Abstract
The changes in the translational repression and variation in mRNA degradation induced by micro RNA are important aspects of tumorigenesis. The association of microRNA-21 with clinicopathologic features and expression of programed cell death 4 (PDCD4) in Iraqi female’s with breast tumors has not been studied. MicroRNAs were extracted from a set of 60 breast tumor tissues and blood samples of females with breast cancer and benign breast lesions obtained after breast-reductive surgery, and only blood samples from 30 normal volunteers. These extracts were evaluated for miR-21 expression by quantitative RT-PCR. Analysis of PDCD4 protein expression was carried out as miR-21 target gene by immunohistochemical tests and correlating the results with patients’ clinicopathological features. Significant overexpression of miRNA-21 was found in breast cancer group. The fold increase in the miR-21 gene expression was significantly higher in circulating exosomes and breast tissues of breast cancer patients as compared to other groups (P < 0.001). Overexpression of miR-21 was also significantly associated with the advanced tumor stage and histological grade. In breast cancer patients, PDCD4 protein expression was decreased to about 70% of the level in the control group. The delta Ct of exosomal and breast tissue miRNA-21 was negatively associated with PDCD4 expression. In conclusion, the translational repression of the PDCD4 induced by the high expression of miR-21 promotes breast cell transformation and development of breast tumor, and circulating miR-21 level could be applied to the screening panels for early detection of women breast cancer.
Electronic supplementary material
The online version of this article (10.1007/s12291-017-0710-1) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Benign breast lesion, Programed death cell 4, MicroRNA-21
Introduction
The threat of increasing breast cancer incidence in women is of high public concern. One out of eight women is diagnosed with breast cancer during her lifetime, with an estimated 1.57 million new cases in 2012 [1]. The incidence of women breast cancer in Iraq increased from 30/100,000 to 40/100,000 in latest Iraqi cancer registry report and this unimodal increase is distributed homogenously in different geographical regions [2, 3]. Al-Janabi et al. [4] reported that the incidence of breast cancer in the first half of year 2015 increased to exceed half of that for 2014 in Kerbala city in Southern Iraq. Breast cancer worldwide exhibits biological heterogeneity in terms of its prognosis and sensitivity to anti-cancer agents. Numerous studies are currently being conducted in an attempt to identify markers of cell growth and differentiation which are associated with breast cancer formation and progression [5]. MicroRNAs (miRNAs) are small RNA molecules that function in apoptosis, cell proliferation, and cell differentiation [6–8]. Several cancer-related miRNAs and their target genes have been identified to be either oncopromotors or oncosupressers in tumor cells [9–11]. MiR-21 is one of the most significantly upregulated miRs in human breast cancer, and its expression contributes to tumor progression and bad prognosis [12–14]. Evidence suggests that miR-21 targets and inhibits programmed cell death 4 (PDCD4) and other tumor-related genes. Several reports have described the regulation of PDCD4 by miRNA-21 [15]. Asangani et al. [16] found a conserved potential site for miRNA-21 within the 3−UTR of PDCD4 mRNA. Lu et al. [17] and Zu et al. [18] demonstrated that the implication of the 3−UTR site as well as the regulation of PDCD4, and that the initiation of invasion and metastasis are promoted by high expression of miRNA-21. MiRNAs are released into the blood by budding or apoptotic and necrotic cells or by active secretion in small particles [19], and thus, exist in exosomes that are important mediators of cell–cell crosstalk. Lipids, proteins, miRNAs and mRNAs are transferred from one cell to another by exosomes [20]. The tumor cell-derived exosomes have an emerging role in tumor progression and metastases [21]. In this study, and for the first time in Iraq, measured the fold change in both breast tissue and circulating tumor cell-derived (exosomal) miR-21 and its possible impact on the expression of neoplastic transformation inhibitor “PDCD4” and further correlated the expression level of miR-21 with the invasiveness of female breast cancer.
Materials and Methods
The study was conducted on 60 female patients (23–67 years) with breast tumors (30 with breast cancer and 30 with benign breast lesions) after getting the approval of the Institutional Review Board at College of Medicine, Al-Nahrain University/Baghdad/Iraq. Blood samples were also taken from thirty age-matched women, who were enrolled in the study as a control group. Written consent was taken from all participants. Patient demographic details, including age, BMI, menopausal status, family history of breast tumors or other malignancies are shown in Table 1 (Supplemented materials).
Five milliliters of blood was aspirated and the sera was fractionated into small tubes for the determination of serum CA 15-3 levels and a portion of the blood was kept in an EDTA tube, centrifuged at 3500×g and the plasma was re-centrifuged at 10,000×g to remove platelets and cell debris and then stored in a liquid nitrogen at − 190 °C for subsequent miRNA extraction and molecular analyses.
Breast Tissue Specimens
Resected breast tumors (benign and malignant) and the corresponding apparent normal tissues away from the tumor site were obtained at theatre from those who are scheduled for surgical mastectomy by senior surgeons. Fresh samples were immediately stored in liquid nitrogen for RNA extraction. Breast cancer patients with previous mastectomy, preoperative radiotherapy or chemotherapy were excluded from this study. The whole breast or resected mass was sent to histopathology laboratories for the accurate diagnosis, TNM classification and staging of breast tumors according to the American Joint Committee on Cancer AJCC [22]. Paraffin embedded blocks of these specimens were sectioned for the subsequent immunohistochemical staining and analysis of PDCD4 protein expression.
Determination of Serum Cancer Antigen 15-3 (CA 15-3)
The level of serum CA 15-3 was determined using 50 μl serum samples from each patient and the control group and an Elisa kit for quantitative measurement of human serum CA 15-3 (Human, Germany).
Molecular Analyses
RNA Isolation from Circulating and Tissue Samples
Exosomes were isolated from 600 μl of plasma using the miRCURY™ Exosome Kit (Exiqon, no. 300110, Denmark), that is based on capturing of water molecules which otherwise form the hydrate envelope of particles in suspension. By mixing the sample with the precipitation buffer, dehydration of the particles will favor the precipitation of the subcellular particles below 100 nm at low centrifugation speed. The total RNA and miRNA were extracted from the isolated exosomes using the miRCURY™ RNA Isolation Kit (Biofluids, Exiqon, no. 300112, Denmark), and the RNA was eluted from the column in a final volume of 15 μl.
