Abstract
Compelling evidence for the inherited nature of essential hypertension has led to extensive research in rats and humans. Rats have served as the primary model for research on the genetics of hypertension resulting in identification of genomic regions that are causally associated with hypertension. In more recent times, genome-wide studies in humans have also begun to improve our understanding of the inheritance of polygenic forms of hypertension. Based on the chronological progression of research into the genetics of hypertension as the “structural backbone,” this review catalogs and discusses the rat and human genetic elements mapped and implicated in blood pressure regulation. Furthermore, the knowledge gained from these genetic studies that provide evidence to suggest that much of the genetic influence on hypertension residing within noncoding elements of our DNA and operating through pervasive epistasis or gene-gene interactions is highlighted. Lastly, perspectives on current thinking that the more complex “triad” of the genome, epigenome, and the microbiome operating to influence the inheritance of hypertension, is documented. Overall, the collective knowledge gained from rats and humans is disappointing in the sense that major hypertension-causing genes as targets for clinical management of essential hypertension may not be a clinical reality. On the other hand, the realization that the polygenic nature of hypertension prevents any single locus from being a relevant clinical target for all humans directs future studies on the genetics of hypertension towards an individualized genomic approach.
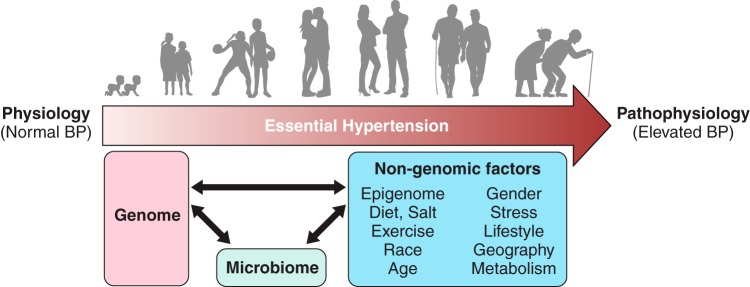
I. BLOOD PRESSURE AS A COMPLEX POLYGENIC TRAIT
The focus on the role of arterial blood pressure as an important reporter of cardiovascular health has deep historical roots dating back to the 18th century. In 1733, the British clergyman Stephen Hales is documented to have made the first published measurement of blood pressure in several animals by inserting fine tubes into arteries and measuring the height to which the column of blood went up (86, 88, 215). In 1836, Richard Bright found an association between renal disease and left ventricular hypertrophy (23). He suggested that left ventricular hypertrophy could be a consequence of high blood pressure. Clinical features of essential hypertension were studied by T. C. Allbutt (6), who then went on to coin the term hyperpiesis to describe essential hypertension, but the term was not popular. Eberhard Frank is believed to have replaced it with the term Essentielle Hypertonie, the English translation of which is Essential Hypertension, i.e., elevated high blood pressure for no known causes (189). Despite these documented observations on the very nature of essential hypertension being noticeable in humans, the clinical significance of hypertension was, interestingly, a discovery attributed to keen observations of individuals from a nonscientific background. As early as in 1906, long before the clinical concept of hypertension as a risk factor became accepted, several insurance companies required medical examiners to document applicants’ blood pressures (44, 190). The extent of blood pressure being a good predictor of serious illness from stroke, heart failure, or renal disease was first recognized by L.I. Dublin and A. J. Lotka, who were demographers working for a life insurance firm in the United States (71, 72). Soon thereafter, the medical community followed with evidence from the studies in populations such as the Framingham study to conclude that essential hypertension is a condition qualitatively distinct from normotension and that elevated blood pressure was not just in those able to afford life insurance (189). There was however the question of whether there was a “dividing line” between normal blood pressure and hypertension. Further clarification that arterial blood pressure did not have a dividing line between normal blood pressure and hypertension was provided by Pickering and co-workers (273, 280, 281) who studied larger populations to study the frequency distributions of arterial blood pressure in populations and emphasized that there is a continuous relationship between arterial pressure and mortality over a full range of arterial pressure. This definition changed the perception of hypertension being a qualitatively different feature from normal blood pressure to a “quantitative” feature, i.e., a trait that varies continually from low to high in a population. The recognition of this feature was in itself a milestone in the understanding of hypertension, but many questions remained as to what caused this variation in populations.
II. EVIDENCE FOR INHERITANCE
Pickering and co-workers (125–127, 283) were among the first to recognize familial correlation of blood pressure (BP). They collected BP data from large groups of normotensive and hypertensive subjects along with their relatives and plotted frequency distribution curves after adjustment for age and gender. BP of relatives of hypertensive subjects were significantly higher than that of the relatives of normotensives (127). There was a linear relationship between the level of BP of individual subjects with their relatives (282). This led to the important conclusion that what was inherited was not only hypertension, but the level of BP regardless of whether it was high or normal. To provide some perspective, this study was published in 1954, which is over half a century after Mendel’s ideas on inheritance were applied to medicine by Archibald Garrod (110). Beginning in 1902, Garrod had studied alkaptonuria, cystinuria, and albinism in relatives of patients and found that genetics explained these inborn errors of metabolism as Mendelian heritable traits, but he also speculated that genetics may play a role in common diseases (110–114).
More precise evidence for genetic factors to influence BP came from correlative observations of a history of hypertension in parents of hypertensive subjects in various populations in Europe and North America (158, 251, 333, 334, 378). Additional evidence was obtained through population studies wherein a greater concordance of blood pressure was observed within families than between families (226), but such studies also raised the question of additional shared nongenetic, environmental factors such as food habits within a family that could influence blood pressure. Twin studies wherein a greater similarity of blood pressures was documented with monozygotic than dizygotic twins (87, 207) suggested that genetic factors strongly influenced the incidence of hypertension. Other studies suggest that genetic effects on BP are evident early in life by demonstrating a significant sib-pair and maternal-children correlation of BP in infants (133, 212) and in children between 2 and 14 yr of age (409). However, a clarification on environmental factors overriding the genetic factors was inferred based on studies of BP variance in a Montreal adoption study of French-Canadian families with both adopted and natural children (8, 9, 19, 20, 252). The study pointed to the highest correlations of BP within children in a shared household environment, regardless of whether they were adopted or not. This correlation between the children was more significant than correlations between the BP of parents and their natural children, leading to the inference that nongenetic, environmental factors contribute more than genetic factors to the extent of BP; however, the identities of both genetic and environmental factors remained enigmatic. Another metric that is used to measure familial resemblance of a trait and hence its genetic component is heritability. The higher the heritability, the stronger is the correlation between phenotype and genotype. From twins and family studies, the heritability of BP ranges from 15 to 40% for the office BP and around 51–69% for night time blood pressure obtained from ambulatory BP monitoring (131, 192, 207). In general, low heritability estimates indicate that genetic mapping would be difficult for that phenotype.
III. WHY AND HOW TO STUDY THE GENETICS OF BLOOD PRESSURE CONTROL
Hypertension being inherited in families serves as an important clue for the presence of susceptibility factors on our genomes that predispose some of us but not others to develop high BP. It is interesting to note that most of the above-mentioned correlative observations were made in the first half of the 20th century well ahead of 1953, the year in which Watson and Crick made the landmark discovery of the molecular structure of DNA (376, 377). This discovery marked the dawn of the era of Molecular Biology and paved the way for a series of monumental discoveries to follow that gave fundamental insights into key concepts that sealed our current understanding of DNA as the genetic material that is inherited. DNA was therefore accepted as the “molecule of life,” and the following three decades since 1953 revolutionized our understanding of the structure-function relationship between the inheritance of DNA and the role it plays in determining the structure of proteins. While the definitions of what constitutes a “gene” were being worked on in the 1950s to the 1980s by molecular biologists, during the same period, physiologists interested in the etiology of hypertension began looking into dietary factors such as the components of the Kempner rice diet influencing the extent of hypertension (186, 375, 383). Salt being an important component influencing BP was inferred by studying various populations around the globe (55). It was apparent that dietary salt intake was directly proportional to the extent of BP observed (55).
IV. RATS AS MODELS TO STUDY THE GENETICS OF HYPERTENSION
To further address the question of whether salt was responsible for the development of hypertension, Lewis Kitchener Dahl et al. (56) conducted experiments with a population of Sprague-Dawley rats, wherein BP of the rats was recorded following their dietary intake of a high salt (8% NaCl) diet. If a high dietary salt intake caused an elevation in BP, all the rats should have developed higher BP. However, this was not the case. Only a subset of rats developed higher BP in response to dietary salt. The fundamental factor driving some but not all rats in the population to develop hypertension in response to dietary salt is now interpretable as genetic susceptibility. By selectively breeding rats with hypertension in response to a high salt intake, not only was further evidence obtained for the inheritance of hypertension, but also the model that ensued came to be known as the Dahl salt-sensitive (S) rat (56, 57), the inbred version of which continues to be one of the most popular models used to study the genetics of hypertension to date. In addition to this model developed in the United States, there are several other models of genetically hypertensive rats that were developed in other regions of the world. These include the spontaneously hypertensive rat (SHR) (272) and the spontaneously hypertensive rats stroke prone (SHRSP) from Japan (261, 272), DOCA salt-sensitive (SBH) rat from Israel (13), Fawn-hooded hypertensive (FHH) rat from The Netherlands (204), Lyon hypertensive (LH) rat from France (75), Milan hypertensive strain (MHS) from Italy (15), Prague hypertensive rat (PHR) from the Czech Republic (132), genetically hypertensive (GH) rat and the albino surgery (AS) rat from New Zealand (141, 343), and inherited stress-induced arterial hypertension (ISIAH) rat from Russia (236). The origins of these models are described in greater detail in a review by Rapp (298). These inbred rats served as models to test the hypothesis that there are regions, referred to as quantitative trait loci (QTLs), on the genome that cause an elevation in BP of these inbred rats.
To identify such genomic regions, comparisons of inbred hypertensive strains had to be made at the genomic DNA level with that of inbred normotensive strains. However, there was a significant roadblock to study genomic DNA, because during the 1970s and early 1980s there were few genetic markers and the technology for genotyping by amplification of DNA through the polymerase chain reaction (PCR) had not yet been developed. The experimental design used to find DNA regions related to BP was to breed hypertensive rats to their normotensive counterparts to generate filial generation 1 (F1) rats. The F1 rats were intercrossed to obtain a filial generation 2 (F2) population. The BP of F2 rats were recorded. Initially, Mendelian genetic markers were used to detect differences between regions of the genomes of hypertensive and normotensive rats. Such polymorphisms were used to track the cosegregation of marker genes with BP. The first example of this marker gene approach was demonstrated in 1972 by Yamori et al. (391) using an electrophoretic protein polymorphism within the renal esterase gene as a genetic marker to study the cosegregation of BP in populations derived from SHR and Wistar-Mishima rats. In the same year, another cosegregation study of BP with a Mendelian polymorphism in adrenal steroid biosynthesis was reported using the Dahl rats (302). In this case the genetic marker turned out to be due to variants in the gene actually causing the BP changes (see sect. IVA). A DNA restriction fragment length polymorphism (RFLP) was also exploited as a marker to demonstrate cosegregation of a segment of rat chromosome 13 containing the renin gene with hypertension in the Dahl rats (311).
During the mid 1980s, largely driven by the discovery of the PCR technology, methods were quickly evolving to improve the detection of DNA polymorphisms on the genome. RFLP was soon replaced by the relatively more efficient method of using polymorphic microsatellite markers at the scale of the entire genome for BP linkage analysis. Such experiments were first reported in 1991 by Jacob et al. (163) and Hilbert et al. (142). A major BP QTL on rat chromosome 10 was thus identified by both these groups (142, 163, 298). This method called as “linkage mapping” or “linkage analysis” and the subsequent method called “substitution mapping” were extensively applied to the study of inheritance of hypertension in rat genetic models (298). The results obtained through these efforts were that specific regions on the rat genome were identified as BP QTLs, meaning regions that were flanked by specific microsatellite markers (genotyped using PCR), were linked to the inheritance of BP in segregating populations (usually F2). Following the studies in 1991–92, when the first such mapping studies were reported using rats (78, 142, 163), several such BP QTLs were either identified solely by linkage analysis or further validated by substitution mapping using congenic or consomic strains. Previous review articles by Rapp (298) and Cowley Jr (45) are highly recommended for a detailed understanding of linkage analysis and substitution mapping as applied to the study of the inheritance of BP using rat models of hypertension. For a quick review on the technique to make congenic strains, which are used for substitution mapping, see Rapp (298). The following section provides an update to these mapping efforts of rat BP QTLs, which are important and significant given the rapid rise in sequencing technology since the review by Rapp in 2000 (298).
The dawn of the 21st Century ushered in the new era of whole genome sequencing, which served as the next wave of opportunity to query genomes in further detail. Following the sequencing of the human genome, the rat genome was sequenced in 2004 (116). The genome sequence obtained was from the Brown Norway (BN) rat, which is a relatively normotensive strain compared with the hypertensive S rat or the SHR. The BN rat strain is highly polymorphic, implying that there are a large number of sequence variants of this strain compared with any other rat strain. While the sequence of this strain served as a general reference for comparison with the sequence data of other rat strains, nonavailability of whole genome sequences of other, especially, genetically hypertensive strains, was an impediment. However, the availability of an assembled, complete rat genome sequence served several purposes: 1) provided the means to define BP loci on the physical map of the rat genome, whereby BP QTLs could be represented in physical locations on a chromosome in kilobases or megabases as units as opposed to genetic distances calculated using recombinations with centimorgans as units; 2) served to improve the resolutions of the already identified BP loci because with the genome sequence, additional markers could be located. For example, additional polymorphic microsatellite markers (which are tandemly repeated nucleotide sequences that range in length from two to five nucleotides repeated in different numbers in different rat strains) could be located and exploited to further delimit the genomic region encompassing a BP locus. 3) Sequences of candidate genes within the BP QTL could be determined in any strain by designing primers to amplify genome DNA of any chromosome.
These enhanced features resulted in high-resolution mapping of the identified BP QTL. A summary of all linkage mapping studies, which identified large rat genomic regions of interest is provided in TABLE 1. The main points to note from these linkage analyses are as follows:
All rat chromosomes harbor BP QTLs, implying that BP causal genes are not limited to any particular chromosome and that the overall genetic control of BP could be facilitated by a large number of randomly distributed genetic elements.
Some of the BP QTLs, for example, on rat chromosomes 1, 2, and 10 are consistently and recurrently detected in multiple genetic linkage studies conducted despite the differences in the hypertensive strains used for the studies. The density of QTLs is not proportional to the size of the chromosomes. Chromosomes 1 and 2 are the longest, but chromosome 10 is among the shorter rat chromosomes.
The context of genetic background wherein the QTL alleles are presented in a segregating population has an important effect on whether a given QTL will be detected. For example, a BP QTL was detected on RNO7 in the backcross population generated with the Dahl S rat, but not in an F2 population.
BP QTLs are not always gender independent. Some are gender specific.
BP QTLs are not always salt sensitive.
Table 1.
Summary of BP QTL linkage mapping studies conducted using the rat as a model organism
|
QTL Location (on Rnor 6.0) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RNO | Hypertensive Model | Linkage Analysis | Diet | Sex | BP Method | Epistatic Interaction | From Marker | To Marker | From base pair # | To base pair # | References |
| 1 | FHH | F1 (FHH X ACI) X FHH | Not mentioned | M | I | N | D1Mit17 | D1Mit5 | 133,795,442 | 215,712,024 | Brown et al. (24) |
| 1 | FHH | F2 (FHH X ACI) | Not mentioned | M | I | N | D1Wox6 | Mt1pa | 137,787,261 | 197,963,072 | Shiozawa et al. (337) |
| 1 | HTG | F2 (HTG X LEW) | LS | B | D | N | D1Rat171 | D1Mgh12 | 166,577,232 | 263,271,840 | Ueno et al. (363) |
| 1 | MHS | F2 (MHS X MNS) | Not mentioned | B | B | N | D1Rat5 | D1Mit9 | 10,259,237 | 49,547,474 | Zagato et al. (398) |
| 1 | MHS | F2 (MHS X MNS) | Not mentioned | B | B | N | D1Rat76 | D1Mit14 | 244,992,467 | 280,632,620 | Zagato et al. (398) |
| 1 | MWF /Fub* | F1 (MWF/Fub X LEW/Fub) X MWF/Fub | LS | M | I | N | D1Rat136 | 43,579,208 | Unmapped | Schulz et al. (328) | |
| 1 | SBH | F2 (SBH X SBN) | LS | M | I | N | D1Mgh2 | D1Mit11 | 23,406,428 | 108,057,505 | Yagil et al. (389) |
