Abstract
Breast cancer is recognized for its different clinical behaviors and patient outcomes, regardless of common histopathological features at diagnosis. The heterogeneity and dynamics of breast cancer undergoing clonal evolution produces cells with distinct degrees of drug resistance and metastatic potential. Presently, single cell analysis have made outstanding advancements, overshadowing the hurdles of heterogeneity linked with vast populations. The speedy progression in sequencing analysis now allow unbiased, high-output and high-resolution elucidation of the heterogeneity from individual cell within a population. Classical therapeutics strategies for individual patients are governed by the presence and absence of expression pattern of the estrogen and progesterone receptors and human epidermal growth factor receptor 2. However, such tactics for clinical classification have fruitfulness in selection of targeted therapies, short-term patient responses but unable to predict the long-term survival. In any phenotypic alterations, like breast cancer disease, molecular signature have proven its implication, as we aware that individual cell’s state is regulated at diverse levels, such as DNA, RNA and protein, by multifaceted interplay of intrinsic biomolecules pathways existing in the organism and extrinsic stimuli such as ambient environment. Thus for complete understanding, complete profiling of single cell requires a synchronous investigations from different levels (multi-omics) to avoid incomplete information produced from single cell. In this article, initially we briefed on novel updates of various methods available to explore omics and then we finally pinpointed on various omics (i.e. genomics, transcriptomics, epigenomics, proteomics and metabolomics) data and few special aspects of circulating tumor cells, disseminated tumor cells and cancer stem cells, so far available from various studies that can be used for better management of breast cancer patients.
Keywords: Single cell omics, Genomics, Transcriptomics, Proteomics, Molecular sub-typing of breast cancer
Introduction and Significance of Single Cell Omics
Breast cancer is one of the prime health complications in the United States and worldwide. The American Cancer Society projected 232,340 new cases of invasive breast cancer and 64,640 new cases of ductal carcinoma in situ (DCIS) to be diagnosed amongst women in the United States in 2013, of which 29% were invasive breast cancer [1]. Amongst the Indian women, Breast cancer is the second most common cancer. As per the current data gathered for ‘Atlas of Cancer in India project’ which is evaluating nationwide patterns of cancer incidence across urban and rural parts of India, suggests that breast cancer is most prevalent cancer in metropolitan cities and is predicted to be the most common type of cancer in the coming decade. The incidence is higher in some cities according to data from the Atlas project (for example, Chandigarh 39.5 per 100,000; North Goa 36.8 per 100,000) as compare to those reported by the population-based registry in New Delhi (28.9 per 100,000). The age-adjusted incidence rate in Bangalore, Chennai, Delhi, Mumbai, and Kolkata, 30.9, 33.0, 31.4, 29.3, and 20.6 per 100,000, respectively [2]. A recent report by the Indian Council of Medical Research forecasts the number of breast cancer cases in India may rise from 106,124 in 2015 to 123,634 in 2020 (Cancer Incidence Rates 1982–2005). According to the National Cancer Registry Programme projections, there is an increasing trend in number of breast cancer deaths in India and have risen to 106,124 in 2015 and predicted to further increase to 123,634 in 2020 (Cancer Incidence Rates 1982–2005) [2].
Study of single cell genomics can potentially facilitate development of new schemes for cell characterization, distinguish cellular transition states, unearth hidden biological features and map molecular markers. These can be used for new classifications of here to fore apparently uniform populations. To achieve this goal, innovative methods are necessary to address the high level of clatter characteristic in single cell genomics, related to technical complications of extremely low amounts of input material and biological issues such as transitory bursts of RNA transcription. Combining single cell studies and hypothetical gene expression analyses can reveal both qualitative and quantitative characteristics of gene regulation, which otherwise would remain undetermined in population averaging gene expression studies [3, 4]. In order to advance single cell studies, microfluidic devices are now being developed to do next generation sequencing (NGS) on suitable linear mRNA after amplification and bar-coding, in lesser time than prior generation of genome experiments.
Breast cancer is a diverse disease characterized by different pathological features, incongruent response to therapeutics, and extensive inconsistencies in long-term patient survival [5]. The heterogeneity observed among breast cancers indicates now well-accepted notion that there is not just one disease with a few alternative subtypes, instead breast cancer exemplifies a group of distinct neoplastic diseases of the breast and the cells composing the breast [5]. Further, the distinct nature and character of these disorders can be appreciated through traditional pathological examination (i.e., in terms of disease morphology) but the true extent of variety among breast cancers can be realized only through molecular analyses. Almost 80% of the invasive breast cancers are represented by invasive ductal carcinoma and invasive lobular carcinoma is the next most common, representing approximately 10% of invasive breast cancers. The less frequent subtypes comprise mucinous, cribriform, inflammatory, papillary, medullary, metaplastic, tubular and micropapillary carcinomas. These subtypes can be further subdivided into classifications based on their molecular signatures (i.e., expression of protein biomarkers or gene expression profiles).
Methods for Isolating and Analyzing Multiple Types of Molecules from a Single Cell
Standardization of an effective method for making cell suspension that could be used in isolation of high-quality RNA or flow cytometer analysis from limiting amount of human origin tissue samples is very challenging. As we are familiar that tissue obtained during gun-biopsy per core is not more than 100 mg so to utilize this less amount of tissue in RNA isolation and for characterizing diverse antibodies in flow cytometer, is the key challenge for molecular biologists.
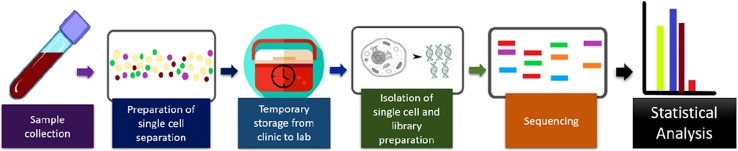
Last 30 years has seen insistent progress in the field of molecular biology and molecular techniques leading to new vistas in study of Omics like genomics, transcriptomics and proteomics which could be employed in solving the conundrum of chronic and untreatable diseases [5–8]. DNA, RNA and proteins are the fundamental molecules required for such diverse branches and, these molecules inhabit within the tissue, cells and cell membranes, enclosing their cytoplasm and genomic contents. These molecules have shown efficiency for use not only as a biomarkers but also in tracing various disease associated pathways [9–13]. The Fig. 1 represents outline of the process of applying single cell sequencing to patient derived tumour samples.
Fig. 1.
Overview of the process of applying single cell sequencing to patient derived tumor samples
For sample preparation to study single cell biology from any tissue, standard methodology should be used. For example, harsh sample treatment could affect nucleic acid quality, quantity along with production of a lot of debris which may affect further gene expression, flow cytometer analysis. Therefore, it is important to standardize the tissue disruption methods to obtain RNA, or single cell suspension from plentiful as well as from minute tissue representatives obtained during surgery or during core biopsy. Enzyme digestion method (Trypsin/Collagenase based method) are better than homogenization based method for single cell preparation. Mostly labs including ours follow, Trypsin based digestion and is most favorable for flow-cytometer analysis since it reduces debris in comparison of mechanical process of shearing [14].
Whole Genome Amplification Methods from Single Cells
Obtaining enough amount of molecules, including DNA or RNA, from single cell is a great challenge. For example, for single cell sequencing, limited amount of DNA or cDNA molecules need to be amplified with higher fidelity and less bias. Several Whole-Genome Amplification (WGA) methods have been used to obtain sufficient DNA for sequencing. Here we quickly sum up a few of these methods and their features.
DOP-PCR
Previously degenerate oligonucleotide-primed Polymerase Chain Reaction (DOP-PCR), was extensively used to amplify genome from single cell. It has vital usages in genome mapping and in identification of the source of markers, determine copy number variations (CNVs), and map translocation breakpoints on a significant genomic scale [15]. However, DOP-PCR has low genome coverage, high amplification prejudices, and high drop-out rate owing to the exponential amplification nature of PCR [16].
MDA
Yet another method is Multiple Displacement Amplification (MDA). It amplifies DNA in single-cell whole-genome analyses. This technique causes, within few hours circular DNA templates amplification by 10,000-fold, using random primers and Phi29 DNA polymerase [17]. MDA gives much higher genome coverage than DOP-PCR, with a drawback of giving rise to chimeric reads and introduces huge amplification bias due to its exponential amplification process [18]. Furthermore, such sequence-dependent bias of MDA is not reproducible along the genome from cell to cell.