Total RNA of the breast tumor tissues and the corresponding normal breast tissues were isolated using the miRCURY™ RNA Cell and Plant Isolation Kit (Exiqon, no. 300110, Denmark) and for serum extraction using Biofluid kit (Exiqon, no. 300111, Denmark). For optimal isolation of RNA from breast tissues that have high lipid content, we used Lysis Additive (Exiqon, no. 300121, Denmark) during extraction as suggested by the manufacturer. Tissue homogenization was carried out using a micro pestle motor-driven hand homogenizer on 20 mg of frozen tissue specimen. The homogenate was then passed 5–10 times through a 25 gauge needle attached to a 5 ml syringe to shear genomic DNA prior to loading to the column.
RNA concentration and purity were analyzed using a UV spectrophotometer at A260 and A280 nm and RNA integrity was checked by ethidium bromide stained − 2% agarose gel electrophoresis (Figs. 1, 2-Supplemented materials).
One Step: Reverse Transcription (RT) and Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Each template RNA sample isolated from a tissue or plasma sample was adjusted to a final concentration of 5 ng/μl with nuclease free water and were reverse transcribed to cDNA in 10 μl total reaction volume using Exiqon Universal cDNA synthesis kit (no. 20330, Denmark). These were incubated in a thermal cycle (Clever, UK) that was programed for 42 °C for 60 min and 5 min at 95 °C, and then maintained at 4 °C. Second step RT-qPCR was performed using the ExiLENT SYBR Green master mix (Exiqon no. 203403, Denmark) and LNA™ PCR miR-21 and miR-16 primer sets (Exiqon, Denmark) as reference gene. Four microliters of cDNA diluted to 1:80 (for tissue) or 1:50 for circulating exosomal miRNA were used as templates in a 10 μl total reaction volume using the following conditions: 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s, 60 °C for 1 min, and a melting curve analysis was carried out at 95 °C for 1 min, 55 °C for 30 s, 95 °C for 30 s. All reactions were carried out on the stratagene (Mx3005P) real-time PCR amplification-detection system (Agilent Technology, Germany). The expression levels of miR-21 were normalized with the miRNA-16 as reference gene. The relative circulating miRNA quantity was determined using the comparative Ct method. The ΔCt was calculated by subtracting the Ct values of miRNA-16 from the Ct values of the miRNA-21.The ΔΔCt was used for the determination of fold difference in the miRNA expression which equals to the 2−ΔΔCt [23]. The efficiency of the of miRNA-21 expression was 93.43% with a slope of − 3.493 and a correlation coefficient of 0.992 whereas the amplification plot and standard curve of miR-16 showed a correlation coefficient of 0.999 and a slope of − 3.55 and PCR efficiency of 91.2% (Figs. 3–5-Supplemented materials).
Immunohistochemical Analysis of PDCD4
Immunohistochemical staining for PDCD4 was performed on 5 μm thick formalin fixed paraffin sections of breast tissue specimens after incubation at 37 °C for one night and subsequent deparaffinization in xylene and rehydration through different percentages of alcohol. The slides were then treated for blocking endogenous peroxidase activity and nonspecific binding. Afterwards, the sections were incubated at 4 °C in a humidified chamber with a monoclonal antibody to PDCD4 at a dilution of 1:200. A two-step technique using universal anti-mouse/anti-rabbit secondary antibody (Abcam, Inc. USA) was used for visualization with the chromogen diaminobenzidine. Finally, sections were counterstained with haematoxylin. Negative controls were performed by replacing the primary antibody with phosphate-buffered saline (PBS). The normal rat liver tissue sample was used as a positive control for PDCD4 (Fig. 6. Supplemented materials). PDCD4 slides showing brown cytoplasmic and nuclear stains were considered positive. The percentage of cells expressing positive IHC staining for the PDCD4 was calculated by counting 2000 tumor cells in the most positive areas in at least 10 high power fields for each sample. The grading of percentage of positive staining were as follows: Grade 0: < 10%, Grade + 1: 10–25%, Grade + 2: 26–50%, Grade + 3: > 50%.
The staining intensity for PDCD4 in malignant breast cells was assessed as being weak, moderate or strong when compared to the staining intensity observed in benign breast cells [22].
Statistical Analysis
All statistical analyses were carried out using version 21.0 statistical software (SPSS Inc., Chicago IL, USA). P values < 0.05 were considered statistically significant. All data were expressed as the mean ± standard error of the mean (SEM). The Student’s t test was used to compare between groups’ ∆Ct values of microRNAs expression levels. Mann–Whitney-U test was applied for the nonparametric data analysis and the Kruskal–Wallis test was used for more than two independent variables. The diagnostic accuracy of biochemical and molecular markers was performed using the receiver operating characteristic (ROC) curve analysis. The cut off values and area under the ROC curve (AUC) were then determined.
Results
Results of the Pathological Characteristics of the Patients with Breast Tumors
Table 1 summarizes data for the histological type, TNM stage, grade, lymph node metastasis, ER, PR and human epidermal growth factor-2 (Her2/neu) status. In this table, of the 30 newly diagnosed breast cancers, twenty (66.7%) samples were having invasive ductal carcinoma, eight (26.5%) with ductal carcinoma in situ (DCIS) and only two (6.7%) showed invasive lobular carcinoma (ILC). The highest percentages of tumors were in the stage II (43.3) and III (43.3%) and 10% were in stage I and 3.3% having stage IV. Regarding the degree of differentiation of the tumor or histological grading, 30% were of grade I, 43.4% of grade II and only 26.7% of tumors were of grade III. The tumor size was less than 2 cm in 56.3% of patients. The lymph node (LN) involvement positive percent was 66.7%, the Her2/neu receptor detection test was negative in 66.7% of the patients, the estrogen receptor (ER) was either positive or negative in 56.7 and 43.3%, respectively, and the progesterone receptor tests were negative in 43.3% and positive in 56.7% of tumor samples.
Table 1.
