Abstract
There is an ever-increasing recognition that bile acids are not purely simple surfactant molecules that aid in lipid digestion, but are a family of molecules contributing to a diverse range of key systemic functions in the host. It is now also understood that the specific composition of the bile acid milieu within the host is related to the expression and activity of bacterially-derived enzymes within the gastrointestinal tract, as such creating a direct link between the physiology of the host and the gut microbiota. Coupled to the knowledge that perturbation of the structure and/or function of the gut microbiota may contribute to the pathogenesis of a range of diseases, there is a high level of interest in the potential for manipulation of the gut microbiota-host bile acid axis as a novel approach to therapeutics. Much of the growing understanding of the biology of this area reflects the recent development and refinement of a range of novel techniques; this study applies a number of those techniques to the analysis of human samples, aiming to illustrate their strengths, drawbacks and biological significance at all stages. Specifically, we used microbial profiling (using 16S rRNA gene sequencing), bile acid profiling (using liquid chromatography-mass spectrometry), bsh and baiCD qPCR, and a BSH enzyme activity assay to demonstrate differences in the gut microbiota and bile metabolism in stool samples from healthy and antibiotic-exposed individuals.
Keywords: microbiota, metabonome, bile, antibiotics, 16S rRNA gene sequencing, qPCR
1. Introduction
1.1. Overview
The last few years have been associated with a rapid increase in understanding of the profound contribution of the gut microbiota to the health of the host, as well as its potential roles in the onset and maintenance of a range of diseases. Much initial interest in the gut microbiota has focused on observational studies which defined changes to the structure of the microbiota in different scenarios (e.g. different disease states, impact of diet or antibiotics, etc). However, more recent emphasis has moved away from solely defining the structure of the microbiota, but refocused upon better defining its function, and specifically the many complex routes of communication (including metabolic pathways, immune axes, etc) between the gut microbiota and the host [1]. Given that a key regulator of the composition of the bile acid pool within mammals is the action of bacterially-derived enzymes within the gastrointestinal tract [2], an improved understanding of the close interplay between the gut microbiota and the host’s bile acid metabolism is an area of particular interest.
1.2. Gut microbiota-bile acid interactions in vivo
Primary bile acids (BA) are synthesised from cholesterol in the liver, where they are conjugated with glycine or taurine. These conjugated bile acids subsequently enter the gallbladder, and are released into the duodenum following the intake of food. Once in the small bowel, the bile acids undertake one of their key physiological roles, the emulsification and solubilisation of dietary lipids. Bile acids will continue along the small intestine, towards the terminal ileum; whilst approximately 95% of bile acids will be reabsorbed via the enterohepatic circulation pathway, the remaining 5% (~400-800 mg per day) are not recovered, and will continue through the distal gut of the terminal ileum and on to the colon [3].
It is within the small intestine that bile acid modification by the gut microbiota is initiated, driven by enzymes that are produced and secreted by gut microbiota members, but which are not produced by the mammalian host. The first stage of bile acid modification by the gut microbiota is from the enzymes named bile salt hydrolases (BSHs). These enzymes deconjugate the taurine and glycine groups from conjugated bile acids via a hydrolysis reaction, and therefore reform the primary bile acids cholate (CA) and chenodeoxycholate (CDCA). BSHs are found mainly within the bacterial phyla Firmicutes and Bacteroidetes, but are widely-distributed throughout most major bacterial divisions and archaeal species of the human gut microbiota [4]. At least eight different bsh genes exist (see Supplementary Figure 1), with each form having specific properties relating to optimal pH, specificity for taurine- or glycine-conjugated bile acids and gene size [4]. The secondary enzymatic steps are 7-α-dehydroxylation. In these steps, the hydroxyl group of C-7 is removed, thus converting primary bile acids to secondary bile acids. Specifically, in humans, this includes the conversion of cholate to deoxycholate (DCA), and the conversion of chenodeoxycholate to lithocholate (LCA), along with the biosynthesis of other secondary bile acids. 7-α-dehydroxylation is a complex, multi-step process, and only performed by strictly anaerobic bacteria with the bile acid-inducible (bai) operon. Based on current microbial genomic annotation, it is estimated that only a very small percentage of gut microbiota members possess 7-α-dehydroxylation activity, with those organisms that do predominantly belonging to the genera Clostridium clusters XIVa and XI [5], [6]. Generation of secondary bile acids creates a more hydrophobic bile acid pool, facilitating the elimination of these bile acids within faeces. A range of other gut microbial metabolic actions against bile acids are also described, including the epimerisation of CDCA to synthesise ursodeoxycholic acid, as well as other pathways that result in the generation of iso-, allo- and oxo-/keto-bile acids [2].
There is now increasing recognition of the diverse roles of bile acids within the host, in particular via their role as endogenous ligands for host cell receptors. These include the nuclear receptor farnesoid X receptor (FXR), and the G protein-coupled plasma membrane bile acid receptor TGR5, all exhibiting varying affinities for different bile acids and their moieties [2]. Bile acids as FXR and TGR5 agonists contribute to a wealth of host physiological processes including the modulation of lipid, glucose and energy homeostasis, as well as the regulation of bile acid synthesis, conjugation and transport. To add to the complexity, there is also evidence that bile acids influence microbiota composition, both via direct and indirect actions [2]. Collectively, the growing evidence for the multiple functions of bile acids within the host – coupled with evidence demonstrating the complex interplay between bile acid metabolism and the gut microbiota – highlights that this axis is a key mechanism by which the gut microbiota directly influences a range of aspects of host physiology.
Two of the most important questions in gut microbiome research are “who is there?” and “what are they doing?”. In the context of bile metabolism we can describe changes in the gut microbiota at several different levels: we can use microbial DNA to define the composition of the gut microbiota and quantify the amount of bile metabolising genes, we can look at the amount of bile metabolising proteins expressed by measuring their enzymatic activity, and we can look at the metabolites being produced by characterising the quantity and composition of bile acid metabolites. In this study we use a set of stool samples from individuals exposed to antibiotics and non-antibiotic-exposed controls to demonstrate how researchers can apply a wide variety of techniques to more fully characterise microbiota-bile interactions in the gut. These techniques include 16S rRNA gene sequencing, liquid chromatography-mass spectrometry-based bile acid profiling, BSH and 7-a-dehydroxylase qPCR, and a BSH enzyme activity assay. In addition, we correlated metataxonomic and metabonomic data to gain a better understanding of the modulation of the bile acid pool by the gut microbiota.