MALBAC
Multiple Annealing And Looping-Based Amplification Cycle (MALBAC) method reduces the sequence-dependent bias introduced by exponential amplification, since it includes quasi-linear amplification through looping-based amplicon protection into PCR [19]. MALBAC uses primers in the initial reaction, designed to share common sequences forming loops and inhibiting the repeated (potentially biased) priming from their ends. This technique offers high stability across the genome. Such amplified DNA on sequencing can accomplish 93% genome cover-age ≥ 1× for a single human cell at a mean sequencing depth of 25× [20].
LIANTI
Linear Amplification via Transposon Insertion (LIANTI) combines Tn5 transposition and T7 in vitro transcription for single-cell genomic analyses facilitating further reduction in amplification bias and errors [21]. During this process Tn5 transposition first randomly fragments and then inserts T7 promoter sequence into genomic DNA. Further, T7 RNA polymerase facilitates generation of amplified antisense RNA. After c-DNA synthesis and second strand synthesis, double-stranded LIANTI amplicons are organized for DNA library preparation and high throughput sequencing. Thus, LIANTI efficiently reduces PCR’s errors and biases (induced by nonspecific priming and exponential amplification) by replacing PCR with in vitro transcription.
The ‘Omics’ Application Based on Single Cell
Brisk technological development has improved the understanding of single cell omics. Single-cell sequencing of genomes, and transcriptomes, is now well-known and largely advantageous, and the primary methods for mapping single-cell epigenomes, proteomes, and metabolomes, are now becoming accessible. Table 1 is representing the various single cell methods and their application to omics of cancer.
Table 1.
Summary of relevant single-cell methods and their applications to cancer [22]
| Experimental method type | Specific methods | Application to cancer “omics” |
|---|---|---|
| Single-cell whole genome amplification | DOP-PCR, MDA, MALBAC | Used in conjunction with next-generation sequencing to detect intra-tumor CNVs and SNPs |
| Single-cell spatial genomics | STAR-FISH | Detects the spatial distribution of intra-tumor CNVs and SNPs. Can be combined with longitudinal analysis to reveal migratory cells |
| Single-cell transcriptome amplification | Smart-seq, Tang et al. method, | single-cell qPCR Identifies cancer-specific gene expression signatures, cancer cell types, alternative-splicing events |
| Single-cell spatial transcriptomics | smFISH, SeqFISH, MERFISH, FISSEQ, TIVA | Can provide spatially-resolved gene expression signatures in tumors. Has potential applications in tracing cell migratory paths and locating tumor-like stem cells |
| Single-cell DNA methylomics | scRRBS, PBAT | Enables the discovery of differential methylation in cancer cells. Potential for broadening understanding of phenotypic plasticity of cancer cells |
| Single-cell chromatin accessibility | ATAC-seq, Pico-Seq | Can give insight into the differential binding of transcription factors in cancer cells |
| Chromosome conformation capture | Hi-C, ChIP-seq | Potential for understanding the mechanisms of cancer heterogeneity through mapping transcription factor-regulatory element interactions |
| Simultaneous multiple single-cell omics | G&T-seq, scTrio-seq, | Provides an integrated view of intra-tumoral heterogeneity through measuring direct interactions between genomic, transcriptomic, epigenetic, and proteomic variation |
| Computational methods Single-cell spatial transcriptomic inference | Seurat, Achim et al. method | Infers cell location through scRNA-seq data and an in situ RNA reference map of several landmark genes, enabling mapping of intra-tumor spatial heterogeneity |
| Pseudo-time ordering | Monocle, TSCAN, Waterfall, SCUBA, Wanderlust, Wishbone | Projects gene expression values from a single time-point to a continuous trajectory over cell differentation. Potential use in understanding differentiation from stem-like cancer cell to matured cancer cell |
| Rare cell-type detection | RaceID, StemID, GiniClust | Potential use in the detection of circulating tumors cells and stem-like cancer cells |
| Clonal evolution inference | SCITE, OncoNEM | Builds lineage trees for understanding evolutionary events such as the development of therapy resistance |
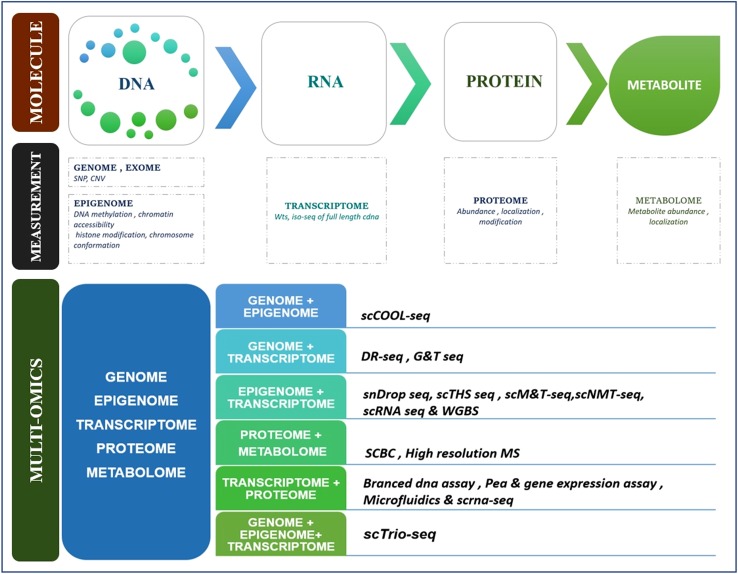
Amalgamating many of these technologies into integrated multi-omic assays of the same single cells will yield exceptional visions in fundamental biology and biomedicine. The Fig. 2 representing the theoretical diagram of multi-omics in single cell.
Fig. 2.
Representing the theoretical diagram of multi-omics in single cell
Single Cell Genomics and Breast Cancer
Single cell genome sequencing (SCGS) has been used to explain mutations, structural variations, aneuploidies, and recombination in the genome, and to study the diversity, evolution and role of genetic mosaicism [23]. SCGS is critical for unveiling genetic heterogeneity and cell-lineage communications between normal and diseased tissues [24]. Since a precise appraisal of prognosis is critical in conceiving a efficacious treatment strategy for cancers, single-cell technology has permitted many novel prognostic factors to be detected and confirmed. For example, this technique was applied to detect and map the source of disseminated tumor cells in breast cancer [25].
Single-cell sequencing is of paramount significance for appreciating the genomic heterogeneity of cancer cells. The earliest amplifications for single cell genomics encompass whole genome amplification, delivering adequate amounts of DNA for successive sequencing. DOP-PCR has low coverage but even amplification of genome and is appropriate for CNV detection [26]. SNP recognition can be done using multiple displacement amplification (MDA), a linear amplification method proficient of higher coverage via the use of Phi-29 polymerase [27]. Multiple annealing and looping based amplification cycles (MALBAC) pools MDA and PCR for a high coverage, and turns out to be a uniform amplification technique, appropriate for either CNV or SNP detection [19]. Intra-tumor CNVs and SNPs in various cancers have been characterized using these techniques. However, one key shortcoming of the above-mentioned methods is that three-dimensional information is lost as soon as single cells are isolated, which is essential to insight about relations of the cell with its micro-environment and may prove important for evaluating drug responsiveness. Lately, a new tool, exclusive-to-allele PCR-FISH (STAR-FISH) [28], has been recognized. This technique can perceive the spatial distribution of both SNPs and CNVs via combination of in situ PCR and FISH. PCR primers targeting mutant and wild type mRNAs, one gene at a time are used. Amplification is followed by hybridization of fluorophores to a 50 over-hang built into each probe. A frequently detected mutation of Her2+ breast cancer before and after chemotherapy is His1047Arg mutation in PIK3CA and ERBB2 (commonly known as HER2). Janiszewska et al. used their method to study this commonly reported amplification. They acknowledged changes in mutational frequency of mutated cells, which enabled a perception of the development of drug resistance in HER2+ breast cancer. In combination with longitudinal analysis, this method was made use of to locate migratory cells [28]. Currently, the technology can only be employed to locate known mutations. The introduction of such spatial methods to single-cell cancer genomics allows genomic heterogeneity to be plotted in space. This presents innovative prospects in studying cell-to-cell interactions, and in recognizing migratory cancer cells and their roles in metastasis.
