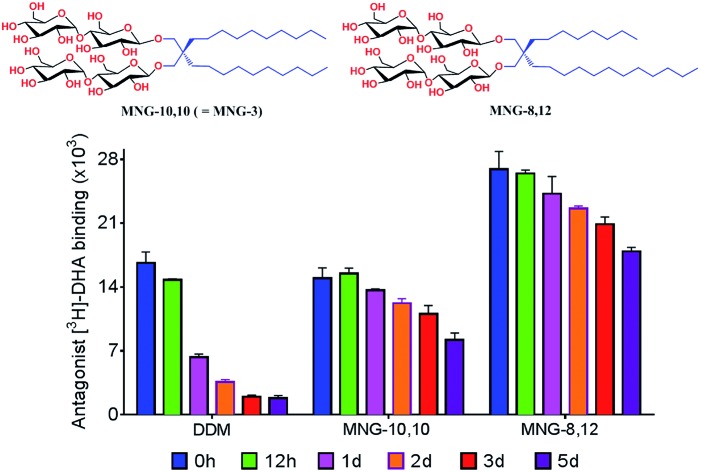
 An asymmetric MNG, MNG-8,12, provided enhanced stability to human G protein-coupled receptors (GPCRs) compared to the symmetric MNG, MNG-3.
An asymmetric MNG, MNG-8,12, provided enhanced stability to human G protein-coupled receptors (GPCRs) compared to the symmetric MNG, MNG-3.
Abstract
Maintaining protein stability in an aqueous solution is a prerequisite for protein structural and functional studies, but conventional detergents have increasingly showed limited ability to maintain protein integrity. A representative novel agent, maltose neopentyl glycol-3 (MNG-3), has recently substantially contributed to membrane protein structural studies. Motivated by the popular use of this novel agent, we prepared asymmetric versions of MNG-3 and evaluated these agents with several membrane proteins including two G protein-coupled receptors in this study. We found that some new MNGs were significantly more effective than MNG-3 at preserving protein integrity in the long term, suggesting that these asymmetric MNGs will find a wide use in membrane protein studies. In addition, this is the first study addressing the favorable effect of detergent asymmetric nature on membrane protein stability.
Introduction
The number of membrane proteins with known structures has been steadily growing since the first structure determination of the light harvesting complex in 1985.1 However, the number of membrane proteins with known structures remains much smaller than that of soluble proteins. Detergents, amphipathic agents, serve as essential tools for the structural study of these bio-macromolecules. These agents are used not only to extract membrane proteins from the membranes, but also to maintain protein integrity over the course of protein purification, necessary for downstream characterization.2 Conventional detergents such as OG (n-octyl-β-d-glucoside), DM (n-decyl-β-d-maltoside), DDM (n-dodecyl-β-d-maltoside), and LDAO (lauryldimethylamine-N-oxide) have been widely used for this purpose.2,3 However, many membrane proteins, particularly eukaryotic proteins and protein complexes with multiple subunits, have the tendency to aggregate/denature over time.2 Maintaining membrane integrity in detergent micelles is challenging presumably due to the inherently dynamic nature of these micellar assemblies. Because of the planar architecture, biological membranes are much less dynamic than detergent micelles and additionally exert lateral pressure on membrane proteins, thereby effectively preventing protein degradation.4 However, in the physiological environment of the native membrane, integral membrane proteins are not compatible for structural and functional characterization. As a consequence, detergent micelles are most widely used in membrane protein research.5 Membrane proteins are implicated in various human diseases such as cystic fibrosis and cancer and thus represent major drug targets.6 Thus, it is of great importance to develop new micellar systems with the ability to efficiently extract membrane proteins from the membranes and to effectively maintain the integrity of the extracted proteins in the long term.2,7
Over the past two decades, many efforts have been made to develop new micellar systems with enhanced efficacy toward membrane protein solubilization and stabilization.8 Most efforts were made by designing novel classes of amphiphiles with distinct architectures from conventional detergents. Different from conventional detergents built by the direct connection between a large head group and a single flexible alkyl chain, most novel amphiphiles developed so far contain multiple head and tail groups, as exemplified by tripod amphiphiles (TPAs),9a,b norbornane-based amphiphiles (NBMs),9c resorcinarene-based glucoside amphiphiles (RGAs),9d xylene-linked maltoside amphiphiles (XMAs),9e neopentyl glycol-based amphiphiles (glucose neopentyl glycols (GNGs), maltose neopentyl glycols (MNGs) and neopentyl glycol-derived triglucosides (NDTs))9f–i and pentasaccharide-bearing amphiphiles (PSEs).9j Single large lipophilic groups such as cholesterol and diosgenin were also used as detergent hydrophobic groups instead of multiple alkyl chains (e.g., chobimalt10a and glyco-diosgenin (GDN)10b). As expected by the successful use of peptides, polymers and dendrimers in many other applications, these scaffolds have been successfully included in novel amphiphile architectures. Lipopeptide detergents (LPDs)11a and β-peptides (BPs)11b are representatives of peptide-based amphiphiles, while amphipols (Apols)12a,b and styrene-maleic acid (SMA)-based lipodisqs12c are primary polymer-based inventions. Very recently, a dendronic structure (e.g., dendronic trimaltosides (DTMs)) was utilized as a detergent lipophilic group.13 It is noteworthy that membrane-mimetic systems with non-micellar architectures have also been developed. For instance, nanodiscs (NDs)14a and bicelles14b consist in a patch of lipid bilayer stabilized by membrane scaffold proteins (MSPs) and detergent molecules, respectively.
Among these agents, a representative novel amphiphile is MNG-3 (aka, LMNG), which has facilitated the elucidation of 40 new membrane protein structures including the β2 adrenergic, acetylcholine and opioid G-protein coupled receptors in the last seven years.15a–m This agent has been increasingly used since its invention in 2010 and has now become one of the most popular agents in membrane protein studies, indicating that the MNG scaffold is highly compatible with membrane protein structural studies. Previously, we reported several MNG derivatives with two identical alkyl groups, but all those ‘symmetric’ agents were inferior to MNG-3 for membrane protein stabilization.9h In the current study, by utilizing the privileged MNG-3 structure, we firstly prepared asymmetric amphiphiles, asymmetric MNGs (A-MNGs), containing two alkyl chains with different chain lengths; all multiple alkyl chain-bearing amphiphiles developed to date are symmetrical in this regard. The newly developed A-MNGs tend to form significantly smaller and more stable micelles than MNG-3. In evaluation with membrane proteins, a couple of A-MNGs were more effective than MNG-3 at stabilizing the tested membrane proteins here. Thus, this is the first study assessing the favourable effect of detergent asymmetricity on membrane protein stability.
Results and discussion
Detergent structures and physical characterization
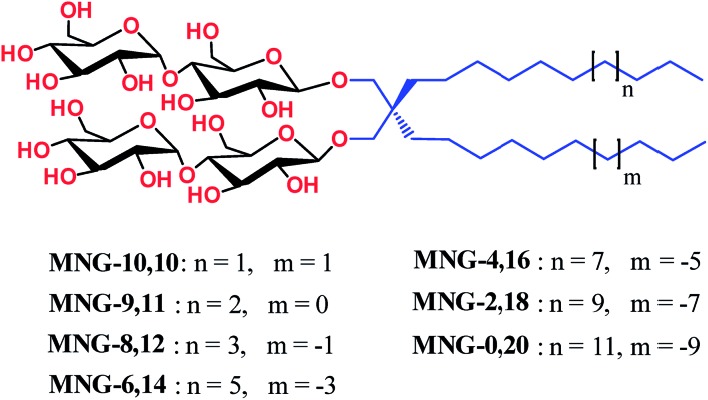
New MNG agents share a branched dimaltoside head group with MNG-3, but differ in terms of symmetricity of the lipophilic groups. MNG-3 has two identical C10 alkyl chains (‘symmetric’) while the new agents have two alkyl chains with different chain lengths and are thus denoted as asymmetric MNGs (A-MNGs). In the new MNGs, one chain length was increased from C11 to C20 while the other chain length was decreased from C9 to C0. In this variation, we maintained the total number of carbon units in detergent alkyl chains as C20, making MNG-3 and the A-MNGs constitutional isomers of each other. Thus, the A-MNGs basically have the same hydrophile–lipophile balance (HLB) as MNG-3, enabling us to compare detergent efficacy based only on variation in the detergent architecture.16 The chain lengths of the two alkyl groups were used for detergent designation. For example, MNG-3 with two C10 alkyl chains was denoted as MNG-10,10 and an A-MNG derivative with C6 and C14 alkyl chains as MNG-6,14. As for a systematic variation in detergent asymmetricity, we prepared MNG-9,11, MNG-8,12, MNG-6,14, MNG-4,16, MNG-2,18, and MNG-0,20 having two, four, eight, twelve, sixteen and twenty-carbon unit differences, respectively. Detergent lipophilic group asymmetricity is proportionally correlated with the chain length difference between the two alkyl groups. As a result, MNG-10,10 is completely symmetric (no difference) while MNG-0,20 is maximally asymmetric (C20 difference). Because of the presence of a single eicosanyl chain, MNG-0,20 is a monopod agent, while all the other A-MNGs and MNG-10,10 are dipod amphiphiles. It is noteworthy that asymmetric amphiphiles were previously reported in other applications,17 but there is no study reporting how detergent asymmetricity influences membrane protein solubilisation and stabilization.
