells at a given point in time. Studies of changes in GE can be used to identify patients at higher risk for various medical conditions, a higher symptom burden, and/or the adverse consequences associated with various treatments. The aims of this article are as follows: (1) to describe the different types of RNA transcripts, (2) to describe the processes involved in GE (i.e., RNA transcription, epigenetics, and posttranscriptional modifications), (3) to describe common sources of variation in GE, (4) to describe the most common methods used to measure GE, and (5) to discuss factors to consider when choosing tissue for a GE study. This article begins with an overview of the mechanisms involved in GE. Then, the factors that can influence the findings from GE experiments (e.g., tissue specificity, host age, host gender, and time of sample collection) are described and potential solutions are presented. This article concludes with a discussion of how the types of tissue used in GE studies can affect study findings. Given that the costs associated with the measurement of changes in GE are decreasing and the methods to analyze GE data are becoming easier to use, nurse scientists need to understand the basic principles that underlie any GE study.
Keywords: gene expression, transcription, epigenetics, quantitative polymerase chain reaction, microarray, ribonucleic acid sequencing
Gene expression (GE) is the synthesis of a functional gene product using the information provided by deoxyribonucleic acid (DNA; Perdew, Vanden-Heuvel, & Peters, 2006). Ribonucleic acid (RNA) is synthesized from DNA through the process of transcription, which is part of the process of GE. Asan organism develops or responds to changes in its environment, cells can adjust the type and amount of GE. Hence, studies of GE provide insights into cellular responses at a given point in time. Over 40 years ago, King and Wilson (1975) demonstrated that major phenotypic differences between organisms could be explained by small changes in the regulatory mechanisms associated with alterations in GE. Clinical research has taken advantage of this property by measuring differences in GE between groups of individuals. These studies have advanced our understanding of disease progression (Koleck & Conley, 2016), differences in symptom severity (Kober et al., 2016; Wright et al., 2017), and the identification of drug targets to treat breast cancer (Dowsett & Dunbier, 2008).
Research on changes in GE has increased for a number of reasons. First, the number of clinical samples available from tissue repositories and the availability of new methods to measure GE from a variety of tissues are growing (Shabihkhani et al., 2014). Second, the costs associated with the measurement and analysis of GE data have decreased by more than 50% (Kukurba & Montgomery, 2015). Third, large databases of experimental GE data are publicly available (e.g., the Gene Expression Omnibus). Fourth, the most current technologies for measuring GE (e.g., RNA sequencing [RNA-Seq]) are becoming more affordable and have broader applicability (Hou et al., 2015). Finally, newer methods to analyze GE data are more accessible and easier to use (Kukurba & Montgomery, 2015).
As the biological materials, GE data, and analytic approaches become more available and accessible, nurse scientists need increased knowledge of the fundamental mechanisms that underlie GE as well as factors that need to be considered when one designs a GE study. Therefore, the purposes of this article are as follows: (1) to describe the different types of RNA transcripts, (2) to describe the processes involved in the regulation of GE (i.e., RNA transcription, epigenetics, and posttranscriptional modifications), (3) to describe common sources of variation in GE, (4) to describe the most common methods used to measure GE, and (5) to discuss factors to consider when choosing tissue for a GE study.
Types of RNA
RNA is a polymeric linear molecule composed of a ribose sugar, a phosphate group, and nitrogenous bases (i.e., adenine, guanine, cytosine, and uracil). Researchers measure the different types of RNA (i.e., coding and noncoding) in a cell to determine GE. In general, RNA transcripts are classified as protein coding or noncoding (Claverie, 2005). The human genome contains approximately 20,000 protein-coding genes and at least the same number of noncoding RNA genes (Byron, Keuren-Jensen, Engelthaler, Carpten, & Craig, 2016; Chen & Weiss, 2015).
As the name suggests, protein-coding genes synthesize messenger RNAs (mRNAs) that, through the processes of translation, produce protein products (Gibson & Muse, 2009, chapter 2). mRNA is distinguished from other types of RNA in that most of it is polyadenylated (poly[A]) at the end of the transcript. One exception is histone mRNA that, instead of a poly(A) tail, has a conserved 3′ stem loop (Dominski & Marzluff, 1999). While mRNA is widely studied, it comprises only 1–5% of the total RNA (Gibson & Muse, 2009, chapter 2).
As shown in Table 1, noncoding RNAs make up the majority of the RNA species present in a cell. There are two main types of noncoding RNAs, namely, the small noncoding RNAs and the long noncoding RNAs. The small noncoding RNAs include microRNA (miRNA), transfer RNA (tRNA)-derived small fragments, P-element Induced Wimpy (PIWI) protein-interacting RNA, small nucleolar RNA, and small interfering RNA. The long noncoding RNAs include promoter-associated long RNA, transcribed ultraconserved regions, tRNA, circular RNA (circRNA), small nuclear RNA (snRNA), pseudogenes, and antisense RNA (Byron et al., 2016; Ng et al., 2016). Ribosomal RNA (rRNA) is the most abundant type of RNA in the intracellular matrix and comprises about 80% of the total RNA. It belongs to neither the small nor the long noncoding RNA categories (Gibson & Muse, 2009, chapter 2). We describe the functions of and detection methods for the noncoding RNAs in Table 1.
Table 1.
Noncoding RNAs.
| Type | Subtype | Function | Detection Method |
|---|---|---|---|
| Small noncoding RNA (20–35 nucleotides) | miRNA (18–20 nucleotides) | Posttranscriptional silencing of complementary mRNA | RNA-Seq and microarray |
| tRF | Controls viral replication, modulates cell proliferation, and inhibits protein synthesis | RNA-Seq | |
| piRNA (26–30 nucleotides) | Interacts with PIWI protein and maintains genome integrity by silencing transposon element in the germ line at transcriptional and posttranscriptional level | RNA-Seq | |
| snoRNA (60–140 nucleotides) | Ribosome biogenesis and rRNA modification | RNA-Seq | |
| siRNA | Binds to complementary sequences in mRNA and causes degradation of mRNA. Binds to complementary DNA sequence and cause methylation for short-term silencing | RNA-Seq | |
| Long noncoding RNA (>200 nucleotides) | PAR | Binds to RNA-binding protein called translocated in liposarcoma and represses transcription | RNA-Seq |
| T-UCR | Plays a role in proper functioning of cell differentiation and proliferation. Changes in level of T-UCR expression is associated with carcinogenesis | Microarray and RNA-Seq | |
| tRNA | Carries an amino acid molecule and docks on the ribosome bound to an mRNA molecule during protein synthesis. Each amino acid binds to a specific tRNA and is required for translation | Northern blotting | |
| circRNA | Regulates splicing and transcription | RNA-Seq | |
| snRNA (100–300 nucleotides) | Plays a role in RNA splicing and processing. Localized in the nucleus | RNA-Seq | |
| Pseudogenes | Regulates tumor suppressors and oncogenes | RNA-Seq | |
| asRNA | Plays a role in gene silencing and regulation of gene expression | RNA-Seq | |
| rRNA | 28S | Combines with proteins to form the ribosomal structure. Binds to the mRNA and tRNA molecules to enable translation | Northern blotting and qPCR |
| 5.8S | |||
| 5S | |||
| 18S |
Note. asRNAs = antisense RNAs; bp = base pair; circRNA = circular RNA; mRNA = messenger RNA; miRNA = micro RNA; PAR = promoter-associated long RNA; piRNA = PIWI-interacting RNA; qPCR = quantitative polymerase chain reaction; RNA = ribonucleic acid; rRNA = ribosomal ribonucleic acid; RNA-Seq = ribonucleic acid sequencing; siRNA = small interfering RNA; snRNA = small nuclear RNA; snoRNA = small nucleolar RNA; tRF = tRNA-derived small fragment; tRNA = transfer RNA; T-UCR = transcribed ultra-conserved regions.
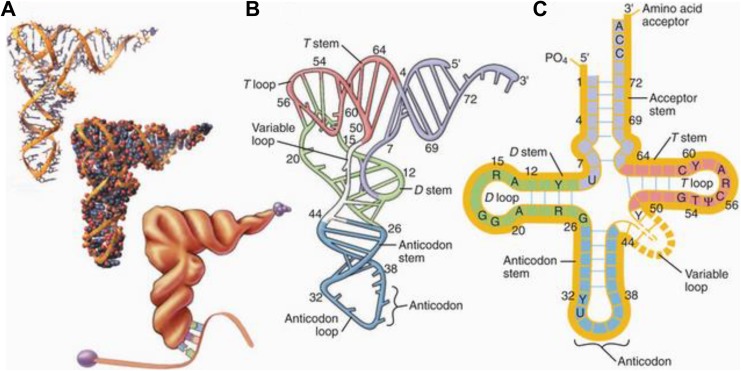
Both tRNA and circRNA play important roles in protein synthesis. As shown in Figure 1, tRNAs fold into cloverleaf structures as a result of short stretches of base-pairing. Each tRNA carries the specific amino acid that is needed to extend the protein during translation of the mRNA. On a ribosome, tRNA binds to mRNA through a three-nucleotide anticodon sequence. The circRNAs have covalent bonds between the 5′and the 3′ ends (Byron et al., 2016) and play a role in transcription, regulation of posttranscriptional RNA splicing (Gibson & Muse, 2009, chapter 2), and sequestration and suppression of miRNA activity (Hansen et al., 2013). miRNA regulates the expression of several genes, while regulation of miRNA function by circRNA highlights the increased complexity of noncoding RNA-mediated regulatory pathways (Hansen, Kjems, & Damgaard, 2013).
