Abstract
Background
PD-L1 immunohistochemistry (IHC) has been traditionally used for predicting clinical responses to immune checkpoint inhibitors (ICIs). However, there are at least 4 different assays and antibodies used for PD-L1 IHC, each developed with a different ICI. We set to test if next generation RNA sequencing (RNA-seq) is a robust method to determine PD-L1 mRNA expression levels and furthermore, efficacy of predicting response to ICIs as compared to routinely used, standardized IHC procedures.
Methods
A total of 209 cancer patients treated on-label by FDA-approved ICIs, with evaluable responses were assessed for PD-L1 expression by RNA-seq and IHC, based on tumor proportion score (TPS) and immune cell staining (ICS). A subset of serially diluted cases was evaluated for RNA-seq assay performance across a broad range of PD-L1 expression levels.
Results
Assessment of PD-L1 mRNA levels by RNA-seq demonstrated robust linearity across high and low expression ranges. PD-L1 mRNA levels assessed by RNA-seq and IHC (TPS and ICS) were highly correlated (p < 2e-16). Sub-analyses showed sustained correlation when IHC results were classified as high or low by clinically accepted cut-offs (p < 0.01), and results did not differ by tumor type or anti-PD-L1 antibody used. Overall, a combined positive PD-L1 result (≥1% IHC TPS and high PD-L1 expression by RNA-Seq) was associated with a 2-to-5-fold higher overall response rate (ORR) compared to a double negative result. Standard assessments of sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) showed that a PD-L1 positive assessment for melanoma samples by RNA-seq had the lowest sensitivity (25%) but the highest PPV (72.7%). Among the three tumor types analyzed in this study, the only non-overlapping confidence interval for predicting response was for “RNA-seq low vs high” in melanoma.
Conclusions
Measurement of PD-L1 mRNA expression by RNA-seq is comparable to PD-L1 expression by IHC both analytically and clinically in predicting ICI response. RNA-seq has the added advantages of being amenable to standardization and avoidance of interpretation bias. PD-L1 by RNA-seq needs to be validated in future prospective ICI clinical studies across multiple histologies.
Electronic supplementary material
The online version of this article (10.1186/s40425-018-0489-5) contains supplementary material, which is available to authorized users.
Keywords: Atezolizumab, Avelumab, cancer immunotherapy, Durvalumab, Nivolumab, Pembrolizumab, PD-L1, Biomarker
Background
Five trial-evaluated immunohistochemistry (IHC) assays for the assessment of CD274 (best known as programmed death ligand-1, PD-L1) expression in formalin-fixed paraffin-embedded (FFPE) samples have been developed as companion and complementary diagnostics alongside immune checkpoint inhibitors (ICIs) targeting PD-L1 and its main receptor (programmed cell death 1, PDCD1, best known as PD-1) [1–5]. While these tests measure PD-L1 protein levels, they differ by antibody clone, staining platform, and scoring system. For instance, while assessment of PD-L1 expression in advanced gastric cancer or gastroesophageal junction adenocarcinoma samples by the PD-L1 IHC 22C3 pharmDx assay uses a “combined positive score” or CPS [6], testing metastatic non-small cell lung cancer (NSCLC) patient samples relies on a “tumor proportion score” or TPS [7]. This variability in scoring methods has contributed to confounding results across clinical trials and in clinical practice, leading to uncertainty about the universal value of PD-L1 expression levels as a biomarker across tumor types [8–10]. The “Blueprint PD-L1 IHC Comparability Project” was an effort to compare the concordance of five antibody clones commonly used for assessing PD-L1 expression by IHC [11, 12]. The two-phase study revealed a good concordance for three of the five antibody clones employed, but suggests that interchanging assays and cutoffs would lead to “misclassification” of PD-L1 status for some patients. Furthermore, the use of FFPE archival tumor tissues with non-standardized fixation and storage methods may be a source of unpredictable and unintended results for adequate PD-L1 antigen retrieval, potentially increasing the heterogeneity of IHC intensity, extent and topography of staining. All these factors complicate the use of PD-L1 status as assessed by IHC for predicting patient clinical response to ICIs [13, 14].
RNA-based assays on FFPE tissues are currently used in the clinic to classify or predict recurrence risk in patients affected by various tumor types, These assays include DecisionDX-Melanoma (Castle Biosciences), Prosigna® (Nanostring Technologies), MammaPrint® (Agendia), Afirma® Thyroid FNA Analysis (Veracyte), and OncoType DX® (Genomic Health) [15–19]. Most of these tests are microarray- or quantitative reverse transcription (qRT)-PCR-based assays specific for a small panel of cancer-related genes. Recently, RNA-seq has emerged as powerful tool to evaluate mRNA expression in the clinic [20–23]. The use of highly-specific primers that target stably expressed genes provides a high level of specificity and sensitivity, allowing for the simultaneous measurement of several targets including genes for sample quality control purposes. Gene expression profiling by RNA-seq has minimal input requirements and has the potential to be far more cost-effective than IHC methods given the scalability of next-generation sequencing. Further, strong concordance between platforms, including gene expression microarrays, qRT-PCR and IHC has demonstrated the analytic validity of RNA-seq, even for challenging FFPE tumor samples [24]. By digitally counting target molecules, RNA-seq enables precise transcriptome quantification that provides a continuum measurement across a large dynamic range of expression.
The objective of this study was to compare RNA-seq to IHC for the assessment of PD-L1, at both analytical and clinical levels, with the intent to validate RNA-seq as a predictor of response in 209 patients with multiple tumor types treated with ICIs. To demonstrate the linearity and sensitivity of PD-L1 RNA-seq as a standalone assay, we tested several tumor samples across multiple dilutions. We then used objective response criteria (RECISTv1.1) to compare measurements of PD-L1 by IHC versus RNA-seq to assess clinical utility.
Methods
Patients and clinical data
Eight collaborating institutions obtained approval by their respective institutional review boards (IRBs) to submit existing de-identified specimens and associated clinical data for use in this study. Patients were identified for inclusion of electronic pharmacy records indicated they received at least one dose of checkpoint inhibition therapy in the course of standard care, had adequate pre-treatment FFPE tissue (minimum 10% tumor nuclei, maximum 50% necrosis) collected within 2 years of first dose, were evaluable for response by RECIST v.1.1, and had known overall survival from first dose of checkpoint blockade. A total of 209 patients were included, encompassing renal cell carcinoma (RCC, n = 45; 7 responders, 38 non-responders), metastatic cutaneous melanoma (n = 76; 32 responders, 44 non-responders), and NSCLC (n = 88; 17 responders, 71 non-responders). Our primary clinical endpoint for analysis was objective response rate (ORR), defined as patients with complete responses (CR) or partial responses (PR), and patients with progressive disease (PD) or stable disease (SD) classified as non-responders (Additional file 1: Table S1) [25].