| 1 | SBH | F2 (SBH X SBN) | IS | B | I | N | D1Mit2 | D1Mgh8 | 140,953,686 | 163,796,432 | Yagil et al (389), |
| 1 | SHR | F2 (SHR X WKY) | Not mentioned | M | I | N | Sa | 189,514,503 | Iwai et al. (161) | ||
| 1 | SHR | F2 (SHR X WKY) | LS | M | B | N | Sa | Mt1pa | 189,514,503 | 197,963,072 | Samani et al. (323) |
| 1 | SHR | RI- SHR X BN | LS | M | D | N | KAL | 100,133,276 | Pravenec et al. (290) | ||
| 1 | SHR | F1 (SHR X F344) X SHR | LS | B | I | Y | D1Rat43 | D1Mgh11 | 144,634,295 | 221,753,518 | Ohno et al. (271) |
| 1 | SHR | F1 (S X SHR) X S | LS | M | I | N | D1Rat189 | D1Rat158 | 90,804,143 | 161,321,256 | Garrett et al. (100) |
| 1 | SHR | F2 (S X SHR) | HS | M | I | N | D1Rat1 | D1Rat335 | 10,065,314 | 67,227,947 | Siegel et al. (339) |
| 1 | SHR/Mol | F1 (SHR/Mol X BB/OK) X SHR/Mol, F1 (SHR/Mol X BB/OK) X BB/OK | LS | B | I | N | Igf2 | D1Mgh12 | 215,828,102 | 263,271,840 | Kovacs et al. (197) |
| 1 | SHRSP | F2 (W.S.10 X SHRSP) | IS | B | D | N | Scnn1b | 191,829,555 | Kreutz et al. (200) | ||
| 1 | SHRSP/izm | F2 (SHRSP/Izm x WKY/Izm) | IS | M | I | N | D1Wox29 | Mt1pa | 130,779,148 | 197,963,072 | Mashimo et al. (237) |
| 1 | SHRSP/izm | F2 (SHRSP/Izm x WKY/Izm) | IS | B | I | N | D1Mgh5 | Mt1pa | 79,689,548 | 197,963,072 | Kato et al. (178) |
| 1 | SHRSP/izm | F1 (SHRSP/Izm x WKY/Izm) X SHRS/Izm | % not found | M | I | N | D1Wox29 | D1Wox10 | 130,779,148 | 236,763,528 | Kato et al. (178) |
| 1 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D1Wox1 | Igf2 | 50,508,884 | 215,828,102 | Garrett et al. (101) |
| 1 | SS/Jr | F1 (S X SHR) X S | LS | M | I | N | D1Rat189 | D1Rat158 | 90,804,143 | 161,321,256 | Garrett et al. (100) |
| 1 | SS/JrHSDMCwi | F2 (S X BN) | HS | F | D | N | D1Rat295 | D1Rat301 | 227,107,736 | 249,206,417 | Moreno et al. (255) |
| 1 | SS/JrHSDMCwi | F2 (S X BN) | HS | F | D | N | D1Rat265 | D1Rat183 | 94,364,073 | 137,084,126 | Moreno et al. (255) |
| 1 | SS/Rkb | F2 (S X SHR) | HS | M | I | N | D1Rat1 | D1Rat335 | 10,065,314 | 67,227,947 | Siegel et al (339) |
| 1 | LH | F2 (LH X LN) | LS | M | D | N | D1Rat278 | 175,980,731 | Bilusic et al. (17) | ||
| 1 | LH | F2 (LH X LN) | LS | M | D | N | D1Rat278 | 175,980,731 | Bilusic et al. (17) | ||
| 1 | ISIAH | F2(ISIAHXWAG) | Not mentioned | M | I | N | D1Rat54 | D1Rat81 | 174,905,700 | 264,802,994 | Redina et al. (313) |
| 1 | ISIAH | F2(ISIAHXWAG) | Not mentioned | M | I | N | D1Rat54 | D1Rat117 | 174,905,700 | 233,490,237 | Redina et al. (313) |
| 1 | ISIAH | F2 (ISAH X WAG) | Not mentioned | M | I | N | D1Rat168 | D1Rat76 | 217,372,257 | 244,992,610 | Redina et al. (314) |
| 1 | ISIAH | F2 (ISAH X WAG) | Not mentioned | M | I | N | D1Rat54 | D1Rat168 | 174,905,700 | 217,372,467 | Redina et al. (314) |
| 1 | SS/Rkb | F2 (S X SHR) | HS | M | I | N | D1Rat1 | D1Rat335 | 10,065,314 | 67,227,947 | Siegel et al. (339) |
| 1 | SS/Hsd | F2 (S x R) | HS | B | D | D1Rat45 | 153,834,077 | Herrera et al. (139) | |||
| 1 | SS/Hsd | F2 (S x R) | HS | M | D | D1Mgh11 | 221,753,409 | Herrera et al. (139) | |||
| 2 | AS | F2 (S X AS) | HS | M | I | N | D2Uia17 | D2Mco25 | 12,666,921 | 33,718,888 | Garrett et al. (102) |
| 2 | GH | F2 (GH X BN) | LS | B | B | N | Gca | 189,840,403 | Harris et al. (130) | ||
| 2 | LH | F2 (LH X LN) | LS | M | D | N | D2Rat270 | 54,436,698 | Bilusic et al. (17) | ||
| 2 | LH | F1 (LH X LN) X LH | LS | M | D | N | D2Mit5 | D2Wox20 | 66,828,049 | 188,448,205 | Vincent et al. (365) |
| 2 | SHR | F2 (SHR X WKY) | LS | M | I | N | D2Wox24 | D2Mgh12 | 189,854,122 | 217,498,710 | Samani et al. (322) |
| 2 | SHR | F2 (SHR X BN) | IS | Not mentioned | D | Y | Mt1pb | Gca | 11,261,631 | 189,856,090 | Schork et al. (327) |
| 2 | SHR | RI- SHR X BN | LS | M | D | N | D2N35 | 153,799,585 | Pravenec et al. (287) | ||
| 2 | SHR/Mol | F1 (SHR X Wild) X SHR | Not mentioned | B | I | N | Fgg | 181,987,217 | Kloting et al. (185) | ||
| 2 | SHRSP/Glasgow | F2 (SHRSP X WKY) | IS | B | D | N | D2Mit5 | Cpb | 66,828,049 | 105,089,659 | Clark et al. (42), |
| 2 | SHRSP/Heidelberg | F2 (SHRSP X WKY) | IS | B | D | Y | Gca | 189,840,403 | Jacob et al. (163) | ||
| 2 | SS/Hsd | F2 (S X R) | HS | M | D | N | D2Mit10 | D2Mit14 | 157,914,311 | 204,585,731 | Herrera et al. (140) |
| 2 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D2Mit1 | D2Mit6 | 3,127,441 | 78,466,260 | Garrett et al. (101) |
| 2 | SS/Jr | F2 (S X MNS) | HS | M | I | N | Fgg | Camk2d | 181,987,217 | 231,132,039 | Deng et al. (69), Deng et al. (65) |
| 2 | SS/Jr | F2 (S X WKY) | HS | M | I | N | Fgg | Camk2d | 181,987,217 | 231,132,039 | Deng et al (69), Deng et al. (65) |
| 2 | BN | F2 (BN X GH) | HS | M | B | Y | D2Mgh7 | D2Mgh11 | 158,159,186 | 204,022,555 | Bilusic et al. (18) |
| 2 | SS/Hsd | F2 (SS/Hsd x R) | HS | F | D | EA4 | Unmapped | Herrera et al. (139) | |||
| 2 | SS/Hsd | F2 (SS/Hsd x R) | HS | F | D | D2Rat143 | 106,156,724 | Herrera et al. (139) | |||
| 2 | Ss/Hsd | F2 (SS/Hsd x R) | HS | M | D | D2Mgh11 | 204,022,334 | Herrera et al. (139) | |||
| 2 | SS/Jr | F2 (S X AS) | HS | M | I | N | D2Uia17 | D2Mco25 | 12,666,921 | 33,718,888 | Garrett et al. (102) |
| 3 | hHTg | F2 (hHTg X BN) | LS | M | D | N | D3Rat126 | 125,753,279 | Klimes et al. (184) | ||
| 3 | HTG | F2 (HTG X LEW) | LS | B | D | N | D3Wox3 | D3Rat17 | 30,846,101 | 127,023,997 | Ueno et al. (363) |
| 3 | SHR | F2 (S X SHR) | HS | M | I | N | D3Rat53 | D3Rat45 | 13,126,914 | 41,510,346 | Garrett et al. (108) |
| 3 | SHR | F2 (S X SHR) | HS | M | I | N | D3Mgh9 | D3Rat75 | 3,285,929 | 55,279,027 | Siegel et al. (339) |
| 3 | SHRSP/Glasgow | F2 (SHRSP X WKY) | IS | B | D | N | D3Mit10 | D3Wox2 | 13,152,311 | 50,533,259 | Clark et al. (42) |
| 3 | SHRSP/Izm | F2 (SHRSP/Izm x WKY/Izm) | IS | M | I | N | D3Mgh16 | D3Mgh8 | 6,000,748 | 26,684,263 | Mashimo et al. (237) |
| 3 | SHRSP/Izm | F2 (SHRSP/Izm x WKY/Izm) | IS | B | I | N | D3Mgh8 | D3Wox10 | 6,363,336 | 51,821,835 | Kato et al. (177) |
| 3 | SHRSP/Izm | F1(SHRSP x WKY) x SHRSP | LS | M | I | N | D3Mit9 | 34,394,121 | Kato et al. (178) | ||
| 3 | SS/Hsd | F2 (SS/Hsd X R) | HS | M | I | Y | D3Rat18 | D3Rat6 | 124,580,247 | 153,412,619 | Herrera et al. (135) |
| 3 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D3Wox3 | D3Mco21 | 30,846,101 | 72,672,468 | Garrett et al. (101) |
| 3 | SS/Jr | F2 (S X BN) | HS | M | I | N | D3Wox20 | D3Wox1 | 129,787,213 | 174,632,112 | Kato et al. (176) |
| 3 | SS/Jr | F1 (S X R) X S | HS | B | I | N | D3Mco16 | D3Rat100 | 25,633,106 | 39,248,617 | Cicila et al. (37) |
| 3 | SS/Rkb | F2 (S X SHR) | HS | M | I | N | D3Mgh9 | D3Rat75 | 3,285,929 | 55,279,027 | Siegel et al. (339) |
| 4 | AS | F2 (S X AS) | HS | M | I | N | D4Uia1 | D4Rat160 | 119,130,374 | Unmapped | Garrett et al. (102) |
| 4 | MHS | F2 (MHS X MNS) | Not mentioned | B | B | N | Add2 | 117,743,710 | Bianchi et al. (16) | ||
| 4 | MWF /Fub | F1 (MWF/Fub X LEW/Fub) X MWF/Fub | LS | M | I | N | D4Rat41 | 97,758,884 | Schulz et al. (328) | ||
| 4 | SHR | F2 (SHR X WKY) | LS | B | D | N | Npy | 79,573,998 | Katsuya et al. (181) | ||
| 4 | SHR | F2 (SHR X BN) | IS | Not mentioned | D | Y | Npy | 79,573,998 | Schork et al. (327) | ||
| 4 | SHR | RI- SHR X BN | LS | M | D | N | Il6 | 3,043,231 | Pravenec et al. (287) | ||
| 4 | SHR/Mol | F1 (SHR X BB/OK) X BB/OK | LS | M | I | N | D4Mit2 | D4Mit24 | 55,791,564 | 79,575,658 | Kovacs et al. (196) |
| 4 | SHR/Sankyo | F2(SHR x WKY) | LS | B | D | N | Npy | 78 | Takami et al. (352) | ||
| 4 | SHRSP/Izm | F2 (SHRSP/Izm x WKY/Izm) | IS | M | I | N | D4Mit2 | Spr | 55,791,564 | 116,916,236 | Mashimo et al. (237) |
| 4 | SHRSP/Izm | F2 (SHRSP/Izm x WKY/Izm) | IS | B | I | N | D4Mgh7 | Try1 | 136,351,734 | 70,779,249 | Kato et al. (177) |
| 4 | SS/Jr | F2 (S X AS) | HS | M | I | N | D4Rat160 | D4Uia1 | Unmapped | 119,130,374 | Garrett et al. (102) |
| 5 | hHTg | F2(hHTg x BN) | LS | M | D | N | D5Mgh9 | 172,402,477 | Klimes et al. (184) | ||
| 5 | HTG | F2 (HTG X LEW) | LS | B | D | N | D5Rat77 | D5Rat105 | 75,995,687 | 154,794,907 | Ueno et al. (363) |
| 5 | MWF /Fub* | F1 (MWF/Fub X LEW/Fub) X MWF/Fub | LS | M | I | N | D5Rat41 | 155,051,873 | Schulz et al. (328) | ||
| 5 | SHR | F2 (SHR X WKY) | LS | B | D | N | D5Mgh14 | 149,568,795 | Zhang et al. (401) | ||
| 5 | SHR | F2 (SHR X WKY) | LS | B | D | N | D5Mgh5 | D5Rat180 | 44,404,276 | 165,718,386 | Ye P et al. (395) |
| 5 | SHR/Mol | F1 (SHR X Wild) X SHR | Not mentioned | B | I | N | Slc2a1 | D5Mgh9 | 138,154,673 | 172,402,610 | Kloting et al. (185) |
| 5 | SHR/NCrlBr | F2 (SHR X BN) | IS | B | D | N | D5Rjr1 | 134,502,121 | Stec et al. (348) | ||
| 5 | SHRSP/izm | F2 (SHRSP/Izm x WKY/Izm) | IS | B | I | N | D5Mgh2 | D5Rat4 | 17,064,231 | 48,722,188 | Kato et al. (177) |
| 5 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D5Mit5 | D5Mco2 | 108,092,659 | 147,641,079 | Garrett et al. (101) |
| 5 | BN | F2 (BN X SHR) | IS | D | N | R589 | Unmapped | Soler et al. (345) | |||
| 5 | SS/Hsd | F2 (S x R) | HS | F | D | D5Rat106 | 156,443,753 | Herrera et al. (139) | |||
| 5 | Ss/Hsd | F2 (S x R) | HS | F | D | D5Rat23 | 105,924,457 | Herrera et al. (139) | |||
| 6 | SHR | F2 (SHR X LEW) | LS | B | I | Y | D6Mit4 | 60,606,186 | Ramos et al. (297) | ||
| 6 | SHR | RI-SHR X BN, BN X SHR | Not mentioned | Not mentioned | D | N | D6Rat46 | D6Rat84 | 13,122,958 | 33,259,316 | Jaworskiet al (164) |
| 6 | SHR | F1 (S X SHR) X S | LS | M | I | N | D6Rat180 | D6Mit3 | 174,130 | 75,623,393 | Garrett et al. (100) |
| 6 | SHR | F2 (S X SHR) | HS | M | I | N | D6Rat80 | D6Rat108 | 1,120,393 | 16,100,257 | Siegel et al. (338) |
| 6 | SS/Jr | F1 (S X SHR) X S | LS | M | I | N | D6Rat180 | D6Mit3 | 174,130 | 75,623,393 | Garrett et al. (100) |
| 6 | SS/Rkb | F2 (S X SHR) | HS | M | I | N | D6Rat80 | D6Rat108 | 1,120,393 | 16,100,257 | Siegel et al. (338) |
| 6 | BN | BN.GH | HS | M | B | Y | D6Mit12 | D6Mit3 | Unmapped (2,916,444 on RNOr_5.0) | 75,623,393 | Bilusic et al. (18) |
| 7 | SHR/Mol | F1 (SHR X Wild) X SHR | Not mentioned | B | I | N | Igf1 | 28,412,198 | 28,486,609 | Kloting et al. (185) | |
| 7 | SS/Jr | F1 (S X R) X S | HS | B | I | N | Cyp11b1 | 112,977,395 | Cicila et al. (41) | ||
| 8 | AS | F2 (S X AS) | HS | M | I | N | D8Mgh9 | D8Mgh4 | 38,202,434 | 89,058,369 | Garrett et al. (102) |
| 8 | HTG | F2 (HTG X LEW) | LS | B | D | N | D8Rat37 | D8Rat117 | 55,435,004 | 125,428,828 | Ueno et al. (363) |
| 8 | SHR | F2 (SHR X BN) | IS | Not mentioned | D | Y | D5Mit3 | D5Mit5 | 83,646,702 | 108,092,802 | Schork et al. (327) |
| 8 | SHR | F2 (SHR X WKY) | HS | B | D | N | D8Mgh10 | Takami et al. (352) | |||
| 8 | SHR | F2 (S X SHR) | HS | M | I | N | D8Rat36 | D8Rat133 | 58,425,510 | 96,998,640 | Garrett et al. (108) |
| 8 | SHR | RI-SHR X BN, BN X SHR | Not mentioned | Not mentioned | D | N | D8Mit6 | D8Rat40 | 11,373,267 | 50,708,951 | Jaworski et al. (164) |
| 8 | SHRSP/izm | F2 (SHRSP/Izm x WKY/Izm) | LS | B | I | N | D8Mit1 | Acaa | 95,349,621 | 128,036,236 | Kato et al. (177) |
| 8 | SS/Iwai | F2 (S/Iwai X WKY) | HS | B | D | N | D8Mgh10 | Takami et al. (352) | |||
| 8 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D8Mgh9 | D8Wox2 | 38,202,434 | 62,427,969 | Garrett et al. (101) |
| 8 | SS/Jr | F2 (S X SHR) | HS | M | I | N | D8Rat36 | D8Rat133 | 58,425,510 | 96,998,640 | Garrett et al. (108) |
| 8 | SS/Jr | F2 (S X AS) | HS | M | I | N | D8Mgh9 | D8Mgh4 | 38,202,434 | 89,058,369 | Garrett et al. (102) |
| 8 | SHR | F2(SHRXBN) | Not mentioned | M | D | N | Apoa02 | R830(D8Mit12) | Unmapped | 59,087,488 | Silva et al. (340) |
| 9 | SHR | F2 (SHR X WKY) | HS | B | D | N | D9Mit2 | 71,771,288 | Takami et al. (352) | ||
| 9 | SHR | F2 (S X SHR) | HS | M | I | N | D9Mit3 | D9Rat5 | 63,269,904 | 93,442,944 | Siegel et al. (339) |
| 9 | SHR | F2 (S X SHR) | HS | M | I | N | D9Uia10 | D9Rat92 | 31,001,440 | 54,885,226 | Garrett et al. (108) |
| 9 | SHRSP/izm | F2 (SHRSP/Izm x WKY/Izm) | LS | B | I | N | D9Wox18 | D9Mit2 | 25,692,373 | 71,771,476 | Kato et al. (178) |
| 9 | SS/Jr | F2 (S X R) | HS | B | I | N | D9Rat12 | D9Rat4 | 73,334,111 | 98,606,834 | Rapp et al. (305) |
| 9 | SS/Jr | F2 (S X SHR) | HS | M | I | N | D9Uia10 | D9Rat92 | 31,001,440 | 54,885,226 | Garrett et al. (108) |
| 9 | SS/Rkb | F2 (S X SHR) | HS | M | I | N | D9Mit3 | D9Rat5 | 63,269,904 | 93,442,944 | Siegel et al. (339) |
| 9 | SS/Jr | F2(SS.SHR(9)X8A X SS) | IS | M | B | N | D9Mco72 | D9Mco93 | 52,686,874 | 98,164,303 | Toland et al. (356) |
| 10 | GH | F2 (GH X BN) | LS | B | B | N | Ace | 94,170,766 | Harris et al. (130) | ||
| 10 | ISIAH | F2 (ISAH X WAG) | Not mentioned | Not mentioned | D | N | D10Wox16 | 83,390,674 | Redina et al. (312) | ||
| 10 | MHS | F2 (MHS X MNS) | Not mentioned | B | B | N | D10Rat82 | D10Rat73 | 32,942,229 | 46,449,947 | Zagato et al. (398) |
| 10 | SHR | F2 (SHR X WKY) | LS | B | D | N | Ace | 94,170,766 | Zhang et al. (400) | ||
| 10 | SHR/Mol | F2 (SHR/Mol X BB/OK) | LS | B | I | N | Abp | Ppy | 56,219,861 | 90,042,877 | Kovacs et al. (197) |
| 10 | SHRSP//HD | F2 (SHRSP//HD X WKY/HD-0) | LS | B | D | N | Chrnb1 | 56,390,671 | 56,403,188 | Kreutz et al. (199) | |
| 10 | SHRSP//HD | F2 (SHRSP//HD X WKY/HD-0) | LS | B | D | N | Ace | 94,170,766 | Kreutz et al. (199) | ||
| 10 | SHRSP/Heidelberg | F2 (SHRSP X WKY) | IS | B | D | Y | Ace | 94,170,766 | Jacob et al. (163) | ||
| 10 | SHRSP/izm | F2 (SHRSP/Izm x WKY/Izm) | IS | M | I | N | Gh1 | D10Mgh1 | 94,486,205 | 102,748,839 | Mashimo et al. (237) |
| 10 | SS/Jr | F2 (S X BN) | HS | M | I | N | D10Mit4 | 36,584,373 | Kato et al. (176) | ||
| 10 | SS/Jr | F2 (S X WKY) | HS | M | I | N | D10Mgh6 | 64,648,175 | Kato et al. (176), Deng et al. (68) | ||
| 10 | SS/Jr | F2 (S X MNS) | HS | M | I | N | D10Wox11 | D10Wox6 | 53,637,485 | Unmapped | Kato et al. (176), Deng et al. (68) |
| 10 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D10Mco30 | D10Mco15 | 76,420,583 | 97,308,569 | Garrett et al. (101) |
| 10 | SS/Jr | F1 (S X SHR) X S | LS | M | I | N | D10Rat38 | D10Mco66 | 31,643,957 | 81,202,850 | Garrett et al. (100) |
| 10 | TGRmRen2-27 SD | F1 (F344 X LEW) X TGRmRen2-27 | Not mentioned | B | I | Y | D10Wox3 | D10Mit4 | 18,246,394 | 36,584,560 | Kantachuvesiri et al. (174) |
| 11 | SHR/Mol | F1 (SHR X Wild) X SHR | Not mentioned | B | I | N | D11Mgh4 | D11Wox6 | 62,653,194 | 86,714,647 | Kloting et al. (185) |
| 11 | SS/Jr | F1 (S X SHR) X S | IS | M | I | N | D11Rat67 | D11Rat50 | 47,263,866 | 86,994,795 | Garrett et al. (103) |
| 11 | SS/Hsd | F2 (S x R) | HS | M | D | D11Mgh5 | 44,444,112 | Herrera et al. (139) | |||
| 12 | SHR | F2 (SHR X LEW) | LS | B | I | Y | D12Mit3 | 24,294,687 | Ramos et al. (297) | ||
| 12 | SS/Jr | F2 (S X WKY) | HS | M | I | N | D12Wox16 | D12Wox8 | 30,641,840 | 44,098,244 | Kato et al. (176) |
| 12 | R | F2 (R X SS) | HS | F | D | D12Mit6 | 14,419,775 | Herrera et al. (139) | |||
| 13 | SHR | F2 (SHR X WKY) | LS | M | I | N | D13Wox5 | D13Mit3 | 55,560,677 | 80,285,772 | Samani et al. (322) |
| 13 | SHR | F2 (SHR X WKY) | LS | B | B | N | Ren | 50,502,724 | 50,514,151 | Sun et al. (351) | |
| 13 | SHR | F2 (SHR X WKY) | Not mentioned | B | B | N | Ren | 50,502,724 | 50,514,151 | Yu et al. (397) | |
| 13 | SHR | F2 (SHR X LEW) | LS | B | I | N | Ren | 50,502,724 | 50,514,151 | Kurtz et al. (208) | |
| 13 | SHR/Mol | F1 (SHR X BB/OK) X BB/OK | LS | M | I | N | D13Uwm1 | 55,560,677 | 55,560,862 | Kovacs et al. (196) | |
| 13 | SHR/Ola | RI- SHR X BN | LS | M | D | N | Ren | 50,502,724 | 50,514,151 | Pravenec et al. (292) | |
| 13 | SHRSP/izm | F2 (SHRSP/Izm x WKY/Izm) | LS | B | I | N | D13Mgh4 | D13Mgh7 | 42,310,744 | 67,207,219 | Kato et al. (177) |
| 13 | SS/Jr | F2 (S X R) | HS | B | I | N | D13Mgh3 | D13Mit3 | 39,372,926 | 80,285,772 | Zhang et al. (402) |
| 13 | SS/Jr | F2 (S X R) X S | HS | B | I | N | Ren | 50,502,724 | 50,514,151 | Rapp et al. (310) | |
| 13 | SS/JrHSDMCwi | F2 (S X BN) | HS | F | D | N | D12Rat22 | 51,955,573 | 51,955,754 | Moreno et al. (255) | |
| 13 | LH | F2 (LH X LN) | LS | M | D | N | D13Rat120 | 49,549,739 | 49,549,883 | Bilusic et al (17) | |
| 14 | MHS | F2 (MHS X MNS) | Not mentioned | B | B | N | D14Rat90 | D14Rat94 | 73,391,467 | 88,870,994 | Zagato et al. (398) |
| 14 | SS/JrHSDMCwi | F2 (S X BN) | LS | F | D | N | D14Rat12 | 61,783,047 | Moreno et al. (255) | ||
| 15 | SHRSP/izm | F2 (SHRSP/Izm x WKY/Izm) | LS | B | I | N | Ednrb | D15Mgh6 | 88,006,977 | 104,003,672 | Kato et al. (177) |
| 15 | SS/JrHSDMCwi | F2 (S X BN) | HS | F | D | N | D15Rat106 | 106,550,444 | Moreno et al. (255) | ||
| 16 | SHR | F2 (SHR X BN) | IS | Not mentioned | D | Y | D16Mi2 | D16Mit5 | 5,013,802 | 16,482,677 | Schork et al. (327) |
| 16 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D16Wox11 | D16Mit3 | 19,398,181 | 47,346,612 | Garrett et al. (101) |
| 17 | FHH | F2 (FHH X ACI) | Not mentioned | M | I | N | D17Rat54 | 18,538,334 | Shiozawa et al. (337)2000 | ||
| 17 | LH | F2 (LH X LN) | LS | M | D | N | D17Rat98 | 64,047,700 | Bilusic et al. (17) | ||
| 17 | SBH | F2 (SBH X SBN) | IS | B | I | N | D17Mgh3 | D17Mit4 | 43,677,641 | 63,994,435 | Yagil et al. (389) |
| 17 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D17Mit2 | D17Mco3 | 33,303,627 | 64,946,465 | Garrett et al. (101) |
| 17 | TGRmRen2-27 SD | F1 (F344 X LEW) X TGRmRen2-27 | Not mentioned | B | I | Y | D17Mit2 | 33,303,627 | Kantachuvesiri et al (174) | ||
| 17 | LH | F2 (LH X LN) | LS | M | D | N | D17Rat32 | 61,036,503 | Bilusic et al (17) | ||
| 18 | SHR/Mol | F2 (SHR/Mol X BB/OK) | LS | B | I | N | Ttr | D18Mit9 | 15,532,963 | 80,696,375 | Kovacs et al. (197) |
| 18 | SHRSP/Heidelberg | F2 (SHRSP X WKY) | IS | B | D | Y | D18Mit7 | 12,597,179 | Jacob et al. (163) | ||
| 18 | SS/Jr | F2 (S X LEW) | HS | M | I | N | D18Mit1 | D18Mco3 | 15,539,427 | 29,530,769 | Garrett et al. (101) |
| 18 | SS/JrHSDMCwi | F2 (S X BN) | HS | M | D | N | D18Mit1 | D18Mit8 | 15,539,427 | 61,985,812 | Cowley et al. (49) |
| 18 | BN | F2 (BN X GH) | HS | M | B | Y | D18Mgh2 | D18Mgh4 | 61,901,172 | 79,948,741 | Bilusic et al. (18) |
| 19 | SHR | F2 (SHR X WKY) | IS | M | D | N | Agt | 57,321,640 | Lodwick et al. (225) | ||
| 19 | SHR | RI- SHR X BN | LS | M | D | N | D19Mit7 | 47,318,201 | Pravenec et al. (287) | ||
| 20 | GH | F2 (GH X BN) | Not mentioned | B | B | N | Tnf | 4,855,829 | Harris et al. (129) | ||
| 20 | MHS | F2 (MHS X MNS) | Not mentioned | B | B | N | D20Rat44 | D20Rat38 | 32,469,646 | 45,719,230 | Zagato et al. (398) |
| 20 | SHR | RI- SHR X BN | LS | M | D | N | D20Mgh4 | 5,875,339 | Pravenec et al. (288) | ||
| 20 | SHRSP/Izm | F1(SHRSP x WKY) x SHRSP | LS | M | I | N | D20Mgh1 | 51,892,390 | Kato et al. (178) | ||
| 20 | R | F2 (R X SS) | HS | M | D | N | D20Rat37 | Unmapped | Herrera et al. (139) | ||
| X | SBH | F2 (SBH X SBN) | IS | B | I | N | DXRat4 | DXRat15 | 12,370,136 | 65,347,605 | Yagil C et al. (390) |
| X | SHR | F2 (SHR X WKY) | IS | B | D | N | DXMgh5 | DXMit4 | 23,143,255 | 61,398,097 | Hilbert et al. (142) |
| Y | SHR | F2 (SHR X WKY) and reciprocal F1 crosses | Not mentioned | B | I | N | Entire Y | Entire Y | Entire Y | Entire Y | Ely et al. (84), |
| Y | SHRSP | F2 (SHRSP X WKY), reciprocal crosses | IS | B | B | N | Entire Y | Entire Y | Entire Y | Entire Y | Davidson et al. (59) |
Bp QTLs that are reported in the literature are curated by their location on the rat genome and organized by chromosome. For details on strain names, please refer to the Rat Genome Database (www.rgd.mcw.edu). LS, low-salt diet (≤1% NaCl); lS, intermediate-salt diet (1% to <2% NaCl); HS, high-salt diet (≥2% to 8% NaCl); M, male; F, female; B, both genders; l, indirect tail-cuff method for BP measurement; D, direct, telemetry method for BP measurement; B, both tail-cuff method and telemetry for BP measurement. Blank spaces in all columns represent data that are insufficient or not provided in the original publications.