The pathological characteristics of breast cancer patients
| Variables | N = 30 | % |
|---|---|---|
| Age (years): ≤ 55 | 14 | 46.7 |
| > 55 | 16 | 53.3 |
| Histological type | ||
| Invasive ductal carcinoma (IDC) | 20 | 66.7 |
| Ductal carcinoma in situ (DCIS) | 8 | 26.7 |
| Invasive lobular carcinoma (ILC) | 2 | 6.7 |
| TNM stage | ||
| I | 3 | 10 |
| II | 13 | 43.3 |
| III | 13 | 43.3 |
| IV | 1 | 3.3 |
| Histologic grade | ||
| I | 9 | 30 |
| II | 13 | 43.4 |
| III | 8 | 26.7 |
| Tumor size | ||
| ≥ 2 cm | 12 | 40 |
| < 2 cm | 18 | 60 |
| Lymph node | ||
| Positive | 20 | 66.7 |
| Negative | 10 | 33.3 |
| Estrogen receptor | ||
| Positive | 17 | 56.7 |
| Negative | 13 | 43.3 |
| Progesterone receptor | ||
| Positive | 17 | 56.7 |
| Negative | 13 | 43.3 |
| Her2/neu receptora | ||
| Positive | 10 | 33.3 |
| Negative | 20 | 66.7 |
aHuman epidermal growth factor receptor 2
Results of Serum Cancer Antigen (CA15-3)
The mean ± SEM of the serum CA15-3 concentration was 42.0 ± 3.24 U/ml in breast cancer group, 33.6 ± 1.9 U/ml in benign breast lesion group and 31.2 ± 1.8 U/ml in the control group (Table 2). Student t test revealed significant increase in the mean values of CA15-3 concentration in breast cancer patients as compared to levels in the control and benign breast lesion groups (P < 0.01, P < 0.05, respectively), while no significant difference was observed between benign breast lesions and control groups (P > 0.05).
Table 2.
Mean serum ± standard error of mean (SEM) concentration of CA15-3 among different studied groups
| Descriptive statistics | Serum concentration of CA15-3 (U/ml) | ||
|---|---|---|---|
| Breast cancer | Benign breast lesion | Control | |
| Mean | 42.0a**,b* | 33.6 | 31.2 |
| ± SEM | 3.24 | 1.9 | 1.8 |
aStudent t test: breast cancer versus control: **P < 0. 01
bStudent t test: breast cancer versus benign breast lesions: *P < 0.05
Results of Molecular Analyses
The Expression Levels of the Circulating (Exosomal) miRNA-21 and miRNA-16 Among the Studied Groups
The amplification plots of the circulating miRNA-21 and miRNA-16 in control, benign breast lesion and breast cancer groups are presented in Fig. 2-Supplemented materials.
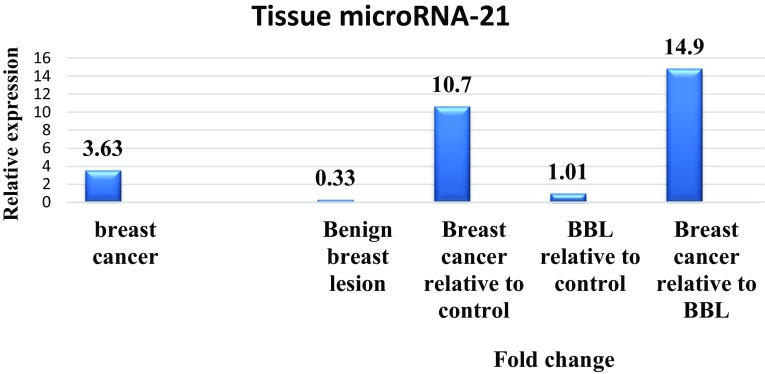
The mean ± SEM threshold cycle of miRNA-21 and miRNA-16, relative expression and median fold change in the circulating plasma miRNA-21 in the studied groups are shown in Fig. 1. The circulating level of miRNA-21 was normalized to that of miRNA-16 as a reference gene. The mean ± SEM relative expression (− ∆Ct) value of miRNA-21 was 3.63 ± 0.33 in the breast cancer samples, 0.33 ± 0.15 in benign breast lesion, and 0.11 ± 0.35 in healthy control group.
Fig. 1.
The relative expression and median of fold change of plasma exosomal microRNA-21 in the breast cancer and benign breast lesion (BBL) groups
Statistical analysis revealed a significant increase in the mean of relative expression in the plasma miRNA-21 in breast cancer patients (P < 0.001) with fold a change of 10.7 as compared with those of the control group, and 14.9-fold change when compared with benign breast lesion (P < 0.001). No statistically significant difference was observed (P > 0.05) between these variables in the benign breast lesion as compared to the control group (Fig. 1).
Result of miRNA-21 and 16 Genes’ Expressions in Breast Cancer and Benign Breast Lesion Tissues
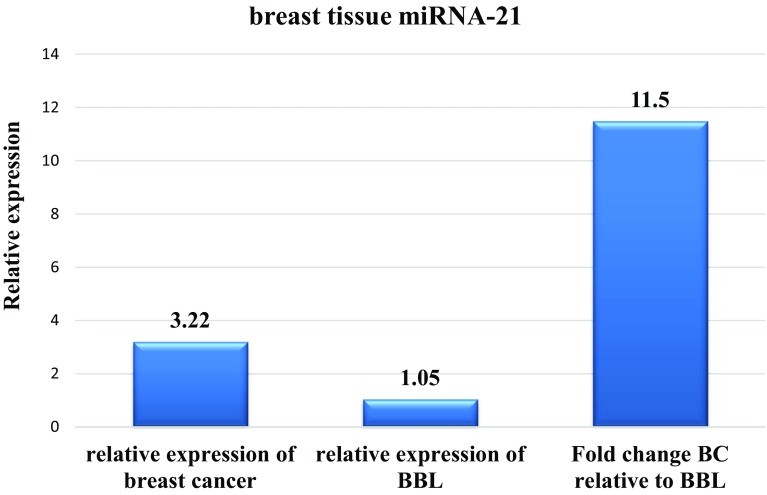
As shown in Fig. 2, the Ct value of miR-21 in breast cancer tissues was lower than that of miR-21 in non-cancer and benign lesion breast tissues, which means that the expression level of miR-21 in breast cancer samples was higher than that in the other groups. The tissue level of miRNA-21 in each tumor sample was normalized to that of a reference gene miRNA-16 and then compared to the non-tumor sites. The mean ± SEM of relative expression (− ΔCt) value of miRNA-21 was 3.22 ± 0.185 in the breast cancer samples, and 1.05 ± 0.183 in benign breast tissue lesion. The mean relative expression level of tissue miR-21 was increased in breast cancer patients with 11.5 fold change when compared to benign breast tissue lesion (P < 0.001, Student’s t test).