2. Material and methods
2.1. Study participants
The study was performed under approval from the UK National Research Ethics Centre (13/LO/1867). Stool samples were collected from a total of eight healthy individuals, and five patients who had recently taken recurrent courses of antibiotics. Antibiotics had been prescribed for a variety of indications, had been used for at least three continuous weeks within the past month, and had last been used between 3 – 6 days prior to sample collection (Supplementary Table 1). Healthy individuals had not used antibiotics or been prescribed regular medications for at least six months prior to sample collection. Stool specimens were put on ice within 15 minutes after collection, transferred to the hospital laboratory, and homogenised and aliquoted within 30 minutes. Samples were frozen to and maintained at -80°C prior to analysis.
2.2. DNA extraction and 16S rRNA gene sequencing
DNA was extracted from 250 mg of stool using the PowerLyzer PowerSoil DNA Isolation Kit (Mo Bio, Carlsbad, CA, USA) following manufacturer’s instructions, with the addition of a bead beating step for 3 minutes at speed 8 in a Bullet Blender Storm (Chembio Ltd, St Albans, UK). DNA was stored at -80°C until it was ready to be used.
Sample libraries were prepared following Illumina’s 16S Metagenomic Sequencing Library Preparation Protocol [7] with two modifications. Firstly, the V1-V2 regions of the 16S rRNA gene were amplified using the primers listed in Table 1. Additionally, the index PCR reactions were cleaned up and normalised using the SequalPrep Normalization Plate Kit (Life Technologies, Paisley, UK). Sample libraries were quantified using the NEBNext Library Quant Kit for Illumina (New England Biolabs, Hitchin, UK). Sequencing was performed on an Illumina MiSeq platform (Illumina Inc., Saffron Walden, UK) using the MiSeq Reagent Kit v3 (Illumina) and paired-end 300bp chemistry.
Table 1.
Primers used for 16S rRNA gene sequencing on the Illumina MiSeq. The forward primer mix was composed of four different forward primers, mixed at a ratio of 4:1:1:1 (28F-YM:28F-Borrellia:28FChloroflex:28F-Bifdo). Bases in bold are the MiSeq adapter sequences.
| Primer name | Primer sequence |
|---|---|
| 28F-YM (forward primer) |
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGAGTTTGATYMTGGCTCAG |
| 28F-Borrellia (forward primer) | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGAGTTTGATCCTGGCTTAG |
| 28FChloroflex (forward primer) | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGAATTTGATCTTGGTTCAG |
| 28F-Bifdo (forward primer) |
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGGTTCGATTCTGGCTCAG |
| 388R (reverse primer) |
GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGCTGCCTCCCGTAGGAGT |
The resulting data was analysed using the Mothur package following the MiSeq SOP Pipeline [8]. The Silva bacterial database was used for sequence alignments (www.arb-silva.de/) and the RDP database reference sequence files were used for classification of sequences using the Wang method [9]. The non-metric multidimensional scaling (NMDS) plot and PERMANOVA p-values were generated using the UniFrac weighted distance matrix generated from Mothur, and analysed using the Vegan library within the R statistical package [10]. Family-level extended error bar plots were generated using the Statistical Analysis of Metagenomic Profiles software package using White’s non-parametric t-test with Benjamini-Hochberg FDR [11]. The α diversity (Shannon diversity index, H’) and richness (total number of bacterial taxa observed, Sobs) were calculated within Mothur and statistical tests (independent t-test and Mann-Whitney U test, respectively) were performed using IBM SPSS Statistics Software version 23. A p-value of 0.05 and a q-value of 0.05 was considered significant.
2.3. Inference of gut microbiota function from 16S rRNA gene sequencing data
To predict the bile-metabolising ability of the microbial communities within the samples, an inferential tool, Piphillin, was applied [12]. This algorithm uses direct nearest-neighbour matching between 16S rRNA gene sequencing datasets and microbial genomic databases to infer the metagenomic content of the samples [12]. In this case, Piphillin was used online [13], using the KEGG May 2017 as reference database, and applying 97% identity cut-off. Inference of gene abundance was assessed for KEGG orthology K01442 (cholylglycine hydrolase, an alternative name for BSH), KEGG orthology K15870 (baiCD, a bacterial gene specific to the 7-α-dehydroxylation pathway) and KEGG pathway ko00121 (corresponding to the secondary bile acid biosynthesis pathway).
2.4. Ultra performance liquid chromatography-mass spectrometry (UPLC-MS) profiling of faecal bile acids
Faecal samples were lyophilized for 24 hours using a VirTis Benchtop BTP 8ZL freeze dryer (BPS, UK). The dried samples were weighed and bile acids were extracted using a 2:1:1 (vol) mixture of water, acetonitrile and 2-propanol in a Biospec bead beater with 1.0 mm Zirconia beads. After centrifugation (16,000 x g, 20 minutes) the supernatant was filtered using 0.45 μm microcentrifuge filters (Costar, Corning).
Quality control (QC) samples were prepared using equal parts of the faecal filtrates. QC samples were used as an assay performance monitor[14], and as a proxy to remove features with high variation. QC samples were also spiked with mixtures of bile acid standards (55 bile acid standards including 36 non-conjugated, 12 conjugated with taurine, seven conjugated with glycine (Steraloids, Newport, RI, USA)) and were analysed along with the stool samples to determine the chromatographic retention times of bile acids and to aid in metabolite identification.
Bile acid analysis of faecal extracts was performed using ACQUITY UPLC (Waters Ltd, Elstree, UK) coupled to a Xevo G2 Q-ToF mass spectrometer equipped with an electrospray ionization source operating in negative ion mode (ESI-), using the method described by Sarafian and colleagues [15].
Waters raw data files were converted to NetCDF format and data were extracted using XCMS (v1.50) package with R (v3.1.1) software. Probabilistic quotient normalisation [16] was used to correct for dilution effects and chromatographic features with coefficient of variation higher than 30% in the QC samples were excluded from further analysis.