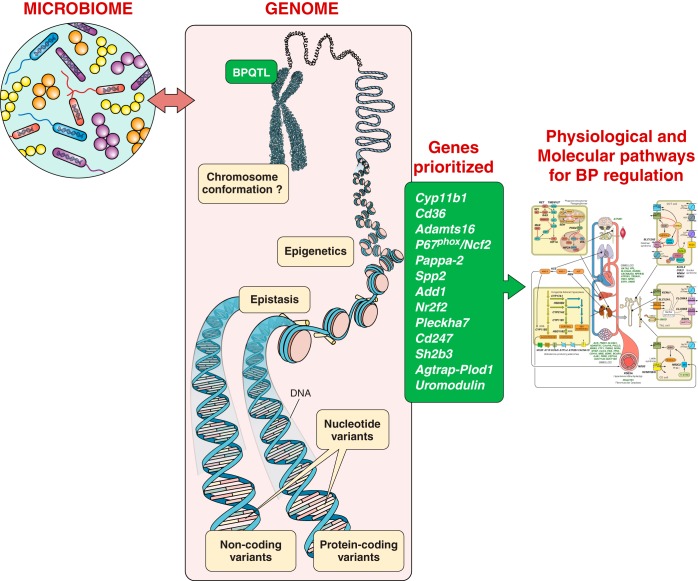
Some but not all of the linkage mapped regions were corroborated with substitution mapping studies. The substitution mapping studies are tabulated and schematically represented in TABLE 2 and FIGURE 1, respectively, and discussed previously (166, 167, 298). As products of such substitution mapping studies, several genes have been prioritized as positional candidates for BP regulation. The genes identified as a result of these mapping efforts are discussed below through section VIII.
Table 2.
Summary of BP QTL identified using rat congenic strains
| QTL Location (on Rnor 6.0) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RNO # | Letter Designated in Figure 1 | Hypertensive Model | Congenic Strain | Diet | Sex | BP Method | Epistatic Interaction | From Marker | To Marker | From base pair # | To base pair # | References |
| 1 | a | SS/Jr | S.LEW | HS | M | D | N | D1Rat211 | D1Rat12 | 33,667,777 | 36,129,328 | Joe et al. (168) |
| 1 | b | SS/Jr | S.LEW/NCrlBR | IS | M | I | N | D1Mgh7 | D1Mco36 | 113,593,576 | 129,209,407 | Saad et al. (319) |
| 1 | c | SS/Jr | S.LEW/NCrlBR | IS | M | I | N | D1Rat35 | D1Rat131 | 130,917,121 | 152,871,103 | Saad et al. (319) |
| 1 | d | SHR | SHR.WKY-Sa | LS | M | I and D | N | D1Wox34 | D1Rat55 | 175,447,029 | Unmapped (184,481,458 on Rnor_5.0) | Frantz et al. (96) |
| 1 | e | SHR | SHR.WKY-Sa | LS | M | I and D | N | D1Rat56 | D1Rat111 | 184,419,946 | 215,097,919 | Frantz S et al. (96) |
| 1 | f | SBH | SBH.SBN | HS | B | I and D | N | D1Rat10 | D1Rat24 | 85,706,847 | 78,434,672 | Yagil et al. (388) |
| 1 | g | SBH | SBH.SBN | HS | B | I and D | N | D1Rat27 | D1Rat74 | 94,201,400 | 241,482,368 | Yagil et al. (388) |
| 1 | h | SHRSP | WKY-1.SHRSP | IS | Not mentioned | D | N | D1Rat29 | D1Rat57 | 108,986,301 | 188,794,576 | Hubner et al. (155) |
| 1 | i | SHR | SHR.BN | IS | M | D | N | D1Rat68 | D1Rat71 | 205,603,081 | 230,420,772 | St Lezin et al. (346) |
| 1 | j | SHRSP/Izm | SHRSP/Izm.WKY.Izm | LS | B | D | N | Klk1 | D1Wox10 | 100,059,967 | 236,763,528 | Kato et al. (178) |
| 1 | k | SHRSP/Izm | .WKY.Izm/ SHRSP/Izm | Not mentioned | M | D | N | D1Wox18 | D1Smu11 | 100,133,276 | 184,185,332 | Cui et al. (54) |
| 1 | l | SHRSP | WKY.SHRSP | LS | M | D | Y (with RNO10) | D1Rat29 | D1Rat57 | 108,986,301 | 188,794,576 | Monti et al. (253) |
| 1 | m | SS/Jr | S.LEW | HS | M | D | Y | Serac-1 | D1Rat19 | 46,942,192 | 49,578,693 | Deng et al. (64) |
| 1 | n | SS/Jr | S.LEW | HS | M | D | Y | D1Uia4 | D1Rat320 | 64,588,516 | 125,875,986 | Deng et al. (64) |
| 1 | o | SS/Jr | S.LEW | HS | M | D | Y | D1Rat320 | D1Mgn32 | 125,875,986 | Unmapped (mentioned as 269,689,xxx in the article) | Deng et al. (64) |
| 1 | p | SS/Jr | S.LEW/NCrlBR | IS | M | I | N | D1Uia8 | D1Rat18 | 49,454,221 | 49,454,378 | Saad et al. (319) |
| 1 | q | SHRSP/izm | WKY.SHRSP | Not mentioned | M | D | N | D1Wox29 | D1Arb21 | 130,779,148 | 199,254,774 | Cui et al. (53) |
| 1 | r | SHRSP/izm | WKY/Izm.SHRSP/Izm | Not mentioned | M | D | Y | D1Smu13 | D1Arb21 | 166,884,926 | 199,254,774 | Xiao et al. (387) |
| 1 | s | SHRSP/izm | SHRSP/Izm.WKY/Izm | Not mentioned | M | D | Y | Apbb1 | D1Arb21 | 170,387,609 | 199,254,774 | Xiao et al. (387) |
| 1 | t | SS/Jr | S.LEW/NCrlBR | IS | M | D | N | D1Mco55 | D1Mco101 | 134,089,429 | 129,209,407 | Mell et al. (244) |
| 1 | u | SS/Jr | S.LEW/NCrlBR | IS | M | D | N | D1Rat200 | D1Mco136 | 133,076,978 | 133,164,521 | Mell et al. (244) |
| 1 | v | SS/Jr | S.LEW/NCrlBR | IS | M | D | N | D1Muo1 | D1Muo28 | 34,894,063 | 35,664,727 | Joe et al. (170) |
| 2 | a | SS/Jr | S.LEW | HS | M | D | N | D2Rat35 | D2Wox18 | 150,211,243 | 181,987,474 | Garrett et al. (106) |
| 2 | b | SS/Jr | S.LEW | HS | M | D | N | D2Wox18 | D2Wox25 | 181,987,474 | 200,585,297 | Garrett et al. (106) |
| 2 | c | SS/Jr | S.LEW | HS | M | D | N | D2Wox25 | D2Rat259 | 200,585,297 | 210,251,018 | Garrett et al. (106) |
| 2 | d | SS/Jr | S.MNS | HS | M | D | N | D2Rat166 | D2Mgh10 | 145,903,536 | 200,453,484 | Dutil et al. (76) |
| 2 | e | SHR/lj | WKY.SHR | LS and HS | M | D | N | D2Rat21 | D2Rat27 | 75,687,495 | 96,556,765 | Alemayehu et al. (5) |
| 2 | f | SHR/lj | WKY.SHR | LS and HS | M | D | N | D2Mgh10 | D2Rat62 | 200,453,324 | 236,318,668 | Alemayehu et al. (5) |
| 2 | g | SHR/lj | SHR.WKY | LS and HS | M | D | N | D2Rat40 | D2Rat50 | 169,852,670 | 207,612,467 | Alemayehu et al. (5) |
| 2 | h | SHR/lj | SHR.WKY | LS and HS | M | D | N | D2Rat161 | D2Mgh10 | 118,446,646 | 200,453,484 | Alemayehu et al. (5) |
| 2 | i | SHR | SHR.BN | LS | M | D | N | D2Rat171 | D2Arb24 | 116,075,644 | 228,737,869 | Pravenec et al. (293) |
| 2 | j | SHRSP | SHRSP.WKY | LS | M | D | N | D2Rat43 | D2Mgh12 | Unmapped (199,443,778 on Rnor_5.0) | 217,498,710 | McBride et al. (241) |
| 2 | k | SS/Hsd | S.R | HS | B | D | N | D2Rat352 | SNP2786652 | 180,909,971 | 187,195,159 | Herrera et al. (137) |
| 2 | l | SHRSP | SHRSP.WKY | IS | M | D | Y (with RNO3) | D2Rat13 | D2Rat157 | 37,861,479 | 241,761,983 | Koh-Tan et al. (187) |
| 2 | m | SHR | SHR.BN | IS | M | D | N | D2Rat226 | D2Rat294 | 177,680,772 | 243,901,375 | Aneas et al. (7) |
| 2 | n | SHR | SHR.BN | IS | M | D | N | D2Rat114 | D2Rat123 | 36,245,223 | 112,175,725 | Aneas et al. (7) |
| 2 | o | SS/Jr | S.LEW | IS | M | D | N | D2Chm277 | Prlr | 56,736,401 | 60,325,692 | Eliopoulos et al. (83) |
| 2 | p | SS/Jr | S.LEW | IS | M | D | N | D2Rat199 | D2Mco17 | 41,179,255 | 42,776,280 | Charron et al. (30), |
| 3 | a | SS/Jr | S.R | HS | B | I | N | D3Mco19 | D3Mco24 | 153,325,515 | 172,890,235 | Cicila et al. (37) |
| 3 | b | SS/Jr | S.LEW | HS | M | D | Y | D3Rat52 | D3Rat130 | 14,090,411 | 48,662,146 | Palijan et al. (277) |
| 3 | c | SS/Jr | S.LEW | HS | M | D | Y | D3Chm63 | D3Rat26 | 55,245,276 | 95,176,874 | Palijan et al. (277) |
| 3 | d | SS/Jr | S.R | HS | M | I | Y | D3Mco81 | D3Mco75 | Unmapped (128,986,468 from Rat genome database) | 173,986,468 | Lee et al. (211) |
| 3 | e | SS/Jr | F2(SREdn3XS) | HS | M | I | Y | D3Mco39 | D3Mco36 | 176,305,697 | 177,366,660 | Lee et al. (211) |
| 3 | f | SHRSP | SHRSP.WKY | IS | M | D | Y | D3Mgh16 | D3Wox28 | 6,000,748 | 156,575,096 | Koh-Tan et al. (187) |
| 3 | g | SS/Jr | S.R | HS | B | I | N | D3Rat61 | D3Rat59 | 153,381,237 | 170,534,769 | Cicila et al. (37) |
| 4 | a | SHR | SHR.BN-Il6/Npy | IS | M | B | N | D4Rat247 | SNP2788971 (labeled as 17.37Mb) | 4,780,787 | Unmapped | Pravenec et al (286) |
| 4 | b | SHR | SHR.BN | IS | M | D | N | D4Rat33 | D4Rat54 | 81,874,073 | 119,546,974 | Aneas et al. (7) |
| 5 | a | SS/Jr | S.LEW/NCrlBR | IS | M | I | Y | D5Uwm31 | D5Rjr1 | 47,842,131 | 134,502,294 | Garrett et al. (107) |
| 5 | b | SS/Jr | S.LEW/NCrlBR | IS | M | I | Y | D5Rat154 | D5Wox39/Lepr | Not mapped (closest marker is D5Mit5 at 108,092,659) | 124,025,214 | Garrett et al. (107) |
| 5 | c | SHR | SHR.BN | LS | M | D | N | D5Wox20 | D5Rat63 | 107,316,809 | 145,726,262 | Pravenec et al. (289) |
| 5 | d | SS/Hsd | S.R | HS | F | D | N | SNP2791496 | SNP2791569 | Unmapped (labeled as 134,909,988) | Unmapped (labeled as 141,744,732) | Herrera et al. (136) |
| 5 | e | SS/Jr | S.LEW/NCrlBR | IS | M | B | Y | D5Mco58 | D5Mco41 | Unmapped (mentioned as 131,853,815) | Unmapped (mentioned as 124,085,611) | Pillai et al. (284). |
| 5 | f | SS/Jr | S.LEW/NCrlBR | IS | M | B | Y | D5Mco42 | D5Mco47 | Unmapped (mentioned as 122,070,175) | Unmapped (mentioned as 117,894,038) | Pillai et al. (284) |
| 7 | a | SS/Jr | S.R | HS | M | D | N | D7Mco19 | D7Mco7 | 115,922,628 | Unmapped (116,060,096 on Rnor_5.0) | Garrett et al. (105) |
| 7 | b | SS/Hsd | S.LEW | IS | M | D | N | D7Chm6 | D7Mgh1 | Unmapped (nearest marker D7Rat73 is at 61,047,589) | Unmapped (nearest marker D7Rat128 is at 124,246,733 | Crespo et al. (51) |
| 8 | a | SHR | SHR.BN-Lx | LS | M | D | N | D8Mit6 | Rbp2 | 11,373,509 | 106,506,834 | Kren et al. (198) |
| 8 | b | SS/Hsd | S.LEW | IS | M | D | N | D8Chm14 | D8Rat16 | 51,844,002 | 101,305,168 | Ariyarajah et al. (10) |
| 8 | c | SS/Hsd | S.LEW | IS | F | D | N | D8Rat51 | D8Rat55 | 30,918,112 | 7,238,999 | Deng et al. (67) |
| 8 | d | S | S.LEW | IS | M | D | N | D8Rat56 | D8Rat51 | 9531047 | 30918267 | Ariyarajah et al. (10) |
| 9 | a | SS/Jr | S.R | HS | M | D | N | D9Mco14 | D9Uia6 | 82,356,030 | 83,686,404 | Meng et al. (247) |
| 9 | b | SS/Jr | S.R | IS | M | B | N | D9Mco95 | D9Mco98 | 81,100,315 | 81,180,041 | Gopalakrishnan et al. (122) |
| 9 | c | SS/Jr | S.R | IS | M | D | N | D9Mco14 | D9Mco61 | 82,356,030 | 89,338,443 | Garrett et al. (104) |
| 9 | d | SS/Jr | S.R | IS | M | D | N | D9Mco14 | Resp18-Intron2 | 82,356,030 | 82,477,136 | Garrett et al. (104) |
| 9 | e | SS/Jr | S.SHR | IS | M | D | N | D9Mco113 | D9Mco124 | 95,430,880 | 97,679,852 | Nie et al. (267), |
| 10 | a | SS/Jr | S.MNS and S.LEW | HS | M | D | N | D10Mco1 | D10Wox23 | 691,236,03 | 836,536,98 | Garrett et al. (109) |
| 10 | b | SS/Jr | S.MNS | HS | M | D | N | D10Mit1 | D10Mco6 | 94,178,327 | 101,482,600 | Garrett et al. (109) |
| 10 | c | SS/Jr | S.LEW | HS | M | D | N | D10M11Mit119 | D10Rat27 | Unmapped (nearest marker D10Wox51 is at 70,428,844 | 75,983,805 | Palijan et al. (278) |
| 10 | d | SS/Jr | S.LEW | HS | M | D | N | D10Rat204 | D10Rat9 | 93,622,786 | Unmapped (99,588,446 on RGSC_v3.4) | Palijan et al. (278) |
| 10 | e | SS/Jr | S.LEW | HS | M | D | N | D10Rat27 | D10Rat93 | 75,983,662 | 80,946,110 | Palijan et al. (278) |
| 10 | f | SHRSP | WKY.SHRSP | LS | M | D | N | Myhse | D10Mit11 | 53,621,375 | 100,759,938 | Monti et al. (254) |
| 10 | g | SHRSP | WKY.SHRSP | LS | M | D | Y | Myh3 | Aldoc | 53,621,375 | 65,590,126 | Monti et al. (253) |
| 10 | h | SS/Jr | S.LEW | IS | M | D | Y | D10Mgh6 | D10Mgh1 | 64,648,175 | 102,748,839 | Crespo et al. (52) |
| 10 | i | SS/Jr | S.LEW | IS | M | D | Y | D10Chm169 | D10Chm147 | 69,808,793 | 70,581,793 | Charron et al. (29) |
| 10 | j | SS/Jr | S.LEW | IS | M | D | Y | D10Chm212 | D10Chm29 | 73,727,917 | 75,099,909 | Charron et al (29) |
| 10 | k | SS/Jr | S.LEW | IS | M | D | Y | D10Got92 | D10Rat127 | 77,055,741 | 86,419,171 | Charron et al. (29) |
| 10 | l | SS/Jr | S.MNS | IS | M | B | N | D10Mco88 | D10Mco89 | 70,647,089 | 71,727,943 | Saad et al. (318) |
| 10 | m | SS/Jr | S.LEW | IS | M | B | N | D10Mco129 | D10Mco147 | 69,910,996 | Unmapped (71,100,513) | Saad et al. (320) |
| 10 | n | SS/Jr | S.LEW | IS | M | B | N | D10Rat58 | D10Mco43 | 70,202,084 | 70,610,989 | Saad et al. (320) |
| 10 | n | SS/Jr | S.LEW | IS | M | B | N | D10Got88 | D10Mco62 | 70,800,238 | Unmapped (70,763,527 on Rnor_5.0) | Saad et al. (320) |
| 10 | o | SS/Jr | S.LEW | HS | M | B | N | SNP marker | SNP marker | 71,028,112 | 71,070,581 | Gopalakrishnan et al. (121) |
| 11 | a | SS/Jr | S.SHR | IS | M | I | N | D11Rat31 | D11Rat50 | 6,673,351 | 86,994,795 | Garrett et al. (103) |
| 13 | a | SS/MCW | SS.BN | HS | M | D | N | Consomic (whole chromosome) | Consomic (whole chromosome) | 1 | 114,033,958 | Cowley et al. (48) |
| 13 | b | SS/MCW | SS.BN | HS | B | D | N | D13Rat7 | D13Rat60 | 14,279,081 | 26,919,398 | Moreno et al. (257) |
| 13 | c | SS/MCW | SS.BN | HS | B | D | N | D13Rat111 | D13Rat101 | 35,301,263 | 53,264,877 | Moreno et al. (257) |
| 13 | d | SS/MCW | SS.BN | HS | F | D | N | D13Rat88 | D13Rat91 | 53,264,698 | 50,799,665 | Moreno et al. (257) |
| 13 | e | SS/MCW | SS.BN | HS | F | D | N | D13Rat178 | D13Got51 | 62,788,897 | 75,026,713 | Moreno et al. (257) |
| 13 | f | SS/MCW | SS.BN | HS | B | D | N | D13Rat111 | D13Rat88 | 35,301,263 | 46,444,796 | Moreno et al. (257) |
| 13 | g | SS/MCW | SS.BN | HS | B | D | N | D13Rat39 | D13Mit2 | 37,352,631 | 65,008,446 | Moreno et al. (257) |
| 13 | h | SS/MCW | SS.BN | HS | B | D | N | D13Rat91 | D13Got45 | 50,799,478 | 72,031,708 | Moreno et al. (257) |
| 13 | i | SS/MCW | SS.BN | HS | B | D | N | D13Rat101 | D13Got45 | 53,264,877 | 72,031,708 | Moreno et al. (257) |
| 13 | j | SS/MCW | SS.BN | HS | M | D | N | D13Rat7 | D13Rat20 | 14,279,081 | 42,155,682 | Moreno et al. (257) |
| 13 | k | SS/MCW | SS.BN | HS | M | D | N | D13Rat7 | D13Rat101 | 14,279,081 | 53,264,877 | Moreno et al. (257) |
| 13 | l | SS/MCW | SS.BN | HS | M | D | N | D13Rat111 | D13Rat101 | 35,301,263 | 53,264,877 | Moreno et al. (257) |
| 13 | m | SS/MCW | SS.BN | HS | M | D | N | D13Rat115 | D13Rat101 | 39,639,775 | 53,264,877 | Moreno et al. (257) |
| 13 | n | SS/MCW | SS.BN | HS | M | D | N | D13Rat88 | D13Rat91 | 46,444,570 | 53,264,877 | Moreno et al. (257) |
| 13 | o | SS/MCW | SS.BN | HS | M | D | N | D13Rat88 | D13Got51 | 46,444,570 | 75,026,713 | Moreno et al. (257) |
| 13 | p | SS/MCW | SS.BN | HS | M | D | N | D13Rat178 | D13Got51 | 62,788,897 | 75,026,713 | Moreno et al. (257) |
| 13 | q | SS/MCW | SS.BN | HS | M | D | N | D13Rat20 | D13Hmgc98 | 42,155,543 | 44,730,157 | Moreno et al. (258) |
| 13 | r | SS/MCW | SS.BN | HS | M | D | N | D13Hmgc755 | D13Hmgc5885 | 76,000,378 | 77,381,370 | Cowley et al. (46) |
| 13 | s | SS/MCW | SS.BN | HS | M | D | N | D13Hmgc1048 | D13Hmgc1050 | Unmapped (mentioned as 81,011kb in the article) | Unmapped (mentioned as 81,717 kb in the article) | Cowley et al. (50) |
| 14 | a | MHS | MNS.MHS | LS | M | D | Y | D14Rat43 | D14Wox15 | 78,446,303 | 81,093,349 | Tripodi et al. (360) |
| 16 | a | SS/Jr | S.LEW | HS | M | D | N | D16Rat88 | D16Rat21 | 1,550,330 | Unmapped (11,086,530 on Rnor_5.0) | Moujahidine et al. (258a) |
| 16 | b | SS/Jr | S.LEW | HS | M | D | N | D16Chm48 | D16Chm60 | 2,471,921 | 3,525,217 | Crespo et al. (52a) |
| 16 | c | SHR | SHR.BN | IS | M | D | N | D16Rat87 | D16Mgh1 | 4,136,355 | 45,905,331 | Aneas et al. (7) |
| 16 | d | SS/Jr | S.LEW | HS | M | D | N | D16Rat12 | D16Chm66 | 1,090,164 | 3,439,525 | Moujahidine et al. (258b) |
| 17 | a | SS/Jr | S.LEW | IS | M | D | N | D17Rat181 | D17Rat97 | 33,209,117 | 58,467,778 | Grondin et al. (123a) |
| 18 | a | SHR | SHR.BN | IS | M | D | N | D18Rat113 | D18Rat99 | 3,719,547 | 32,487,870 | Johnson et al. (170a) |
| 18 | b | SHR | SHR.BN | IS | M | D | N | D18Rat40 | D18Rat82 | 61,499,531 | 73,016,546 | Johnson et al. (170a) |
| 18 | c | SS/Jr | S.LEW | IS | M | D | Y | D18Wox7 | D18Rat101 | 15,539,551 | 27,743,236 | Charron et al. (30) |
| 18 | d | SS/Jr | S.LEW | IS | M | D | Y | D18Rat101 | D18Chm56 | 27,743,024 | 48,499,517 | Charron et al. (30) |
| 18 | e | SS/Jr | S.LEW | IS | M | D | Y | D18Rat55 | D18S481 | 54,108,375 | Unmapped | Charron et al. (30) |
| 19 | a | SHR/Ola | SHR.BN-Agt | LS | M | D | N | D19Mit7 | D19Rat57 | 47,318,314 | 60,220,451 | St Lezin et al. (346a) |
| 20 | a | SHR | SHR.BN | LS | M | D | N | D20Cebr215s7 | D20Rat23 | Unmapped (nearest mentioned Tnfα maps to 4,857,203) | 21,569,567 | Pausova et al. (279a) |
| X | a | SHR | SHR.BB/OK | unknown | B | I | N | Ar-Mysc-Pfkb1 | DXMgh3 | 17,823,554 (location of Mysc) | 11,969,489 | Klöting et al. (185a) |
| Y | a | SHR | SHR.BN | IS | M | D | N | Consomic (entire chromosome) | Consomic (entire chromosome) | 1 | 3,310,458 | Kren et al. (198a) |
| Y | b | SHRSP | SHRSP.WKY and WKY.SHRSP | IS | M | D | N | Consomic (entire chromosome) | Consomic (entire chromosome) | 1 | 3,310,458 | Negrin et al. (236a) |
BP QTLs that are reported in the literature are curated by their location on the rat genome and organized by chromosome. For details on strain names, please refer to the Rat Genome Database (www.rgd.mcw.edu). LS, low-salt diet (≤1% NaCI); IS, intermediate-salt diet (1% to <2% NaCI); HS, high-salt diet (≥2% to 8% NaCI); M, male; F, female; B, both genders; I, indirect tail-cuff method for BP measurement; D, direct, telemetry method for BP measurement; B, both tail-cuff method and telemerty for BP measurement.