SNPs and Mutations and Breast Cancer
While the degree of analysis of tumor cells is indeed of the quintessence, it is also vital to amplify the information that is retrieved from each cell. For genome-wide DNA analysis of mutation and copy number profiling, two whole-genome amplification (WGA) approaches have been commonly accepted for single-cell DNA-seq: multiple displacement amplification (MDA) [29], and the multiple-annealing, looping-based amplification-based cycle method (MALBAC). Choice of method depends primarily on the question of interest. Each method is related with artefacts introduced owing to allelic drop out (ADO), false discovery of mutations due to amplification or sequencing errors and preferential amplification of specified genomic sites. Overall, MDA displays greater steadiness in comparison with MALBAC. On the contrary, MDA false-negative rates are also higher due to lower genome coverage and low consistency of coverage. Rates of ADO differ greatly, with percentages as low as 1% reported for MALBAC and as high as 65% for MDA [30], even though ~ 10% ADO can perhaps be expected on average for most samples. Considering this, MALBAC has been the method worthwhile for copy number aberration, however it is significant to note that reproducibility of results for smaller copy number aberrations remains low [31].
Furthermore, precise molecular variants of breast cancer were found to be linked to specific genetic modifications. For example, HER2+ and basal like breast cancers exhibit a high rate of somatic mutation in the TP53 tumor suppressor gene (72–80%), whereas other molecular variants display TP53 gene mutations relatively less frequently (12–29%). Luminal A, luminal B, and HER2+ subtypes presented significant percentages of mutation in the PIK3CA gene (45%, 29%, and 39%, respectively), but basal-like breast cancers are only rarely related with mutation of this gene (9%). It is vital that very limited genes were perceived to be mutated at more than 10% frequency within or across the molecular subtypes of breast cancer, but several genes (including at least 177 cancer-associated genes) were mutated in lesser numbers of cancer (> 20,000 non silent somatic mutations among 510 breast cancers). CNVs showing gene deletions and amplifications were found to influence several genes and gene regions, comprising amplifications in PIK3CA and ERBB2 chromosomal regions and deletions in TP53 and MAP2K4 chromosomal regions, among many [32].
Single Cell Epi-genomics and Breast Cancer
Epigenetics plays a crucial role in regulating gene expression in cancer, and exploring the heterogeneity of epigenetic patterns may contribute in understanding causal transcriptomic heterogeneity. As a dynamic process, it may add to the phenotypic plasticity of cancer cells, for example aiding in the differentiation of cancer stem cells.
Studies have exhibited remarkably low levels of global DNA methylation along with hyper-methylation of definite regions, such as tumor suppressor gene promoter regions, providing robust indication for the role of epigenetic abnormalities in cancer dissemination. The characterization of intra-tumor epigenetic heterogeneity has been explored less extensively due to its technological challenges. However, multiple epigenetic tactics have recently been customized for single-cell purposes. Establishing DNA methylation patterns has conventionally been performed by bisulfite sequencing methods, nevertheless bulk techniques have not performed well in the single-cell setting owing to DNA degradation occurring during bisulfite conversion. Some methods have modified bisulfite sequencing for single-cell, including scRRBS (reduced representation bisulfite sequencing) [33], and PBAT (post-bisulfite adapter-tagging) wherein a modified version of bisulfite sequencing is employed to each cell individually. ScRRBS relieves the problem of high DNA loss by substituting the several purification steps preceding bisulfite sequencing with a single-tube reaction. A restriction enzyme recognizing CpG islands is exploited to cut the genome, selecting CpG island regions for subsequent conversion and sequencing. By sequencing particularly these regions, this method offers low-cost but low-coverage sequencing [33]. This technique has been utilized in human hepatocellular carcinoma tissue along with transcriptome sequencing. Methylation levels at all CpG sites were established and consequently used to cluster the tissue into two subpopulations through unsupervised graded clustering. An immense heterogeneity was obtained between and within these sub-populations. Remarkably, when the same clustering method was harnessed using CNV patterns, a matching clustering was found [34]. PBAT is a rather unprejudiced whole-genome method, which focusses on the issue of bisulfite-conversion-induced DNA degradation and achieves suitable library preparation after bisulfite sequencing. Typically, adapter-tagging is done prior to bisulfite conversion and sequencing templates are degraded, but changing the order of these events impedes this problem [35]. While using PBAT, differential methylation of distal regulatory elements was unravelled in mouse embryonic stem cells [35]. These elements cannot usually be captured by scRRBS, making it look good for higher-coverage cancer methylation studies.
Another important regulator of gene expression is chromatin structure. Largely transcription factors can only bind to open chromatin regions, however a small number of pioneer factors perhaps bind to closed chromatin, initiating it to open up so that other factors can bind. The genome-wide landscape of chromatin openness can be evaluated by using either ATAC-seq (assay for transposase-accessible chromatin) or DNase-seq [36]. These methods differ in the DNA-cutting enzymes, analogous to Tn5 and DNase I, respectively. Both methods have been fitted to single-cell analysis. Two single-cell methods have modified ATAC-seq. A combinatorial indexing tactic labels nuclei with unique barcodes to ease their grouping and processing together. Groups of nuclei are positioned in wells, barcoded, and then sent on through a second set of wells and barcoded again. Assuming that each nuclei is highly probable to pass through an exclusive combination of wells, the barcoding is enormously cell-specific. In a microfluidic tactics, cells are captured and analyzed separately. This technique has been used to dig up inconsistency of transcription factor motif accessibility in cancer cell lines. For DNase-seq, a single-cell method called Pico-Seq [36], separates cells using FACS prior to DNase I treatment. To evade a large damage of digested DNA during subsequent library preparation, a circular carrier DNA is added after digestion. The carrier DNA will not be amplified in the ensuing PCR due to its mismatch with the adaptor ligation process. Notably, the authors used their method to formalin-fixed paraffin-embedded follicular thyroid cancer patient tissue and, observed a SNV in one of the patient samples that inhibits the binding of tumor suppressor protein p53 [36]. The above-mentioned methods have started to deliver new systematic revelations into cancer heterogeneity. Besides, two more single-cell methods, Hi-C and ChIP-seq, have been lately developed and show potential for use in forthcoming cancer epigenetic studies. A type of chromosome conformation capture that quantifies interactions between genomic loci, Hi-C can be utilized to locate trans-regulatory elements and their targets [37], ChIP seq, which differentiates interactions between DNA and DNA-binding proteins, can determine transcription factor regulatory element interactions. Results associated to examinations of gene expression patterns, gene mutations, DNA copy number, DNA methylation, and miRNA expression patterns in a huge cohort of approximately 800 breast cancers were published in 2012 by the Cancer Genome Atlas Network [32]. This study distinctly revealed that breast cancer is a heterogeneous disease with multiple discrete molecular variants and also there is great variety among the well-known major molecular subtypes. This suggests that the molecular progressions influencing the pathogenesis of breast cancers of a particular molecular subtype can vary, involving varied procedures for gene activation or inactivation and different genes signifying positive and negative mediators of neoplastic development and progression, and that, therefore, no singular molecular mechanism of breast cancer pathogenesis exists [32].
Global hypo methylation and gene-specific hyper-methylations are Cancer-associated modifications in DNA methylation. Present-day evidence advocates that these epigenetic processes play a key role in breast carcinogenesis. Genes that have been recognized to be directly quietened by DNA methylation in breast cancer encompass cell-cycle control genes (CDKN2A), steroid receptor genes [ESR1 (alias Era, NR3A1), PGR (alias PR, NR3C3), and RARB (alias HAP, NR1B2, RRB2)], tumor suppressor genes (BRCA1), genes associated with cancer metastasis (CDH1, TIMP3), and many others [38]. Loss of expression of ER is often related with hyper methylation of the ESR1 gene. Moreover hyper methylation of specific genes, hypo-methylation influencing large chromosomal regions can be merged with aberrant or inappropriate expression of genes that contribute to cancer development and progression. Additionally, genome-wide demethylation promotes chromosomal instability by destabilizing peri-centromeric regions of certain chromosomes. Thus, epigenetic processes effective in breast cancer may contribute to transformed expression of specific genes, altered expression of genes positioned in common chromosomal regions, and/or genetic instability bring about copy number alterations.