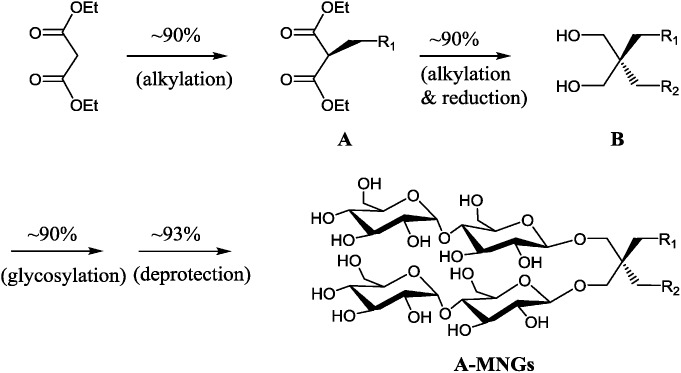
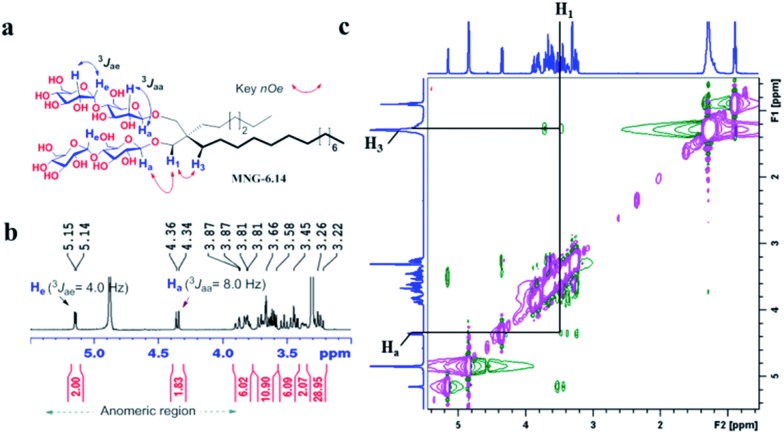
The preparation of an asymmetric amphiphile is generally substantially more difficult than that of its symmetric counterpart as introduction of two different alkyl chains into the detergent scaffold usually requires differentiation of two reaction sites with identical reactivity. Consequently, screening of one reaction site via a protecting group is necessary for the preparation of an asymmetric amphiphile, often resulting in an additional synthetic step and a decrease in the synthetic efficiency/yield of a target product. As synthetic accessibility is an important detergent feature for a widespread use in membrane protein studies, the aim of this study was to enhance detergent efficacy for membrane protein stability without impacting its practicability. In this respect, the A-MNGs introduced here seem optimal as, without introducing a protection group, two different alkyl chains could be introduced stepwise into the MNG scaffold efficiently using diethylmalonate as the starting material (Scheme 1). The first alkyl group was attached to diethylmalonate with a high yield (∼90%) using DMF-THF and K2CO3 as the solvent and base, respectively. Under these mild conditions utilizing a weak base (K2CO3), the formation of the di-alkylated product was impeded due to steric hindrance experienced by the mono-alkylated product (A), but the second alkylation was facilitated by the use of a strong base (NaH) in DMF in the subsequent step. Furthermore, the second alkylation and reduction by LiAlH4 was combined to afford a di-alkylated malonate-derived diol (B) with a two-step yield of ∼90%. Lastly, a maltose head group was attached to each alcohol of the di-alkylated diol via glycosylation and deprotection (∼90 and ∼93%, respectively). The synthetic protocol comprised of four high-yielding steps, as efficient as that used for the synthesis of MNG-10,10.9g The glycosylation was conducted using AgOTf as a promoter and perbenzoylated maltosylbromide as a glycosyl donor in the presence of a weak proton scavenger (i.e., 2,4,6-collidine). Due to the presence of a carbonyl-containing protecting group (i.e., a benzoyl group) in the vicinity of anomeric carbon, the stereochemistry of new glycosidic bonds was expected to be β (anchimeric assistance). The selective β-linkage formation was supported by the 1H NMR spectra of the A-MNGs (Fig. 1 & S1†). All new agents produced a doublet peak at 4.35 ppm with vicinal coupling constants (J) of 8.0 Hz. These chemical shift (δ) and coupling constant (J) are typical for β-glycosidic bonds. The peak corresponding to the α-glycosidic bond, appearing at δ = 5.17 ppm with J = 4.0 Hz, was also observed in the NMR spectra as maltoside is made of connection of two glucose units via an α-linkage. Detergent asymmetricity, defined as the chain length difference between the two alkyl chains in this study, did not affect the chemical shifts of the NMR peaks corresponding to the anomeric protons (Fig. S1†). Similarly, no noticeable difference in the chemical shift between the methyl protons (CH3) at the two alkyl chain terminals was observed for most A-MNGs; MNG-10,10, MNG-9,11, MNG-8,12 and MNG-6,14 all showed the same chemical shift of 0.90 ppm for these protons (Fig. S1†). This result indicates that, in the MNG scaffold, detergent asymmetricity is not well reflected by the methyl NMR peaks. However, the effect of detergent asymmetricity was detected in the methyl peaks of the highly asymmetric A-MNGs (MNG-4,16 and MNG-2,18). The methyl protons of the short chains in these agents (butyl for MNG-4,16 and ethyl for MNG-2,18) showed chemical shifts of 0.89 and 0.83 ppm, respectively, which were slightly and substantially different from that of the long chain methyl protons (0.90 ppm) (Δδ = 0.01 and 0.06 ppm, respectively) (Fig. S1†). The detergent structure was further characterized by 2D NOESY experiments using MNG-6,14 as a representative (Fig. 2c). A NOE correlation signal was observed between two hydrogens (H1 and H3) neighbouring the central quaternary carbon. An additional correlation signal was observed between the anomeric proton (Ha) and H1. These NOE signals are consistent with the chemical structure of this A-MNG (Fig. 2a).
Scheme 1. Synthetic scheme of A-MNGs using diethylmalonate as a starting material. Two different alkyl groups (R1CH2- and R2CH2-) were sequentially introduced into diethylmalonate to stepwise give mono-alkylated malonates (A) and di-alkylated propane-1,3-diol (B) in high yields (∼90% for each step). Two maltose units were attached to the resulting di-alkylated diol derivatives (B) via β-selective glycosylation and global deprotection with a high yield.
Fig. 1. Chemical structures of asymmetric MNGs (A-MNGs) and previously developed MNG-10,10. MNG-10,10 contains two identical decyl (C10) chains while the new agents have two different alkyl chains as the lipophilic groups, but the total numbers of carbon units (CH2) are all the same as C20 for the alkyl chains of A-MNGs or MNG-10,10. The A-MNGs were designated to include the number of carbon units of each alkyl chain.
Fig. 2. (a) Chemical structure of MNG-6,14 is shown to illustrate anomeric protons (Ha), their couplings with the neighbouring protons, and a set of protons of interest responsible for key NOE correlation signals. (b) Partial 1H NMR spectrum in the anomeric region for MNG-6,14 showing its high anomeric purity. The spectrum of MNG-6,14 showed a doublet at 4.35 ppm, with a vicinal axial–axial coupling constant (3Jaa) of 8.0 Hz, a typical peak characteristic of β-anomeric proton (Ha). The peaks for the anomeric protons with α-linkage (He) appeared at ∼5.15 ppm with a vicinal axial–equatorial coupling constant (3Jae) of 4.0 Hz. (c) Partial 2D NOESY NMR spectrum of MNG-6,14 in CD3OD. Only key 1H–1H NOE correlation signals are assigned. MNG-6,14 showed an NOE correlation signal between two protons of Ha and H1. Because of a close proximity in space, an NOE correlation signal between H1 and H3 was additionally observed in the spectrum.