Fig. 2.
The relative expression and median of fold change of breast tissue microRNA-21 in the breast cancer and benign breast lesion (BBL) groups
Results of Correlation Between Circulating Exosomal and Tissue miRNA-21 and PDCD4 in the Study Groups
Figure 3 shows positive significant correlation of tissue miRNA-21 relative expression with circulating plasma levels of miRNA-21 in the breast cancer group (r = 0.44, P = 0.016). In relation to PDCD4, we registered significant inverse correlation (P < 0.01) between Ct of tissues (r = − 0.517) and circulating miRNAs-21 (r = − 0.59) gene expression and PDCD4 level in the breast cancer group. Loss or reduction of PDCD4 expression was significantly associated with increased levels of circulating and tissues miRNA-21 in breast cancer patients.
Fig. 3.
The correlation between a circulating miRNA-21 and tissue miRNA-21, b tissue miRNA-21 and tissue programed death cell 4, c circulating miRNA-21 and PDCD4 in patients with breast cancer
The Diagnostics Performance of the Biomarkers in the Studied Groups
Receiver operator characteristics curve (ROC) analysis of tissue miRNA-21 (Fig. 4) in breast cancer patients when compared with benign breast lesion group (Fig. 4a) displayed 100% sensitivity and specificity and the best cut off of the relative expression reading was 0.87. MiRNA-21in plasma of breast cancer patients (Fig. 4b) recorded lowest sensitivity (93.3%), specificity (83.3%) at cut off value of 1.7. The sensitivity, specificity and best cut off values of relative expression of circulating miRNA-21 in the breast cancer patients when compared with healthy control group were 90, 86.7%, and 1.2, respectively. ROC curve analysis of serum CA 15-3 in malignant breast tumors revealed a low sensitivity and specificity (70 and 56.7%) compared to controls at cut off value 32.5 U/ml whereas in comparison to benign breast lesion, the sensitivity and specificity of serum CA 15-3 were 60 and 50%, respectively at cut off value 33.5 U/ml (Fig. 4c).
Fig. 4.
The receiver operating characteristic (ROC) curve showing cut off value, sensitivity, specificity and area under curve for a ROC of tissue miRNA-21 in BC group relative to the BBL group (cut off value = 0.87, sensitivity = 100%, specificity = 100%, and area under curve = 1). Plasma miRNA-21 in breast cancer (BC) group relative to the healthy control (HC) group (cut off value = 1.2, sensitivity = 90%, specificity = 86%, and area under curve = 0.93), and in BC group relative to the benign breast lesion (BBL) group (cut off value = 1.7, sensitivity = 93.3%, specificity = 83.3%, and area under curve = 0.94). c serum CA 15-3 in breast cancer with control (cut off value = 32.5, sensitivity = 70%, specificity = 56%, and area under curve = 0.69) and benign breast lesion groups (cut off value = 33.5, sensitivity = 60%, specificity = 50%, and area under curve = 0.639)
Relationship Between MiRNA-21 and CA15-3 in Breast Cancer Patients and Their Clinicopathological Features
Table 3 shows no statistically significant difference in the mean plasma and tissue expression levels of miRNA-21 and serum CA15-3 in breast cancer patients in relation to age at presentation. Twenty invasive ductal carcinoma (NOS) tumors showed highly significant increases in circulating and tissue miRNA-21 expression levels from both ductal carcinoma in situ (DCIS) and invasive lobular carcinoma (ILC) groups (P < 0.05 and P < 0.01), also the mean serum CA15-3 was higher in invasive ductal carcinoma (NOS) P = 0.05 (Kruskal–Wallis test). The up-regulated expression of miRNA-21 (plasma and tissue) was associated with advanced TNM stage (P = 0.01 for tissue miRNA-21). In Her-2-positive breast tumors, there were a statistical trend towards decreased levels of circulating miRNAs 21 as compared to Her-2-negative tumor samples (P < 0.05). Tissue levels of miRNA-21 showed no difference in Her-2 positive and Her-2 negative breast cancer specimens. Whereas, the mean level of serum CA15-3 revealed no significant association and change with the Her-2 status.
Table 3.