The relative intensities of the features were corrected to the dry weight of the faecal samples.
2.5. Integration of 16S rRNA gene sequencing data and bile acid mass spectrometry data
Correlations between two “omic” datasets acquired from the same set of samples were determined using regularised Canonical Correlation Analaysis (rCCA). rCCA modelling of metataxonomic (16S rRNA gene sequencing) and metabonomic (bile acid mass spectrometry) data was employed in the mixOmics library within the R statistical package [17], [18]. The regularisation parameters were determined using the shrinkage method. The rCCA similarity scores between the variables were plotted as heatmaps using the clustered image maps (cim) function. Hierarchical clustering (complete linkage, Euclidean distance) was used to obtain the order of the variables. The correlation circle plot was generated using the plotVar function, which plots strong correlations between variables (plots variables with a correlation above 0.5 outside of the inner circle).
2.6. Abundance and activity of bile-metabolising enzymes
2.6.1. Real-time PCR for the quantification of BSH and baiCD gene abundance
qPCR was performed using extracted DNA to quantify gene abundance. Gene abundance was quantified for i) specified groups of bsh (using degenerate primer sets previously designed and optimised by our group (Table 2)) and ii) baiCD (using primers previously described in the literature [19]).
Table 2.
Primers sequence and PCR conditions for bsh and baiCD qPCR.
| Group | Primer Sequence (5’-3’) | F/R | Cycling Conditions | Expected Product Size (bp) |
|---|---|---|---|---|
| 1a | CACATATTGTGGCACGAACAATHGAR TGGGG | F | 95°C for 10 min, (95°C for 15 sec, 55°C for 1 min) × 40 cycles | 570 |
| CTGTGCCCGGATACAGATTAACRTAR TTRTT | R | |||
| 1b | CGGCGTTCCGCATTTYTAYGARAA | F | 95°C for 10 min, (95°C for 15 sec, 55°C for 1 min) × 40 cycles | 318 |
| GTTCAATGCCAATCGGAATATCRAAR TTRTT | R | |||
| 3c/e | TTTTGGCCGAACACTGGAYTAYGARTT | F | 95°C for 5 min, (95°C for 15 sec, 54°C for 30 sec, 72 for 10 min) × 40 cycles | 774 |
| TCAACGGAGCCCAGAATATGRAARA AYTG | R | |||
| baiCD | GGWTTCAGCCCRCAGATGTTCTTTG | F | 94°C for 2 min, (94°C for 20 sec, 52°C for 30 sec, 69°C for 90 sec) × 35 cycles, 68°C for 10 min | 1300 |
| GAATTCCGGGTTCATGAACATTCTKCKAAG | R |
A total reaction volume of 25µl was used for each reaction, consisting of 20µl master mix and 5µl diluted DNA (12.5ng total per reaction). All DNA was diluted in buffer EB (Qiagen, Hilden, Germany). A standard master mix consisting of 5.5µl PCR grade water (Roche, Penzberg, Germany), 12.5µl of 2x SYBR green master mix (ThermoFisher Scientific, Waltham, Massachusetts, USA), 1µl of 10µM forward primer (Eurofins Genomics, Wolverhampton, UK) and 1µl of 10µM reverse primer (Eurofins Genomics) was used. One bacterial strain from the relevant reference group was selected as a standard for each primer set (bsh group 1a – Bacteroides plebius; bsh group 1b – Bacteroides ovatus; bsh group 3c/e – Blautia obeum; baiCD – Clostridium scindens (DSMZ 5676, Braunschweig, Germany) (Supplementary Methods). Serial dilutions of each isolate were used to create a standard curve. Thermocycling conditions for each primer set are summarised in Table 2. A melt curve stage was performed post-cycling to confirm primer specificity. Products were also visualised using the 2200 Tapestation System (Aligent Technologies, Santa Clara, California, USA) in combination with D1000 Reagents and D100 Screentapes (Aligent Technologies), following the manufacturer’s protocol.
Copy number was calculated from qPCR data using the following formula: gene abundance = (quantity (ng) x 6.022 x 1023 (gene copy number/mol)) / (length of product x 1 x 109 (ng/g) x 660 (g/mol)). A mean copy number for each set of triplicates was calculated and divided by the total DNA per reaction to obtain average copy number per ng DNA.
2.6.2. Bile salt hydrolase enzyme activity assay
Faecal water was prepared and total faecal protein quantified using a similar method to that previously-described by Morris and Marchesi [20], but with the addition of bacterial and mammalian protease inhibitor cocktails (G Biosciences, St Louis, MO, USA), as well as DTT to 1mM final concentration (Roche, Welwyn Garden City, UK) to minimise enzyme oxidation [21].
The BSH assay itself was an adaptation of a precipitation-based assay [21]–[23]. The assay was performed in a clear flat-bottomed 96-well microtitre plate and incubated at 37°C at pH 5.8 for up to 8 hours. In a total volume of 200μl, 500μg of faecal protein was incubated with sodium phosphate buffer (pH 5.8, final concentration of 0.02mM), and taurodeoxycholic acid (Merck, Damstadt, Germany) (at final concentration 1mM). To prevent evaporation during incubation, wells were overlaid with 50μl of light paraffin oil (0.85g/ml; PanReac AppliChem, Barcelona, Spain) [23]. Samples were assayed in triplicate, with precipitation of insoluble deoxycholic acid monitored by absorbance measurement at 600nm (A600) using a microplate reader (MultiSkan Go, Thermo Scientific, Dartford, UK). Faecal protein incubated with phosphate-buffered saline served as a negative control, and faecal protein incubated with varying concentrations of deoxycholic acid (Merck) was used to establish a standard curve to quantify precipitate formation.
2.6.3. Statistical analysis
A Mann-Whitney U test was used to compare the BSH activity and the BSH and baiCD gene abundance data between the antibiotic treated and healthy cohorts. A p-value of <0.05 was considered significant.