Single Cell Transcriptomics and Breast Cancer
Similar to single-cell genome evaluation, the first efforts in single-cell transcriptomics were of the amplification of the transcriptome to enable for quantification and sequencing of the transcriptome. Whole transcriptome amplification techniques comprise poly-A tailing methods and template-switching approaches for example Smart-seq [39]. Targeted gene expression profiles can also be determined with high sensitivity by multiplexing qPCR [40]. In amalgamation with single-cell RNA sequencing and qPCR, these techniques have been used in several cancer studies. Cancer-specific gene expression signatures and alternative-splicing events have been recognized for melanoma [40]. Such signatures have led to the perception of cancer cell types, such as cancer stem cells. The relative impacts of clonal evolution and multi-lineage disparity in transcriptomic heterogeneity have been studied in the perspective of colon cancer [41]. Current technologies have been developed to compute gene expression levels in situ, thereby maintaining spatial information. Here we review recent single-cell spatial transcriptomic methods and their potential for future use in cancer studies. These methods share the same basic principle as single-molecule fluorescence in situ hybridization (smFISH), whereby fluorescently-labeled DNA oligonucleotide probes are hybridized to their complementary target mRNA, and are then spotted via fluorescence microscopy. The latest practices described below have greatly enhanced detection proficiency and output. SeqFISH (sequential FISH) is a deviation of smFISH that utilizes sequential hybridization to permit for multiplexing. Each mRNA is attributed a unique sequence of fluorophores which create a barcode, that facilitates decoding of each mRNA. In the first round of this process, probes aiming the same mRNA are labeled with the same fluorophore. Furthermore, these probes are hybridized, imaged, and then purged. Subsequently, the same probes are labeled with a different fluorophore, and the same sequence of steps is followed. Multiple such rounds creates a unique barcode of colors unique for a specific mRNA. Thus each probe set targeting a particular mRNA gets labeled with a unique barcode. For F fluorophores and N hybridization rounds, this means FN mRNAs can be visualized. As this number surges quickly with an increasing number of fluorophores and hybridization rounds, this technique can potentially be used to sequence all known genes with restrained numbers of fluorophores and hybridization cycles. Initially this method was employed to immobilized yeast cells and mouse embryonic stem cells, but has been since encompassed to deep tissues such as the brain. MERFISH (multiplexed error-robust FISH) is a similar approach that also allows for error rectification by using a smart choice of barcodes. Precisely, barcode sequences are chosen to include only those which are separated by a certain Hamming distance (Hamming distance = number of changes in a barcode sequence required to transform one sequence into another). Because not all possible barcodes encode a specific mRNA, this encoding scheme suggests a measures to error detection and correction. The authors have engaged this approach to synchronously measure 1001 genes in human fibroblast cells. Two fluorophores and 14 hybridization rounds authorize all encoding sequences to be split by a Hamming distance of 2 [42]. Of note, these authors illustrate that their barcode design helps reduce the error rate significantly. FISSEQ is another in situ technique based on sequencing. RNA is first reverse-transcribed and amplified [43]. The amplicons are crosslinked to the cellular matrix and sequenced by using the SOLiD SBL (sequencing-by-ligation) method. This has been used to a simulation of the wound healing response in primary fibroblasts and the authors witnessed differentially expressed genes midst migrating cells and contact-inhibited cells [43]. Such a process could similarly be employed to find differentially expressed genes in migratory versus non-migratory tumor cells. Besides, in vivo quantification of transcriptomic profiles can also be by using a technology called TIVA (transcriptome in vivo analysis) in which a photo-activatable biotin-labeled TIVA-tag is inserted into live cells, attached to mRNA upon selective photo-activation, and recaptured via streptavidin beads and captured mRNA is eventually sequenced. TIVA was applied on live mouse and human brain tissue, as well as mouse brain cells in culture and a comparison of live and culture mouse brain cells displays notable differences in gene expression levels, emphasizing that cells removed from their natural environment may not be representative of the same cells in vivo [44]. The above referred techniques give increasingly multiplexed ways of spatially mapping gene expression patterns. While most of the applications to date have been restrained to cell culture, we believe that soon they will be valid for tissue samples. If they can be customized to tumor cross-sections, these methods will have great impression on investigating the cancer progression path. For example, the position of tumor-like stem cells could be mapped within the tumor. If longitudinal measurements are taken, cell migratory paths may also be discovered. Gene expression profiling computes the activity of thousands of genes to create a global picture of cellular function. Some gene expression profiling tests utilize fresh tissue while others use tissue preserved in paraffin. Using tumor samples from a biopsy or other surgical procedure, gene expression profiling measures the specific messenger RNA that the genes code. The mRNA in a tumor is measured to appraise which genes in a cell are active. Identifying which genes are active can reveal a tumor’s uniqueness that will determine clinical outcome in breast cancer patients.
Computer analysis of the results facilitated the assessment of relationships between the gene expression patterns and information from the women’s medical records (i.e., cancer recurrence and whether the woman benefited from treatment such as chemotherapy). Researchers then developed numbering scales or scores for predicting how likely cancer will relapse or how likely a woman would benefit from treatment. MammaPrint® and Oncotype DX® are two well-known gene expression profiling tests for breast cancer, which compute gene expression levels within the tumor to produce number scores and its probability of the risk of distant disease recurrence. These tests can be utilized together with data such as a woman’s age, whether her cancer has spread, and whether the tumor tests positive for hormone receptors to guide treatment decisions.
MammaPrint® is a 70-gene signature assay obtained from the investigation of 78 frozen samples from lymph node-negative breast cancers, smaller than 5 cm, from patients younger than 55 years, treated at the Netherlands Cancer Institute. A prognostic signature was recognized comprising 70 genes, by matching the expression profiles of tumors from patients who developed distant metastasis within 5 years with those who did not. Oncotype DX® is a 21-gene RT-PCR test for estrogen receptor (ER)-positive breast cancer, which was based upon prospective selection of 250 genes related with cancer pathology and prognosis, from the published literature and genomic databases. Comparing the aforesaid assays, MammaPrint® uses fresh-frozen tumor tissue which is more frequently available from procedures done in Europe, whereas Oncotype DX® uses tissues preserved in paraffin, a typical procedure in the United States. Lack of direct comparison of the two methods, keeps the issue unanswered as to which test is better than the other at predicting what the best treatment will be or whether a breast cancer will return.
In a prospective, randomized phase III clinical trial called the MINDACT, the 70-gene unique assay is presently being evaluated. This phase III trial will enroll 6000 patients and will compare MammaPrint® to Adjuvant Online, a popular clinical-pathological prognostic tool, for selecting patients for adjuvant chemotherapy in node-negative breast cancer. Yet another phase III trial is TAILORx where the Oncotype DX® test is being weighed in another prospective [45]. This phase III trial will enroll over 10,000 women at 900 sites in the United States and Canada and investigate if genes frequently associated with recurrence can be used to ascribe early breast cancer patients to the most fitting and effective treatment. Both Oncotype DX® and MammaPrint® are useful for women who have hormone receptor-positive early breast cancer. These two assays have only one gene in common and this lack of overlap in genes might be due to the way tumor tissues are prepared for the tests and other differences in procedures that the laboratories use. Also, the heterogeneous nature of breast cancer means that many genes are implicated in the cancer molecular pathway. MammaPrint® has been approved by the US Food and Drug Administration (FDA) for its prognostic value (i.e., predicting the likelihood that breast cancer will return). However, it is not endorsed for predicting the response to treatment. That question is still under investigation. Oncotype DX has yet to be approved by the FDA, although the safety and performance standards of the test conducting labs are regulated under the Clinical Laboratory Improvements Amendments (CLIA). Although these two tests are extensively available for use by physicians, more corroboration is needed to apprehend how clinically useful the tests are.
