Abstract
GEMIN4 is a member of the GEMIN gene family which is involved in multiple pathologies including cancer. It is located on Chr17p13.3, the most notorious chromosome and a hotspot for various carcinomas. We therefore intend to find genetic variants of GEMIN4 gene associated with renal cell carcinoma risk (RCC). This study comprised 100 patients and 225 controls. Genotyping of GEMIN4 gene variants was done using Taqman® assay. The association of GEMIN4 variants and risk prediction of RCC was done by statistical analysis. Haplotype analysis was done to see the combined effect of variants on RCC. Patients carrying variant genotype, CC of GEMIN4 T/C rs7813 showed significant association whereas in case of GEMIN4 G/C rs910925 variant genotype, CC significant risk was found. GEMIN4 rs7813 T/C variant genotype, CC showed risk with smoking (p = 0.034). Our study gives a substantive support for the association between the GEMIN4 gene variants and RCC risk.
Keywords: Renal cell carcinoma (RCC), GEMIN4 gene, Cancer susceptibility
Introduction
Renal cell carcinoma (RCC) is the third most common genitourinary malignancies. RCC accounts for more than 87% of all renal malignancies and is the most common neoplasm of the kidney. Renal carcinoma is heterogeneous being comprised of variety of histological variants having different survivals, genetics and response to the treatment given. The most common pathology for RCC is clear cell renal cell carcinoma (ccRCC). The rate of occurrence of RCC varies worldwide being the highest in Europe and North America and lowest in Asia and South America [1]. The ratio for RCC occurrence M:F is 2:1 [1]. The precise cause of RCC is poorly understood, but some specific lifestyle factors such as smoking can play an important etiological factor.
MicroRNAs (miRNAs) are about 20 nucleotide long non-coding RNA molecules which plays a role in post-transcriptional regulation [2, 3]. MiRNAs play a regulatory role in many human genes. Any misregulation in post transcriptional events could lead to cancer occurrence and progression [4]. Some studies reported the association of pre-miRNA gene variants to prostate cancer, supporting the potential interdependence between alterations in the miRNA pathway and the development of prostate cancer [5].
Gemin4 protein is a key member of GEMIN protein family which is involved in various physiological processes [6]. SNPs in the GEMIN4 gene affects DNA repair in liver cancers and hence lead to the development of liver cancers in Chinese population [7]. Studies have shown association of GEMIN4 SNPs with the clinical outcome of bladder cancer [8], renal cancer [9, 10], ovarian cancer [11], lung cancer [12], prostate cancer [6] and esophageal cancer [13]. Any truncation of Gemin4 protein will affect the differential expression of some miRNAs which could be related to the malignant tumors. There is no consensus about the effects of GEMIN4 gene variants in the pathogenesis of RCC among North Indians. Therefore, the present pilot study is designed to evaluate the association between common polymorphisms in the GEMIN4 gene and the risk of RCC in North Indian population.
Materials and Methods
Ethical Statement
The study design was approved by the Institutional Ethics Committee. Duly signed informed consents were taken from each study subject.
Study Subjects
The present study of RCC was conducted in the Department of Urology and Renal Transplantation. 100 histologically confirmed RCC patients (M/F 81/19; mean age 49.93 + 11.39 years) were enrolled for the study. Exclusion criteria for patient selection was chemotherapy and radiation therapies. 225 unrelated controls (mean age 55.08 years, and M/F ratio as 202/23) were recruited randomly from the unrelated individuals. 6 ml of peripheral blood was collected from every patient with 3 ml of blood each in different vials containing 0.5 M EDTA (pH 8.0) as an anti-coagulant and stored at − 80 °C for further analysis. At the time of enrolment patients and controls were interviewed for demographic details and lifestyle details like smoking. The RCC patients were grouped further into non-smokers and smokers.
Clinical Data Collection
The basic detail and characteristics of patients are demonstrated in Table 1. The pathological details of the tumor and therapy, relapse etc. were provided by concerned personnels. American Joint Committee on Cancer’s TNM staging system was used to classify the tumor. 79 of 100 patients had clear cell RCC (ccRCC) type and rest had non-clear cell RCC.
Table 1.
