Abstract
Background & Aims
Faecal microbiota transplantation (FMT) is effective for treating recurrent Clostridioides difficile infection (CDI), but there are concerns regarding its long-term safety. Understanding the mechanisms of the effects of FMT could help us design safer, targeted therapies. We aimed to identify microbial metabolites that are important for C difficile growth.
Methods
We used a CDI chemostat model as a tool to study the effects of FMT in vitro. The following analyses were performed: C difficile plate counts, 16S rRNA gene sequencing, 1H-NMR spectroscopy, and UPLC mass spectrometry bile acid profiling. FMT mixtures were prepared using fresh faecal samples provided by donors enrolled in the FMT Programme. Results from chemostat experiments were validated using human stool samples, C difficile batch cultures, and C57BL/6 mice with CDI. Human stool samples were collected from 16 patients with recurrent CDI and 5 healthy donors participating in an FMT trial.
Results
In the CDI chemostat model, clindamycin decreased valerate and deoxycholic acid concentrations and increased C difficile total viable counts (TVC) and valerate precursors, taurocholic acid, and succinate concentrations. After we stopped adding clindamycin, levels of bile acids and succinate recovered, whereas levels of valerate and valerate precursors did not. In the CDI chemostat model FMT increased valerate concentrations and decreased C difficile TVC (94% reduction), spore counts (86% reduction), and valerate precursor concentrations—concentrations of bile acids were unchanged. In stool samples from patients with CDI, valerate was depleted before FMT, but restored after FMT. C difficile batch cultures confirmed that valerate decreased vegetative growth, and that taurocholic acid is required for germination but had no effect on vegetative growth. C difficile TVC were decreased by 95% in mice with CDI given glycerol trivalerate compared to phosphate-buffered saline.
Conclusions
We identified valerate as a metabolite that is depleted with clindamycin and only recovered with FMT. Valerate is a target for a rationally designed recurrent CDI therapy.
Keywords: bacteria, stool transplant, gut microbiome, pathogen
Introduction
Clostridioides difficile (formerly Clostridium difficile) is an anaerobic, spore-forming, Gram-positive bacterium that causes opportunistic infections in the human colon, usually after antibiotic exposure. C difficile infection (CDI) can lead to diarrhoea, pseudomembranous colitis, toxic megacolon, intestinal perforation, multi-organ failure, and death.1 A recent study showed that the incidence of recurrent CDI has disproportionately increased relative to CDI overall, therefore the demand for recurrent CDI therapies may be rising.2
The principle behind faecal microbiota transplantation (FMT) is to use stool from a healthy donor to replace the microorganisms and ecosystem functions that are depleted in the gut of recurrent CDI patients. While FMT is highly effective at treating recurrent CDI,3 it lacks a detailed mechanism of action and it is unclear whether all the microbes included in the preparations are required to resolve disease. There are concerns regarding the long-term safety, reproducibility, composition, and stability of FMT preparations,4 and potential risks include transmission of infections, invasive administration routes, and concerns treating high-risk individuals (frail/elderly or immunosuppressed patients). In addition, as more studies describe the role of the gut microbiota in disease, it is unclear whether FMT could result in the transfer of a gut microbiota which later contributes to disease (e.g. colorectal cancer, obesity, inflammatory bowel disease, etc).
C difficile causes disease after germination, where cells change from their dormant spore state to their active vegetative state.5 Previous studies have suggested that exposure to antibiotics alters the composition and functionality of the gut microbiota, changing the global metabolic profile to an environment that supports C difficile germination and vegetative growth.6 Studies have shown that antibiotic exposure and FMT alters many microbial metabolic pathways, including bile acid metabolism7 and succinate metabolism.8 In fact, Ott and colleagues showed that sterile faecal filtrate from healthy stool donors was able to cause remission from recurrent CDI in a preliminary investigation of five patients with the condition.9 It is possible the sterile faecal filtrate contained bacterial metabolites or enzymes that were sufficient to inhibit C difficile spore germination and vegetative growth.
Mechanistic studies are challenging to conduct in vivo due to the wide variety of factors which influence the composition and functionality of the gut microbiota. Firstly, samples from recurrent CDI patients prior to FMT are usually collected while they are still on suppressive vancomycin. Therefore, it is difficult to determine whether changes in specific bacteria or metabolites following FMT are due to the FMT administration, or whether these changes could have occurred in the absence of FMT due to recovery of the gut microbiota following cessation of antibiotic treatment. Changes in diet can also cause profound changes in the composition and functionality of the gut microbiota, especially short chain fatty acid production,10 and diet is especially difficult to control in human studies. Recurrent CDI patients may eat differently before and after receiving FMT, and may eat differently from healthy controls. Studies on diet, the gut microbiota, and short chain fatty acid production have often relied on fermentation data in vitro and animal data due to the challenges associated with human studies.11 Therefore, human studies could lead to “false positives” for mechanisms of C difficile pathogenesis.
Data collected from chemostat studies can be used to complement microbiome data collected from human and animal studies to more easily determine a mechanism of action for specific disease states or interventions.12 Chemostat models are artificial systems that mimic some of the spatial, temporal, and environmental conditions found in the human gut.13 Chemostats have many advantages over human and animal studies, which have been discussed in detail previously.12 Bacterial communities cultured in these models are highly reproducible, stable, complex, and representative of the bacterial communities found in vivo.14,15 This means researchers can perform longitudinal studies in these systems that can directly link changes in the gut microbiota structure and function to an experimental intervention. Chemostats have previously been used to model CDI and test the effects of several treatments on C difficile growth and pathogenesis (e.g. antibiotics,16–19 bacteriophages,20 and lactoferrin21).
We used a twin-vessel single-stage distal gut chemostat model as a tool to study CDI and the effects of FMT under tightly-controlled conditions in vitro. We hypothesised that exposure to antibiotics kills bacteria that perform important functions in the gut microbial ecosystem, resulting in a “metabolic dysbiosis” where the loss or reduction of specific microbial metabolic pathways creates an environment that promotes C difficile germination and growth. We also hypothesised that FMT administration would reverse these effects by restoring the bacteria responsible for performing these key metabolic functions. Our aim was to identify these metabolites so we could propose new therapeutic approaches to treat recurrent CDI that are well-defined, effective, and safe.
Materials and Methods
Chemostat model of CDI
The chemostat models used in this study were two identical Electrolab FerMac 200 series bioreactor systems (Electrolab, Tewkesbury, UK). Chemostat inoculum and growth medium were prepared and vessels were inoculated and operated as previously described (see Supplementary Methods).14 Stool samples were collected under approval from the UK National Research Ethics Centres (13/LO/1867). We performed three separate twin-vessel chemostat experiments. In each experiment, two identical vessels (“VA” receiving saline vehicle control and “VB” receiving FMT preparation) were inoculated with a 10% (w/v) faecal slurry prepared using fresh faeces from a healthy donor not exposed to antibiotics within the previous 2 months (Run 1= male in his 40's; Run 2= male in his 60’s; Run 3= male in his 80’s). We used clindamycin and C difficile spores to induce CDI in our chemostat model following a modified version of the methods previously described by Freeman and colleagues (see Table 1 and Supplementary Methods).22 After stopping clindamycin dosing, microbial communities were allowed to stabilise before administering the FMT preparation or saline vehicle control. FMT mixtures were prepared using fresh faecal samples provided by donors enrolled within Imperial’s FMT Programme.23 Stool samples from these faecal donors have previously been used to successfully treat recurrent CDI patients.
