Abstract
As a significant component of monocot cell walls, (1,3;1,4)-β-glucan has conclusively been shown to be synthesized by the cellulose synthase-like F6 protein. In this study, we investigated the synthetic activity of other members of the barley (Hordeum vulgare) CslF gene family using heterologous expression. As expected, the majority of the genes encode proteins that are capable of synthesizing detectable levels of (1,3;1,4)-β-glucan. However, overexpression of HvCslF3 and HvCslF10 genes resulted in the synthesis of a novel linear glucoxylan that consists of (1,4)-β-linked glucose and xylose residues. To demonstrate that this product was not an aberration of the heterologous system, the characteristic (1,4)-β-linkage between glucose and xylose was confirmed to be present in wild type barley tissues known to contain HvCslF3 and HvCslF10 transcripts. This polysaccharide linkage has also been reported in species of Ulva, a marine green alga, and has significant implications for defining the specificity of the cell wall content of many crop species. This finding supports previous observations that members of a single CSL family may not possess the same carbohydrate synthetic activity, with the CSLF family now associated with the formation of not only (1,3)- and (1,4)-β-glucosidic linkages, but also (1,4)-β-glucosidic and (1,4)-β-xylosidic linkages.
Short abstract
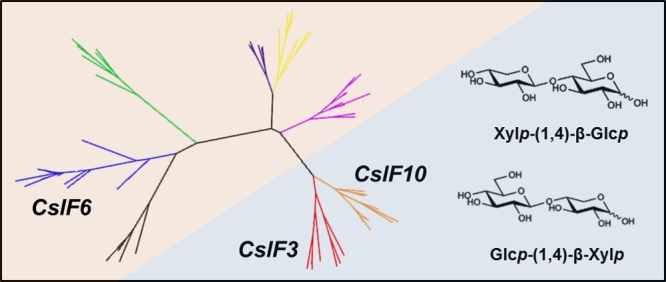
Characteristic Xylp-(1,4)-β-Glcp and Glcp-(1,4)-β-Xylp polysaccharide linkages found associated with cellulose synthase-like F3 and F10 gene expression in barley.
Introduction
Cell walls are essential extracellular matrices of plants, providing the structural integrity required for cell growth, division, and differentiation.1 The cell walls of the world’s most economically important crop species, such as barley, wheat, rice and maize, have been intensively studied for their dietary benefits and applications in industry. Plant cell walls consist of cellulose microfibrils embedded in a gel-like three-dimensional matrix of noncellulosic polysaccharides.2 In members of the Poaceae family, the noncellulosic polysaccharides are predominantly mannan, heteroxylan, and (1,3;1,4)-β-glucan, with lower amounts of xyloglucan and pectin. Proportions of the individual cell wall components vary substantially across different species, tissues, and cell types, influencing cell wall physicochemical properties and potential downstream applications. One of the approaches for characterizing the composition of plant cell walls is to use biochemical methods to determine the types of linkages between the monomers of constituent polysaccharides.3 The polysaccharide composition of a given wall is estimated based on prior knowledge of the relative proportions of particular components and linkages therein. However, this can be accurate only if we have a good understanding of the possible linkages likely to be present within the target species.
The cellulose synthase gene superfamily encodes enzymes of the glycosyltransferase (GT) family 2.4 In addition to the cellulose synthase (CesA) clade there are 11 separate cellulose synthase-like (CslA-M) clades that are considered to be involved in the synthesis of noncellulosic polysaccharides,5,6 although experimental evidence has yet to be obtained for most gene products. Functional characterization has linked the synthesis of (1,4)-β-glucan to CesA genes,7 mannan and glucomannan to CslA genes,8,9 xyloglucan to CslC,10−12 and (1,3;1,4)-β-glucan to CslF, CslH, and CslJ genes.6,13,14 There is some debate regarding the role of the CslD genes in the synthesis of mannan15 or cellulose,16−19 stemming from conflicting results in different systems.
The barley genome (Hordeum vulgare) contains 10 CslF family members, which have expanded in number through a series of recent duplication events originating from HvCslF6 and HvCslF7, to form three additional phylogenetic clades (HvCslF4, HvCslF11, and HvCslF13; HvCslF8, HvCslF9, and HvCslF12; HvCslF3 and HvCslF10).20,21HvCSLF4 and HvCSLF6 have been demonstrated to synthesize (1,3;1,4)-β-glucan in heterologous systems devoid of (1,3;1,4)-β-glucan.13,22 However, the existence of a “β-glucanless” HvCslF6 mutant indicates that the CSLF6 protein is responsible for the synthesis of the majority of the (1,3;1,4)-β-glucan in the barley cell wall.23 At this stage, the mechanism of (1,3;1,4)-β-glucan synthesis by CSLF6 is unknown, but mutation studies of the CSLF6 transmembrane and catalytic regions suggest that the CSLF6 enzyme is able to catalyze the formation of both the (1,3)- and (1,4)-β-glucosidic linkages present in (1,3;1,4)-β-glucan chains.24,25
In this study, we further investigated the synthetic activity of the barley CslF gene family and determined which members are capable of synthesizing (1,3;1,4)-β-glucan in a heterologous expression system (Nicotiana benthamiana), with the exception of HvCslF11 and HvCslF12 that have been previously tested.21 A novel linear glucoxylan was synthesized in the heterologous host, and its presence in native barley tissues was confirmed. The biochemical evidence provided in this study reveals a new function of CslF genes in barley.
Results
Each member of the barley CslF gene family was introduced into N. benthamiana leaves using Agrobacterium infiltration and expressed constitutively under the control of the CaMV35S promoter. Leaf tissues were harvested after 6 days and screened for the presence of (1,3;1,4)-β-glucan using lichenase hydrolysis assays.26 The characteristic oligosaccharides released from (1,3;1,4)-β-glucan by the lichenase were observed for leaves infiltrated with HvCslF6, HvCslF7, HvCslF8, and HvCslF9 (data not shown). When calculated against well-characterized oligosaccharide standards, the (1,3;1,4)-β-glucan levels in N. benthamiana plants expressing HvCslF7, HvCslF8, or HvCslF9 (<0.1%) were much lower than that produced by plants transformed with HvCslF6 (1.6%), and consistent with previous results comparing the amount of (1,3;1,4)-β-glucan synthesized by the products of the HvCslH(14) and HvCslJ(6) genes. No (1,3;1,4)-β-glucan derived oligosaccharide products could be detected in N. benthamiana plants expressing the remaining HvCslF family members, i.e., HvCslF3, HvCslF4, HvCslF10, and the empty vector control.