Association of circulation (exosomal), tissue miRNA-21 expression and serum level of CA15-3 with clinical and pathological features of breast tumors
| Variable | Exosomal miRNA-21 (− ΔCt) | Tissue miRNA-21 (− ΔCt) | Serum CA-153 (U/ml) | |
|---|---|---|---|---|
| N | Mean ± SEM | Mean ± SEM | Mean ± SEM | |
| Age distribution | ||||
| Age ≤ 55 | 14 | 3.64 ± 0.4 | 3.27 ± 0.28 | 44.9 ± 5.45 |
| > 55 | 16 | 3.61 ± 0.59 | 3.27 ± 0.26 | 39.4 ± 3.8 |
| Histological type | ||||
| Invasive ductal carcinoma (NOS) | 20 | 4.29 ± 0.33*a | 3.39 ± 0.23*a | 49.5 ± 12.5*a |
| Ductal carcinoma in situ (DCIS) | 8 | 2.75 ± 0.60*a | 3.05 ± 0.34*a | 32.8 ± 1.78 |
| Invasive lobular carcinoma (ILC) | 2 | 0.512 ± 0.71 | 2.26 ± 0.26 | 32.3 ± 3.72 |
| TNM stage | ||||
| I | 3 | 0.3 ± 0.29 | 2.06 ± 0.57 | 32.2 ± 3.5 |
| II | 13 | 3.4 ± 0.35*a | 2.84 ± 0.25**a | 33.7 ± 1.86 |
| III | 13 | 3.57 ± 0.44*a | 3.77 ± 0.21**a | 36.67 ± 8.9 |
| IV | 1 | 4.26 | 4.42 ± 0.00**a | 44 |
| Histologic grade | ||||
| I | 9 | 3.28 ± 0.71 | 2.73 ± 0.41 | 35.3 ± 3.25 |
| II | 13 | 4.23 ± 0.46 | 3.56 ± 0.23 | 32.3 ± 3.6 |
| III | 8 | 3.1 ± 0.59 | 3.08 ± 0.29 | 33.8 ± 2 |
| Tumor size | ||||
| ≥ 2 cm | 12 | 4.02 ± 0.41 | 3.39 ± 0.26 | 31.9 ± 2.0 |
| < 2 cm | 18 | 3.76 ± 0.365 | 2.97 ± 0.23 | 36.1 ± 3.6 |
| Lymph node | ||||
| Positive | 20 | 3.76 ± 0.37 | 3.30 ± 0.24 | 33 ± 2.0 |
| Negative | 10 | 3.35 ± 0.7 | 3.06 ± 0.29 | 34.8 ± 4.0 |
| Estrogen receptor | ||||
| Positive | 17 | 3.67 ± 0.38 | 3.04 ± 0.25 | 32.8 ± 2.16 |
| Negative | 13 | 3.57 ± 0.61 | 3.46 ± 0.27 | 34.0 ± 3.43 |
| Progesterone receptor | ||||
| Positive | 17 | 3.34 ± 0.37 | 3.26 ± -0.28 | 33.76 ± 2.08 |
| Negative | 13 | 3.99 ± 0.6 | 3.17 ± 0.23 | 33.3 ± 3.6 |
| Her2/neu receptorc | ||||
| Positive | 10 | 2.46 ± 0.55*b | 3.05 ± 0.3 | 35.5 ± 1.78 |
| Negative | 20 | 4.21 ± 0.36 | 3.31 ± 0.24 | 32.4 ± 3.7 |
aKruskal–Wallis test: *P < 0.05; **P < 0.01
bMann–Whitney test: *P < 0.05
cHuman epidermal growth factor receptor 2
Histological Findings for Programed Cell Death 4 (PDCD4) in Malignant and Benign Breast Tissue Lesions
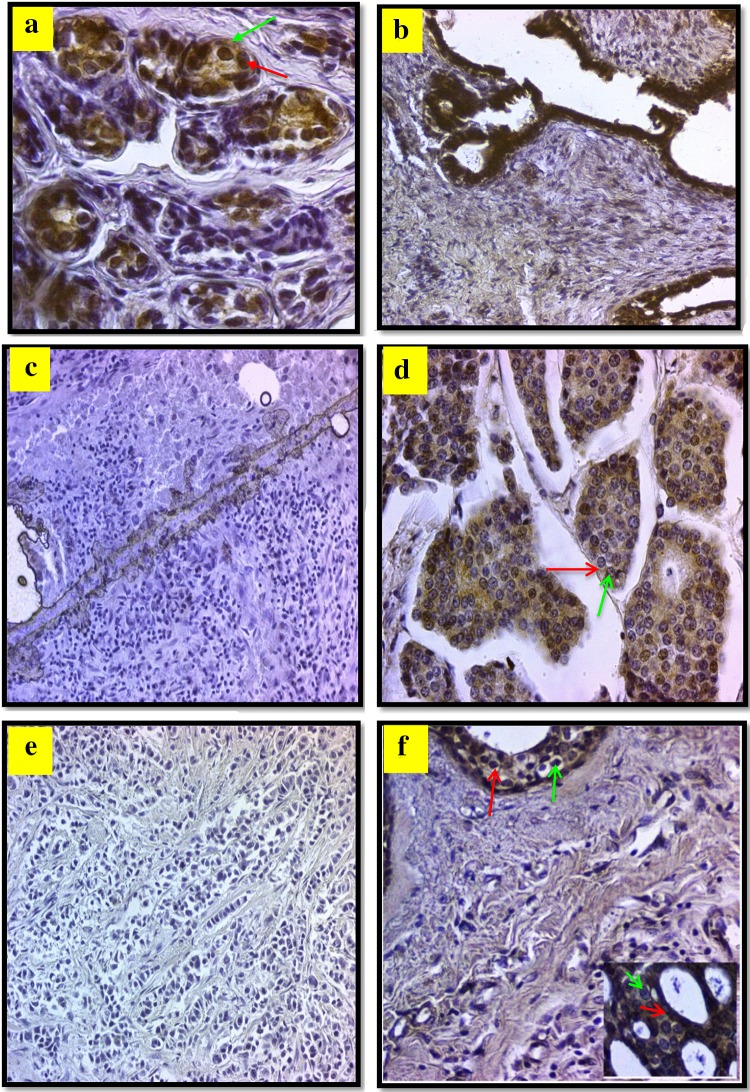
PDCD4-specific staining was positive in all cases of Benign breast tissue samples and the protein was found in both nuclear and cytoplasmic portions (Fig. 5a–f). In contrary, the PDCD4 specific staining was focused in the cytoplasm of breast cancer cells. The cytoplasmic staining was not as intense as nuclear staining in most breast cancer tissues. Figure 5a, b depict moderate and strong nuclear staining intensities for PDCD4. The invasive ductal carcinoma grade 0 showed negative reaction in the nucleus and cytoplam of less than 10% of cells and loss of positive nucleocytoplasmic reaction (Fig. 5e). Negative reaction is displayed in the nucleus and cytoplam of invasive ductal carcinoma grade 0 (Fig. 5c). Staining intensities of 25–50% in grade II invasive ductal carcinoma are shown in Fig. 5d. Loss of positive reaction was observed in the nucleo-cytoplasmic regions of less than 10% of cells invasive lobular carcinoma grade 0 and in ductal carcinoma in situ grade II (Fig. 5f). PDCD4 was expressed at varying levels in twenty (66.7%) of invasive ductal carcinoma, eight (26.7%) of ductal carcinoma in situ (DCIS) and two (6.7%) cases of Invasive Lobular carcinoma (ILC). Some differences in the distribution and intensity of staining were observed between the groups. In the breast cancer specimens for patients with invasive ductal carcinoma, the immunohistochemical reaction for the detection PDCD4 showed positive expression mainly in cytoplasmic region with few positive expressions in the nucleus (Fig. 5c, d). Figure 5c revealed negative expression of PDCD4 in both cytoplasm and nucleus.