Table 1. Time periods used for CDI chemostat experiments to compare the effects of FMT to saline.
| Vessel | Days 0-17 | Days 18-24 | Day 24 | Days 25-31 | Days 32-42 | Day 42 | Days 43-54 |
|---|---|---|---|---|---|---|---|
| Control vessel (VA) | Stabilisation period (no intervention) | Stable communities (no intervention) | Added C difficile spores | Added C difficile spores (day 25) and clindamycin every 12 hrs (days 25-31) | Stabilisation period (no intervention) | Added saline | No intervention |
| Test vessel (VB) | Stabilisation period (no intervention) | Stable communities (no intervention) | Added C difficile spores | Added C difficile spores (day 25) and clindamycin every 12 hrs (days 25-31) | Stabilisation period (no intervention) | Added FMT | No intervention |
16S rRNA gene sequencing (metataxonomics)
DNA extraction is described in the Supplementary Methods. Sample libraries amplifying the V3-V4 region of the 16S rRNA gene were prepared following Illumina’s 16S Metagenomic Sequencing Library Preparation Protocol,24 with a few modifications. First, we used the SequalPrep Normalization Plate Kit (Life Technologies, Carlsbad, USA) to clean up and normalise the index PCR reactions. Also, we used the NEBNext Library Quant Kit for Illumina (New England Biolabs, Ipswich, USA) to quantify the sample libraries. Sequencing was performed on an Illumina MiSeq platform (Illumina Inc, San Diego, USA) using the MiSeq Reagent Kit v3 (Illumina) and paired-end 300 bp chemistry. The resulting data were pre-processed and analysed as described in the Supplementary Methods.
1H-NMR spectroscopy
Chemostat culture supernatants were prepared for 1H-NMR as described in the Supplementary Methods. One-dimensional 1H-NMR spectra were acquired from chemostat culture supernatants at 300 K on a Bruker DRX 600 MHz NMR spectrometer or a Bruker AVANCE III 600 MHz NMR spectrometer (Bruker Biospin, Germany). A standard one-dimensional NMR pulse sequence [RD-90°-t1-90°-tm-90°-acq] was used with a recycle delay (4 s) and mixing time (100 ms). The 90° pulse length was around 10 μs and 32 scans were recorded. Metabolites concentrations were quantified from spectra using the Chenomx NMR suite software (Chenomx Inc, Edmonton, Canada).25
To confirm the identity of key metabolites in chemostat culture supernatants a series of NMR spectra including 1D 1H NOESY, 2D 1H−1H TOCSY and 1H−1H COSY of a chemostat culture supernatant and a metabolite standard were recorded (see Supplementary Methods).
Ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) bile acid profiling
Bile acids were extracted from 50 µL of chemostat culture by adding 150 µL of cold methanol, followed by incubation at -30°C for 2 hours. Tubes were centrifuged at 9500 x g and 4°C for 20 min and 120µL of supernatant was loaded into vials. Bile acid analysis was performed using an ACQUITY UPLC (Waters Ltd, Elstree, UK) coupled to a Xevo G2 Q-ToF mass spectrometer. The MS system was equipped with an electrospray ionization source operating in negative ion mode, using methods previously described by Sarafian and colleagues.26 Data pre-processing and analysis are described in the Supplementary Methods.
Gas chromatography-mass spectrometry (GC-MS) analysis of human stool samples
Human stool samples were collected from recurrent CDI patients (n=16) and healthy donors (n=5) as part of a randomised clinical trial comparing the efficacy of capsulized and colonoscopic FMT for the treatment of recurrent CDI, as previously described.27 Pre-FMT samples were collected from recurrent CDI patients while on suppressive vancomycin. Post-FMT samples were collected 1, 4, and 12 weeks after FMT treatment. All patients were successfully treated following a single FMT.
A targeted GC-MS protocol was used to identify and quantify short chain fatty acids from human stool samples as previously-described.28 Samples were analysed on an Agilent 7890B GC system, coupled to an Agilent 5977A mass selective detector (Agilent, Santa Clara, CA). Data analysis was performed using MassHunter software (Agilent).
C difficile batch cultures
We tested the effects of valerate (Fisher Scientific) on the vegetative growth of three C difficile ribotypes (010, 012, and 027) as well as several gut commensal bacteria (Bacteroides uniformis, Bacteroides vulgatus, and Clostridium scindens) (see Supplementary Methods). We centrifuged an overnight culture of the test isolate at 3000 x g for 10 minutes and resuspended the cells in brain heart infusion broth (Sigma-Aldrich) (supplemented with 5 mg/mL yeast extract (Sigma-Aldrich), and 0.1% L-cysteine (Sigma-Aldrich)), containing varying concentrations of valerate (0, 1, 2, 3, 4, 5, 10, and 20 mM, pH of broth adjusted to 6.8) in triplicate. The OD600 was measured at time zero and cultures were incubated at 37°C in an ElectroTek AW 400TG Anaerobic Workstation (ElectroTek, West Yorkshire, UK). Additional OD600 measurements were taken at 2, 4, 6, and 8 hours post-inoculation. We plotted the changes in OD600 (from a time point during the exponential phase) against each concentration of valerate tested. We used ANOVA and Tukey post hoc test to determine whether the concentration of valerate tested affected the growth of the test isolate compared to batch cultures grown in the absence of valerate.
We also tested the effects of taurocholic acid (TCA) on C difficile germination and vegetative growth using batch cultures (see Supplementary Methods).
Mouse model of CDI
Mouse experiments were performed under the authority of the UK Home Office outlined in the Animals (Scientific Procedures) Act 1986 after ethical review by Imperial College London Animal Welfare and Ethical Review Body (PPL 70/7969). We adhered to standards articulated in the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
Wild-type C57BL/6 mice (8-10-week-old; female) were purchased from Envigo (UK) and acclimatised for 1 week prior to use. Mice were housed five per cage in individually ventilated cages with autoclaved food (RM1, Special Diet Services), bedding (Aspen chip 2 bedding), and water (provided ad libitum). Mice were subjected to a 12 h light and 12 h dark cycle at 20–22°C.
