Abstract
In recent years protein therapeutics have seen increasing use in the therapeutic arena. As with traditional small molecule drug substances, one is obligated to ensure purity and stability of the various dosage forms. With these higher molecular weight therapeutics a common approach for analytical characterization is enzymatic digestion followed by gradient elution liquid chromatography with mass spectrometry detection to create a peptide map (bottom-up protein analysis). Due to the difficult to separate mixtures frequently encountered, there is the need for advanced chromatographic systems featuring increased resolution and/or peak capacity that can be operated in the gradient elution format. Presently we describe an extreme ultra-pressure liquid chromatography (XUPLC) system that has been implemented as an in-house add-on to a commercial ultra-pressure chromatography system. This add-on allows operation at the 38 Kspi range, accommodates the use of capillary columns in excess of one meter packed with sub-2 μm particles and can be operated in the gradient elution format. To evaluate the utility of this system, rat growth hormone was used as a model protein and was exposed to light (λ 254 nm) to create a stress environment. When enzymatic digests of control and stressed protein were analyzed with the XUPLC system using MS detection, greater than 92% peptide coverage was achieved, including the identification some peptides where pre-oxidation of Met residues had occurred, as well as chemistry specifically related to the photolysis of protein disulfide linkages. When the same samples were analyzed by commercial UPLC and compared to the XUPLC results, the utility of the increased peak capacity available with the XUPLC was apparent as previously co-eluting peaks were now well resolved. In particular one specific degradation route was identified where a pair of isobaric cis/trans diastereomerically related peptides were well resolved by XUPLC while they were unresolved by UPLC. Clearly the use of this system operating at the higher pressure regime with long capillary columns is and will be useful in continued investigations of protein stability, especially in cases where only subtle differences in the amino acid residues have occurred during degradation.
Keywords: Photochemical mediated protein degradation, XUPLC, Meter capillary columns, Sub-2 μm particles
Introduction
Therapeutic Proteins and Photoinduced Degradation
Apparently the first therapeutic protein was the pancreatic extract, insulin, which was used in the treatment of diabetes by Grant and Macleod, who subsequently received the 1923 Nobel Prize in Physiology or Medicine [1]. In past several decades, therapeutic proteins have acquired a significant role in the treatment of various pathologies such as various cancers and inflammatory conditions [2]. As of 2012 there were 100 of these agents approved for clinical use in the European Union and the USA [3]. As with traditional small molecule pharmaceuticals, protein therapeutics must be prepared in high purity as stable dosage forms. Being composed of constituent amino acids, protein therapeutics have available numerous chemical pathways of degradation such as deamidation, hydrolysis, elimination, rearrangements, oxidation, photochemical reactions and photo-mediated oxidations; reactions that are frequently facile due to the presence of higher-order structure resulting in proximity of the reactive functional moieties [4].
Pharmaceutical scientists and regulatory authorities have recognized the susceptibility of therapeutic proteins to light mediated degradation and accordingly have established guidelines for photostabiliy testing. In particular residues in proteins that are subject to primary photooxidation include the aromatic amino acids tryptophan, tyrosine, phenylalanine, and the thiol containing amino acids cysteine/cystine, with reaction schemes of the ensuing chemical pathways being illustrated in a recent publication [5]. As regards methionine residues, the formation of methionine sulfoxide, resulting of the creation of an additional stereocenter, is known to occur by interaction with metal-ions in the presence of molecular oxygen and by a multi-step process initiated by electron transfer from excited state tryptophan to form superoxide anion [6], subsequently forming hydrogen peroxide, which can interact with methionine to eventually result in formation of the sulfoxide [7].
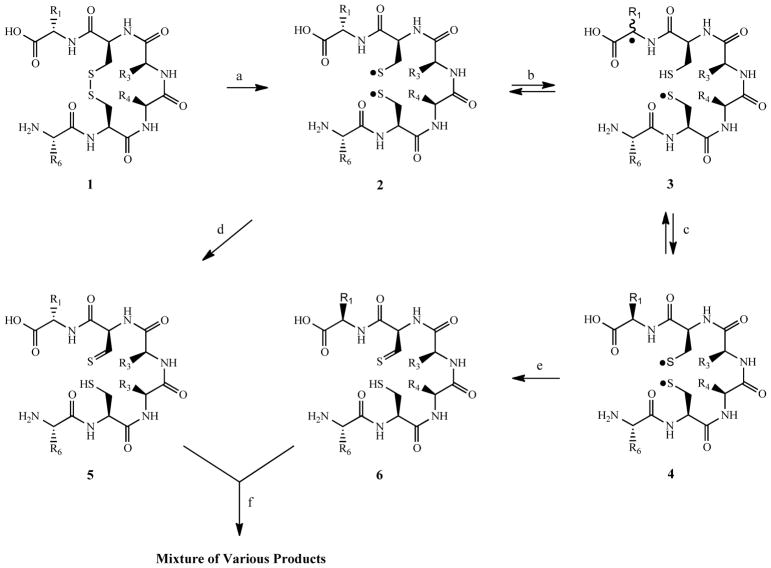
Recognizing the potential for photoinduced degradation of protein therapeutics, for the past several years and continuing to the present, the Schöneich group together with various collaborators have initiated a systematic investigation of such processes, with particular interest in the disulfide linkage. The investigations began with small molecular weight substances, with continuing efforts focusing on more structurally complex higher molecular weight substances [8–15]. Emerging from these efforts is an appreciation of various reactions pathways and products that are subsequent to photoinduced homolytic bond cleavage of disulfide bonds to form the corresponding thiyl radicals. The initially ensuing reactions include hydrogen atom exchange with the peptide backbone and/or sulfur radical disproportionation, processes which result in residue racemization and the formation of a thiol-thioaldehyde combination, respectively [10–13]. These initially formed sulfur bearing residues have been found to undergo a series of reactions ultimately leading to the formation of products such as hemidithioacetals, cyclic thioethers and cyclic vinylthioethers, with the ultimate products formed being dependent on whether the disulfide bond was an inter- or intra-molecular linkage [10–13].
In the case of small peptides possessing an intramolecular disulfide linkage, the initial steps of photo-induced disulfide cleavage are illustrated in Fig. 1. Species 2–4 represent initial thiol radical formation and residue racemization, with products 5–6 being formed from a subsequent disproportionation process. Of note, products 5 and 6 represent only one of the plausible thiol-thioaldehyde pairs. These initially formed products are reactive species that have been shown to undergo a series of reactions leading to the numerous other products [10–15]. As previously noted, the various degradation pathways result in a mixture of products bearing differing, but often structurally similar functional groups. Importantly, numerous members of these mixtures are chemically related as diastereomers, which possess similar chemical-physical properties.
Fig. 1.
Reactions illustrating the initial steps in photoinduced degradation of a cyclic peptide.
(a) light induced formation of cys thiyl radicals; (b) intramolecular H atom abstraction by thiyl radical; (c) H atom transfer resulting in stereocenter racemization; (d) disproportionation reaction leading to thiol and thioaldehyde formation (for non-symmetrical disulfide peptides the alternative product pair may be formed); (e) disproportionation reaction of racemized peptide; (f) continuing degradation processes per various secondary processes [11].
Analysis of Protein Degradation Products
Liquid chromatography (LC) electrospray ionization (ESI) time-of-flight (TOF) mass spectrometry (MS) has served as the predominant analytical approach for the analysis of photoinduced degradation of various model peptides [9, 11, 12]. For proteins, due to their larger mass imparting inefficient chromatographic and mass spectral properties, the analytical workflow typically includes a proteolytic digestion step to provide a mixture of peptides [16]. The various reactions previously noted were first delineated using model peptides and latter confirmed in higher molecular weight substances such as insulin [10] and monoclonal antibodies [13–15]. While LC-MS has been essential in these investigations, examination of the various chromatograms frequently reveals the presence of partially or coeluting peaks, thus placing a high burden on the MS dimension to elucidate product structure, and in certain situations potentially leading to an oversimplification and/or incorrect conclusions regarding the nature of products formed. While presenting details of all such possibilities is beyond the scope of the present publication, examination of structures 5 and 6 in Fig. 1 illustrates such an issue. In this case, the two peptides are diastereomerically related due to racemization of the stereocenter at the alpha-carbon of the first residue. In such situations one would expect each peptide to undergo substantially identical collision-induced dissociation (CID) MS fragmentation, and if chromatographic coelution occurred, the transformation of the L- to D-configuration would go undetected. In contrast complete chromatographic resolution would indicate the presence, but not the absolute identity, of at least two products. In such situations full characterization can be accomplished by employing a combination of chemical, chromatographic and detection techniques that together allow for the determination of the individual D- and L-amino acids [12, 15].
Practical Aspects of High Peak Capacity Chromatographic Systems
As one considers the previously noted types of products that result from photoinduced protein degradation, including the reaction pathways presented in Fig. 1 and the resultant analytical challenges, it is clear that chromatographic systems possessing high peak capacity (PC) (high resolution) would be beneficial in profiling complex degradation profiles. In principle the achievement of increased peak capacity would appear to be straightforward through the use of long columns packed with small particles operated in a gradient elution format. For gradient elution separations exhibiting unit resolution (Rs = 1.0), PC capacity is related to various parameters by equation 1 where tG is the gradient time, and W the 4σ peak width [17].
| (1) |
In linear solvent strength (LSS) GE, as is commonly encountered in reversed-phase liquid chromatography (RPLC), peak width is represented by equation 2 where ke is the column retention value at the time of elution, to the column dead time and N* gradient elution efficiency [18].
| (2) |
Combing these relationships leads to equation 3, where it is seen that PC is directly proportional to the square root of column length and inversely related to plate height equations such as the Knox [17] or van Deemter equations [19].
| (3) |
Column pressure drop (ΔP) is related to various parameters as described by equation 4, with ϕ, η and u representing the column resistance factor, mobile phase viscosity and linear velocity, respectively [17].
| (4) |
Comparison of these relationships (equations 3 and 4) indicates that PC ∝ ΔP1/2, the square root of length and reciprocal of particle size. While the use of long columns packed with sub-2 micron particles appears to provide a straightforward approach for the achievement of high PC, inherent to this is the unavoidable limitation of the need for substantially higher operating pressure that will quickly exceed the 6 Kpsi limit of so-called legacy chromatography systems. Another option, but ultimately limited, would be an increase in the gradient time. However, there are practical and performance issues to such an approach that can be appreciated by inspection of equation 5, where k* represents the instantaneous retention factor at mid-column in LLS-GE (F the volumetric flow rate, Vm the column dead time, Δϕ the gradient solvent range and S = (d(log k)/dϕ), a characteristic of a particular analyte in combination with the retention phase [18].
| (5) |
While PC may be increased by an increase in tG, (equation 3), this is not without issue as continued increase in gradient time will lead to a separation exhibiting a substantially non-optimal k* value and associated diminishing detection sensitivity. Alternatively, if one balances this effect by a decrease in F, then the diminishment in linear velocity will likewise result in non-optimal operating conditions as governed by band dispersion equations and a concomitant decrease in PC [20].