Single Cell Proteomics and Breast Cancer
Single-cell proteomics tactics, although still limited in comparison of genomics methods, but now developing quickly. Currently, the commonly applied single-cell proteomics methods based on targeting specific proteins by means of tagged antibodies. Fluorescence-based screening of protein by fluorescence-activated cell sorting (FACS) or fluorescence microscopy, as well as single-cell Western blotting, permit protein assessment in single cells with a low level of multiplexing (approximately 10–15 proteins in total), though other multiplexed methods, including the practice of oligonucleotide-labelled antibodies followed by qPCR metal-tagged antibodies followed by mass cytometry and single-cell mass spectrometry are developing. Though, these primary methods are still limited to the revealing of tens to hundreds of proteins per cell.
A cell’s proteome correlates genotype to phenotype by sensing various internal and external stimuli. Like, tumor suppressor protein p53 is central in many cancers. High magnification and resolution based single-cell analyses exposed that the results of the previous cell based studies not able to expose p53’s exact dynamic response [46]. In place of reduced magnitude, each and every cells display series of equal p53 pulses with static amplitude and duration, independent of the intensity of external stimuli. The misrepresentative mean outcomes from the majority of cell studies are associated to a decreased cell number and loss of synchronization between single cells at later times. So, single-cell proteomics will provide dynamic and comprehensive knowledge of genetic heterogeneity in their responses to drugs and other external stimuli.
Single Cell Metabolomics and Breast Cancer
Metabolomics, when combined with genomics, transcriptomics and proteomics, provides us a comprehensive opinion to fully understand the functionality of each individual cell. Within a single cell, the transcripts derived from DNA are translated into proteins, which act as enzymes to catalyze intermediate products of metabolism. Therefore, metabolites act as a connection between genotype and phenotype on single cell level, providing a coherent view on single cell’s behavior. Numerous in silico studies have established the correlation of the various metabolites by clustering approach. Like one study by Tang et al., 2014 has classified 399 metabolites into distinct groups by hierarchical clustering (cluster 3.0) and then showed with TreeView. They found that many groups of metabolites were highly clustered and correlated. These groups comprise metabolites that are documented to be in the same metabolic pathways as well as surprising correlation between metabolites in different metabolic pathways. Metabolites from varied pathways clustered in the same groups might represent two diverse metabolic pathways which are controlled by the same genetic change or influenced similarly by the metabolic reprogramming. Like, they demonstrated a cluster of metabolites comprising many intermediates of various lipids associated with glycerophosphocholines. They also reported two separate clusters of amino acids and di-amino acids (glycine-proline, glutamate-leucine, and alanine–tyrosine) and N-acetyl-amino acids (N-acetyl-aspartate, N-acetyl-ornithine and N-acetyl-aspartyl-glutamate). Both clusters may represent products of protein degradation and catabolism and can be used to identify breast tumors with higher protein catabolism [47].
Breast Cancer Sub-typing and Molecular Characterization
Earlier studies of transcriptomics accomplished on DNA microarrays documented many molecular subtypes of breast cancer. Subgrouping of breast cancers were documented using computational methods that assessed similarities in the gene expression profiles created for individual breast cancers among large cohorts of breast cancer samples. Clusters were characterized by typical gene expression patterns recognized by overexpressed genes. The initial study of this type recognized four major molecular subtypes of breast cancer: (1) ER+/luminal, (2) HER2+ (HER2-enriched), (3) basal-like, and (4) normal-like, [48]. Several transcription profiling studies of invasive breast cancer validated that these molecular subtypes are dissimilar and robust between breast cancer cohorts and using different gene sets for cluster analysis [49]. The newly classified molecular subtypes of breast cancer are (1) luminal A (ER+), (2) luminal B (ER+/HER2-enriched), (3) HER2+ (HER2-enriched), (4) basal-like, (5) claudin-low, and (6) normal-like [50].
Additionally, the molecular subtypes of breast cancer as demonstrated by transcriptomic analysis are linked with diverse clinical outcomes [51], the robustness of molecular subtype category of breast cancer based on transcriptomics analysis has been verified by various study. Though breast cancer grouping methods display decent reproducibility, suggesting that these are reproducible biological subtypes, breast cancers that are not classifiable by conventional approaches are now recognized with regular frequency.
Luminal A and Luminal B Breast Cancers
ER+ breast cancers are seen commonly and comprise of two major molecular classifications: luminal A and luminal B. Luminal A breast cancers are the most common, with a frequency of 28–31%. Luminal B breast cancers are known by ER positivity escorted by amplification and/or overexpression of the HER2 gene. Luminal B types are not so frequent, it represents nearly 20% of patients [51], and the expression status of proliferation linked genes is one of most central factor which make difference between luminal A and luminal B breast cancers. Moreover, the two ER+ breast cancer subtypes, luminal A and luminal B, are accompanied with a good prognosis and admirable long-term survival (approximately 80–85% 5-year survival), whereas the ER negative subtypes (HER2+ and basal-like) are challenging to manage and are linked with poor prognosis (approximately 50–60% 5-year survival). Survival of patients with ER+ breast cancers directs the existence of effective targeted therapy which is available in the form of anti-estrogen treatment (like, tamoxifen). Moreover, among the ER+ breast cancers, the luminal B subtypes are linked with a considerably worse prognosis, when compared with the luminal A subtype. The differences in patient survival are mainly due to differences in response of luminal A and luminal B breast cancers to anti-estrogen treatment [52].
HER2+ Breast Cancers
HER2 belongs to human epidermal growth factor receptor family, which includes EGFR (alias HER1), HER3, and HER4. HER2 acts as an oncogene in several cancers including breast cancers, exhibits its effect of carcinogenesis mostly by over expression or due to gene amplification. Recently, this subtypes of breast cancers was discovered through transcriptomic analyses that recognized a cluster of breast cancers with strong expression of the ERBB2 proto-oncogene. HER2+ breast cancers occurs nearly 17% of all breast cancers, with a frequency of 12–21% across different populations. HER2 overexpression (HER2+) in breast cancer is linked with poor clinical outcomes [53], but is also have a positive therapeutic responses to anti-HER2 drugs (e.g., trastuzumab). HER2+ breast cancers are usually ER+, so therapy for these cancers does not comprise of anti-estrogenic hormonal treatment. Instead, treatments for the HER2+ breast cancers are centered on combinations of targeted drugs (e.g., trastuzumab) and cytotoxic chemotherapy [54].
Basal-Like and Claudin-Low Breast Cancers
Both, the basal-like and claudin-low molecular subtypes indicate subsets of triple-negative breast cancers (as classified by immunohistochemistry),that is without expression of ER and PR (ER+/PR+) and also missing amplification of ERBB2 (HER2+) [55]. The basal cell phenotype of breast cancer was initially observed in immune-histochemical studies, and since then it has been re-explored by various transcriptomic analyses. The basal-like subtype is characteristically HER2+ and reveals some characteristics of breast myoepithelial cells [56]. Basal-like breast cancers indicates about 15% of all breast cancers. The basal-like breast cancers have unusual high capability of cell proliferation and worst clinical outcomes. It has been shown that Basal-like breast cancers respond to preoperative chemotherapy [57].
However, the observation it was perceived that the patients with basal-like breast cancers have a very poor prognosis, also shown a higher likelihood of relapse/recurrence in these patients where the drug response was not achieved [57]. Claudin-low breast cancers have shown about 10% of all breast cancers. These breast cancers are augmented for markers of epithelial-to-mesenchymal transition and stem cell-like and/or tumor-initiating cell features [58]. Like, basal-like cancers, claudin-low breast cancers well respond to some chemotherapeutic agents, but patients have poor recurrence-free and overall survival outcomes. This observation may reveal the information that these cancers display mesenchymal features and may show resistance to standard chemotherapy treatment [58]
Normal-Like Breast Cancers
The normal-like breast cancers are so named because they incline to cluster closely with normal breast epithelium in microarray studies. It is still not clear whether this is a different molecular subtype of breast cancer or merely a grouping of breast cancers that are not otherwise classifiable because of its spread into normal epithelium. Nevertheless, this subset of breast cancer is regularly conveyed in gene expression studies.