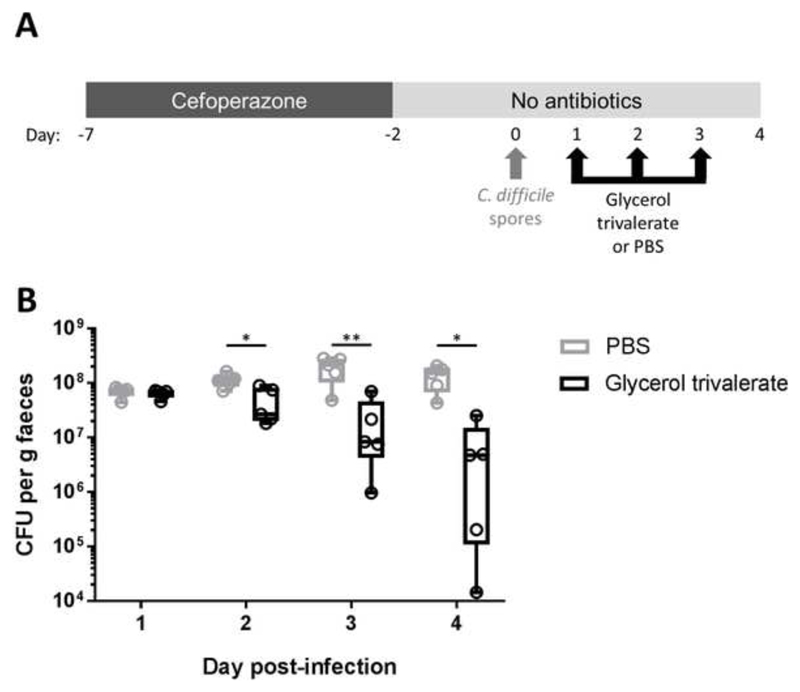
We used a previously published mouse model of C difficile infection as described by Winston and colleagues (Figure 6A).29 Briefly, mice were given 0.5 mg/ml cefoperazone in their drinking water for 5 days (from day -7 to day -2), followed by autoclaved antibiotic-free water for the remainder of the experiment. C difficile spores were prepared and enumerated (see Supplementary Methods) and mice were challenged with 105 C difficile spores by oral gavage on day 0. Mice were orally gavaged with 200 µl of 15 mM glycerol trivalerate (n=5) or 200 µl of PBS (n=5) on days 1, 2, and 3. Faecal samples were collected on days 1, 2, 3, and 4 and C difficile total viable counts (TVC) were quantified (see Supplementary Methods). Mice were not fasted before oral gavages and all interventions were performed during the light cycle.
Figure 6.
Glycerol trivalerate significantly reduced faecal C. difficle total viable counts in a CDI mouse model. (A) Experimental design. Mice were given cefoperazone in their drinking water for 5 days, then switched to antibiotic-free autoclaved water for the remainder of the experiment. On day 0 mice were orally gavaged with 105 CFU of C difficile spores. On days 1, 2, and 3 mice were orally gavaged with glycerol trivalerate (n=5) or PBS (n=5). (B) C difficile TVC were quantified from mouse faecal pellets on days 1, 2, 3, and 4. The box excludes the upper and lower 25% (quartiles) of data, and the lines go to maximum and minimum values. All data points are marked with “o” in the plot. Independent t-test of log10-transformed C difficile TVC, *p<0.05, ** p<0.01.
Statistical analysis and data integration
Statistical tests were performed using IBM SPSS Statistics Software version 23 (paired t-test, independent t-test, ANOVA) or GraphPad Prism version 7.03 (Mann-Whitney U test, Friedman test). Short AsyNchronous Time-series Analysis (SANTA) was used to determine whether there were significant changes in data trajectories at several time periods over the course of the chemostat experiments (see Supplementary Methods).30 Spearman's rho statistic and p-values were calculated for C difficile TVC and metabolite data using the cor and cor.test functions, respectively, within the stats base library within R. A p-value less than 0.05 was considered significant. We used regularised Canonical Correlation Analysis (rCCA) to correlate metataxonomic and metabolomic data using the mixOmics library within R (see Supplementary Methods).31
Results
C difficile total viable counts and spore counts from chemostat culture samples
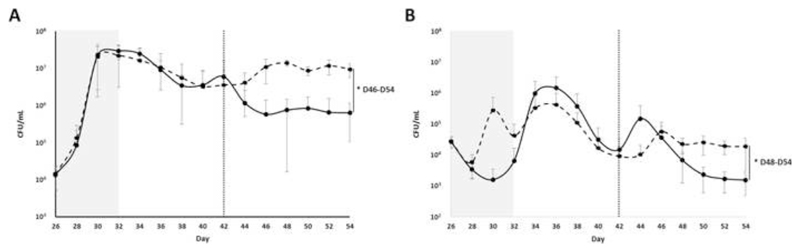
Following the addition of C difficile spores to each vessel C difficile TVC and spore counts were enumerated every other day until the end of the experiment (Figure 1). We found an increase in C difficile TVC during the clindamycin-dosing period (p<0.001). We also found a 94% reduction in C difficile TVC (p=0.025) and an 86% reduction in C difficile spore counts (p=0.034) in FMT-treated cultures compared to saline-treated cultures.
Figure 1.
Average C difficile plate counts taken from VA (saline-treated cultures, black dashed line) and VB (FMT-treated cultures, black solid line) over the course of the experiment. (A) Average C difficile total viable counts. (B) Average C difficile spore plate counts. The grey shaded box indicates the clindamycin-dosing period, while the vertical dotted line indicates the day of FMT or saline dosing. Error bars represent the mean ± standard deviation (*, p<0.05 by SANTA analysis).
Bacterial community composition of chemostat culture samples
We found that clindamycin dosing of chemostat cultures altered the composition and the biomass of the bacterial communities (Figure S1). Clindamycin dosing caused an initial decrease in the total bacterial biomass one day after starting clindamycin dosing (p<0.001), however the total bacterial biomass increased over time to reach pre-clindamycin levels by the end of the clindamycin dosing period (p>0.05). Clindamycin dosing also resulted in a decrease in bacterial diversity (p=0.002), richness (the number of species present, p=0.001) and evenness (how evenly distributed each species is, p=0.004) (Figure S2). There was a significant increase in richness following FMT treatment (p=0.035), however diversity (p>0.05) and evenness (p>0.05) remained unchanged.