The era of ultrahigh-pressure liquid chromatography (UHPLC) was ushered in with the demonstration of a research instrument operating at 100 Kpsi by the Jorgenson research group in 1997 [21]. Realizing the attributes of operation at higher pressures, the initial commercial UHPLC system, with a pressure limit of 15 Kpsi, was introduced in 2004 with numerous manufacturers soon following suit [22]. From a practical and operational perspective, the use of capillary columns is advantageous in terms of detection and/or sample volume as these parameters are scaled to column dimension as the reciprocal of radius squared. In larger bore columns operated at higher pressures there is the potential for sufficient heat generation to cause unwanted dispersive processes; however with the increased surface to volume ratio of capillary column diameters of less than 250 μm this issue is avoided [23]. Importantly, packing techniques for the fabrication of long capillary columns packed with sub-2 micron columns [24, 25] have been established. Due to these various factors it is clearly advantageous to design a high PC chromatographic system based on utilization of capillary columns.
Recently, in a collaboration with the Jorgenson group at UNC Chapel Hill, we implemented a capillary-based chromatography system with operational capability over the 35–40 Kpsi range. As in the initial description of such a system [26], a constant pressure add-on chromatographic system was fabricated, albeit with certain design differences. The attributes of operation at constant pressure versus constant flow have been recently noted [27 and references provided therein], together with an updated version of the initially described system that is presently being utilized by the Jorgenson group.
Herein we provide a description of the system being used at The University of Kansas, which we term an extreme ultra-pressure liquid chromatography (XUPLC) system. In the present publication emphasis is placed on its application in profiling the photoinduced degradation of the biologically relevant protein rat growth hormone (rGH) (Fig. 2) rather than extensive chromatographic evaluation, which recently has been reported by our collaborators [27]. Of note, in profiling the rGH degradants, we have compared results obtained with the XUPLC system to those obtained using a commercial capillary column and obtained significantly increased resolution of various critical peak groups with the former technique.
Fig. 2.
Sequence of rGH utilized in the present investigation. Met residues are highlighted in red and the two intra-chain disulfide linkages are highlighted in green (53C-C164) and blue (181C-C189), respectively. The underlined residues indicate coverage of the various peptides (some residues are overlapping, refer to Table 2 data) obtained from LCMS analysis of the digested protein
Experimental
Chemicals and Solvents, Devices, Hardware and Tubing
Optima grade water, Optima grade 0.1% formic acid in water, Optima grade 0.1% formic acid in acetonitrile, Optima grade formic acid and HPLC grade acetonitrile were obtained from Fisher Scientific (Fairlawn, NJ). HPLC grade acetone was obtained from Sigma-Aldrich (St. Louis, MO). Formamide (98%) was obtained from Aldrich (Milwaukee, WI) and Kasil 1 (Kasil) was obtained from PQ Corporation (Valley Forge, PA). An Impulse Stripall® Thermal Wire Stripper (model #TWC-1) with a Thermal Controller and mounting was obtained from Telodyne (San Diego, CA). Polyimide coated fused-silica (FS) tubing of various dimensions (15 μm – 180 μm id x 360 μm od) was obtained from Polymicro Technologies (Phoenix, AZ). Unless otherwise noted the various valves, fittings and stainless steel (SS) tubing were obtained from Valco Instruments (Houston, TX).
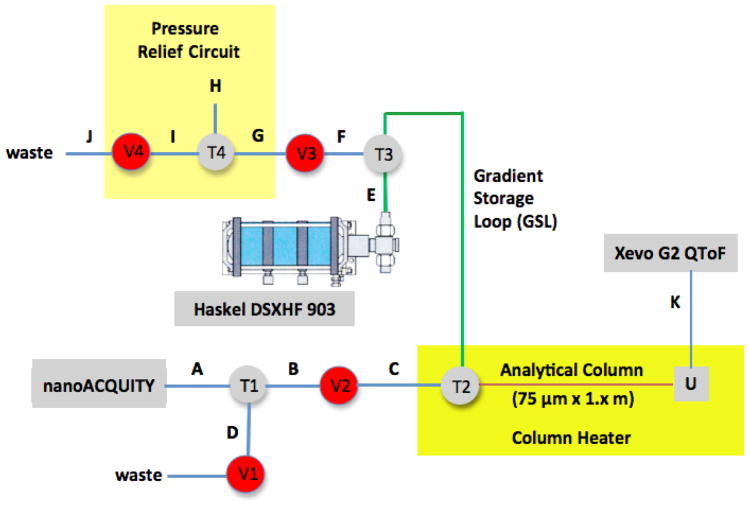
XUPLC Liquid Path Design and Assembly
The liquid flow path for the XUPLC system (Fig. 3) was assembled around a nanoAcquity UPLC and XEVO G2 QToF mass spectrometer (Waters Corporation, Milford, MA) fitted with a nano-electrospray interface (nESI). The various connections were as follows: T1 and T4, 1/16″ micro-volume connectors; T2 and T3, 1/32″ micro-volume connectors; U, PicoClear Union for 360 μm od tubing (New Objective, Woburn, MA); A, FS tubing with frit, 25 μm id x 360 od x 10″ (Waters, #430001570); B, FS tubing with frit, 25 μm id x 360 μm od x 18″ (Waters, #430002242); C, Ni-Clad FS tubing, 50 μm id x 1/32″ od x 25 cm; D, Ni-Clad FS tubing, 50 μm id x 1/16″ od x 25 cm; E, FS tubing, 15 μm id x 360 μm od x 3′; F, SS tubing, 0.005″ id x 1/32″ od x 10 cm; G, SS tubing, 0.005″ id x 1/32″ od x 30 cm; H, FS tubing, 15 μm id x 360 μm od x 2.5 m (Polymicro); I, SS tubing, 0.005″ id x 1/32″ od x 30 cm; J, PTFE tubing, 0.010″ id x /1/16″ od x 2′; K, FS tubing, 20 μm id x 360 od x 50 cm. V1, 10 Kpsi air actuated on/off valve, 1/16″, 0.50 mm bore; V2–V4, 40 Kpsi air actuated on/off valve, 1/16″, 0.15 mm bore. The gradient storage loop (GSL) was assembled using SS tubing, 0.005 ″ id x 1/32″ od x 130′. The various connections involving 1/32″ SS tubing (0.005″ id) were set to the various valves and tees using commercially available SS ferrules and nuts. Connection of FS tubing (360 μm od) to 1/32″ internal ports required a combination of commercial and in-house parts. A PEEK fused silica adapter (1/32″ for 0.20 ≤ 0.25 mm tubing) was fitted to the FS followed by PEEK tubing sleeve (1/32″ od x 0.015″ id x 1″ length). The connection was completed by use of a 1/32″ nut-collet-collar assembly. The pneumatic amplified liquid pump (PALP) with an actual amplification ratio of 1038:1 and an operational limit of 75 Kpsi enabled constant pressure chromatography at 37.4 Kpsi (Model DSXHF 903, Haskel International, Inc., Burbank, CA).
Fig. 3.
Schematic of the liquid flow-path for the In-House XUPLC Add-On Based on a nanoAcquity UPLC and Xevo G2 ESI QToF MS
T1–T4, micro-volume connectors; U, capillary tubing union; A, B, E, F and H, fused silica tubing; C–D, Ni-Clad fused silica tubing; G, I, stainless steel tubing; J, PTFE tubing; V1, 10 Kpsi air actuated on/off valve; V2–V4, 40 Kpsi air actuated on/off valves. Additional details are provided in the experimental section.
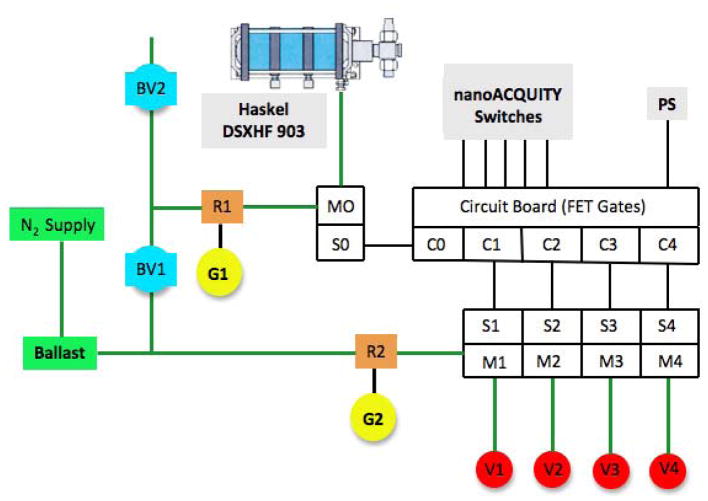
Electrical and Pneumatic Control Circuits
Control of the XUPLC fluid flow path was controlled the electrical and pneumatic circuits that are depicted in Fig. 4. The nanoAcquity back panel on/off switches were used to actuate FET gates (C0–C4) of the circuit board (minor modification of original UNC in-house design). Power was provided for the circuit board and the various solenoids by a 24V direct current power supply. S1–S4 (Humphrey, #HG010LE1 24VDC) and M1–M4 (Humphrey, #HGO10M6A) are solenoid values and the associated manifolds that allow control of the high-pressure on/off values (noted in the previous section). S0 and M0 represent an integrated solenoid-manifold (Humphrey, #320 24VDC) used to control the PALP. The Humphrey solenoids and manifolds were obtained from Skarda Equipment Company, Inc., Omaha, NE. BV1 and BV2 are on-off values (McMaster-Carr, #47865K21, ¼″ NPT female connections, bass ball valve) used to control nitrogen gas access. R1-G1 and R2-G2 are regulators (McMaster-Carr, #4959K21) allowing for final control of nitrogen input pressure; however, gauge G1 was upgraded to an ultra-high accuracy test gauge (McMaster-Carr, #4009K5843, 60 psi limit) to allow precise control of nitrogen pressure to the PALP. The ballast tank (Andy Mark, #am2478, 500 ml capacity, ¼″ NPT ports) was used to buffer inlet nitrogen pressure for the various components. N2 gas was supplied using a liquid nitrogen Dewar. For the primary gas distribution lines polyethylene tubing (0.170″ id, ¼″ od) was used in conjunction ¼″ push-to-connect fittings (McMaster-Carr); however, gas connections to the high-pressure on-off valves (S-M series) were completed using polyurethane tubing (1/8″ id, 3/16″ od, McMaster-Carr #5195T62).