3. Results
3.1. 16S rRNA gene sequencing
16S rRNA gene sequencing analysis showed patients taking recurrent antibiotics had altered compositions of their gut microbiotas compared to healthy controls (Figure 1A, p < 0.01, PERMANOVA). Patients taking recurrent antibiotics had lower microbial community diversity (Figure 1B, p < 0.001, independent t-test) and richness (Figure 1C, p < 0.01, Mann-Whitney U test) compared to healthy controls. Statistical analysis showed that the altered microbiota in patients taking recurrent antibiotics were due to decreases in the relative abundances of the families Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, and Oscillospiraceae, and increases in the relative abundance of the family Enterobacteriaceae compared to healthy controls (Figure 1D).
Figure 1.
Antibiotics alters the gut microbiota composition in patients taking recurrent antibiotics compared to healthy controls. (A) Nonmetric multidimensional scaling (NMDS) plot showing the difference in gut microbiota composition of patients taking recurrent antibiotics and healthy controls (p < 0.01, PERMANOVA). (B) α diversity was decreased in patients taking recurrent antibiotics compared to healthy controls (*** p < 0.001, independent t-test). (C) Richness (total number of bacterial taxa observed) was decreased in patients taking recurrent antibiotics compared to healthy controls (** p < 0.01, Mann-Whitney U test). (D) Extended error bar plot comparing the differences in the mean proportions of significantly altered families and the difference in the proportions of the means (White’s non-parametric t-test with Benjamini-Hochberg FDR). Plot only shows families where the difference between the proportions was greater than 1%.
3.2. Inference of gut microbiota function from 16S rRNA gene sequencing data
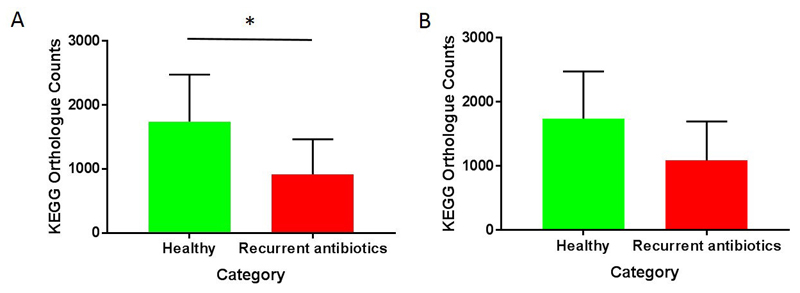
Results from Piphillin analysis are shown in Figure 2. Predicted gene abundance for bsh (KEGG orthologue K01442) was significantly reduced in patients who had taken recurrent antibiotics (Figure 2A, p < 0.05, Mann-Whitney U test). It was not possible to predict gene abundance counts for all samples for baiCD (KEGG orthologue K15870) at the cut-off of 97% identity used, implying very low counts. Predicted secondary bile acid biosynthesis (ko00121) trended lower in patients with recurrent antibiotic use compared to controls, but this was not significant (Figure 2B, p = 0.08).
Figure 2.
Inference of bile-metabolising function from 16S data using Piphillin. (A) Bile salt hydrolase KEGG orthologue counts (K01442) (* p < 0.05, Mann-Whitney U test). (B) Secondary bile acid biosynthesis KEGG orthologue counts (ko00121) (p > 0.05, Mann-Whitney U test).
3.3. Multivariate statistics analysis of UPLC-MS profiling data
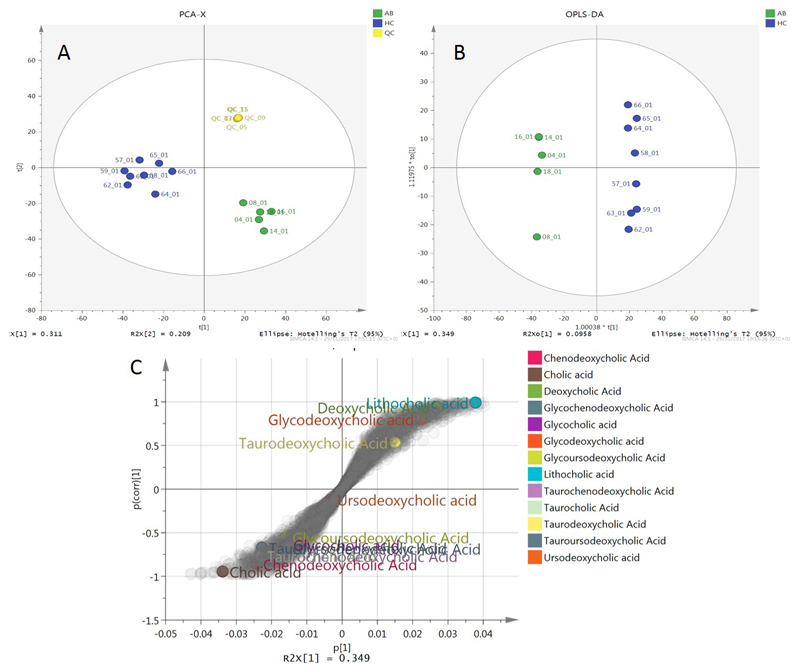
The data table produced by XCMS after normalization to the dry weight of the samples was introduced to SIMCA 14.1 (MKS Umetrics AB). Principal component analysis (PCA) was performed to visualise clustering of samples and assess the quality of the run using the QC samples (Figure 3A). Furthermore, supervised OPLS-DA was performed (Figure 3B) to reveal the features that were responsible for the discrimination between the recurrent antibiotic-treated and healthy control groups. This feature identification was achieved using the S-plot presented in Figure 3C, where feature in the edges of the S-shaped cloud of features were responsible for the separation. Features on top right were higher in the healthy control group, and in bottom left higher in the group treated with recurrent antibiotics. Annotated bile acids are highlighted in the plot.
Figure 3.
Multivariate analysis of UPLC-MS bile acid profiling data. (A) PCA scores plot (B) OPLS-DA scores plot (C) OPLS-DA S-plot, showing the contribution of bile acids to the separation of the two groups. AB: recurrent antibiotic treated patients; HC: healthy controls; QC: quality controls.
Univariate analysis for differences in specific bile acids between healthy participants and people treated with recurrent antibiotics was also performed; data are presented in Supplementary Figure 2.