Global metabolic profiling of chemostat culture samples by 1H-NMR spectroscopy
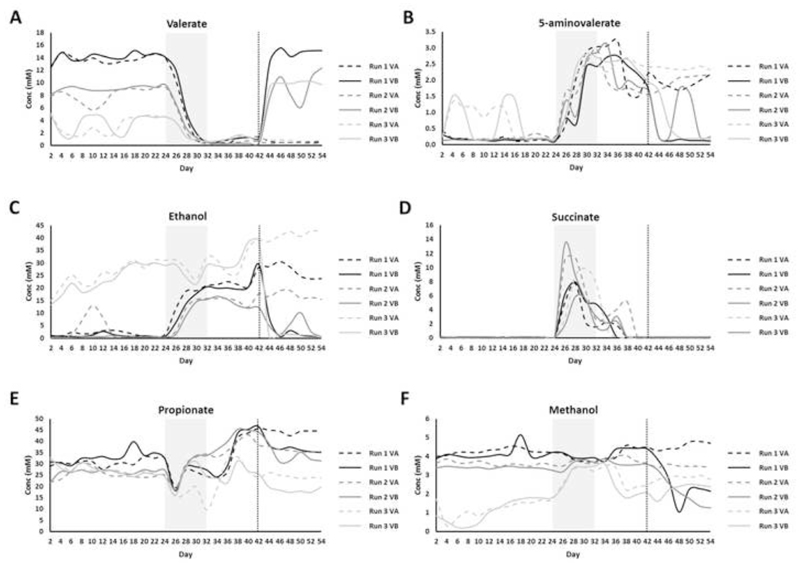
We performed 1H-NMR spectroscopy as an exploratory technique to generate global metabolite spectral profiles for samples collected over the course of the chemostat experiments. There was a significant decrease in valerate (p=0.004), butyrate (p=0.004), and acetate (p=0.013) and a significant increase in 5-aminovalerate (p=0.004), ethanol (p=0.021), succinate (p=0.004), and isobutyrate (p=0.004) during the clindamycin dosing period compared to the steady state period (Figures 2 and S3). rCCA modelling was used to determine correlations between 16S rRNA gene sequencing data and metabolite data that correspond to clindamycin dosing. The unit representation plot showed a clear separation between steady state cultures and cultures sampled during and after the clindamycin dosing period (Figure S4A). The correlation circle plot showed that the separation between the cultures was due to decreases in the levels of valerate and acetate, and increases in the levels 5-aminovalerate, succinate, isobutyrate, and ethanol during and after the clindamycin dosing period (Figure S4B). This plot also showed strong correlations between bacterial genera and these metabolites. During the clindamycin-dosing period there were significant correlations between C difficile TVC and valerate (rS=-0.59, p=0.005), 5-aminovalerate (rS=0.54, p=0.010), and succinate (rS=-0.49, p=0.022).
Figure 2.
1H-NMR metabolites that changed following clindamycin treatment and with FMT (VA= saline-treated cultures, dashed line; VB= FMT-treated cultures, solid line). (A) valerate, (B) 5-aminovalerate, (C) ethanol, (D) succinate, (E) propionate, and (F) methanol. The shaded grey box indicates the clindamycin-dosing time period, while the vertical dotted line indicates the day of FMT or saline dosing. SANTA analysis with Benjamini-Hochberg FDR was used to compare the following: steady state cultures to clindamycin-treated cultures, steady state cultures to post-clindamycin cultures, and FMT-treated cultures to saline treated cultures.
Following the end of the clindamycin-dosing period the levels of succinate, butyrate, acetate, and isobutyrate recovered to pre-antibiotic levels, and these levels were not affected by FMT treatment (Figures 2 and S3). However, after stopping clindamycin dosing the levels of valerate (p=0.009) were still significantly decreased and the levels of 5-aminovalerate (p=0.009) and ethanol (p=0.013) were still significantly increased compared to pre-antibiotic levels (Figure 2), indicating the levels of these metabolites did not recover after stopping clindamycin dosing. Moreover, after stopping clindamycin dosing the levels of isovalerate decreased (p=0.009) and the levels of propionate increased (p=0.036) compared to pre-antibiotic levels (Figures 2 and S3).
There was a significant increase in valerate (p=0.032), and significant decreases in 5-aminovalerate (p=0.032), ethanol (p=0.032), propionate (p=0.032), and methanol (p=0.039) in FMT-treated cultures compared to saline-treated cultures (Figure 2). rCCA modelling was also used to determine correlations between 16S rRNA gene sequencing data and metabolite data that correspond to FMT or saline treatment. The unit representation plot showed a clear separation between FMT-treated cultures and saline-treated cultures along the second canonical variate (Figure S4C). The correlation circle plot showed that the separation between FMT- and saline-treated cultures was due to increases in the levels of valerate and decreases in the levels of 5-aminovalerate, ethanol, and methanol in FMT-treated cultures (Figure S4D). This plot also showed strong correlations between bacterial genera and these metabolites. Following FMT or saline treatment there was a significant strong negative correlation between C difficile TVC and valerate (rS=-0.67, p=1.48x10-4), and significant strong positive correlations between C difficile TVC and 5-aminovalerate (rS=0.76, p=6.07x10-6), ethanol (rS=0.81, p=1.72x10-6), and methanol (rS=0.72, p=2.98x10-5).
The identity of the proposed valerate peaks was confirmed in the chemostat culture supernatants by performing 1D-1H-NMR spectroscopy of a sample before and after valerate spike-in, as well as 2D-1H-NMR spectroscopy of a valerate-containing sample (Figures S5 and S6, Supplementary Results). The identities of other metabolites were also confirmed in the chemostat culture supernatants by performing 2D-1H-NMR and STOCSY (Figures S7 and S8).
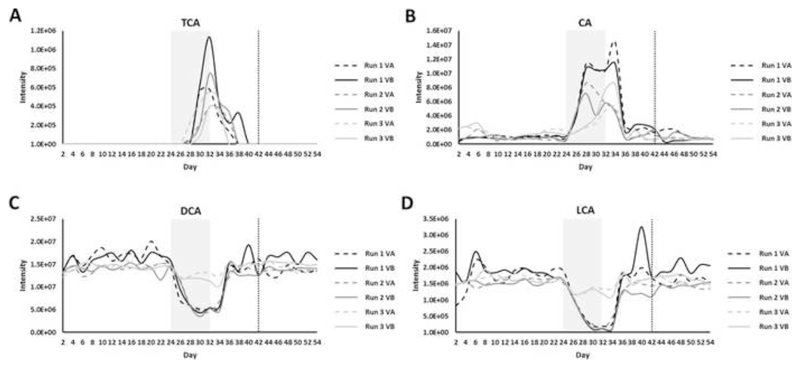
Bile acid UPLC-MS profiling of chemostat culture samples
Due to the important role that bile acids play in C difficile spore germination and vegetative growth,5 we also measured changes in bile acids over the course of the chemostat experiments. During the clindamycin dosing period there was a significant increase in the conjugated primary bile acids taurocholic acid (TCA, p=0.004), glycocholic acid (GCA, p=0.005), and glycochenodeoxycholic acid (GCDCA, p=0.005), in the unconjugated primary bile acids cholic acid (CA, p=0.004) and chenodeoxycholic acid (CDCA, p=0.037), and in the conjugated secondary bile acids taurodeoxycholic acid (TDCA, p=0.045) and glycodeoxycholic acid (GDCA, p=0.004) (Figures 3 and S9). During the clindamycin dosing period there was a significant decrease in the secondary bile acids deoxycholic acid (DCA, p=0.006), lithocholic acid (LCA, p=0.005), and ursodeoxycholic acid (UDCA, p=0.037) (Figures 3 and S9). Following the end of the clindamycin dosing period the levels of these bile acids recovered to steady state levels (before clindamycin dosing), and these levels were not affected by FMT treatment.
Figure 3.