Immunohistochemical studies
In melanoma samples, PD-L1 expression was assessed using the Dako Omnis platform (Agilent, Santa Clara, CA) and the 28–8 pharmDx antibody (Agilent, Santa Clara, CA), which is the FDA-approved complementary diagnostic for nivolumab. For RCC and NSCLC samples, the 22c3 pharmDx antibody (Agilent, Santa Clara, CA) was employed on Autostainer Link 48 (Agilent, Santa Clara, CA), which is the FDA-approved companion diagnostic for pembrolizumab. Established cutoffs for the diagnostics in each histologic type were used to score PD-L1 IHC tumor proportion score (TPS) and immune cell staining (ICS) as follows: melanoma TPS, 1%, [13] NSCLC TPS, 50 and 1%, [7] RCC TPS, 1%; RCC ICS, 1%.
RNA-seq profiling
RNA was extracted from each sample following microscopic tissue review by an anatomical pathologist and selection of specimen representing tumor cells and associated microenvironment. Gene expression was evaluated by targeted RNA-seq of 384 immune transcripts using an analytically validated assay [23]. Absolute reads were generated using Torrent Suite’s plugin immuneResponseRNA (v5.2.0.0) and further normalized to yield normalized reads per million (nRPM), using previously described methods [23]. For all 394 genes including PD-L1, nRPM values were subsequently ranked (gene expression rank) from 0 to 100 based on expression of these genes in a reference population representing a wide range of gene expressions in various tumor types, as previously described [23]. A subset of samples with varying PD-L1 expression levels were serially diluted to demonstrate sensitivity and linearity of detection.
Data analysis
To demonstrate the linearity of PD-L1 mRNA detection, coefficient of determination (R2) was calculated for the absolute reads generated across various library dilutions. To investigate the relationship between PD-L1 expression by targeted RNA-seq and IHC, IHC TPS and ICS results were categorized as either high or low using the previously described FDA-approved complementary and companion diagnostic scoring guidelines and one-way ANOVA and Tukey honest significant difference (HSD) was performed for all PD-L1 values across all samples. To compare IHC versus RNA-seq for prediction of response, values of TPS ≥1% for melanoma, TPS ≥1% and ≥ 50% for NSCLC, and TPS and ICS ≥1% for RCC were compared to RNA-seq expression interpretations of high (rank ≥75) and not-high (rank < 75), relative to a reference population. To compute sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy, a positive result was considered as IHC TPS of ≥1% for melanoma, TPS of ≥1% and ≥ 50% for NSCLC, and TPS and ICS ≥1% for RCC, and high value for RNA-seq expression (rank ≥75). A negative result was considered as IHC TPS of < 1% for melanoma, TPS of < 1 and < 50% for NSCLC, and TPS and ICS < 1% for RCC, and a moderate or low value for RNA-seq expression. Logistic regression was then performed to evaluate the prediction of response based on tumor type, IHC result, and RNA-seq result.
Results
Linearity of PD-L1 assessment by RNA-seq
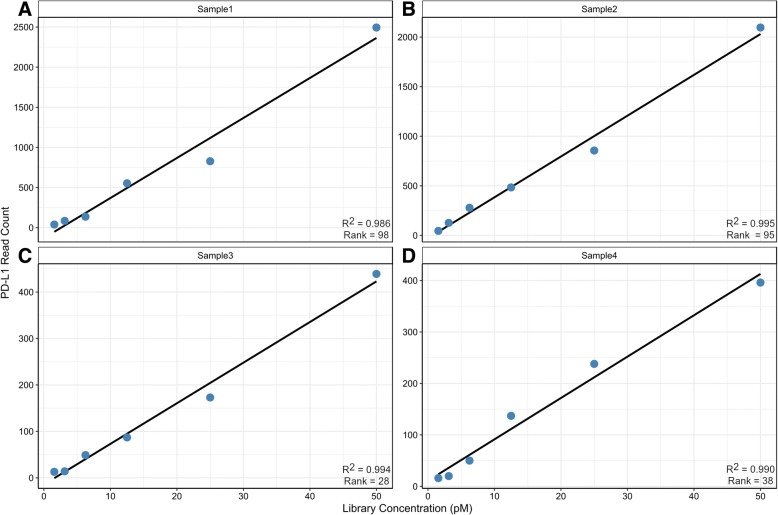
Linearity of PD-L1 assessment by RNA-seq was determined by comparing the absolute reads relative to an input of 1.5625, 3.125, 6.25, 12.5, 25, and 50 pM RNA library for tumor samples representing diverse levels of expression (Fig. 1; Additional file 1: Table S2). Samples #1 and #2 represent high expressors (PD-L1 > 75 rank), while samples #3 and #4 represent moderate expressors (PD-L1 = 25–75 rank). For samples #1 and #2, PD-L1 transcript detection values ranged from 0 to > 2400 absolute reads, demonstrating a robust positive linear correlation (R2 > 0.98) for clinical specimens expressing high PD-L1 levels. For samples #3 and #4, PD-L1 transcript detection values ranged from 0 to < 450 absolute reads, demonstrating a positive linear correlation (R2 > 0.98) for clinical specimens expressing low-to-moderate PD-L1 levels. Overall, these results demonstrate that detection of PD-L1 mRNA levels in FFPE samples by RNA-seq is consistent across a dynamic range of expression, and that PD-L1 transcripts can be reliably quantified by a continuous variable of absolute transcript reads down to values approaching background.
Fig. 1.