Demographical details of renal cell cancer patients and healthy controls
| Variables | Patients n = 100 n (%) |
Controls n = 225 n (%) |
#p value |
|---|---|---|---|
| Sex | |||
| Female | 19 (19.0) | 23 (10.2) | 0.229 |
| Male | 81 (81.0) | 202 (89.8) | |
| Age (years) | |||
| Mean age ± SD | 49.93 ± 11.39 | 55.08 ± 10.14 | 0.313 |
| Smokinga | |||
| Non-smokers | 32 (34.4) | 161 (73.9) | < 0.001 |
| Smokers | 61 (65.6) | 57 (26.1) | |
| Stage | |||
| T1 | 25 (25.0) | – | – |
| T2 | 43 (43.0) | – | |
| T3 | 12 (12.0) | – | |
| T4 | 20 (20.0) | ||
| Grade | |||
| G1 | 20 (20.0) | – | – |
| G2 | 42 (42.0) | – | |
| G3 | 38 (38.0) | – | |
| Histology | |||
| Clear cell RCC | 79 (79.0) | ||
| Non-clear cell RCC | 21 (21.0) | ||
The statisticaly significant values are shown in bold
BCG i + m Bacillus Calmette-Guerin induction + maintenance
aThe sum could not add up to the total due to some missing values
#Student t test was used to determine the p value
SNPs Selected
SNPs were selected based on available literature and their role in various carcinomas. SNP info and F-SNP were used for Tag SNPs selection. Three SNPs of GEMIN4 viz. GEMIN4 C/T rs3744741, GEMIN4 T/C rs7813 and GEMIN4 G/C rs910925 were selected for the present study. Details of selected SNPs are given in Table 2.
Table 2.
Characteristic of candidate SNPs of GEMIN-4 Gene
| SNP ID | Major/minor allele | MAF (%) |
|---|---|---|
| rs3744741 | C/T | 28 |
| rs7813 | T/C | 29 |
| rs910925 | G/C | 29 |
SNP single nucleotide polymorphism, MAF minor allele frequency
DNA Extraction and Genotyping
Blood DNA was extracted using salting-out method [14]. Polymorphisms in GEMIN4 C/T rs3744741, GEMIN4 T/C rs7813 and GEMIN4 G/C rs910925 were genotyped using TaqMan SNP (Applied Biosystems, USA) genotyping assay using 96-well plate Real time PCR system along with positive and negative controls each assay, and 10% of the samples were randomly selected and run in duplicates with 100% concordance.
Statistical Analysis
The power of the study was calculated using Quanto software, version 1.0 (available from: http://hydra.usc.edu/gxe) To analyse basal characteristics, we used Chi square tests for the categorical data. Associations between GEMIN4 gene polymorphisms and RCC risk were estimated using adjusted odds ratios (AORs) and 95% confidence intervals (95% CIs) from multivariate logistic regression, which was used to adjust the effect factor (i.e., age, gender and smoking). The statistical analysis was done using the Statistical Package for Social Sciences software, version 20.0 (SPSS, Chicago, IL), and p < 0.05 was considered statistically significant.
In Silico Analysis
Functional effects in GEMIN4 gene was determined using F-SNP (http://compbio.cs.queensu.ca/F-SNP/) [15].
Results
Demographic Details
The characterization of patients and controls are demonstrated in Table 1. There was no significant difference between the patients and controls on age (p = 0.313) and gender (p = 0.229). Number of patients with smoking habit (65.6%) are higher than controls (p value = 0.001). 79% of patients had conventional clear cell carcinoma. Patients having other histological forms of carcinoma were 21%. Approximately 25% of patients were in stage I, 43, 12, and 20% was found to be in stage II, III, and IV, respectively. Genotype distributions for all the SNPs in the control group were in concordance with Hardy–Weinberg equilibrium (p = 0.855).