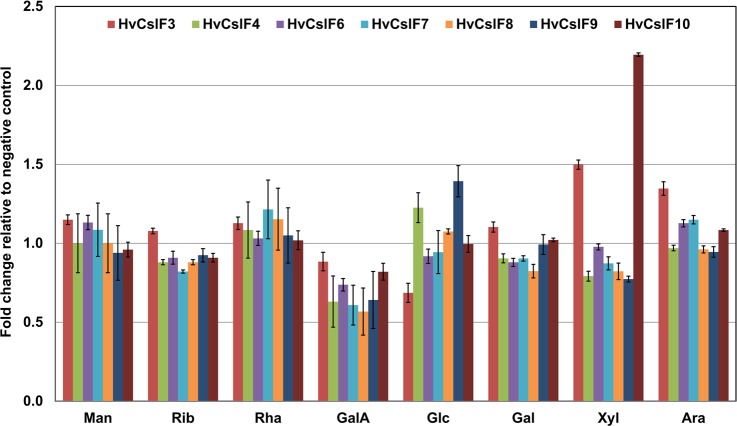
A monosaccharide analysis was performed to determine if there were any other detectable changes to the cell walls of the N. benthamiana leaves infiltrated with the HvCslF genes (Figure 1). Analysis of monosaccharides released by acid hydrolysis showed an increase in glucose content relative to the N. benthamiana negative control (AGL1) for HvCslF4 and HvCslF9, and a decrease for the HvCslF3 samples with high levels of variation between sample replicates. This phenomenon is often observed when expressing HvCslF6 in the N. benthamiana system as the synthesis of (1,3;1,4)-β-glucan appears to interfere with normal cell wall synthesis resulting in a variability of measurable glucose.24 However, there was a consistent increase in xylose content in the leaf samples expressing HvCslF3 and HvCslF10 of 0.3 ± 0.02% (w/w) and 0.65 ± 0.01% (w/w), respectively, corresponding to 1.5 and 2.2 fold higher levels than the negative control. This suggested that any glucan products synthesized by HvCSLF3 or HvCSLF10 were being substituted with xylose to form xyloglucan, or that there was an increase in another product containing xylose as a result of HvCslF3 and HvCslF10 expression.
Figure 1.
Monosaccharide analysis of N. benthamiana leaf samples expressing members of the HvCslF family. Values are presented as fold change relative to a negative control infiltrated with Agrobacterium containing an empty expression vector (AGL-1). Man (mannose), Rib (ribose), Rha (rhamnose), GalA (galacturonic acid), Glc (glucose), Gal (galactose), Xyl (xylose), and Ara (arabinose). Error bars indicate standard error of the mean normalized against the original value prior to calculation of fold change, N = 3.
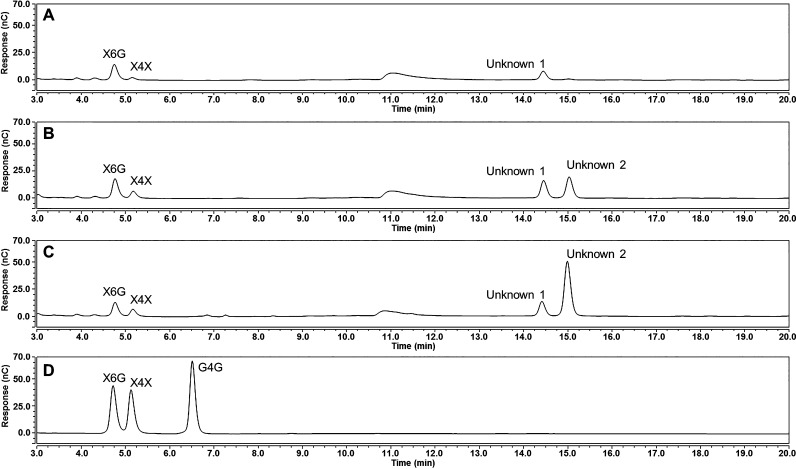
A Driselase treatment was performed to determine if there was an increase in xylan or xyloglucan in the leaves expressing HvCslF3 or HvCslF10.27 The Driselase enzyme mixture is capable of hydrolyzing most sugar linkages present in N. benthamiana tissues, including the hydrolysis of (1,4)-β-xylan into xylose and xylobiose, and (1,4)-β-glucan into glucose and cellobiose. However, the hydrolytic enzyme mixture cannot hydrolyze the (1,6)-α-linkage between the glucan backbone and xylose substitutions found in xyloglucan. If xyloglucan is present in the leaves then the characteristic isoprimeverose disaccharide (xylopyranosyl-α-(1,6)-glucopyranose) should be observed when using high-performance anion-exchange chromatography (HPAEC). Isoprimeverose, xylobiose, and an unidentified oligosaccharide (Unknown 1, Figure 2A), which eluted at a longer retention time, were observed in the empty vector control sample. However, there was no evidence of an increase in isoprimeverose in the HvCslF3 and HvCslF10 expressing leaves relative to the empty vector control (Figure 2B,C). The only changes observed were increases in xylobiose and Unknown 1, and the occurrence of another unknown oligosaccharide (Unknown 2, Figure 2B,C).
Figure 2.
HPAEC-PAD traces of oligosaccharides produced post-Driselase hydrolysis of N. benthamiana leaf tissue overexpressing an empty vector control (A), HvCslF3 (B), and HvCslF10 (C). Standards for cellobiose (G4G), xylobiose (X4X), and isoprimeverose (X6G) are included (D). Unknown 1 and 2 refer to peaks identified for further characterization. x-axis, time; y-axis, abundance.
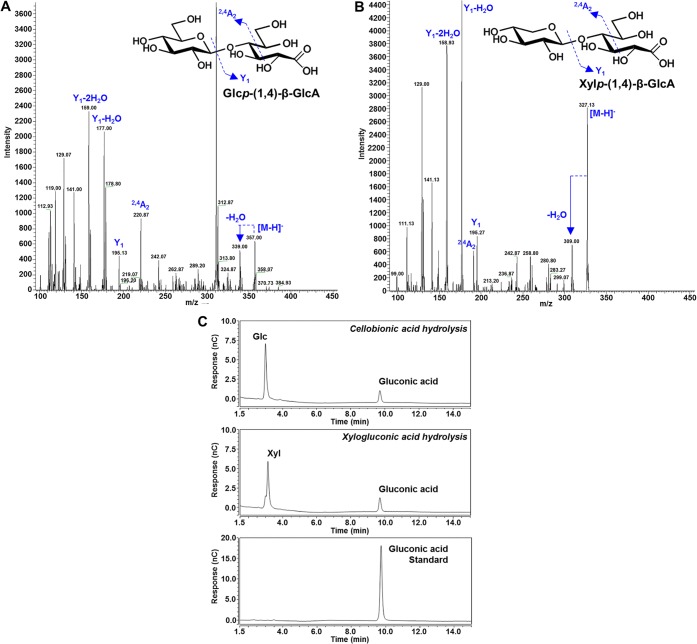
Mass spectrometric analysis (MS) with negative ion electrospray was used to determine the nominal mass of these oligosaccharides and of another unidentified product that could be released by acid hydrolysis from both of the unknown oligosaccharides. The first oligosaccharide (Unknown 1, Figure 2) was identified as a disaccharide, called cellobionic acid [glucopyranosyl-(1,4)-β-gluconic acid, (m/z 357.00, Figure 3A)], that can be hydrolyzed by acid to form glucose and gluconic acid (Figure 3C). The identity of gluconic acid (m/z 195.0510, C6H12O7) was confirmed through chromatographic comparison with a commercial standard (Figure 3C) under two sets of elution conditions. Oxidative enzymes present in commercial enzyme cocktails, such as Driselase, lead to the formation of cellobionic acid as a product of the oxidation of cellobiose.28 The presence of cellobionic acid in all our N. benthamiana samples was a product of cellulose hydrolysis. Thus, other polysaccharides containing several segments of consecutive 1,4-β-linked glucosyl residues, such as (1,3;1,4)-β-glucan, should also release cellobionic acid. This was confirmed by Driselase hydrolysis of cellulose and mixed-linked β-glucan standards (data not shown).