Fig. 5.
Microphotograph view of the immunohistochemical staining of programed cell death 4 PDCD4 in breast tissue with a fibrocystic disease (Grade III) with positive reaction mainly in the nucleus (green arrow) and cytoplamic (red arrow) of more than 50% of cells with modrate intensity, ×40. b Fibroadenoma (Grade III) with positive reaction in the nucleus and cytoplamic of more than 50% of cells with strong intensity, ×20. c Invasive ductal carcinoma (Grade 0) negative reaction in the nucleus and cytoplam of less than 10% of cells, ×20. d Invasive ductal carcinoma (Grade II) positive reaction mainly in the cytoplam (red arrow) and less in nucleus (green arrow) of less than 50% of cells of moderate intensity, ×40. e Invasive lobular carcinoma (Grade 0) loss positive reaction in the nucleus and cytoplam of less than 10% of cells, ×20. f Ductal carcinoma in situ (Grade II) positive reaction in the cytoplam (red arrow) and nucleus (green arrow) of less than 50% of cells strong intensity, ×20 (inset ×40)
Breast cancer specimens from the patients with DCIS showed less than 50% positive expression in nucleus and cytoplasm (Fig. 5f), whereas patients specimens with ILC showed negative PDCD4 expression (Fig. 5e).
As seen in Table 4, 90% of specimens from Benign breast tissues exhibit moderate to strong PDCD4 protein expression. Seven out of 30 (23.3%) breast cancer tissues had no detectable PDCD4 protein expression, while 11 out of 30 (36.7%) malignant breast tissues exhibited weak PDCD4 expression and 40% (12/30) showed moderate or strong expression. The overall expression of PDCD4 in breast cancer was significantly lower as compared to Benign breast tissues (P < 0.01).
Table 4.
Distribution of the expression of programed cell death 4 (PDCD4) in benign and malignant breast tumors
| Breast tumor | PDCD4 expressiona | Total | Reaction grade | ||
|---|---|---|---|---|---|
| Weak | Moderate | Strong | |||
| No. % | No. % | No. % | No. % | ||
| Malignant (n = 30) | Positive | 23 (66.7) | 11 (36.7) | 9 (30) | 3 (10) |
| Negative | 7 (23.3) | – | – | – | |
| Benign (n = 30) | Positive | 30 (100) | 3 (10) | 8 (26.7) | 19 (63.3) |
| Negative | 0 (0%) | – | – | – | |
aIndependent t test: *P < 0.01
Discussion
The present study showed that the highest frequency of breast cancer occurs in Iraqi women with a mean-age of 52 years, while the mean age for women with benign cases and for healthy control was (40 years old). These results were similar to those found by other studies conducted in Iraq; documenting that the malignant breast lesions occur mostly in mean age of 45–51 years [24–26] and similar mean age prevalence was found in other published reports [27, 28]. The peak age of breast cancer in Asian countries is between 40 and 50 but this still lower than those observed in most developed countries like USA and UK which have a peak age between 60 and 70 years [29]. The high incidence of breast cancer at lower ages than those of developed countries is due to increased exposure to pollution from extensive use of gasoline toward electric generators and the remnants of war and weapons, decreased birth rate, the increase in the age of marriage and probably the westernization of Iraqi diet.
The results of the present study show that the expression level of plasma miRNA-21 was significantly higher (fold change = 10.7, P < 0.001) in breast cancer patients than in controls and is 14.9 times higher than in the benign breast lesion group. This finding is in line with those reported by Asaga et al. [30] and Mar-Aguilar et al. [31], who reported that the levels of miR-21 were significantly higher in the sera of breast cancer patients than in the control group and that high-circulating miR-21 concentrations were correlated significantly with the presence and extent of breast cancer. Han et al. [32] found that the serum concentrations of miR-21 was significantly higher in patients with breast cancer as compared to healthy controls. Chen et al. [33] observed that the levels of plasma miR-21 and miR-152 were increased in breast and lung cancer patients.
The expression of miR-21 in breast tissues of breast cancer patients is found to be statistically higher relative to the benign breast lesion group with a median fold change of 11.5. This finding is similar to the results of several other studies that revealed miR-21 expression in breast cancer tissue is significantly higher than those in the normal breast tissue, suggesting that miR-21 functions as an oncogene in cancer development [34, 35].
Yan et al. [36] found that miR-21 was significantly up-regulated in breast cancer. Breast cancer patients with elevated miR-21 expression have the worst prognosis and the high levels of miR-21 appear to be significantly interrelated with the advance of the clinical stage, lymph node metastases, and bad prognosis. Toraih et al. [37] reported high expression of miR-21 in tissues and sera of 30 patients with breast cancer, and 30 cancer-free participants with risk factors for developing breast cancer. The higher levels of serum miR-21 were associated with of high tumors grade, more nodal involvement and advanced clinical stages. Li et al. [38] found that miR-21 was overexpressed in breast cancer tissue in comparison with para-carcinoma site. The circulating miRNA may also play a role in in cell to cell crosstalk and facilitate processes including antigen presentation and hemostasis. Exosomes can release their contents following plasma membrane fusion at target cells. These findings further suggest that exosomes containing miRNAs may have the potential for promoting tumor metastasis and are probable therapeutic targets to treat breast malignancy under diverse pathological conditions [39–41].
The sensitivity and specificity of circulating miRNA-21 in breast cancer were found to be 90, 86.7%, respectively, in reference to the control group, and 93, 83.3%, respectively, in relation to the benign breast lesion group. In the study of Gao et al. [42], the circulating miR-16 and miR-21 were amplified and quantified by real-time PCR in 89 breast cancer patients and 55 healthy controls and they reported a significant increase in circulating miR-21 in breast cancer patients compared to controls (P < 0.001). The sensitivity and specificity of miR-21 were 87.6 and 87.3%, respectively with an absence of correlation with the status of ER, PR and clinical stages. Meta analyses [43] on eleven studies with a total of 918 breast cancer patients and 613 controls reported that sensitivity, specificity and diagnostic odds ratio with their 95% confidence intervals (CIs) were 0.72 (95% CI 0.69–0.75), 0.80 (95% CI 0.77–0.83, respectively. The area under the curve of ROC was 0.85.