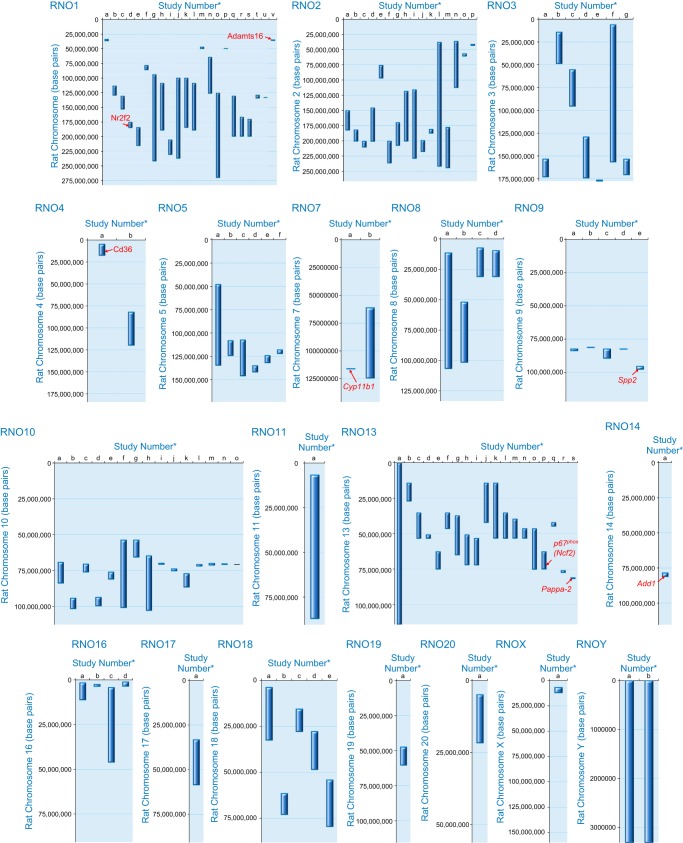
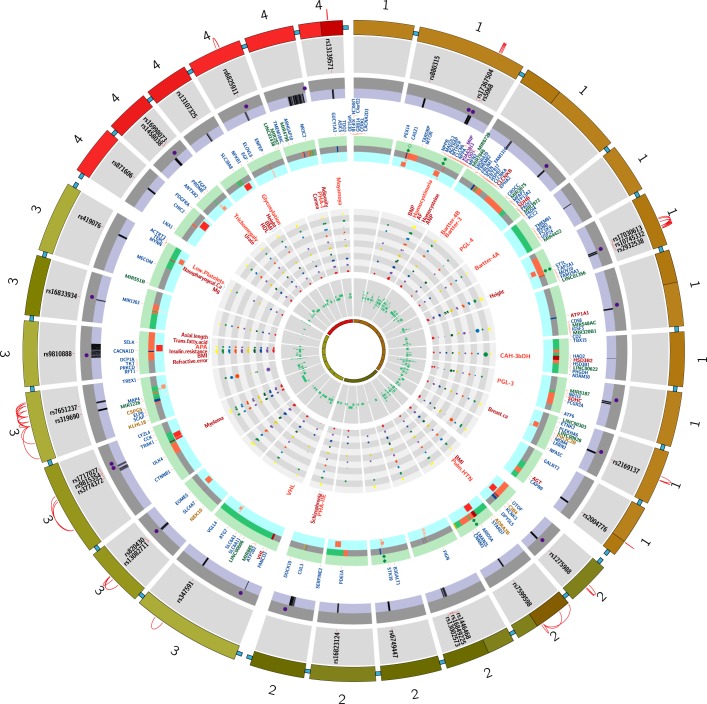
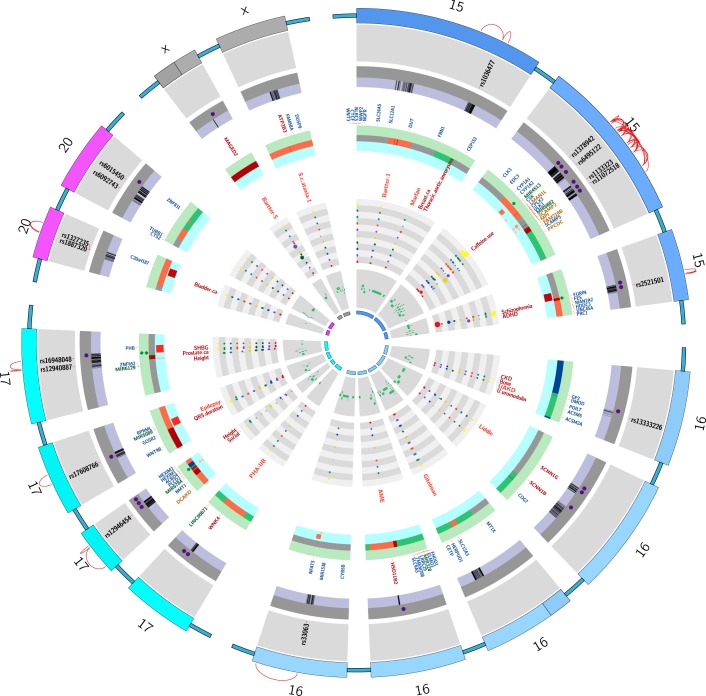
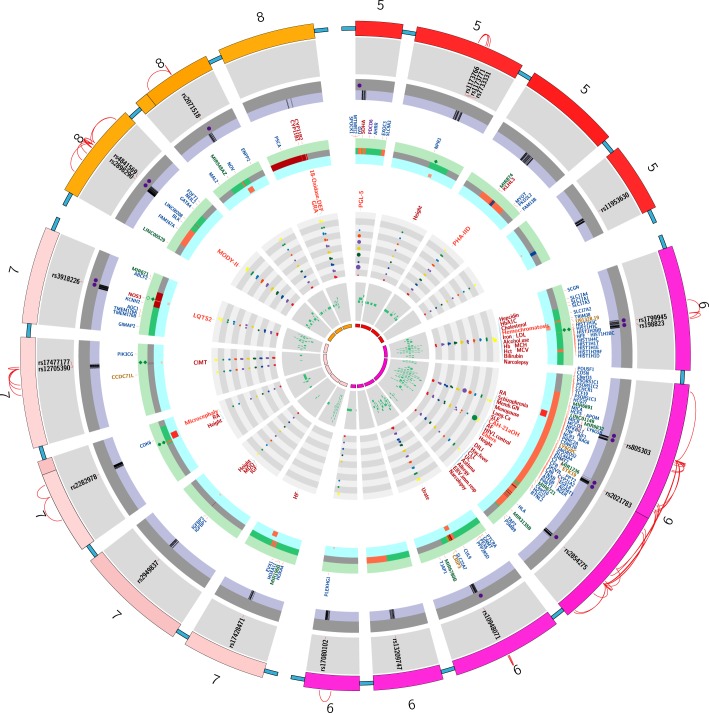
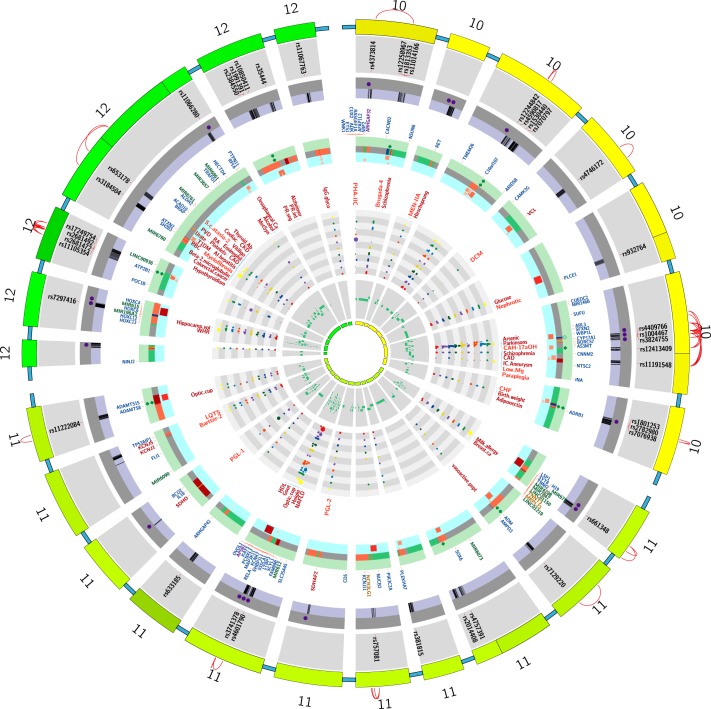
FIGURE 1.
Regions on the rat genome mapped using substitution mapping. *The letters on the x-axis are studies listed with the same alphabet in Table 2. The y-axis of each panel in this figure represents the length of a single rat chromosome. Bars represent the locations of BP QTLs. These locations were determined by searching the Ensembl data (www.ensembl.org) and the rat genome data (www.rgd.mcw.edu) for the rat genome version 6.0. In cases where the locations of the end markers were not available on the rat genome version 6.0, other closest markers or other versions of the rat database were used. Red arrows with names of genes indicate locations on congenic strains where the evidence for the actual gene accounting for the QTL is “strong.” Data with mapping ambiguity for positioning markers on the rat genome map are not featured in this diagram.
A. Cyp11b1
With the use of the mapping approach, the first locus to be deduced as a bona fide rat BP QTL was the gene Cyp11b1, which codes for both 18- and 11β-hydroxylation of adrenal steroids. The premise for this locus to be tested as a BP QTL was the observation of a striking difference in adrenal steroidogenesis between Dahl S and R rats. Compared with R rats, S rats had an increased ability to 18-hydroxylate 11-deoxycorticosterone (DOC) to form 18 hydroxy-11-deoxycorticosterone (18OH-DOC) and a reduced ability to 11β-hydroxylate DOC to form corticosterone (300–302, 304). These biochemical changes resulted in higher circulating levels of the weak mineralocorticoid 18OH-DOC in S rats (309), which was interpreted to cause increased BP especially on a high-salt diet (304). These steroid profiles were shown to be due to a single genetic locus (302) and the enzyme responsible (Cyp11b1) was identified in 1976 based on strain differences in Warburg’s partition constant [inhibition by carbon monoxide (CO)] (303). Further genetic proof was obtained through the characterization of a congenic strain constructed by introgressing ~22 centimorgans (cM) of rat chromosome 7 including the Cyp11b1 gene from the R rat into the S rat. The resultant congenic strain had a significantly lower BP and increased survival compared with the S rats (38). This locus was defined by further substitution mapping to be within 177 kb (FIGURE 1, RNO7) (105). Five S and R allelic single nucleotide polymorphisms (SNPs) were identified within the coding region of Cyp11b1, all of which were nonsynonymous substitutions (41, 238). Further specific evidence was obtained through site-directed mutagenesis and experiments conducted with artificially constructed chimeric genes of S and R rats to demonstrate that the strain-specific steroid patterns were due to the substitutions in exon 7 coding for amino acid residues 381 and 384 which probably alters the structure of the steroid binding site of the Cyb11b1 enzyme (238, 269).
Mutations in the CYP11B1 human gene are known to cause rare monogenic forms of inherited hypertension (381, 382). In a limited study of essential hypertension with 160 subjects (12), eight novel missense heterozygous mutations were identified in the CYP11B1 gene that alters the encoded amino acids: R43Q, L83S, H125R, P135S, F139L, L158P, L186V, and T196A. None of these mutations accounted for hypertension; however, in vitro testing indicated that the variants L158P and L83S severely impaired while R43Q, F139L, P135S, and T196A increased the enzymatic activity of 11β-hydroxylase, suggesting the importance of these affected residues to enzyme function (12), which may point to lack of power for detecting associations. Further evidence for the association of CYP11B1 in humans, albeit modest, was obtained through a resequencing approach in 560 individuals with extreme systolic BP belonging to the GenNet cohort with European American and African American ancestry (266). Association of CYP11B1 was detectable only after pooling all coding and noncoding variants at evolutionarily conserved sites (266). An interesting relationship between CYP11B1 and the neighboring gene CYP11B2, encoding aldosterone synthase, was discovered by haplotypic analysis. The pattern of variation across the entire CYP11B locus was determined by sequencing 26 normotensive subjects homozygous for the −344 and intron conversion variants within CYP11B2. Four common haplotypes with 83 variants associated with −344 and intron conversion were identified confirming strong linkage disequilibrium across the region. Two novel CYP11B1 polymorphisms upstream of the coding region (−1889 G/T and −1859 A/G) were identified as contributing to the common haplotypes. Hypertensive subjects (n = 512) from the British Genetics of Hypertension Study population were genotyped for these polymorphisms, and the study strongly suggested that the impaired 11β-hydroxylase efficiency associated previously with the CYP11B2 −344 and intron conversion variants was due to linkage with these polymorphisms in CYP11B1 (12). Overall, the data from all these studies points to both coding and noncoding variants of CYP11B1 as imparting a modest but significant effect (average allelic effect <1 mmHg, P = 0.005), on BP regulation via regulation of steroidogenesis in humans.
V. BEYOND HIGH-RESOLUTION MAPPING: POSITIONAL CLONING OF INHERITED LOCI FOR BLOOD PRESSURE REGULATION
Substitution mapping using congenic strains has been highly successful to resolve BP QTLs from large intervals of a few megabases to, in some cases, very short intervals encompassing less than a megabase or in some cases, a few kilobases (104, 105, 120, 122, 268, 318). However, there are hundreds of variations presenting as candidate quantitative trait nucleotides within each of these highly resolved loci. Further substitution mapping alone is highly unlikely to uncover the precise genetic elements imparting the change in BP because substitution mapping relies on naturally occurring meiotic recombinations, the frequency of occurrence of which is inversely proportional to the size of the genomic segment. In other words, the shorter the congenic interval, the rarer it is for recombinations to occur within that interval. Given this impediment, other complimentary approaches were needed to move the field forward from mapping to positional cloning of BP QTLs.
VI. IDENTIFICATION OF MAPPED BP QTLs WITH COMPLIMENTARY APPROACHES
A. Transcriptome Analysis
Soon after the sequencing of the human genome, prompted by the availability of the microarray technology for quantitatively evaluating gene expression on a large scale, transcriptomic analyses were superimposed with the substitution mapping approach to assess whether candidate genes with QTLs were differentially expressed between a parental strain and a congenic strain. Although this method did not directly test the genomic variation within the QTL segment causing the BP effect, the hypothesis for such studies was that variation within the QTL segment within regulatory regions such as promoters or enhancers could influence the level of expression of a BP QTL gene. The contemplation of applying this technology was prompted by the application of such studies at that time to studies on yeast mutants (349), in mice (79, 175), and in rats (3). With the use of the combinatorial approach of mapping and microarrays, complement factor was identified as a susceptibility locus in a murine model of allergic asthma, Cr2 as a susceptibility locus for murine systemic lupus erythematosus, and Cd36 as a locus causing defective fatty acid and glucose metabolism in the SHR (3, 79, 175).
One of the first genes identified using a combinatorial approach of mapping and microarray analyses is a fatty acid transporter called Cd36. Aitman et al. (3) were mapping genes for metabolic syndrome using the SHR rat. They used congenic strains with genomic segments introgressed from the normotensive Brown Norway rat into the SHR background and observed partially reduced insulin resistance of the SHR. The QTL interval affecting glucose and fatty acid metabolism was thereby located to a genomic segment spanning 36 cM on rat chromosome 4 with Cd36 as a candidate gene at the peak of the QTL (3). The SHR rat had lower expression of Cd36 and a functional fatty acid transporter deficiency in both adipose tissue and heart. The lower expression of Cd36 in SHR rats was traced to a genomic deletion within the 3′-untranslated region, the only region represented on the microarray. Congenic substitution of chromosome 4 (including Cd36) from the Brown Norway (BN) rat onto the SHR also caused reductions in BP and ameliorated dietary-induced glucose intolerance, hyperinsulinemia, and hypertriglyceridemia. These results demonstrated that a single chromosome region could influence a broad spectrum of cardiovascular risk factors including hypertension and metabolic syndrome. The interpretation that Cd36 was a genetic determinant of BP was thwarted by the analysis of Cd36 genotypes in the stroke-prone SHR strain, which, despite being hypertensive, did not inherit the deletion variant of Cd36 carried by the SHR. So, for a while, it was inferred that the deletion polymorphism of Cd36 was not important to the hypertensive phenotype of the SHR (294).
Later however, definitive proof for Cd36 as a BP QTL was obtained by focusing on the renal expression of Cd36. With the use of an integrated renal whole transcriptome profiling experiment coupled with linkage analysis in a BXH/HXB panel of rat recombinant inbred strains developed from SHR and BN rats, Cd36 was identified as a potential expression QTL (eQTL) linked to BP (156). This evidence was strengthened by the observation that the renal quantitative expression of Cd36 correlated inversely with arterial BP in these recombinant inbred strains (286). Furthermore, another SHR.BN congenic strain (SHR-Chr.4a subline) with a shorter introgressed segment carrying the wild-type allele for Cd36 also demonstrated a decrease in BP (FIGURE 1, RNO4) (286). To investigate whether selective lack of wild-type Cd36 in the kidney is sufficient to promote increased BP, renal transplantation experiments were conducted using donor kidneys from either the SHR progenitor that lacks wild-type Cd36 or from a SHR-TG19 transgenic strain with robust renal expression of wild-type Cd36 into recipient SHR.BN congenic rats. Rats receiving kidneys from SHR (with mutant Cd36) had significantly higher BP compared with rats that received kidneys from SHR-TG19 transgenic strain (expressing wild-type Cd36) (286). These data provided compelling evidence for Cd36 as a genetic determinant of BP. This conclusion with rat models is also supported with data from Cd36 knockout mice, which develop hypertension (188). It is to be noted that humans with CD36 deficiency exist, with prevalence of 2–3% reported in Asian and African populations (143, 209). BP of Japanese individuals with CD36 deficiency was reported to be elevated compared with BP in age-matched controls (250). SNPs of CD36 are also reported to be associated with essential hypertension (222) and ischemic stroke (404) among Chinese populations.
CD36 is referred to as a scavenger receptor, but has been demonstrated to transport fatty acids and facilitate the uptake of long-chain fatty acids and oxidized lipids (43, 182, 210). Although the findings from molecular genetic studies clearly demonstrate that a primary defect in fatty acid transport can promote disordered carbohydrate metabolism in the SHR (291) and evidence is mounting (36, 117) for the involvement of CD36 in metabolic dysfunction via signaling pathways such as the c-Jun N-terminal kinase (JNK) activation and Toll-like receptors (182), association of CD36 with endothelial dysfunction (335), and the recognition of Cd36 as a multifunctional immune-metabolic receptor with many ligands (2), the critical question of the precise molecular mechanism impacted by CD36 in the kidney, heart, or blood vessels, to regulate BP, remains unknown.