Molecular Biomarkers Expression in Various Subtype of Breast Cancer
Recent cutting edge technological developments have allowed the simultaneous evaluation of multiple RNAs (DNA micro-arrays) or proteins (tissue arrays) in tumour samples [59]. These studies have revealed that the breast tumors could be classified into few classes by the over-expression of exclusive groups of genes/proteins [60]. As per past’s studies, nearly two-thirds of tumors have characteristics reminiscent of the luminal epithelial component of the breast. These lesions are commonly obtainable with well differentiated, low grade and display relatively high levels of steroid receptors, cytokeratins KRT8, KRT18 and KRT19, BCL2, CDH1, MUC1, the transcription factors GATA3, FOXA1, XBP1 [61], TFF1, TFF3, SLC39A6, CDKN1A, CDKN1B and CCND1. In comparison of the ‘luminal epithelial-like’ lesions, about 15% of tumors have a low level of the above mentioned markers, whereas they express relatively high levels of cytokeratins KRT5 and KRT17, CDH3, EGFR, FOXC1, KIT, SERPINB5, TRIM29, GABRP, MMP7, SLPI and various proliferation markers. Most of these ‘basal/myoepithelial-like’ tumors are not well differentiated and have a high grade [62]. Partly they may be associated with the rare medullary carcinomas122 and mutations in the familial cancer susceptibility BRCA1 gene [63]. Tumors overexpressing ERBB2 due to gene amplification may be organized into a separate class (ERBB2 subtype), more closely related to the ‘basal/myoepithelial-like’ than to the ‘luminal epithelial-like’ lesions. Moreover, the ‘luminal epithelial-like’, ‘basal/myoepithelial-like’ and ‘ERBB2’ classes are also found in breast cancer cell lines [64], most of which are derived from DTC (obtained in most cases from pleural effusions).
Additionally, it has been perceived that among the markers stated above, many are relatively associated to a specific class. EGFR, SERPINB5 and GABRP are frequently expressed by ‘basal/myoepithelial-like’ tumors, while high ERBB2 levels are markedly expressed in lesions of the ‘ERBB2’ class. ESR1, TFF1 and TFF3, the expression of which is exactly linked, are found at high levels only in ‘luminal epithelial-like’ tumors. Other markers associated to this well-differentiated, low-grade class are the secreted proteins PIP, SCGB2A1, SCGB2A2 and SCGB2D1, as well as the mucins MUC1 and SBEM, the transcription factor SPDEF and ANKRD30A characterize stable portrait of breast cancer during progression, despite increasing heterogeneity. The breast tumor classes differentiated by gene/protein signatures put forward that any tumor biology conveys to a large extent the biology of the cell of origin at the time of initiation. Tumors initiating from more undifferentiated epithelial cells have a hasty growth pattern and more aggressive behavior and outcome compared with those beginning in a more differentiated epithelial cells. Therefore, the description of tumors seems to be stable during progression.
It is now recognized on the behalf of past studies and a number of data regarding breast cancer biology, pathology and genetics that exists during progression to metastasis, although undergoing increasing genetic alterations, most breast tumors largely maintain their portrait (luminal epithelial-like, basal/myoepithelial like, ERBB2). In fact, the grade (I–III) and the expression of markers, such as ESR1, PGR, TFF1, EGFR, ERBB2, P53 and various proliferation markers, etc. are generally concordant between primaries and metastases [65, 66].
Special Aspects of Single Cell Omics
Liquid Biopsy: CTCs (Circulating Tumor Cells) and DTCs (Disseminating Tumor Cells) in Breast Cancer
Existence of circulating tumor cells (CTCs) in peripheral blood and disseminated tumor cells (DTCs) in bone marrow of tumor patients has become an vibrant area of translational cancer research, with numerous groups developing new diagnostic assays and more than 250 clinical trials incorporating CTC counts as a biomarker in patients with various types of solid tumors. CTC investigation could play a role as a “liquid biopsy,” which will permit physicians to follow cancer changes over time and mold precise treatment, and it signifies a potential new diagnostic field for advanced-stage patients; the sensitive CTC detection platforms allow monitoring of disease and treatment efficacy, thus it is helping in tailoring precision medicine.
CTCs are tumor cells released into the peripheral blood from a primary lesion or metastatic lesion by natural behavior, diagnosis operation or treatment operation. The existence of CTCs means that tumor cells are not limited to the primary lesion, but can develop into distant metastases. Further, CTCs have been shown to appear in the bloodstream astonishingly early, before metastatic lesions could be observed by histologic analysis [67]. Numerous studies have established that CTCs are significant in forecasting disease progression and survival [68], watching the complex tumor genomes, diagnosing tumor recurrence and metastasis and guiding therapeutic management [69]. Thus, CTCs lend themselves well as non-invasive biomarkers that can be painlessly accepted by patients. Circulating tumor DNA (ctDNA) encodes tumor specific sequences that can be used as another form of liquid biopsy, which can be noninvasively repeated during treatment and follow-up. Studies have presented that ctDNA can reveal genotype information of the tumor, indicating that ctDNA analysis could effectively substitute tumor biopsy. Moreover, advances in sequencing technology has meant that ctDNA can now timely monitor tumor progression and therapeutic responses of various solid cancers.
The opinion of a non-invasive liquid biopsy that could expose metastatic mechanisms makes CTCs is an active area of cancer research. Further, researchers have scanned the various blood markers and identified transcripts that were organ-specific by deep comparative transcriptome analysis across forty or more different organs in humans and mice.
A list of bio-markers that have been utilized in assays to discover disseminated tumour cells (DTCs) by antibody or nucleic acid-based techniques also summarized in Table 2 which are commonly used and further, Table 3 showed Clinical significance of CTCs detection in breast cancer.
Table 2.
A list of markers used to detect disseminated tumour cells by antibody or nucleic acid-based techniques [66]
| Marker (gene) name | Gene locus | Standard name | Other frequently-used names |
|---|---|---|---|
| ANKRD30A | 10p11.21 | Ankyrin repeat domain 30A | Breast cancer antigen NY-BR-1; B726P |
| B305D | 21q11.1-q11.2 | Antigen B305D | B305D, isoform A (B305D-A) B305D, isoform C (B305D-C) |
| CD44 | 11p13-pter | Antigen CD44 | Hermes antigen; PGP1 |
| CDH1 | 16q22.1 | Cadherin-1 (epithelial) | E-cadherin; Uvomorulin |
| KRT19 | 17q21-q22 | Keratin 19 | Cytokeratin 19 (CK19) |
| KRT7 | 12q12-q14 | Keratin 7 | Cytokeratin 7 (CK7); Sarcolectin (SCL) |
| GABRP | 5q32-q33 | γ-Aminobutyric acid type A receptor pi subunit | GABA receptor A, pi polypeptide (GABARAP); GABAA receptor, pi polypeptide (GABA A(pi)) |
Table 3.