All A-MNGs were water soluble up to 20%. Aggregation behaviours of the new agents were investigated in terms of the critical micelle concentration (CMC) and micelle size. Individual CMCs were estimated by micellar encapsulation of a hydrophobic fluorescent dye, diphenylhexatriene (DPH).18 The micelle size was determined in terms of the hydrodynamic radius (Rh) by dynamic light scattering (DLS) experiments. The results are summarized and compared with MNG-10,10 and DDM in Table 1. The CMC values of the new agents tend to decrease with increasing difference in the chain length of the two alkyl groups (i.e. detergent asymmetricity). For instance, MNG-0,20 with the largest chain length difference showed a CMC value five-fold smaller than MNG-10,10 with two identical alkyl chains (∼2 vs. ∼10 μM). As all A-MNGs have the same number of carbon units (C20) in the alkyl chain region, the variation in detergent CMC is not due to change in hydrophobicity of the lipophilic group but due to geometrical variation of the A-MNGs. Detergent micelles have a central core congested with multiple alkyl chains and thus the presence of a bulky group at the alkyl chain tip significantly decreases detergent tendency to self-aggregate.19 Because of the presence of the two identical alkyl groups, the lipophilic group terminal of MNG-10,10 is likely bulkier than those of the monopod MNG (i.e., MNG-0,20) and the other A-MNGs, thereby giving the higher CMC. The bulkiness of the lipophilic group terminal likely decreases with increasing detergent asymmetricity (i.e., the length difference between the two lipophilic chains) in the MNG architecture, explaining the trend observed for the CMCs of the A-MNGs; the CMCs of these agents decreased with increasing detergent asymmetricity.
Table 1. Molecular weights (MWs) and critical micelle concentrations (CMCs) of novel agents (A-MNGs), MNG-10,10 and a conventional detergent (DDM), and hydrodynamic radii (Rh; n = 4–5) of their micelles at room temperature.
| Detergent | MW a | CMC (μM) | CMC (wt%) | R h b (nm) |
| MNG-10,10 | 1005.20 | ∼10 | 0.001 | 9.8 ± 0.2 |
| MNG-9,11 | 1005.20 | ∼8 | 0.0008 | 8.3 ± 1.0 |
| MNG-8,12 | 1005.20 | ∼8 | 0.0008 | 6.0 ± 0.2 |
| MNG-6,14 | 1005.20 | ∼6 | 0.0006 | 3.8 ± 0.1 |
| MNG-4,16 | 1005.20 | ∼4 | 0.0004 | 3.5 ± 0.2 |
| MNG-2,18 | 1005.20 | ∼2 | 0.0002 | 4.1 ± 0.2 |
| MNG-0,20 | 1005.20 | ∼2 | 0.0002 | 3.9 ± 0.1 |
| DDM | 510.62 | 170 | 0.0087 | 3.4 ± 0.0 |
aMolecular weight of detergents.
bHydrodynamic radius of detergents measured at 1.0 wt% by dynamic light scattering experiments.
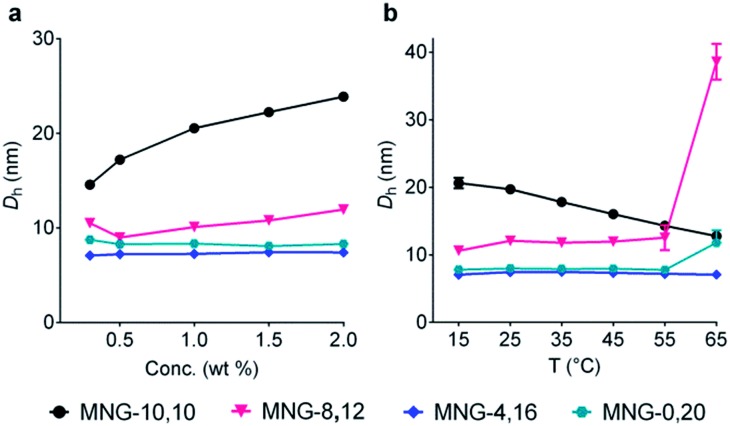
The A-MNGs were also different from MNG-10,10 in terms of micelle size. Micelles formed by the A-MNGs were substantially smaller than those formed by MNG-10,10 with a hydrodynamic radius (Rh) of 9.8 nm (Table 1). The size difference compared to MNG-10,10 micelles tended to increase with increasing detergent asymmetricity. For example, MNG-9,11 and MNG-8,12 having chain length differences of two and four-carbon units between two alkyl groups, respectively, formed detergent micelles with an Rh of 8.3 and 6.0 nm. When the chain length difference was further increased to 8 and 12-carbon units, the micelle size (Rh) continued to decrease to 3.8 (MNG-6,14) and 3.5 nm (MNG-4,16), respectively. This trend of micelle size depending on detergent asymmetricity is likely due to a gradual change in detergent geometry from a cylindrical to a conical shape when detergent asymmetricity was increased. Interestingly, a further increase in chain length difference up to 16/20-carbon units resulted in enlarged micelles, as can be seen in the Rh values of MNG-2,18 (4.1 nm) and MNG-0,20 (3.9 nm), indicative of the engagement of another factor (i.e., the length of the detergent lipophilic group) in determining the detergent micelle size; a long alkyl chain detergent tends to form larger micelles than a short chain agent. Thus, the micelle size of the A-MNGs appeared to be determined mainly by detergent asymmetricity, with a small contribution from the length of the detergent lipophilic group. The differences in self-aggregation behaviours between the A-MNGs and MNG-10,10 (CMC and micelle size) observed here indicate that detergent asymmetricity substantially influences detergent–detergent interactions in a micellar environment.
Detergent micelles were further analyzed in terms of size distribution to investigate micellar homogeneity. When the micellar distributions were represented by number-weighted DLS profiles, all A-MNGs tested here showed a single set of populations, while the intensity-weighted DLS profiles indicated the presence of additional large aggregates in the case of some MNGs (MNG-9,11, MNG-8,12 and MNG-6,14) (Fig. S2†). As scattered light intensity is known to be proportional to D6 where D represents micelle diameter,20 the relative populations of the large aggregates formed by these agents were calculated to be very small (less than one in 106), calculated from the fact that the big aggregates are >10-fold larger than the small micelles (Fig. S2b†). This result indicates high micellar homogeneity for all the A-MNGs. The detergent micelle size was further studied by the variation in the detergent concentration. The symmetric MNG (i.e., MNG-10,10) showed a gradual increase in micelle size with increasing detergent concentration (Fig. 3a), which was also seen in the intensity-weighted DLS profile of this agent (Fig. S3a†). Similarly, micelles formed by MNG-8,12 appeared to steadily enlarge with increasing detergent concentration (Fig. 3a). However, the intensity-weighted DLS profiles indicate that the apparent increase in the MNG-8,12 micelle size is due to a gradual increase in the relative proportion of large aggregates compared to small micelles rather than an actual increase in micelle size (Fig. S3b†). MNG-4,16 and MNG-0,20 with rather high asymmetricity were almost invariable in micelle size with changing detergent concentration, which can also be seen in the intensity-weighted DLS profiles of these agents (Fig. S3c and d†). We also investigated the micelle sizes of these selected MNGs (MNG-10,10, MNG-8,12, MNG-4,16 and MNG-0,20) with increasing solution temperature from 15 °C to 65 °C. In the case of MNG-10,10, we found a gradual decrease in micelle size with increasing solution temperature (Fig. 3b). In contrast, the micelles formed by the other three A-MNGs (MNG-8,12, MNG-4,16 and MNG-0,20) exhibited little change in micelle size up to 55 °C. However, an abrupt increase in micelle size was notable for MNG-8,12 or MNG-0,20 at 65 °C, which likely to be the result of an increase in the relative proportion of large aggregates (MNG-8,12) or the appearance of large aggregates (MNG-0,20) at this high temperature, with concomitant decreases in the proportions of small micelles (Fig. S4b and d†). Thus, there were no actual substantial changes in the micelle sizes of these detergents (MNG-8,12 and MNG-0,20) in this range of temperature variation (15–65 °C). Combined together, all tested A-MNGs (MNG-8,12, MNG-4,16 and MNG-0,20) showed rather invariant micelle size with the change in detergent concentration or solution temperature whereas MNG-10,10 with the symmetric lipophilic group showed substantial variation in micelle size with the same environmental changes. In addition, of the four tested MNGs, micelles formed by MNG-4,16 appeared to be the most stable, reflected by no change in its DLS profiles with the variation of detergent concentration or solution temperature. The detergent CMCs, micelle size and micellar stability observed here indicate that the asymmetric MNGs are quite different from their symmetric counterpart in terms of self-assembly behaviours despite the common features of chemical structure and HLB.