B. Targeted Gene-Editing Approaches
In 2003, a set of guidelines was suggested (1) to confirm candidates within QTLs beyond the fine-mapping stage to be recognized as bona fide quantitative trait genes (QTGs). Mentioned among these guidelines is the use of knockout and knock-in models (1). While knockout (or null alleles) of a candidate QTL can be used for deficiency-complementation testing, knock-ins can serve the purpose of direct testing of replacement of one allele with another at the candidate QTL to alter BP. Prior to 2009, on one hand, fine-mapping of BP QTLs was steadily progressing, but on the other hand, specific gene-targeting technology was lagging in the rat (162). In 2009, a major breakthrough was achieved by the creation of the world’s first targeted knockout rat using zinc-finger nucleases (115). This discovery paved the way for the assessment of several prioritized genes within fine-mapped BP QTLs to be further examined for their effect on BP in targeted gene deletion models. Although not directly a candidate gene for a BP QTL, as a proof-of-principle for this method to work, renin, a major gene involved in BP regulation, was targeted using the zinc-finger nuclease system and the resultant Ren−/− rats, which had a 10-bp deletion in exon 5, resulting in a frameshift mutation, were demonstrated to have almost 50 mmHg lower BP compared with the heterozygous Ren+/− rats (256).
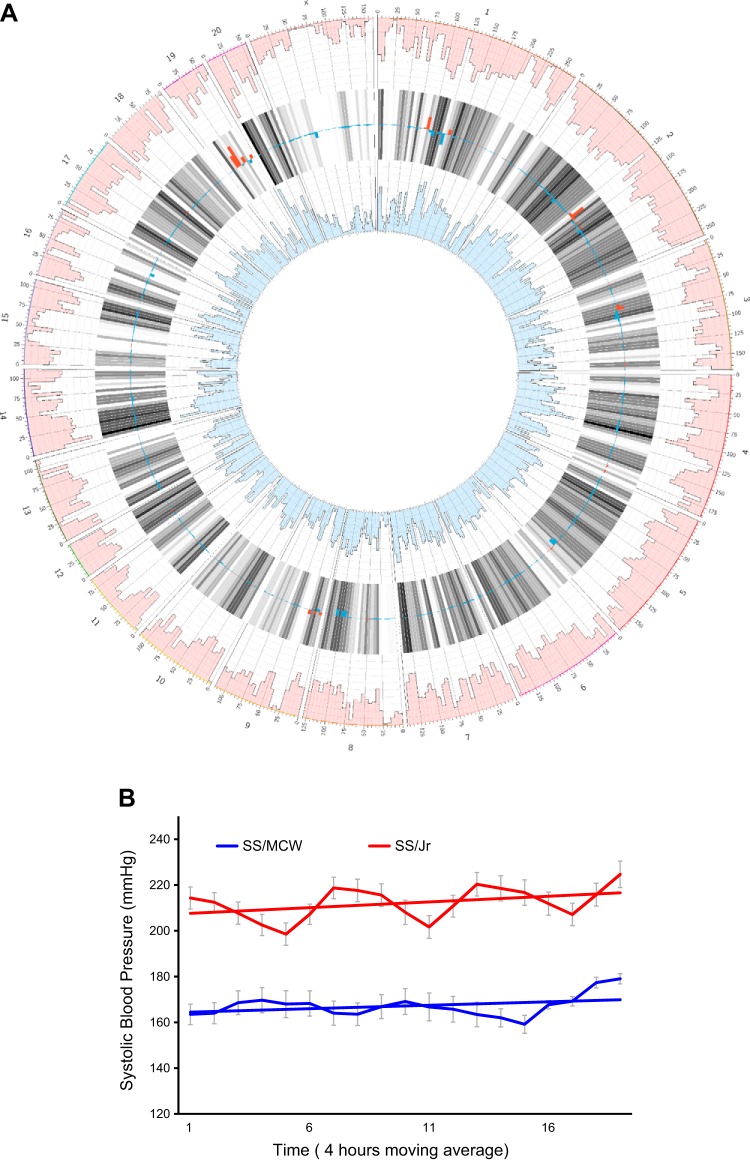
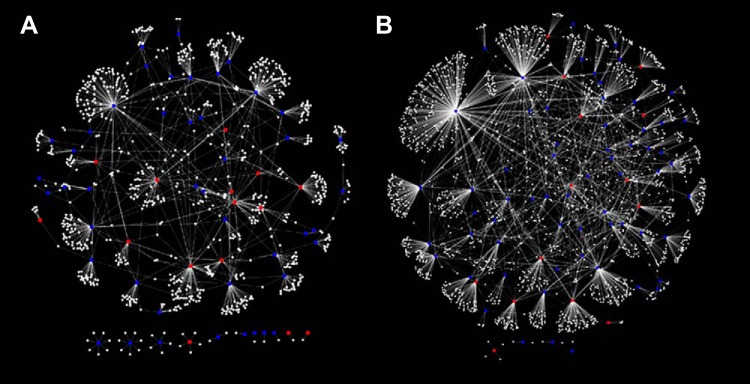
These studies were conducted by researchers in the Medical College of Wisconsin using the SS/MCW rat. Since then, it is important to point out that the Medical College of Wisconsin led a program for creating knockout genes on the background of the SS/MCW rat. The SS/MCW rat originated from the Dahl S rat and was rederived from (genetically contaminated) SS/JrHsd rats originally obtained as a congenic control strain from Dr. T. Kurtz (University of California-San Francisco) in 1991 (257). At the genomic level, SS/MCW is different from the original colony of inbred Dahl SS/Jr rats maintained at the University of Toledo by ~1,353,492 base pairs (FIGURE 2A obtained from the Rat Genome Database, www.rgd.mcw.edu). To assess whether these genetic differences had any effect on BP, BP readings from SS/MCW and SS/Jr rats were both measured under identical housing and dietary conditions in experiments conducted at the University of Toledo. The data presented in FIGURE 2B illustrate that these genetic differences cause the systolic BP of SS/MCW rat to be 40 mmHg lower than that of the authentic inbred SS/Jr rat. The situation is particularly unfortunate since in general larger effects of any genetic or environmental manipulations to alter BP in the rat are best accomplished on a very permissive genetic background. The SS/MCW rat is relatively less permissive compared with the SS/Jr based on its BP level. So obviously one has to be careful about choosing “control” strains for genetically engineered SS rats. To be specific, a targeted gene-edited model (disruption, knockout, or knock-in strain) made in the SS/MCW rat should not be compared with SS/Jr rats but only to SS/MCW and a targeted gene-edited model made in the SS/Jr rat should not be compared with SS/MCW rats.
FIGURE 2.
A: Circos plots of genomic variant densities between SS/Jr (or S/Jr) and SS/MCW strains of genetically hypertensive rat strains. The outermost ring of numbers 1–20 and X indicate rat chromosomes. The numbers beneath the outer ring that are labels of tick marks represent locations on each chromosome in megabases. The pink-colored outer circumference of Manhattan plots represents the histogram of variants of the SS/MCW strain compared with the rat reference sequence from Brown Norway rat. The blue innermost circumference of Manhattan plots is the histogram of variants of the SS/Jr strain compared with the rat reference sequence from Brown Norway rat. The histograms consist of 10 levels wherein each level represents 2,500 variants. The gray bars in between the 2 histograms (pink and blue) show the average of the 2 densities with darker bars for higher densities. The key data with regard to SS/MCW and SS/Jr strains in this plot are the overlaid red and blue bars over the gray bars. The density of variants is overlaid on the gray average bars with red bars for higher SS/MCW density and blue bars for higher SS/Jr density. The range of density difference from the bottom to the top of a gray bar is minus 12,500 to plus 12,500 variants. This Circos plot was drawn and based on the requested analysis conducted by the Rat Genome Database (www.rgd.mcw.edu). Comparisons of blood pressure (BP) readings from SS/MCW are lower than that of the SS/Jr rats (see FIGURE 2B). Thus this genetic difference could be one of the reasons for this observed relatively lower BP reported for the SS/MCW strain compared with the BP of the SS/Jr strain. B: radiotelemetry measurements of systolic BP of male SS/Mcw and SS/Jr rats. Studies were conducted as per IACUC approved protocols at the University of Toledo. Rats were weaned at 28–30 days of age and fed a low-salt (0.3% NaCl) Harlan Teklad diet. At 40–42 days of age, all rats were placed on a 2% NaCl diet and maintained on this diet for 24 days. While on the high-salt diet, rats were surgically implanted with C40 BP radiotelemetry transmitters. Their BP was monitored on day 25 post the high-salt diet regimen by radiotelemetry. The average systolic BP data plotted in this graph were collected from 4 independent BP studies for SS/MCW (total n = 30) rats and 3 independent BP studies from SS/Jr rats (total n = 21). Data points are 4-h moving averages ± SE. The straight lines through the BP data are trend lines.
1. A disintegrin-like metalloproteinase with thrombospondin motifs 16 (Adamts16)
The first reported application of the use of the ZFN method for tracking down a BP QTL candidate gene was for the gene Adamts16 (118). Adamts16 was a primary candidate gene within a BP QTL located on rat chromosome 1. The QTL was initially detected through a linkage analysis of an F2 population derived from SS/Jr (or S) and normotensive LEW rats (101). By substitution mapping, linkage was confirmed and demonstrated to be due to three independent BP QTLs named QTL1a, QTL1b, and QTL2 (170, 319) (FIGURE 1, RNO1). QTL2 was further mapped in multiple iterations and located within 804.6 kb. The primary congenic strain [labeled as S.LEW (D1Mco4) in Ref. 319] had a BP lowering effect of −30 mmHg and the final two congenic strains (labeled as D1Mco4x1x3Bx1 and D1Mco4x1x3Bx2), both spanning 804.6 kb, had BP lowering effects of −14 and −18 mmHg. These strains contained two genes, one of which was Adamts16, which harbored two nonsynonymous variants and five synonymous variants (170). Adamts16 was therefore prioritized as a candidate BP QTL and tested using the ZFN gene-targeting approach. The Adamts16 gene-edited S rat contained a 17-bp deletion in the first exon, which introduced a stop codon in the transcript. The BP of this Adamts16−/− S rat was significantly lower than that of the S rat, providing additional evidence for the involvement of Adamts16 in BP regulation (118). Other physiological observations pointed to the vasculature as a potential site of action of Adamts16 to lower BP. Adamts16−/− rats exhibited significantly lower aortic pulse wave velocity and vascular media thickness compared with S rats. The mechanosensory cilia of vascular endothelial cells from the Adamts16−/− rats were longer than that of the S rats. Furthermore, Adamts16−/− rats survived longer when compared with the S rats. Further studies will be required for a deeper molecular mechanistic understanding of how Adamts16 impacts BP. The translational significance of this locus as a BP QTL was indicated by the detection of linkage to BP in the Quebec Family Study of a region on human chromosome 5 encompassing ADAMTS16 under the QTL LOD plot (170). While ADAMTS16 was only one of several candidate genes in the linkage analysis, specific SNPs (rs2086310 in particular) within ADAMTS16 were tested and confirmed for association with BP not only in the Quebec Family Study, but also replicated in another cohort, GenNet (170).
2. p67phox, a cytosolic subunit of NAD(P)H oxidase
Through mapping experiments conducted using the Dahl SS/MCW and BN rats, four independent genomic regions ranging from 4.5 to 16 Mega basepairs (Mb) were identified on chromosome 13 (257). Each of these BP QTL regions significantly lowered salt sensitivity in the SS/MCW rat (TABLE 2 and FIGURE 1, RNO13) (46). The largest effect among these QTLs was found in a region that overlapped with the original renin gene region first mapped by Rapp (311). The largest region however, spanning 16 Mb, was mapped using a congenic strain SS.13BN26, which was reported to be mapped to 12.2 Mb containing p67phox, a cytosolic subunit of NAD(P)H oxidase, as a candidate gene. The mRNA expression of p67phox was significantly higher in the renal outer medulla of SS/MCW rats compared with SS.13BN26 congenic rats (89). SS/MCW rats had a 204 bp deletion and 4 SNPs within the promoter region of p67phox and the promoter activities tested through transfection and luciferase reporter studies in immortalized rat medullary thick ascending limb cells further supported these genetic variances in the promoter region to contribute to higher mRNA expression of p67phox in the SS/MCW compared with SS.13BN26 (89). To further examine the functional relevance of p67phox in salt-sensitive hypertension, a ZFN-based p67phox null mutant (p67phox−/−) rat was generated and tested for BP along with the SS/MCW rat. The p67phox−/− rats had a 5 bp deletion in their genomic sequence. p67phox being a subunit of NADPH oxidase, an enzyme critical for macrophage respiratory burst activity, macrophage activation in response to phorbol myristate acetate was examined. Respiratory burst activity was completely abolished in the p67phox null mutant (p67phox−/−) rat compared with controls. Also, salt-sensitive hypertension renal oxidative stress and renal glomerular injury were significantly attenuated in the p67phox null mutant (p67phox−/−) rat compared with SS/MCW rats (89). These phenotypes were associated with a significantly lower H2O2 concentrations in the medullary interstitial fluid of p67phox null mutant (p67phox−/−) rats. Collectively, authors interpreted these data to suggest that p67phox plays a crucial role in the development of salt-sensitive hypertension, which at least in part is due to its effects noted in the kidney. From a mapping standpoint, the region containing p67phox is large and cannot be ignored for the presence of other genetic determinants of BP, but the data on p67phox are compelling to speculate that it is indeed a genetic determinant of BP. Validation studies will be required to assess the naturally occurring polymorphisms of p67phox as the precise quantitative nucleotide variants causing alterations in BP and renal phenotypes. Authors also indicate that SNPs in human p67phox have not been found in human genetic association studies, but suggest that stratification of populations based on salt sensitivity may serve as a better design to uncover any association of p67phox with human salt-sensitive hypertension.
VII. OTHER GENES PRIORITIZED THROUGH MAPPING STUDIES
A. Pappalysin-2 (Pappa2)
Also known as pregnancy-associated plasma protein-A2, Pappa2 is a metalloproteinase in the metzincin superfamily. Pappa2 was prioritized through substitution mapping on rat chromosome 13, using SS/MCW and BN rats (46, 50, 257). The highest resolution reported within which Pappa2 is mapped is 0.71 Mb (chromosome 13, 81.01–81.72 Mb) containing two other annotations, a protein coding gene Astn1 and a microRNA miR-488 (50). BN alleles in this genomic segment lowered salt-sensitive mean arterial BP by 24 mmHg (50). Although no nonsynonymous variants were observed, eight SNPs, five insertions, and two deletion polymorphisms are reported within a 25 kb 5′ upstream sequence of Pappa2. Expression of Pappa2 was eight times higher in the renal cortex of the SS/MCW.BN congenic strain compared with S. By the observation of immuno-costaining of Pappa2 with a Na+-K+-Cl2 cotransporter (Nkcc2), Pappa2 was more precisely located in the cortical thick ascending limbs of renal proximal tubules (50). Exactly how the upregulation of Pappa2 in the congenic strain protected from elevated BP is not clear. The authors (50) point out that Pappa2 is known as a protease that can cleave Igfbp-5 (insulin-like growth factor binding protein 5) (274, 392) and thereby may regulate hydroxyapatite and insulin-like growth factor (IGF)-1 binding. Clinical associations between Pappa2, IGF-1, hypertension, and cardiovascular risk have also been reported (50, 173, 330, 385). To further support the candidacy of Pappa2, validation experiments with gene-edited models to assess the naturally occurring polymorphisms of Pappa2 will be required.
B. Secreted Phosphoprotein 2 (Spp2)
Substitution mapping of BP QTLs on rat chromosome 9 (FIGURE 1, RNO9) led to the prioritization of Spp2 as the sole gene with a nonsynonymous variation. The QTL was mapped using congenic strains developed from the introgression of SHR alleles onto the genome of the S rat, which demonstrated significant lowering of BP and proteinuria (268). A nonsynonymous G/T polymorphism was detected in the Spp2 gene between the S and S.SHR congenic rats. Interestingly, the T allele was rare, being detected from among 45 rat strains, only in substrains of SHR and WKY. Importantly, this polymorphism was potentially pleiotropic, because, in addition to improved cardiovascular and renal function, high salt fed congenic animals carrying the SHR T variant of Spp2 demonstrated a significantly lower bone mass and altered bone microarchitecture. In addition to Spp2, the mapped congenic segment contains two other genes, Arl4c and Trpm8, but neither of these genes had any nonsynonymous variants (268). The deduced inferences from the current observations on the candidacy of Spp2 are nevertheless translationally interesting because a GWAS study reports an association to systolic BP responses to cold pressor test (34, 243) and salt-sensitivity (243) on human chromosome 2q37 to a region close to SPP2 and TRPM8. An interesting feature to note is that the BP QTL containing Spp2 is mapped between a relatively salt-insensitive hypertensive rat, the SHR, and a salt-sensitive hypertensive strain, the S rat. To formally test these genes, further mapping and application of targeted gene-edited models will be required. If indeed Spp2 presents as a valid BP QTL, targeting this gene will require caution as the protective allele for BP has adverse effects on bone mass.
C. Adducin
A linkage analysis using an F2 population of Milan hypertensive and normotensive rats detected a BP QTL on rat chromosome 14 including the gene Adducin 1 (Add1) (398). Adducin is a heterodimer of α- and β-subunits that functions to promote the assembly of actin with spectrin. Milan hypertensive and normotensive rats harbor polymorphisms at the α-adducing locus wherein an A to T polymorphism results in a Y316F substitution and at the β-adducin locus wherein a G to A polymorphism results in a R529Q amino acid substitution in the Milan normotensive strain. These polymorphisms cosegregate with a significant increment in BP of the Milan hypertensive strain (16). Congenic strain analyses (FIGURE 1, RNO14) as well as human associations (14, 360) have confirmed that adducin is an important gene implicated in BP regulation. To assess mechanisms, renal function was studied. Polymorphisms of adducin affect the development of glomerular lesions by modulating the expression of podocyte proteins (90). In humans, three polymorphisms within the ADD genes [ADD1 (Gly460Trp-rs4961), ADD2 (C1797T-rs4984), and ADD3 (IVS11+386A>G-rs3731566)] are reported to influence brachial arterial diameter, distensibility, and compliance (331). Physiological interaction between the ADD1 and WNK1-NEDD4L pathways influences the effects of variants in all of these genes on BP and renal sodium handling (234). There was also a strong correlation between endogenous ouabain levels, renal sodium handling in hypertension, and α-adducin polymorphisms (232, 235). Also, adducin polymorphisms in both rats and humans are demonstrated to affect renal cellular endocytosis and Na/K pump activity (359). Much research was since conducted by the Bianchi group (81) that has resulted in clinical trial for rostafuroxin, a selective inhibitor of Src-SH2 interaction with mutant adducin for sodium handling in hypertensives (347). Unfortunately, the results were not encouraging as rostafuroxin did not reduce BP at any dose (347). Polygenic nature of hypertension is perhaps not permissive enough for a single gene effect to be promising. A personalized therapeutic approach is proposed for future approaches to results from single candidate gene analysis for hypertension (233).
VIII. PRIORITIZING CANDIDATE GENES BASED ON HUMAN GENOME-WIDE ASSOCIATION STUDIES
Technological advancements in targeted gene-editing approaches in model organisms were not only useful to validate fine-mapped rat BP loci, but were also favored as an approach to validate genes that were prioritized through genome-wide association studies (GWAS) in humans. GWAS, wherein SNPs associated with disease are detected on a genome-wide scale, are different from linkage studies in that inheritance is not a consideration, because associations are reported among unrelated individuals. Nevertheless, GWAS came into prominence as the next step after the human genome sequence was deciphered. The first genome-wide association study for hypertension was reported in 2007 by the Wellcome Trust Case Control Consortium (WTCCC) (380). The study included ∼2,000 cases of essential hypertensives and ∼3,000 controls, which, at that time, represented the single largest human association study for hypertension. Surprisingly, no major association was detected with any SNP for hypertension. However, six single-nucleotide polymorphisms (SNPs) were reported as being associated with hypertension with marginal significance (380). Comparative mapping revealed that the homologous locations of four out of these six human SNPs (rs2820037, rs6997709, rs11110912, and rs2398162 on human chromosomes 1q43, 8q24, 12q23, and 15q26) map within regions of the rat genome identified as BP quantitative trait loci (QTL) on rat chromosomes 17, 7, 7, and 1, respectively (101). It is also interesting that among the many strain comparisons that were used for mapping BP QTLs, all four regions are reported as BP QTL containing regions originally identified from a single linkage analysis between the hypertensive Dahl Salt-sensitive (S) rat and the Lewis (LEW) rat (101).
A. Nuclear Receptor 2, Factor 2 (Nr2f2)
The human 15q26 region containing the gene NR2F2 was tabulated in the supplementary data section of the first human GWAS conducted for hypertension, the WTCCC study (380). The homologous rat chromosome 1 region containing Nr2f2 was identified as a BP QTL within a 13 Mb region (169, 319) (FIGURE 1, RNO1 and TABLE 2), whereby Nr2f2 was also a positional candidate locus (one of many genes within the region) for BP control. The candidacy of NR2F2 in human hypertension was further corroborated in humans through a haplotypic analysis of the WTCCC data (26). All these different studies in both humans and rats collectively pointed to Nr2f2 as a plausible BP QTL, but fell short of providing conclusive evidence. The targeted gene-editing approach was applied to address the question of whether disruption of Nr2f2 impacts BP of the S rat (206). A ZFN-based Nr2f2 mutant rat generated for this purpose had a 5-amino acid deletion (Δ amino acid residues159–163) in the hinge region of the Nr2f2 protein. BP of the Nr2f2 mutant rats were significantly lower than that of the S rats. An enhanced binding of the mutant Nr2f2 with the transcription factor Friend of GATA 2 (Fog2) was noted. Furthermore, by chromatin immunoprecipitation, an enhanced binding of the mutant form of Nr2f2 to the promotor of atrial natriuretic factor (Anf) was observed. Anf is a direct target gene influenced by the Nr2f2-Fog2 interaction (157) and a vasorelaxant (183, 203). Mesenteric arteries from Nr2f2 mutant rats relaxed better than the S rats. Conclusions from this study were that the extent of BP is inversely linked to the extent of interaction between Nr2f2 and Fog2 through the hinge region of Nr2f2 and through this interaction, transcription of Anf, is regulated.