Genotyping and Association of GEMIN4 C/T rs3744741, GEMIN4 T/C rs7813 and GEMIN4 G/C rs910925 with RCC
The gene variants of GEMIN4 viz. GEMIN4 C/T rs3744741, GEMIN4 T/C rs7813 and GEMIN4 G/C rs910925 were evaluated and their genotype distribution in RCC patients is detailed in Table 3. No significant association of GEMIN4 C/T rs3744741 was seen in case of RCC risk (CT type; OR 1.18, CI 0.65–2.14, p = 0.582, TT type; OR 2.16, CI 0.85–5.53, p = 0.106). We did not find any association in the dominant model (CT + TT; OR 1.31, CI 0.75–2.28, p = 0.339) or at allelic level (T allele; OR 1.26, CI 0.88–1.82, p = 0.206) of GEMIN4 C/T rs3744741 in RCC. Although the other two variants of GEMIN4 were found to be associated with manifold risk in developing RCC. In case of GEMIN4 T/C rs7813, significant association was observed in variant genotype (CC type; OR 3.37, CI 1.44–7.89, p = 0.005), dominant model (TC + CC; OR 1.94, CI 1.11–3.41, p = 0.021) as well as at allelic level in variant allele (C allele; OR 1.66, CI 1.17–2.35, p = 0.004). While in the third variant of GEMIN4, i.e. GEMIN4 G/C rs910925 significant risk was seen only in variant genotype (CC type; OR 2.37, CI 1.12–4.99, p = 0.024) and variant allele (C allele; OR 1.56, CI 1.11–2.19, p = 0.010).
Table 3.
Genotype and allelic frequency distribution of GEMIN4 C/T (rs3744741), GEMIN4 T/C (rs7813) and GEMIN4 G/C (rs910925) gene polymorphism in RCC patient and healthy controls
| Genetic model | Genotypes | Controls n = 225 n (%) |
Patients n = 100 n (%) |
#p value | ORa (95%CI) |
|---|---|---|---|---|---|
| GEMIN4 C/T rs3744741 | |||||
| Additive | CC | 123 (54.7) | 51 (51.0) | Ref | Ref |
| CT | 86 (38.2) | 36 (36.0) | 0.582 | 1.18 (0.65–2.14) | |
| TT | 16 (7.1) | 13 (13.0) | 0.106 | 2.16 (0.85–5.53) | |
| Dominant | CC | 123 (54.7) | 51 (51.0) | Ref | Ref |
| CT + TT | 102 (45.3) | 49 (49.0) | 0.339 | 1.31 (0.75–2.28) | |
| Multiple | C | 332 (73.8) | 138 (69.0) | Ref | Ref |
| T | 118 (26.2) | 62 (31.0) | 0.209 | 1.26 (0.88–1.82) | |
| GEMIN4 T/C rs7813 | |||||
| Additive | TT | 115 (51.1) | 39 (39.0) | Ref | Ref |
| TC | 89 (39.6) | 41 (41.0) | 0.918 | 1.03 (0.56–1.90) | |
| CC | 21 (9.3) | 20 (20.0) | 0.005 | 3.37 (1.44–7.89) | |
| Dominant | TT | 115 (51.1) | 39 (39.0) | Ref | Ref |
| TC + CC | 110 (48.9) | 61 (61.0) | 0.021 | 1.94 (1.11–3.41) | |
| Multiple | T | 319 (70.9) | 119 (59.5) | Ref | Ref |
| C | 131 (29.1) | 81 (40.5) | 0.004 | 1.66 (1.17–2.35) | |
| GEMIN4 G/C rs910925 | |||||
| Additive | GG | 100 (44.4) | 37 (37.0) | Ref | Ref |
| GC | 93 (41.3) | 35 (35.0) | 0.693 | 0.88 (0.47–1.65) | |
| CC | 32 (14.2) | 28 (28.0) | 0.024 | 2.37 (1.12–4.99) | |
| Dominant | GG | 100 (44.4) | 37 (37.0) | Ref | Ref |
| GC + CC | 125 (55.5) | 63 (63.0) | 0.062 | 1.72 (0.97–3.04) | |
| Multiple | G | 293 (65.1) | 109 (54.5) | Ref | Ref |
| C | 157 (34.9) | 91 (45.5) | 0.010 | 1.56 (1.11–2.19) | |
The statistically significant values are shown in bold
OR odds ratio, CI confidence interval
aOR (95%CI); Age, gender and smoking adjusted odds ratio and 95% confidence interval
#Student’s t test was used to determine p value
Association of GEMIN4 Genotypes with Smoking as a Risk Factor of RCC
After analysing the data at genotypic and allelic level we tried to relate the association with smoking, which is a great risk factor in causing RCC. For this study, we stratified the patients amongst two groups: smokers and non-smokers, and calculations were done accordingly. We found significant association in variant genotype of GEMIN4 T/C rs7813 with smoking in RCC (CC; OR 3.51, CI 1.16–4.29, p = 0.034). Whereas, the other gene variants GEMIN4 C/T rs3744741 and GEMIN4 G/C rs910925 did not show any association with smoking risk factor in RCC. The detailed data is shown in Table 4.