Fig. 4.
Schematic of the electrical and pneumatic circuits used to control the fluidic flow path
Throughout, green lines represent gas connections and black lines electrical connections. C0–C4 are electrical circuits than are controlled by the nanoAcquity real panel on/off switches, while PS represents circuit board direct current power supply. S0–S4 represents solenoid valves that work in conjunction with the gas flow manifolds M0–M4, while BV1and BV2 are on-off ball values. G1 (ultra-high accuracy) and G2 are pressure gauges with R1 and R2 being the associated pressure regulators. Further details are provided in the experimental section.
Capillary Column Packing and Fabrication
The long capillary columns, typical dimensions of 360 μm OD, 75 μm id x ≥ 1.0 m length) were packed in house using CSHTM C18 1.7 μm particles (Waters). The outlet frit was fabricated by the addition of formamide (30 μL) to Kasil (90 μL) contained in a 1.5 mL microcentrifuge tube. The solution was immediately subjected to vortex mixing (~10 s) followed by dipping one end of a blank capillary column into the solution for a period of ~2 s, which resulted in ~ 2–3 mm of the now polymerizing solution to be contained in the capillary. Typically the capillary was heated overnight at 60° C. The terminal length of the capillary was then cut to leave about 1–2 mm of frit material, which was then ready for packing. Packing was accomplished by transferring an acetone slurry (80 mg/mL) of particles (previously sonicated for 10 min) to a packing bomb, subsequently attaching a fritted blank capillary and initially applying 3–4 Kpsi of pressure (acetone used as the packing solvent) until the first 2–3 cm of the column was filled. At this time the pressure was increased to 40 Kpsi for the duration of the packing process, which for 1.3-meter columns typically required about 1–1.25 hours. The remainder of the fabrication process consisted of several steps [25, 28]: a) slow decompression of the packing system, b) connection to a second station where washing was accomplished using 1-1 acetonitrile-water at 50 Kpsi, c) slow decompression, d) recompression to 10 Kpsi, e) formation of a temporary thermal inlet frit using the Teledyne Thermal Wire Stripper, and f) slow decompression), all of which are previously described procedures [25, 28], with the exception of the inlet frit fabrication. Prior to inlet frit formation, the packed capillary was typically set-aside for one to several days, allowing the solvents near column terminus to evaporate. For inlet frit formation, the column was dipped into a solution of Kasil-formamide (1.5–1, v/v), for ~15 s, which resulted in the solution being drawn 1–2 mm into the column. At this point the inlet only was subjected to heating for about 2 h, with the column then being set aside ready for use.
XUPLC Solvent Program, Operation Stages and Chromatography
The XUPLC add-on requires loading the GSL with a solvent program that serves various functions. Mobile phase A (MPA) was Optima grade water with 0.1% formic acid and mobile phase B (MPB) was Optima grade acetonitrile with 0.1% formic acid. Of note, the solvent program operated in a last in first out (LIFO) format. The exact nature of the composition and volume of solvent loaded onto the GSL is presented in Figure 5, where the various stages of the solvent program as associated with the operation stages of a separation are delineated. A discussion of system operation and the role of these various linked operations will be presented in the results and discussion section. The column was housed in a column heating device held at 45 °C, with a sample mass of 0.20 μg injected (2 μL of 4.5 μM protein digest).
Fig 5.
XUPLC separation of control rGH digest, with insets (A) and (B) showing pre-irradiation oxidation of Met residues
UPLC Utilizing a Commercial Column
For comparison purposes, samples determined by XUPLC were additionally analyzed by a commercial UPLC system (Waters nanoAcquity), with the system operated in a non-trapping format. Plumbing from the nanoAcquity injection valve (2 μl loop) to the commercial UPLC column (CSH C18, 75 μm x 25 cm, 1.7 μm particles, Waters), was accomplished using FS tubing with frit, 25 μm id x 360 od x 10″ (Waters, #430001570) from the injection value to a nano-tee (#289002576, Waters) to which the column was attached. The injection loop was filled with 2 μL of sample then the following solvent program was initiated using a flow rate of 0.30 μL/min. Time zero to 0.1 min, 3% MPB; 0.1–50 min, 3–35% MPB; 50–70 min, 35–95% MPB; 70–80 min, 95% MPB; 8–83 min, 95–3% MPB; 83–109 min, 3% MPB. The column was housed in a column heater device held at 45 °C, with a sample mass of 0.20 μg injected (2 μL of 4.5 μM protein digest).
Nano electrospray Ionization Time-of-Flight MS and MS/MS Analysis
The commercial column was connected directly to the nano electrospray interface while the in-house columns connected to the interface as indicated in Fig. 3, with details previously described in a previous section. A stainless steel emitter (#ES523, 50 mm, 30 μm id x 150 mm od, Thermo Scientific) fitted with a 180 μm id x 360 μm sleeve was utilized. Mass spectrometry was performed on a Waters Xevo-G2 (Waters Corporation, Milford, MA) operating in the positive mode. The desolvation gas flow and the desolvation temperature were set to 1000 L/h and 100° C, respectively. The cone gas flow was set to 25 L/h and a source temperature of 100° C. The capillary voltage and cone voltage were set to 2,700 and 45 V, respectively. The Xevo-G2 acquisition rate was set to 0.5 s with a 0.0 s interscan delay. Argon was used as the collision gas. The instrument was operated in the MSE mode. The instrument was operated with the first resolving quadruple in a wide pass mode with the collision cell operating with different alternating energies. To acquire the non-fragmented MS1 spectrum, the collision cell was operated at 3 eV. Fragmented MS2 ion spectra were acquired by ramping the collision cell energies from 18 to 45 eV. The MS and MS/MS spectra obtained by the Q-TOF G2 were analyzed using MassLynx program V4.1 (Waters), with further LC-MS/MS data analysis accomplished using Byonic software (Protein Metrics, San Carlos, CA). The rGH digests were searched against a database with the amino acid sequence of the protein using 10 ppm and 20 ppm mass tolerance parameters for precursor and fragment ions respectively. An observed peak capacity (separation window from first identified peptide till end of data acquisition) was calculated from the peak full width at half maximum (FWHM) for a number of peptides that were fully resolved spanning the early to late elution region. The FWHM values were multiplied by 1.7 to provide the 4σ peak width, with peak capacity then calculated by dividing the separation window (estimated from the time of first to last identified peptides) by the 4σ peak width [18].
Protein production
The rGH plasmid, provided by GenScript (New Jersey, USA) was utilized by KanPro Research Inc. for rGH expression. The plasmid was transfected into BL21 (DE3), a type of E. coli cells, which is mainly used for expression of various proteins [29]. In brief, the E. coli cells were cultured in 10 ml lysogeny broth (LB) culture media with the antibiotics (kanamycin and chloramphenicol) for around 12 Hr. This 10.0 ml culture was added to 1.0 L LB media and was placed in a shaking incubator at 37° C and 200 rpm with the antibiotics (kanamycin and chloramphenicol), until optical density of the media reached a value of 0.6. Isopropyl β-D-1-thiogalactopyranoside (IPTG) (1.0 mM) was added to the LB media and incubation was continued for 4 Hr. Cells were centrifuged at 4,000 rpm for 10 min and the cell pellet was suspended in the lysis buffer (50 mM tris hydrochloride, 500 mM sodium chloride, pH 8). The mixture was then sonicated until it was no longer viscous. Inclusion bodies of rGH were isolated by centrifuging at 19,500 rpm for 30 min followed by a water wash. Inclusion bodies were dissolved in the lysis buffer containing 6 M urea and were refolded by following proprietary procedures and dialyzed in a phosphate buffer saline (PBS) containing 88 mM mannitol.
Photoirradiation
The photoirradiation was carried out in a Rayonet photoreactor RPR-200 [Southern New England Ultra Violet Company (Branford, CT)] equipped with four 35 W low-pressure mercury lamps (RPR-2537Å) at λ = 254 nm. Each sample (200 μL) contained 45 μM (1 mg/mL) of rGH in 25mM pH 7.2 sodium phosphate buffer. Samples were placed in quartz tubes and saturated with Argon (Ar) prior to photoirradiation for a period of 30 minutes.
Digestion Protocol
For peptide mapping of the rat growth hormone (rGH), 200 μL of either the control or the stressed samples were mixed with 100μL of 50mM ammonium bicarbonate buffer (pH 8.2), 25 μL of guanidine hydrochloride solution (6 M, in ammonium bicarbonate buffer, 50 mM, pH 8.2) and 50 μL dithiothreitol solution (25 mM) for the reduction of the disulfide bonds. The mixtures were incubated at 50° C for 30 min. The thiolate groups of the reduced cysteine residues were then alkylated by the addition of 50 μL of iodoacetamide (100 mM). After 1 hour of incubation in the dark at 37° C, the protein was precipitated with 500μL of 0.5 M perchloric acid, and centrifuged at 14,000 g for 10 min at 4°C. The resulting pellet was washed twice with milliQ water then reconstituted in 200 μL of ammonium bicarbonate buffer (50 mM, pH 8.2). Digestion was performed by the addition of 10 μg of trypsin/chymotrypsin with a ratio [protein]: [enzyme] = 20 for each sample. After overnight incubation at 37°C, peptides were purified using Amicon ultra 0.5 centrifugal devices equipped with 10 kDa membranes in order to separate undigested protein and trypsin/chymotrypsin from the proteolytic peptides.
Results and Discussion
Aspects of XUPLC Operation and Solvent Program
Mass Lynx files controlled the XUPLC system, which consists of a series of values, a pneumatic amplified liquid pump (PALP), a gradient storage loop (GSL) a capillary chromatography column and a series of tees, connectors and tubing as described in the experimental section (Fig 3). Gradient storage loops have been previously described in liquid chromatography, an early application described by Knox [30] for gradient formation prior to the availability of suitable two-pump system controllers, with more recent examples in capillary HPLC [31] and UPLC [26, 27 and references cited therein]. The system illustrated in Fig. 3 represents some modifications as compared to previous versions [26, 27]. Early designs [26] were absent T4 and V4 and as a result at the end of a chromatographic run valve V3 was opened resulting in rapid system depressurization, thus resulting in stress on the inlet Kasil frit. In an effect to circumvent this situation, the present design (Fig. 4) implements these elements, with the tubing segment H being fused silica of 15 μm id x 2.5 m in length. At the end of a run, V3 is opened for a period of 3.7 min, with the now accessible 15 μm id capillary providing a significant resistance to liquid flow, thus slowing the depressurization and minimizing the associated stress on the inlet frit. While the rate of depressurization was not monitored, the electrospray process was observed to halt over approximately a two-minute period.