Figure 1.
A two-dimensional and three-dimensional representation of transfer RNA (tRNA) cloverleaf structure. tRNA carries an amino acid molecule at its 3′ end, and its anticodon end binds to the codon on the messenger RNA molecule for that amino acid during protein synthesis. Source: This figure was published in Pollard, Earnshaw, and Lippincott-Schwartz (2007, fig. 17-3, p. 300). Copyright 2008 by Elsevier. Reprinted with permission.
Regulation of GE
The processes involved in the regulation of GE include transcription, a number of epigenetic processes, and posttranscriptional modifications. Regulation of GE is cell-specific and involves a number of complex biochemical processes that are essential for the development of the organism as well as for the organism’s ability to respond to changes in the environment (e.g., response to injury; Perdew et al., 2006). In order to be able to interpret GE data, it is important to understand the mechanisms that regulate GE.
Transcription
Transcription is the process by which a functional gene product (i.e., RNA) is synthesized. This product, or transcript, may be a precursor to a protein (through translation), a subunit for a larger molecule (e.g., ribosome), or a functional molecule in and of itself (e.g., miRNA). Measurement of GE is the quantification of the transcribed gene product. The transcriptome is the complete set of RNA transcripts present in a cell at a specific developmental stage or physiological condition. Transcription involves the unwinding of DNA, the attraction and binding of transcription factors, and the action of the transcriptional machinery to produce RNA from the DNA template (Gibson & Muse, 2009, chapter 4; Perdew et al., 2006). Regulatory regions within DNA and transcription factors are the fundamental units involved in the transcription process.
Regulatory Regions
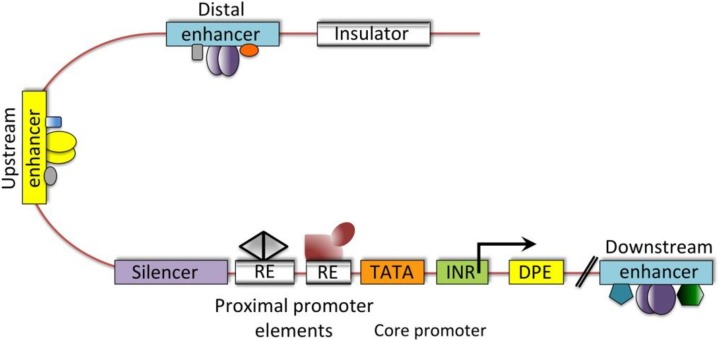
The specific regions of DNA that are associated with transcription and the regulation of GE in eukaryotes are the promoter, enhancer, silencer, and insulator (Maston, Evans, & Green, 2006). As shown in Figure 2, regulatory regions of DNA such as promoters and enhancers are called cis-regulatory elements (Levine & Tjian, 2003). These cis-regulatory elements form the cis-regulatory module (CRM) also called a transcription factor binding site (TFBS). Transcription factors bind to a TFBS to regulate GE (Liu, Yu, Zack, Zhu, & Qian, 2008).
Figure 2.
The cis-regulatory elements on the deoxyribonucleic acid that regulate gene expression in mammals. The cis-regulatory elements are fundamental regions for transcription initiation and its regulation. DPE = downstream promoter element; INR = initiator sequence; RE = response element; TATA box = promoter region where transcription begins. Source: Reprinted with permission from Macmillan (Levine & Tjian 2003). Copyright 2003 by Nature.
The promoter region of a gene is located upstream from its transcription initiation site. Typically, the promoter region has a conserved sequence of 25–35 base pairs upstream from the transcription initiation site that contains a motif of TATA repeats (i.e., the “TATA box,” a conserved sequence in the promoter region where transcription begins). Transcription factors bind to the promoter region of a gene and facilitate the binding of RNA polymerase, which initiates transcription. The promoter sequence defines the direction of transcription. The DNA strand that is transcribed is called the sense strand (Gibson & Muse, 2009, chapter 2).
The enhancer regions of DNA are required for precise regulation of tissue-specific GE. Enhancers activate transcription independent of their location, distance from, or orientation to the promoter. Enhancers contain multiple TFBSs and can be transcribed into noncoding RNAs. These noncoding RNAs, together with a protein complex called cohesin, stabilize long-distance enhancer–promoter interactions and facilitate transcription (Ong & Corces, 2011).
Silencers are regions of DNA that have the opposite effect of enhancers. Transcriptional repression is achieved through two kinds of silencers, namely, silencer elements and negative regulatory elements (NREs). Silencer elements are position independent and direct active repression of transcription. NREs are position-dependent and direct passive repression of transcription. Silencers function in association with the promoter and determine the mechanism of transcription repression. They can be an intrinsic part of the promoter region. Interactions between silencers and enhancers and other transcriptional elements are important for the regulation of GE (Ogbourne & Antalis, 1998).
Insulators are DNA sequences that protect an expressing gene from its surrounding environment. The two types of DNA insulator sequences are the barrier element and the enhancer-blocking element. The barrier element binds a protein complex that prevents DNA methylation. In contrast, an enhancer-blocking element interferes with interactions between the enhancer and promoter regions of DNA. Enhancer blocking occurs when an insulator is present between an enhancer and a promoter. This insulator element prevents the enhancer from activating GE of an adjacent gene (Ghirlando et al., 2012).
Transcription Factors
Transcription factors are proteins that initiate and regulate transcription. Fewer than 2,000 transcription factors control GE (Holdt et al., 2016). Transcription factors share common structural motifs such as a zinc finger, a leucine zipper, and helix-loop-helix structures. Inducible transcription factors are activated by protein kinases to bind to target response elements. For example, an increase in the levels of a serum hormone can activate specific cell-surface receptors, which induces a cascade of protein kinase activated cell-signaling pathways that lead to the activation of specific transcription factors (Gibson & Muse, 2009, chapter 2). Any changes in the expression of transcription factors, hormone levels, or cell-surface receptors can change the levels of GE.
Transcription factors control the levels of GE in a cell through selective transcription of a subset of genes. For example, in the case of an embryonic stem cell, while more than 1,200 genes encode for transcription factors, only a limited number of genes need to be expressed to reprogram cells into pluripotent stem cells (Messina, Glasscock, Gish, & Lovett, 2004). Overexpression of transcription factors can lead to morphological changes. For example, overexpression of c-Myc can cause cancer and is associated with increased aggressiveness of certain cancers as well as with poorer clinical outcomes (Hoffman & Liebermann, 2008).
Fundamental Mechanisms of GE in Mammals
Mammalian GE is regulated through complex interactions among multiple processes and occurs primarily at the initiation of transcription (Beyersmann, 2000). The regulatory structure of a mammalian gene consists of a coding sequence for RNA, a proximal upstream promoter region that binds general transcription factors, a distant enhancer sequence that binds inducible transcription factors, as well as insulator and silencer regions. Unlike the general eukaryotic transcription mechanism, the mammalian transcription mechanism requires unique transcriptional enhancers that control the expression of over 2,000 protein-coding genes to maintain cell-type-specific functions. Gene regulatory elements and their target genes occur in chromosomal loop structures formed by the interaction between two DNA sites that are bound by a CCCTC-binding factor (CTCF) protein and occupied by the cohesion complex (Dixon et al., 2012). These interactions are essential for normal gene activation and repression in humans (Zuin et al., 2014). These chromosomal scaffolds are preserved throughout development and can be perturbed in disease states by genetic and epigenetic factors (Hnisz, Day, & Young, 2016).
Epigenetic Regulation
Epigenetic regulation allows for changes in GE in response to the environment. Epigenetic regulation of GE can occur as a result of DNA methylation, histone modifications, or noncoding RNA expression (Stephens, Miaskowski, Levine, Pullinger, & Aouizerat, 2012).
DNA methylation occurs primarily at the cytosine base of the molecule that is adjacent to guanine (i.e., CpG site). Cytosine is converted to 5-methylcytosine by DNA methyltransferase. DNA methylation of a CpG island in the promoter region of a gene can repress GE by blocking the binding of transcription factors to the methylated promoter site (Phillips, 2008). For example, hypermethylation of DNA can lead to silencing of tumor suppressor genes like BRCA1, which results in tumorigenesis (Chakravarthi, Nepal, & Varambally, 2016). In certain cases, CpG islands activate transcription (Spruijt & Vermeulen, 2014). For example, CpG-binding transcription factors (e.g., Kruppel like factor 4) can activate transcription and cause mature human cells to produce induced pluripotent stem cells.