Fig. 3. Micelle size variation of the selected MNGs (MNG-10,10, MNG-8,12, MNG-4,16 and MNG-0,20) with increasing detergent concentration from 0.3 wt% to 2.0 wt% (a) or with elevating solution temperature from 15 °C to 65 °C (b). For concentration variation experiments, the solution temperature was kept at 25 °C, while the detergent concentration was maintained at 1.0 wt% for temperature variation experiments. Micelle sizes of the individual MNGs were represented by hydrodynamic diameters (Dh; z-average), which were calculated from the intensity-weighted DLS data shown in the ESI. Error bars, SEM; n = 4–5.
Detergent evaluation with diverse membrane proteins
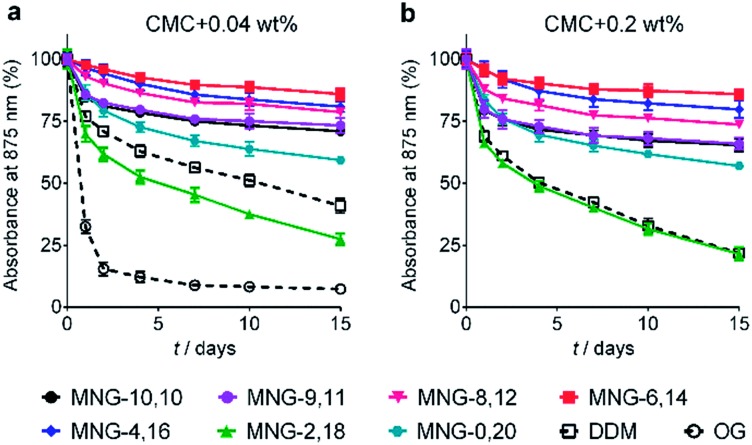
The A-MNGs were first evaluated with a photosynthetic superassembly from Rhodobacter (R.) capsulatus to investigate detergent efficacy toward membrane protein stabilization.21 The photosynthetic assembly comprises the light-sensitive light harvesting complex I (LHI) and a reaction centre complex (RC).9b Because of the presence of several cofactors (e.g., chlorophyll and carotenoids) embedded in the complex interior, the native complex gives a strong absorption signal at 875 nm. Upon protein denaturation, the cofactors dissociate from the complex, resulting in a decrease in the absorption peak intensity at 875 nm. Thus, the structural integrity of the LHI-RC could be conveniently assessed by monitoring absorbance at 875 nm.9b For detergent evaluation, the LHI-RC complex was first extracted with 1.0 wt% DDM from the membranes and the extracted complex was purified in 1xCMC DDM via a Ni2+-NTA affinity column. The collected DDM-purified LHI-RC was diluted into buffer solutions containing the individual A-MNGs, DDM or OG to give final detergent concentrations of CMCs + 0.04 wt%. Protein stability was monitored over the course of a 20-day incubation at room temperature. Consistent with a previous result, the OG-solubilized LHI-RC rapidly lost integrity over time, while DDM-solubilized complexes lost integrity much more gradually.9f As expected, MNG-10,10 was substantially more effective than DDM at maintaining complex integrity in the long term (Fig. 4a). After the 20-day incubation, MNG-10,10 retained approximately 70% protein integrity while use of DDM resulted in approximately 40% retention of protein integrity. Of the A-MNGs, MNG-2,18 was the poorest at stabilizing the complex. The monopod MNG (MNG-0,20) was better than DDM, but worse than MNG-10,10. MNG-9,11 with the smallest asymmetricity was similar to MNG-10,10 at retaining complex integrity, while the other A-MNGs (MNG-8,12, MNG-6,14 and MNG-4,16) were superior to MNG-10,10 under the conditions tested, with the best efficacy observed for MNG-6,14, followed by MNG-4,16 and MNG-8,12. The best A-MNG (i.e., MNG-6,14) was markedly effective at stabilizing the complex, resulting in more than 85% retention of protein integrity after the 20-day incubation. When the detergent concentration was increased to CMC + 0.2 wt%, a similar detergent efficacy order was obtained (Fig. 4b). Again, MNG-2,18 was the worst of the tested detergents, while the efficacies of MNG-9,11 and MNG-0,20 were comparable to and worse than that of MNG-10,10, respectively. The other A-MNGs (MNG-8,12, MNG-6,14 and MNG-4,16) were superior to MNG-10,10, with the best performance again found for MNG-6,14. Thus, we identified three new MNG derivatives (MNG-8,12, MNG-6,14 and MNG-4,16) that are more effective than the original MNG (MNG-10,10) at preserving complex integrity in the long term.
Fig. 4. Long-term stability of LHI-RC complexes solubilized in an A-MNG (MNG-9,11, MNG-8,12, MNG-6,14, MNG-4,16, MNG-2,18, or MNG-0,20), MNG-10,10 and a conventional detergent (DDM or OG). Two different detergent concentrations were used: (a) CMCs + 0.04 wt% and (b) CMCs + 0.2 wt%. Because of the very poor protein stability observed at a detergent concentration of CMC + 0.04 wt%, OG was not included in the experiment using the detergents at CMC + 0.2 wt%. The native complexes strongly absorb 875 nm light, which was utilized to assess protein stability over time. Protein stability was monitored at regular intervals during a 15-day incubation at room temperature. Error bars, SEM; n = 2.
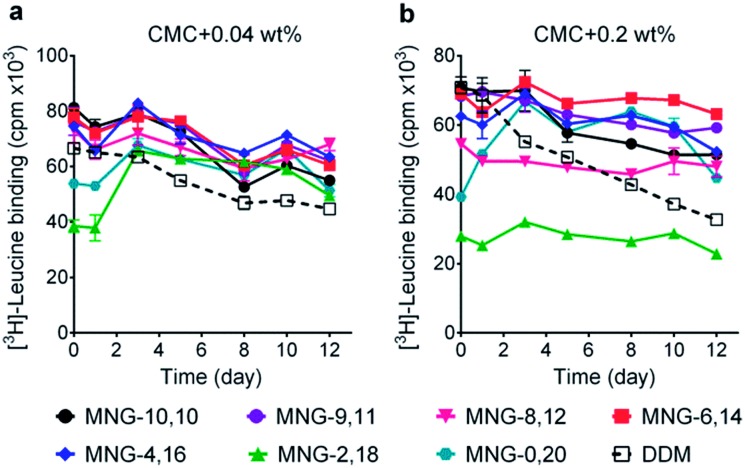
We next turned to the leucine transporter (LeuT) from the bacteria Aquifex aeolicus to further evaluate the ability of these agents to stabilize membrane proteins.22 After extraction from the membrane by treatment with 1.0 wt% DDM, the transporter was purified in 0.05% of the same detergent, and this was used for sample dilution in the next step. The final concentrations of the individual A-MNGs and MNG-10,10 were CMCs + 0.04 wt%. LeuT stability was assessed by monitoring the ability of the transporter to bind the radio-labelled substrate ([3H]-leucine (Leu)) via a scintillation proximity assay (SPA).23 The substrate binding ability was measured at regular intervals during a 12-day incubation at room temperature. Consistent with a previous result, MNG-10,10 was better than DDM at retaining transporter activity (Fig. 5a).9g,h The transporter solubilized in the long alkyl chain A-MNGs (MNG-2,18 and MNG-0,20) gave rather low activity at day 0 compared to DDM, but transporter activity was fully recovered following a 3-day incubation. This restoration in transporter activity could be ascribed to a slow detergent exchange following the sample dilution. Because of the presence of the long alkyl chains (C18 and C20), these agents (MNG-2,18 and MNG-0,20) may need more time to find an optimal position/conformation around the transporter in the process of detergent exchange. The other A-MNGs outperformed DDM and, more importantly, were at least comparable to MNG-10,10, with the best performance being detected for MNG-4,16. A similar detergent efficacy order was observed when the detergent concentration was increased to CMC + 0.2 wt%, with detergent efficacy difference being slightly more pronounced (Fig. 5b). MNG-2,18 again gave low transporter activity compared to DDM at day 0, but retained that low activity of the transporter until the end of incubation. A similar behaviour was observed for MNG-8,12, although the transporter in this MNG started at a higher baseline activity. MNG-0,20 also gave a rather low activity of the transporter at day 0, but transporter activity was fully recovered over a 3-day incubation; the same result was obtained at CMC + 0.04 wt% for this detergent. MNG-10,10 outperformed DDM, a gold standard conventional detergent, but some A-MNGs such as MNG-6,14 and MNG-4,16 were even better than this symmetric MNG. Notably, MNG-6,14 was highly effective at preserving transporter activity during the 12-day incubation. Collectively, the A-MNGs with intermediate asymmetricity (MNG-6,14 and MNG-4,16) were superior to MNG-10,10 in retaining transporter stability in the long term.