Table 4.
Association of GEMIN4 C/T (rs3744741), GEMIN4 T/C (rs7813) and GEMIN4 G/C (rs910925) gene variants with smoking habit in RCC susceptibility
| Genotype | Patients Non smokers n = 32; n (%) |
Patients Smoker n = 61; n (%) |
p value | ORa (95% CI) |
|---|---|---|---|---|
| GEMIN4 C/T rs3744741 | ||||
| CC | 16 (50.0) | 30 (49.2) | Ref | Ref |
| CT | 10 (31.2) | 24 (39.3) | 0.721 | 0.80 (0.23–2.71) |
| TT | 6 (18.8) | 7 (11.5) | 0.129 | 0.31 (0.07–1.41) |
| GEMIN4 T/C rs7813 | ||||
| TT | 11 (45.8) | 35 (50.7) | Ref | Ref |
| TC | 8 (33.4) | 26 (37.7) | 0.227 | 2.09 (0.63–6.91) |
| CC | 5 (20.8) | 8 (11.6) | 0.034 | 3.51 (1.16–4.29) |
| GEMIN4 G/C rs910925 | ||||
| GG | 9 (37.5) | 26 (37.7) | Ref | Ref |
| GC | 7 (29.2) | 25 (36.2) | 0.507 | 1.53 (0.43–5.42) |
| CC | 8 (33.3) | 18 (26.1) | 0.587 | 1.46 (0.37–5.85) |
Statistically significant values are shown in bold
OR odds ratio, CI confidence interval
aOR (95%CI) Age, gender adjusted odds ratio and 95% confidence interval
Association of Histological Cell Type of RCC with GEMIN4 Gene Variants
Tumor histology plays a major role in detection of cancer stage as well as grade. If any gene is associated with the cellular histology of tumor tissue, we can detect the stage by detecting that gene. Here we tried to correlate the GEMIN4 gene variants at genotypic level with RCC cell type to find any significant association. No significant association of any of the three gene variants of GEMIN4 with RCC risk was observed. No association of RCC tumor with GEMIN4 gene variants could be due to small number of samples. (Table 5).
Table 5.
Genotype frequency of GEMIN4 C/T (rs3744741), GEMIN4 T/C (rs7813) and GEMIN4 G/C (rs910925) of RCC patients based on RCC cell type
| Genotype | Cell type | p value | ORa (95%CI) | |
|---|---|---|---|---|
| Non-clear RCC (n = 21) | Clear cell RCC (n = 79) | |||
| GEMIN4 C/T rs3744741 | ||||
| CC | 12 (57.1) | 39 (49.4) | Ref | Ref |
| CT | 6 (28.6) | 30 (38.0) | 0.483 | 0.650 (0.219–1.932) |
| TT | 3 (14.3) | 10 (12.7) | 0.973 | 0.975 (0.230–4.129) |
| GEMIN4 T/C rs7813 | ||||
| TT | 9 (42.9) | 30 (38.0) | Ref | Ref |
| TC | 9 (42.9) | 32 (40.5) | 0.904 | 0.938 (0.328–2.678) |
| CC | 3 (14.3) | 17 (21.5) | 0.469 | 0.588 (0.140–2.472) |
| GEMIN4 G/C rs910925 | ||||
| GG | 9 (42.9) | 28 (35.4) | Ref | Ref |
| GC | 7 (33.3) | 28 (35.4) | 0.660 | 0.778 (0.254–2.379) |
| CC | 5 (23.8) | 23 (29.1) | 0.531 | 0.676 (0.199–2.301) |
aOR (95%CI) Age, gender adjusted odds ratio and 95% confidence interval
OR odds ratio, CI confidence interval
Association of GEMIN4 C/T rs3744741, GEMIN4 T/C rs7813 and GEMIN4 G/C rs910925 Haplotypes with RCC Risk
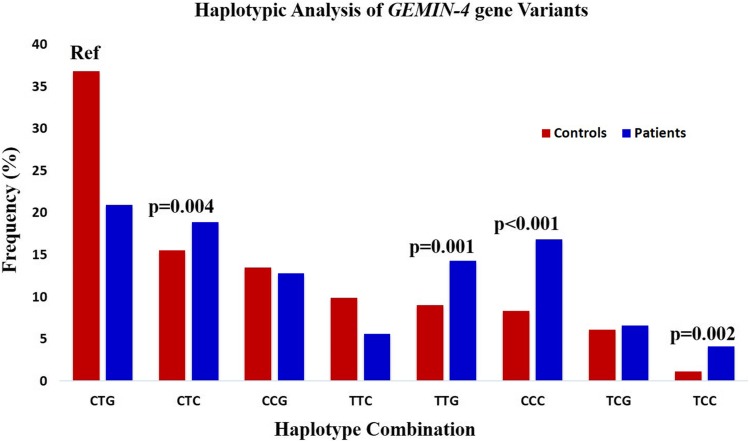
Haplotype analysis (Combinatorial effect of SNPs) may be more manifesting in predicting the association compared to single nucleotide polymorphism, as individual polymorphism is likely to confer modest effects to the risk of RCC. We constructed haplotype sets for GEMIN4 gene polymorphisms, wherein CTG was taken as a reference. We demonstrate significant association in 3 sets of haplotype combinations (TTG, CCC and TCC) with RCC risk, the significant p values and OR are shown in Fig. 1.