3.4. Integration of metataxonomic and metabonomic data
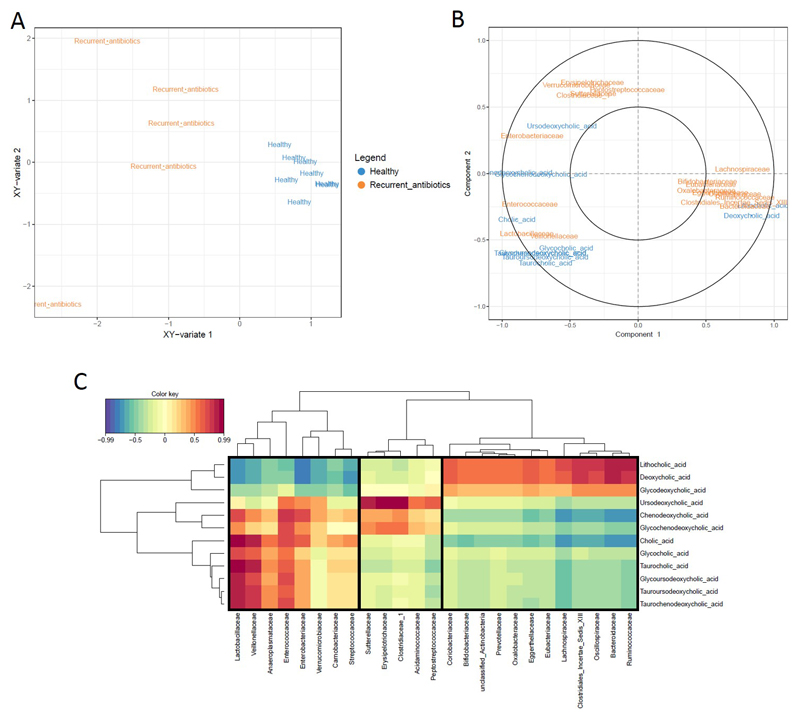
rCCA modelling was used to determine correlations between metataxonomic (16S rRNA gene sequencing) and metabonomic (bile acid mass spectrometry) data (Figure 4). We found that correlations between bacterial families and bile acids clustered into three distinct groups (Figure 4). Group 1 consisted of correlations where bacterial families were positively associated with conjugated and unconjugated primary bile acids, and negatively correlated with secondary bile acids DCA and LCA. Group 2 consisted of families positively correlated with ursodeoxycholic acid. Group 3 consisted of families positively correlated with secondary bile acids DCA and LCA, and negatively associated with unconjugated primary bile acids CA and CDCA. Enterobacteriaceae, which increased in the recurrent antibiotics group, clustered in group 1. Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, and Oscillospiraceae, which decreased in the recurrent antibiotics group, clustered in group 3.
Figure 4.
Regularized CCA (rCCA) modelling of metataxonomic (16S rRNA gene sequencing data, family-level) and metabonomic data (bile acid data). (A) The representation of units for the first two canonical variates showing the correlations between variables in patients receiving recurrent antibiotics and healthy controls. (B) Correlation circle plot showing strong correlations between metataxonomic and metabonomic data (plot only shows variables with a correlation above 0.5). Variables projected in the same direction from the origin have a strong positive correlation, and variables projected in opposite directions form the origin have strong negative correlations. Variables that are at a farther distance from the origin have a stronger correlation. (C) Heatmaps of the rCCA similarity scores between metataxonomic and metabolomic data. Bacterial families outlined in black boxes clustered according to correlations with distinct groups of bile acids.
3.5. Abundance and activity of bile-metabolising enzymes
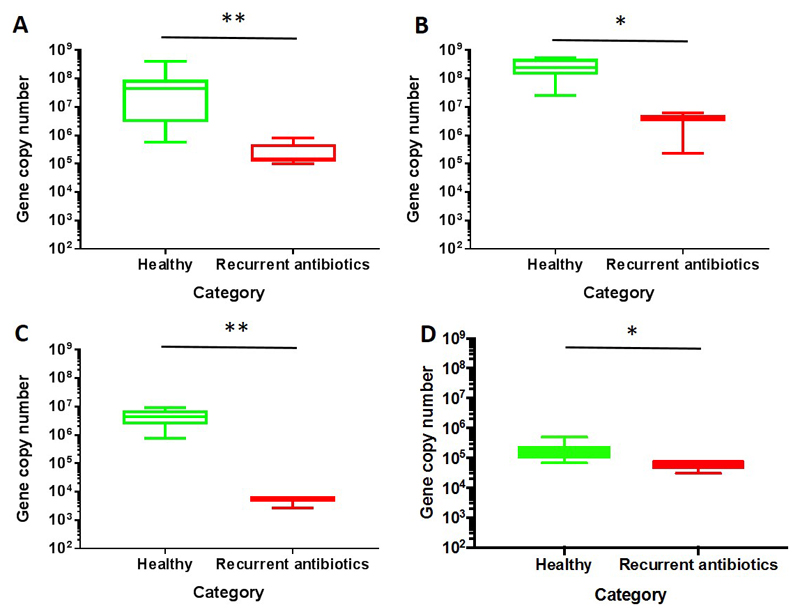
Results from qPCR assays are displayed in Figure 5. Recurrent antibiotic use was associated with a significantly reduced abundance of bsh genes for all BSH groups tested compared to healthy control participants. Specifically, after recurrent antibiotic use, there was reduced abundance of the genes of bsh group 1a gene (p < 0.01, Mann-Whitney U test), bsh group 1b gene (p < 0.05, Mann-Whitney U test), and bsh group 3c/e gene (p < 0.01, Mann-Whitney U test). baiCD gene abundance also significantly reduced after recurrent antibiotic use (p < 0.05, Mann-Whitney U test).
Figure 5.
qPCR to quantify gene abundance of bile metabolising genes. (A) bsh group 1a gene (** p < 0.01, Mann-Whitney U test); (B) bsh group 1b gene (* p < 0.05, Mann-Whitney U test); (C) bsh group 3c/e (** p < 0.01, Mann-Whitney U test); (D) baiCD gene (p < 0.05, Mann-Whitney U test).