Bile acids that changed following clindamycin treatment and correlated with C difficile TVC (VA= saline-treated cultures, dashed line; VB= FMT-treated cultures, solid line). (A) taurocholic acid (TCA), (B) cholic acid (CA), (C) deoxycholic acid (DCA), and (D) lithocholic acid (LCA). The shaded grey box indicates the clindamycin-dosing period, while the vertical dotted line indicates the day of FMT or saline dosing. Steady state cultures were compared to clindamycin-treated cultures using SANTA analysis with Benjamini-Hochberg FDR.
rCCA modelling was used to determine correlations between 16S rRNA gene sequencing data and bile acid data during the clindamycin-dosing period. The unit representation plot showed separation between cultures sampled before and during the clindamycin-dosing period along the first canonical variate, but no separation between cultures sampled before and after the clindamycin-dosing period (Figure S10A). The correlation circle plot showed that the separation between cultures sampled before and during clindamycin dosing was due to increases in the levels of TCA, CA, CDCA, GCA, GDCA, and GCDCA, and decreases in the levels of DCA, LCA, and UDCA during the clindamycin-dosing period (Figure S10B). This plot also showed strong correlations between bacterial genera and bile acids.
There were significant strong correlations between C difficile TVC and several bile acids during the clindamycin dosing period, including TCA (rS=0.68, p=6.29x10-4), CA (rS=0.61, p=0.003), DCA (rS=-0.75, p=8.86x10-5), and LCA (rS=-0.76, p=6.36x10-5).
GC-MS analysis of human stool samples
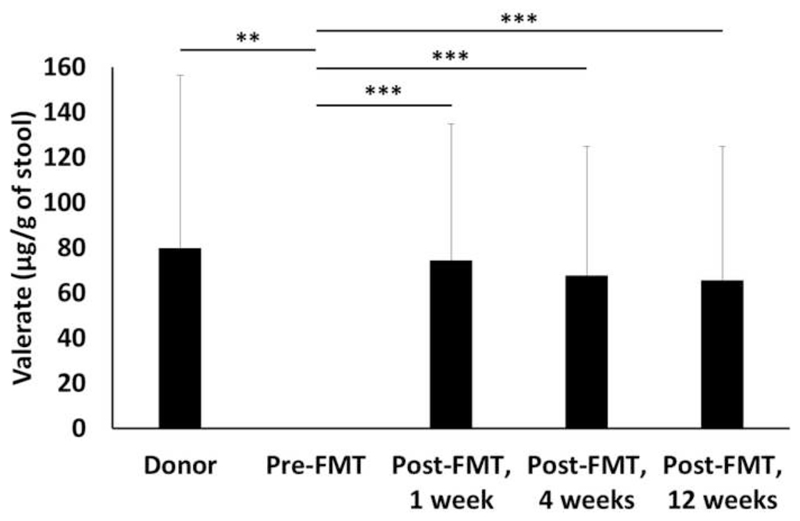
To confirm the findings from our chemostat experiments we measured the levels of valerate in human stool samples from healthy FMT donors, recurrent CDI patients pre-FMT and at several time points post-successful FMT (1, 4, and 12 weeks after FMT treatment) (Figure 4). Valerate was depleted in stool samples from recurrent CDI patients pre-FMT compared to healthy donors (p=0.0075). Valerate levels were significantly increased in CDI patients post-FMT compared to pre-FMT (p=0.0007 at 1 week, 4 weeks, and 12 weeks). There were no significant differences in the levels of valerate in stool samples from healthy donors compared to any of the time points from CDI patients collected post-FMT (p>0.05 for all comparisons).
Figure 4.
Effect of FMT on the concentration of valerate in stool from healthy FMT donors (n=5) and recurrent CDI patients pre-FMT (n=16) and at several time points post-FMT (n=16). Mann-Whitney U test for donors vs. pre-FMT, Friedman test for pre-FMT vs. post-FMT. ** p<0.01, *** p<0.001.
C difficile batch culture experiments with valerate and TCA
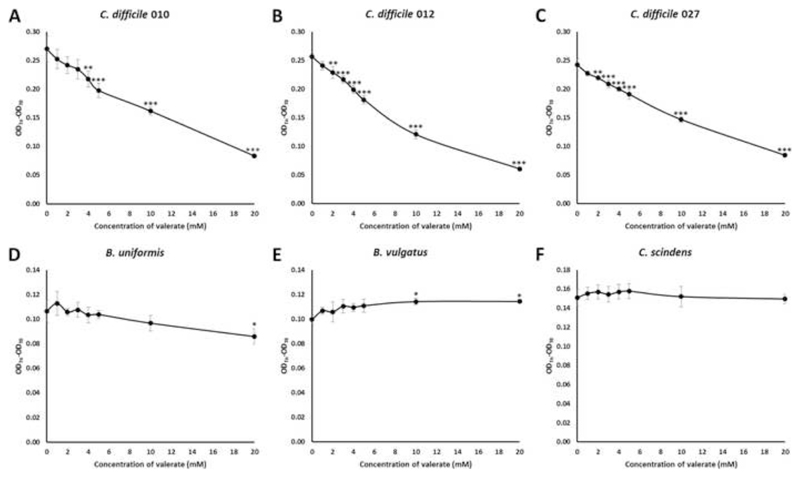
Batch culture experiments were performed to directly study the effects of specific metabolites of interest on C difficile germination and vegetative growth and to confirm the findings from our chemostat experiments. These experiments showed that valerate inhibited the vegetative growth C difficile ribotype 010 at concentrations ≥ 4 mM (p=0.008), ribotype 012 at concentrations ≥ 2 mM (p=0.003), and ribotype 027 at concentrations ≥ 2 mM (p=0.008) (Figure 5). The concentration of valerate in FMT-treated chemostat culture supernatants remained above 4 mM for all samples, whereas the concentration of valerate in saline-treated cultures remained below 2 mM for all samples. As a control, we also tested the effects of valerate on commensal gut isolates (B uniformis and B vulgatus, two representatives of Bacteroidetes, and C scindens, a representative of Firmicutes). B uniformis was only inhibited in broth containing 20 mM valerate (p=0.026). B vulgatus was not inhibited at any concentration of valerate that was tested, and showed more growth in broths containing 10 mM (p=0.020) or 20 mM (p=0.019) valerate. C scindens was not inhibited at any concentration of valerate tested (p>0.05 for all concentrations of valerate tested).
Figure 5.
Valerate inhibits C difficile vegetative growth in batch cultures. Vegetative cells were inoculated into supplemented brain heart infusion broth containing varying concentrations of valerate (0, 1, 2, 3, 4, 5, 10, and 20 mM) and OD600 measurements were taken at 0, 2, 4, 6, and 8 hours. The change in OD600 (from a time point during the exponential phase) was plotted against the concentrations of valerate tested. (A) C difficile ribotype 010, (B) C difficile ribotype 012, (C) C difficile ribotype 027, (D) B. uniformis, (E) B. vulgatus, (F) C. scindens. Error bars represent the mean ± standard deviation, * p<0.05, ** p<0.01, *** p<0.001.
Batch culture experiments also confirmed previous findings showing that TCA is required for C difficile spore germination but had no effect on vegetative growth (Figure S11).