The overall operation of the XUPLC system can be divided into nine distinct stages, as delineated in Table 1. As noted previously, utilizing a GSL results in a last in first out (LIFO) format. Per the solvent program used, this results in a 10 μL of 7% MPB flowing through the capillary column prior to encountering the gradient program (7%–45% MPB). The remaining solvent program entails an isocratic hold at 45% MPB, a pulse of 70% MBP and a return to 7% MPB. Following the chromatographic step, the previously noted decompression stage occurs followed by reloading the GSL with 7% MPB that is used for re-equilibration over a 35 minute period. Of note, data acquisition occurs over a 275 minute period (stopped at this point due to file size limitations) followed by continued operation of the system for a total of 295 minutes; however, with an observed flow rate of 0.22 μL/min (measured with 50% MPB) this is insufficient time to playback the entire solvent program. Aspects of gradient playback in terms of the observed chromatography will be discussed in a subsequent section.
Table 1.
Detailed description of fluids loaded into the Gradient Storage Loop together with a summary of the various aspects of fluidic control (refer to Fig. 3 for description of fluid connections, valves and the pneumatic pump).
| Time (min) | Relative Time | Pneumatic Pump | Valve 1 | Valve 2 | Valve 3 | Valve 4 | MPB % | Flow (a) (μL/min) | Volume (μL) |
|---|---|---|---|---|---|---|---|---|---|
| Gradient Method Loading | |||||||||
|
| |||||||||
| 0.00 | −32.50 | Off | Closed | Open | Open | Open | 7 | 4.00 | − |
| 0.80 | −31.70 | Off | Closed | Open | Open | Open | 7 | 4.00 | 3.20 |
| 0.90 | −31.60 | Off | Closed | Open | Open | Open | 70 | 4.00 | 0.40 |
| 6.40 | −26.10 | Off | Closed | Open | Open | Open | 45 | 4.00 | 22.00 |
| 6.50 | −26.00 | Off | Closed | Open | Open | Open | 45 | 4.00 | 0.40 |
| 29.50 | −3.00 | Off | Closed | Open | Open | Open | 7 | 4.00 | 92.00 |
| 30.00 | −2.50 | Off | Closed | Open | Open | Open | 7 | 4.00 | 2.00 |
| Sample Loading | |||||||||
|
| |||||||||
| 30.00 | −2.50 | Off | Closed | Open | Open | Open | 7 | 4.00 | − |
| 32.00 | −0.50 | Off | Closed | Open | Open | Open | 7 | 4.00 | 8.00 |
| Switch to XUPLC Ready State | |||||||||
|
| |||||||||
| 32.00 | −0.50 | Off | Open | Closed | Closed | Closed | 50 | 4.00 | (b) |
| 32.50 | 0.00 | Off | Open | Closed | Closed | Closed | 50 | 4.00 | (b) |
| Initiate XUPLC and Data Acquisition | |||||||||
|
| |||||||||
| 32.50 | 0.00 | On | Open | Closed | Closed | Closed | (c) | 0.22 | - |
| 307.50 | 275.00 | On | Open | Closed | Closed | Closed | (c) | 0.22 | 60.50 |
| Data Acquisition Terminates | |||||||||
|
| |||||||||
| 307.50 | 275.00 | On | Open | Closed | Closed | Closed | (c) | 0.22 | - |
| 327.50 | 295.00 | On | Open | Closed | Closed | Closed | (c) | 0.22 | 4.40 |
| Switch to Depressurization Mode | |||||||||
|
| |||||||||
| 327.50 | 295.00 | Off | Closed | Closed | Open | Closed | - | - | - |
| 330.50 | 298.00 | Off | Open | Closed | Open | Open | - | - | - |
| 331.00 | 298.50 | Off | Open | Open | Open | Open | - | - | - |
| 331.20 | 298.70 | Off | Closed | Open | Open | Open | - | - | - |
| Equilibration Method Loading (d) | |||||||||
|
| |||||||||
| 332.50 | 300.00 | Off | Closed | Open | Open | Open | 7 | 4.00 | - |
| 338.55 | 306.05 | Off | Closed | Open | Open | Open | 7 | 4.00 | 24.20 |
| Switch to XUPLC Equilibration (d) | |||||||||
|
| |||||||||
| 338.90 | 306.40 | On | Open | Closed | Closed | Closed | 50 | 4.00 | - |
| 339.00 | 306.50 | On | Open | Closed | Closed | Closed | 50 | 4.00 | - |
| Column Equilibration | |||||||||
|
| |||||||||
| 339.00 | 306.50 | On | Open | Closed | Closed | Closed | 7 | 0.22 | - |
| 374.00 | 341.50 | On | Open | Closed | Closed | Close | 7 | 0.22 | 7.70 |
| Return to Gradient Loading Method | |||||||||
4.00 μL/min flow is from the nanoAcquity and 0.22 μL/min from XUPLS system with 50% MPB
Washing lines and venting to waste per Fig 3.
Gradient program playback
System pause prior to initiation of next stage
XUPLC System Robustness
An advantage of the XUPLC system is that it can largely be assembled from off-the-shelf commercial components; however, a major concern is the trouble free function of the various components on a day-to-day basis. Experience in these laboratories has shown the most frequently encountered operational failure lies in the integrity of on/off valve V2 (Fig. 4). Subsequent to each V2 failure, examination of the column inlet revealed a void over the first few millimeters. Clearly catastrophic failure of the inlet Kasil frit has occurred with pressure cycling leading to particles exiting the column and lodging in the valve mechanism. Apparently over time this results in surface abrasion and eventual valve failure and to date there have been no exceptions to this scenario. In further support of this conclusion, V3 is subject to the same high pressures but was not observed to fail at pressures up to 40 Kpsi over an extended period of time. These results were obtained with the initially used base-system, which did not contain the pressure relief circuit (PRC) shown in Fig. 3. Due to this continued problematic aspect, the basic compromised was modified by adding the PRC, consisting of V4 and the resistance capillary H as illustrated in Fig 3. It was postulated that this modification would provide for a diminished decompression rate subsequent to a run, creating less stress on the inlet frit and concomitantly serving to protect V2 (circuit dimension and operation details provided in the previous section). In an effort to further mitigate this issue, the capillary columns were installed in a reverse fashion, as compared to the packing configuration (refer to the experimental section), with the Kasil outlet (2–3 mm) being used as the inlet in the chromatographic system. Due to the heating process utilized in the fabrication of this frit, it was posited that an increased ruggedness would result. The combination of these two factors did result in an increased column integrity, thus allowing for routine operation for periods from several weeks to 3 months; however, as yet there is no parameter identified that allows prediction of the column inlet frit lifetime.
The use of Kasil (solution of potassium silicate) frits in packed bed capillary liquid chromatography has been established for many years, with an early example being that of Cortes et al. [32]. In a typical example, formamide is added to a solution of potassium silicate, which initiates a polymerization sequence (polysiloxane bond formation), eventually leading to the formation of a highly interconnected porous gel [33]. In a packed capillary column this process occurs in the void space around the particles leading to an entrapping network that functions as a “chemical frit”. When silicates undergo polymerization in the absence of formamide, a poorly interconnected micro-porous gel forms, in contrast to the result occurring in the presence of formamide (thought to undergo hydrolysis to formic acid and ammonia) where a gel structure is formed that exhibits increased pore radius and specific pore volume or a mesoporous material with good chromatographic permeability [32, 33]. Unfortunately the skeleton of such mesoporous gels will constitute a smaller fraction of the overall volume and will be more susceptible to mechanical stress. In contrast to silica-based bonded phases, these bare silica chemical frits possess surface sites highly accessible to chemical attack and thus create skeletal defects, which over time will additionally weaken the gel structure. In the initial capillary chromatography applications, these chemical frits were not subject to the much higher decompression stress as encountered in the present work. Due to this apparent weak point, the present system design incorporating the aforementioned pressure relief circuit was implemented and has provided for an improved but not indefinite column lifetime.
XUPLC Separation of rGH Digest Control Samples
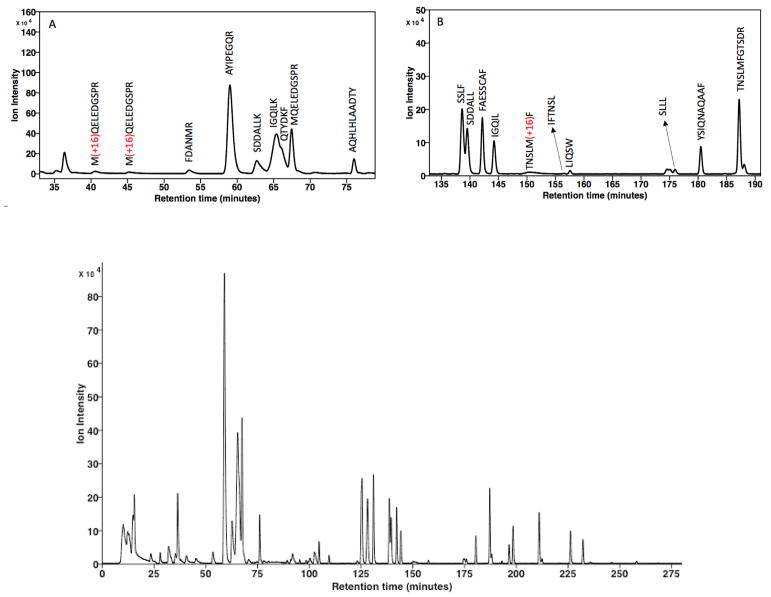
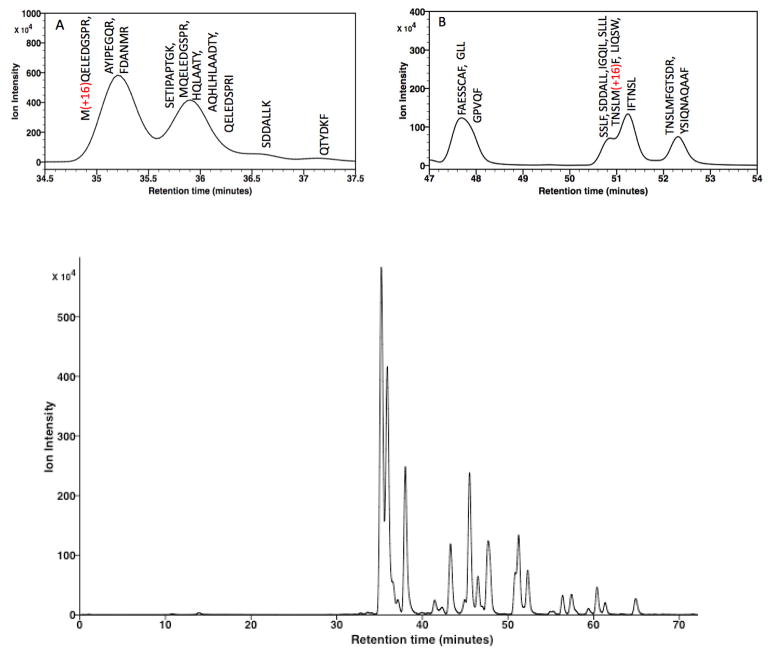
As noted previously, playback of the contents of the GSL (Table 1) occurred over a 295 min period, with a flow rate of 0.22 μL/min this constitutes a total volume of 64.9 μL. A typical chromatogram for a control rGH digest is shown in Fig. 5 where the first identified peptide is observed near 41 min (Fig. 5, inset A), with all identified peaks listed in Table 2. The observed retention values are highly reproducible with and with RSD < 0.15 (n = 3), except for the arginine containing peptide 4 (RSD 0.34). None of the peaks eluting prior to this peptide have been identified and appear to be late eluting substances from a previous injection; however data analysis with the Byonic software program indicated > 92% coverage.