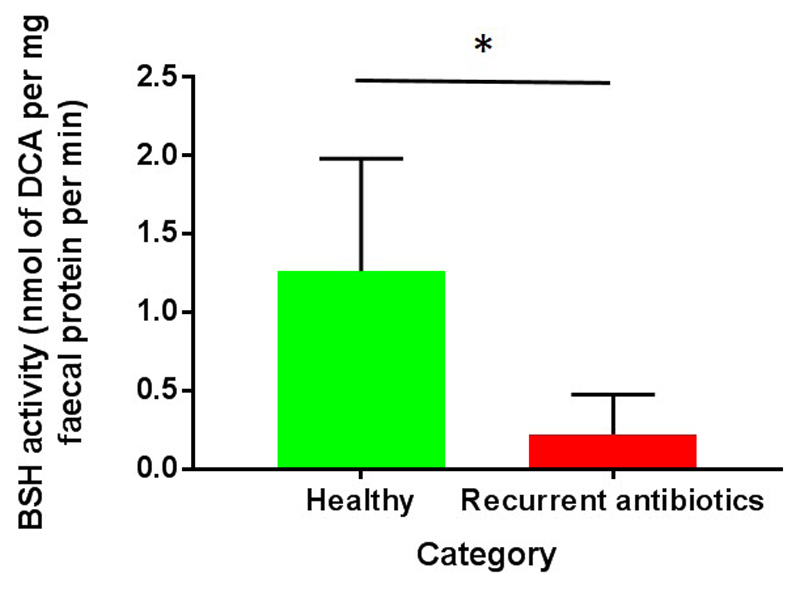
Use of recurrent antibiotics is associated with marked reduction in BSH enzyme activity within faecal samples (Figure 6, p < 0.01, Mann-Whitney U test).
Figure 6.
Bile salt hydrolase (BSH) enzyme activity assay. Taurodeoxycholic acid was used as the substrate for the enzyme assay, and results are therefore expressed as rate of deoxycholic acid formation (* p < 0.05, Mann-Whitney U test).
4. Discussion and Conclusions
In this study, we performed a range of analyses upon stool samples taken from healthy participants and people with recent antibiotic use as a means of demonstrating a range of techniques that may be applied to delineate gut microbiota-host bile acid interactions.
We found that patients taking recurrent antibiotics had gut microbiotas with reduced proportions of known bile-metabolising enzyme function, including the families Bacteroidaceae, Lachnospiraceae and Ruminococcaceae. Consistent with this, recurrent antibiotic use was associated with enrichment of stool primary bile acids (both conjugated and unconjugated) and loss of secondary bile acids. Correlation analysis showed a distinct clustering of bacterial families and bile acids into three groups, where Enterobacteriaceae was positively correlated with unconjugated primary bile acids, and Bacteroidaceae, Lachnospiraceae, Ruminococcaeceae and Oscillospiraceae were positively correlated with secondary bile acids. Further analysis demonstrated a loss of BSH gene abundance and enzyme activity within the gut of antibiotic-treated patients, coupled with a loss of 7-α-dehydroxylase baiCD gene abundance related to antibiotic use. Most fundamentally, these results emphasise the close and complex interplay between the gut microbiota and bile acid metabolism, and reinforce that any perturbation of the gut microbiota (in this case by antibiotics) may result in marked changes to host physiology. These findings are consistent with other comparable work within this area, including the demonstration that early life antibiotic exposure is associated with a long lasting reduction in bile salt hydrolase function [24]. Furthermore, it has also been recognised that Clostridium difficile infection (a gastrointestinal infection occurring predominantly in patients with antibiotic-associated gut dysbiosis) is associated with perturbation of host bile acid profiles, possibly mediated through alteration of gut bile metabolising enzyme functionality [25]–[27].
We used 16S rRNA gene sequencing to determine the differences in the composition of the gut microbiota between patients taking recurrent antibiotics and healthy controls. We found an increase in the relative abundance of Enterobacteriaceae and a decrease in the relative abundance of Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, and Oscillospiraceae in the recurrent antibiotic group compared to healthy controls. However, it is important to note that we are reporting changes in the relative abundances of these groups, not the absolute abundances. The total read numbers per sample does not provide information on the total number of 16S rRNA gene copies in the sample [28]. This is especially important in samples where a change in the total bacterial biomass occurs, for example with antibiotic treatment (as is the case in this study). While it is possible that the absolute abundance of Enterobacteriaceae increases after recurrent antibiotics, it is also possible that the absolute abundance of Enterobacteriaceae has remained unchanged, and there was a decrease in the total biomass due to a decrease in the absolute abundances of Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, and Oscillospiraceae. Studies can account for these changes in bacterial biomass by performing 16S rRNA gene qPCR, and weighting their relative abundance data to get a more informative representation of the microbial community composition.
Whilst 16S rRNA gene sequencing data provides information on the bacterial composition of the sample, it does not provide information regarding the potential functional capabilities of the bacteria and subsequent interactions with the host. Metagenomic sequencing provides information on the collection of genomes in a sample, followed by assembly or mapping to a reference database which allows gene annotation. However, metagenomic sequencing is more expensive than metataxonomics, and the data analysis can be more challenging. In this study, we used Piphillin [12] to indirectly infer the abundance of functional genes as a straightforward and cost-free addition to the study. Piphillin has certain advantages compared to other inferential software tools (including its ease of use, speed of output and the ability to select a reference database of interest [12]), but has not to our knowledge been applied before now for analysis of human gut metataxonomic data. Our intention was to use this method as an exploratory technique, to later confirm with additional methods of analysis (qPCR, LC-MS, and an enzyme assay). The Piphillin results here predicted a reduced bsh gene abundance in the recurrent antibiotic group compared to healthy controls, and our qPCR data and enzyme assay were consistent with this. Whilst Piphillin predicted a trend towards reduced secondary bile acid biosynthesis within the recurrent antibiotic group, it was not able to specifically predict baiCD gene abundance, and we used qPCR to explore this instead. Our experience here and in other work with inferential algorithms is that whilst they may be a helpful and broadly accurate additional tool to start exploring the function of the microbiota, the current limitations in metagenomic annotation mean that results obtained in this way must be interpreted with caution. However, the constant improvements in metagenome annotation are likely to make such tools ever-more accurate over time.