Glycerol trivalerate intervention study in a CDI mouse model
Next, we determined whether valerate could inhibit C difficile growth when administered as an intervention in a CDI mouse model (Figure 6A). To avoid the rapid uptake of valerate and obtain sufficient release in the gastrointestinal tract, valerate was administered to the mice in the form of glycerol trivalerate (which is hydrolysed by lipases to release valerate32). One-day post-infection (prior to glycerol trivalerate or PBS administration) there was no significant difference in the levels of C difficile TVC in mouse faeces in each group (p=0.584). Glycerol trivalerate or PBS was administered to C difficile-infected mice by oral gavage 1, 2, and 3 days post-infection (n=5 per group). There was a significant decrease in C difficile TVC in glycerol trivalerate-treated mice compared to PBS-treated mice (Figure 6B, p=0.027 on day 2, p=0.007 on day 3, and p=0.024 on day 4). After only 3 doses of glycerol trivalerate or PBS, glycerol trivalerate-treated mice had an average of 95% less C difficile TVC per gram of faeces compared to PBS-treated mice. No adverse effects were noted in glycerol trivalerate-treated mice over the course of the experiment. These results corroborate the other findings presented in this study and support the therapeutic use of valerate to treat CDI.
Discussion
We used several ‘omic’ techniques to study the effects of FMT on CDI in a chemostat model under tightly controlled conditions. The aim of our study was to directly link changes in C difficile counts to changes in the structure and function of the cultured faecal microbiota. We confirmed our findings by analysing human stool samples, performing C difficile batch culture experiments, and performing an interventional study in a CDI mouse model. A summary of the key findings and proposed interactions between C difficile, valerate, and TCA are outlined in Figure S12.
In our study we found valerate significantly inhibited the growth of C difficile, both in vitro and in vivo. Valerate is a short chain fatty acid produced via amino acid fermentation by members of the gut microbiota.33 Valerate was significantly depleted in chemostat culture samples during clindamycin dosing and did not recover after clindamycin dosing was stopped. Valerate significantly increased with FMT treatment and had a strong negative correlation with C difficile TVC. These findings were corroborated with human data which showed valerate was depleted in recurrent CDI patient stool but was restored following successful FMT. Batch culture experiments confirmed that valerate directly inhibited the vegetative growth of several C difficile ribotypes, but had minimal effects on the growth of other commensal gut bacteria tested. Moreover, we showed that glycerol trivalerate significantly decreased C difficile TVC in a CDI mouse model. We hypothesise that maintaining or restoring the levels of valerate in the gut microbiota of CDI patients will inhibit the vegetative growth of C difficile. This restoration could be accomplished by directly supplying the gut with valerate (e.g. in the form of glycerol trivalerate) or by administering bacteria capable of transforming valerate precursors into to valerate.
There are several different metabolic pathways that lead to valerate production. 5-aminovalerate is a product of the anaerobic degradation of protein hydrolysates by members of the gut microbiota, in particular Clostridium species.34 In this pathway, proline is reduced to 5-aminovalerate by proline reductase in a Stickland-type fermentation.35–37 5-aminovalerate is then fermented to valerate in a series of reactions mediated by gut bacteria.34,38 We showed that 5-aminovalerate was increased in chemostat cultures following clindamycin dosing and remained increased after clindamycin dosing was stopped. This finding is supported by data from a patent submitted by Savidge and Dann, who proposed CDI patients can be distinguished from non-infected subjects by measuring elevated levels of 5-aminovalerate in the patient’s stool, urine, or blood.39 Moreover, Fletcher and colleagues showed that 5-aminovalerate was elevated in the caecal content of C difficile infected mice.40 In another metabolic pathway, some Clostridium species can ferment ethanol and propionate to valerate.41 We found ethanol and propionate increased after clindamycin dosing and decreased following FMT treatment. Taken together, changes in the levels of valerate and valerate precursors (5-aminovalerate, ethanol, and propionate) over the course of our chemostat experiments supports our hypothesis that disruption of the valerate pathway due to antibiotics results in an environment that permits C difficile vegetative growth.
In this study we performed an intervention experiment to test the effects of valerate (in the form of glycerol trivalerate) in a CDI mouse model. For the first time we showed that glycerol trivalerate-treated mice had significantly reduced C difficile TVC compared to PBS-treated control mice (95% reduction in glycerol-trivalerate treated mice after 3 doses). The results from our study are consistent with the previously published study by Theriot and colleagues, who briefly mention that antibiotic-exposed mice (who were susceptible to CDI) had a 66-fold decrease in valerate compared to non-antibiotic controls (who were resistant to CDI).6 They also showed that antibiotic-exposed mice had increased levels of amino acids required for C difficile growth (including proline, a precursor to 5-aminovalerate) compared to non-antibiotic controls. Six weeks later, antibiotic-exposed mice were fully-resistant to CDI following exposure to C difficile spores. As shown in the supplementary material of their paper, Theriot and colleagues found that valerate levels had partially recovered six weeks later, and were only 4.9-fold decreased in antibiotic-exposed mice compared to non-antibiotic controls.
C difficile spores can persist following the cessation of antibiotics and germinate to vegetative cells which initiate disease relapse. In our study we found that TCA, a known potent germinant for C difficile spores,5 was increased during clindamycin dosing, remained elevated for several days after stopping clindamycin, and had a strong positive correlation with C difficile TVC. However, after stopping clindamycin and allowing chemostat communities to recover and stabilise, we found that bile acid levels recovered to pre-clindamycin levels and did not change with FMT treatment. We hypothesise that the transient increase in TCA found during and shortly after stopping clindamycin dosing stimulated C difficile spore germination, resulting in vegetative growth. Once clindamycin dosing stopped and the levels of TCA decreased to pre-clindamycin levels, C difficile spores had already germinated and were growing in their vegetative state which is no longer affected by the presence of TCA (as shown by Sorg and Sonnenshein5, and confirmed in our study using batch cultures). These results are also consistent with the previously published study by Theriot and colleagues, who showed that antibiotic-exposed mice (who were susceptible to CDI) had a 15-fold increase in TCA compared to non-antibiotic controls (who were resistant to CDI).6 Six weeks later (when antibiotic-exposed mice were fully-resistant to CDI) the levels of TCA in antibiotic-exposed mice recovered to the levels found in non-antibiotic controls, suggesting there was insufficient levels of TCA to stimulate C difficile germination. Further discussion of bile acid data is included in the Supplementary Discussion.
The first line of therapy for an initial episode of CDI is vancomycin/metronidazole therapy. These antibiotics kill C difficile vegetative cells, but C difficile spores can persist.42 If vancomycin/metronidazole therapy is stopped while these C difficile spores are present, the elevated levels of TCA (present following the cessation of antibiotics) will cause C difficile spore germination, and low levels of valerate will allow C difficile vegetative growth and therefore recurrent disease. To prevent CDI initiation and relapse following the cessation of antibiotics, it is also important to maintain low levels of TCA in the gut. One way to accomplish this would be to ensure the maintenance of bile salt hydrolase enzymes during and after antibiotic exposure. These enzymes are produced by commensal gut bacteria and are responsible for deconjugating tauro- and glyco-conjugated bile acids. It has been proposed that antibiotics kill bile salt hydrolase-producing bacteria, resulting in the accumulation of TCA in the gut. Therefore, re-inoculation of bile salt hydrolase-producing bacteria with FMT may be responsible for degrading TCA present following the cessation of antibiotics, and prevent the germination of C difficile spores. A more controlled method of restoring bile salt hydrolase activity in the gut microbiomes of CDI patients could be to avoid the administration of live microorganisms by administering purified bile salt hydrolase enzyme preparations.