PD-L1 transcript detection across serial dilutions of 4 tumor samples. PD-L1 transcript detection across serial dilutions of 4 tumor samples. Results demonstrate high, moderate, and low PD-L1 expression and can be reliably quantified by a continuous variable of absolute transcript reads. a Sample 1: Melanoma with high expression. b Sample 2: Melanoma with high expression. c Sample 3: RCC with moderate expression. d Sample 4: RCC with moderate expression
Analytical comparison of IHC and RNA-seq results
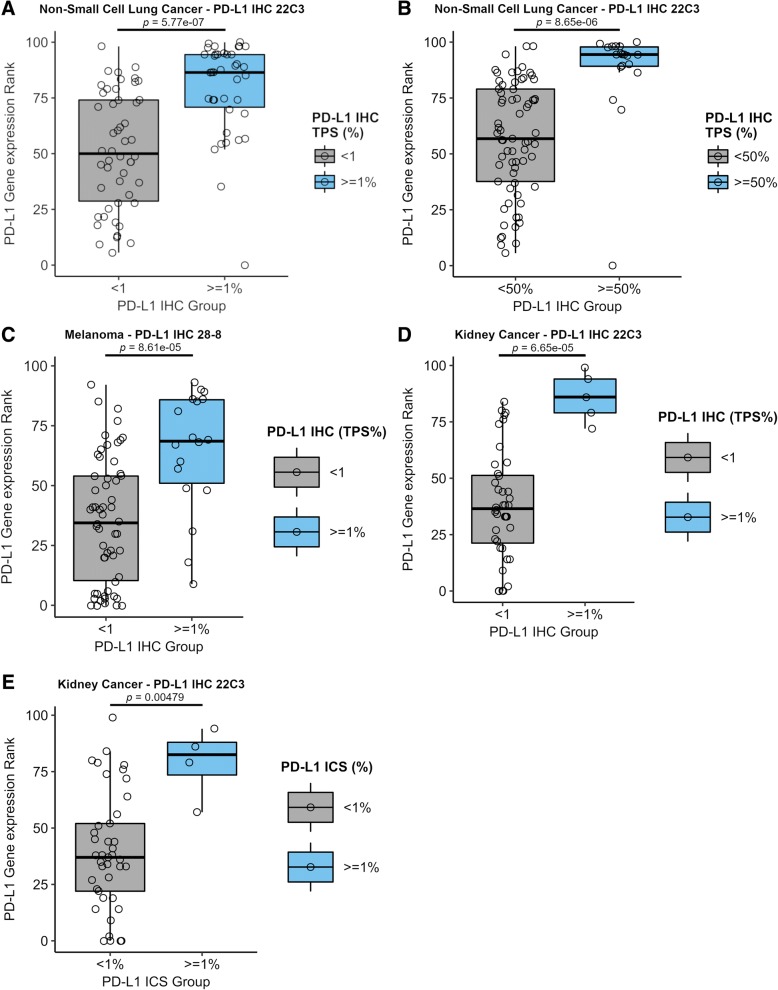
For the 209 samples evaluated, the highest rate of a positive result, defined as IHC TPS of ≥1% for melanoma, TPS of ≥1% and ≥ 50% TPS for NSCLC, and TPS or ICS ≥1% for RCC, or RNA-seq rank ≥75, was observed with NSCLC samples for both IHC and RNA-seq (Table 1). One-way ANOVA demonstrated a statistically significant correlation between PD-L1 RNA-seq rank and IHC (TPS or ICS, p < 2e-16) across the three tumor types. The null hypothesis, which specified that the means of PD-L1 gene expression ranks in each IHC group would not differ, was rejected. Therefore, we performed Tukey HSD for multiple pairwise-comparisons between the means of the IHC TPS/ICS high and not-high groups. Ad-hoc Tukey’s HSD comparisons of NSCLC mean TPS at < 1% (Fig. 2a), NSCLC TPS at < 50% (Fig. 2b), melanoma TPS at < 1% (Fig. 2c), and RCC TPS < 1% (Fig. 2d) or ICS < 1% (Fig. 2e), demonstrated significant differences (p < 0.01) between the various groups that were consistent with RNA-seq ranks.
Table 1.
PD-L1 IHC and RNA-seq results for 209 samples
| Test Result | Test | RCC | Melanoma | NSCLC |
|---|---|---|---|---|
| ≥1% TPS | IHC | 5 (11%) | 19 (25%) | 38 (43%) |
| < 1% TPS | IHC | 40 (89%) | 57 (75%) | 50 (57%) |
| ≥50% TPS | IHC | NA | NA | 19 (22%) |
| < 50% TPS | IHC | NA | NA | 69 (78%) |
| ≥1% ICS | IHC | 4 (9%) | NA | NA |
| < 1% ICS | IHC | 41 (91%) | NA | NA |
| > 75 rank (high) | RNA-seq | 9 (20%) | 11 (14%) | 35 (40%) |
| 25–75 rank (moderate) | RNA-seq | 24 (53%) | 39 (51%) | 42 (48%) |
| > 25 rank (low) | RNA-seq | 12 (27%) | 26 (35%) | 11 (12%) |
| Total | 45 | 76 | 88 |
Fig. 2.
Ad-hoc Tukey’s HSD test comparing PD-L1 expression by RNA-seq (Y axis) with IHC (X axis). Box plots show concordance of the two measurements across multiple clinical cutoffs for IHC and different tumor types. a NSCLC mean TPS at < 1%. b NSCLC mean TPS at < 50%. c Melanoma TPS mean at < 1%. d RCC mean TPS < 1% or E) ICS < 1%
Objective response rates
To investigate the clinical utility of a positive result for PD-L1 by RNA-seq, IHC or both, we evaluated ORR in RCC (n = 45), melanoma (n = 76), and NSCLC (n = 88) patients receiving an FDA-approved ICI (Additional file 1: Table S1). For these comparisons, RNA-seq results ≤75 rank were combined into a single RNA-seq “not high” group, enabling a more direct binary comparison to IHC. The ORR for patients stratified by PD-L1 IHC levels was consistent with previously published values for each tumor type [26–30] (Table 2), supporting that our study population was not biased in selection, and allowing for comparison of the IHC results to those for RNA-seq.
Table 2.