Histone modification is another vehicle for epigenetic regulation of GE, DNA is bound to histone proteins to form a nucleosome. Nucleosomes are arranged in a compact chromatin structure in the nucleus of the cell. Certain amino acids on the histone protein can be modified by acetylation, phosphorylation, or methylation. Histone modifications can affect GE through two mechanisms. First, histone modifications can result in a less compact DNA structure, which makes it more accessible for transcription. Second, proteins can bind to the modified amino acid on the histone protein and alter the transcription of DNA (Bannister & Kouzarides, 2011).
Finally, epigenetic regulation of GE can be mediated by noncoding RNA expression. Noncoding RNAs play important roles in the regulation of transcriptional and posttranscriptional processes (discussed below). For example, miRNAs regulate GE by repressing the translation or promoting the degradation of mRNA (Flowers, Froelicher, & Aouizerat, 2013). miRNAs can bind to the mRNA molecule and inhibit protein synthesis. The expression of these miRNAs varies over time depending on changes in the intracellular and external environments (Radom-Aizik, Zaldivar, Oliver, Galassetti, & Cooper, 2010).
Posttranscriptional Processes
Posttranscriptional processes convert noncoding RNA into a functional gene product and prepare mRNA for translation. Posttranscriptional processes involve 5′ capping of pre-mRNA, removal of intron sequences from RNA by splicing, alternative splicing of pre-mRNA, addition of a poly(A) tail to the pre-mRNA, gene fusion transcript processing, and modulation of mRNA stability (Perdew et al., 2006). Among these processes, removal of intron sequences from RNA by splicing, alternative splicing, and gene fusion transcript processing events can affect the regulation of GE.
Splicing of intron sequences from pre-mRNA is catalyzed by RNA-protein complexes called small nuclear ribonucleoprotein particles (snRNPs). The RNAs found in snRNPs are a type of long noncoding RNA. There are five types of snRNAs: U1, U2, U4, U5, and U6. The U1 RNA has a sequence complimentary to the 5′ end of the intron. U2 RNA recognizes sequences close to the 3′ end. The U4, U5, and U6 RNAs form a complex called the splicesome, which removes the intron region and joins the two exons together (Perdew et al., 2006).
Alternative splicing occurs when the same gene gives rise to multiple different transcripts. Alternative splicing of a pre-mRNA transcript can lead to changes in GE. The coding region of a gene is called the exon. On average, a human gene contains 10–15 exons. Through alternative splicing, these exon sequences can form different combinations that encode different versions of a protein. Therefore, mRNA transcripts from the same gene can differ as a result of the different combinations of exon sequences that can get incorporated into the mature mRNA transcript (Gibson & Muse, 2009, chapter 2; Lee & Young, 2013).
As in the case of alternative splicing, gene fusion events can occur when two noncontinuous genomic regions join to form a single transcript. The resulting transcripts are called fusion transcripts. In some cases, these gene fusion events change transcript levels in cells and cause a disease state (Gibson & Muse, 2009, chapter 2). For example, fusion of the transmembrane protease serine 2 gene (TMPRSS2) with the transcription factor gene v-ets avian erythroblastosis virus E26 oncogene homologue (ERG) results in the TMPRSS2-ERG fusion gene. This fusion gene transcribes a fusion transcript that results in the synthesis of a chimeric protein that causes an epithelial malignancy (Mertens, Johansson, Fioretos, & Mitelman, 2015). Intron splicing, alternative splicing, and gene fusion events can be measured and analyzed as part of a GE study.
Common Sources of Variation in GE
A number of factors can influence GE. Researchers can control for some of these factors during the statistical analysis of GE data, and they need to consider them in the design of a GE study, so that they collect appropriate data for subsequent analyses. Tissue specificity, host age, host gender, time of day or season, environment, and heritable variations are all factors that can influence GE and should be considered in the design of GE studies.
Tissue Specificity
GE varies from cell to cell and tissue to tissue (Byron et al., 2016; Font-Tello et al., 2015). For example, expression of perilipin1 (PLIN1) occurs in adipocytes but not in fibroblasts, peripheral nerves, or chondrocytes (Human Protein Atlas, n.d.). Compared to GE in mature cells, GE in embryonic stem cells is completely different. Because these cells are actively dividing, about 60% of the coding genes are transcribed into mRNA, and a minority of the genes are cell-specific (Lee & Young, 2013). In most tissues, a subset of RNA transcripts are expressed nonspecifically at a constant level and are termed housekeeping genes. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), for example, is a housekeeping gene expressed across most human tissues (Barber, Harmer, Coleman, & Clark, 2005). As discussed below, expression of housekeeping genes serves as a check for cell function, and researchers use it for quality control of GE data.
The Tissue-Specific Gene Expression and Regulation (TIGER) online database provides information on the depth and breadth of tissue-specific GE (Liu et al., 2008). The TIGER database contains three types of data on unique levels of regulation of GE: (1) tissue-specific GE profiles, (2) tissue-specific combinatorial gene regulation with multiple transcription factors, and (3) CRM detection. Across 30 tissues, 7,261 tissue-specific genes have been identified and approximately 9,060 tissue-specific transcription-factor interactions have been predicted (n ∼ 300 per tissue; Liu et al., 2008; Su et al., 2004).
Prior knowledge of variations in background levels of GE in resting cells is beneficial when investigators are undertaking studies to determine associations between tissue-specific changes in GE and a particular phenotype. A common example of tissue-specific GE is the various cellular components of peripheral blood. In blood, GE occurs in monocytes, dendritic cells, natural killer cells, CD4+ T lymphocytes, CD8+ T lymphocytes, and B lymphocytes (Cole, Hawkley, Arevalo, & Cacioppo, 2011). Depending on the phenotype under investigation, relevant changes in GE can occur in a single or multiple cell types.
For example, in a study that compared chronically lonely individuals to a healthy control group, 98 genes were differentially expressed between the two groups (Cole et al., 2011). While upregulated GE was found predominantly in dendritic cells, downregulated GE occurred in dendritic cells and monocytes. Upregulated genes were involved in leukocyte activation and inflammation, and downregulated genes were involved in Type I interferon antiviral responses. The identification of these cellular-specific pathways provides insights into the biological processes that may contribute to loneliness (Lee & Young, 2013). For GE studies of peripheral blood, investigators can use appropriate analytic procedures (e.g., tissue-of-origin analysis; Cole et al., 2011) to evaluate the relative contributions of various cell types to changes in GE and the particular phenotype of interest.
Finally, heterogeneity in GE within a population of similar cells (e.g., monocytes) can be a source of unexpected variation. Factors associated with variations in GE within a specific cell type include cell-cycle stage, the presence or absence of microbes, and differences in the microenvironment. Researchers should thus consider sources of variations in GE among similar populations of cells in a tissue when conducting GE analyses. Single-cell transcriptome profile sequencing can be done to detect variability in GE within cells and between cells of the same tissue sample. It allows for expression-based clustering of cell types in a tissue, detection of altered transcription in matched cell types, and discovery of new cell types (Sandberg, 2014).
Host Age
GE varies with age. In a meta-analysis (de Magalhaes, Curado, & Church, 2009), the authors found that as the age of humans increased, 56 genes were consistently overexpressed and 17 genes were underexpressed. Changes in GE with age are likely caused by DNA damage (Bahar et al., 2006; Lu et al., 2004). Age-related changes in GE appear to be gradual (Peters, 2006). In any GE study, researchers should record the participants’ age. If investigators are using tissue banked samples, they should match the specimens on age. If a study reveals changes in the expression of genes that are known to change their levels of expression as humans age, researchers should control for participants’ age in the statistical analyses.
Host Gender
Gender influences GE in whole blood. In one study that enrolled 41 healthy males and 36 healthy females (Whitney et al., 2003), 46 genes (i.e., 35 in females and 11 in males) showed gender-associated differences in GE. In microarray experiments, researchers found significant differences in GE by gender for both autosomal and sex chromosome genes (Whitney et al., 2003; Xu et al., 2013). For example, while investigators found no gender-associated differences in neutrophil counts in one study (Whitney et al., 2003), they did find a small number of genes that were highly expressed in the neutrophils of females. Depending on the phenotype under investigation, if a study reveals differences in expression in genes whose expression is known to be influenced by gender, then researchers may need to control for the participants’ gender in the statistical analyses.
Time of Sample Collection
Researchers have observed variations in GE in multiple tissues over a 24-hr period (Storch et al., 2002) and across different seasons (Dopico et al., 2015). For example, GE levels in whole blood (Whitney et al., 2003), human parotid saliva (Hardt et al., 2005), and in the liver and heart (Storch et al., 2002) exhibit diurnal variations. If possible, researchers should collect blood, saliva, and other tissues at the same time of day in all participants. If this approach is not possible, then they should record the time the sample was collected.
In addition, seasonal variations in GE can occur. In one study (Dopico et al., 2015), more than 4,000 protein-coding mRNAs in white blood cells and adipose tissue exhibited seasonal changes in GE. Again, when designing a GE experiment, researchers should record the date and time of day at which they collected the specimen. If investigators find that genes with known temporal variations in GE are differentially expressed, having the date and time of the collection may allow for the control of this potential source of variability in subsequent analyses.