Figure 3.
LC-ESI-qTOF MS/MS analysis of unknown oligosaccharides (Figure 2) fractionated from Driselase digests of N. benthamiana leaf tissue. Unknown 1, cellobionic acid (A), and unknown 2, xylogluconic acid (B). x-axis, m/z; y-axis, intensity. HPAEC-PAD traces of monosaccharides produced postacid hydrolysis (C) of unknown 1 (glucose and gluconic acid) and unknown 2 (xylose and gluconic acid) with gluconic acid standard. x-axis, time; y-axis, abundance. x-axis, m/z; y-axis, intensity.
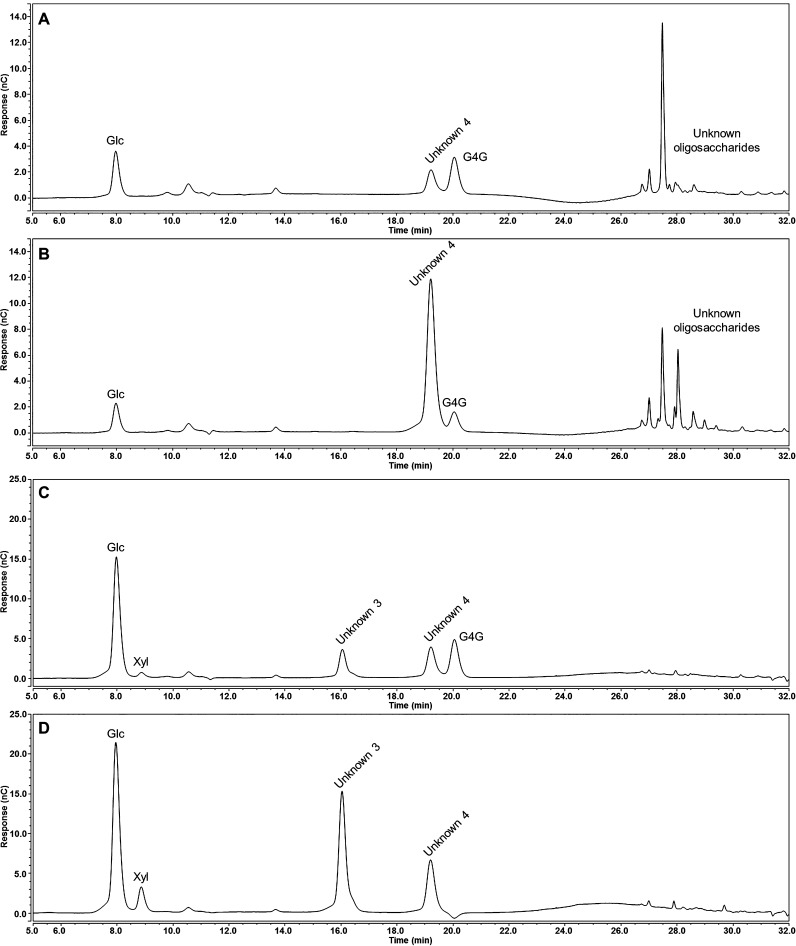
The second oligosaccharide (Unknown 2, Figure 2) was identified as a disaccharide (m/z 327.13, Figure 3B) that can be hydrolyzed by acid to form xylose and gluconic acid (Figure 3C). It was proposed that cellobionic acid and the second oligosaccharide (Unknown 2) originated from a polysaccharide containing xylose and glucose in a linear chain of (1,4)-β-linked residues. To test this hypothesis, the HvCslF3 and HvCslF10 expressing samples were incubated with two cellulase enzymes with differing substrate specificities: a cellulase from Aspergillus niger (E-CELAN), which has a preference for (1,4)-β-linkages in cellulose and (1,3;1,4)-β-glucan, and a cellulase from Trichoderma longibrachiatum (E-CELTR), which has a broader specificity including (1,4)-β-xylan29 and lower molecular weight glucan oligosaccharides.30 Relative to the empty vector controls, hydrolytic reactions with E-CELAN resulted in an increase of cellobiose, an unknown disaccharide (Unknown 4, Figure 4), and a number of unknown oligosaccharides eluting at a longer retention time for both HvCslF3 (Figure 4A) and HvCslF10 (Figure 4B) expressing samples. As the buffers used for each enzyme led to a baseline artifact in the HPAEC chromatograms (Figure S4), visualization of the oligosaccharides with a longer elution time was achieved by subtraction of the chromatogram corresponding to the negative control from N. benthamiana leaf tissue transformed with the empty vector. Therefore, any visible peaks are a direct increase in signal relative to the controls and not the total signal from each sample. However, it is clear that there were no background peaks of Unknown 4 visible in the negative control.
Figure 4.
HPAEC-PAD traces of oligosaccharides produced posthydrolysis with cellulolytic enzymes of N. benthamiana leaf tissue overexpressing of HvCslF3 and HvCslF10. Results have been normalized by the subtraction of the negative control spectra from N. benthamiana leaf tissue overexpressing an empty vector control (original traces available in Figure S4, Supporting Information). Oligosaccharides released from HvCslF3 (A) and HvCslF10 (B) expressing tissue by a cellulase from Aspergillus niger (E-CELAN), which has a preference for (1,4)-β-linkages in cellulose and (1,3;1,4)-β-glucan. Oligosaccharides released from HvCslF3 (C) and HvCslF10 (D) expressing tissue by a cellulase from Trichoderma longibrachiatum (E-CELTR), which has a broader specificity. Unknown 3 and 4 refer to peaks identified for further characterization. x-axis, time; y-axis, abundance.
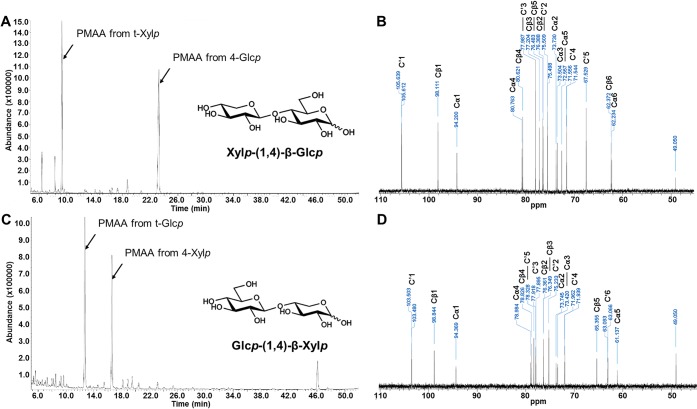
Incubation with E-CELTR resulted in cellobiose, xylobiose, two unknown disaccharides (Unknowns 3 and 4, Figure 4), and a considerably smaller number of peaks most likely related to higher molecular weight compounds for both HvCslF3 (Figure 4C) and HvCslF10 (Figure 4D) expressing samples. One of the unknown peaks arising from the E-CELTR treatment had the same retention time as the unknown peak from the E-CELAN condition (Unknown 4). Isolation of each unknown peak and analysis of the monosaccharides released following acid hydrolysis indicated that both disaccharides were composed of xylose and glucose at a 1:1 ratio (data not shown). Permethylation glycosidic linkage analysis (Figure 5A,C) confirmed that the disaccharides were xylopyranosyl-(1,4)-glucopyranose (Unknown 4) and glucopyranosyl-(1,4)-xylopyranose (Unknown 3). 13C NMR analysis (Figure 5B,D) demonstrated that the sugars were β-linked, and equivalent to the previously characterized synthetic disaccharides, xylopyranosyl-(1,4)-β-glucopyranose and glucopyranosyl-(1,4)-β-xylopyranose.31
Figure 5.