ORR across tumor type and individual biomarker result
| Disease | Test | PD-L1 result | Responders | Non-responders | Total | ORR |
|---|---|---|---|---|---|---|
| Melanoma | IHC TPS | ≥1% | 10 | 8 | 18 | 55.60% |
| < 1% | 22 | 36 | 58 | 37.90% | ||
| RNA-seq | High | 8 | 3 | 11 | 72.70% | |
| Moderate & low | 24 | 41 | 65 | 46.20% | ||
| NSCLC | IHC TPS | ≥1% | 10 | 28 | 38 | 26.30% |
| < 1% | 7 | 43 | 50 | 14.00% | ||
| IHC TPS | ≥50% | 8 | 11 | 19 | 42.10% | |
| < 50% | 9 | 60 | 69 | 13.00% | ||
| RNA-seq | High | 10 | 25 | 35 | 28.60% | |
| Moderate & low | 7 | 46 | 53 | 11.90% | ||
| RCC | IHC TPS | ≥1% | 2 | 3 | 5 | 40.00% |
| < 1% | 5 | 35 | 40 | 12.50% | ||
| IHC ICS | ≥1% | 1 | 3 | 4 | 25.00% | |
| < 1% | 6 | 35 | 41 | 14.60% | ||
| RNA-seq | High | 3 | 6 | 9 | 33.30% | |
| Moderate & low | 4 | 32 | 36 | 8.30% |
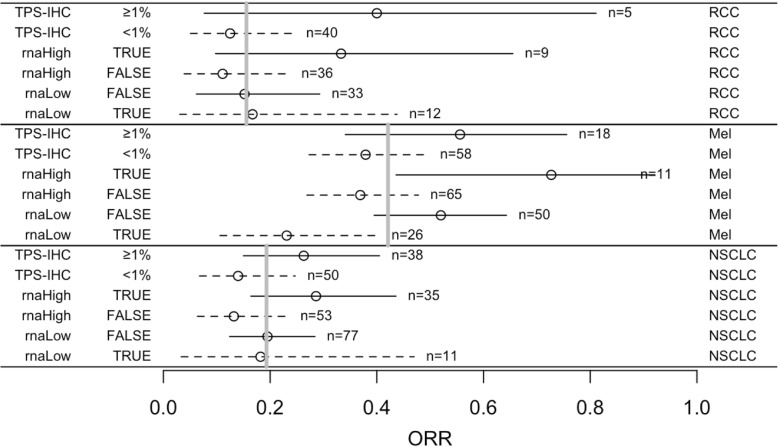
ORR was 42.1% for melanoma, 15.6% for RCC, and 19.3% for NSCLC patients (Fig. 3 and Additional file 1: Table S3). ORR, as measured by various PD-L1 measurements, ranged from 37.9% (IHC < 1% TPS) to 72.7% (RNA-seq high) for melanoma, 11.9% (RNA-seq low) to 42.1% (IHC > 50% TPS) for NSCLC, and 8.3% (RNA-seq low) to 40.0% (IHC > 1% TPS) for RCC. For positive PD-L1 assessments across tumor types, the maximum ORR was 72.7% (RNA-seq) and 55.6% (IHC), 28.6% (RNA-seq) and 42.1% (IHC), and 33.3% (RNA-seq) and 40% (IHC) for melanoma, NSCLC and RCC, respectively. Conversely, negative PD-L1 assessments resulted in ORR of 46.2% (RNA-seq) and 37.9% (IHC) for melanoma, 11.9% (RNA-seq) and 13–14% (IHC) for NSCLC, and 8.3% (RNA-seq) and 12.5–14.6% (IHC) for RCC, with the latter two histologies’ ORR dependent on IHC cutoff and staining interpretation. One important implication of assessing PD-L1 levels is using negative results to support clinical decision making against the administration of ICIs [29]. For RCC, the PPVs for IHC TPS of ≥1% and for RNA-seq high were rather uncertain due to the small population size. In melanoma, only a low RNA-seq result had a notably high NPV (1.0–0.23 = 0.77). For NSCLC samples, neither test had much predictive power for response (Fig. 3).
Fig. 3.
Proportions of responses in subgroups defined by tests for PD-L1 expression. Objective response rate (ORR) was 42.1% for melanoma (Mel), 15.6% for renal cell carcinoma (RCC), and 19.3% for non-small cell lung carcinoma (NSCLC) (grey bars). Each complementary pair of subsets corresponds to positive predictive value (PPV, solid line) and 1 – negative predictive value (NPV, dashed line) (circles). The intervals are 90% confidence intervals. TPS-IHC = PD-L1 tumor proportion score (TPS) by IHC, rnaHigh TRUE = PD-L1 RNA-seq expression is high, rnaHigh FALSE = PD-L1 RNA-seq expression is low or moderate (considered “negative”), rnaLow TRUE = PD-L1 RNA-seq expression is low, rnaLow FALSE = PD-L1 RNA-seq expression is moderate or high (considered “positive”)
An important comparison of these two methodologies is the ORR for concordant or discordant results when combining RNA-seq and IHC PD-L1 expression results. Concordant negative results (IHC/RNA-seq −/−) were associated with the lowest observed ORR across all three tumor types. Conversely, concordant positive results (IHC/RNA-seq +/+) were not consistently associated with the highest ORR. Although only four cases, discordant result between IHC and RNA-seq (IHC/RNA-seq −/+) were associated with 75% ORR in melanoma, (3 responders and 1 non-responder), the highest ORR of any tumor. A high ORR was also documented amongst NSCLC patients with discordant results (IHC/RNA-seq +/−) at a ≥ 50% TPS cutoff (ORR = 66.7%), but not at a ≥ 1% value (ORR = 20%) (Additional file 1: Table S4). There were 34 patients treated with ipilimumab, 22 with ipilimumab + nivolumab, and 153 with either single-agent pembrolizumab, nivolumab, or atezolizumab (anti-PD-1). For melanoma, a 67% response rate was observed for patients with RNA-seq high treated with an anti-CTLA4 agent, which increased to 80% when analyzed for anti-PD-1 therapy alone. RCC (n = 10) and NSCLC (n = 2) had limited number of patients treated with anti-CTLA4 agents, but RNA-seq high was associated with the only RCC response to ipilimumab + nivolumab, as well as non-response for all RNA-seq low patients (Additional file 1: Table S5).
Clinical utility of PD-L1 IHC versus RNA-seq
Standard parameters of sensitivity, specificity, PPV, NPV, and accuracy were used to compare the clinical utility of PD-L1 assessment with IHC versus RNA-seq (Table 3). RNA-seq in melanoma samples had the lowest sensitivity (25%) and the highest PPV (72.7%) of all test results. The highest sensitivity at 58.8%, shared by both IHC with TPS ≥1% and RNA-seq high in NSCLC samples, was coupled with the lowest PPV at 26.3 and 28.6%, respectively. High NPV (> 85%) was seen with IHC TPS ≥50% and RNA-seq in NSCLC samples, as well as with IHC TPS ≥1% and RNA-seq in RCC samples. However, PPV was suboptimal for all these biomarkers. Sensitivity, specificity, PPV, and NPV for a double positive result (IHC/RNA-seq +/+), as compared to a single positive result, was minimally different from direct comparisons and offered little advantage. The results of these analyses epitomize the typical trade-off of sensitivity for specificity, and vice versa, with an overall less than optimal performance of the tests to predict responders.
Table 3.