Mass spectrometric techniques are the workhorse of bile acids analysis due to their sensitivity and specificity compared to other assays. High resolution time-of-flight mass spectrometry using a soft ionization method (electrospray ionization, ESI) coupled with ultra-performance liquid chromatography is our analytical method of choice as it can provide comprehensive coverage of bile acids and lipids species from complex biological samples needing minimal sample pre-treatment [15]. In our study, we found that antibiotic exposure had a significant impact upon the composition of the bile acid pool, which could have implications on host physiology. In order to develop interventions that target the bile acid metabolic pathway, researchers need to be able to identify specific bacterial taxa responsible for these bile acid conversions.
One difficulty with ‘omics’ methodologies is the complexity of the datasets generated, often with very large numbers of variables. Software packages such as mixOmics offer researchers useful exploratory approaches to highlight important correlations between bacteria and metabolites. Integration of metataxonomic and metabonomic data can provide researchers with information on the potential roles of microorganisms with in an ecosystem, however it is important to remember that correlation does not equal causation. Strong correlations between bacteria and metabolites must be confirmed with further experiments, such as assays in vitro where researchers can assess the direct effects of a substrate/metabolite on the growth or activity of a microorganisms of interest. Examples of assays in vitro which may be used include batch cultures, mammalian cell line assays, enzyme assays, etc. It is also important to note that there is no consensus on which data integration method is the best method to integrate metataxonomic and metabonomic data sets, as this is an actively developing field of research.
bsh qPCR primer sets were designed to quantify the differences in bsh gene abundance in our samples. We found a statistically significant decrease in bsh group 1a, group 1b, and group G3c/e gene abundance, together with a significant reduction in that of baiCD, associated with antibiotic use. Even though these primers were optimised by us to target a select group of BSH-producing bacteria and were confirmed to not cross-react between groups, the bacterial strains used from each group during the optimisation stage were subject to availability. Therefore, it is reasonable to suggest that, due to their degenerate nature, the primers could also target the bsh gene in other bacterial species within a group which were not tested during the optimisation stage, thereby potentially providing a more comprehensive assessment of bsh gene abundance within the faecal samples. DNA sequencing would be required to categorically confirm the BSH-producing bacterial species targeted by these primer sets. We also performed qPCR of the baiCD operon; whilst this operon is not found in all bacteria with 7-α-dehydroxylating ability, it is present within the two bacterial species with high activity of this enzyme, Clostridium scindens and Clostridium hiranonis, and most strains of these species will be amplified by this PCR [19]. Furthermore, Clostridium scindens is particularly of interest within this context, since its loss from the gut microbiota in association with antibiotic use has been associated with altered gut bile acid metabolism and a potential vulnerability to Clostridium difficile infection [27]. Whilst this qPCR will not amplify certain bacteria with low secondary bile acid biosynthesis functionality (including Clostridium leptum and Clostridium sordeii), good correlation has been noted between baiCD PCR assay results and 7-α-dehydroxylase activity in an in vitro assay, demonstrating that this is still a highly useful assay [19].
Whilst qPCR of bacterial genes is useful, similar to metataxonomic data, there are concerns that what is being assessed relates to which bacterial genes are present, rather than if those genes are being actively transcribed and the resultant functional effects. As such, metatranscriptomics – the sequencing of RNA from within a microbial community – is of great interest for its ability to more directly establish gene transcription and therefore microbiota functionality. However, there remain certain practical difficulties in undertaking such studies, including the considerable cost, the computational complexity, and the difficulties in high-quality RNA extraction and sequencing given its relative instability compared to DNA. Furthermore, whilst protocols have been described that aim to simplify collection of stool and preserve samples for subsequent streamlined combined metagenomic and metatranscriptomic analysis (e.g. via the addition of ethanol or RNAlater to samples) [29], the implications of these preservatives upon the quality of the metabolic profile obtained from the sample remain undefined.
The quantification of gene abundance using qPCR data, metagenomic data, and Piphillin data cannot categorically confirm gene expression and functionality in vivo. Therefore, we developed an enzyme activity assay to measure the amount of BSH activity in each sample through substantial adaptation of a plate-based precipitation assay [21]. Other groups have used a ninhydrin assay to measure BSH activity [23], [30]; however, these studies used pure bacterial strains, and in our experience, this assay is not sensitive enough to detect BSH activity within faecal water. Whilst BSH activity does not require strict anaerobic conditions, 7-α-dehydroxylation does [33], complicating development of a similar activity assay. However, an assay applying thin layer chromatography and radiolabelled cholic acid to human caecal aspirate or stool obtained after enema use to assess 7-α-dehydroxylase activity has been described [33], [32].
In this study, we compared healthy people with patients taking antibiotics, and did not match the participants for other demographics. There are a variety of variables that have been shown to influence the composition and/or functionality of the gut microbiota, which (in addition to antibiotics/microbial infections) include diet, age, surgery, stress, BMI, and pregnancy[34]–[36]. As such, we are unable to say if the differences seen between our groups related purely to antibiotic use, or if there was a contribution from other factors. Where studies compare healthy and diseased groups in attempting to generate novel hypotheses regarding the contribution of gut microbiota-bile acid interactions to the disease process, regard for these factors must be taken to ensure that control groups are appropriate.
Future challenges regarding methodology within this area remain. The relationship between the gut microbiota, bile acid metabolism and the host is complex and bidirectional, and methodologies that further delineate this relationship are required. Development of standardised pipelines for analysing these complex datasets – coupled with more standardised methods for integration of different data sets – are key immediate challenges. At present, whilst there is growing sophistication in our ability to define and correlate gut microbial and bile acid profiles, there is little work (particularly within humans) that has linked this back to systemic host effects. Given the growing recognition that bile acids are signalling molecules with complex systemic effects upon the host, it is clearly of interest and importance to be able to link microbial and bile acid interplay to host physiological function, in relation to health and disease.
Supplementary Material
Acknowledgements
Infrastructure support was provided by the National Institute for Health Research (NIHR) Imperial Biomedical Research Centre (BRC).
Funding: BHM is the recipient of a Medical Research Council Clinical Research Training Fellowship (grant reference: MR/R000875/1). The Division receives financial support from the National Institute of Health Research (NIHR) Imperial Biomedical Research Centre (BRC) based at Imperial College Healthcare NHS Trust and Imperial College London.