Structural analysis of the disaccharides produced posthydrolysis with E-CELTR from N. benthamiana leaves overexpressing HvCslF3 and HvCslF10. Total ion current (TIC) chromatogram following methylation linkage analysis (A) and the 13C NMR spectra (B) of the disaccharide Xylp-(1 → 4)-Glcp. Total ion current (TIC) chromatogram of partially methylated alditol acetates (C) and the 13C NMR spectra (D) of the disaccharide Glcp-(1 → 4)-Xylp. x-axis, time; y-axis, abundance. C′1–6 correspond to the carbons of the nonreducing end of the disaccharide, and Cα and Cβ refer to the α and β anomeric carbons of the reducing end of the disaccharide. MS fragmentation patterns of the partially methylated alditol acetates in parts A and C are available in Figure S1.
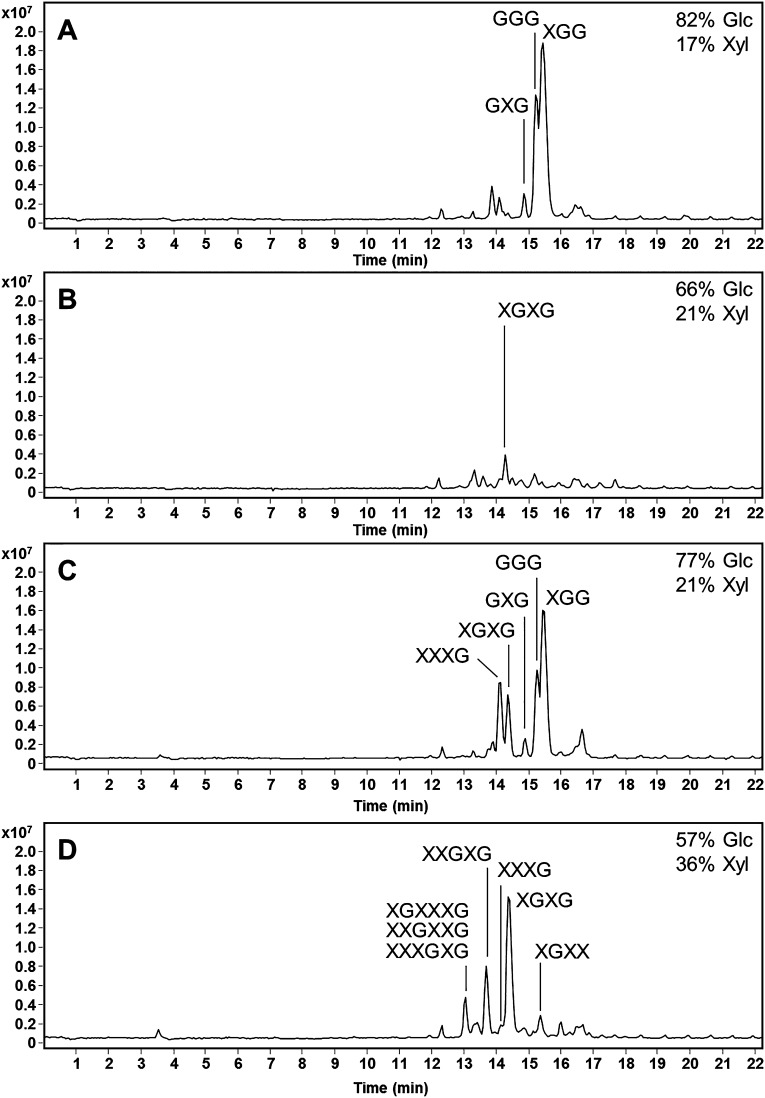
The later eluting oligosaccharides produced postcellulase (E-CELAN) hydrolysis of N. benthamiana leaves expressing HvCslF3 and HvCslF10 were further characterized following fractionation using Carbon SPE (Bond Elut, Agilent Technologies, Singapore). Monosaccharides and disaccharides were removed by eluting with acetonitrile in water up to 10%, and then fractions containing the larger oligosaccharides were collected with 15% acetonitrile and 55% acetonitrile. These fractions were analyzed using HPAEC-PAD, monosaccharide analysis, and LC-MS2 as their PMP derivatives (Figure 6A–D). Monosaccharide analysis indicated that each fraction was predominantly composed of glucose and xylose with higher concentrations of glucose. The Glc:Xyl ratio was lower in the 55% acetonitrile fractions of both HvCslF3 and HvCslF10 samples (Figure 6B,D), which contained higher molecular weight oligosaccharides. A higher number of higher molecular weight oligosaccharides were observed in the HvCslF10 samples (Figure 6C,D) supporting the larger xylose increase visible in Figure 1. Analysis of the MS2 fragmentation patterns (Figure S5) and the assumption (based on molecular weight) that hexose and pentose sugars corresponded to glucose and xylose, respectively, allowed structural predictions of each oligosaccharide to be made (Figure 6A–D). Oligosaccharides containing between three and six sugars were detected with various combinations of glucosyl and xylosyl residues. Up to three consecutive xylosyl residues could be observed in some oligosaccharides.
Figure 6.
Structural analysis of the oligosaccharides produced postcellulase (E-CELAN) hydrolysis of Nicotiana benthamiana leaves expressing HvCslF3 and HvCslF10. Extracted ion chromatograms (EIC 700–1500) of the PMP derivatives from the HvCslF3 15% (A) and 55% (B) acetonitrile oligosaccharide fractions and the HvCslF10 15% (C) and 55% (D) acetonitrile oligosaccharide fractions are presented. Monosaccharide contents of each fraction are indicated with the total mol % of the glucose and xylose in the top right corner. MS fragmentation patterns used to calculate the structure of each oligosaccharide labeled in parts A–D are available in Figure S5. G = glucose, X = xylose.