Table 2.
XUPLC reproducibility for control samples of rGH digest (n = 3)
| # | Peptide | Residue | m/z | RT (min) | SD (min) | %RSD |
|---|---|---|---|---|---|---|
| 1 |
|
124-133 | 1117.52 | 41.27 | 0.112 | 0.11 |
| 2 |
|
124-133 | 1117.52 | 45.39 | 0.107 | 0.11 |
| 3 | FDANMR | 145-150 | 753.33 | 53.59 | 0.181 | 0.34 |
| 4 | AYIPEGQR | 35-42 | 933.47 | 59.00 | 0.108 | 0.18 |
| 5 | SETIPAPTGK | 55-64 | 1000.53 | 65.39 | 0.061 | 0.09 |
| 6 | QELEDGSPRI | 125-134 | 1030.48 | 65.39 | 0.072 | 0.11 |
| 7 | SDDALLK | 151-157 | 648.69 | 66.14 | 0.127 | 0.19 |
| 8 | IGQILK | 135-138 | 671.80 | 66.14 | 0.115 | 0.17 |
| 9 | MQELEDGSPR | 124-133 | 1161.49 | 67.42 | 0.072 | 0.11 |
| 10 | HQLAADTY | 22-29 | 918.40 | 70.75 | 0.111 | 0.16 |
| 11 | QTYDKF | 140-145 | 801.87 | 70.84 | 0.075 | 0.11 |
| 12 | AQHLHQLAADTY | 18-29 | 1367.61 | 75.94 | 0.042 | 0.06 |
| 13 | TNSLM | 98-102 | 565.26 | 78.18 | 0.139 | 0.18 |
| 14 | GLL | 160-162 | 302.20 | 102.99 | 0.171 | 0.17 |
| 15 | RFAESSCAF | 183-191 | 1074.46 | 104.79 | 0.087 | 0.08 |
| 16 | TDMELLR | 71-77 | 877.44 | 125.65 | 0.110 | 0.09 |
| 17 | GPVQF | 88-92 | 547.28 | 125.65 | 0.135 | 0.11 |
| 18 |
|
53-64 | 1307.60 | 128.29 | 0.114 | 0.09 |
| 19 | SIQNAQAAF | 44-52 | 949.47 | 131.36 | 0.122 | 0.09 |
| 20 | SSLF | 8-11 | 453.51 | 138.84 | 0.167 | 0.12 |
| 21 | SDDALL | 151-156 | 633.31 | 139.75 | 0.168 | 0.12 |
| 22 |
|
184-191 | 918.34 | 142.35 | 0.123 | 0.09 |
| 23 | IGQIL | 134-138 | 543.35 | 144.48 | 0.164 | 0.11 |
| 24 |
|
98-103 | 728.33 | 152.39 | 0.197 | 0.11 |
| 25 | IFTNSL | 96-101 | 694.37 | 156.92 | 0.112 | 0.07 |
| 26 | LIQSW | 82-86 | 646.35 | 157.77 | 0.137 | 0.09 |
| 27 | SLLL | 79-82 | 445.58 | 176.30 | 0.246 | 0.14 |
| 28 | YSIQNAQAAF | 43-52 | 1188.56 | 180.80 | 0.174 | 0.10 |
| 29 | TNSLMFGTSDR | 98-108 | 1228.56 | 187.56 | 0.193 | 0.10 |
| 30 | DLHKAETY | 168-175 | 975.46 | 191.20 | 0.127 | 0.07 |
| 31 | TDMELL | 71-75 | 721.34 | 196.96 | 0.250 | 0.13 |
| 32 | LGPVQF | 87-92 | 660.37 | 198.36 | 0.193 | 0.10 |
| 33 | SCF | 163-165 | 413.37 | 198.78 | 0.302 | 0.15 |
| 34 | FSLL | 78-81 | 479.28 | 211.50 | 0.217 | 0.10 |
| 35 | DLEEGIQAL | 115-123 | 987.49 | 226.67 | 0.214 | 0.09 |
| 36 | EEGIQAL | 117-123 | 760.38 | 232.67 | 0.217 | 0.09 |
| 37 | LIQSWL | 82-87 | 759.44 | 232.67 | 0.217 | 0.09 |
| 38 | MFPAMPL | 1-7 | 806.39 | 258.63 | 0.261 | 0.10 |
It is instructive to review the observed chromatogram in context with the solvent program and in particular the volumes of the various components of the fluid flow path from the GSL to the MS. Unfortunately a system dead-time marker was not utilized; however since the dimensions of the various components are known, the various volumes and associated dead time for an unretained substance can be estimated. From column inlet through electrospray emitter the system volume is calculated to be 3.65 μL or in terms of time 16.1 min (column porosity εT 0.65 was assumed). The next section of the GSL contained the sample plus loop wash volume (2 μL + 6 μL). This represents times of 9.01 min and 27.27 min, respectively, with the time now totaling ~53 min where elution occurs under isocratic conditions. For unretained sample components, the system volume would be ~25 min, thus is appears the first identified peak (elution time at 41 minutes, Table 2) was minimally retained (est. k′ 0.5). The next solvent segment was a 2.0 μL volume (9.01 min) isocratic segment (7% MPB), which brings the total volume to 13.65 μL (62.06 min) prior to the start of the gradient solvent program. Playback of the gradient solvent program occurred over the period of 62–295 minutes (data collection stopped at 275 min due to file size limitations) or 46.8 μL (data acquisition) and 51.2 μL (total playback). Since the gradient solvent program of 92 μL ranges from 7% – 45% MPB, the solvent program during data collection ranged from 7% to 26.3% MPB, while the continuing playback ranged from 26.3% to 28.2% MPB. At this point, the system entered the decompression mode, followed by a subsequent initial state equilibration solvent (24.2 μL of 7% MPB) being loaded into the GSL, followed by an equilibration period of 35 min (7.7 μL), which represents ~2.3 column volumes. While this may seem as a minimal time (volume) for equilibration, it should be noted that the sample and initially encountered solvent program are of the same composition, effectively representing a continuing isocratic re-equilibration volume and resulting in excellent peak retention reproducibility (Table 2).
Referring to the chromatograms shown in Fig. 5 there are several points of interest. The first two peptides identified (inset A, tR 41.3 and 45.4 min) elute under isocratic conditions and represent pre-irradiation oxidation of residue 124 (peptides 1 and 2, sequence 124–133), with another such site being observed during gradient elution (inset B, tR 152.4 min) at residue 102 (peptide 24, sequence 98–103). Since the oxidation of Met residues result in the formation of a new stereocenter of either R or S configuration, the transformation of residue 124 results in diastereomerically related peptides that are resolved in this case. When the sample was subjected to UPLC these products co-eluted (data presented and discussed in a subsequent section). A similar situation exists for the later eluting peptide; however, only a single peak was observed despite the potential for diastereomer formation. One may speculate that in the former situation the oxidation site lies at the N-terminus and perhaps is more accessible for interaction with the retention phase as compared to the latter case where the oxidation site lies within the peptide sequence. Additional XUPLC chromatograms are provided in Supplemental Data.
XUPLC Peak Capacity, Gradient Capacity Factor and the Solvent Program
Calculation of what is presently termed an observed peak capacity (PCobs) was performed for a series of peptides that were fully resolved over the entire chromatogram shown in Fig. 5, with these data summarized in Table 3. The separation window is under a combination of isocratic and gradient elution conditions (isocratic solvent program until 62 min, noted in previous section), and since the first the eluting peptides (Table 3, peptides 1, 2 and 4) were not subject to gradient peak compression, it is instructive to examine these data in more detail. The diminished PCobs for peptides 1 and 2 (tailing peak shape) is readily attributed to the injection process where a large injection volume (2μL sample) is being introduced onto the column relative to the column dead time (est. 3.39 μL). Peptide 4 also exhibits diminished PCobs and of note it undergoes elution while the solvent program is still in the isocratic mode. The remaining peptides, which elute under gradient conditions, all have significantly larger PCobs values, with the exception of peptide 18. Examining structural aspects of peptides 1, 2, 4, and 18, one immediately notes the presence of arginine and lysine residues. In the acidic mobile phase employed each of these peptides exhibits positive charges sites on the R-groups, with hydrophobic methylene groups separating these sites from the peptide backbone, thus potentially providing for a combination of an electrostatic and hydrophobic interaction with the retention phase and residual silanol groups to result in diminished chromatographic efficiency.
Table 3.