Clinical significance of CTCs screening in breast cancer [66]
| Method | Marker | CTC detection rate | Clinical significance |
|---|---|---|---|
| Early breast cancer | |||
| Nested RT-PCR | CK-19 | 44 of 148 (30%) | DFI: p = 0.001, OS: p = 0.014 |
| RT-qPCR | CK-19 | Node negative 36 of 167 (21.6%) | DFI: p < 0.001, OS: p = 0.008 |
| RT-qPCR | CK-19, mammaglobin HER-12 | CK-19: 72 of 145 (41%) | DFI: CK-19 (p < 0.001), OS: CK-19 (p = 0.044) |
| Mammaglobin: 14 of 175 (8%) | DFI: mammaglobin (p = 0.011), OS: mammaglobin (p = 0.034) | ||
| HER-2: 50 of 175 (29%) | DFI: HER-2 (p < 0.001) | ||
| RT-qPCR | CK-19, ER | 181 of 444 (41%) | DFI: CK-19 and ER- (p = 0.001) OS: CK-19 and ER- (p = 0.001) |
| RT-qPCR | CK-19 | After adjuvant therapy: 179 of 437 (41%) | DFI: p < 0.001, OS: p = 0.003 |
| RT-qPCR | CK-19 | Before adjuvant therapy: 91 of 165 (55.2%) | Before adjuvant therapy: DFI: p = 0.081, OS: p = 0.024 |
| After adjuvant therapy: 79 of 162 (48.8%) | After adjuvant therapy: DFI: p = 0.057, OS: p = 0.128 | ||
| RT-qPCR CellSearch |
CK-19 Pan-CK |
99 of 133 (31.7%) Before and/or after neoadjuvant chemotherapy: 32 of 118 (27%) |
DFI: p = 0.001 and OS: p = 0.001) DFI: ps0.013 |
| CellSearch | Pan-CK | Before chemotherapy therapy: 95 of 115 (82.6%) | Before chemotherapy: DFI: p = 0.007, OS: p = 0.0006 |
| After chemotherapy: 85 of 115 (73.9%) | After chemotherapy: DFI: p = 0.04, OS: p = 0.02 | ||
| CellSearch | Pan-CK | Before chemotherapy: 140 of 1489 (9.4%) | Before chemotherapy: DFI: p < 0.0001, OS: p = 0.023 |
| After chemotherapy: 129 of 1489 (8.7%) | After chemotherapy: DFI: p = 0.054, OS: p = 0.154 | ||
| ICC | CK | 47 of 71 (66%) | OS: ps0.071,DFI: p = 0.052 |
| RT-PCR | CK-19, HER-2, P1B, PS2, epithelial glycoprotein 2 | 43 of 72 (60%) | DFI: ps0.031, OS: p = 0.03 |
| ICC | CK and HER-2 | 17 of 35 (49%) | DFI: p < 0.005, OS: p < 0.05 |
| Nested RT-PCR | Mammaglobin | 14 of 101 (13.9%) | DFI: p = 0.020, OS: p = 0.009 |
| Metastatic breast cancer | |||
| Cell Search | Pan-CK | 87 of 177 (49%) | DFI: p < 0.001, OS: p < 0.001 |
| Cell Search | Pan-CK | 43 of 83 (52%) | DFI: p = 0.0014, OS: p = 0.0048 |
| CellSearch | Pan-CK | 92 of 195 (47.2%) | DFI: p = 0.0122, OS: p = 0.0007 |
| CellSearch | Pan-CK | 35 of 138 (25%) | OS: p < 0.0001 |
DFI disease-free interval, OS overall survival
Besides CTCs and blood proteins as tumor biomarkers, circulating DNAs, mRNAs and microRNAs from tumor cells are being explored as substitute tumor biomarkers and for monitoring cancer recurrence. Further, studied evidence suggest that CTCs may exhibit phenotypes distinct from their corresponding primary tumors. Lately, laboratory results in collaboration with the BioMEMS for CTC research laboratory at the University of Michigan has jointly described isolated CTCs by using a highly-sensitive microfluidic capture device and noted HER2 positive CTCs from the blood of metastatic breast cancer patients had HER2 negative primary tumors [70]. This provides a potential description for the surprising finding that HER2 blockade in the adjuvant setting benefits women whose breast tumors do not display HER2 gene amplification. Additionally, study on other cancers, like in prostate cancer patients, researchers examined the functional diversity of viable, single CTCs for clonal comparison and mapping of heterogeneity. They informed that only a rare subset of isolated CTCs were resistant to anoikis within blood circulation, showing metastatic characteristics such as invasiveness and producing proteases in patients with late-stage, metastatic castration-resistant prostate cancer (mCRPC).
The various findings suggests that CTCs alone may be insufficient to fully clarify the metastatic potential of tumor cells in the circulation of cancer patients [71]. Additionally, disseminated tumor cells (DTCs) in the bone marrow of breast cancer patients have also been noted in tumor metastasis. In addition to CTCs, DTCs from breast cancer patients have also been explored as an independent prognostic factor using whole-genome amplification (WGA) followed by NGS and described a clear difference in the copy number between the DTCs and matched primary tumors, indicating that the DTC underwent further evolution at the copy number level. Therefore, single cell analyses of CTCs and DTCs are an important tool for explaining tumor heterogeneity as well as complexity of the cancer genome.
Breast Cancer Stem Cells (BCSCs)
Teamwork and collaborations between translational labs and biotechnology companies including Fluidigm Corporation (San Francisco, CA) and Denovo Sciences (Plymouth, MI) are in progress for developing and/or optimizing microfluidic devices to explore the heterogeneity of breast CSCs and circulating tumor cells (CTCs) at the single cell level. In an early attempt to explore heterogeneity of CSCs and CTCs, researchers group has screened the gene expression signature of the CD44+/CD24-, ALDH+ sorted CSC populations and bulk cells from breast cancer cell lines and patient derived xenografts at the single cell level using Fluidigm’s C1 and BioMark HD platforms. These three sorted fractions show distinct patterns of gene expression from one other, but also noticeably show heterogeneity within each sorted population of CSCs. This observed heterogeneity would otherwise be masked using conventional gene expression methods based on average population studies [72]. Our outcomes and other researchers in the field believe that single cell analysis will soon become a transformational technology in cancer biology as well as in clinical cancer practice. Upcoming studies combining thousands of single cancer cells using these advanced technologies and others for assay preparations along with the novel computational methods will enable researchers to better rebuilt intracellular networks, re-evaluate cell types and states and transform our knowledge about the process of decision making in individual cells at the genomic level.
Breast cancer is perfect model for studying tumor heterogeneity, because it is hierarchically organized, similar to the normal mammary epithelium and the disease process is identified to be highly heterogeneous. Our research and along with others have shown that hierarchically organized breast cancer is driven by a small fraction of tumor cells that display stem cell properties [13, 73]. This small population of breast CSCs also termed breast cancer initiating cells was first identified among solid tumors by characteristics of their expression of the cell surface markers EpCAM+/CD24-/CD44+ and their capability to recapitulate heterogeneous populations of tumor cells [74]. Additionally, it has been shown that both normal and malignant breast CSCs express high levels of the enzyme aldehyde dehydrogenase (ALDH) that serves as a predictor of poor clinical outcome in breast cancers [75]. These two types of breast CSCs are anatomically distinct, one with EMT (epithelial-to-mesenchymal transition) and one with MET (mesenchymal-to-epithelial transition) gene expression profiles. Moreover, they vibrantly show transition between the mesenchymal and epithelial-like states reflective of their normal counterparts in the mammary epithelial hierarchy [76]. Remarkably, this plasticity of breast CSCs from a quiescent mesenchymal state to a proliferative epithelial-like state plays a critical role for these cells to create sizable metastatic nodules in distant organs. Indeed, there is increasing experimental evidence to suggest that such transition, termed colonization, is vital for development of successful macrometastasis.
The usefulness of single-cell omics has conveyed great vision into our understandings of dissimilar biological processes with extensive inferences for both basic and clinical research that have formerly impossible to decide from bulk population cells. Although, certain hurdles like procedure of single cell isolation, whole genome amplification, library construction, sequencing, bioinformatics analysis, and data integration are hampering the path that needs to be resolve. In conclusion, even with difficulties, we are certain that single cell omics will open new vistas in field of diagnosis and therapeutics, thus it will help in better management of breast cancer patients.
Acknowledgements
Funding was provided by Science and Engineering Research Board (Grant No. PDF/2015/000322).
Conflict of interest
All Authors declare that there is no conflict of interest.
Ethical Approval
This article is review article, so does not contain any studies with animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Raina V, Tyagi BB, Manoharan N. Two year report of the population based cancer registries, 2004–2005. Incidence and distribution of cancer. New Delhi: National Cancer Registry Programme, Indian Council of Medical Research; 2009. p. 63–5. https://canceratlasindia.org.