Clinical utility comparison of IHC TPS and RNA-seq rank results
| Prediction Method | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| Melanoma IHC ≥1% | 31.3% | 81.8% | 55.6% | 62.1% |
| Melanoma RNA-seq > 75 | 25.0% | 93.2% | 72.7% | 63.1% |
| Melanoma IHC ≥1% & RNA-seq > 75 | 20.8% | 94.6% | 71.4% | 64.8% |
| NSCLC IHC ≥1% | 58.8% | 60.6% | 26.3% | 71.4% |
| NSCLC IHC ≥50% | 47.1% | 84.5% | 42.1% | 87.0% |
| NSCLC RNA-seq > 75 | 58.8% | 64.8% | 28.6% | 86.8% |
| NSCLC IHC ≥1% & RNA-seq > 75 | 63.6% | 68.0% | 30.4% | 89.5% |
| NSCLC IHC ≥50% & RNA-seq > 75 | 46.2% | 83.9% | 37.5% | 88.1% |
| RCC IHC ≥1% | 28.6% | 92.1% | 40.0% | 87.5% |
| RCC RNA-seq > 75 | 42.9% | 84.2% | 33.3% | 88.9% |
| RCC IHC ≥1% & RNA-seq > 75 | 33.3% | 93.9% | 50.0% | 88.6% |
Sensitivity = TP/(TP + FN)
Specificity = TN/(TN + FP)
Positive predictive value (PPV) = TP/(TP + FP)
Negative predictive value (NPV) = TN/(TN + FN)
To evaluate RNA-seq as the gold standard and determine whether IHC adds predictive value, a logistic regression model was employed to evaluate the prediction of response to treatment based on tumor type, PD-L1 by IHC, and PD-L1 levels by RNA-seq (Table 4). As expected, the model shows that melanoma patients (p = 0.0026) have a higher response rate than patients with RCC and NSCLC, and that the expression rank interpretation (“RNA-seq”) has a significant linear (“RNA-seq.L”) relationship to response (equally spaced scoring from low to moderate, and from moderate to high). The RNA-seq.L estimate, 0.96, is the increase in the log odds of response moving from low to moderate, or from moderate to high (odds increased by a factor of 2.6, and by 6.8 going from Low to High). There is no indication of a further quadratic effect (“RNA-seq.Q”), but the sample size is small for detecting such a departure from equal spacing. A model examining whether the relationship between RNA-seq and ORR varies by tumor type showed no significant interaction (p = 0.80). A model examining whether the PD-L1 RNA-seq and IHC results interact in predicting response also showed no significant interaction (p = 0.45). Among the three tumor types analyzed in this study, the only non-overlapping confidence interval for predicting response was for “RNA-seq low vs high” in melanoma.
Table 4.
Logistic regression for predicting response category “CR or PR” versus “SD or “PD”
| Estimate | Std. Error | z value | Pr(>|z|) | |
|---|---|---|---|---|
| TumorType RCC | (reference) | – | – | – |
| TumorType Melanoma (versus RCC) | 1.50 | 0.50 | 3.01 | 0.0026 |
| TumorType NSCLC (versus RCC) | −0.13 | 0.53 | −0.25 | NS |
| IHC TPS ≥1% | 0.41 | 0.40 | 1.03 | NS |
| RNA-seq.L | 0.96 | 0.40 | 2.43 | 0.015 |
| RNA-seq.Q | 0.21 | 0.28 | 0.76 | NS |
Discussion
PD-L1 appears as one of the most controversial biomarkers to be introduced into clinical practice. Despite prior evidence demonstrating that both technologic and histologic variability limit clinical utility [31, 32], four IHC based tests are currently approved for guiding treatment decisions in patients with multiple tumor types. Clinicians, patients and other stakeholders seeking reliable PD-L1 diagnostic assays are subjected to the lack of IHC standardization and face real implications to clinical care. Recently, the Blueprint Working Group was established with cooperation from the FDA, IHC platform vendors, professional oncology organizations and the pharmaceutical industry to provide a comparison of different PD-L1 IHC tests in NSCLC, including scoring methods for expression [11] . The initial findings showed comparable results for 22C3, 28–8, and SP263 antibody clones, but reduced sensitivity for the SP142 clone, when evaluating staining in tumor cells. In the phase 1 study, only 50% of the cases demonstrated concordant positive staining above the antibody specific cutoffs. These results are alarming given that the Blueprint study involved pathologists with considerable expertise in NSCLC and the utilization of large resection specimens (n = 38) that are typically easier to evaluate. While ‘no’ and ‘very high’ PD-L1 expression were mostly concordant, the Blueprint study demonstrated that the ‘low’ to ‘intermediate’ expression levels seen in most NSCLC patients can result in discrepancy. In these instances, a continuous measure, rather than using a specific TPS cutoff, may better predict ICI treatment efficacy. The more recent phase 2 study [12] confirms the previously reported differences in sensitivity between the IHC assays, and reports reliability among pathologists ranging from very strong for TPS scoring to poor for immune cell scoring. Given the challenges associated with PD-L1 IHC, it is surprising the paucity of information that exists for PD-L1 measurement in FFPE tumors by other methods.
In contrast to the Blueprint study, our study explores measuring PD-L1 by a single alternative methodology, RNA-seq by direct comparison between FDA-approved PD-L1 IHC assays and a laboratory developed RNA-seq test. Although previous studies demonstrate PD-L1 as measured by IHC is a predictive biomarker of response to ICIs [33], it was unclear if an alternative methodology would validate PD-L1 utility as a predictive biomarker. It was not the intent of this study to debate the clinical utility of PD-L1 IHC, but to assess the clinical utility of PD-L1 by RNA-seq. Unlike IHC, RNA-seq quantitates the number of expressed mRNA transcripts in the entire tumor microenvironment without subjective scoring methods and cell type discrimination. When performed in a CLIA laboratory setting with a validated protocol [23], our data demonstrates that RNA-seq is a highly sensitive and robust assay for measuring PD-L1 across a continuum of expression levels.
Our study employed a considerably large (n = 209) cohort of samples from multiple institutions treated with one or more FDA-approved ICIs. The specimens were of variable tumor mass, including a large fraction of needle core biopsies and FNA cell blocks. Furthermore, multiple tumor types were evaluated and the pathologist reading IHC slides was not a renowned expert in any one particular disease. In total, the study reflects a real-world clinical scenario in which archival specimens representing several commonly tested histologies are evaluated for PD-L1 expression.
Data obtained from this study revealed that PD-L1 expression as measured by RNA-seq is highly correlated to IHC both analytically and clinically. Overall, ICI response varied between tumor types but as expected, each demonstrated highest ORR with either a PD-L1 positive RNA-seq or IHC. We acknowledge that the relatively small number of PD-L1 positive cases, especially RCC, may limit the evaluation of RNA-seq as a predictive assay for the tumor types evaluated, but ORR for patients stratified by PD-L1 IHC levels was consistent with previously published values. Even with these limitations however, RNA-seq high and low results for melanoma demonstrates significant PPV and NPV, respectively, and resulted in an overall 73% ORR compared to 56% ORR by PD-L1 IHC. Although the combined positive (IHC/RNA-seq +/+) samples were associated with an overall higher ORR than the combined negative (IHC/RNA-seq −/−) samples, the highest ORR of any tumor type were associated with discordant melanoma IHC and RNA-seq results (IHC/RNA-seq −/+). Given the small number of samples (n = 4) in this grouping, a larger cohort with this phenotype is required to understand if the predictive power is tumor type specific or a direct result of PD-L1 quantitation in melanoma by RNA-seq. NSCLC samples, which had the largest number of PD-L1 positive RNA-seq and IHC cases, did not however demonstrate predictive power for either test. Most interesting is the ORR for combined positive (IHC/RNA-seq +/+) and negative (IHC/RNA-seq −/−) results. While a combined positive did not always demonstrate the highest ORR across tumor types, the combined negative result did have the lowest ORR, except for RCC IHC/RNA-seq +/− which was represented by only one case.