Fig. 5. Effect of asymmetric MNGs on LeuT stability. A DDM-purified transporter was diluted in a buffer containing an asymmetric MNG (MNG-9,11, MNG-8,12, MNG-6,14, MNG-4,16, MNG-2,18 or MNG-0,20) at a concentration of CMC + 0.04 wt% (a) or CMC + 0.2 wt% (b) and detergent efficacy for transporter stability were compared with DDM and MNG-10,10. LeuT activity was assessed by [3H]-leucine binding using the scintillation proximity assay (SPA) at designated time points over a 12-day incubation period at room temperature. Data points are means ± SEM (error bars); n = 2.
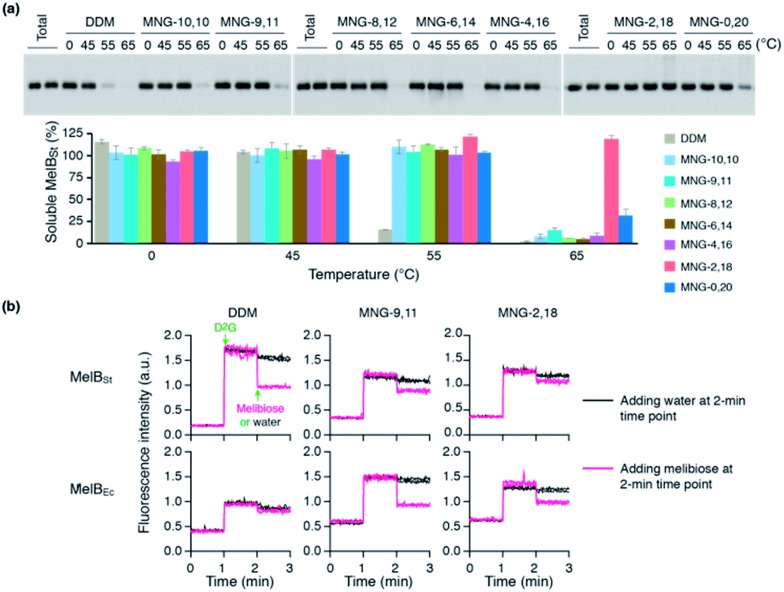
The encouraging results of the A-MNGs with LHI-RC and LeuT prompted us to further test these agents with melibiose permease of Salmonella typhimurium (MelBSt).24 In this experiment, the individual MNGs were directly used to extract MelBSt from the membranes. Therefore, we could exclude the effect of DDM on protein stability via this direct extraction protocol. MelBSt extraction was carried out at four different temperatures (0, 45, 55 and 65 °C) and the amount of soluble MelBSt under each condition was analysed by SDS-PAGE and Western blotting following ultracentrifugation of the extracted samples. As can be seen in Fig. 6a, MNG-10,10 at 0 °C was as efficient as DDM at extracting MelBSt from the membranes, consistent with a previous result.24d Similar efficiencies were observed for all the individual A-MNGs. When the incubation temperature was increased to 45 °C, MelBSt extracted with DDM or the individual MNGs showed full water-solubility, indicating that all these agents are good at stabilizing this transporter. However, a further increase of incubation temperature to 55 °C led to a large difference in the amount of soluble MelBSt between DDM and the tested MNGs. DDM gave an almost complete loss in soluble MelBSt from the solution, presumably due to significant aggregation or denaturation of the proteins under these conditions. In contrast, all A-MNGs including MNG-10,10 turned out to be fully effective at retaining MelBSt solubility at this temperature. The efficacy of the A-MNGs for MelB thermo-stability was differentiated by a further increase of incubation temperature to 65 °C. At this high temperature, most A-MNGs failed to yield soluble MelBSt; however, MNG-2,18 was still effective at maintaining soluble MelBSt and MNG-0,20 yielded a small amount of soluble protein. Based on this thermo-stability analysis of MelBSt, MNG-9,11 and MNG-2,18 were selected to test their effects on MelB function. The functionality of MelB extracted with these two detergents was assessed via a galactoside binding assay utilizing a fluorescent sugar 2′-(N-dansyl)aminoalkyl-1-thio-β-d-galactopyranoside (D2G). Upon addition of D2G, functional MelB effectively binds to this fluorescent ligand, leading to a strong fluorescence emission via Förster resonance energy transfer (FRET) from MelB tryptophan (Trp) residues to the dansyl moiety on D2G. This strong fluorescence emission is partially reversed by addition of an excess amount of melibiose as this displaces the bound D2G. Thus, monitoring fluorescence emission intensity by the sequential addition of D2G and melibiose is a good estimation for MelBSt functionality. MNG-9,11- or MNG-2,18-solubilized MelBSt was less responsive to this assay than the transporter in DDM (Fig. 6b), as reported previously for MNG-10,10.9h,24d As observed for MNG-10,10,24d the strong binding of these MNGs to the transporter could be mainly responsible for the decreased responsiveness of the transporter here as the strong detergent association may reduce protein conformational dynamics. When a less stable MelB homologue, MelB from Escherichia coli (MelBEc), was used, DDM failed to produce a functional transporter whereas two A-MNGs (MNG-9,11 and MNG-2,18) were effective at preserving MelBEc in a functional state (Fig. 5b). Thus, the A-MNGs were superior to DDM in retaining MelB in a soluble/functional state.
Fig. 6. (a) Thermo-stability of MelBSt solubilized in A-MNGs, MNG-10,10 or DDM. E. coli membranes containing MelBSt were incubated with 1.5 wt% individual detergents for 90 min at four different temperatures (0, 45, 55, and 65 °C). The protein samples solubilized by the individual detergents were analysed by SDS-PAGE and Western blotting. The amount of soluble MelBSt in each sample was estimated from gel image analysis and expressed as a percentage of the total amount of MelBSt in the untreated membrane (Total) in the histogram. Error bars, SEM; n = 2–4. (b) MelB functional assay. Right-side-out (RSO) membrane vesicles containing MelBSt or MelBEc were treated with DDM, MNG-9,11, or MNG-2,18 and the resulting MelB extracts after ultracentrifugation were then subjected to a functional assay (i.e., melibiose reversal of FRET from Trp to dansyl-2-galactoside (D2G)). Changes in fluorescence emission intensity were monitored following additions of D2G and an excess amount of melibiose at the 1-min and 2-min time points, respectively (magenta line). Addition of water instead of excess melibiose at the 2-min time point was used for control data (black line).