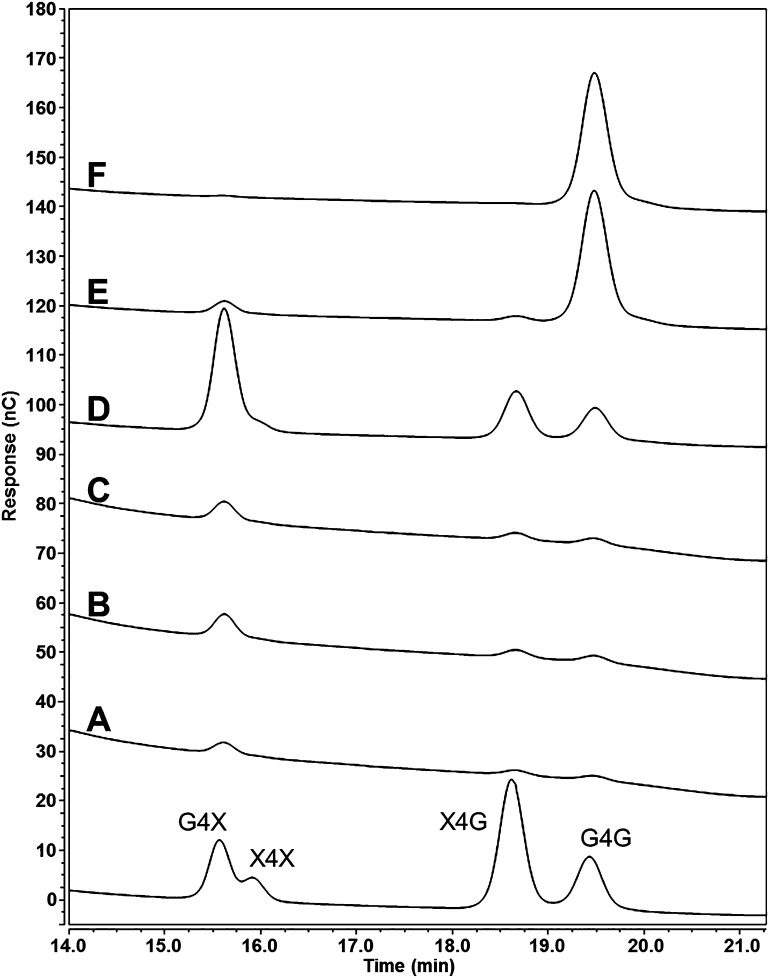
The solubility of the glucoxylan polysaccharide synthesized by HvCslF10 in N. benthamiana was investigated using a sequential series of solvent extractions, including water at 100 °C, dimethyl sulfoxide (DMSO) at 50 and 100 °C, and DMSO with increasing concentrations of the ionic liquid 1-ethyl-3-methylimidazolium acetate (EmimAc, 2%, 6%, and 20%) at 60 °C. At the right concentration and temperature, EmimAc is capable of solvating one of the least water soluble and most recalcitrant polysaccharides, cellulose.32 Each fraction was screened for the presence of the diagnostic disaccharides following hydrolysis with the cellulase enzyme, E-CELTR. Xylp-(1,4)-β-Glcp and Glcp-(1,4)-β-Xylp disaccharides were observed at low levels in the water (Figure 7A) and DMSO fractions (Figure 7B,C). However, the addition of 2% EmimAc solubilized the majority of the glucoxylan polysaccharide (∼80%, Figure 7D). Cellobiose also appeared in the 2% EmimAc fraction, either released from the glucoxylan or cellulose, with the majority of cellobiose appearing in the 6% EmimAc (Figure 7E) and 20% EmimAc (Figure 7F) fractions that contained low levels of the Xylp-(1,4)-β-Glcp and Glcp-(1,4)-β-Xylp disaccharides.
Figure 7.
HPAEC-PAD traces of oligosaccharides produced posthydrolysis with E-CELTR from solvent soluble fractions extracted from N. benthamiana leaves overexpressing HvCslF10; water soluble fraction (A), 50 °C DMSO soluble fraction (B), 100 °C DMSO soluble fraction (C), 60 °C DMSO/EmimAc (98:2) soluble fraction (D), 60 °C DMSO/EmimAc (94:6) soluble fraction (E), and 60 °C DMSO/EmimAc (80:20) soluble fraction (F). Standards for Glc-(1,4)-β-Xyl (G4X), xylobiose (X4X), Xyl-(1,4)-β-Glc (X4G), and cellobiose (G4G) are included. x-axis, time; y-axis, abundance.
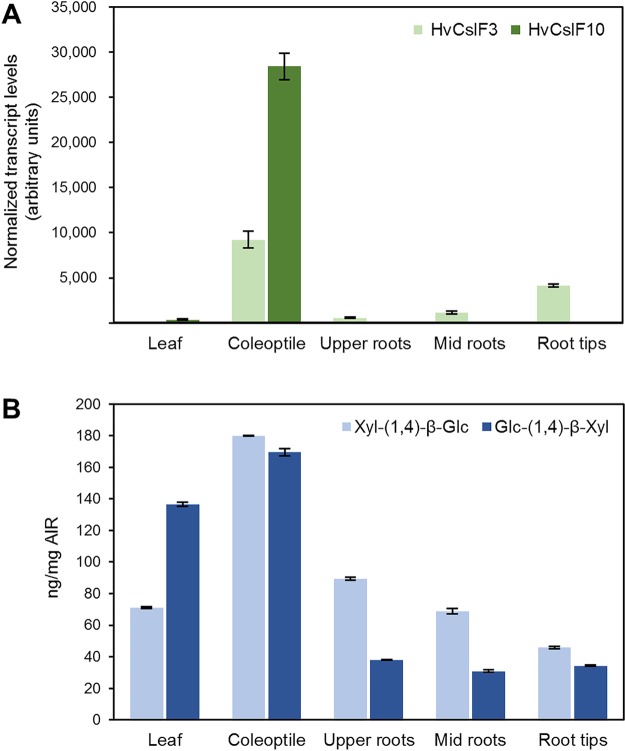
Given that glucoxylan had only been observed in a heterologous expression system, we next determined whether (1,4)-β-linked glucose and xylose occur in barley tissues expressing HvCslF3 or HvCslF10. Barley seedlings were grown for 7 days, harvested, and divided into leaf and root sections for analysis of HvCslF3 and HvCslF10 transcript levels. HvCslF10 transcript was observed only in leaf and coleoptile tissues, at a low level in the former, and substantially higher level in the latter (Figure 8A). HvCslF3 transcript was observed at its highest levels in the coleoptile tissue and was detected in all root tissues, increasing toward the root tip. Each tissue was screened for the presence of the diagnostic disaccharides following hydrolysis with E-CELTR. Analysis by liquid chromatography electrospray-ionization quadrupole time-of-flight mass spectrometry (LC-ESI-qTOF-MS) clearly showed that the enzymatic hydrolysis of the barley tissues released the diagnostic disaccharides with the same retention times compared to those observed in the heterologous expression system (Figure S2A–H). The MS and MS/MS profiles of each peak were matched with the corresponding disaccharide standards. The amounts of Xylp-(1,4)-β-Glcp and Glcp-(1,4)-β-Xylp disaccharides in tissues of barley seedlings were quantified by measuring the LC-ESI-qTOF peak area compared against a standard curve of purified disaccharides (Figure 8B). The highest concentrations of each disaccharide were found in the coleoptiles, which also contained the highest HvCslF3 and HvCslF10 transcript levels (Figure 8A). High levels were also detected in tissues with lower levels of HvCslF3 and HvCslF10 transcripts. This is not unexpected as the abundance of the disaccharides within each tissue is measured from a single time point and would be a culmination of HvCslF3 and HvCslF10 transcription, translation, activation, and turnover during the development of the tissue.
Figure 8.
Normalized transcript levels of HvCslF3 and HvCslF10 genes in tissues of barley seedlings, cv. Golden Promise (A). Error bars indicate standard deviation. Quantification of Xyl-(1,4)-β-Glc and Glc-(1,4)-β-Xyl disaccharides produced postcellulase (E-CELTR) hydrolysis of tissues of barley seedlings (B). Error bars indicate standard deviation.