Unexpectedly, we found that melanomas with PD-L1 IHC TPS > 1% had a significantly improved response to all ICI monotherapies, including anti-PD-1 and anti-CLTA-4 monotherapy. We were also surprised to find that elevated PD-L1 by RNA-seq quantification was an even better predictor of response to the same monotherapies. This is a provocative finding given that the PD-1/PD-L1 axis is currently thought to be entirely distinct and not interacting with the CTLA-4 axis [34]. However, we do acknowledge that our melanoma data set is limited in size and this finding needs to be confirmed in future studies.
In addition to the inherent advantages of standardized methods for PD-L1 assessment by RNA-seq, this technology is also convenient as it enables highly multiplexed testing of several patients in a single run, with per sample costs approaching those of IHC when performed in batch sizes greater than twenty. By measuring multiple transcripts simultaneously, RNA-seq is well suited to characterize the functional state of immune cells in the tumor microenvironment for biomarkers of antigen presentation, IFN-γ signaling, T-cell active cytokines and other biological features that are responsive to PD-1 checkpoint blockade. It is beyond the scope of this study to report data for the focused set of nearly 400 other genes included in the transcriptome panel, however evaluating RNA for immune gene expression in addition to PD-L1 has been shown to be predictive of efficacy to anti-PD-1 therapy across multiple tumor types with more accuracy than PD-L1 IHC [35–37]. For example, a T-cell inflamed signature based on IFN-γ genes was associated with response to anti-PD-1 therapy in multiple tumor types [35], and an algorithmic approach which combines gene expression profiling with tumor mutational burden (TMB) and PD-L1 IHC improves prediction of response to ICIs in melanoma [37]. These multi-marker approaches contribute more comprehensive information to the cancer immunity cycle [38] than a single analyte and could improve personalized combination immunotherapy treatment options in patients that have failed prior immunotherapy by targeting over-expressed immunomodulatory factors, including LAG-3, GITR, ICOS, TIM-3, and OX40 [39] across multiple tumor types [40, 41]. Co-overexpression of PD-L1 and PD-L2 (another PD-1 ligand) in the same tumor, as well as overexpression of other co-inhibitory or co-activatory molecules can reliably indicate whether checkpoint blockade is a significant factor in a specific case [42, 43]. Though not presented here, RNA-seq may enable a qualitative and quantitative analysis of tumor infiltrating lymphocytes that inform on whether the tumor is immunologically “hot” or “cold”, and provide data to explore new biomarker opportunities [32].
For analytical purposes, the expression of other genes has value relative to PD-L1 for use as an endogenous control for PD-L1 normalization. It is feasible that the various subjective IHC interpretation requirements for estimating percentages of either tumor, immune or combined cell staining could be replaced by normalization of PD-L1 RNA-seq by cell specific markers. Normalization against one or several markers, such as an immunohistochemical stain which the surgical pathologist determines to be highly specific for neoplastic cells in a given case, CD45 for hematopoietic cells, CD3 for T-cells, CD8 for cytotoxic T-cells, or CD68 for macrophages could result in an objective qualitative PD-L1 result that can be automated to reproducibly report PD-L1 expression relative to specific cell types in the tumor microenvironment. With proper tissue review and selection, RNA-seq analysis of the tumor and associated microenvironment is designed to provide a PD-L1 score minimally influenced by sample selection bias and tumor heterogeneity, and to minimize the current PD-LI testing variability which may impact clinical decisions and the uptake of precision immune oncology treatments [44].
A major limitation of our study linked to development of RNA-seq as a standardized measurement of PD-L1 expression, is a lack of external standards. In our study, RNA-seq measurements have a proprietary method of normalization that influence the results for all genes in the panel. Our control samples and genes with a similar impact are also proprietary. Additionally, the interpretation of these measurements by rank is derived via comparison to a unique non-public reference database. Although these are significant issues, the standardization of PD-L1 measurement by RNA-seq is much more attainable than in the case of IHC. External, publicly available standards could be developed and shared similarly to how minimal residual disease for BCR-ABL has become standardized [45]. Another limitation of this study is its retrospective nature of testing archival specimens to assess the presence of a dynamically upregulated biomarker which can change during the disease course. Therefore, PD-L1 status by RNA-seq needs to be further validated in future prospective ICI clinical trials.
The confluence of the Blueprint study and our work supports that alternative measurements of PD-L1 expression beyond IHC, such as RNA-seq, should be considered for clinical use to improve response prediction in patients being considered to receive ICI treatments.
Conclusions
In summary, our study shows that measurement of PD-L1 mRNA expression by RNA-seq is comparable to PD-L1 expression by IHC assays, both analytically and clinically, with evidence that for melanoma samples RNA-seq may be superior to IHC. At minimum, mRNA expression by RNA-seq provides another layer of PD-L1 detection which can exploited to predict tumor response to ICI. The predictive performance of RNA-seq to measure PD-L1 expression (in comparison to IHC) is affected to some extent by histologic factors, but the overall results suggest that moving forward with this technology is a viable approach for this dynamic biomarker.
Additional file
Table S1. Samples (n=209) sorted within tumor type by PD-L1 rank (RNA-seq). Table S2. PD-L1 (CD274) RNA-seq dilution series. Table S3. Responders and non-responders across tumor types and biomarker result. Table S4. ORR across tumor type and combined biomarker results for all ICI therapies. Table S5. Responders and non-responders across tumor types and individual biomarker result. (XLSX 39 kb)
Acknowledgements
The following individuals participated with the collection of biospecimens and associated clinical data including Larson Hsu from Roswell Park Cancer Institute, Ryan Winters from the Biosample Repository Facility at Fox Chase Cancer Center, Mary Shields and Ashley Gibbs from Northwest Oncology, Rosemary Makar and Amy Fricke from the Oregon Health & Science University Knight BioLibrary, Pearl Abernathy from Mission Health System, Stephanie Kaufman acting as honest broker from OmniSeq, Inc.