Environment
GE is influenced by environmental factors. For example, in one study of changes in GE as a result of exposure to traffic-related pollutants (Chu et al., 2016), a core set of 25 transcripts were differentially expressed. These 25 genes were implicated in pathways associated with cancer, heart disease, and chronic lung disease. Particularly for disease susceptibility studies, researchers may need to collect data on a number of environmental factors (e.g., stress, air pollutants, diet) depending on the phenotype under consideration (Armenise et al., 2017; Bouchard-Mercier et al., 2013; Kundakovic & Jaric, 2017; Mancini et al., 2017; Ruegsegger et al., 2017).
Inherited Variation
GE is influenced by inherited (i.e., genetic) variations. A large body of research has explored how genetic variations influence GE and risk for certain diseases (Gibson, Powell, & Marigorta, 2015). For example, in one seminal study of obesity traits in a large population-based cohort (Emilsson et al., 2008), researchers evaluated differences in GE in adipose tissue and blood associated with obesity-related traits. While the differences in GE in adipose tissue were highly correlated with obesity-related traits, the investigators did not find these associations for blood. In addition, the investigators evaluated the genetic component to GE and found a significant heritable contribution to the observed variations in GE in both tissue types. Changes in GE of inflammatory- and immune-response genes were causally associated with obesity. This example highlights the genetic (i.e., heritable) contributions to variations in GE. When designing a GE study, researchers may want to account for the genetic component of GE during data analysis.
Transcript Level and Serum Protein Levels May Not Correlate
Although protein-coding gene transcript levels may change, the transcript levels for a specific mRNA may not correlate with the serum protein product levels (Li & Xie, 2011). As a result, knowledge of the level of GE does not provide a reliable estimate of protein levels. Actual transcript and protein levels can fluctuate in cells based on the characteristics of the extracellular and intracellular environments. Transcription of mRNA may not translate into a protein product because of rapid degradation of mRNA, degradation of the protein product, a reservoir effect, or other factors (Li & Xie, 2011). If the purpose of a research study is to evaluate the correlation between changes in GE and serum protein levels, researchers should measure protein levels in addition to transcription.
Common Methods for Measurement of GE
The most common laboratory methods used to measure GE levels are Northern blotting, quantitative polymerase chain reaction (qPCR), DNA microarray, and RNA-Seq. In this section, we describe these methods and compare the benefits and challenges of each of these methods.
Northern Blotting
Northern blot analysis is a standard method used to determine the size and quantity of a specific RNA in a sample. The RNA is size fractionated by polyacrylamide or agarose gel electrophoresis and transferred to a nitrocellulose membrane. RNA molecules form covalent bonds with the nitrocellulose membrane. Probes that are complimentary to the transcript are labeled prior to hybridization with a chemiluminescence dye. The nitrocellulose membrane with the bound RNA is hybridized with one or more specifically labeled probes. The hybridized nitrocellulose membrane is exposed overnight to an X-ray film, which is processed for detection of signal to determine the presence or the absence of the transcript (Josefsen & Nielsen, 2011). Northern blotting is often used to measure tRNA or a specific set of mRNAs (Janssen, Diner, & Hayes, 2012). As shown in Table 2, the advantages of Northern blotting include the simplicity of the procedure and low cost. Limitations of this technique are that it is time-consuming, only a small number of samples can be analyzed at one time, and it requires a large amount of starting material and stringent oligonucleotide hybridization (Streit, Michalski, Erkan, Kleeff, & Friess, 2008).
Table 2.
Techniques for RNA Measurement.
| Method | Technique | Initial RNA Processing Step | Strengths | Limitations |
|---|---|---|---|---|
| Northern blotting | Hybridization-based assay | mRNA, tRNA labeled, and bound to nitrocellulose paper |
|
|
| qPCR | PCR quantification–based assay | mRNA reverse transcribed to cDNA |
|
|
| Microarray | Multistep workflow for target preparation hybridization-based assay | mRNA reverse transcribed to cDNA |
|
|
| RNA-Seq | Adapter ligation-, PCR amplification-, and sequencing-based assay | mRNA reverse transcribed to cDNA or labeling of miRNA, tRNA, and rRNA |
|
|
Note. cDNA = complementary deoxyribonucleic acid; GWAS = genome-wide association studies; miRNA = microribonucleic acid; mRNA = messenger ribonucleic acid; PCR = polymerase chain reaction; qPCR = quantitative polymerase chain reaction; RNA = ribonucleic acid; rRNA = ribose ribonucleic acid; RNA-Seq = RNA sequencing; tRNA = transfer ribonucleic acid.
qPCR
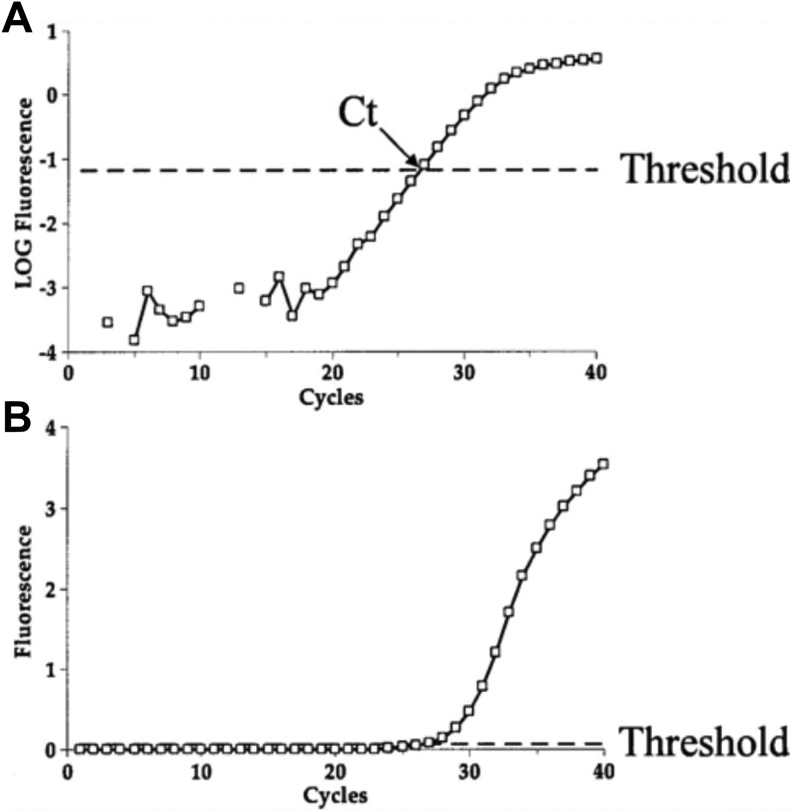
qPCR is a method to quantify GE in real time (VanGuilder, Vrana, & Freeman, 2008). The quantity is measured using a spectrophotometer. mRNA is often used as the template for the qPCR reaction. During qPCR, the mRNA template is converted to complimentary DNA (cDNA) using reverse transcriptase. Then, the single-stranded cDNA is synthesized into double-stranded DNA using DNA polymerase. The reaction proceeds exponentially as the double-stranded DNA replicates. The amount of DNA is measured after each round of amplification. A fluorescent label is added to the reaction mixture so that, as the amplified DNA molecules accumulate, the fluorescent values are recorded at the end of each cycle. The level of the fluorescent signal is directly proportional to the amount of amplified DNA that is present. As shown in Figure 3, the threshold cycle (Ct) value for a particular DNA is the cycle number when the fluorescence signal of the amplifying DNA molecule is first detected above the baseline threshold. The Ct value depends on the amount of mRNA present at the start of qPCR and provides an estimate of the level of GE (VanGuilder et al., 2008).
Figure 3.
The end result of real-time polymerase chain reaction (PCR) is the amplification plot. The x-axis is the PCR cycle, whereas the y-axis is the fluorescence that increases throughout the reaction. (a) Amplification plot on a logarithmic scale, demonstrating appropriate threshold. (b) Amplification plot on a linear scale, demonstrating the same threshold. Note that on a logarithmic scale, the linear phase appears to be a straight line. CT = crossing threshold. Source: Reprinted with the permission of Springer from Peirson and Butler (2007, fig. 3). Copyright 2007 by Humana Press.
A serial dilution of a control DNA of a known concentration is prepared to generate a standard curve for absolute quantification. The standard curve measures the exact amount of template in the sample. More than one transcript can be detected by using a method called multiplex qPCR. Multiplex qPCR includes probes for genes labeled with different reporter dyes in the same reaction mix. This approach allows for the detection of the levels of RNA from multiple genes in the same qPCR reaction (VanGuilder et al., 2008).
As shown in Table 2, the advantages of qPCR include ease of use, the relatively short period of time for quantifying mRNA transcripts (8–12 hr), and the ability to detect multiple mRNA transcripts using a multiplex approach (Smith & Osborn, 2009). However, several limitations warrant consideration, namely, one needs a priori knowledge of the sequence of the target transcript that is to be quantified and only a small number of transcripts can be quantified in each reaction, which limits the throughput of this method.