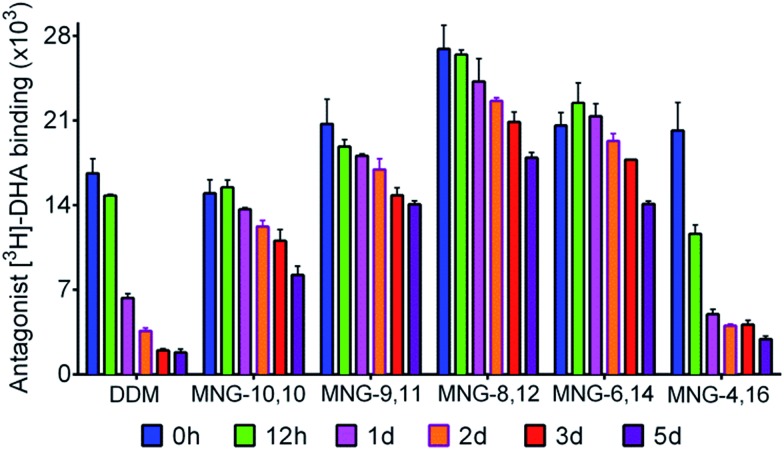
In order to investigate the compatibility with G protein-coupled receptors (GPCRs), the A-MNGs were evaluated with the human β2 adrenergic receptor (β2AR).25 The receptor was purified with 1% DDM and the resulting receptor was diluted into buffer solutions supplemented with the individual MNGs to give final detergent concentrations of CMCs + 0.2 wt%. Receptor stability was assessed by monitoring the ability of the receptor to bind a radioactive antagonist ([3H]-dihydroalprenolol (DHA)).26 After a 30-min detergent dilution, specific receptor activity was measured as a preliminary assessment of detergent efficacy for β2AR stabilization. We found that all the MNGs except MNG-2,18 and MNG-0,20 gave higher receptor activity than DDM (Fig. S5†). Some A-MNGs (MNG-8,12, MNG-6,14 and MNG-4,16) were even better than MNG-10,10 in this regard. Based on this result, we selected MNG-9,11, MNG-8,12, MNG-6,14, and MNG-4,16 to further evaluate detergent efficacy for long-term receptor stability. Receptor activity was monitored at regular intervals during a 5-day incubation at room temperature. Consistent with previous results, MNG-10,10 was superior to DDM at maintaining receptor stability in the long-term (Fig. 7).9g,h Of the selected A-MNGs, MNG-4,16 with the longest alkyl chain was worse than MNG-10,10 and more or less comparable to DDM in this regard. In contrast, the other A-MNGs (MNG-9,11, MNG-8,12, and MNG-6,14) were superior to MNG-10,10 in stabilizing the receptor in the long term, with the best performance observed for MNG-8,12. The best A-MNG (i.e., MNG-8,12) gave initial receptor activity nearly two-fold higher than MNG-10,10 and showed markedly enhanced receptor stability over the whole period of the incubation (Fig. 7).
Fig. 7. Long-term stability of β2AR solubilized in DDM, MNG-10,10, or an A-MNG (MNG-9,11, MNG-8,12, MNG-6,14, or MNG-4,16). The DDM-purified receptor was diluted into individual detergent-containing buffer solutions to give final detergent concentrations of CMCs + 0.2 wt%. The specific ligand binding ability of the receptor was measured using the radiolabeled antagonist ([3H]-dihydroalprenolol (DHA)) following 30-min dilution (0 h) and was further monitored at regular intervals during a 5-day incubation at room temperature. Error bars, SEM; n = 3.
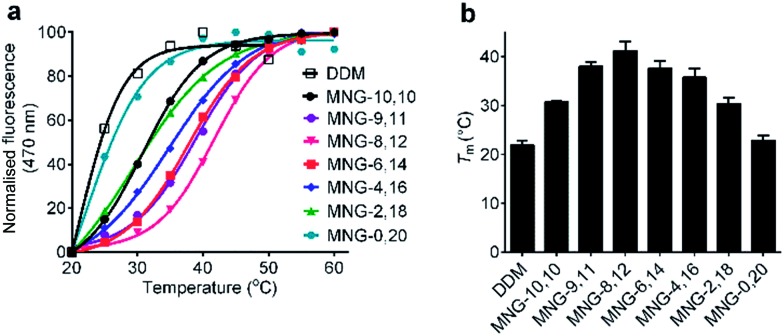
The A-MNGs were also evaluated with another GPCR, the human μ-opioid receptor (MOR).27 For this study, a DDM-purified receptor was incubated with buffer solutions containing 1.0 wt% individual detergents. Sample solutions were prepared with a final detergent concentration of 0.5 wt% by dilution. MOR stability was assessed by estimating melting temperature (Tm), utilizing a N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl] maleimide (CPM) assay.28 The results are summarized in Fig. 8 and Table S1.† MOR solubilized in DDM gave a low Tm value of 21.9 °C, indicating that the receptor is relatively unstable in this conventional detergent. When we used the MNG agents, the Tm of the receptor substantially increased, except for MNG-0,20 (22.9 °C). As expected, MNG-10,10, a significantly optimized novel agent for GPCR stability, gave a Tm value of the receptor of 30.8 °C, higher than DDM by 8.9 °C. The use of the A-MNGs further increased Tm values of the receptor, with the best performance observed for MNG-8,12 with intermediate asymmetricity. The use of MNG-8,12 resulted in a Tm value of 41.1 °C, higher than that of MNG-10,10 by 10.3 °C. MNG-9,11 and MNG-6,14 that have one-step less and more detergent asymmetricity than MNG-8,12, respectively, gave receptor Tm values of 38.0 and 37.6 °C. Further increase in hydrophobic asymmetricity reduced detergent efficacy for receptor stabilization; MNG-4,16- and MNG-2,18-solubilized receptors gave Tm values of 35.8 and 30.4 °C, respectively, still higher than or comparable to MNG-10,10. Thus, this result further supports the assumption that detergent asymmetricity plays a crucial role in determining detergent efficacy for GPCR stabilization. Of note, MNG-8,12 was the best of the A-MNGs for both GPCRs (β2AR and MOR), indicating that this A-MNG has potential for GPCR structural studies.
Fig. 8. Representative thermostability profile (a) and melting temperature (Tm) (b) of MOR dissolved in MNG-10,10 or an A-MNG (MNG-9,11, MNG-8,12, MNG-6,14, MNG-4,16, MNG-2,18, or MNG-0,20). DDM was used as a positive control. The DDM-purified receptor was mixed into individual detergent-containing buffer solutions to give final detergent concentrations of 0.5 wt%. Receptor stability was assessed by Tm obtained from the CPM assay. Error bars, SD; n = 2–4.
Asymmetric MNGs were evaluated with diverse membrane proteins and their efficacy was compared with a symmetric MNG (i.e., MNG-10,10). The A-MNGs introduced here were prepared in four high-yielding synthetic steps and are comparable to the original MNG (MNG-10,10) in terms of synthetic convenience.19 The large reactivity difference between the first and second alkylation steps enabled us to introduce two different alkyl chains into the MNG scaffold in a stepwise manner. Thus, the MNG scaffold offered a unique opportunity to prepare asymmetric amphiphiles with synthetic accessibility. Detergent synthetic accessibility is essential for a widespread use in membrane protein research. The A-MNGs also showed promising results in detergent evaluation with the multiple membrane proteins. Of the A-MNGs, MNG-6,14 was the most effective for LHI-RC and LeuT while MNG-2,18 and MNG-8,12 were the best for MelBSt and β2AR/MOR stability, respectively. Thus, the optimal detergent asymmetricity for protein stability tends to be protein-specific. Despite this protein-specific nature, MNG-6,14 with intermediate asymmetricity was superior to MNG-10,10 in stabilizing LHI-RC, LeuT, β2AR and MOR while MNG-8,12 and MNG-4,16 with a little low and high asymmetricity, respectively, were more effective than the symmetric MNG at stabilizing LHI-RC/β2AR/MOR and LHI-RC/LeuT, respectively. These results indicate the presence of optimal detergent asymmetricity universally applicable to membrane protein stability, as exemplified by MNG-6,14 with a C8 chain length difference between the two alkyl chains. A little deviation from this optimal detergent asymmetricity appeared to be fine, as seen in the results for MNG-8,12 and MNG-4,16 with C4 and C12 chain length differences, respectively. Notably, via asymmetric MNG invention, we identified a few MNGs as new chemical tools that are more effective than MNG-10,10 at stabilizing the membrane proteins tested here. As MNG-10,10 has proved effective for membrane protein structural studies, particularly for GPCRs, these A-MNGs, particularly MNG-8,12, displaying enhanced stabilization efficacy are expected to find wide use in GPCR structural studies.