Funding
This research was funded by OmniSeq, Inc. (Buffalo, NY).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author’s contributions
MK, GD, KGM, KS, KD, LJT, JZ, ML, DM, SJM, JMC, DJG, TZ, MZ, PG, IAF, LdlCM, AS, BG, AM, JT, RS, RJ, DH, and NS collected and submitted deidentified patient data with corresponding clinical samples from their respective institutions with IRB approval. JMC, SP, MN, STG, YW, RD and CM contributed to the experimental design of this analysis. JMC, SP, MN, STG, APS, BB, JA, VG, YW, FLL, WB, LG, MG and CM prepared and analyzed patient datasets and corresponding clinical samples and were major contributors in writing and revising the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Existing deidentified specimens and associated clinical data was submitted to OmniSeq from Roswell Park Comprehensive Cancer Center (Buffalo, NY), Fox Chase Cancer Center (Philadelphia, PA), Duke University Medical Center (Durham, North Carolina), Dartmouth-Hitchcock Medical Center (Lebanon, NH), Medical College of Wisconsin (Milwaukee, WI), Meharry Medical College (Nashville, Tennessee), Northwest Oncology (Munster, IN), and Hospital Universitario Virgen Macarena (Sevilla, Spain) with institutional review board approval from each individual institution. OmniSeq’s analysis utilized deidentified data that was considered non-human subjects research under IRB-approved protocol (BDR #080316) at Roswell Park Comprehensive Cancer Center (Buffalo, NY).
Consent for publication
Not applicable.
Competing interests
Disclosures: KGM, KS, KD, LJT, JZ, ML, DM, SJM, JMC, DJG, TZ, MZ, PG, IAF, LdlCM, AS, BG, AM, JT, RS, RJ, DH, NS and RD have no conflicts of interest to disclose. JMC, SP, MN, STG, APS, BB, JA, VG, YW, FLL, MG and CM are all employees of OmniSeq, Inc. (Buffalo, NY) and hold restricted stock in OmniSeq, Inc. MK, GD, WB, CM, STG, and JMC are employees of Roswell Park Comprehensive Cancer Center (Buffalo, NY). Roswell Park Comprehensive Cancer Center is the majority shareholder of OmniSeq, Inc. LG provide remunerated consulting to OmniSeq, Inc.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dako . PD-L1 IHC 22C3 pharmDx. 2015. pp. 1–12. [Google Scholar]
- 2.Dako . PD-L1 IHC 28–8 pharmDx. 2017. [Google Scholar]
- 3.Ventana Medical Systems . VENTANA PD-L1 (SP142) Assay. 2016. pp. 1–8. [Google Scholar]
- 4.Ventana Medical Systems. VENTANA PD-L1 (SP263) Assay. 2017;1. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf16/P160046B.pdf
- 5.Teixidó C, Vilariño N, Reyes R, Reguart N. PD-L1 expression testing in non-small cell lung cancer. Ther Adv Med Oncol. 2018 11;10(14):175883591876349. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29662547. [DOI] [PMC free article] [PubMed]
- 6.Dako. PD-L1 IHC 22C3 pharmDx: Gastric or Gastroesophageal Junction Adenocarcinoma [interpretation manual]. Santa Clara, CA: Dako; 2017. Available from: https://www.agilent.com/cs/library/usermanuals/public/29219_pd-l1-ihc-22C3-pharmdx-gastric-interpretation-manual_us.pdf
- 7.Dako. PD-L1 IHC 22C3 pharmDx: Non-Small Cell Lung Cancer [interpretation manual]. Santa Clara, CA: Dako; 2015. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf15/P150013c.pdf
- 8.Paxton A. Making the best of PD-L1 IHC testing. CAP Today. 2016 Jul;1–5. Available from: http://www.captodayonline.com/making-best-pd-l1-ihc-testing/
- 9.Hansen AR, Siu LL. PD-L1 Testing in Cancer. JAMA Oncol. 2016 Jan 1;2(1):15. Available from: 10.1001/jamaoncol.2015.4685%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/26562503%0Ahttp://www.ncbi.nlm.nih.gov/pubmed/26562503%0Ahttp://oncology.jamanetwork.com/article.aspx?doi=10.1001/jamaoncol.2015.4685 [DOI] [PubMed]
- 10.Bhaijee F, Anders RA. PD-L1 Expression as a Predictive Biomarker. JAMA Oncol. 2016 1;2(1):54. Available from: http://oncology.jamanetwork.com/article.aspx?doi=10.1001/jamaoncol.2015.3782 [DOI] [PubMed]
- 11.Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol. 2017 1 [cited 2017 Dec 15];12(2):208–22. Available from: http://www.sciencedirect.com/science/article/pii/S1556086416335365?via%3Dihub [DOI] [PubMed]
- 12.Tsao MS, Kerr KM, Kockx M, Beasley M-B, Borczuk AC, Botling J, et al. PD-L1 immunohistochemistry comparability Study in real-life Clinical samples: results of blueprint Phase 2 project. J Thorac Oncol 2018 Sep;13(9):1302–11. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1556086418306269 [DOI] [PMC free article] [PubMed]
- 13.Dako . PD-L1 IHC 28–8 pharmDx Interpretation Manual Melanoma. 2017. [Google Scholar]
- 14.Mathew M, Safyan RA, Shu CA. PD-L1 as a biomarker in NSCLC: challenges and future directions. Ann Transl Med 2017 Sep;5(18):375–375. Available from: http://atm.amegroups.com/article/view/16060/16633 [DOI] [PMC free article] [PubMed]
- 15.Cook RW, Middlebrook B, Wilkinson J, Covington KR, Oelschlager K, Monzon FA, et al. Analytic validity of DecisionDx-Melanoma, a gene expression profile test for determining metastatic risk in melanoma patients. Diagn Pathol. 2018;13(1):1–8. doi: 10.1186/s13000-018-0690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh PS, Wilde JI, Tom EY, Reynolds JD, Chen DC, Chudova DI, et al. Analytical performance verification of a molecular diagnostic for cytology-indeterminate thyroid nodules. J Clin Endocrinol Metab 2012 Dec;97(12):E2297–306. Available from: https://academic.oup.com/jcem/article-lookup/doi/10.1210/jc.2012-1923 [DOI] [PubMed]
- 17.Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics. 2015;8(1):1–14. doi: 10.1186/s12920-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beumer IJ, Persoon M, Witteveen A, Dreezen C, Chin S, Sammut S, et al. Prognostic value of MammaPrint ® in invasive lobular. Breast Cancer. 2016:139–46. [DOI] [PMC free article] [PubMed]
- 19.Baehner FL. The analytical validation of the oncotype DX recurrence score assay. Ecancermedicalscience. 2016;10:1–11. doi: 10.3332/ecancer.2016.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penland SK, Keku TO, Torrice C, He X, Krishnamurthy J, Hoadley KA, et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Investig 2007 Apr 12;87(4):383–91. Available from: http://www.nature.com/doifinder/10.1038/labinvest.3700529 [DOI] [PubMed]
- 21.Li P, Conley A, Zhang H, Kim HL. Whole-Transcriptome profiling of formalin-fixed, paraffin-embedded renal cell carcinoma by RNA-seq. BMC Genomics. 2014 11;15(1):1087. Available from: http://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-15-1087 [DOI] [PMC free article] [PubMed]