Microarray
Microarrays have been used to measure GE for over 15 years (Schulze & Downward, 2001). Microarray technology employs the principle of nucleic acid hybridization of cDNA strands to quantify a large number of transcripts in a single experiment (Sinicropi, Cronin, & Liu, 2006). Two types of microarrays can be used for GE. The in situ synthesized oligonucleotide microarray uses oligonucleotides that are 25 bases in length and are attached to a chip surface by a light-directed method (Fodor et al., 1991). The cDNA microarray uses a single-stranded cDNA that is reverse transcribed from a single strand of mRNA. The single-stranded cDNA is converted into double-stranded DNA through PCR amplification. Then, the PCR product is immobilized on the array with each PCR product spot about 80–200 μm in diameter. The double-stranded PCR product is denatured to form a single strand before use. This single-stranded DNA represents the mRNA template.
While microarray designs differ among vendors and laboratories (Petersen et al., 2005), a microarray experiment is always a multistep process. First, RNA is extracted from the tissue and reverse transcribed to cDNA. Then, cDNA is processed for labeling. The labeled nucleic acids are transferred to a microarray chip for hybridization with immobilized probes. The microarray chips are hybridized in a temperature-controlled chamber. The microarray chips are washed after the hybridization step in salt buffer, and the hybridized, tagged, fluorescent-labeled nucleic acid sequences remain on the microarray chip. Then, the hybridized microarray chip is scanned to read the fluorescent excitation signals. The intensity of the fluorescent signals detected is directly proportional to the amount of transcribed RNA. Finally, various software packages and statistical approaches are used to analyze the GE data (Trevino, Falciana, & Barrera-Saldana, 2007).
As shown in Table 2, the advantages of microarray quantitation are that a large number of transcripts can be quantified in a single experiment, tens of thousands of transcripts can be measured simultaneously, the costs are relatively low, and a prior knowledge of transcript sequences is not required. The limitations include the fact that multiple tissue samples cannot be tested in one assay; a control and a test tissue sample need to be prepared separately, which takes more time and may contribute to increases in the variance of output data; and RNA quantification is determined by image processing that requires specialized equipment and software (Sinicropi et al., 2007).
RNA-Seq
RNA-Seq quantifies the levels of various types of RNA in a sample by sequencing the RNA directly and counting the number of sequences. This approach is different from Northern blotting, which quantifies RNA through gel electrophoresis; qPCR, which quantifies RNA through amplification and dye intensity; and microarray, which quantifies RNA through template hybridization and dye intensity. While several methods for sequencing RNA exist (Hrdlickova, Toloue, & Tian, 2017), they all share a similar overall process. First, the target RNA is extracted and purified from the sample. The type of RNA sequenced depends on the objective of the study. For example, total RNA-Seq attempts to measure all of the expressed RNA. Coding RNA can be enriched by poly(A) capture techniques, and small RNAs can be enriched through size selection and gel electrophoresis. In addition, input RNA can be enriched for a particular RNA species of interest through removal of unwanted species. For example, rRNA and globin RNA account for a large proportion of total RNA and are often removed through ribosomal and globin depletion products prior to sequencing library preparation. Alternatively, if the RNA sequences are known, RNA capture techniques can isolate specific types of RNA using complementary probes (Hrdlickova et al., 2017).
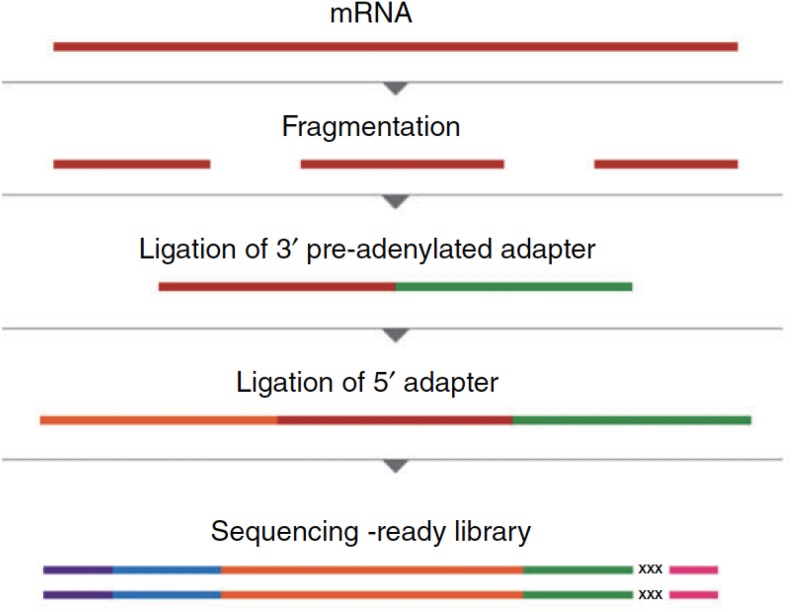
As shown in Figure 4, the RNA sample is prepared as a library prior to sequencing. The RNA is typically fragmented, and fragments of 100–500 bases are isolated. The input RNA is ligated to specific sequence linkers called adapter molecules, which may include primers for subsequent amplification steps. The fragmented and adapter-ligated RNA molecules are typically 150–550 base pairs in length. The adapter-ligated RNA fragments are amplified and sequenced in parallel multiple times (Hrdlickova et al., 2016).
Figure 4.
RNA library preparation procedure for sequencing RNA. The mRNA molecule is fragmented. Adapter molecules with known sequences are ligated to the 3′ and 5′ end of the fragmented mRNA molecules. Ligating adapters with unique barcode sequences to fragmented mRNA molecules mediates processing full-length mRNA sequence. mRNA = messenger RNA; RNA = ribonucleic acid. Source: Reprinted with permission from Hrdlickova, Toloue, and Tian (2017). Copyright 2016 by Wiley Periodicals.
Several advantages of sequencing and quantitation using RNA-Seq exist. Massive parallel sequencing of transcripts allows for the detection of underlying genomic alterations at single-nucleotide resolution. RNA-Seq has a greater dynamic range to quantify transcripts compared to microarray technology. Thousands of differentially expressed genes, tens of thousands of differentially expressed gene isoforms, mutations and germ-line variations in thousands of expressed genetic variants, and transcript isoforms and splice variants can be detected (Wang, Gerstein, & Snyder, 2009). Compared to microarray, RNA-Seq can detect 30% more differentially expressed genes. However, the limitations of RNA-Seq include a higher cost per sample relative to microarray and a higher computational and data-storage burden for downstream analyses. Fortunately, cheaper assays with increased sensitivity to detect different types of RNA are evolving. In addition, data-storage procedures and data-analysis tools are becoming easier to use (Byron et al., 2016).
Factors to Consider When Choosing Tissue to Measure GE
GE studies can be performed on a wide range of human tissues (e.g., whole blood, breast biopsies, and gut microbiome; Agus et al., 2016; Bondar et al., 2014; Chakravarthi et al., 2016). However, a number of issues regarding tissue selection warrant consideration.
Tissue Type
First, it is important to determine which tissue (e.g., blood vs. a muscle biopsy) is the most appropriate one to use to answer the research question. Given its widespread use, some considerations regarding the use of blood versus other types of tissue for GE studies warrant discussion. The advantages of using blood are that it is relatively easy to collect and can be stored for long-term use. However, as noted above, blood is a heterogeneous tissue, and GE levels differ among cell types. Whole blood includes red blood cells, white blood cells, and platelets suspended in plasma. Peripheral blood mononuclear cells (PBMCs) include monocytes and lymphocytes (e.g., T cells and B cells) that can be isolated from whole blood. For some studies, RNA transcript levels should be collected in more homogenous cell populations, such as T or B lymphocytes, that may have potential biomarkers for a specific phenotype. However, additional processing is required to isolate specific cell types (Keating & Hartmann, 2017).
The level of RNA transcript detected in blood depends on the cell type and the cell-isolation method used. The PAXgene (Qiagen, Valencia, CA) method captures RNA transcripts from all of the cell types in whole blood. No cell-isolation procedure is done prior to RNA isolation. Alternatively, the Ficoll method was developed to isolate a subset of white blood cells such as PBMCs prior to RNA extraction. A downside of this method is that 8 ml of blood are required to extract RNA in contrast to the 2.5 ml needed for the PAXgene method (Min et al., 2010).
Due to the susceptibility of RNA molecules to degradation, researchers must take care when extracting RNA from tissue. In particular, they should perform RNA extractions under RNase-free laboratory conditions. Fortunately, commercial RNA extraction packages are available, and in most situations only standard molecular laboratory equipment (e.g., a microcentrifuge, micropipettes, and vacuum manifold) is required. RNA extraction from peripheral blood takes approximately 2 hr.