- 3.Dwivedi S, Chikara G, Samdariya S, Pareek P, Sharma P, Khattri S, et al. Molecular biotechnology for diagnostics. In: Khan MS, Khan IA, Barh D, et al., editors. Applied molecular biotechnology: the next generation of genetic engineering. New Delhi: CRC Press, Taylor & Francis Group, Inc; 2016. pp. 303–333. [Google Scholar]
- 4.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polyak K. Breast cancer: origins and evolution. J Clin Investig. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dwivedi S, Sharma P. Prospects of molecular biotechnology in diagnostics: step towards precision medicine. Indian J Clin Biochem. 2017;32(2):121–123. doi: 10.1007/s12291-017-0650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dwivedi S, Purohit P, Misra R, Pareek P, Goel A, Khattri S, et al. Diseases and molecular diagnostics: a step closer to precision medicine. Indian J Clin Biochem. 2017;32(4):374–398. doi: 10.1007/s12291-017-0688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dwivedi S, Shukla KK, Gupta G, Sharma P. Non-invasive biomarker in prostate carcinoma: a novel approach. Indian J Clin Biochem. 2013;28(2):107–109. doi: 10.1007/s12291-013-0312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dwivedi S, Goel A, Mandhani A, Khattri S, Sharma P, Misra S, et al. Functional genetic variability at promoters of pro-(IL-18) and anti-(IL-10) inflammatory affects their mRNA expression and survival in prostate carcinoma patients: five year follow-up study. Prostate. 2015;75(15):1737–1746. doi: 10.1002/pros.23055. [DOI] [PubMed] [Google Scholar]
- 10.Dwivedi S, Goel A, Khattri S, Mandhani A, Sharma P, Misra S, et al. Genetic variability at promoters of IL-18 pro- and IL-10 anti-inflammatory gene affects susceptibility and their circulating serum levels: an explorative study of prostate cancer patients in North Indian populations. Cytokine. 2015;74(1):117–122. doi: 10.1016/j.cyto.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Dwivedi S, Goel A, Khattri S, Mandhani A, Sharma P, Pant KK. Tobacco exposure by various modes may alter pro-inflammatory (IL-12) and anti-inflammatory (IL-10) levels and affects the survival of prostate carcinoma patients: an explorative study in North Indian population. Biomed Res Int. 2014;2014:158530. doi: 10.1155/2014/158530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma P, Dwivedi S. Nutrigenomics and nutrigenetics: new insight in disease prevention and cure. Indian J Clin Biochem. 2017;32(4):371–373. doi: 10.1007/s12291-017-0699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dwivedi S, Shukla S, Goel A, Sharma P, Khattri S, Pant KK. Nutrigenomics in breast cancer. In: Barh D, editor. Omics approaches in breast cancer. New Delhi: Springer; 2014. pp. 127–151. [Google Scholar]
- 14.Dwivedi S, Purohit P, Misra R, Pareek P, Vishnoi JR, Sharma P, et al. Methods for isolation of high quality and quantity of RNA and single cell suspension for flow-cytometry from cancer tissue: a comparative analysis. Indian J Clin Biochem. 2017 doi: 10.1007/s12291-017-0719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter NP, Bebb CE, Nordenskjo M, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-K. [DOI] [PubMed] [Google Scholar]
- 16.Paunio T, Reima I, Syvänen A-C. Preimplantation diagnosis by whole-genome amplification, PCR amplification, and solid-phase minisequencing of blastomere DNA. Clin Chem. 1996;42:1382–1390. [PubMed] [Google Scholar]
- 17.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasken RS. Single-cell sequencing in its prime. Nat Biotechnol. 2013;31:211–212. doi: 10.1038/nbt.2523. [DOI] [PubMed] [Google Scholar]
- 19.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genom Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, et al. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI) Science. 2017;356:189–194. doi: 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsoucas D, Yuan GC. Recent progress in single-cell cancer genomics. Curr Opin Genet Dev. 2017;42:22–32. doi: 10.1016/j.gde.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome as-sembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer pa-tients. Proc Natl Acad Sci. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demeulemeester J, Kumar P, Møller EK, Nord S, Wedge DC, Peterson A, et al. Tracing the origin of disseminated tumor cells in breast cancer using single-cell se-quencing. Genome Biol. 2016;17:250. doi: 10.1186/s13059-016-1109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janiszewska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA, et al. In situ single-cell analysis identifies heterogeneity for PIK3 CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet. 2015;47:1212–1219. doi: 10.1038/ng.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, et al. Whole genome multiple displacement amplification from single cells. Nat Protoc. 2006;1:1965–1970. doi: 10.1038/nprot.2006.326. [DOI] [PubMed] [Google Scholar]
- 30.Van Loo P, Voet T. Single cell analysis of cancer genomes. Curr Opin Genet Dev. 2014;24:82–91. doi: 10.1016/j.gde.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H, Zhu P, Wu X, Li X, Wen L, Tang F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528:142–146. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Widschwendter M, Berger J, Müller HM, Zeimet AG, Marth C. Epigenetic downregulation of the retinoic acid receptor-beta2 gene in breast cancer. J Mammary Gland Biol Neoplasia. 2001;6:193–201. doi: 10.1023/A:1011360724350. [DOI] [PubMed] [Google Scholar]
- 39.Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.TAILORx trial. http://www.cancer.gov/clinicaltrials/digestpage/TAIL.
- 46.Batchelor E, Loewer A, Lahav G. The ups and downs of p53: understanding protein dynamics in single cells. Nat Rev Cancer. 2009;9:371–377. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang X, Lin C-C, Spasojevic I, Iversen ES, Chi J-T, Marks JR. A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Res BCR. 2014;16:415. doi: 10.1186/s13058-014-0415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 49.Sørlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a populationbased study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 54.Mukai H. Treatment strategy for HER2-positive breast cancer. Int J Clin Oncol. 2010;15:335–340. doi: 10.1007/s10147-010-0107-0. [DOI] [PubMed] [Google Scholar]
- 55.Carey LA. Breast cancer: HER2ea good addiction. Nat Rev Clin Oncol. 2012;9:196–197. doi: 10.1038/nrclinonc.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wetzels RH, Holland R, van Haelst UJ, Lane EB, Leigh IM, Ramaekers FC. Detection of basement membrane components and basal cell keratin 14 in noninvasive and invasive carcinomas of the breast. Am J Pathol. 1989;134:571–579. [PMC free article] [PubMed] [Google Scholar]
- 57.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 58.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dwivedi S, Purohit P, Mittal P, Goel A, Verma R, Khattri S, et al. Genetic engineering: towards gene therapy and molecular medicine. In: Barh D, Azevedo V, et al., editors. Omics technologies and bio-engineering: towards improving quality of life. Cambridge: Academic Press; 2017. pp. 507–530. [Google Scholar]
- 60.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 2006;27(7):96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 62.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 63.Bertucci F, Finetti P, Cervera N, Charafe-Jauffret E, Mamessier E, Adélaïde J, et al. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancer. Cancer Res. 2006;66:4636–4644. doi: 10.1158/0008-5472.CAN-06-0031. [DOI] [PubMed] [Google Scholar]
- 64.Lacroix M, Leclercq G. Hereditary breast cancer: an update on genotype and phenotype. In: Yao PA, editor. New breast cancer research. New York: Nova Science Publishers; 2006. pp. 27–51. [Google Scholar]
- 65.Charafe-Jauffret E, Ginestier C, Monville F. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 66.Dwivedi S, Goel A, Sadashiv Verma A, Shukla S, Sharma P, et al. Molecular diagnosis of metastasizing breast cancer based upon liquid biopsy. In: Barh D, et al., editors. Omics approaches in breast cancer. New Delhi: Springer; 2014. pp. 425–459. [Google Scholar]
- 67.Dwivedi S, Purohit P, Misra R, Pareek P, Goel A, Khattri S, et al. Diseases and molecular diagnostics: a step closer to precision medicine. Indian J Clin Biochem. 2017;32:374–398. doi: 10.1007/s12291-017-0688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20(8):897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 70.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nanotechnol. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao X, Choudhury AD, Yamanaka YJ, Adalsteinsson VA, Gierahn TM, Williamson CA, et al. Functional analysis of single cells identifies a rare subset of circulating tumor cells with malignant traits. Integr Biol (Camb) 2014;6:388–398. doi: 10.1039/C3IB40264A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levitin HM, Yuan J, Sims PA. Single-cell transcriptomic analysis of tumor heterogeneity. Trends Cancer. 2018;4(4):264–268. doi: 10.1016/j.trecan.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 74.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dwivedi S, Sharma P. Stem cell biology: a new hope in regenerations and replenishments therapy. Ind J Clin Biochem. 2018;33(4):368–370. doi: 10.1007/s12291-018-0792-4. [DOI] [PMC free article] [PubMed] [Google Scholar]