XUPLC peak capacity for selected peptides spanning the retention window
| # | Peptide | Residue | m/z | RT (min) | PW (min) | PC |
|---|---|---|---|---|---|---|
| 1 |
|
124-133 | 1117.52 | 41.27 | 1.22 | 192 |
| 2 |
|
124-133 | 1117.52 | 45.39 | 1.11 | 211 |
| 4 | AYIPEGQR | 35-42 | 933.47 | 59.00 | 0.69 | 342 |
| 12 | AQHLHQLAADTY | 18-29 | 1367.61 | 75.94 | 0.32 | 734 |
| 18 |
|
53-64 | 1307.60 | 128.29 | 0.62 | 378 |
| 19 | SIQNAQAAF | 44-52 | 949.47 | 131.36 | 0.36 | 656 |
| 22 |
|
184-191 | 918.34 | 142.35 | 0.51 | 458 |
| 23 | IGQIL | 134-138 | 543.35 | 144.48 | 0.43 | 543 |
| 28 | YSIQNAQAAF | 43-52 | 1188.56 | 180.80 | 0.35 | 673 |
| 31 | TDMELL | 71-75 | 721.34 | 196.96 | 0.43 | 551 |
| 32 | LGPVQF | 87-92 | 660.37 | 198.36 | 0.38 | 615 |
| 34 | FSLL | 78-81 | 479.28 | 211.50 | 0.41 | 580 |
| 35 | DLEEGIQAL | 115-123 | 987.49 | 226.67 | 0.35 | 679 |
| 36 | EEGIQAL | 117-123 | 760.38 | 232.67 | 0.35 | 673 |
| 37 | LIQSWL | 82-87 | 759.44 | 232.67 | 0.35 | 673 |
Considering the subset of peptides in Table 3 (peptides 12–37) that elute under gradient elution conditions it is instructive to calculate the instantaneous gradient retention factor k* (equation 5). Using the mean molecular weight (MWmean 886.9) of these peptides, with all the other needed parameters being readily available, k* 8.35 was calculated, which is generally considered to be within an optimum range from a resolution and time perspective [34]. In HPLC, gradients are frequently implemented and adjusted from a %B/min perspective, with typical values being 1–2 %B/min being widely adopted. Examination of the XUPLC gradient from this perspective reveals the gradient program was 0.091% MPB/min, seemingly a very shallow program; however, this view is in terms of a conventional dimension HPLC column of 4.6 mm id x 150 mm in length. To appreciate the gradient program for the presently used capillary column, it is useful to examine equation 6, where the parameter tG•F/Vm represents the gradient volume in terms of column volume (Vm). Expressing the gradient program in terms of % MPB in terms of the number of column volumes (#Cvol) leads to equation 6 where the present numerical values are shown (VG, gradient volume during the period for peak capacity calculation).
| (6) |
In the case of conventional HPLC, utilizing a typical gradient program (Δ B% 1.0, F 2.0 mL/min, tG 50 min, column dimensions as previously noted), this calculation leads to 1.62% MPB/Cvol, a similar value to the presently used XUPLC gradient program. These calculations illustrate the importance of viewing the gradient elution program in terms of solvent strength change per column volume regardless of the column geometry. In the case of capillary columns, characterization of the solvent program in terms of %B per time is highly misleading due to the proportionally larger time required per column volume. In any case, the XUPLC k* value does lie within a generally accepted optimal range. From the measured flow rate and the estimated column porosity, the linear velocity was calculated to be 1.89 mm/s or in terms of reduced velocity 7.72 (diffusion coefficient was estimated for the median mobile composition and temperature used during gradient elution, MWmean, and dp 1.9 μm [35, 36]). While not established in the present research, in terms of typical van Deemter plots, these values lie in an optimal region with respect a performance with respect to time (linear velocity) [23].
UPLC Separation of rGH Digest Control Samples
For comparison, separations of the rGH control digest samples were subjected to commercial UPLC, with the resulting chromatograms shown in Fig. 6 and the data summarized in Table 4. The UPLC separations were conducted in the non-trapping model, which is not time efficient but was thought to serve as a better format for a comparison to the XUPLC separations. When the fluid flow path volume was estimated, from the injection loop to the emitter tip, one arrives at a volume 3.25 μL requiring 10.83 minutes or to column inlet 7.56 minutes. Elution of the initial identified peptides (peptides 1 and 2 co-elute but are the same as the XUPLC experiment, refer to Table 3) occurred at 32.33 min. Of note, the solvent composition at the initiation of the gradient program and for the sample was 3% MPB, weaker than that of the XUPLC starting point. While no substance was included as a void volume marker, it is clear that these initially eluting peptides are subject to chromatographic retention. Upon inspection of the chromatograms displayed in Fig. 6, one notes an overall lack of chromatographic resolution as compared to the previously presented XUPLC separation. Of particular note are the insets A and B where the pre-irradiated oxidation peaks are identified, but along with numerous other peptides constitute co-eluting bands. These various substances were identified from the MS fragmentation data; however, the chromatograms clearly illustrate the need for high PC chromatographic systems. Analysis of the gradient program using MWave to estimate the S value (as for the XUPLC calculation) one can calculate k* 8.06, 0.64% MPB/min, and 1.53% MPB/Cvol. These values are similar to those of the XUPLC separation with a slight increase of the gradient program steepness based on Δ %MPB per column volume. Review of the peptide retention data of Table 4 reveals that all of the previously identified peptides were again found; however, in numerous cases there is a change in retention order or gradient selectivity. While not within the scope of the present research, such selectivity differences in gradient elution are commonplace for irregular samples [18] when there are even small changes in the effective steepness as in the present situation. The diminished resolution for the 25 cm commercial column compared to the 118 cm in-house fabricated XUPLC column is to a large extent explained by the large increase in theoretical plates that are available in the latter case. Additional UPLC chromatograms are included in Supplemental Data.
Fig 6.
UPLC separation of control rGH digest, with insets (A) and (B) showing pre-irradiation oxidation of Met residues
Table 4.
UPLC reproducibility for control samples of rGH digest (n = 3), with reference to the observed XUPLC retention order
| # | Ref | Peptide | Residue | m/z | RT (min) | SD (min) | %RSD |
|---|---|---|---|---|---|---|---|
| 1 | 1 |
|
124-133 | 1117.52 | 34.72 | 0.134 | 0.29 |
| 2 | 2 |
|
124-133 | 1117.52 | 34.72 | 0.134 | 0.29 |
| 3 | 4 | AYIPEGQR | 35-42 | 933.47 | 35.20 | 0.195 | 0.55 |
| 4 | 3 | FDANMR | 145-150 | 753.33 | 35.20 | 0.169 | 0.48 |
| 5 | 5 | SETIPAPTGK | 55-64 | 1000.53 | 35.91 | 0.193 | 0.54 |
| 6 | 9 | MQELEDGSPR | 124-133 | 1161.49 | 35.91 | 0.197 | 0.55 |
| 7 | 10 | HQLAADTY | 22-29 | 918.40 | 35.91 | 0.191 | 0.53 |
| 8 | 12 | AQHLHQLAADTY | 18-29 | 1367.61 | 35.91 | 0.218 | 0.67 |
| 9 | 6 | QELEDGSPRI | 125-134 | 1030.48 | 35.91 | 0.179 | 0.50 |