Tissue Availability
Depending on the research question and tissue availability, researchers can collect the tissue from the patient directly or obtain it from a biobank. Large repositories of tissue samples are available for research use. Some examples include the following: the National Institutes of Health (NIH) NeuroBioBank (https://neurobiobank.nih.gov/), the Mayo Clinic Biobank (http://www.mayo.edu/research/centers-programs/mayo-clinic-biobank/overview), and the Global Biobank Directory, Tissue Bank, and Biorepositories (http://specimencentral.com/biobank-directory/). Consent is obtained from the participant at the time of tissue collection for research use.
For the purposes of a research study, advantages of obtaining samples from a biobank include a streamlined process for sample acquisition, no need for consent from the donor at the time tissue is ordered, availability of these samples at low cost, and easy accessibility. However, one disadvantage of using samples from a biobank is that the quality of RNA may be compromised. Research showed that RNA stored at −80 °C degrades within 5 years (Shabihkhani et al., 2014). The tolerance of RNA for freeze–thaw events is low. Before requesting samples from a biobank, researchers should examine the quality control and quality assurance procedures the biobank employs.
Tissue Management
Care must be taken to properly preserve RNA during and after sample collection. RNA is a single-stranded molecule and is susceptible to fragmentation by ubiquitous RNase. For peripheral blood, special collection tubes (e.g., PAXgene tubes) are used to stabilize the RNA (Takeda et al., 2015). RNA samples can be stored at 4 °C for a day after collection and then must be placed in a −80 °C freezer for long-term storage.
Fresh tissue samples that are to be stored long term for pathology studies or RNA extraction for GE experiments are treated with a formaldehyde solution diluted with water and transferred to small containers called cassettes for processing. After processing, the tissue is put in a mold with hot paraffin wax. After the wax cools, the tissue becomes a block (i.e., formalin-fixed paraffin-embedded [FFPE]). The fixation of these samples with formalin causes cross-linking of nucleic acids with protein molecules, covalent modification of RNA, and fragmentation of RNA transcripts. In the past, because of the RNA degradation that occurred, researchers did not use FFPE samples for GE studies. However, numerous methods are now available to isolate RNA from FFPE samples (Zhou, Sahin, & Myers, 2015).
Alternatively, biopsies can be frozen without processing. A fresh biopsy should be frozen soon after collection in liquid nitrogen to preserve the RNA (i.e., fresh frozen [FF] sample). In one study (Ripoli et al., 2016), levels of GE from FF and FFPE samples showed only 63% concordance. Compared to FF samples, FFPE samples showed lower levels of GE. Whether to use FF or FFPE samples will depend on the availability of these samples and the specific research question. While some studies showed that the use of an RNA-stabilizing reagent called RNAlater (Ambion, Foster City, CA) improved preservation of RNA (Freidin et al., 2012; Martin et al., 2017) researchers in other studies did not observe this effect (Micke et al., 2006; Mutter et al., 2004).
Quality Control
Regardless of the source for extraction, researchers must perform quality-control procedures to determine the quality and quantity of the RNA and to check for degradation before the start of a GE experiment. Processing of degraded RNA in test samples can lead to spurious results (Fleige & Pfaffl, 2006). Some RNAs may be preferentially degraded in a sample while others are stable (Mayne, Shepel, & Geiger, 1999). For example, in blood samples, while miRNA remains stable, other RNA species may be degraded (Eikmans, Rekers, Anholts, Heidt, & Claas, 2013).
Quality control procedures used to determine the quality of RNA in a sample are well described elsewhere (Zhou et al., 2015). For example, one method to check the quality of RNA is to determine the expression levels of housekeeping genes (e.g., GAPDH, actin, or 18S rRNA). For this method, a qPCR procedure is performed and fluorescent signal intensity values are evaluated for housekeeping gene transcripts. mRNA stability of housekeeping genes can be affected by cell treatment conditions and cell types. Some housekeeping gene transcripts are more stable than others in a specific cell type. It is important to validate the expression level of different housekeeping genes before use to normalize target GE. If degradation of housekeeping gene transcripts is detected in a test sample compared to a control sample, then researchers should eliminate that sample from the study (Julian, de Oliveira, Perry, Tufik, & Chagas, 2014).
Conclusions
GE is a complex, multifaceted process with intricate regulatory mechanisms. An evaluation of changes in GE may help to explain differences in various phenotypes. For example, GE studies can be used for the following: to diagnose specific diseases (Lapuk et al., 2010), evaluate differences in disease prognoses (Koleck & Conley, 2016), evaluate the mechanisms that underlie differences in symptom severity (Kober et al., 2016), aid in treatment decisions, and identify patients at increased risk for adverse events from specific treatments (Cardoso et al., 2016; Koleck & Conley, 2016). GE studies provide insights into the interplay among pathways that may explain underlying biological mechanisms involved in the transition from a healthy to a disease state, the occurrence and severity of symptoms, and interindividual differences in responses to pharmacologic and nonpharmacologic interventions. Increased access to tissue specimens, technological advances in the measurement of GE, reduced costs associated with acquisition and measurement of GE, and the development of more robust analysis procedures have enabled more research groups to design and conduct GE studies. GE data in combination with other “omics” data (e.g., genome, proteome, and methylome) can provide important information to predict, diagnose, and treat common health conditions.
Footnotes
Author Contributions: Komal P. Singh contributed to conception and design, acquisition, and interpretation; drafted the manuscript; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Christine Miaskowski contributed to conception, design, and interpretation; drafted the manuscript; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Anand A. Dhruva contributed to design and interpretation, critically revised the manuscript, gave final approval, and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Elena Flowers contributed to design and interpretation, critically revised the manuscript, gave final approval, and agrees to be accountable for all aspects of work ensuring integrity and accuracy. Kord M. Kober contributed to conception and design, acquisition, and interpretation; drafted the manuscript; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by a grant from the National Cancer Institute (NCI, CA134900). Dr. Christine Miaskowski is an American Cancer Society Clinical Research Professor and is supported by a grant from NCI (CA168960). Komal P. Singh is supported by a grant from the American Cancer Society and by a T32 grant from the National Institute of Nursing Research (NR016920).
References
- Agus A. Denizot J. Thevenot J. Martinez-Medina M. Massier S. Sauvanet P. … Barnich N. (2016). Western diet induces a shift in microbiota composition enhancing susceptibility to adherent-invasive E. coli infection and intestinal inflammation. Scientific Reports, 6, 19032 doi:10.1038/srep19032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenise C. Lefebvre G. Carayol J. Bonnel S. Bolton J. Di Cara A. … Valsesia A. (2017). Transcriptome profiling from adipose tissue during a low-calorie diet reveals predictors of weight and glycemic outcomes in obese, nondiabetic subjects. American Journal of Clinical Nutrition, 106, 736–746. doi:10.3945/ajcn.117.156216 [DOI] [PubMed] [Google Scholar]
- Bahar R. Hartmann C. H. Rodriguez K. A. Denny A. D. Busuttil R. A. Dollé M. E. … Vijg J. (2006). Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature, 441, 1011–1014. [DOI] [PubMed] [Google Scholar]
- Bannister A. J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Research, 21, 381–395. doi:10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber R. D., Harmer D. W., Coleman R. A., Clark B. J. (2005). GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiological Genomics, 21, 389–395. [DOI] [PubMed] [Google Scholar]
- Beyersmann D. (2000). Regulation of mammalian gene expression. Experientia Supplementum, 89, 11–28. [DOI] [PubMed] [Google Scholar]
- Bondar G. Cadeiras M. Wisniewski M. Maque M. Chittoor J. Chang E. … Deng M. (2014). Comparison of whole blood and peripheral blood mononuclear cell gene expression for evaluation of the perioperative inflammatory response in patients with advanced heart failure. PLoS One, 9, e115097 doi:10.1371.journal.pone.0115097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard-Mercier A., Paradis A. M., Rudkowska I., Lemieux S., Couture P., Vohl M. C. (2013). Associations between dietary patterns and gene expression profiles of healthy men and women: A cross-sectional study. Nutrition Journal, 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron S. A., Keuren-Jensen K. R. V., Engelthaler D. M., Carpten J. D., Craig D. W. (2016). Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nature Review Genetics, 17, 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso F. van’t Veer L. J. Bogaerts J. Slaets L. Viale G. Delaloge S. … Piccart M. (2016). 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. New England Journal of Medicine, 375, 717–729. doi:10.1056/NEJMoa1602253 [DOI] [PubMed] [Google Scholar]