Fig. 1.
Haplotypic analysis of GEMIN4 gene variants in RCC patients and controls
In silico Analysis for the Functionality of GEMIN-4 Gene Variants
The location of SNPs of GEMIN4 gene i.e. rs3744741, rs7813 and rs910925 is described in Table 6 and the results of In-silico analysis showed change in transcriptional regulation for all the candidate SNPs (Table 6).
Table 6.
In silico analysis of GEMIN4 gene polymorphisms by F-SNP
| SNPs of GEMIN-4 gene | Functional category | Prediction tool | Prediction result | FS score | Location |
|---|---|---|---|---|---|
| rs3744741 | Protein coding | SNP effect | Deleterious | 1 | Exonic |
| Splice regulation | ESE finder | Changed | |||
| Post translational | OGPET | Exist | |||
| rs7813 | Protein coding | SNP effect | Deleterious | 0.451 | 3′UTR |
| Splice regulation | ESE finder | Changed | |||
| Post translational | OGPET | Not exist | |||
| rs910925 | Protein coding | SNP effect | Benign | 0.264 | Exonic |
| Splice regulation | ESE finder | Changed | |||
| Post translational | OGPET | Not exist |
Discussion
GEMIN4 gene is located on chromosome 17p13.3, a hotspot for various melanomas, and is a protein coding gene. GEMIN4 is an important gene of miRNA machinery. The expression of many human genes is regulated by miRNA processing. [6, 16]. Aberrations in the regulatory pathway of the miRNA could lead to altered miRNA transcription, splicing, and transcriptional regulation of cell proliferative and apoptotic genes. Therefore, polymorphisms in the miRNA pathways may contribute to cancer progression [5]. In this study, significant associations were found in SNPs of GEMIN4 gene and RCC risk. The results of this study would lead to the further concept of the mechanism of expression of miRNAs and their relation with genetic variants in the miRNA regulatory pathway genes viz. GEMIN4 and the susceptibility of RCC. Our findings are consistent with the results that these two polymorphisms, GEMIN4 G/C rs910925 and GEMIN4 T/C rs7813, have potential roles in carcinogenesis, such as renal cell carcinoma [10], bladder cancer [9], and ovarian cancer [17] in other studies. No significant association with the third polymorphism and RCC risk was observed in our study, suggestive of either no role in the pathogenesis of RCC, or perhaps related to the limited sample size. So, case–control studies with large sample size and of different ethnicity should be performed to validate our results. In this study, we found significant associations between SNPs in miRNAs biogenesis pathway and the risk of RCC. Recent studies have shown that, disrupting miRNAs processing through the knockdown of DROSHA, DGCR8, and DICER1, could accelerate cellular transformation and tumorigenesis [8].
Thomson et al. [18] have shown that the repression of mature miRNAs is not consistent with the reductions in the primary miRNAs transcripts, suggesting the existence of altered regulations of miRNAs processing in human cancers. These lines of evidence are in concordance with the recent profiling of miRNAs expression, which showed the general repression of miRNAs in a variety of tumors and cancer cell lines [6, 8, 16].