B. Pleckstrin Homology Domain Containing Family A Member 7 (Plekha7)
Five independent GWAS in multiple populations associated the human SNP rs381815 in intron 1 of Pleckha7 with systolic BP (93, 144, 145, 213, 220). To test this association, the rat Plekha7 gene was mutated by applying the ZFN approach in the SS/MCW rat (85). Plekha7 mutant S rats had blunted hypertension and renal disease, which the authors attribute to improvements in vascular function because in vitro assays revealed enhanced endothelium-dependent and flow-mediated dilation in the mutant vessels compared with wild-type (WT) controls, which was associated with augmented intracellular calcium release in endothelial cells and an increased bioavailability of the vasodilator nitric oxide. Plekha7 encodes an adherens 84 junction protein (295) and is a cytoplasmic protein reported to stabilize cadherins and nectins at adhesion junctions (124, 193, 195, 279, 332). More recently, PLEKHA7 is reported to regulate cellular behavior via miRNAs by associating with the microprocessor complex at the apical zonula adherens (193, 194). Through a proteomic approach, proteins associating with PLEKHA7 were identified to be cytoskeletal-related and RNA-binding proteins. Loss of PLEKHA7 activates the actin regulator cofilin by regulating the levels and associating with PP1alpha, a phosphatase responsible for cofilin activation (193, 194). Interestingly, cofilin1 is involved in hypertension-induced renal damage (370). Therefore, it is possible that Plekha7 affects hypertension-induced renal function in addition to the alterations in vascular function detailed in the Plekha7 mutant SS/MCW rat.
C. Cd247
CD247 encodes the zeta chain of the T-cell receptor. T cells are increasingly being recognized as important in the etiology of salt-sensitive hypertension (60, 134, 366, 386). A large genome-wide human linkage analysis in the GenNet Network of the Family Blood Pressure Program (FBPP) identified a highly significant linkage peak on human chromosome 1q spanning a 100 cM region (354). By genotying 1,569 SNPs within this linkage peak in 2,379 individuals, associations were found for both systolic and diastolic BP in or near 2 genes, GPA33 and CD247. In CD247, a single-nucleotide polymorphism variant in intron 1 is associated with diastolic and systolic BP in hypertensive black and European American subjects (82). Given these associations, Cd247 was tested as a candidate gene for hypertension through ZFN-mediated deletion of Cd247 in the SS/MCW rat (316). These mutant Cd247 rats had no functional T cells and demonstrated significantly lower hypertension, renal disease, and lower infiltration of T cells in to the kidneys in response to dietary high salt (316). Thus authors conclude that Cd247 is an important link between T cell biology and development of hypertension and renal disease (316).
D. Sh2b3
SH2B adaptor protein 3 also known as lymphocyte-specific adapter protein Lnk is a gene nominated as a candidate gene by GWAS of hypertension and renal disease (21, 150, 151, 213). Sh2b3 is an intracellular adaptor protein expressed in hematopoietic and endothelial cells. To test the contribution of Sh2b3 to hypertension in the Dahl SS/MCW rat, a ZFN mutant rat with a 6-bp deletion in the Sh2b3 gene was generated (317, 408). As a result, the native proline-leucine-glutamic acid sequence is replaced with a single glutamine in the Sh2b3em1Mcwi mutant rat, resulting in modification of the phosphotyrosine-peptide binding pocket of the SH2 domain. BP of the Sh2b3 mutant Dahl SS/MCW rat was attenuated. Associated with the lowering of BP was reduced renal damage and blunted infiltration of immune cells into the kidney (317). Transplanting bone marrow from Sh2b3em1Mcwi mutants to Dahl SS/MCW rats fed a high (2% NaCl) salt diet resulted in a significant decrease in mean arterial pressure and kidney injury (317). Although not proven to be an inherited factor for susceptibility to develop hypertension, studies in Sh2b3−/− mice also confirm these functions of Sh2b3, whereby Sh2b3 is recognized as a key gene implicated in the development of hypertension (58).
E. GWAS-Nominated Agtrap-Plod1 Locus
GWAS not only detect single gene associations, but also present with associations to multiple SNPs clustered by proximity within multiple genes in linkage disequilibrium, thus depriving clarity on which of the closely linked genes are likely to be significant determinants of BP. An example of this in the case of a GWAS for BP in a genomic region on human chromosome 1:11,736,084–11947845bp, containing six genes, Agtrap, Mthfr, Clcn6, Nppa, Nppb, and Plod1, reported in 11 human studies (35, 98, 165, 171, 179, 213, 221, 264, 265, 357, 403). To overcome this limitation of GWAS and to determine the relative contributions of each of these genes to BP regulation, ZFN-based mutant rats were individually constructed and characterized on the Dahl S background (92). The results demonstrated that five of the six genes influence BP and/or renal function (92), indicating that multiple genes within a short genomic segment can individually modulate BP.
IX. HAS RAT GENETICS ADVANCED THE SEARCH FOR INHERITANCE OF HYPERTENSION IN HUMANS?
Taking a broad perspective, it is well to recognize that the fact that rats can be bred for hypertension and for susceptibility to salt-induced hypertension is a profound statement of what phenomena are also likely in human hypertension. While this is easily demonstrated in rats because of the freedom to breed and change the environment for the life of the rat, similar manipulation in humans is impossible. And of course the genetic and salt susceptibility data available from humans are compatible with what is unequivocal in rats. At the time the initial work in rats was done, such results were far from given.
This article began with documenting the quest for inherited factors for the development of hypertension and then cataloged a large number of BP QTLs on almost every chromosome of the rat. These BP QTLs have yielded only a handful of genes that have varied tiers of experimental proof to suggest that inheritance of allelic forms of these protein-coding genes impact BP. In other words, these genes are putative BP quantitative trait genes in rats and perhaps in humans as well. The number of loci mentioned above is relatively small and by no means representative of the entire blueprint of the genetic/inherited factors responsible for BP regulation. What are we missing? Observations made during the course of the mapping studies that are beyond the discovery of SNPs within protein-coding genes are worth noting as they support the reasoning that much remains to be discovered. These observations are listed below.
The numbers of BP QTLs identified in either rats or humans far exceeds the number of causative genes identified in these QTLs to date (listed in TABLE 1).
Variants within protein-coding genes are not the sole variants present within positionally mapped BP QTLs. There are a larger number of variants within any mapped segment that are outside of protein-coding genes or other annotations. These may have unknown effects.
Further mapping of BP QTLs that were originally identified as single LOD peaks have demonstrated that underneath each of these single LOD peaks is a cluster of independently operating BP QTLs.
Many BP QTLs have also been detected which do not operate independently, but require allelic interactions with other QTLs to impart a change in BP. These observations are mainly in rat genetic studies and provide important clues to the genomic architecture governing the pathophysiology of hypertension in humans. They suggest that inheritance of the level of BP of an individual attributed to major effects of a small number of genes is perhaps a gross underestimation.
Ideally the discovery of new genetic inputs to BP control in the rat would lead to new therapies for hypertension in humans. The fact that the work on Adducin did not result in a successful treatment for hypertension (347) does not abrogate the desirability for further work to truly understand the complex genetics of hypertension. In addition, if the same result turns out to be true for all the genes found to be causative for hypertension in the rat on a very permissive (artificially constructed by selective breeding) genetic background, then we need to know that therapeutic targeting of singe genes on a nonpermissive outbred genetic background is an unworkable approach. If that turns out to be true for the genes found using the rat models, then the same situation is likely to be true for any causative genes for hypertension discovered by GWAS in humans.
X. EPISTASIS OF BP QTLs
Epistasis or gene-gene interactions occur when the effect of alleles at one locus depends on the alleles at another locus. In the context of BP studied in rats, such interactions can be discovered in segregating populations by comparing the effect on BP of genotypes at one marker versus the effect on BP of genotypes at another marker. This is done by a two-way factorial analysis of variance and finding a significant interaction term. Note that in an F2 population this analysis will include all three possible genotypes at each marker.
In working with congenic strains for BP QTL on the background of a given strain, epistasis can be defined as the nonadditive effect (positive or negative) when combining two congenic strains into a new double congenic strain. In this case it is important to appreciate that only homozygous genotypes are being compared at the two QTLs involved. The BP of four strains (the background strain, two congenic strains, and the double congenic strain) is required. The analysis of BP again involves a two-way factorial analysis of variance on the genotypes at the QTLs being tested (306). Epistasis is present if this analysis yields a significant interaction term.
One of the early reports of such an epistasis between two BP QTLs was weak statistical evidence for epistasis between two BP QTLs on rat chromosomes 2 and 10 (69). Subsequently, separate congenic strains were constructed on the S background for the appropriate regions of Chr 2 (D2Mit5-D2Mit8) or 10 (D10Mco1-D10Mco6) introgressed from a normotensive strain (66, 73). Evidence for individual effects of these two segments was obtained by the observation that each congenic strain had lower BP than the S rats. However, when a double congenic strain was constructed containing low BP QTL alleles from both chromosomes 2 and 10 and tested for BP, the sum effect of the two regions was smaller than the additive effects of decreasing BP relative to S. This provided definitive evidence for a strong epistatic interaction on BP of the QTL on chromosomes 2 and 10. At that time, this was the only documentation of an interaction on a quantitative trait in a mammalian system (306). Unfortunately, the identities of the two genes in epistasis remain unknown as there are no further reports of mapping of the QTLs involved. Epistasis has been reported for a number of other BP QTLs (29–31, 33, 39, 63, 77, 187, 253, 277, 284, 326, 355). Furthermore, a theoretical modeling study of epistasis of BP QTLs through analysis of data obtained by substitution mapping using congenic rats is reported by Rapp (299). In the case of human hypertension, detections of epistatic relationships are largely limited with the current approach of querying for individual loci that are associated with BP.
In working with Dahl rats in particular, the usual practice is to make congenic strains introgressing QTL segments of chromosome from the normotensive strain into the S-rat genetic background. There is a good reason for this. It is simply that in early work the S-rat genetic background was found to be permissive for finding genetic effects but the R-rat genetic background was not (40, 41, 310). The same is true for the LEW rat background. In this case the LEW background has been studied extensively using congenic strains, by introgressing S rat QTL chromosomal segments (associated with increasing BP) on the LEW background. No or minimal effects on BP were observed (30, 51, 52) even when multiple S rat QTL chromosomal segments were introgressed into the same congenic strain (52). Additional work suggests that a segment on rat chromosome 18 may be important in maintaining low BP in the LEW rat (52). The result of this is that one should not expect the relatively large observable effects of introgressed chromosomal segments on the S genetic background to translate into effects on other genetic backgrounds. Such a profound influence of genetic background is presumably due to epistatic effects.
An attempt has been made to place interacting pairs of QTLs into two “epistatic modules” wherein pairs of QTLs within a module show epistatic effects but pairs of QTLs from different modules show only additive effects (32). Whether such modules are actually a valid concept has been questioned on the basis of biased sampling and lack of a statistical analysis to establish their existence (307). Such objections could be overcome by the systematic collection of further data, but for now the concept, albeit interesting, remains to be adequately supported.
There are some interesting effects of epistasis on dominance in comparing Dahl S rats to LEW rats. An F1 cross between S and LEW has a BP equal to LEW (52), that is, the LEW phenotype is dominant. Interestingly, the LEW alleles at 9 out of 10 BP QTL when studied individually on the S background were dominant to the S alleles (74). This strong dominance is lost in an F2 population derived from S and LEW (101), suggesting that dominance of LEW QTL alleles is highly dependent on genetic background.
When repeated backcrosses of LEW were made to S, it took three backcrosses (after making the initial F1 cross) for the BP of the backcross population to increase only modestly (52), and it was still well below that of S rats. After backcross 3, the population would be expected to have only 6.25% of LEW alleles. This again demonstrates the powerful effect of the LEW background to lower BP. This is surprising because the inbred LEW rats used were never selectively bred for low BP.
The focus on genetic interactions between pairs of QTL is an oversimplification because: 1) a given gene may interact pairwise with several other genes; and 2) higher order interactions exist (128, 353) but have not been addressed in hypertensive rats. A study on higher order QTL interactions involving insulin-like growth factor-1 in mice (128) is especially interesting because the three-locus and four-locus interactions were considerably larger than the component two-locus interactions. One can speculate that S rats might accumulate QTL alleles leading to higher-order interactions increasing BP during selective breeding while the LEW rat would not accumulate (by chance) QTL alleles leading to higher-order interactions decreasing BP because inbred LEW rats were never selectively bred for BP. Thus substitution of a LEW allele on the S background at a QTL that was part of a haplotype creating a higher-order interaction would be expected to disrupt the higher-order interaction and show a disproportionately large effect. In contrast, the reciprocal substitution of an S allele on the LEW background at one of the QTLs in such a complex would not disrupt a higher order interaction that did not exist and would, therefore, be expected to have a lesser effect.
It is anticipated that the genes underlying BP QTL exist in metabolic pathways which eventually impinge on BP and that any given pair is either in series or in parallel. A model for pairwise QTL interactions using congenic strains has been constructed assuming the QTL act like switches that can be either on or off (299). The switches themselves are not imaginary, but are based on the basic biochemical construct of binding curves between two molecules of any type. The binding curves (and therefore the switches) have allelic and metabolic inputs.
FIGURE 3 gives actual data from the literature showing epistatic patterns. It is often convenient in this work to express the effect of a congenic strain as the deviation from the S strain. Thus S rat BP is always at zero, and if a congenic strain decreases (or increases) BP compared with S, the deviation is negative (or positive). This convention is used in FIGURE 3, but the reader is advised that sometimes the double congenic is used as a baseline in the literature. This results in a purely technical artifact causing what looks like positive epistasis in one frame of reference to look like negative epistasis in the other, and vice versa (299).
FIGURE 3.
Epistatis in rat models. The data in A are redrawn from Pillai et al. (284), and the data in B are redrawn from Figure 1D of Chauvet et al. (39). In each panel BP is given for two congenic strains each for a different QTL and for a double congenic strain which has the introgressed chromosomal segments from both of the original congenic strains. All congenic strains are on the Dahl S genetic background. BP of each congenic strain is expressed as a deviation from Dahl S rats which is defined as the zero point. Blue arrows on the right of the figures show the effect expected if the two QTL effects were additive, and the red arrows show the difference between expected and observed. In both panels the epistatic effects were reported to be significant (P < 0.05). The pattern in A is compatible with two QTL in parallel metabolic pathways, and the pattern in B is compatible with two QTL in series in the same metabolic pathway (299).
The data in FIGURE 3A (284) are identical to the pattern predicted if two QTL switches are in parallel, and in both congenic strains the S allelic switch (an on switch) is replaced by a LEW allelic switch (an off switch). The data in FIGURE 3B (32) are identical to the pattern predicted if two QTL switches are in series, and in both congenic strains the S allelic switch (an on switch) is replaced by a LEW allelic switch (an off switch). Other more common patterns can be simulated by assigning leak properties to the switches under certain logical constraints, but such patterns do not always result in a unique solution as to the relative position of the QTL in metabolic pathways, possibly because most of the QTL involved are not due to a single genetic factor. The point is, however, that the epistatic patterns vary and contain information that can be extracted (299).
Note in FIGURE 3A that the congenic strains are both on the same chromosome (chromosome 5) and that individually they would be undetectable. The complex was of course found when a QTL on chromosome 5 was dissected by construction of nested congenic substrains from either end of the original congenic segment, resulting in the finding that the effect of the original QTL disappeared when its components were separated (107). The structure and its epistatic effect have been confirmed and the location of the components better defined with the component QTL separated by ~2 Mb (284). If two such subcomponents, each without a BP effect of its own, were on different chromosomes, they could only be discovered by testing genetic markers for interactions throughout the genome in a segregating population. As an aside, the merits of dissecting QTL using nested or contiguous congenic strains has been discussed (308).
Another property of the QTL switches is that one of the inputs is the concentration of one of the ligands, and this of course can vary. This lends itself to inputs subject to metabolic changes induced by environmental challenges or, for example, by sex hormones. The quintessential environmental factor is of course dietary sodium chloride and the changes it induces in the renin-angiotensin-aldosterone system. Dietary salt does increase rat [Na+] in cerebrospinal fluid (153). One example of a salt-sensitive QTL that alters BP, heart rate, and renal sympathetic nerve activity in response to changes in cerebrospinal fluid [Na+] is on chromosome 10 (152). Dietary salt also markedly influences the mRNA for the strong candidate gene Pappa2 on chromosome 13 noted above (50). Suppression of aldosterone by dietary salt is also necessary for the steroid changes resulting from the Cyp11b1 polymorphism between S and R rats to be effective (304). Dietary factors other than salt are also known to influence BP in the Dahl S rat (239, 240), and sex-dependent QTL are also known in the this model (67, 138, 139).
One thing that has not been done intensively enough in work with hypertensive rats is to look systematically for two-way interactions between genetic markers throughout the genome, similar to studies in other model organisms (154, 229, 358), although the techniques used in some model organisms are not always applicable to the rat. Higher order interactions on BP in the rat also need to be sought. In the original study of an F2 population derived from S and LEW, several QTL interactions were found by testing markers against each other, but this was not extensive enough to uncover larger scale networks of interactions (101). Programs for this that would be applicable to the rat have been developed (329, 362), and one was used successfully in a time study of urinary albumin excretion (UAE) using a backcross population F1(SxSHR)xS. In this study 11 pairwise interactions were observed, most of which were age dependent (100). Viewed in the context of variable environmental and metabolic inputs into the statistical existence and presumed associated functional importance of QTL, the expression of QTL and interaction networks can be viewed as somewhat ephemeral.
Most of the QTL in the Dahl S rat that have been extensively dissected by substitution mapping using congenic strains have (as predicted) turned out to be compound (contain multiple loci harboring alleles with both positive, negative, and interactive effects on BP). Having dissected the QTL into components, we now need to understand why clusters of interacting loci influencing BP (that is, the observable QTL) in a genome scan of a segregating population exist in the first place. It is interesting that it was relatively easy to obtain Dahl S rats with hypertension by only several generations of selection (56) in spite of the fact that many genes are involved. This implies, although it seems unlikely, that haplotypes favoring high (or low) BP in relatively few chromosomal segments (equates to already known compound QTL) should exist in the outbred Sprague-Dawley rats from which Dahl rats were selected. Or, more likely, it could be that compound QTL are in a sense an artifact of the selection process, during which highly interactive alleles for increased BP (in the case of the S rat) are assembled relatively quickly by chromosomal crossover events into hypertensive haplotypes where the loci involved happen to be loosely linked. This would explain why essentially all observable BP QTL turn out to be compound QTL. The same perspective could be applied to compound QTL found in studies by crossing any selectively inbred strains for any trait.
It hardly seems likely that an essential hypertensive human is genetically assembled by the chance association of alleles each with a BP effect of about +1 mmHg at 25 or more loci (as GWAS studies would lead us to contemplate) and that this could result in a 15–20% prevalence of hypertension in human populations. In any case it appears that we are missing some fundamental genetic phenomenon in rats and humans. That “missing something” is probably our lack of an adequate understanding of the complex architecture of genetic interactions.
Unlike in rat models, genetic background is not taken into consideration as a critical factor in human studies, so candidate genes in epistasis are not detected, let alone validated in any other species. The status of human research into finding loci linked or associated with hypertension is captured in the following sections describing studies conducted in humans that encompass investigations on monogenic forms of hypertension and more recent queries on a genome-wide scale for individual loci that are associated with BP.
XI. GENETIC STUDIES OF HUMAN HYPERTENSION AND HYPOTENSION
Diseases or traits can be broadly classified as common (complex, polygenic) or simple (Mendelian, monogenic). Simple traits are attributed to dysfunction of a single gene, whereas a complex interplay of genetic and environmental factors contribute to the pathogenesis of common diseases (sometimes referred to as multifactorial inheritance). Genetic variants with large effect sizes manifest as Mendelian or single-gene disorders, accounting for only a small fraction of cases in the population and manifest in infancy and early adulthood. Mendelian disease mutations are highly penetrant and under a very strong selection, which ensures lower frequencies in populations. Tracing patterns of genetic segregation in families have been used successfully in Mendelian diseases as exemplified by discoveries using linkage analysis.
Susceptibility variants involved in common (complex) diseases/traits, unlike monogenic traits, do not have high penetrance, are not under a strong selection, and present with lower allelic heterogeneity. Nevertheless, previous selection can be a factor in hypertension, which is a modern disease and could very well be an undesirable pleiotropic effect of a preserved genotype that was perhaps optimized for fitness in the ancient environment. Compared with populations of European ancestry, susceptibility to develop hypertension is higher in people of African ancestry (27). Ancestral sodium-conserving alleles are more prevalent in people of African origin, who also demonstrate increased rates of hypertension and sodium sensitivity (262, 379, 396). These variants may have exerted significant adaptive phenotypic effects in the past, for example, in the time of low salt intake, and now under changed environmental circumstances may have become maladaptive.
The role of genetics in human BP regulation and hypertension was established through family and twin studies which showed a significantly higher risk of hypertension among subjects with one or two hypertensive parents, and a greater correlation of BP levels in monozygotic twins compared with dizygotic twins (178, 281) (see sect. II). The distribution of BP in the population shows a normal unimodal pattern which supports a complex multifactorial basis of BP regulation. GWAS are the standard methods for genetic dissection of complex traits, which is based on the common disease/common variant hypothesis stated as follows: “the genetic variants underlying complex traits occur with a relatively high frequency (>1%), have undergone little or no selection in earlier populations and are likely to date back to >100,000 yr ago” (216). The spectrum of genetic variants and their target tissues that affect blood pressure regulation are summarized in FIGURE 4.
FIGURE 4.
Mutations altering blood pressure in humans and GWAS loci linked to plausible genes. The figure shows the circulatory system and pathways that modulate blood pressure containing genes with known mutations that cause monogenic high or low blood pressure syndromes. In addition, plausible genes linked GWAS loci are shown based on multiple lines of evidence (eQTL or nonsynonymous SNPs or genome-wide 3D proximity maps) (374).