MiR-21 is considered an ‘oncomir’ leading to tumorous cell proliferation, apoptosis repression and augmentation of migration, invasion, angiogenesis, and metastasis. The increased miR-21 expression level significantly correlated with tumor grade, tumor size, lymph nodal involvement, lympho-vascular invasion in patients with a worse prognosis. Therapeutic inhibition of miRNAs using a specific antisense oligonucleotide of miR-21 may still be beneficial for a large number of resistant cancers [44].
PDCD4 gene has the potential of tumor suppression and the PDCD4 can inhibit cell proliferation and promote cell apoptosis through Sp1 transcription factor [45]. The present study demonstrated that the IHC staining for PDCD4 is expressed in both nuclear and cytoplasm fraction of cells but it is abundantly expressed as a cytoplasmic protein in benign breast lesion (90% moderately to strong) cells. Whereas, lower expression of PDCD4 is found more in malignant breast cancer (overall 66.7% weak to strong). The expression of PDCD4 was observed within the epithelial cells of normal breast tissue and in the nucleo-cytoplasmic fractions. These findings are in agreement with those previously recorded by Wen et al. [46], who observed a primarily nuclear localization of PDCD4 with decreased protein expression in DCIS compared to normal breast tissues where the PDCD4 expression was predominantly cytoplasmic. PDCD4 is able to the nucleo-cytoplasmic shuttling, and modulate the function of DAXX and therefore it promote DNA damage [47]. The PDCD4 gene product was reported to be the transcription regulator of specific genes through modulating the activity of transcription factors c-Jun, Sp1and p53 [45, 47]. Modelska et al. [48] reported a correlation of PDCD4 with ER expression in breast tumors, specifically the nuclear PDCD4 (ρ = 0.035). In ER-positive tumors, we observed that the intense PDCD4 positive immunohistochemical staining was associated with lower grade, smaller size and less nodal metastasis in female’s breast cancer. In estrogen-positive breast cancers, downregulation of PDCD4 was found to be mediated by upregulation of Her2/neu via the MAPK and Akt signaling pathways, as well as miR-21 in aromatase inhibitor-resistant breast cancer cells [49, 50].
Furthermore, we recorded low protein expressions of PDCD4 in breast cancer tissues compared to benign breast lesion tissues that was also associated with the increase in the expression of breast cancer tissue and exosomal miR-21. Similar expression of miR-21 and suppression of PDCD4 has been previously reported by Qi et al. [51] who revealed a positive staining of miR-21 in 15 of 17 of IDC specimens with low miR-21 expression in normal tissue (13%), and in flat epithelial atypia, FEA was (47%), 75% in DCIS, and 88% in IDC. The PDCD4 expression was observed in the cytoplasm and nuclei of luminal cells. The expression of PDCD4 was found predominantly in nuclei in normal breast tissue (13 vs. 73% in nucleus, respectively) and FEA (13 vs. 93% in nucleus, in an ordered manner), whereas in DCIS and IDC positive intense cytoplasmic staining was found in nearly 50% of the cases with reduced nuclear staining intensity. However, the nuclear PDCD4 and the miR-21 staining patterns for normal and IDC cases were inversely interrelated. For FEA and DCIS histological types such a relation was not obvious.
The finding of a strong correlation between exosomal and breast tissue miRNA-21 expression in breast tumors (P < 0.01) pinpoint to the origin of this circulating miRNA-21 and the usefulness of exosomal miRNA determination in screening for breast tumors and as a prognostic marker in these patients.
In conclusion, the increased expression and the interrelation of both breast tissue and exosomal miR-21 in DCIS, and IDC and their inverse correlation with the foremost nuclear expression of PDCD protein signifies the oncogenic role of miR-21 in pathogenesis and the spread of breast cancer in these patients, as well as its potential use as a biomarker for disease screening and response to therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors would express their sincere appreciation to Professor Robert C. Benjamin, University of North Texas, USA for revising and editing this article prior to submission to the journal.
Author Contribution
Professor Dr. NAH has conceived, designed the experiments and share in writing the paper. Dr. MMA has collected the samples, performed the molecular experiments under the supervision of Professor Dr. NAH, contributed reagents/materials/analysis tools and analysed the data. Professor Dr. AGH has performed the immunohistochemical analyses and interpretation of the results.
Compliance with Ethical Standards
Conflict of interest
All authors declare that they have no conflict of interest.
Contributor Information
Meena M. Abdulhussain, Email: khalid_abood99@yahoo.com
Najat A. Hasan, Email: onlynajat@yahoo.com, Email: onlynajat@gmail.com, Email: dr.najat.ar.hassan@colmed-alnahrain.edu.iq
Alaa G. Hussain, Email: alaa-ghani@colmed-alnahrain.edu.iq
References
- 1.International Agency for Research on Cancer: Early Detection and Prevention. Globocan 2012. Available from: http://www.iarc.fr/en/research-groups/sec10/index.php. Accessed 13 June 2017.
- 2.Alsaraj M, Alsaed SJ. Iraqi cancer registry 2011. Baghdad: Iraqi Cancer Board, Ministry of Health; 2011. pp. 25–38. [Google Scholar]
- 3.Majid RA, Hassan HA, Muhealdeen DN, Mohammed HA, Hughson MD. Breast cancer in Iraq is associated with a unimodally distributed predominance of luminal type B over luminal type A, surrogates from young to old age. BMC Womens Health. 2017;17:27. doi: 10.1186/s12905-017-0376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AL-Janabi AA, Naseer ZH, Hamody TA. MEpidemiological study of cancers in Iraq-Karbala from 2008 to 2015. Intern J Med Res Health Sci. 2017;6(1):79–86. [Google Scholar]
- 5.The Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumors. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Iorio MV, Croce CM. MicroRNA involvement in human cancer. Carcinogen. 2012;33(6):1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, et al. High miR-expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117:13–40. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 11.Sioud M, Cekaite L. Profiling of miRNA expression and prediction of target genes. Methods Mol Biol. 2010;629:257–271. doi: 10.1007/978-1-60761-657-3_16. [DOI] [PubMed] [Google Scholar]
- 12.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. MiR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 13.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 14.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 15.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21(miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 17.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 18.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 20.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 21.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 23.Yang M, Chen J, Su F, Yu B, Lin L, Liu Y, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alwan, N. Iraqi initiative of a regional comparative breast cancer research project in the Middle East. J Cancer Biol Res. 2014. Available from: https://www.jscimedcentral.com/CancerBiology/cancerbiology-spidbreastcancer-1016.php. Accessed 13 June 2017.