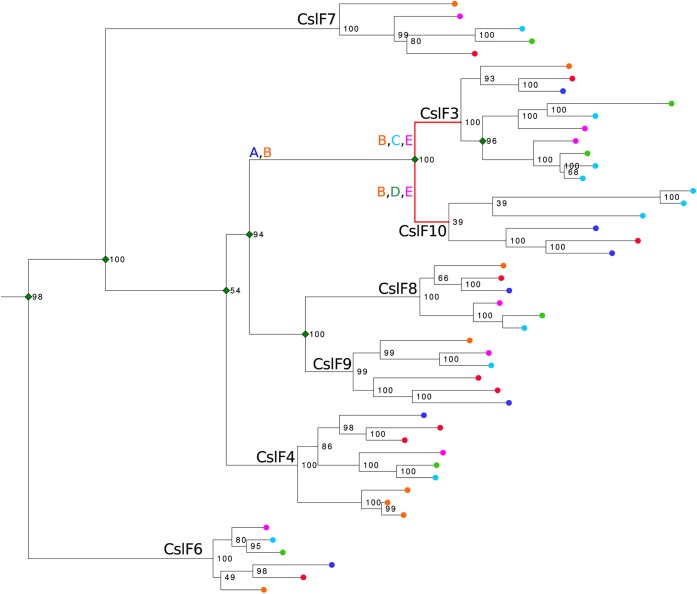
We addressed the question of CslF3 and CslF10 evolution compared to other CslF sequences in six fully sequenced Poaceae species by reconstructing a phylogeny using RAxML. Seven clades that existed prior to the divergence of extant species were identified with CslF3 and CslF10 forming a monophyletic grouping (Figure 9), concordant with previous observations.1,20,21 To determine evolutionary rates and how selection has operated on the CslF3 and CslF10 sequences we used the BUSTED model as implemented in HYPHY33 to test for diversifying episodic selection. Five hypotheses were tested: (A) A burst of episodic selection occurred in the ancestral branch of the CslF3 and CslF10 group. (B) There had been sustained selection in the ancestral group and following the CslF3 and CslF10 split. There was selection independently in the branches leading to CslF3 (C) and CslF10 (D). Finally, (E) there was selection in both branches leading to the CslF3 and CslF10 groups. Diversifying episodic selection was detected in hypotheses B (p = <0.005), C (p = <0.005), D (p = <0.005), and E (p = <0.005). BUSTED reports selection if one site in one branch of the “foreground” group is detected. As selection was not detected in hypothesis A, wherein a burst of episodic selection occurred in the ancestral branch of the CslF3 and CslF10 group, we concluded that only the branches leading to CslF3 and CslF10 were under significant diversifying episodic selection.
Figure 9.
Maximum likelihood tree of 48 CslF sequences from Hordeum vulgare (red ●), Brachypodium distachyon (dark blue ●), Oryza sativa (orange ●), Setaria italica (purple ●), Sorghum bicolor (light blue ●), and Zea mays (green ●). Bootstrap support values are indicated on nodes in black. Green ◆ indicates gene duplication events that have occurred prior to the emergence of extant species. Letters A–E represent the evolutionary hypotheses tested using BUSTED as implemented in HyPhy.
Discussion
The analysis of N. benthamiana tissues transiently expressing HvCslF3 and HvCslF10 genes driven by the 35S promoter indicated that both of the corresponding proteins are unable to synthesize detectable levels of the (1,3;1,4)-β-glucan that has been observed following expression of other members of the CslF family. Enzymatic treatment of HvCslF3 or HvCslF10 expressing tissue with E-CELTR, a cellulase with broad specificity that is able to hydrolyze Glcp-(1,4)-β-Glcp and Xylp-(1,4)-β-Xylp linkages,29 released the Xylp-(1,4)-β-Glcp and Glcp-(1,4)-β-Xylp disaccharides. Enzymatic hydrolysis with E-CELAN, a cellulase with strong specificity toward (1,4)-β-glucan chains greater than and equal to cellopentose,30 released the Glcp-(1,4)-β-Xylp disaccharide along with a series of higher molecular weight oligosaccharides consisting of single or consecutive (1,4)-β-xylosyl residues within the (1,4)-β-glucan chain.
The Xylp-(1,4)-β-Glcp and Glcp-(1,4)-β-Xylp disaccharides have been observed previously in the cell wall from species of Ulva, a marine alga (Ulvales, Chlorophyta). Three main polysaccharides have been found in the green seaweed Ulva rigida.34 The major polysaccharide was composed of sulfated glucuronorhamnoxylans (ulvan) along with two noncellulosic fractions consisting of glucuronans and glucoxylans. The glucoxylan could be separated into four fractions, based on solubility. The range of solubilities was proposed to result from variable Glc:Xyl ratios and the potential for the glucoxylans to self-associate or associate with cellulose through hydrogen bonding. While the fractions with a lower Glc:Xyl ratio (less than 1:1) were extracted relatively easily, the fourth fraction resisted treatment with 1 and 4 M KOH before finally being extracted along with α-cellulose, following a cold acidic chlorite treatment and another 4 M KOH treatment. This is most likely due to the higher Glc:Xyl ratio (5:1) and longer stretches of (1,4)-β-glucan that can form intermolecular alignments with adjoining chains. Preliminary attempts to solubilize the glucoxylan synthesized by the HvCslF10 gene product (Figure 7) required the application of ionic liquids to facilitate solubility in DMSO, suggesting that the glucoxylan product produced by HvCSLF3 and HvCSLF10 may contain a higher ratio of glucose to xylose. This is partially supported by the analysis of the higher molecular weight oligosaccharides (Figure 6); however, for this to be confirmed, the polysaccharide products of HvCSLF3 and HvCSLF10 will need to be fractionated away from the background N. benthamiana cell wall components and analyzed further using conventional biochemical methods to determine the linkage types between the monosaccharide components and the ratios of each therein. The physicochemical properties of the isolated polysaccharide will determine if the presence of the xylose residues within the (1,4)-β-glucan chain affects the ability of the polysaccharide to form intermolecular alignments with adjoining chains. If the native barley glucoxylan has similar physicochemical properties, then the polysaccharide could play a more structural role in the cell wall compared to the conventional soluble (1,3;1,4)-β-glucan produced by HvCSLF6.
The production of glucoxylan by HvCSLF3 and HvCSLF10 challenges the concept that members of a single CSL family possess the same carbohydrate synthetic activity, with the CSLF family now demonstrated to catalyze the formation of not only (1,3)- and (1,4)-β-glucosidic linkages, but also (1,4)-β-glucosidic and (1,4)-β-xylosidic linkages. The production of different polysaccharides has been suggested previously within the CSLC family, which is involved in the synthesis of (1,4)-β-glucosidic linkages in the xyloglucan backbone in the Golgi.35 Members of the CSLC family are also proposed to play a role in cellulose synthesis; however this is thought to be due to synthesis of the (1,4)-β-glucan backbone at the plasma membrane in the absence of xylosyltransferases and not because of alternative linkages formed by the synthase.35 The production of different polysaccharides has been demonstrated within the CSLA family, which is involved in the synthesis of 1,4-β-mannan and glucomannan backbones.8,9 Perhaps the finding of another Csl clade with multiple carbohydrate synthetic activity will prompt a broader screen of each clade within the cellulose synthase superfamily.