It is difficult to know a precise reason why the A-MNGs displayed favourable behaviours for membrane protein stability compared to MNG-10,10 as detergent–protein interaction remains elusive. Despite this difficulty, plausible explanations for enhanced detergent efficacy observed here are described below by taking into account key differences between MNG-10,10 and the A-MNGs in terms of detergent–protein or detergent–detergent interactions. First, the length of the detergent lipophilic group affects detergent–protein interactions. As membrane proteins have a narrow range of hydrophobic thickness of around 30 Å, the length of the detergent lipophilic group needs to be compatible with the hydrophobic dimensions of a target protein. In this regard, MNG-10,10 with a C10 alkyl chain seems a little short while the long alkyl chain A-MNGs (e.g., MNG-2,18 and MNG-0,20) will be too long. Thus, the A-MNGs with an intermediate chain length (MNG-8,12 (C12), MNG-6,14 (C14) and MNG-4,16 (C16)) would have a range of alkyl chain lengths optimal for protein stability. Interestingly, the alkyl chain lengths of these A-MNGs (MNG-8,12, MNG-6,14 and MNG-4,16) are similar to those of phospholipids in biological membranes. Furthermore, because of the variation of the alkyl chain length from C12 to C16, this set of A-MNGs could be useful for structural/functional studies of membrane proteins with varied hydrophobic thickness (28–32 Å). An advantageous effect of the A-MNGs on protein stability can also be attributed to favourable detergent–detergent interactions in the micellar environment. The low CMCs of these agents compared to that of MNG-10,10 are good indications of favourable detergent–detergent interactions, associated with enhanced micelle and protein stability. Considering spherical micelle formation driven by a hydrophobic effect, we conceive that an asymmetric detergent with high hydrophobic density is optimal for stable micelle formation as it allows detergent alkyl chains to effectively pack in the micelle interior. In this regard, MNG-0,20 with a large branched dimaltoside head group and a single thin chain (C20) would be suboptimal as micelles formed by this agent would contain large empty spaces in their interior, particularly in a hydrophobic region close to the hydrophilic surface. An ethyl pendant of MNG-2,18 will help increase the hydrophobic density of detergent micelles by occupying the empty spaces in the micellar interior, but is probably too small. The C4 and C6 alkyl chains of MNG-4,16 and MNG-6,14, respectively, are likely to be large enough to sufficiently fill the micellar empty spaces, and therefore favourable for detergent–detergent interactions. At this stage, it is hard to know a priori which alkyl chain length is the best for fitting the empty spaces in order to maximize hydrophobic detergent–detergent interactions, but the well-accommodated C4 and C6 alkyl chains in the micellar empty spaces are supported by the fact that no significant change in micelle size was observed with increasing the alkyl chain length from C2 to C4 to C6; with this increase in alkyl chain length, the micelle size (Rh) only marginally varied from 4.1 (MNG-2,18) to 3.5 (MNG-4,16) to 3.8 nm (MNG-6,14). A substantial increase in micelle size was detected for MNG-8,12 (6.0 nm), but this apparent micelle enlargement turned out to be due to the appearance of the large aggregates rather than an actual increase in detergent micelle size, as described above (Fig. S2b†). In contrast, the alkyl chains of MNG-9,11 and MNG-10,10 (i.e., C9 and C10, respectively) appeared to be too large in this regard, as indicated by substantial and actual increases in micelle size for these A-MNGs (8.3 and 9.8 nm, respectively) (Fig. S2b†). Thus, the micelle expansion observed for MNG-9,11 or MNG-10,10 is likely to be a natural consequence of accommodation of the large alkyl chain (C9/C10) in the micelle interior. This micelle enlargement decreases alkyl chain density (i.e., hydrophobic density) in the micelle interior, a less effective arrangement for favourable interactions with a target membrane protein. By recognizing multiple structural differences between MNG-10,10 and the A-MNGs associated with detergent–protein or detergent–detergent interactions, we reasoned that the length, asymmetricity and hydrophobic density of the detergent lipophilic group are responsible for the favourable behaviours of the A-MNGs with intermediate asymmetricity (e.g., MNG-4,16, MNG-6,14 or MNG-8,12) compared to MNG-10,10. Based on this discussion, it is clear that multiple factors are interactively involved in determining detergent efficacy for membrane protein stabilization and enhanced detergent efficacy can be attained only by a detergent molecule with an optimal range of individual factors. Such multiple optimal detergent aspects appear to be achieved in MNG-6,14 as this agent was effective at stabilizing all the tested membrane proteins here (LHI-RC, LeuT, MelBSt, β2AR, and MOR).
Conclusions
By preparing new MNGs (A-MNGs) with a range of detergent asymmetricity and evaluating them for membrane protein solubilisation/stabilization, we identified a few detergents (MNG-4,16, MNG-6,14 and MNG-8,12) that displayed enhanced protein stability compared to MNG-3 (i.e., MNG-10,10). This result is notable in considering that the symmetric MNG (MNG-3) is popularly used in structural studies of GPCRs and other membrane proteins. The new agents were prepared via a straight-forward protocol comprising four high-yielding steps, which facilitates widespread utility in membrane protein research. Importantly, we firstly compared the symmetric vs. asymmetric detergents in terms of protein stabilization efficacy, a comparison which highlighted the importance of detergent asymmetricity for protein stability. This study not only introduces the biochemical tools useful for protein structural studies, but also provides insights into detergent structure–efficacy relationships based on detergent asymmetricity.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grants 2016R1A2B2011257 and 2018R1A6A1A03024231 to P.S.C.) and by the National Institutes of Health (grants R01GM122759 and R21NS105863 to L.G).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8sc02560f
References
- Deisenhofer J., Epp O., Miki K., Huber R., Michel H. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- (a) Serrano-Vega M. J., Magnani F., Shibata Y., Tate C. G. Proc. Natl. Acad. Sci. U. S. A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Newstead S., Ferrandon S., Iwata S. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) He Y., Wang K., Yan N. Protein Cell. 2014;5:658–672. doi: 10.1007/s13238-014-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newstead S., Hobbs J., Jordan D., Carpenter E. P., Iwata S. Mol. Membr. Biol. 2008;25:631–638. doi: 10.1080/09687680802526574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglin T. C., Conboy J. C. Biophys. J. 2008;95:186–193. doi: 10.1529/biophysj.107.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) White S. H., Wimley W. C. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; (b) Bowie J. U. Curr. Opin. Struct. Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]; (c) Lacapere J. J., Pebay-Peyroula E., Neumann J. M., Etchebest C. Trends Biochem. Sci. 2007;32:259–327. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]; (d) Philips R., Ursell T., Wiggins P., Sens P. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Drake M. T., Shenoy S. K., Le owitz R. J. Circ. Res. 2006;99:570–582. doi: 10.1161/01.RES.0000242563.47507.ce. [DOI] [PubMed] [Google Scholar]; (b) Lappano R., Maggiolini M. Nat. Rev. Drug Discovery. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- (a) Garavito R. M., Ferguson-Miller S. J. Biol. Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]; (b) Privé G. G. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- (a) Chae P. S., Wander M. J., Cho K. H., Laible P. D., Gellman S. H. Mol. BioSyst. 2013;9:626–629. doi: 10.1039/c3mb25584k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang Q., Tao H., Hong W. X. Methods. 2011;55:318–323. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) McQuade D. T., Quinn M. A., Yu S. M., Polans A. S., Krebs M. P., Gellman S. H. Angew. Chem., Int. Ed. 2000;39:758–761. [PubMed] [Google Scholar]; (b) Chae P. S., Cho K. H., Wander M. J., Bae H. E., Gellman S. H., Labile P. D. Biochim. Biophys. Acta. 2014;1838:278–286. doi: 10.1016/j.bbamem.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Das M., Du Y., Ribeiro O., Hariharan P., Mortensen J. S., Patra D., Skiniotis G., Loland C. J., Guan L., Kobilka B. K., Byrne B., Chae P. S. J. Am. Chem. Soc. 2017;139:3072–3081. doi: 10.1021/jacs.6b11997. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Hussain H., Du Y., Tikhonova E., Mortensen J. S., Ribeiro O., Santillan C., Das M., Ehsan M., Loland C. J., Guan L., Kobilka B. K., Byrne B., Chae P. S. Chem.–Eur. J. 2017;23:6724–6729. doi: 10.1002/chem.201605016. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) H Cho K., Du Y., Scull N. J., Hariharan P., Gotfryd K., Loland C. J., Guan L., Byrne B., Kobilka B. K., Chae P. S. Chem.–Eur. J. 2015;21:10008–10013. doi: 10.1002/chem.201501083. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Chae P. S., Rana R. R., Gotfryd K., Rasmussen S. G. F., Kruse A. C., Cho K. H., Capaldi S., Carlsso E., Kobilka B., Loland C. J., Gether U., Banerjee S., Byrne B., Lee J. K., Gellman S. H. Chem. Commun. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Chae P. S., Rasmussen S. G. F., Rana R. R., Gotfryd K., Chandra R., Goren M. A., Kruse A. C., Nurva S., Loland C. J., Pierre Y., Drew D., Popot J. L., Picot D., Fox B. G., Guan L., Gether U., Byrne B., Kobilka B., Gellman S. H. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Cho K. H., Husri M., Amin A., Gotfryd K., Lee H. J., Go J., Loland C. J., Guan L., Byrne B., Chae P. S. Analyst. 2015;140:3157–3163. doi: 10.1039/c5an00240k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Sadaf A., Mortensen J. S., Capaldi S., Tikhonova E., Hariharan P., Ribeiro O., Loland C. J., Guan L., Byrne B., Chae P. S. Chem. Sci. 2016;7:1933–1939. doi: 10.1039/c5sc02900g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Ehsan M., Du Y., Scull N. J., Tikhonova E., Tarrasch J., Mortensen J. S., Loland C. J., Skiniotis G., Guan L., Byrne B., Kobilka B., Chae P. S. J. Am. Chem. Soc. 2016;138:3789–3796. doi: 10.1021/jacs.5b13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Howell S. C., Mittal R., Huang L., Travis B., Breyer R. M., Sanders C. R. Biochemistry. 2010;49:9572–9583. doi: 10.1021/bi101334j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chae P. S., Rasmussen S. G. F., Rana R. R., Gotfryd K., Kruse A. C., Nurva S., Gether U., Guan L., Loland C. J., Byrne B., Kobilka B. K., Gellman S. H. Chem.–Eur. J. 2012;18:9485–9490. doi: 10.1002/chem.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) McGregor C.-L., Chen L., Pomroy N. C., Hwang P., Go S., Chakrabartty A., Privé G. G. Nat. Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; (b) Tao H., Lee S. C., Moeller A., Roy R. S., Siu F. Y., Zimmermann J., Stevens R. C., Potter C. S., Carragher B., Zhang Q. Nat. Methods. 2013;10:759–761. doi: 10.1038/nmeth.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Tribet C., Audebert R., Popot J.-L. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Popot J. L. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]; (c) Dörr J. M., Scheidelaar S., Koorengevel M. C., Dominguez J. J., Schäfer M., van Walree C. A., Killian J. A. Eur. Biophys. J. 2016;45:3–21. doi: 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaf A., Du Y., Santillan C., Mortensen J. S., Molist I., Seven A. B., Hariharan P., Skiniotis G., Loland C. J., Kobilka B. . K., Guan L., Byrne B., Chae P. S. Chem. Sci. 2017;8:8315–8324. doi: 10.1039/c7sc03700g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Nath A., Atkins W. M., Sligar S. G. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]; (b) Ujwal R., Bowie J. U. Methods. 2011;55:337–341. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Rosenbaum D. M., Zhang C., Lyons J., Holl R., Aragao D., Arlow D. H., Rasmussen S. G. F., Choi H. J., DeVree B. T., Sunahara R. K., Chae P. S., Gellman S. H., Dror R. O., Shaw D. E., Weis W. I., Caffrey M., Gmeiner P., Kobilka B. K. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Haga K., Kruse A. C., Asada H., Kobayashi T. Y., Shiroishi M., Zhang C., Weis W. I., Okada T., Kobilka B. K., Haga T., Kobayashi T. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kruse A. C., Ring A. M., Manglik A., Hu J., Hu K., Eitel K., Hubner H., Pardon E., Valant C., Sexton P. M., Christopoulos A., Felder C. C., Gmeiner P., Steyaert J., Weis W. I., Garcia K. C., Wess J., Kobilka B. K. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Suzuki H., Nishizawa T., Tani K., Yamazaki Y., Tamura A., Ishitani R., Dohmae N., Tsukita S., Nureki O., Fujiyoshi Y. Science. 2014;344:304–307. doi: 10.1126/science.1248571. [DOI] [PubMed] [Google Scholar]; (e) Dickson V. K., Pedi L., Long S. B. Nature. 2014;516:213–218. doi: 10.1038/nature13913. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Hauer F., Gerle C., Fischer N., Oshima A., Shinzawa-Itoh K., Shimada S., Yokoyama K., Fujiyoshi Y., Stark H. Structure. 2015;23:1769–1775. doi: 10.1016/j.str.2015.06.029. [DOI] [PubMed] [Google Scholar]; (g) Yin J., Mobarec J. C., Kolb P., Rosenbaum D. M. Nature. 2015;519:247–250. doi: 10.1038/nature14035. [DOI] [PubMed] [Google Scholar]; (h) Kang Y. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Perez C., Gerber S., Boilevin J., Bucher M., Darbre T., Aebi M., Reymond J. L., Locher K. P. Nature. 2015;524:433–438. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]; (j) Taniguchi R., Kato H. E., Font J., Deshpande C. N., Wada M., Ito K., Ishitani R., Jormakka M., Nureki O. Nat. Commun. 2015;6:8545. doi: 10.1038/ncomms9545. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Dong Y. Y. Science. 2015;347:1256–1259. doi: 10.1126/science.1261512. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Paulsen C. E., Jean-Paul A., Gao Y., Cheng Y., Julius D. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Schmidt H. R., Zheng S., Gurpinar E., Koehl A., Manglik A., Kruse A. C. Nature. 2016;532:527–530. doi: 10.1038/nature17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Chae P. S., Kruse A. C., Gotfryd K., Rana R. R., Cho K. H., Rasmussen S. G. F., Bae H. E., Chandra R., Gether U., Guan L., Kobilka B. K., Loland C. J., Byrne B., Gellman S. H. Chem.–Eur. J. 2013;19:15645–15651. doi: 10.1002/chem.201301423. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cho K. H., Hariharan P., Mortensen J. S., Du Y., Nielsen A. K., Byrne B., Kobilka B. K., Loland C. J., Guan L., Chae P. S. ChemBioChem. 2016;17:2334–2339. doi: 10.1002/cbic.201600429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Wu G., Wang Z., Zhang X. Langmuir. 2014;30:1531–1535. doi: 10.1021/la405000a. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A., London E. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- Li Y., Reeve J., Wang Y., Thomas R. K., Wang J., Yan H. J. Phys. Chem. B. 2005;109:16070–16074. doi: 10.1021/jp0523874. [DOI] [PubMed] [Google Scholar]
- Plum M. A., Steffen W., Fytas G., Knoll W., Menges B. Opt. Express. 2009;17:10364–10371. doi: 10.1364/oe.17.010364. [DOI] [PubMed] [Google Scholar]
- Laible P. D., Kirmaier C., Udawatte C. S., Hofman S. J., Holten D., Hanson D. K. Biochemistry. 2003;42:1718–1730. doi: 10.1021/bi026959b. [DOI] [PubMed] [Google Scholar]
- Deckert G. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- Quick M., Javitch J. A. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Guan L., Nurva S., Ankeshwarapu S. P. J. Biol. Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ethayathulla A. S., Yousef M. S., Amin A., Leblanc G., Kaback H. R., Guan L. Nat. Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hariharan P., Guan L. J. Gen. Physiol. 2017;149:1029–1039. doi: 10.1085/jgp.201711788. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Amin A., Hariharan P., Chae P. S., Guan L. Biochemistry. 2015;54:5849–5855. doi: 10.1021/acs.biochem.5b00660. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Maehrel C., Cordat E., Mus-Veteau I., Leblanc G. J. Biol. Chem. 1998;273:33192–33197. doi: 10.1074/jbc.273.50.33192. [DOI] [PubMed] [Google Scholar]
- Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Bobilka B. K. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- (a) Yao X., Parnot C., Deupi X., Ratnala V. R. P., Swaminath G., Farrens D., Kobilka B. Nat. Chem. Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]; (b) Swaminath G., Steenhuis J., Kobilka B., Lee T. W. Mol. Pharmacol. 2002;61:65–72. doi: 10.1124/mol.61.1.65. [DOI] [PubMed] [Google Scholar]
- Manglik A., Kruse A. C., Kobilka T. S., Thian F. S., Mathiesen J. M., Sunahara R. K., Pardo L., Weis W. I., Kobilka B. K., Granier S. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Alexandrov A., Mileni M., Chien E. Y., Hanson M. A., Stevens R. C. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]; (b) Hanson M. A., Cherezov V., Griffith M. T., Roth C. B., Jaakola V.-P., Chien E. Y. T., Velasquez J., Kuhn P., Stevens R. C. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.