We also found that succinate transiently increased in chemostat cultures with clindamycin dosing, but negatively correlated with C difficile TVC. We believe the negative correlation between C difficile TVC and succinate is because C difficile uses succinate for growth. A study by Ferreyra and colleagues found that succinate was present at low levels in the guts of healthy mice, but transiently increased with antibiotics or motility disturbance.8 They found that C difficile can metabolise succinate to butyrate, and it exploits the increase in succinate to grow in perturbed gut microbiota. Again, these findings are consistent with our findings from chemostat experiments. Members of Bacteroidetes and the Negativicutes class of Firmicutes can degrade succinate to propionate, and as such succinate does not usually accumulate to high levels in the guts of healthy humans.43 Another strategy to give C difficile vegetative cells a competitive disadvantage would be to maintain succinate metabolism during antibiotic exposure by administering succinate-degrading enzymes, so succinate is no longer available for C difficile growth.
The findings from our study support the hypothesis that antibiotic exposure causes a depletion of specific metabolic pathways normally found in healthy gut microbiotas, resulting in a metabolite environment that favours C difficile germination and growth. It also supports our hypothesis that FMT administration reverses these effects by restoring the bacteria responsible for performing these key metabolic functions. These findings have the potential to directly impact clinical practice in the foreseeable future by developing targeted treatments for CDI by different routes, alone or in combinations: (1) directly supplement the gut with valerate (to inhibit C difficile vegetative growth); (2) directly supplement the gut with bile salt hydrolase enzymes (to degrade taurocholic acid and prevent C difficile spore germination); and (3) directly supplement the gut with succinate-degrading enzymes (to degrade succinate and give C difficile vegetative cells a competitive disadvantage). These proposed interventions are well-defined and represent safer options that will avoid all the risks involved with administering live microorganisms to patients and will not promote antimicrobial resistance.
The advantage of using valerate (instead of live microorganisms or relatively undefined FMT preparations) is that valerate is a well-defined small molecule that is normally present in the healthy gut. Valerate may be a preferred treatment for CDI in immunocompromised patients where administration of bacteria may increase the risk of a subsequent infection, or in CDI patients that require further antibiotic treatment for other purposes (where the antibiotic treatment would kill bacteria present in FMT preparations and render the treatment less effective). Valerate could also be delivered using more patient-friendly methods that do not require the need to preserve live microorganisms. In this study glycerol trivalerate was orally gavaged to mice, but it has also been included as an additive in animal feed. Therefore, valerate has the potential to be administered to patients via a less invasive route compared to FMT. This promising new therapy merits further evaluation in prospective studies in vivo.
Supplementary Material
Lay Summary.
The short chain fatty acid valerate is depleted from the gut following antibiotics and restored with FMT. Valerate significantly decreased C. difficile growth in an interventional mouse model of infection.
Editor's Note.
Background and Context: Although FMT is effective at treating recurrent CDI there are concerns regarding its long-term safety. Mechanistic understanding of how FMT works is required to design targeted, well-defined, and safer therapies.
New Findings: Antibiotics depleted valerate from the gut (permitting C. difficile growth) while FMT restored valerate (decreasing C. difficile growth). Valerate significantly decreased C. difficile growth in an interventional CDI mouse model.
Limitations: We have yet to identify the minimum dose of glycerol trivalerate required to eliminate C. difficile in mouse faeces.
Impact: Valerate can be used as a safer, microorganism-free method to treat CDI, especially in vulnerable patients (e.g. immunocompromised or paediatric patients) or patients that require further antibiotic treatment.
Grant support
The Division of Integrative Systems Medicine and Digestive Disease at Imperial College London receives financial support from the National Institute of Health Research (NIHR) Imperial Biomedical Research Centre (BRC) based at Imperial College Healthcare NHS Trust and Imperial College London. This article is independent research funded by the NIHR BRC, and the views expressed in this publication are those of the authors and not necessarily those of the NHS, NIHR, or the Department of Health. BHM is the recipient of a Medical Research Council (MRC) Clinical Research Training Fellowship (grant reference: MR/R000875/1). DK received research funding from Alberta Health Services and University of Alberta Hospital Foundation. TBC is a Sir Henry Dale Fellow jointly funded by the Wellcome Trust and Royal Society (Grant Number 107660/Z/15Z).
Abbreviations
- 1H-NMR
proton nuclear magnetic resonance
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- CDI
Clostridioides difficile infection
- COSY
correlation spectroscopy
- DCA
deoxycholic acid
- FDR
false discovery rate
- FMT
faecal microbiota transplantation
- GC-MS
gas chromatography-mass spectrometry
- GCA
glycocholic acid
- GCDCA
glycochenodeoxycholic acid
- GDCA
glycodeoxycholic acid
- LCA
lithocholic acid
- NOESY
nuclear Overhauser enhancement spectroscopy
- OD600
optical density at 600 nm
- OTU
operational taxonomic unit
- PBS
phosphate buffered saline
- rCCA
regularised Canonical Correlation Analysis
- SANTA
Short AsyNchronous Time-series Analysis
- STOCSY
statistical total correlation spectroscopy
- TCA
taurocholic acid
- TDCA
taurodeoxycholic acid
- TOCSY
total correlation spectroscopy
- TVC
total viable counts
- UDCA
ursodeoxycholic acid
- UPLC-MS
ultra-performance liquid chromatography-mass spectrometry
- VA
vessel A
- VB
vessel B
Footnotes
Disclosures: DK has received research funding from Rebiotix. All other authors disclose no conflicts.