The findings of our study indicate that genetic alterations of miRNAs regulation might be associated with cancer development and progression. We analyzed the haplotype combination of GEMIN4 gene variants with their three polymorphic sites and RCC risk. We found significant association in three sets with more than 1.5 folds’ risk in RCC. Three SNPs of the GEMIN4 (rs7813 and rs2740348) and GEMIN3 gene (rs197412) were found to be associated with altered RCC risk [10].
Nonetheless, we tried to get more powerful comprehension about the influence of these SNPs on RCC risk using a pathway-based polygenic approach and identify a pattern toward an increasing RCC risk with an increasing number of unfavourable genotypes. Yang and group in one of their study have discussed about genetic variants in miRNA genes and their biogenesis pathway and their susceptibility to bladder cancer. In the same study, they have elaborated about 41 functional miRNA related SNPs, among all, 7 SNPs (rs910924, rs2740348, rs7813, rs910925, rs3744741, rs1062923, rs4968104) of GEMIN-4 gene were discussed [8]. One SNP of GEMIN-4 gene i.e. rs7813 was found to be significantly associated with bladder cancer risk [8], hepatocellular carcinoma [7], which complies to our results. Yang et al. [8], also found the SNPs to have physiological effects and tumorigenesis of bladder cancer. F-SNP is a collection of all the SNPs having functional effects on various carcinomas. The functional effects of GEMIN4 C/T rs3744741, GEMIN4 T/C rs7813 and GEMIN4 G/C rs910925 is discussed in Table 6, rs3744741 and rs7813 have deleterious effect in bladder cancer indicating a predisposition of these SNPs in risk of bladder cancer (http://compbio.cs.queensu.ca/F-SNP/).
This finding thus can contribute to prove the fact that RCC is polygenic process and thus miRNAs machinery genes may have a greater ability to characterize high-risk populations. Further studies in a larger population are required to validate these results. The GEMIN-4 gene, one of the miRNAs regulatory pathway genes, contributes to increased risk of RCC.
Limitations of the Study
The sample size of our study was relatively small; therefore, studies with larger sample sizes with sufficient large subgroups are warranted to validate our findings.
Acknowledgements
This study was funded by Department of Science and Technology (DST) [SR/SO/HS-120/2007], New Delhi. The assistance of relevant clinical information of the patients by the Urologists and Pathologist is duly acknowledged.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Archana Verma, Email: archanavermakv@gmail.com.
Vibha Singh, Email: vibhasngh03@gmail.com.
Praveen Kumar Jaiswal, Email: praveenj.bbau@gmail.com.
Rama D. Mittal, Phone: +91 522 2494116, Email: ramamittal@gmail.com
References
- 1.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, et al. Up-regulation of dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, Lu C, Ye Y, Chang J, Yang H, Lin J, et al. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum based chemotherapy. Pharmacogenet Genomics. 2008;18:955–965. doi: 10.1097/FPC.0b013e32830efdd4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Liu J, Wei M, He Y, Liao B, Liao G, et al. Genetic variants in the microRNA machinery gene GEMIN4 are associated with risk of prostate cancer: a case-control study of the Chinese Han population. DNA Cell Biol. 2012;31(7):1296–1302. doi: 10.1089/dna.2011.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan D, He M, Wang J, Qiu X, Zhou W, Luo Z, et al. Two variants of the human hepatocellular carcinoma-associated HCAP1 gene and their effect on the growth of the human liver cancer cell line Hep3B. Genes Chromosomes Cancer. 2004;39(1):48–58. doi: 10.1002/gcc.10293. [DOI] [PubMed] [Google Scholar]
- 8.Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 9.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31:1805–1812. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang D, Meyer L, Chang DW, Lin J, Pu X, Ye Y, et al. Genetic variants in Micro RNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765–9776. doi: 10.1158/0008-5472.CAN-10-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang X, Yin Z, Li X, Xia L, Zhou B. Polymorphisms in GEMIN4 and AGO1 genes are associated with the risk of lung cancer: a case-control study in Chinese female non-smokers. Int J Environ Res Public Health. 2016;13(10):E939. doi: 10.3390/ijerph13100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1(6):460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucl Acids Res. 2008;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some micro-RNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 17.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 18.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20(16):2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]