A. Monogenic Pathways of BP Regulation and Hypertension
Discovery of the monogenic Mendelian forms of hypertension has mainly been through positional cloning using large family pedigrees, with multiple members of the family showing a clear inheritance pattern. Patients with these types of disorders represent <1% of the hypertensive population and are considered to have secondary hypertension. Mutations causing monogenic hypertension are characterized by being rare with a major defect that usually disrupts a single pathway. Despite the complexity and the presence of several systems and physiological pathways that control BP, the majority of mapped monogenic hypertension syndromes are due to mutations in genes that play key roles in renal-sodium handling (228, 369), which have contributed enormously to our understanding of salt and water homeostasis and blood pressure regulation. TABLE 3 summarizes the different forms of monogenic hypertension, their key features, and causal genes.
Table 3.
Monogenic disorders of blood pressure regulation with characteristic clinical features and treatment
| Syndrome | Subtypes | Inheritance | Locus | Gene | BP | Renin | Aldosterone | Serum K+ | Catecholamines | Treatment |
|---|---|---|---|---|---|---|---|---|---|---|
| Liddle’s syndrome | MIM 177200 | AD | 16p12.2 | SCNN1B-SCNN1G | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Amiloride or triamterence. |
| Gitelman syndrome | MIM 263800 | AR | 16q13 | SLC12A3 | ↓↓ | ↑↑ | — | ↓↓ | — | Oral potassium and magnesium supplementation with adequate salt and water. |
| Bartter syndrome | Type 1 “antenatal” MIM 601678 | AR | 15q21.1 | SLC12A1 | ↓↓ | ↑↑ | ↑↑ | ↓↓ | — | Potassium supplementation and use of cyclooxygenase inhibitors, angiotensin converting enzyme (ACE) inhibitors, and potassium sparing diuretics. |
| Type 2 “antenatal” MIM 241200 | AR | 11q24.3 | KCNJ1 | |||||||
| Type 3 “antenatal” | AR | 1p36.13 | CLCNKB | |||||||
| Type 4a MIM 602522 | AR | 1p32.3 | BSND | |||||||
| Type 4b (digenic) MIM 613090 | AR | 1p36.13 | CLCNKA, CLCNKB | |||||||
| Type 5 antenatal MIM 300971 | AR | Xp11.21 | MAGED2, MAGED, BARTS5 | |||||||
| Familial hyperaldosteronism (FH) | FH type 1, or glucocorticoid remediable aldosteronism MIM 103900 | AD | 8q24.3 | CYP11B1 | ↑↑ | ↓↓ | ↑↑ | ↓ | - | Dexamethasone |
| FH type 2 MIM 605635 | AD | 7p22.3—7p22.1 | ||||||||
| FH type 3 MIM 613677 | AD | 11q24.3 | KCNJ5 | |||||||
| Apparent mineralocorticoid excess (AME) | MIM 218030 | AR | 16q22.1 | HSD11B2 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Low sodium diet and spironolactone |
| Pseudohypoaldosteronism (PHA) | PHA1A MIM 177735 | AD | 4q31.2 | NR3C2 | ↓↓ | ↑↑ | ↑↑ | ↑↑ | — | Thiazide diuretics, prostaglandin inhibitors, alkalizing agents, and potassium-binding resins. |
| PHA2B “Gordon syndrome” MIM 614491 | AD | 17q21.2 | WNK4 | ↑↑ | ↓↓ | ↑ | ↑ | — | ||
| PHA2C “Gordon syndrome” MIM 614492 | AD | 12p12.3 | WNK1 | |||||||
| PHA2D “Gordon syndrome” MIM 614495 | AD/AR | 5q31.2 | KLHL3 | |||||||
| PHA2E “Gordon syndrome” MIM 614496 | AD | 2q36.2 | CUL3 | |||||||
| Sporadic aldosterone-producing adenoma (APA), or primary aldosteronism | AD | 11q24.3 | KCNJ5 | ↑↑ | ↓↓ | ↑↑ | ↓ | — | Surgery, aldosterone anta gonists. | |
| AD | 1p31.1 | ATP1A1 | ||||||||
| AD | 3p21.3 | CACNA1D | ||||||||
| Xq28 | ATP2B3 | |||||||||
| Hypertension exacerbation in pregnancy | MIM 605115 | AD | 4q31.2 | NR3C2 | ↑↑ | ↓↓ | ↓↓ | ↓ | — | Spironolactone contraindicated; sodium chloride treatment. |
| 11β-Hydroxylase | MIM 202010 | AR | 8q21 | CYP11B1 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Glucocorticoid therapy. |
| 3β-Hydroxysteroid dehydrogenase | MIM 613890 | AR | 1p12 | HSD3B2 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Glucocorticoid therapy. |
| 17α-Hydroxylase deficiency | MIM 202110 | AR | 10q24.3 | CYP17A1 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Glucocorticoid therapy, potassium sparing diuretics. |
| 21-Hydroxylase deficiency | MIM 201910 | AR | CYP21A2 | ↑↑ | ↓↓ | ↓↓ | ↓↓ | — | Glucocorticoid therapy. | |
| Hypertension and brachydactyly syndrome | Bilginturan syndrome MIM 112410 | AD | 12p12.2 | PDE3A | ↑↑ | — | — | — | — | Possible role for PDE3 inhibition |
| Paragangliomas (PGL) | Paragangliomas 1 MIM 168000 | AD | 11q23.1 | SDHD | ↑↑ | — | — | — | ↑↑ | Surgery, adrenergic blockers (alpha-blockade followed by beta-blockade). |
| Paragangliomas 2 MIM 601650 | AD | 11q12.2 | SDHAF2 | |||||||
| Paragangliomas 3 MIM 605373 | AD | 1q23.3 | SDHC | |||||||
| Paragangliomas 4 MIM 115310 | AD | 1p36.13 | SDHB | |||||||
| Paragangliomas 5 MIM 614165 | AD | 5p15.3 | SDHA | |||||||
| von Hippel–Lindau syndrome | MIM 193300 | AD | 3p25.3 | VHL | ↑↑ | — | — | — | ↑↑ | |
| Multiple endocrine neoplasia, type IIA | MIM 171400 | AD | 10q11.2 | RET | ↑↑ | — | — | — | ↑↑ | |
| NOS3-pregnancy-induced hypertension | MIM 163729 | AD | 7q36.1 | NOS3 | ↑↑ | — | — | — | ↑↑ |
↑, Increase; ↓, decrease; —, no effect.
B. Polygenic Pathways of BP Regulation and Hypertension
Several GWAS have been conducted using BP as a quantitative trait, or by using a binary definition of hypertension. All the significant GWAS signals are summarized in TABLE 4, and additional unpublished efforts are ongoing to discover more common variants associated with BP using larger and larger population samples. The first GWAS was a case-control design from the WTCCC, published in 2007 (380). The study examined seven common complex diseases using 2,000 cases each and 3,000 shared controls. The study genotyped ~500,000 SNPs using the 500 K Affymetrix SNP chip and reported a total of 24 significant disease-SNP association signals (P < 5.0 × 10−7). Hypertension was the only trait without any significant association signal across the genome. Following this a series of large collaborative GWAS (95, 160, 180, 213, 214, 227, 276, 321, 367) with increasing sample sizes and in different ancestries resulted in the discovery of several common SNPs associated with BP and hypertension. The largest of these was the International Consortium for Blood Pressure genome-wide association studies (ICBP-GWAS) which conducted a GWAS meta-analysis for systolic and diastolic BP in >69,899 European individuals, followed by validation in 132,000 individuals (160). The study identified 29 independent SNPs at 28 loci. Although the majority of SNPs identified by ICBP were intragenic, some loci were in gene desert regions or in genomic regions that has no gene encoding protein with a biological plausible effect on BP.
Table 4.
Summary of all GWAS results for BP and hypertension in different ancestries
| Coded Allele Frequency |
BP Effect |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP | Genotype | Coded Allele | European | Asian | African | European | Asian | African | Nearest Gene(s) |
| 1p36.2 | rs880315 | C/T | C | 0.35 | 0.59 | 0.16 | ↑ | ↑ | CASZ1 | |
| 1p36.22 | rs17367504 | A/G | G | 0.17 | 0.10 | 0.06 | ↓ | ↓ | MTHFR, CLCN6, NPPA, NPPB | |
| rs5068 | C/T | C | 0.07 | 0.00 | 0.01 | ↓ | ||||
| 1p13.2 | rs2932538 | C/T | C | 0.73 | 0.80 | 0.85 | ↓ | SLC16A1, CAPZA1, ST7L, MOV10 | ||
| rs17030613 | A/C | C | 0.19 | 0.45 | 0.05 | ↑ | ||||
| rs10745332 | A/G | A | 0.74 | 0.81 | 0.77 | ↑ | ||||
| 1q32.1 | rs2169137 | C/G | G | 0.74 | 0.94 | 0.80 | ↑ | MDM4 | ||
| 1q42.2 | rs2004776 | A/G | A | 0.26 | 0.67 | 0.54 | ↑ | AGT | ||
| 2p23.2 | rs1275988 | A/G | A | 0.60 | 0.23 | 0.08 | ↓ | KCNK3 | ||
| 2q11.2 | rs7599598 | A/G | A | 0.57 | 0.63 | 0.09 | ↓ | FER1L5 | ||
| 2q24.3 | rs1446468 | A/G | A | 0.53 | 0.47 | 0.96 | ↓ | FIGN | ||
| rs13002573 | A/G | G | 0.25 | 0.40 | 0.11 | ↓ | FIGN | |||
| rs16849225 | C/T | C | 0.75 | 0.59 | 0.94 | ↑ | FIGN | |||
| rs6749447 | G/T | G | 0.28 | 0.72 | 0.58 | ↑ | STK39 | |||
| 2q32.1 | rs16823124 | A/G | A | 0.23 | 0.51 | 0.10 | ↑ | PDE1A | ||
| 3p25.3 | rs347591 | G/T | G | 0.33 | 0.23 | 0.51 | ↓ | HRH1-ATG7 | ||
| 3p24.1 | rs13082711 | C/T | T | 0.80 | 0.94 | 0.96 | ↓ | SLC4A | ||
| rs820430 | C/T | T | 0.64 | 0.40 | 1.00 | ↑ | ||||
| 3p22.1 | rs9815354 | A/G/T | A | 0.23 | 0.12 | 0.17 | ↑ | ↑ | ULK4 | |
| rs3774372 | C/T | T | 0.77 | 0.87 | 0.81 | ↓ | ||||
| rs1717027 | C/T | T | 0.22 | 0.12 | 0.66 | ↑ | ||||
| 3p21.31 | rs319690 | A/G | A | 0.51 | 0.75 | 0.41 | ↑ | MAP4 | ||
| rs7651237 | A/G | G | 0.64 | 0.94 | 0.88 | ↑ | ||||
| 3p21.1 | rs9810888 | G/T | G | 0.53 | 0.59 | 0.46 | ↑ | CACNA1D | ||
| 3q26.1 | rs16833934 | A/G | G | 0.37 | 0.17 | 0.65 | ↓ | MIR1263 | ||
| 3q26.2 | rs419076 | G/T | T | 0.48 | 0.13 | 0.57 | ↑ | MECOM | ||
| 4q12 | rs871606 | A/G | A | 0.87 | 0.78 | 0.76 | ↑ | ↑ | CHIC2 | |
| 4q21.21 | rs16998073 | A/T | T | 0.19 | 0.30 | 0.05 | ↑ | ↑ | FGF5 | |
| rs1458038 | A/G | A | 0.27 | 0.34 | 0.05 | ↑ | ||||
| 4q24 | rs13107325 | A/C/T | T | 0.10 | 0.00 | 0.00 | ↓ | SLC39A8 | ||
| 4q25 | rs6825911 | C/T | C | 0.20 | 0.48 | 0.54 | ↑ | ENPEP, PITX2 | ||
| 4q32.1 | rs13139571 | A/C | C | 0.74 | 0.68 | 0.88 | ↑ | GUCY1A3-GUCY1B3 | ||
| 5p13.3 | rs1173771 | C/T | C | 0.51 | 0.57 | 0.81 | ↑ | NPR3-C5orf23 | ||
| rs7733331 | C/T | T | 0.50 | 0.42 | 0.42 | ↓ | ||||
| rs1173766 | C/T | C | 0.52 | 0.59 | 0.62 | ↑ | ||||
| 5q33.3 | rs11953630 | A/C/T | T | 0.34 | 0.06 | 0.15 | ↓ | EBF1 | ||
| 6p22.2 | rs1799945 | C/G | G | 0.18 | 0.04 | 0.00 | ↑ | ↑ | HFE | |
| rs198823 | G/T | T | 0.65 | 0.23 | 0.60 | ↓ | ||||
| 6p21.33 | rs805303 | C/T | C | 0.70 | 0.57 | 0.30 | ↑ | BAG1 | ||
| rs2021783 | C/T | C | 1.00 | 0.81 | 1.00 | ↑ | CYP21A2 | |||
| 6p21.32 | rs2854275 | G/T | T | 0.08 | 0.03 | 0.08 | ↓ | HLA-DQB1 | ||
| 6p21.1 | rs10948071 | C/T | T | 0.70 | 0.67 | 0.05 | ↓ | CRIP3 | ||
| 6q22.33 | rs13209747 | C/G/T | T | 0.45 | 0.48 | 0.12 | ↑ | ↑ | ↑ | RSPO3 |
| 6q25.1 | rs17080102 | C/G | C | 0.06 | 0.01 | 0.09 | ↓ | ↓ | ↓ | PLEKHG1 |
| 7p15.2 | rs17428471 | G/T | T | 0.08 | 0.05 | 0.14 | ↑ | ↑ | ↑ | EVX1-HOXA |
| 7p12.3 | rs2949837 | A/T | A | 0.23 | 0.61 | 0.00 | ↑ | IGFBP3 | ||
| 7q21.2 | rs2282978 | C/T | C | 0.36 | 0.06 | 0.43 | ↑ | CDK6 | ||
| 7q22.3 | rs17477177 | C/T | T | 0.72 | 0.91 | 0.93 | ↓ | PIK3CG | ||
| rs12705390 | A/G | G | 0.72 | 0.91 | 0.93 | ↓ | ||||
| 7q36.1 | rs3918226 | C/T | T | 0.10 | 0.00 | 0.00 | ↑ | NOS3 | ||
| 8p23.1 | rs4841569 | A/G | G | 0.57 | 1.00 | 0.89 | ↑ | BLK-GATA4 | ||
| rs2898290 | C/T | C | 0.58 | 0.02 | 0.55 | NR | ||||
| 8q24.12 | rs2071518 | C/T | T | 0.20 | 0.19 | 0.60 | ↑ | NOV | ||
| 10p12.31 | rs11014166 | A/T | A | 0.63 | 0.97 | 0.89 | ↑ | ↓ | CACNB2 | |
| rs1813353 | A/G | A | 0.65 | 0.92 | 0.85 | ↑ | ||||
| rs4373814 | C/G | G | 0.63 | 0.53 | 0.43 | ↓ | ||||
| rs12258967 | C/G | C | 0.64 | 1.00 | 0.73 | ↑ | ||||
| 10q21.2 | rs1530440 | C/T | T | 0.16 | 0.20 | 0.02 | ↓ | c10orf107 | ||
| rs4590817 | C/G | G | 0.82 | 1.00 | 0.82 | ↑ | ||||
| rs12244842 | G/T | T | 0.25 | 0.19 | 0.35 | ↓ | ||||
| rs7070797 | A/G | A | 0.15 | 0.00 | 0.00 | ↓ | ||||
| 10q22.2 | rs4746172 | C/T | C | 0.23 | 0.56 | 0.19 | ↑ | VCL | ||
| 10q23.33 | rs932764 | A/G | G | 0.43 | 0.58 | 0.15 | ↑ | PLCE1 | ||
| 10q24.32 | rs1004467 | C/T | T | 0.92 | 0.67 | 0.81 | ↑ | CYP17A1-NT5C2 | ||
| rs11191548 | C/T | T | 0.92 | 0.72 | 0.99 | ↑ | ↑ | |||
| rs12413409 | A/G | G | 0.92 | 0.72 | 0.98 | ↑ | ||||
| rs4409766 | C/T | T | 0.93 | 0.78 | 0.82 | ↑ | ||||
| rs3824755 | C/G | C | 0.07 | 0.23 | 0.17 | ↓ | ||||
| 10q25.3 | rs2782980 | C/T | T | 0.27 | 0.15 | 0.44 | ↓ | ADRB1 | ||
| rs7076938 | C/T | C | 0.33 | 0.17 | 0.45 | ↓ | ||||
| rs1801253 | C/G | G | 0.32 | 0.15 | 0.41 | ↓ | ||||
| 11p15.5 | rs661348 | C/T | C | 0.45 | 0.58 | 0.11 | ↑ | LSP1-TNNT3 | ||
| 11p15.4 | rs7129220 | A/G | G | 0.89 | 1.00 | 0.95 | ↓ | ADM | ||
| 11p15.1 | rs381815 | A/C/T | T | 0.30 | 0.25 | 0.18 | ↑ | PLEKHA7 | ||
| rs757081 | C/G | G | 0.63 | 0.66 | 1.00 | ↑ | PIK3C2A, NUCB2, NCR3LG1 | |||
| 11p15.2 | rs2014408 | C/T | T | 0.20 | 0.21 | 0.03 | ↑ | ↑ | ↑ | SOX6 |
| rs4757391 | C/T | C | 0.18 | 0.17 | 0.23 | ↑ | ||||
| 11q13.1 | rs4601790 | A/G | G | 0.25 | 0.42 | 0.07 | ↑ | EHBP1L1 | ||
| rs3741378 | A/G | A | 0.15 | 0.38 | 0.36 | ↓ | RELA | |||
| 11q22.1 | rs633185 | C/G | G | 0.68 | 0.55 | 0.82 | ↓ | FLJ32810-TMEM133 | ||
| 11q24.3 | rs11222084 | A/T | T | 0.40 | 0.07 | 0.26 | ↑ | ADAMTS8 | ||
| 12q13.13 | rs7297416 | A/C | C | 0.25 | 0.62 | 0.38 | ↓ | HOXC4 | ||
| 12q21.33 | rs11105354 | A/G | G | 0.12 | 0.42 | 0.11 | ↓ | ATP2B1 | ||
| rs2681492 | A/G | A | 0.88 | 0.58 | 0.84 | ↑ | ||||
| rs2681472 | C/T | T | 0.88 | 0.58 | 0.89 | ↑ | ↑ | |||
| rs17249754 | A/G | G | 0.88 | 0.59 | 0.84 | ↑ | ↑ | |||
| 12q24.12 | rs3184504 | C/T | T | 0.45 | 0.00 | 0.00 | ↑ | SH2B3 | ||
| rs653178 | A/G | A | 0.56 | 1.00 | 1.00 | ↓ | ||||
| 12q24.13 | rs11066280 | A/T | T | 1.00 | 0.75 | 1.00 | ↑ | RPL6-ALDH2 | ||
| 12q24.21 | rs35444 | C/T | T | 0.59 | 0.73 | 0.55 | ↑ | TBX5-TBX3 | ||
| rs2384550 | A/G | A | 0.37 | 0.09 | 0.34 | ↓ | ||||
| rs10850411 | C/T | T | 0.72 | 0.46 | 0.66 | ↑ | ||||
| rs1991391 | C/T | C | 0.64 | 0.91 | 0.58 | ↑ | ||||
| rs11067763 | A/G | A | 0.89 | 0.61 | 0.65 | ↑ | MED13L | |||
| 15q21.1 | rs1036477 | A/G | G | 0.10 | 0.42 | 0.61 | ↓ | FBN1 | ||
| 15q24.1 | rs6495122 | A/C | A | 0.38 | 0.78 | 0.78 | ↑ | CYP1A1-ULK3 | ||
| rs1378942 | G/T | G | 0.32 | 0.79 | 1.00 | ↑ | ||||
| 15q24.2 | rs11072518 | C/T | T | 0.34 | 0.42 | 0.44 | ↑ | COX5A | ||
| rs1133323 | A/G | A | 0.59 | 0.17 | 0.00 | ↓ | ||||
| 15q26.1 | rs2521501 | A/T | T | 0.63 | 0.93 | 0.80 | ↑ | FURIN-FES | ||
| 16p12.3 | rs13333226 | A/G | G | 0.18 | 0.05 | 0.38 | ↑ | UMOD | ||
| 16q22.1 | rs33063 | A/G | A | 0.19 | 0.15 | 0.00 | ↑ | NFAT5 | ||
| 17q21.31 | rs12946454 | C/T | T | 0.71 | 0.62 | 0.80 | ↑ | PLCD3 | ||
| 17q21.32 | rs17608766 | C/T | T | 0.91 | 1.00 | 1.00 | ↓ | GOSR2 | ||
| 17q21.33 | rs12940887 | C/T | T | 0.41 | 0.09 | 0.03 | ↑ | ZNF652 | ||
| rs16948048 | A/G | G | 0.42 | 0.09 | 0.42 | ↑ | ||||
| 20p12.2 | rs1327235 | A/G | G | 0.52 | 0.48 | 0.53 | ↑ | JAG1 | ||
| rs1887320 | A/G | A | 0.58 | 0.43 | 0.54 | ↑ | ||||
| 20q13.32 | rs6015450 | A/G | G | 0.07 | 0.00 | 0.22 | ↑ | GNAS-EDN3 | ||
| rs6092743 | A/G | A | 0.06 | 0.00 | 0.06 | ↑ | C20orf174 | |||
| rs12244842 | G/T | T | 0.25 | 0.19 | 0.35 | ↓ | ||||
| rs7070797 | A/G | A | 0.15 | 0.00 | 0.00 | ↓ | ||||
BP, blood pressure; ↑, increase; ↓, decrease.
While most of the GWAS for BP have taken the quantitative route studying BP as a continuous variable, two studies analyzed hypertension as a binary trait (276, 321). These studies identified a novel locus located in the promoter region of Uromodulin gene (UMOD), which is exclusively expressed in the kidney and may influence BP by a novel sodium homeostatic pathway (276) and another locus in the promoter region of NOS3 (321) which codes for nitric oxide synthase 3 implicated in vascular smooth muscle relaxation, angiogenesis, and renal tubular response to salt loading.
GWAS for populations other than European descent were also performed with the aim of replicating the variants identified in European populations, and also finding new population-specific loci (95, 180, 227). The success of replicating the previously reported loci for European population in the other population suggests that the physiological effects of these loci may be generalized across populations with diverse genetic backgrounds. Yet, identifying novel loci also suggests that populations with different genetic background may have unique genetic factors as a result of differences in allele frequencies or population-specific factors that interact with genes to influence BP.