The CslF gene family evolved after the Graminids and Restiids split from the other Poales. The family originated from a gene duplication in either a clade nested within the CslD clade, its closest relative in the CesA superfamily, or their immediate common ancestor. Subsequent gene duplication events following the evolution of the Poaceae (grasses) created the seven CslF subfamilies currently recognized.6 The majority of these events occurred in a paralogous cluster syntenic in all currently sampled grasses (Schwerdt et al., 2015). Figure 9 shows the CslF3 and CslF10 families forming a monophyletic grouping that is, along with the CslF8/F9 family, the most recently diverged CslF lineage. A common hypothesis is that the redundancy of gene duplications facilitates the evolution of novel enzyme function. The gene duplications that created the paralogous clustered CslF genes in the Poaceae may have reduced purifying selection, facilitating the evolution of (1,4)-β-xylosidic linkage synthesis.
The evolution of novel protein function can be accompanied by a directional shift in substitution rates. Episodic diversifying selection was tested for during the evolution of the CslF3 and CslF10 families since the CslF3/F10 and CslF8/F9 groups split. As seen in Figure 9, selection was detected in the branches leading to the CslF3 and CslF10 families, but not in the CslF3/F10 ancestral branch. If we assume that synthesis of (1,4)-β-xylosidic linkages is restricted to CSLF3 and CSLF10, then perhaps this function was acquired prior to their separation and subsequently lost in sister groups. Alternatively, considering three highly diverged GT2 families (CslF6, CslH, and CslJ) independently evolved to synthesize (1,3)-β-glucosidic linkages from presumably an ancestral (1,4)-β-glucan synthase (Little et al., 2018), there could be latent functional variation in the CslF family. Furthermore, shifts in substitution rates may not be reflective of selection for neofunctionalization but of protein fold stability or tissue specific codon usage.36
The analysis of cell wall composition not only is a difficult chemical process, but also relies on an estimation of how the jigsaw puzzle fits together without knowing what the complete picture should be, which leads to reliance on previous knowledge of what relative proportions of linkages are present in a particular polysaccharide and within the target species. Previously in barley, (1,4)-linked-glucose could be assigned to cellulose, (1,3;1,4)-β-glucan, xyloglucan, or starch, and (1,4)-linked-xylose could be assigned to heteroxylan. The finding of a new polysaccharide containing both (1,4)-linked glucose and xylose will complicate the assignment of these monosaccharides to a specific polymer and the interpretation of the linkage data. Given that there has been a relative surge in finding new polysaccharides and new gene families over recent years,6,37 there may be more that have not been found because the right methods have not been used to observe them. This highlights the need for linkage analysis to be conducted parallel to complementary methods, such as oligosaccharide mapping, to guide how the monomers are assembled into larger polysaccharides. The assignment of these linkages could potentially change the estimated fine structure and solubility of cellulose, heteroxylan, and (1,3;1,4)-β-glucan, with significant downstream effects for the predicted benefits of plant materials selected for biofuel saccharification or as sources of dietary fiber. Now that we have a simple assay to identify the diagnostic glucoxylan disaccharides, it will be interesting to see how widespread the polysaccharide is in different plant species and to determine the physicochemical properties associated with its presence in the wall.
Acknowledgments
We would like to acknowledge Philip Clements from the University of Adelaide for assistance with the 13C NMR analysis, Natoiya Lloyd from the Australian Wine Research Institute (AWRI) Metabolomics Facility for assistance in the analysis of gluconic acid, and Kylie Neumann for maintenance of plants in the glasshouse.
Glossary
Abbreviations
- Csl
cellulose synthase-like
- DMSO
dimethyl sulfoxide
- EmimAc
1-ethyl-3-methylimidazolium acetate
- GT
glycosyltransferase
- GH
glycosylhydrolase
- HPAEC
high-performance anion-exchange chromatography
- KOH
potassium hydroxide
- LC-ESI-qTOF-MS
liquid chromatography electrospray-ionization quadrupole time-of-flight mass spectrometry
- NMR
nuclear magnetic resonance spectroscopy
- Q-PCR
quantitative polymerase chain reaction
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00568.
Author Contributions
A.L. and J.L. conceived and designed the experiments. A.L., J.L., D.W.J., S.F.K., N.J.S., J.G.S., X.X., M.H., and R.A.B. performed the experiments. A.L., J.L., D.W.J., N.J.S., J.G.S., X.X., M.H., and V.B. analyzed the data. V.B. and R.A.B. contributed reagents/materials/analysis tools. A.L. J.G.S., and V.B. wrote the paper. A.L., J.G.S., D.W.J., N.J.S., R.A.B., and V.B. edited the paper.
This work was supported by the Australian Research Council Centre of Excellence in Plant Cell Walls (CE110001007).
The authors declare no competing financial interest.
Notes
Safety statement: no unexpected or unusually high safety hazards were encountered.
Supplementary Material
References
- Burton R. A.; Jobling S. A.; Harvey A. J.; Shirley N. J.; Mather D. E.; Bacic A.; Fincher G. B. The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 2008, 146 (4), 1821–1833. 10.1104/pp.107.114694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R. A.; Gidley M. J.; Fincher G. B. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 2010, 6 (10), 724–732. 10.1038/nchembio.439. [DOI] [PubMed] [Google Scholar]
- Pettolino F. A.; Walsh C.; Fincher G. B.; Bacic A. Determining the polysaccharide composition of plant cell walls. Nat. Protoc. 2012, 7 (9), 1590–1607. 10.1038/nprot.2012.081. [DOI] [PubMed] [Google Scholar]
- Lombard V.; Golaconda Ramulu H.; Drula E.; Coutinho P. M.; Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42 (D1), D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T.; Somerville C. The cellulose synthase superfamily. Plant Physiol. 2000, 124, 495–498. 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little A.; Schwerdt J. G.; Shirley N. J.; Khor S. F.; Neumann K.; O’Donovan L. A.; Lahnstein J.; Collins H. M.; Henderson M.; Fincher G. B.; Burton R. A. Revised phylogeny of the Cellulose Synthase gene superfamily: insights into cell wall evolution. Plant Physiol. 2018, 177 (3), 1124–1141. 10.1104/pp.17.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear J. R.; Kawagoe Y.; Schreckengost W. E.; Delmer D. P.; Stalker D. M. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U. S. A. 1996, 93 (22), 12637–12642. 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A. H.; Wilkerson C. G.; Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (6), 2221–2226. 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga K. S.; Barreiro R.; Whitten B.; Stecca K.; Hazebroek J.; Randhawa G. S.; Dolan M.; Kinney A. J.; Tomes D.; Nichols S.; Anderson P. Guar seed β-mannan synthase is a member of the cellulose synthase supergene family. Science 2004, 303 (5656), 363–366. 