- Chakravarthi B. V., Nepal S., Varambally S. (2016). Genomic and epigenomic alterations in cancer. American Journal of Pathology, 186, 1724–1735. doi:10.1016/j.ajpath.2016.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Weiss W. A. (2015). Alternative splicing in cancer: Implications for biology and therapy. Oncogene, 34, 1–14. [DOI] [PubMed] [Google Scholar]
- Chu J. H., Hart J. E., Chhabra D., Garshick E., Raby B. A., Laden F. (2016). Gene expression network analyses in response to air pollution exposures in the trucking industry. Environmental Health, 15, 101 doi:10.1186/s12940-016-0187-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J. M. (2005). Fewer genes, more noncoding RNA. Science, 309, 1529–1530. doi:10.1126/science.1116800 [DOI] [PubMed] [Google Scholar]
- Cole S. W., Hawkley L. C., Arevalo J. M. G., Cacioppo J. T. (2011). Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proceedings of the National Academy of Sciences of the United States of America, 108, 3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes J. P., Curado J., Church G. M. (2009). Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics, 25, 875–881. doi:10.1093/bioinformatics/btp073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R. Selvaraj S. Yue F. Kim A. Li Y. Shen Y. … Ren B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 485, 376–380. doi:10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Marzluff W. F. (1999). Formation of the 3’ end of histone mRNA. Gene, 239, 1–14. [DOI] [PubMed] [Google Scholar]
- Dopico X. C. Evangelou M. Ferreira R. C. Guo H. Pekalski M. L. Smyth D. J. … Todd J. A. (2015). Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nature Communications, 6, 7000 doi:10.1038/ncomms8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett M., Dunbier A. K. (2008). Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clinical Cancer Research, 14, 8019–8026. doi:10.1158/1078-0432.CCR-08-0974 [DOI] [PubMed] [Google Scholar]
- Eikmans M., Rekers N. V., Anholts J. D., Heidt S., Claas F. H. (2013). Blood cell mRNAs and microRNAs: Optimized protocols for extraction and preservation. Blood, 121, e81–e89. [DOI] [PubMed] [Google Scholar]
- Emilsson V. Thorleifsson G. Zhang B. Leonardson A. S. Zink F. Zhu J. … Stefansson K. (2008). Genetics of gene expression and its effect on disease. Nature, 452, 423–428. doi:10.1038/nature06758 [DOI] [PubMed] [Google Scholar]
- Fleige S., Pfaffl M. W. (2006). RNA integrity and the effect on the real-time qRT-PCR performance. Molecular Aspects of Medicine, 27, 126–139. [DOI] [PubMed] [Google Scholar]
- Flowers E., Froelicher E. S., Aouizerat B. E. (2013). Measurement of microRNA: A regulator of gene expression. Biological Research for Nursing, 15, 167–178. doi:10.1177/1099800411430380 [DOI] [PubMed] [Google Scholar]
- Fodor S., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. (1991). Light-directed, spatially addressable parallel chemical synthesis. Science, 251, 767–773. [DOI] [PubMed] [Google Scholar]
- Font-Tello A. Juanpere N. de Muga S. Loranzo M. Lorente J. A. Fumado L. … Hernandez S. (2015). Association of ERG and TMPRSS2-ERG with grade, stage, and prognosis of prostrate cancer is dependent on their expression levels. Prostrate, 75, 1216–1226. doi:10.1002/pros.23004 [DOI] [PubMed] [Google Scholar]
- Freidin M., Bhudia N., Lim E., Nicholson A., Cookson W., Moffatt M. (2012). Impact of collection and storage of lung tumor tissue on whole genome expression profiling. Journal of Molecular Diagnostics, 14, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R., Giles K., Gowher H., Xiao T., Xu Z., Yao H., Felsenfeld G. (2012). Chromatin domains, insulators, and the regulation of gene expression. Biochimica et Biophysica Acta, 1819, 644–651. doi:10.1016/j.bbagrm.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G., Muse S. V. (2009). A primer of genome science (pp. 191–258). Sunderland, MA: Sinauer Associates. [Google Scholar]
- Gibson G., Powell J. E., Marigorta U. M. (2015). Expression quantitative trait locus analysis for translational medicine. Genome Medicine, 7, 60 doi:10.1186/s13073-015-0186-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T., Clausen B., Bramsen J., Finsen B., Damgaard C., Kjems J. (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Hansen T. B., Kjems J., Damgaard C. K. (2013). Circular RNA and miR-7 in cancer. Cancer Research, 73, 5609–5612. doi:10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- Hardt M., Witkowska H. E., Webb S., Thomas L. R., Dixon S. E., Hall S. C., Fisher S. J. (2005). Assessing the effects of diurnal variation on the composition of human parotid saliva: Quantitative analysis of native peptides using iTRAQ reagents. Analytical Chemistry, 77, 4947–4954. doi:10.1021/ac050161r [DOI] [PubMed] [Google Scholar]
- Hnisz D., Day D. S., Young R. A. (2016). Insulated neighborhoods: Structural and functional units of mammalian gene control. Cell, 167, 1188–1200. doi:10.1016/j.cell.2016.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B., Liebermann D. A. (2008). Apoptotic signaling by c-MYC. Oncogene, 27, 6462–6472. doi:10.1038/onc.2008.312 [DOI] [PubMed] [Google Scholar]
- Holdt L. M. Stahringer A. Sass K. Pichler G. Kulak N. A. Wilfert W. … Teupser D. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nature Communications, 7, 12429 doi:10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z. Jiang P. Swanson S. A. Elwell A. L. Nguyen B. K. Bolin J. M. … Thomson J. A. (2015). A cost-effective RNA sequencing protocol for large-scale gene expression studies. Scientific Reports, 5, 9570 doi:10.1038/srep09570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlickova R., Toloue M., Tian B. (2017). RNA-Seq methods for transcriptome analysis. Wiley Interdisciplinary Reviews. RNA, 8 doi:10.1002/wrna.1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Protein Atlas. (n.d.) Perilipin1 (PLIN1). Retrieved October 6, 2017, from http://www.proteinatlas.org/ENSG00000166819-PLIN1/tissue/adipose+tissue
- Janssen B. D., Diner E. J., Hayes C. S. (2012). Analysis of aminoacyl- and peptidyl-tRNAs by gel electrophoresis. Methods in Molecular Biology, 905, 291–309. doi:10.1007/978-1-61779-949-5_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsen K., Nielsen H. (2011). Northern blotting analysis. Methods in Molecular Biology, 703, 87–105. doi:10.1007/978-1-59745-248-9_7 [DOI] [PubMed] [Google Scholar]
- Julian G. S., de Oliveira R. W., Perry J. C., Tufik S., Chagas J. R. (2014). Validation of housekeeping genes in the brains of rats submitted to chronic intermittent hypoxia, a sleep apnea model. PLoS One, 9, e109902 doi:10.1371/journal.pone.0109902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating P., Hartmann J. X. (2017). Isolation and purification of Th9 cells for the study of inflammatory diseases in research and clinical settings In Walker J. M. (Series Ed.) Methods in molecular biology (Vol. 1585); Goswami R. (Ed.), Th9 cells: Methods and protocols (pp. 247–255). New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- King M. C., Wilson A. C. (1975). Evolution at two levels in humans and chimpanzees. Science, 188, 107–116. [DOI] [PubMed] [Google Scholar]
- Kober K. M. Dunn L. Mastick J. Cooper B. Langford D. Melisko M. … Aouizerat B. E. (2016). Gene expression profiling of evening fatigue in women undergoing chemotherapy for breast cancer. Biological Research for Nursing, 18, 370–385. doi:10.1177/1099800416629209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleck T. A., Conley Y. P. (2016). Identification and prioritization of candidate genes for symptom variability in breast cancer survivors based on disease characteristics at the cellular level. Breast Cancer (Dove Med Press), 8, 29–37. doi:10.2147/BCTT.S88434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukurba K. R., Montgomery S. B. (2015). RNA sequencing and analysis. Cold Spring Harbor Protocols, 2015, 951–969. doi:10.1101/pdb.top084970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M., Jaric I. (2017). The epigenetic link between prenatal adverse environments and neurodevelopmental disorders. Genes (Basel), 8 doi:10.3390/genes8030104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuk A. Marr H. Jakkula L. Pedro H. Bhattacharya S. Purdom E. … Durinck S. (2010). Exon-level microarray analyses identify alternative splicing programs in breast cancer. Molecular Cancer Research, 8, 961–974. doi:10.1158/1541-7786.MCR-09-0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Young R. A. (2013). Transcriptional regulation and its misregulation in disease. Cell, 152, 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Tjian R. (2003). Transcription regulation and animal diversity. Nature, 424, 147–151. doi:10.1016/j.cell.2013.02.014 [DOI] [PubMed] [Google Scholar]
- Li G. W., Xie X. S. (2011). Central dogma at the single-molecule level in living cells. Nature, 475, 308–315. doi:10.1038/nature10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu X., Zack D. J., Zhu H., Qian J. (2008). TiGER: A database for tissue-specific gene expression and regulation. BMC Bioinformatics, 9, 271 doi:10.1186/1471-2105-9-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Pan Y., Kao S. Y., Li C., Kohane I., Chan J., Yankner B. A. (2004). Gene regulation and DNA damage in the ageing human brain. Nature, 429, 883–891. doi:10.1038/nature02661 [DOI] [PubMed] [Google Scholar]