References
- [1].Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metabolism. 2012 Nov 07;16(5):559–564. doi: 10.1016/j.cmet.2012.10.007. Cell Press. [DOI] [PubMed] [Google Scholar]
- [2].Wahlströ A, Sayin SI, Marschall H-U, Bä Ckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- [3].Hofmann AF. The Continuing Importance of Bile Acids in Liver and Intestinal Disease. Arch Intern Med. 1999 Dec;159(22):2647. doi: 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- [4].Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008 Sep;105(36):13580–5. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kitahara M, Takamine F, Imamura T, Benno Y. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2000 May;50(3):971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- [6].Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006 Feb;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- [7].Illumina. 16S Metagenomic Sequencing Library Preparation. [Accessed: 27-Nov-2017]; [Online]. Available: https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html.
- [8].Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013 Sep;79(17):5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007 Aug;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].R: The R Project for Statistical Computing. [Accessed: 27-Nov-2017]; [Online]. Available: https://www.r-project.org/.
- [11].Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014 Nov;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Iwai S, et al. Piphillin: Improved Prediction of Metagenomic Content by Direct Inference from Human Microbiomes. PLoS One. 2016 Nov;11(11):e0166104. doi: 10.1371/journal.pone.0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Second Genome Therapeutics. Piphillin. [Accessed: 14-Feb-2018]; [Online]. Available: http://secondgenome.com/solutions/resources/data-analysis-tools/piphillin/.
- [14].Sangster T, Major H, Plumb R, Wilson AJ, Wilson ID. A pragmatic and readily implemented quality control strategy for HPLC-MS and GC-MS-based metabonomic analysis. Analyst. 2006 Oct;131(10):1075. doi: 10.1039/b604498k. [DOI] [PubMed] [Google Scholar]
- [15].Sarafian MH, et al. Bile Acid Profiling and Quantification in Biofluids Using Ultra-Performance Liquid Chromatography Tandem Mass Spectrometry. Anal Chem. 2015 Oct;87(19):9662–9670. doi: 10.1021/acs.analchem.5b01556. [DOI] [PubMed] [Google Scholar]
- [16].Veselkov KA, et al. Optimized Preprocessing of Ultra-Performance Liquid Chromatography/Mass Spectrometry Urinary Metabolic Profiles for Improved Information Recovery. Anal Chem. 2011 Aug;83(15):5864–5872. doi: 10.1021/ac201065j. [DOI] [PubMed] [Google Scholar]
- [17].Lê Cao K-A, González I, Déjean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009 Nov;25(21):2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gonzalez I, Déjean S, Martin P, Baccini A. CCA An R Package to Extend Canonical Correlation Analysis. J Stat Softw. 2008 Jan;23(12):1–14. [Google Scholar]
- [19].Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7alpha-dehydroxylating bacteria in human feces. Clin Chim Acta. 2003 May;331(1–2):127–34. doi: 10.1016/s0009-8981(03)00115-3. [DOI] [PubMed] [Google Scholar]
- [20].Morris LS, Marchesi JR. Assessing the impact of long term frozen storage of faecal samples on protein concentration and protease activity. J Microbiol Methods. 2016 Apr;123:31–38. doi: 10.1016/j.mimet.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Smith K, Zeng X, Lin J, Chincholkar S, Mohnl M. Discovery of Bile Salt Hydrolase Inhibitors Using an Efficient High-Throughput Screening System. PLoS One. 2014 Jan;9(1):e85344. doi: 10.1371/journal.pone.0085344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ling WH, Korpela R, Mykkänen H, Salminen S, Hänninen O. Lactobacillus strain GG supplementation decreases colonic hydrolytic and reductive enzyme activities in healthy female adults. J Nutr. 1994 Jan;124(1):18–23. doi: 10.1093/jn/124.1.18. [DOI] [PubMed] [Google Scholar]
- [23].Tanaka H, Hashiba H, Kok J, Mierau I. Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl Environ Microbiol. 2000 Jun;66(6):2502–12. doi: 10.1128/aem.66.6.2502-2512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Korpela K, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. 2016 Jan;7 doi: 10.1038/ncomms10410. 10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Weingarden AR, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. AJP Gastrointest Liver Physiol. 2014 Feb;306(4):G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Allegretti JR, et al. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther. 2016 Jun;43(11):1142–53. doi: 10.1111/apt.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2014 Oct;517(7533):205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gloor GB, Reid G. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol. 2016 Aug;62(8):692–703. doi: 10.1139/cjm-2015-0821. [DOI] [PubMed] [Google Scholar]
- [29].Franzosa EA, et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A. 2014 Jun;111(22):E2329–38. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Z, Zeng X, Mo Y, Smith K, Guo Y, Lin J. Identification and characterization of a bile salt hydrolase from Lactobacillus salivarius for development of novel alternatives to antibiotic growth promoters. Appl Environ Microbiol. 2012 Dec;78(24):8795–802. doi: 10.1128/AEM.02519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 2014 Apr;7(1):12–8. doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- [32].Thomas LA, Veysey MJ, French G, Hylemon PB, Murphy GM, Dowling RH. Bile acid metabolism by fresh human colonic contents: a comparison of caecal versus faecal samples. Gut. 2001 Dec;49(6):835–42. doi: 10.1136/gut.49.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Thomas LA, King A, French GL, Murphy GM, Dowling RH. Cholylglycine hydrolase and 7a-dehydroxylase optimum assay conditions in vitro and caecal enzyme activities ex vivo. Clin Chim Acta. 1997;268(268):61–72. doi: 10.1016/s0009-8981(97)00169-1. [DOI] [PubMed] [Google Scholar]
- [34].DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011 Aug;8(9):523–31. doi: 10.1038/nrgastro.2011.133. [DOI] [PubMed] [Google Scholar]
- [35].Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012 Jun;336(6086):1255–62. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dave M, Higgins PD, Middha S, Rioux KP. The human gut microbiome: current knowledge, challenges, and future directions. Transl Res. 2012 Oct;160(4):246–57. doi: 10.1016/j.trsl.2012.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.