| 10 | 7 | SDDALLK | 151-157 | 648.69 | 36.60 | 0.123 | 0.34 |
| 11 | 11 | QTYDKF | 140-145 | 801.87 | 37.20 | 0.231 | 0.62 |
| 12 | 8 | IGQILK | 135-138 | 671.80 | 38.01 | 0.198 | 0.51 |
| 13 | 15 |
|
183-191 | 1074.46 | 41.42 | 0.181 | 0.44 |
| 14 | 13 | TNSLM | 98-102 | 565.26 | 42.20 | 0.186 | 0.44 |
| 15 | 18 |
|
53-64 | 1307.60 | 43.28 | 0.174 | 0.40 |
| 16 | 16 | TDMELLR | 71-77 | 877.44 | 45.51 | 0.157 | 0.34 |
| 17 | 19 | SIQNAQAAF | 44-52 | 949.47 | 46.49 | 0.141 | 0.30 |
| 18 | 22 |
|
184-191 | 918.34 | 47.68 | 0.122 | 0.26 |
| 19 | 17 | GPVQF | 88-92 | 547.28 | 47.68 | 0.124 | 0.26 |
| 20 | 14 | GLL | 160-162 | 302.20 | 47.68 | 0.174 | 0.36 |
| 21 | 20 | SSLF | 8-11 | 453.51 | 50.82 | 0.143 | 0.30 |
| 22 | 21 | SDDALL | 151-156 | 633.31 | 50.82 | 0.186 | 0.50 |
| 23 | 23 | IGQIL | 134-138 | 543.35 | 50.82 | 0.152 | 0.48 |
| 24 | 27 | SLLL | 79-82 | 445.58 | 50.82 | 0.252 | 0.39 |
| 25 | 24 |
|
98-103 | 728.33 | 51.23 | 0.237 | 0.41 |
| 26 | 26 | LIQSW | 82-86 | 646.35 | 51.23 | 0.247 | 0.26 |
| 27 | 25 | IFTNSL | 96-101 | 694.37 | 51.23 | 0.202 | 0.42 |
| 28 | 30 | DLHKAETY | 168-175 | 975.46 | 51.40 | 0.135 | 0.26 |
| 29 | 29 | TNSLMFGTSDR | 98-108 | 1228.56 | 52.31 | 0.220 | 0.42 |
| 30 | 28 | YSIQNAQAAF | 43-52 | 1188.56 | 52.31 | 0.246 | 0.47 |
| 31 | 32 | LGPVQF | 87-92 | 660.37 | 54.94 | 0.129 | 0.23 |
| 32 | 31 | TDMELL | 71-75 | 721.34 | 57.41 | 0.143 | 0.25 |
| 33 | 33 |
|
163-165 | 413.37 | 57.45 | 0.174 | 0.30 |
| 34 | 35 | DLEEGIQAL | 115-123 | 987.49 | 59.38 | 0.122 | 0.21 |
| 35 | 34 | FSLL | 78-81 | 479.28 | 60.39 | 0.180 | 0.30 |
| 36 | 36 | EEGIQAL | 117-123 | 760.38 | 61.34 | 0.181 | 0.30 |
| 37 | 37 | LIQSWL | 82-87 | 759.44 | 61.34 | 0.175 | 0.29 |
| 38 | 38 | MFPAMPL | 1-7 | 806.39 | 64.92 | 0.197 | 0.30 |
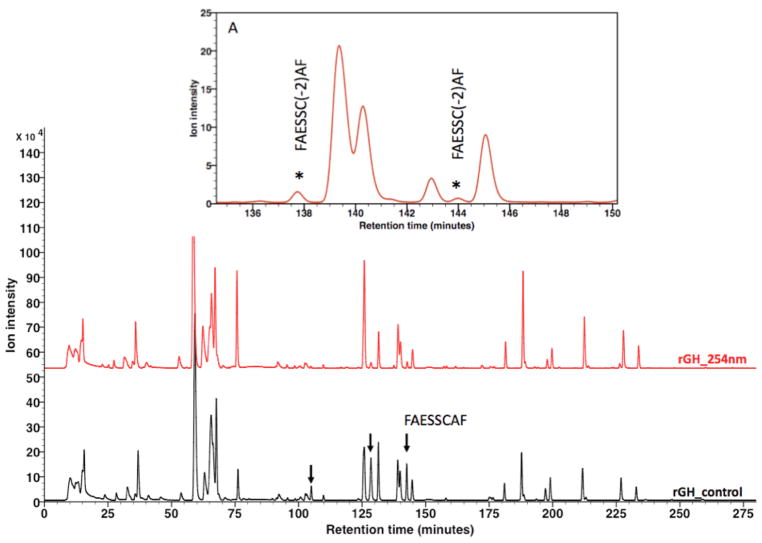
Profiling Photochemical Induced Degradation of rGH
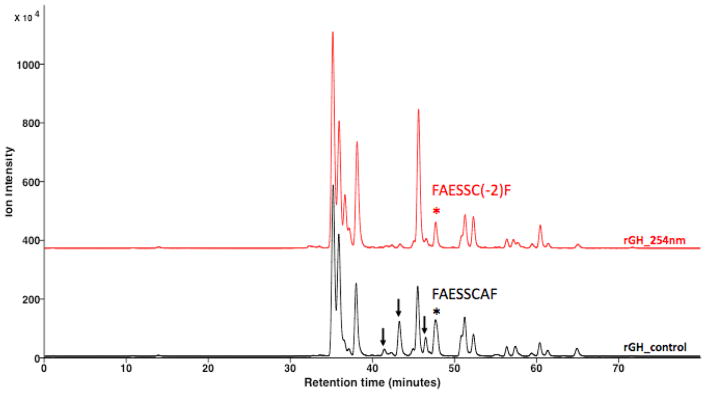
The photochemical mediated degradation of proteins to a variety of first, second and subsequent generation products [9–14] have been established as previously discussed. To explore the utility of XUPLC in profiling protein degradation and identification of various degradants, rGH was selected as a model substance and subjected to 254 nm light, followed by reduction, alkylation and enzymatic digestion (details provided in the experimental section). XUPLC of irradiated rGH subjected to this protocol resulted in the chromatogram of Fig. 7 where one notes that some of the control sample bands have diminished, with newly formed substances appearing in the irradiated sample. Focusing on one band in particular, degradation of peptide FAESSCAF (tR 142.35 min, refer to the structural assignment of peptide 22 in Table 4) results in the formation of two chromatographically resolved products (inset A, tR 137.7 and 142.35 min). The peptide ladder for the parent is shown in Fig. 9, with the same data for the degradants shown in Fig. 10. Based on the present MS data together with the established chemistry for photochemical induced degradation of disulfide linkages (Fig. 1), these products were assigned as diastereomerically related cis/trans isomers; however, with the present data it is not possible to establish specific structural assignments. UPLC separation of these samples lead to the chromatograms shown in Fig. 8 where the parent peptide FAESSCAF (tR 47.6 min) and the associated degradants FAESSC(-2)AF were observed to be unresolved, although structurally identified in the MS dimension.
Fig 7.
XUPLC separation of photochemical induced versus control of rGH digest, inset A indicates identified degradation products that are isobaric. Arrows (↓) indicate several peptides that are diminishing over time when subjected to 254 nm light including a cysteine containing peptide (tR 142.35) that undergoes transformation to an apparent cis-trans pair of ene-thiols (tR 137.7 and 144 min).
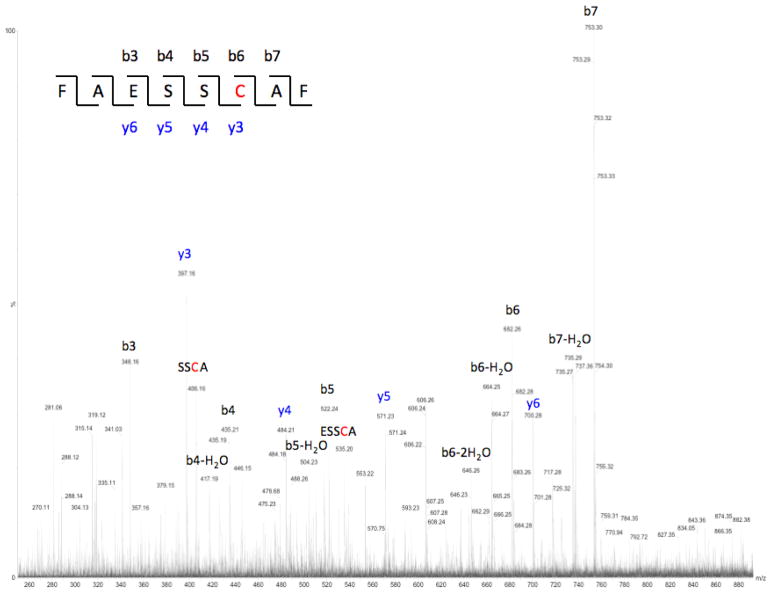
Fig. 9.
CID mass spectrum of a peptide from digestion of a control rGH sample that possesses a cys residue (181C) that is a potential site for photochemical induced degradation.
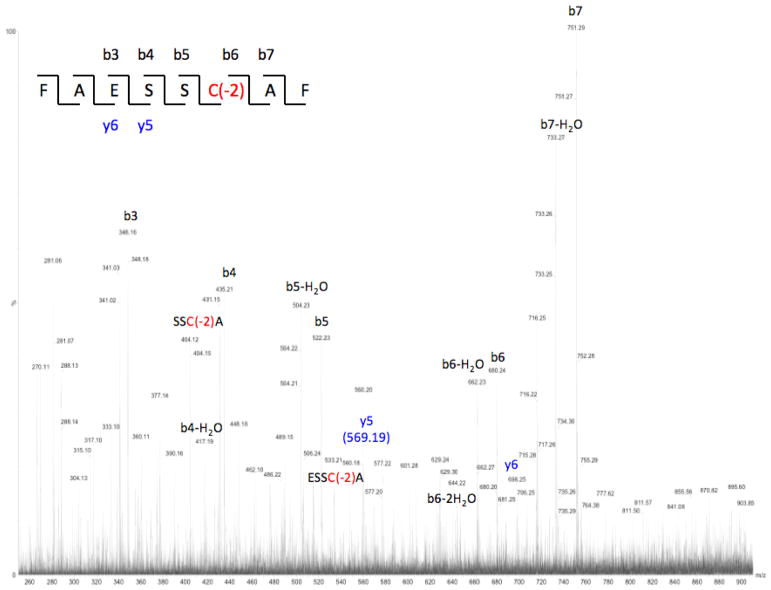
Fig. 10.
CID mass spectrum of a peptide resulting from digestion of rGH possessing a cys residue (181C) that was found to have undergone photochemical induced degradation when subjected to 254 nm light.
Fig. 8.
UPLC separation of photochemical induced versus control of rGH digest showing several peptides that are subject to photochemical mediated degradation (↓) including the peptide FAESSCAF (tR 47.68 min) whose degradation product is unresolved from the parent substance.
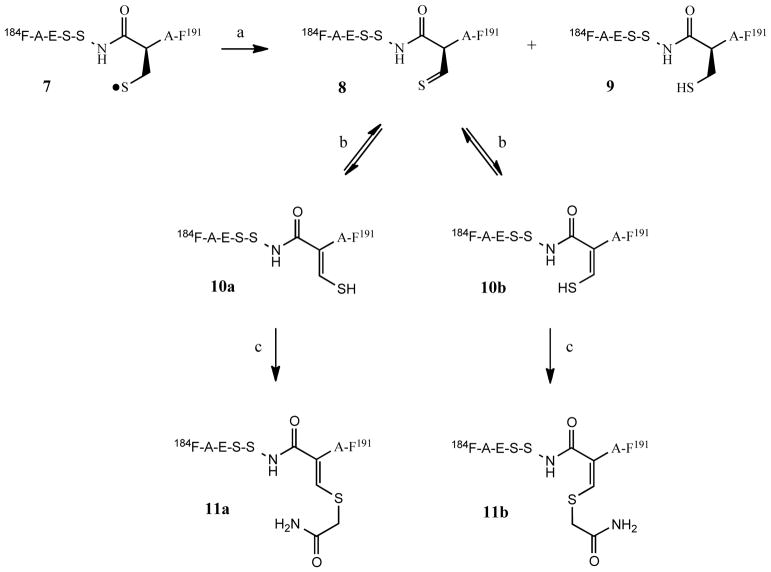
Further to the general reaction scheme of Fig. 1, it is instructive to specifically examine the origin of the identified products. Referring to the primary sequence of rGH (Fig. 2), one can conclude that these products arise from photolysis of the 181C-C190 disulfide linkage near the carboxylic terminus, which is shown by the reaction sequence of Fig. 11. As regards identification of residues near this region, the sequence from 176L-R182 was not identified, thus the fate one of the cysteine residues (C181) of the disulfide bond of interest remained undetected (refer to Table 2). Returning to the reaction scheme of Fig. 11, identification of the reactions responsible for the formation of the degradation products, including the origin of the cis/trans isomers is relatively straightforward. The initially formed thiyl radical (7) undergoes an established disproportionation process leading the formation of the thioaldehyde (8) and the thiol (9), which has previously been identified as peptide 22 (Table 2). Thioaldehyde species are known to participate in tautomeric equilibria with the corresponding ene-thiols (10a and 10b), with no obvious choice of one isomer being preferred over the other in the present case. Once species 10a and 10b undergo alkylation the product no longer possesses a mobile proton and further equilibration between products 11a and 11b would not be expected.
Fig. 11.
Reaction sequence illustrating the formation of rGH degradation products from exposure to light
(a) disproportionation of initially formed thiyl radicals to the corresponding thioaldehyde and thiol; (b) tautomerization of thioaldehyde 8 leading to cis/trans ene-thiols; (c) alkylation with iodoacetamide to form cis/trans products detected by mass spectrometry. Product 9 corresponds to peptide 22 of Table 2.
The presently identified products represent the early stage degradation process for protein disulfide linkages; however, such products are known to undergo a continuing series of reactions leading to numerous products some of which are isobaric [9–14]. Using the present XUPLC system these subsequent generation products have been observed for the related protein human growth hormone (unpublished results) and will be elaborated upon in a separate publication.