- Mancini A. Vitucci D. Labruna G. Imperlini E. Randers M. B. Schmidt J. F. … Buono P. (2017). Effect of lifelong football training on the expression of muscle molecular markers involved in healthy longevity. European Journal of Applied Physiology, 117, 721–730. doi:10.1007/s00421-017-3562-8 [DOI] [PubMed] [Google Scholar]
- Martin N. M., Cooke K. M., Radford C. C., Perley L. E., Silasi M., Flannery C. A. (2017). Time course analysis of RNA quality in placenta preserved by RNAlater or flash freezing. American Journal of Reproductive Immunology, 77, e12637 doi:10.1111/aji.12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston G. A., Evans S. K., Green M. R. (2006). Transcriptional regulatory elements in the human genome. Annual Review of Genomics and Human Genetics, 7, 29–59. [DOI] [PubMed] [Google Scholar]
- Mayne M., Shepel P. N., Geiger J. D. (1999). Recovery of high-integrity mRNA from brains of rats killed by high-energy focused microwave irradiation. Brain Research, 4, 295–302. [DOI] [PubMed] [Google Scholar]
- Mertens F., Johansson B., Fioretos T., Mitelman F. (2015). The emerging complexity of gene fusions in cancer. Nature Reviews Cancer, 15, 371–381. doi:10.1038/nrc3947 [DOI] [PubMed] [Google Scholar]
- Messina D. N., Glasscock J., Gish W., Lovett M. (2004). An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Research, 14, 2041–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micke P., Ohshima M., Tahmasebpoor S., Ren Z. P., Ostman A., Ponten F., Botling J. (2006). Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Laboratory Investigation, 86, 202–211. doi:10.1038/labinvest.3700372 [DOI] [PubMed] [Google Scholar]
- Min J. L. Barrett A. Watts T. Pettersson F. H. Lockstone H. E. Lindgren C. M. … McCarthy M. I. (2010). Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics, 11, 1–14. doi:10.1186/1471-2164-11-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutter G. L., Zahrieh D., Liu C., Neuberg D., Finkelstein D., Baker H. E., Warrington J. A. (2004). Comparison of frozen and RNAlater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics, 5, 88 doi:10.1186/1471-2164-5-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K. W. Anderson C. Marshall E. A. Minatel B. C. Enfield K. S. Saprunoff H. L. … Martinez V. D. (2016). PIWI-interacting RNAs in cancer: Emerging functions and clinical utility. Molecular Cancer, 15, 5 doi:10.1186/s12943-016-0491-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbourne S., Antalis T. M. (1998). Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochemical Journal, 331, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C. T., Corces V. G. (2011). Enhancer function: New insights into the regulation of tissue-specific gene expression. Nature Reviews Genetics, 12, 283–293. doi:10.1038/nrg2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson S. N., Butler J. N. (2007). Quantitative polymerase chain reaction. In Rosato E. (Ed.), Circadian rhythms. Methods in molecular biology (Vol. 362, p. 352). Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Perdew G. H., Vanden-Heuvel J. P., Peters J. M. (2006). Regulation of gene expression—Molecular mechanisms. Totowa, NJ: Humana Press. [Google Scholar]
- Peters R. (2006). Ageing and the brain. Postgraduate Medical. Journal, 82, 84–88. doi:10.1136/pgmj.2005.036665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen D. Chandramouli G. V. Geoghegan J. Hilburn J. Paarlberg J. Kim C. H. … Kawasaki E. S. (2005). Three microarray platforms: An analysis of their concordance in profiling gene expression. BMC Genomics, 6, 63 doi:10.1186/1471-2164-6-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips T. (2008). The role of methylation in gene expression. Nature Education, 1, 116. [Google Scholar]
- Pollard T. D., Earnshaw W. C., Lippincott-Schwartz J. (2007). Cell biology (2nd ed, p. 300). Oxford, England: Elsevier. [Google Scholar]
- Radom-Aizik S., Zaldivar F., Jr, Oliver S., Galassetti P., Cooper D. M. (2010). Evidence for microRNA involvement in exercise-associated neutrophil gene expression changes. Journal of Applied Physiology, 109, 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoli F. L. Mohr A. Conradine Hammer S. Willenbrock S. Hewicker-Trautwein M. Hennecke S. … Nolte I. (2016). A comparison of fresh frozen vs. formalin-fixed, paraffin-embedded specimens of canine mammary tumors via branched-DNA assay. International Journal of Molecular Sciences, 17, 724 doi:10.3390/ijms17050724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegsegger G. N. Grigsby K. B. Kelty T. J. Zidon T. M. Childs T. E. Vieira-Potter V. J. … Booth F. W. (2017). Maternal western diet age-specifically alters female offspring voluntary physical activity and dopamine- and leptin-related gene expression. FASEB Journal, 31, 5371–5383. doi:10.1096/fj.201700389 R [DOI] [PubMed] [Google Scholar]
- Sandberg R. (2014). Entering the era of single-cell transcriptomics in biology and medicine. Nature Methods, 11, 22–24. [DOI] [PubMed] [Google Scholar]
- Schulze A., Downward J. (2001). Navigating gene expression using microarrays—A technology review. Nature Cell Biology, 3, E190–E195. doi:10.1038/35087138 [DOI] [PubMed] [Google Scholar]
- Shabihkhani M. Lucey G. M. Wei B. Mareninov S. Lou J. J. Vinters H. V. … Yong W. H. (2014). The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clinical Biochemistry, 47, 258–266. doi:10.1016/j.clinbiochem.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinicropi D., Cronin M., Liu M. (2006). Gene expression profiling utilizing microarray technology and RT-PCR In Ferrari M., Ozkan M., Heller M. J. (Eds.), BioMEMS and biomedical nanotechnology (pp. 23–46). Boston, MA: Springer. [Google Scholar]
- Smith C. J., Osborn A. M. (2009). Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiology Ecology, 67, 6–20. doi:10.1111/j.1574-6941.2008.00629.x [DOI] [PubMed] [Google Scholar]
- Spruijt C. G., Vermeulen M. (2014). DNA methylation: Old dog, new tricks? Nature Structural & Molecular Biology, 21, 949–954. doi:10.1038/nsmb.2910 [DOI] [PubMed] [Google Scholar]
- Stephens K. M., Miaskowski C. A., Levine J. D., Pullinger C. R., Aouizerat B. E. (2012). Epigenetic regulation and measurement of epigenetic changes. Biological Research for Nursing, 15, 373–381. doi:10.1177/1099800412444785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch K. F., Lipan O., Leykin I., Viswanathan N., Davis F. C., Wong W. H., Weitz C. J. (2002). Extensive and divergent circadian gene expression in liver and heart. Nature, 417, 78–83. doi:10.1038/nature744 [DOI] [PubMed] [Google Scholar]
- Streit S., Michalski C. W., Erkan M., Kleeff J., Friess H. (2008). Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nature Protocols, 4, 37–43. doi:10.1038/nprot.2008.216 [DOI] [PubMed] [Google Scholar]
- Su A. Wiltshire T. Batalov S. Lapp H. Ching K. A. Block D. … Hogenesch J. B. (2004). A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America, 101, 6062–6067. doi:10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M., Funato T., Ikemoto M., Nanmoku T., Urasaki Y., Iwatani Y. (2015). Clinical usefulness of the PAXgene™ bone marrow RNA system for stabilizing total RNA. Journal of Clinical Laboratory Analysis, 29, 61–67. doi:10.1002/jcla.21729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino V., Falciana F., Barrera-Saldana H. A. (2007). DNA microarrays: A powerful tool for biomedical and clinical research. Molecular Medicine, 13, 527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGuilder H. D., Vrana K. E., Freeman W. M. (2008). Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques, 44, 619–626. doi:10.2144/000112776 [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: A revolutionary tool for transcriptomics. Nature Review Genetics, 10, 57–63. doi:10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney A. R., Diehn M., Popper S. J., Alizadeh A. A., Boldrick J. C., Relman D. A., Brown P. O. (2003). Individuality and variation in gene expression patterns in human blood. Proceedings of the National Academy of Sciences of the United States of America, 100, 1896–1901. doi:10.1073/pnas.252784499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. Hammer M. Paul S. M. Aouizerat B. E. Kober K. M. Conley Y. P. … Miaskowski C. (2017). Inflammatory pathway genes associated with inter-individual variability in the trajectories of morning and evening fatigue in patients receiving chemotherapy. Cytokine, 91, 187–210. doi:10.1016/j.cyto.2016.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang F., Liu Y., Yu Y., Gelernter J., Zhang H. (2013). Sex-biased methylome and transcriptome in human prefrontal cortex. Human Molecular Genetics, 23, 1260–1270. doi:10.1093/hmg/ddt516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. H., Sahin A. A., Myers J. N. (2015). Biobanking in genomic medicine. Archives of Pathology & Laboratory Medicine, 139, 812–818. doi:10.5858/arpa.2014-0261-RA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J. Dixon J. R. van der Reijden M. I. Ye Z. Kolovos P. Brouwer R. W. … Wendt K. S. (2014). Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proceedings of the National Academy of Sciences of the United States of America, 111, 996–1001. doi:10.1073/pnas.1317788111 [DOI] [PMC free article] [PubMed] [Google Scholar]