- 22.Blomquist TM, Crawford EL, Lovett JL, Yeo J, Stanoszek LM, Levin A, et al. Targeted RNA-Sequencing with Competitive Multiplex-PCR Amplicon Libraries. Lenburg M, editor. PLoS One . 2013 13;8(11):e79120. Available from: http://dx.plos.org/10.1371/journal.pone.0079120 [DOI] [PMC free article] [PubMed]
- 23.Conroy JM, Pabla S, Glenn ST, Burgher B, Nesline M, Papanicolau-Sengos A, et al. Analytical validation of a next-generation sequencing assay to monitor Immune responses in solid tumors. J Mol Diagnostics 2018 Jan 20;20(1):95–109. Available from: http://linkinghub.elsevier.com/retrieve/pii/S152515781730363X [DOI] [PubMed]
- 24.Paluch BE, Glenn ST, Conroy JM, Papanicolau-Sengos A, Bshara W, Omilian AR, et al. Robust detection of immune transcripts in FFPE samples using targeted RNA sequencing. Oncotarget 2017 Jan 10;8(2):3197–205. Available from: http://www.impactjournals.com/oncotarget/index.php?journal=oncotarget&page=article&op=view&path%5B%5D=13691&path%5B%5D=43523 [DOI] [PMC free article] [PubMed]
- 25.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer. 2009 Jan;45(2):228–47. Available from: 10.1016/j.ejca.2008.10.026 [DOI] [PubMed]
- 26.McDermott DF, Sosman JA, Sznol M, Massard C, Gordon MS, Hamid O, et al. Atezolizumab, an Anti–Programmed Death-Ligand 1 Antibody, in Metastatic Renal Cell Carcinoma: Long-Term Safety, Clinical Activity, and Immune Correlates From a Phase Ia Study. J Clin Oncol. 2016 10;34(8):833–42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26755520. [DOI] [PubMed]
- 27.Daud AI, Wolchok JD, Robert C, Hwu W-J, Weber JS, Ribas A, et al. Programmed Death-Ligand 1 Expression and response to the Anti–Programmed Death 1 Antibody Pembrolizumab in Melanoma. J Clin Oncol 2016 Dec;34(34):4102–9. Available from: http://ascopubs.org/doi/10.1200/JCO.2016.67.2477 [DOI] [PMC free article] [PubMed]
- 28.Long G V., Atkinson V, Cebon JS, Jameson MB, Fitzharris BM, McNeil CM, et al. Standard-dose pembrolizumab in combination with reduced-dose ipilimumab for patients with advanced melanoma (KEYNOTE-029): an open-label, phase 1b trial. Lancet Oncol. 2017 Sep;18(9):1202–10. Available from: 10.1016/S1470-2045(17)30428-X [DOI] [PubMed]
- 29.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of Non–Small-Cell Lung Cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 30.Langer CJ, Gadgeel SM, Borghaei H, Papadimitrakopoulou VA, Patnaik A, Powell SF, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol 2016 Nov;17(11):1497–508. Available from: 10.1016/S1470-2045(16)30498-3 [DOI] [PMC free article] [PubMed]
- 31.Yu J, Wang X, Teng F, Kong L. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. 2016 ;volume 9:5023–39. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27574444. [DOI] [PMC free article] [PubMed]
- 32.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016 Dec;17(12):e542–51. Available from: 10.1016/S1470-2045(16)30406-5 [DOI] [PMC free article] [PubMed]
- 33.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016 May 15;16(5):275–87. Available from: 10.1038/nrc.2016.36%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/27079802 [DOI] [PMC free article] [PubMed]
- 34.Brüggemann C, Kirchberger MC, Goldinger SM, Weide B, Konrad A, Erdmann M, et al. Predictive value of PD-L1 based on mRNA level in the treatment of stage IV melanoma with ipilimumab. J Cancer Res Clin Oncol. 2017 Oct 14;143(10):1977–84. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28616701. [DOI] [PMC free article] [PubMed]
- 35.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN- γ – related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017 Jun 26;127(8):2930–40. Available from: https://www.jci.org/articles/view/91190 [DOI] [PMC free article] [PubMed]
- 36.Prat A, Navarro A, Paré L, Reguart N, Galván P, Pascual T, et al. Immune-Related Gene Expression Profiling After PD-1 Blockade in Non–Small Cell Lung Carcinoma, Head and Neck Squamous Cell Carcinoma, and Melanoma. Cancer Res. 2017 1;77(13):3540–50. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28487385. [DOI] [PubMed]
- 37.Morrison C, Pabla S, Conroy JM, Nesline MK, Glenn ST, Dressman D, et al. Predicting response to checkpoint inhibitors in melanoma beyond PD-L1 and mutational burden. J Immunother Cancer. 2018 9;6(1):32. Available from: https://jitc.biomedcentral.com/articles/10.1186/s40425-018-0344-8 [DOI] [PMC free article] [PubMed]
- 38.Chen DS, Mellman I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity. 2013 Jul 25;39(1):1–10. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23890059. [DOI] [PubMed]
- 39.Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat Rev Clin Oncol. 2016 15;13(5):273–90. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23890059. [DOI] [PMC free article] [PubMed]
- 40.Herbst RS, Soria J, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014 Nov 27;515(7528):563–7. Available from: http://www.nature.com/doifinder/10.1038/nature14011 [DOI] [PMC free article] [PubMed]
- 41.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012 Apr 1;12(4):252–64. Available from: 10.1038/nrc3239%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/22437870 [DOI] [PMC free article] [PubMed]
- 42.Martinez GJ, Pereira RM, Äijö T, Kim EY, Marangoni F, Pipkin ME, et al. The transcription factor NFAT promotes exhaustion of activated CD8 + T cells. Immunity. 2015;42(2):265–278. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 2018 Jun 13;558(7710):454–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29899446. [DOI] [PMC free article] [PubMed]
- 44.Udall M, Rizzo M, Kenny J, Doherty J, Dahm S, Robbins P, et al. PD-L1 diagnostic tests: a systematic literature review of scoring algorithms and test-validation metrics. Diagn Pathol. 2018 9;13(1):12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29426340. [DOI] [PMC free article] [PubMed]
- 45.Network NCC. NCCN Clinical practice guidelines in oncology (NCCN guidelines®) - chronic myeloid leukemia, version 4.2018 [Internet]. Vol. 4.2018. National Comprehensive Cancer Network: Fort Washington, Pennsylvania; 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Samples (n=209) sorted within tumor type by PD-L1 rank (RNA-seq). Table S2. PD-L1 (CD274) RNA-seq dilution series. Table S3. Responders and non-responders across tumor types and biomarker result. Table S4. ORR across tumor type and combined biomarker results for all ICI therapies. Table S5. Responders and non-responders across tumor types and individual biomarker result. (XLSX 39 kb)
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.