References
- 1.Mitu-Pretorian OM, Forgacs B, Qumruddin A, et al. Outcomes of patients who develop symptomatic Clostridium difficile infection after solid organ transplantation. Transplant Proc. 2010;42:2631–2633. doi: 10.1016/j.transproceed.2010.04.057. [DOI] [PubMed] [Google Scholar]
- 2.Ma GK, Brensinger CM, Wu Q, et al. Increasing incidence of multiply recurrent Clostridium difficile infection in the united states: a cohort study. Ann Intern Med. 2017;167:152–158. doi: 10.7326/M16-2733. [DOI] [PubMed] [Google Scholar]
- 3.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 4.Petrof EO, Gloor GB, Vanner SJ, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: 'RePOOPulating' the gut. Microbiome. 2013;1 doi: 10.1186/2049-2618-1-3. 3-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weingarden AR, Chen C, Bobr A, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306:G310–9. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreyra JA, Wu KJ, Hryckowian AJ, et al. Gut microbiota-produced succinate promotes C difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ott SJ, Waetzig GH, Rehman A, et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology. 2017;152:799–811.e7. doi: 10.1053/j.gastro.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 10.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald JAK. In vitro models of the human microbiota and microbiome. Emerging Topics in Life Sciences. 2017;1:373–384. doi: 10.1042/ETLS20170045. [DOI] [PubMed] [Google Scholar]
- 13.Macfarlane GT, Macfarlane S. Models for intestinal fermentation: association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr Opin Biotechnol. 2007;18:156–162. doi: 10.1016/j.copbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 14.McDonald JA, Schroeter K, Fuentes S, et al. Evaluation of microbial community reproducibility, stability and composition in a human distal gut chemostat model. J Microbiol Methods. 2013;95:167–174. doi: 10.1016/j.mimet.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Van den Abbeele P, Grootaert C, Marzorati M, et al. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl Environ Microbiol. 2010;76:5237–5246. doi: 10.1128/AEM.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman J, Baines SD, Jabes D, et al. Comparison of the efficacy of ramoplanin and vancomycin in both in vitro and in vivo models of clindamycin-induced Clostridium difficile infection. J Antimicrob Chemother. 2005;56:717–725. doi: 10.1093/jac/dki321. [DOI] [PubMed] [Google Scholar]
- 17.Baines SD, O'Connor R, Saxton K, et al. Comparison of oritavancin versus vancomycin as treatments for clindamycin-induced Clostridium difficile PCR ribotype 027 infection in a human gut model. J Antimicrob Chemother. 2008;62:1078–1085. doi: 10.1093/jac/dkn358. [DOI] [PubMed] [Google Scholar]
- 18.Chilton CH, Crowther GS, Freeman J, et al. Successful treatment of simulated Clostridium difficile infection in a human gut model by fidaxomicin first line and after vancomycin or metronidazole failure. J Antimicrob Chemother. 2014;69:451–462. doi: 10.1093/jac/dkt347. [DOI] [PubMed] [Google Scholar]
- 19.Chilton CH, Crowther GS, Baines SD, et al. In vitro activity of cadazolid against clinically relevant Clostridium difficile isolates and in an in vitro gut model of C difficile infection. J Antimicrob Chemother. 2014;69:697–705. doi: 10.1093/jac/dkt411. [DOI] [PubMed] [Google Scholar]
- 20.Meader E, Mayer MJ, Steverding D, et al. Evaluation of bacteriophage therapy to control Clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe. 2013;22:25–30. doi: 10.1016/j.anaerobe.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Chilton CH, Crowther GS, Spiewak K, et al. Potential of lactoferrin to prevent antibiotic-induced Clostridium difficile infection. J Antimicrob Chemother. 2016;71:975–985. doi: 10.1093/jac/dkv452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman J, Baines SD, Saxton K, et al. Effect of metronidazole on growth and toxin production by epidemic Clostridium difficile PCR ribotypes 001 and 027 in a human gut model. J Antimicrob Chemother. 2007;60:83–91. doi: 10.1093/jac/dkm113. [DOI] [PubMed] [Google Scholar]
- 23.Mullish BH, Marchesi JR, Thursz MR, et al. Microbiome manipulation with faecal microbiome transplantation as a therapeutic strategy in Clostridium difficile infection. QJM. 2015;108:355–359. doi: 10.1093/qjmed/hcu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Illumina I. 16S Metagenomic Sequencing Library Preparation (Part # 15044223 Rev. B); 2013. 2017 Available at: https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223b.pdf.
- 25.Weljie AM, Newton J, Mercier P, et al. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 26.Sarafian MH, Lewis MR, Pechlivanis A, et al. Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry. Anal Chem. 2015;87:9662–9670. doi: 10.1021/acs.analchem.5b01556. [DOI] [PubMed] [Google Scholar]
- 27.Kao D, Roach B, Silva M, et al. Effect of oral capsule- vs colonoscopy-delivered fecal microbiota transplantation on recurrent Clostridium difficile infection: a randomized clinical trial. JAMA. 2017;318:1985–1993. doi: 10.1001/jama.2017.17077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Villalba R, Gimenez-Bastida JA, Garcia-Conesa MT, et al. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J Sep Sci. 2012;35:1906–1913. doi: 10.1002/jssc.201101121. [DOI] [PubMed] [Google Scholar]
- 29.Winston JA, Thanissery R, Montgomery SA, et al. Cefoperazone-treated mouse model of clinically-relevant Clostridium difficile strain R20291. J Vis Exp. 2016;(118) doi: 10.3791/54850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfer A. SANTA-App: Interactive package for Short AsyNchronous Time-series Analysis (SANTA) in R, implemented in Shiny; 2017. 2017 Available at: https://github.com/adwolfer/SANTA-App.
- 31.Le Cao K, Rohart F, Gonzalez I, et al. mixOmics: Omics Data Integration Project. R package version 6.1.2. 2017 2017 Available at: https://CRAN.R-project.org/package=mixOmics.
- 32.Schwartz B. The effect of temperature on the rate of hydrolysis of triglycerides by pancreatic lipase. J Gen Physiol. 1943;27:113–118. doi: 10.1085/jgp.27.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neis EP, Dejong CH, Rensen SS. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barker HA, D'Ari L, Kahn J. Enzymatic reactions in the degradation of 5-aminovalerate by Clostridium aminovalericum. J Biol Chem. 1987;262:8994–9003. [PubMed] [Google Scholar]
- 35.Seto B, Stadtman TC. Purification and properties of proline reductase from Clostridium sticklandii. J Biol Chem. 1976;251:2435–2439. [PubMed] [Google Scholar]
- 36.Hodgins DS, Abeles RH. Studies of the mechanism of action of D-proline reductase: the presence on covalently bound pyruvate and its role in the catalytic process. Arch Biochem Biophys. 1969;130:274–285. doi: 10.1016/0003-9861(69)90034-4. [DOI] [PubMed] [Google Scholar]
- 37.Seto B. The Stickland reaction. In: Knowles CJ, editor. Diversity of Bacterial Respiratory Systems. II. Boca Raton, FL: CRC Press; 1980. pp. 49–64. [Google Scholar]
- 38.Buckel W. Unusual enzymes involved in five pathways of glutamate fermentation. Appl Microbiol Biotechnol. 2001;57:263–273. doi: 10.1007/s002530100773. [DOI] [PubMed] [Google Scholar]
- 39.Savidge T, Dann S. Methods and uses for metabolic profiling for Clostridium difficile infection. 2013 PCT/US2012/064218. [Google Scholar]
- 40.Fletcher JR, Erwin S, Lanzas C, et al. Shifts in the gut metabolome and Clostridium difficile transcriptome throughout colonization and infection in a mouse model. mSphere. 2018;3 doi: 10.1128/mSphere.00089-18. eCollection 2018 Mar-Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bornstein BT, Barker HA. The energy metabolism of Clostridium kluyveri and the synthesis of fatty acids. J Biol Chem. 1948;172:659–669. [PubMed] [Google Scholar]
- 42.Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2011;9:88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 43.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.