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- Liepman A. H.; Wilkerson C. G.; Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (6), 2221–2226. 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J. C.; Lerouxel O.; Drakakaki G.; Alonso A. P.; Liepman A. H.; Keegstra K.; Raikhel N.; Wilkerson C. G. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (20), 8550–8555. 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A. H.; Cavalier D. The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: recent advances and future perspectives. Front. Plant Sci. 2012, 3, 109. 10.3389/fpls.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R.; Wilson S.; Hrmova M.; Harvey A.; Shirley N.; Medhurst A.; Stone B.; Newbigin E.; Bacic A.; Fincher G. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 2006, 311 (5769), 1940–1942. 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- Doblin M. S.; Pettolino F. A.; Wilson S. M.; Campbell R.; Burton R. A.; Fincher G. B.; Newbigin E.; Bacic A. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (14), 5996–6001. 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y.; Yin L.; Oikawa A.; Scheller H. V. Mannan synthase activity in the CSLD family. Plant Signaling Behav. 2011, 6 (10), 1620–1623. 10.4161/psb.6.10.17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin M. S.; De Melis L.; Newbigin E.; Bacic A.; Read S. M. Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol. 2001, 125 (4), 2040–2052. 10.1104/pp.125.4.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.; Szumlanski A. L.; Gu F.; Guo F.; Nielsen E. A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 2011, 13 (8), 973–980. 10.1038/ncb2294. [DOI] [PubMed] [Google Scholar]
- Dhugga K. S. Biosynthesis of non-cellulosic polysaccharides of plant cell walls. Phytochemistry 2012, 74, 8–19. 10.1016/j.phytochem.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Douchkov D.; Lueck S.; Hensel G.; Kumlehn J.; Rajaraman J.; Johrde A.; Doblin M. S.; Beahan C. T.; Kopischke M.; Fuchs R. The barley (Hordeum vulgare) cellulose synthase-like D2 gene (HvCslD2) mediates penetration resistance to host-adapted and nonhost isolates of the powdery mildew fungus. New Phytol. 2016, 212 (2), 421–433. 10.1111/nph.14065. [DOI] [PubMed] [Google Scholar]
- Schwerdt J. G.; MacKenzie K.; Wright F.; Oehme D.; Wagner J. M.; Harvey A. J.; Shirley N. J.; Burton R. A.; Schreiber M.; Halpin C. Evolutionary dynamics of the Cellulose synthase gene superfamily in grasses. Plant Physiol. 2015, 168 (3), 968–983. 10.1104/pp.15.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M.; Wright F.; MacKenzie K.; Hedley P. E.; Schwerdt J. G.; Little A.; Burton R. A.; Fincher G. B.; Marshall D.; Waugh R.; Halpin C. The barley genome sequence assembly reveals three additional members of the CslF (1,3;1,4)-β-glucan synthase gene family. PLoS One 2014, 9 (3), e90888-1–e90888-9. 10.1371/journal.pone.0090888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R. A.; Collins H. M.; Kibble N. A. J.; Smith J. A.; Shirley N. J.; Jobling S. A.; Henderson M.; Singh R. R.; Pettolino F.; Wilson S. M.; Bird A. R.; Topping D. L.; Bacic A.; Fincher G. B. Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure. Plant Biotechnology Journal 2011, 9 (2), 117–135. 10.1111/j.1467-7652.2010.00532.x. [DOI] [PubMed] [Google Scholar]
- Taketa S.; Yuo T.; Tonooka T.; Tsumuraya Y.; Inagaki Y.; Haruyama N.; Larroque O.; Jobling S. A. Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-ß-D-glucan biosynthesis. J. Exp. Bot. 2012, 63 (1), 381–92. 10.1093/jxb/err285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitroff G.; Little A.; Lahnstein J.; Schwerdt J. G.; Srivastava V.; Bulone V.; Burton R. A.; Fincher G. B. (1, 3; 1, 4)-β-Glucan biosynthesis by the CSLF6 enzyme: position and flexibility of catalytic residues influence product fine structure. Biochemistry 2016, 55 (13), 2054–2061. 10.1021/acs.biochem.5b01384. [DOI] [PubMed] [Google Scholar]
- Jobling S. A. Membrane pore architecture of the CSLF6 protein controls (1–3, 1–4)-β-glucan structure. Science Advances 2015, 1 (5), e1500069 10.1126/sciadv.1500069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClear B. V.; Glennie-Holmes M. Enzymatic quantification of (1,3),(1, 4)-β-glucan in barley and malt. J. Inst. Brew. 1985, 91, 285–295. 10.1002/j.2050-0416.1985.tb04345.x. [DOI] [Google Scholar]
- Gardner S. L.; Burrell M.; Fry S. C. Screening of Arabidopsis thaliana stems for variation in cell wall polysaccharides. Phytochemistry 2002, 60 (3), 241–254. 10.1016/S0031-9422(02)00046-8. [DOI] [PubMed] [Google Scholar]
- Cannella D.; Chia-wen C. H.; Felby C.; Jørgensen H. Production and effect of aldonic acids during enzymatic hydrolysis of lignocellulose at high dry matter content. Biotechnol. Biofuels 2012, 5 (1), 26. 10.1186/1754-6834-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan D.; Cornaggia C.; Liadova A.; McCormack N.; Ivory R.; McKie V. A.; Ormerod A.; McCleary B. V. Novel substrates for the automated and manual assay of endo-1, 4-β-xylanase. Carbohydr. Res. 2017, 445, 14–22. 10.1016/j.carres.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Mangan D.; Cornaggia C.; McKie V.; Kargelis T.; McCleary B. V. A novel automatable enzyme-coupled colorimetric assay for endo-1, 4-β-glucanase (cellulase). Anal. Bioanal. Chem. 2016, 408 (15), 4159–4168. 10.1007/s00216-016-9507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petráková E.; Krupová I.; Schraml J.; Hirsch J. Synthesis and 13 C NMR spectra of disaccharides related to glucoxylans and xyloglucans. Collect. Czech. Chem. Commun. 1991, 56 (6), 1300–1308. 10.1135/cccc19911300. [DOI] [Google Scholar]
- Gericke M.; Liebert T.; Seoud O. A. E.; Heinze T. Tailored media for homogeneous cellulose chemistry: ionic liquid/co-solvent mixtures. Macromol. Mater. Eng. 2011, 296 (6), 483–493. 10.1002/mame.201000330. [DOI] [Google Scholar]
- Murrell B.; Weaver S.; Smith M. D.; Wertheim J. O.; Murrell S.; Aylward A.; Eren K.; Pollner T.; Martin D. P.; Smith D. M. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015, 32 (5), 1365–1371. 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B.; Lahaye M. Cell-wall polysaccharides from the marine green alga Ulva “rigida”(Ulvales, Chlorophyta). Extraction and chemical composition. Carbohydr. Res. 1995, 274, 251–261. 10.1016/0008-6215(95)00138-J. [DOI] [PubMed] [Google Scholar]
- Dwivany F. M.; Yulia D.; Burton R. A.; Shirley N. J.; Wilson S. M.; Fincher G. B.; Bacic A.; Newbigin E.; Doblin M. S. The CELLULOSE-SYNTHASE LIKE C (CSLC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Mol. Plant 2009, 2 (5), 1025–1039. 10.1093/mp/ssp064. [DOI] [PubMed] [Google Scholar]
- Drummond D. A.; Wilke C. O. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 2008, 134 (2), 341–352. 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. W.; Lahnstein J.; Hsieh Y. S.; Xing X.; Yap K.; Chaves A. M.; Scavuzzo-Duggan T. R.; Dimitroff G.; Lonsdale A.; Roberts E. M. Functional characterization of a glycosyltransferase from the moss Physcomitrella patens involved in the biosynthesis of a novel cell wall arabinoglucan. Plant Cell 2018, 30, 1293. 10.1105/tpc.18.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.