Conclusions
An XUPLC system has been assembled that represents a slight design modification to that developed in the Jorgenson laboratory [26, 27]. The present system was routinely operated at pressure 38 Kspi when equipped with capillaries of 118 cm packed with sub-2μm particles. The photochemical mediated degradation of the protein rGH was used to evaluate the capability of this system in supporting investigations of protein degradation. For most of the peptides resulting from enzymatic digestion of rGH, PCobs values ~ 460–735 were observed. Comparison of XUPLC separations to those obtained with a 25 cm commercial UPLC column revealed substantially enhanced resolution, albeit at the expense of time. Chemical degradation of protein disulfide linkages exposed to 254 nm light was demonstrated for rGH, where a well resolved isobaric cis/trans diastereomerically related octa-peptide was observed by XUPLC, but unresolved using the commercial UPLC column. The most serious operational limitation for the presently described XUPLC system is column lifetime, which is related to the integrity of the inlet chemical frit; however, an altered design is currently being implemented that is expected to both preserve inlet frit integrity and allow routine operation for long periods of time. Cleary for complex chemical degradation problems, as encountered in protein chemistry, chromatographic systems utilizing long capillary columns with the associated higher pressures will be invaluable.
Supplementary Material
Acknowledgments
The University of Kansas based investigators gratefully acknowledge the J.R. and Inez Jay funds, awarded to MLF by the Higuchi Biosciences Center at The University of Kansas, for providing partial funding for this research, and the numerous discussions and fabrication assistance from JTS and our collaborators at UNC Chapel Hill.
References
- 1.Nobelprize.org. Nobel Media AB; 2014. The Nobel Prize in Physiology or Medicine 1923. Web. 6 May 2016. http//www.nobelprize.org/nobel_prizes/medicine/laureates/1923. [Google Scholar]
- 2.Hawe A, Wiggenhorn M, Van de Weert M, Garbe JHO, Manler H-C, Jiskoort W. Forced degradation of therapeutic proteins. J Pharm Sci. 2012;101(3):895–913. doi: 10.1002/jps.22812. [DOI] [PubMed] [Google Scholar]
- 3.Dimitrov DS. Therapeutic proteins. Methods Mol Biol. 2012;899:1–26. doi: 10.1007/978-1-61779-921-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siahaan TJ, Schöneich C. In: Pharmaceutical Formulation Development of Peptides and Proteins. 2. Hovgaard L, Frokjaer S, van de Weert M, editors. CRC Press; Boca Raton, FL: 2013. [Google Scholar]
- 5.Kerwin BA, Remmele RL., Jr Protect from light: photodegradation and protein biologics. J Pharm Sci. 2007;96(6):1468–1479. doi: 10.1002/jps.20815. [DOI] [PubMed] [Google Scholar]
- 6.Creed D. The photophysics and photochemistry of the near-UV absorbing amino acids I. Tryptophan and its simple derivatives. Photochem Photobiol. 1984;39(4):537–562. [Google Scholar]
- 7.Vogt W. Oxidation of methionyl residues in proteins: Tools, targets, and reversal. Free Radic Biol Med. 1995;18(1):93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 8.Barrón LB, Waterman KC, Filipiak P, Hug GL, Nauser T, Schöneich C. Mechanism and Kinetics of photoisomerization of a cyclic disulfide, trans-4,5-1,2-dithiacyclohexane. J Phys Chem A. 2004;108:2247–2255. [Google Scholar]
- 9.Mozziconacci O, Sharov V, Williams TD, Kerwin BA, Schöneich Peptide cysteine thiyl radicals abstract hydrogen atoms from surrounding amino acids: the photolysis of a cystine containing model peptide. J Phys Chem B. 2008;112:9250–9257. doi: 10.1021/jp801753d. [DOI] [PubMed] [Google Scholar]
- 10.Mozziconacci O, Williams TD, Kerwin BA, Schöneich C. Reversible intramolecular hydrogen transfer between protein cysteine thiyl radicals and αC–H bonds in insulin: control of selectivity by secondary structure. J Phys Chem B. 2008;112:15,921–15,932. doi: 10.1021/jp8066519. [DOI] [PubMed] [Google Scholar]
- 11.Mozziconacci O, Kerwin BA, Schöneich C. Photolysis of an intrachain peptide disulfide bond: primary and secondary processes, formation of H2S, and hydrogen transfer reactions. J Phys Chem B. 2010;114:3668–3688. doi: 10.1021/jp910789x. [DOI] [PubMed] [Google Scholar]
- 12.Mozziconacci O, Kerwin BA, Schöneich Reversible hydrogen transfer between cysteine thiyl radical and glycine and alanine in model peptides: covalent H/D exchange, radical-radical reactions and L- to D-ala conversion. J Phys Chem B. 2010;114:6751–6762. doi: 10.1021/jp101508b. [DOI] [PubMed] [Google Scholar]
- 13.Mozziconacci O, Kerwin BA, Schöneich C. Exposure of a monoclonal antibody, IgG1, to UV-light leads to protein dithiohemiacetal and thioether cross-links: a role for thiyl radicals? Chem Res Toxicol. 2010;23:1310–1312. doi: 10.1021/tx100193b. [DOI] [PubMed] [Google Scholar]
- 14.Zhou S, Mozziconacci O, Kerwin BA, Schöneich C. The photolysis of disulfide bonds in IgG1 and IgG2 leads to selective intramolecular hydrogen transfer reactions of cysteine thiyl radicals, probed by covalent H/D exchange and RPLC-MS/MS analysis. Pharm Res. 2013;30:1291–1299. doi: 10.1007/s11095-012-0968-1. [DOI] [PubMed] [Google Scholar]
- 15.Mozziconacci O, Schöneich C. Sequence-specific formation of D-amino acids in a monoclonal antibody during light exposure. Mol Pharmaceutics. 2014;11:4291–4297. doi: 10.1021/mp500508w. [DOI] [PubMed] [Google Scholar]
- 16.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Re Biomed En. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 17.Dolan JW, Snyder LR, Djordjevic NM, Hill DW, Waeghe TJ. Reversed-phase chromatographic separation of complex samples by optimizing temperature and gradient time I. Peak capacity limitations. J Chromatogr A. 1999;857:1–20. doi: 10.1016/s0021-9673(99)00765-7. [DOI] [PubMed] [Google Scholar]
- 17.Knox JH. Practical Aspects of LC Theory. J Chromatogr Sci. 1977;15:352–364. [Google Scholar]
- 18.Snyder LR, Dolan JW. High-Performance Gradient Elution: The Practical Application of the Linear-Solvent-Strength Model. Wiley Interscience; Hoboken, NJ: 2007. [Google Scholar]
- 19.Neuw UD. HPLC Columns: Theory, Technology and Practice. Wiley-VCH; New York: 1997. [Google Scholar]
- 20.Snyder LR, Stadalius . In: High-Performance Liquid Chromatography: Advance and Perspectives. Horváth C, editor. Vol. 4. Academic Press; Orlando, FL: 1986. [Google Scholar]
- 21.MacNair JE, Lewis KC, Jorgenson JW. Ultrahigh-Pressure Reversed-Phase Liquid Chromatography in Packed Capillary Columns. Anal Chem. 1997;69:983–989. doi: 10.1021/ac961094r. [DOI] [PubMed] [Google Scholar]
- 22.Dong MW. Myths in Ultrahigh-Pressure Liquid Chromatography. LCGC North America. 2013;31:868–880. [Google Scholar]
- 23.Jorgenson JW. Capillary Liquid Chromatography at Ultrahigh Pressures. Annu Rev Anal Chem. 2010;3:129–50. doi: 10.1146/annurev.anchem.1.031207.113014. [DOI] [PubMed] [Google Scholar]
- 24.Angus PDA, Demarest CW, Catalano T, Stobaugh JF. Aspects of column fabrication for packed capillary electrochromatography. J Chromatogr A. 2000;1126:50–57. doi: 10.1016/s0021-9673(00)00529-x. [DOI] [PubMed] [Google Scholar]
- 25.Franklin EG. Utilization of Long Columns Packed with Sub-2 μm Particles Operated at High Pressures and Elevated Temperatures for High-Efficiency On-Dimensional Liquid Chromatography Separations. The University of North Carolina; Chapel Hill: 2012. [Google Scholar]
- 26.Stobaugh JT. Strategies off Differential Proteomic Analysis by Liquid Chromatography-Mass Spectrometry. The University of North Carolina; Chapel Hill: 2012. [Google Scholar]
- 27.Grinias KM, Godinho JM, Franklin EG, Stobaugh JT, Jorgenson JW. Development of a 45 kpsi ultrahigh pressure liquid chromatography instrument for gradient separations of peptides using long Microcapillary columns and sub-2 μm particles. J Chromatogr A. 2016;1469:60–67. doi: 10.1016/j.chroma.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods JM. Fluorogenic Derivatization of Pro-Quinoidal Species: From Biogenic Amines to Protein Bound 3-Nitrotyrosine; Appendix, Implementation of Chromatographic Systems Which Surpass Commercial Pressure Limits for Enhanced Resolution and Peak Capacity. The University of Kansas Lawrence; 2014. [Google Scholar]
- 29.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:1–17. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knox JH. Gradient storage method for liquid chromatography. 3.981,801 US patent. 1976
- 31.Davis M, Stahl D, Lee T. Low flow high-performance liquid chromatography solvent delivery system designed for tandem capillary liquid chromatography-mass spectrometry. J Am Soc Mass Spectrom. 1995;6:571–577. doi: 10.1016/1044-0305(95)00192-G. [DOI] [PubMed] [Google Scholar]
- 32.Cortes HJ, Pfeiffer CD, Richter BE, Stevens TS. Porous ceramic bed supports for fused silica packed capillary columns used in liquid chromatography. J High Res Chromatogr and Chromatogr Commun. 1987;10:446–448. [Google Scholar]
- 33.Lenza RFS, Vasconcelos WL. Structural evolution of silica sols modified with formamide. Mat Res. 2001;4:175–179. [Google Scholar]
- 34.Robards K, Haddad PR, Jackson PE. Principles and Practice of Modern Chromatographic Methods. Academic Press; London: 1994. [Google Scholar]
- 35.Stadalius MA, Gold HS, Snyder LR. Optimization model for the gradient elution separation of peptide mixtures by reversed-phase high-performance liquid chromatography. J Chromatogr. 1984;296:31–39. [Google Scholar]
- 36.Snyder LR, Kirkland JJ, Glajch JL. Practical HPLC Method Development. 2. John Wiley & Sons, Inc; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.