- 25.AL-wasiti EA, Hasan NA, AL-Salhi AR. Evaluation of markers of oxidative DNA damage in females with breast tumors. Iraqi J Med Sci. 2010;8(1):51–65. [Google Scholar]
- 26.Abed Oun MA, El-Yssin HD, Al-Alwan NA. Prevalence of soluble fas protein in breast cancer patients: correlation with the clinico-pathological parameter. Iraqi Postgr Med J. 2016;15:107–117. [Google Scholar]
- 27.DeSantis C, Siegel R, Bandi P, Jemal A. breast cancer statistics, 2011. CA Cancer J Clin. 2011;61(6):409–418. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 28.Ravdin PM, Cronin KA, Howlader N, Berg CD, Chlebowski RT, Feuer EJ, et al. The decrease in breast-cancer incidence in 2003 in the United States. N Engl J Med. 2007;356(16):1670–1674. doi: 10.1056/NEJMsr070105. [DOI] [PubMed] [Google Scholar]
- 29.Leong SP, Shen ZZ, Liu TJ, Agarwal G, Tajima T, Paik NS, et al. Is breast cancer the same disease in Asian and Western countries? World J Surg. 2010;34(10):2308–2324. doi: 10.1007/s00268-010-0683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 31.Mar-Aguilar F, Mendoza-Ramíreza JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, et al. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.1155/2013/259454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, et al. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. 2017;92(2):55–66. doi: 10.4174/astr.2017.92.2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Liu H, Zou H, Chen R, Dou Y, Sheng S, et al. Evaluation of plasma miR-21 and miR-152 as diagnostic biomarkers for common types of human cancers. J Cancer. 2016;7(5):490–499. doi: 10.7150/jca.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdel-Hamid NR, Mohammed EA, Abbas AH, Badr FM. MicroRNA-21 expression in primary breast cancer tissue among Egyptian female patients and its correlation with chromosome 17 aneusomy. Mol Diagn Ther. 2015;19(6):365–373. doi: 10.1007/s40291-015-0161-4. [DOI] [PubMed] [Google Scholar]
- 35.De Mattos-Arruda L, Bottai G, Nuciforo PG, Di Tommaso L, Giovannetti E, Peg V, et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget. 2015;6(35):37269–37280. doi: 10.18632/oncotarget.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan LX, Liu YH, Xiang JW, Wu QN, Xu LB, Luo XL, et al. PIK3R1 targeting by miR-21 suppresses tumor cell migration and invasion by reducing PI3 K/AKT signaling and reversing EMT, and predicts clinical outcome of breast cancer. Int J Oncol. 2016;48(2):471–484. doi: 10.3892/ijo.2015.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toraih EA, Mohammed EA, Farrag S, Ramsis N, Hosny S. Pilot study of serum MicroRNA-21 as a diagnostic and prognostic biomarker in Egyptian breast cancer patients. Mol Diagn Ther. 2015;19:179–190. doi: 10.1007/s40291-015-0143-6. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Xin S, Yang D, Li X, He Z, Che X, et al. Down-regulation of PDCD4 expression is an independent predictor of poor prognosis in human renal cell carcinoma patients. J Cancer Res Clin Oncol. 2012;138(3):529–535. doi: 10.1007/s00432-011-1121-y. [DOI] [PubMed] [Google Scholar]
- 39.Hornick N, Huan J, Goloviznina NA, Potter A, Kurre P. Hypoxia regulates exosomal microrna content, trafficking and function of key elements in the AML microenvironment. Blood. 2013. http://www.bloodjournal.org/content/122/21/742. Accessed 13 July 2017.
- 40.Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci USA. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W, Lee DY, Ben-David Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int J Physiol Pathophysiol Pharmacol. 2011;3(2):140–155. [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y, Cai Q, Huang Y, Li S, Yang H, Sun L, et al. MicroRNA-21 as a potential diagnostic biomarker for breast cancer patients: a pooled analysis of individual studies. Oncotarget. 2016;7(23):34498–34506. doi: 10.18632/oncotarget.9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J, Zhang Q, Xu J, Guo L, Li X. Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chin J Cancer Res. 2013;25(6):743–748. doi: 10.3978/j.issn.1000-9604.2013.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Badr FM. Potential role of miR-21 in breast cancer diagnosis and therapy. JSM Biotechnol Bioeng. 2016;3(5):1068. [Google Scholar]
- 45.Leupold JH, Yang HS, Colburn NH, Asangani I, Post S, Allgayer H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene. 2007;26:4550–4562. doi: 10.1038/sj.onc.1210234. [DOI] [PubMed] [Google Scholar]
- 46.Wen YH, Shi X, Chiriboga L, Matsahashi S, Yee H, Afonja O. Alterations in the expression of PDCD4 in ductal carcinoma of the breast. Oncol Rep. 2007;18(6):1387–1393. [PubMed] [Google Scholar]
- 47.Kumar N, Wethkamp N, Waters LC, Carr MD, Klempnauer K-H. Tumor suppressor protein Pdcd4 interacts with Daxx and modulates the stability of Daxx and the Hipk2-dependent phosphorylation of p53 at serine 46. Oncogenesis. 2013 doi: 10.1038/oncsis.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, et al. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015 doi: 10.1038/cddis.2014.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Böhm M, Sawicka K, Siebrasse JP, Brehmer-Fastnacht A, Peters R, Klempnauer K-H. The transformation suppressor protein Pdcd4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene. 2003;22:4905–4910. doi: 10.1038/sj.onc.1206710. [DOI] [PubMed] [Google Scholar]
- 50.Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep. 2016;5(4):395–402. doi: 10.3892/br.2016.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi L, Bart J, Tan LP, Platteel I, Sluis TVD, Huitema S, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. doi: 10.1186/1471-2407-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.