In contrast to monogenic syndromes, interpreting the signals from GWAS is not straight forward. The success of GWAS is dependent on three critical factors: 1) sufficiently large sample size drawn from a population of appropriate genetic background, 2) efficient genotyping panel that adequately covers variation within the whole genome, and 3) powerful statistical methods that can reveal genuine association signals. A major drawback is the large number of statistical tests performed in GWAS which increases the chance of type I error. The penalty applied for multiple testing is to use a genome-wide significant P value threshold of 5 × 10−8 or lower which is the Bonferroni-corrected alpha of 0.05 for testing a million markers. As the design of GWAS chips are based on whole genome coverage using linkage disequilibrium, most of the significant SNPs reported by GWAS of all traits lie within noncoding regions. These SNPs are not the causal variant for the trait studied, but are likely to be in linkage disequilibrium with the causal variant nearby. While this approach has been extremely fruitful in discovering novel loci, several challenges exist in the interpretation of GWAS findings. For each SNP mapped by GWAS, it is likely that within ∼100,000 bases of the locus, there exists a causal gene. The biggest challenge of GWAS is not the discovery of significant trait associated SNPs, but the need to understand for each GWAS SNP: 1) the causal variant, 2) the causal gene, 3) the mechanism by which the variant affects the gene function, and 4) the mechanism by which the gene function affects the phenotype. However, as most of the GWAS signals occur in the noncoding regions of the genome, they require complementary mechanistic studies to delineate the physiological mechanisms underlying their genetic association with BP. Around 85–90% of GWAS hits tag noncoding variants only and identifying causal regulatory variants and target gene require a combination of genetic fine mapping, epigenomic profiling, individual reporter assays, expression quantitative trait loci (eQTL) studies in appropriate tissues, or creating isogenic cellular models [e.g., via genome editing (80)]. The putative gene thus identified is then modulated in vitro in primary cell culture or in vivo in animal models to identify its role in determining the original phenotype (259).
The success of GWAS in identifying a multitude of robust signals for BP is summarized in TABLE 4 and illustrated in FIGURES 5–8, which present all the GWAS signals for BP and hypertension along with details of genomic features near the GWAS SNPS including proximate genes that may help identify the causal variant tagged by the GWAS SNP.
FIGURE 5.
Chromosome 1-4: genetic landscape of monogenic and polygenic blood pressure/hypertension syndromes, causal genes, GWAS loci, and information used to prioritize functional genes and variants tagged by the GWAS SNPs (154). The Circos plot tracks from outside inward are as follows: 1) chromosome ideogram; 2) location and ID of GWAS SNPs for BP and hypertension; 3) CpG islands: epigenetic markers such as methylation sites can mark transcriptional activity; 4) DNAse I hypersensitivity sites are open chromatin sites and GWAS variants located in these sites have been shown to control distant genes; 5) genes underlying GWAS SNPs or monogenic BP syndromes: genes in red are monogenic BP genes, while genes in blue are plausible candidate genes that may be causal for the GWAS SNP. For each SNP mapped by GWAS, it is likely that within ∼100,000 bases of the locus, there exists a causal gene. This track just highlights a fraction of the genes present within the selected genetic loci that are most plausible candidates for BP, and the innermost track depicts all the genes that are present within each genetic loci from which the plausible candidates are selected. 6) Regulatory polymorphisms from ORegAnno (Open Regulatory Annotation): location of regulatory regions, transcription factor binding sites, RNA binding sites, regulatory variants, haplotypes, and other regulatory elements from the Open Regulatory Annotation; 7) structural variation (deletion = red, insertion = blue, duplication = yellow): location of structural variation where low frequency and rare variants with intermediate to large effect sizes may lie and require further targeted sequencing studies; 8) multiple alignments of 100 vertebrate species and measurements of evolutionary conservation: highly conserved sites indicating possibly functional sites depicted by size and color intensity of the filled squares; 9) monogenic BP syndromes and GWAS traits for the SNPs and monogenic genes in the outer 2 tracks; 10) GTEX expression data of genes within the selected chromosomal segments: adipose, adrenal, aorta, artery, brain, left ventricle, kidney cortex, liver, muscle, and blood; 11) genes within the chromosome segment. The loops on the top of the ideograms are representation of eQTL locations of GWAS SNPs. Expression quantitative trait loci (eQTL) mapping is performed to find statistical association between a genetic variant and the transcript level of a gene considered as a quantitative trait. eQTL studies can be used as a general method to help identify a set of target genes as many SNPs associated with GWAS traits were shown to be eQTLs.
FIGURE 8.
Chromosome 15-X: genetic landscape of monogenic and polygenic blood pressure/hypertension syndromes, causal genes, GWAS loci, and information used to prioritize functional genes and variants tagged by the GWAS SNPs (154). See legend to Figure 5 for Circos plot track details.
FIGURE 6.
Chromosome 5-8: genetic landscape of monogenic and polygenic blood pressure/hypertension syndromes, causal genes, GWAS loci, and information used to prioritize functional genes and variants tagged by the GWAS SNPs (154). See legend to Figure 5 for Circos plot track details.
FIGURE 7.
Chromosome 10-12: genetic landscape of monogenic and polygenic blood pressure/hypertension syndromes, causal genes, GWAS loci, and information used to prioritize functional genes and variants tagged by the GWAS SNPs (154). See legend to Figure 5 for Circos plot track details.
C. Specific Genetic Pathways of BP Regulation and Dysregulation
We now describe the genetic pathways the perturb BP regulation that manifest as clinical syndromes associated with high or low blood pressure in humans. While the majority of the pathways are rare monogenic syndromes (TABLE 3), we also describe a novel pathway for BP regulation identified through GWAS.
1. Glucocorticoid-remediable aldosteronism or familial hyperaldosteronism type 1
This is a rare autosomal dominant disorder characterized by early-onset hypertension, hyperaldosteronism, variable hypokalemia, low plasma renin activity (PRA), and abnormal production of 18-oxocortisol and 18-hydroxycortisol. This is attributed to a chimeric gene caused by the fusion of the 5′ regulatory sequences of 11β-hydroxylase [CYP11B1; which confers adrenocorticotropic hormone (ACTH) responsiveness] with the distal coding sequences of aldosterone synthase (CYP11B2). This leads to ACTH becoming the main controller of aldosterone secretion, rather than angiotensin II or potassium (218). Low-dose glucocorticoids to suppress ACTH secretion, or amiloride to directly block the epithelial sodium channel (ENaC), or spironolactone to block binding of aldosterone to the mineralocorticoid receptor (MCR) are the specific treatment options for hypertension in these individuals.
2. Apparent mineralocorticoid excess
Hypertension due to apparent mineralocorticoid excess (AME) ensues due to the absence or reduced activity of 11β-hydroxysteroid dehydrogenase (HSD11B2), an enzyme which metabolizes cortisol to prevent its binding to the mineralocorticoid receptor. Reduced activity of HSD11B2 results in cortisol to not be metabolized but acts as if it were a potent mineralocorticoid (28). Patients diagnosed with AME syndrome respond well to low-sodium diet and are treated with spironolactone, which blocks binding of both cortisol and aldosterone to mineralocorticoid receptors.
3. Pseudohypoaldosteronism type II (Gordon’s syndrome)
This is a form of hypertension associated with hyperkalemia, non-anion gap metabolic acidosis, and increased salt reabsorption by the kidney. The WNK (with-no-lysine [K]) kinases play central roles in regulating mammalian BP by initiating a signaling pathway that controls the activity of critical ion cotransporters in the kidney NCC (Na+/Cl− ion cotransporter) and NKCC2 (Na+/K+/2Cl− cotransporter 2). Gordon's syndrome is caused by mutations in WNK1, WNK4, Kelch-like 3 (KLHL3), and Cullin 3 (CUL3) genes. CUL3 and KLHL3 mutations putatively inhibit the ubiquitylation of WNK4, and probably other WNK isoforms, resulting in the the overactivation of NCC/NKCC2 ion cotransporters, and consequently, increased salt retention and hypertension (22, 384). Treatment consists of either a low-salt diet or thiazide diuretics, aimed at decreasing chloride intake and blocking Na+-Cl- cotransporter activity, respectively.
4. Liddle's syndrome
Liddle’s syndrome is an autosomal dominant condition presenting with hypertension and aldosterone excess, but associated with low renin and aldosterone levels. It is caused by mutations in the genes coding the β- or γ-subunits of ENaC, SCNN1B and SCNN1G. The mutations result in deletions of proline-rich regions of the protein products of these ENaC subunits, which are essential for binding of Nedd4–2 (NEDD4L), a regulatory repressor that promotes ENaC channel degradation (336). Increased rates of sodium reabsorption, volume expansion, and hypertension result due to lack of binding activity of β- and γ-subunits to Nedd4, which results in constitutive expression of sodium channels and prolongation of the half-life of ENaCs at the renal distal tubule apical cell surface. Patients with Liddle's syndrome are treated with amiloride or triamterene to lower BP and correct acidosis and hypokalemia.
5. Bartter's syndrome
This is a salt-losing condition characterized by hypokalemic metabolic alkalosis and normal or low BP with increased renin activity and high aldosterone levels. The defective mechanism is located in the thick ascending limb (TAL) of Henle's loop and comprises loss of function of NKCC2 or a group of other proteins which lead to secondary loss of function of NKCC2-ROMK channel, chloride channel Kb (ClC-Kb), Bartin, and calcium sensing receptor (CaSR) (341). Increased urine levels of prostaglandin E2 (PGE2) and diminished susceptibility to the pressor effect of ANG II and norepinephrine are features of Bartter’s syndrome. Patients with Bartter's syndrome are more prone to volume depletion, diarrhea, spasm, fever, and dangerous hypokalemia, which are life-threatening complications, especially during the postnatal period. Chronic treatment of underlying abnormalities of Bartter’s syndrome including features to treat increased prostaglandin synthesis and RAAS activity, which aggravate electrolyte and acid base disturbances, are potassium supplementation and use of cyclooxygenase inhibitors, angiotensin converting enzyme (ACE)-inhibitors, and potassium sparing diuretics.
6. Gitelman's syndrome
In Gitelman's syndrome, the defective mechanism is located in the distal convoluted tubule and comprises loss of function of the sodium-chloride cotransporter (NCC) (342) and is characterized by hypokalemic metabolic alkalosis along with significant hypomagnesemia, low urinary calcium excretion, and low blood pressure. The prevalence of Gitelman's syndrome is very low, estimated to be 1:40,000 in the general population and the prevalence of heterozygotes in the Caucasian population is ~1%. Chronic treatment of patients with Gitelman's syndrome comprises maintaining effective extracellular volume with adequate salt and water consumption along with oral potassium and magnesium supplementation. Indomethacin, amiloride, and eplerenone are also used to treat hypokalemia.
7. Primary aldosteronism
Individuals with primary aldosteronism constitutively produce aldosterone from the adrenal gland, resulting in hypertension with variable hypokalemia and a suppressed circulating renin. A gain-of-function somatic mutation in a K+ channel, KCNJ5, which results in membrane depolarization and enhanced aldosterone production, is a common genetic defect noted for ~40% of aldosterone producing adenomas. Mutations in three other genes, encoding the α1-subunit of Na+-K+-ATPase itself; and ATP2B3, a plasma membrane Ca2+-ATPase 3 homologous to the sarcoplasmic endoplasmic reticulum Ca2+-ATPases (SERCA); and CACNA1D, encoding an L-type Ca2+ channel CaV1.3, are observed in ~7% of the cases (399). Whereas APAs in adrenal zona glomerulosa cells harbor gain-of-function mutations in genes important for the regulation of Na+ and Ca2+, ATP1A1, and CACNA1D, respectively, KCNJ5 mutations are common in APAs resembling cortisol-secreting cells of the adrenal zona fasciculata (25). Adrenalectomy of the affected adrenal gland in APAs cures or ameliorates hypertension in the majority of patients.
8. Phaeochromocytomas and paragangliomas
Rare neuroendocrine tumors of the adrenal glands and the sympathetic and parasympathetic paraganglia are called phaeochromocytomas and paragangliomas, respectively. RET pro-to-oncogene mutations cause autosomal dominantly inherited phaeochromocytomas. Other susceptibility genes include genes that encode succinate dehydrogenase subunits A, B, C, and D (SDHA, SDHB, SDHC, and SDHD, respectively) with heterozygous germline mutations of SDHB, SDHC, and SDHD causing familial pheochromocytoma-paraganglioma syndromes known, respectively, as paraganglioma 4, paraganglioma 3, and paraganglioma 1 and the tumor suppressor gene VHL observed in families with von Hippel-Lindau syndrome (91). Recently discovered predisposing genes for pheochromocytoma/paraganglioma include KIF1Bbeta, PHD2, and SDHAF2 (91).
9. Uromodulin
A GWAS of BP extremes showed the minor G allele of a UMOD promoter SNP, rs13333226, was associated with a lower risk of hypertension and reduced urinary UMOD excretion (276). Uromodulin gene expression is exclusively localized to the thick ascending limb of the loop of Henle (TAL) in the kidney where 25% of the filtered sodium is reabsorbed. An increased localization of the salt-retaining NKCC2 (sodium-potassium-chloride cotransporter 2) in subapical vesicles of TAL cells with reduced phosphorylation, both resulting in reduced cotransporter activity was noted in UMOD knockout mice (260). The resulting effects noted are a greater sodium excretion as compared with wild-type mice, and a 20 mmHg lower BP in the knockout mice at baseline, as measured by radiotelemetry (123). Notably, this difference in BP was exacerbated with salt, whereby the knockout mice were resistant to the hypertensive effects of salt (123). Conversely, UMOD overexpression was associated with an increase in BP (361). The main sodium transporter in TAL is NKCC2, which is blocked by the commonly used loop-diuretic furosemide. Trudu et al. (361) showed furosemide treatment significantly enhanced natriuresis and reduced BP levels both in the transgenic mice and in the hypertensive individuals homozygous for the UMOD increasing allele. Thus GWAS has directed focus on a novel pathway of BP regulation involving altered expression of uromodulin which appear to influence sodium homeostasis and opens an avenue for translational studies to discover or repurpose drugs for treatment of hypertension.
10. Natriuretic peptide
Common SNPs in the chromosomal region containing NPPA and NPPB, the genes encoding the ANP and BNP propeptides, show consistently that alleles associated with increased circulating natriuretic peptide concentrations are also associated with lower systolic blood pressure (264, 265). The GWAS SNP rs5068 lies in the 3′-UTR of the NPPA gene which encodes the pro-peptide of ANP, NT-proANP. Healthy volunteers, which were homozygous for the risk allele of rs5068 showed lower NT-proANP expression possibly mediated through a microRNA miR-425 and provides a putative mechanism to explain how the risk allele reduces ANP level and consequently increases BP (11). The genetic effect of rs5068 on circulating NT-pro-ANP levels is comparable to the environmental change induced by switching from an extremely low-salt diet (230 mg/day) to a diet with salt content typical of a Western diet (4600 mg/day) (11).
XII. EMERGING CONCEPTS
A. Noncoding RNA, Epigenome, and the Inheritance of Hypertension
Variations within the protein-coding genome and epistatic interactions therein do not completely account for the heritability of hypertension. A vast majority of variants detected through human GWAS for hypertension are within noncoding genomic segments. These can be categorized into variants that are within regions that can generate noncoding RNA and variants that are within segments with no evidence of any annotation. Research on the latter is rather difficult and stagnant at this point; however, the hypothesis that variants within noncoding RNA molecules that can influence BP has gained some attention (201). In rat models of hypertension, various classes of noncoding RNAs have been profiled. With the ue of the Dahl SS/MCW rat and a consomic model (SS13.BN), wherein substitution of chromosome 13 of the Dahl SS/MCW rat with that of the BN rat lowered BP, a total of 377 microRNAs were profiled in the renal medulla under the conditions of a high-salt diet (224). In this study, five microRNAs are reported as differentially expressed between the SS/MCW and the SS13.BN strain (224). Of these two microRNAs, Mir214 and Mir29b are located on chromosome 13. miR-214 was readily detectable in the SS/MCW strain, but undetectable in the SS13.BN strain. miR-29b was expressed higher in the SS13.BN strain relative to the SS/MCW strain. The Dahl S rat demonstrates substantial renal medullary interstitial fibrosis, which was hypothesized as at least in part due to the upregulation of miR-29b, which was predicted to regulate genes encoding many proteins of the extracellular matrix. The observation that the expression of nine collagen genes and matrix metalloproteinase 2 (Mmp2), integrin β1 (Itgb1), and other genes related to the extracellular matrix were all downregulated in the renal medulla of SS13.BN provided further proof for the link between miR-29b and associated renal medullary interstitial fibrosis and hypertension. Further evidence was provided through luciferase construct assays wherein miR-29b suppressed the activity of luciferase when the reporter gene was linked to a 3′-untranslated segment of collagen genes Col1a1, Col3a1, Col4a1, Col5a1, Col5a2, Col5a3, Col7a1, Col8a1, Mmp2, or Itgb1. While miR-29b is an attractive candidate as an inherited element that may regulate BP, formal detection of variants within miR-29b or factors that influence its expression differentially would support its candidacy. For now, it appears to be an important mediator of renal fibrosis and related hypertension.
The second class of noncoding RNAs are the long noncoding RNAs. In rats, there are very few investigations that have led to the prioritization of noncoding elements as inherited factors for the development of hypertension partly due to the fact that the rat genome annotation for noncoding RNA genes is limited. There are two reports of profiling for rat long noncoding RNAs (119, 368). These are important starting points for future investigations into the relationships between variants within long noncoding RNAs and hypertension. As far as inheritance of noncoding RNA variants as BP QTLs, there is indirect evidence provided by mapping of a BP QTL on rat chromosome 9 through the congenic approach by introgressing normotensive Dahl R rat alleles onto the genome of the Dahl S rat to a <81.8 kb segment (122). This relatively short genome segment has no protein-coding gene annotations within it, but the congenic strain demonstrated a strong BP lowering effect, thus providing evidence for variants within noncoding regions of the genome to influence BP. A second report is on an even shorter segment of <42.5 kb, which when replaced by LEW rat alleles on the genome of the S rat, accounts for a BP increasing effect. There is a single protein-coding gene within this region called rififylin, but the gene itself does not have coding-sequence variants between S and LEW (121). The expression of rififylin is higher in the congenic strain with the increased BP compared with the S rat, and this increased expression is demonstrated to cause a delay in cellular recycling, a molecular mechanism linked to the observation of not only increased BP, but also shorter QT intervals (121). The expression of rififylin to be higher in the congenic strain is circumstantial to indicate that a variant or variants that are not responsible for coding for a protein are responsible for the observed BP effect. If these sequence variants are noncoding in the rat, it is of interest to note a parallel in humans wherein SNPs within noncoding regions around the rififilyin gene are also associated with aberrant, shorter QT intervals in humans (264).
B. Epigenetics and Hypertension
Epigenetics refers to molecular changes and associated phenotypes that are inherited, but do not involve variations in the DNA nucleotide sequence. Epigenetic mechanisms involve modification of DNA through methylation or histone modification or noncoding RNA, which then contribute to biological regulation by influencing the expression of protein-coding genes. Due to the feature that epigenetic marks such as methylation or histone modification are heritable and can be transmitted either through mitosis or meiosis even without altering the underlying DNA sequence, epigenetics is an inherited factor contributing to the genesis of hypertension. Epigenetic inheritance is an essential mechanism that accounts for the stable propagation of gene activity from one generation of cells to the next. Unlike genomic variants, epigenetic alterations can be difficult to study as they are often tissue or cell-type specific. There are very few studies reported that test the hypothesis that methylation of genomes at CpG sites is associated with hypertension. A study was conducted on a genome-wide scale using DNA from leukocytes from a small cohort of eight African-American hypertensive subjects and eight normotensive age-matched controls, but this initial study lacked power to detect any significant associations (371). Validations were conducted with additional subjects to demonstrate that higher methylation of at least one gene, the sulfatase 1 (SULF1) gene, was associated with hypertension (371). A recent study in humans was conducted using thin-layer chromatography to determine 5-methylcytosine (5mC) levels in blood DNA samples from 60 subjects with essential hypertension and 30 control subjects (344). Lower levels of 5mC were observed in DNA of patients with essential hypertension that correlated with the stage of hypertension (344). Among animal models, a similar approach of genome-wide methylation status analysis is reported using renal outer medullary tissue from the Dahl S hypertensive rat model and the SS.13BN26 congenic strain with a significantly attenuated salt-induced hypertension and renal injury. The SS.13BN26 congenic strain has a 12.9 Mbp segment introgressed from the BN rat. The specific methylations characterized were 5mC and 5-hydroxymethylcytosines (5hmC), both of which were mapped at single-base resolutions (223). The study also examined the effect of increased dietary salt intake and compared the SS rat with the congenic SS.13BN26 rat to investigate the effect of genomic segment substitution on 5mC and 5hmC and the association of 5mC and 5hmC with changes in the disease phenotypes. While there were no detectable alterations in DNA methylation as a result of the chromosome 13 substitution, there was a notable alteration in response to dietary salt in both the strains examined. Nearly 80% of the CpG islands that were differentially methylated in response to salt and associated with differential mRNA abundance were intragenic CpG islands.
Overall, it has become quite clear that a DNA sequence-based approach may only partially explain the involvement of our genome in the genesis of hypertension. Epigenetics is an attractive proposition for explaining some of the “missing heritability” (47, 97, 191, 216, 372, 373) for further studies, which are clearly needed to test and ascertain the causal implications of epigenetics in BP regulation.
C. Microbiome and the Inheritance of Hypertension
Beyond all the features presenting as genomic or epigenomic factors for the inheritance of hypertension, investigations into the realm of the microbiome have opened a new and previously unsuspected role of microbiota in regulating BP (FIGURE 9). Early clues for the involvement of the genome in microbiotal influenced alterations in BP came from the reports on short-chain fatty acid (SCFA) receptor knockout mice (285). Circulating SCFAs originate largely from gut microbiota as they are the end products of bacterial fermentation, which are absorbed into the host circulation. The link between SCFAs produced by the gut microbiota and modulation of BP via these SCFA receptors was demonstrated by using antibiotics to reduce the biomass of the gut microbiota before monitoring BP in these SCFA receptor gene deletion models described below.
FIGURE 9.
Cartoon representation of factors influencing the etiology of hypertension. The red double-sided arrow represents the interactions between the host genome and nongenomic factors that act in concert to influence the transition from normal physiology of BP regulation to the pathophysiological state of elevated BP.
Olfactory receptor 78 and G protein-coupled receptor 41 are both receptors for short-chain fatty acids. Both Gpr41 and Olfr78 are expressed in smooth muscle cells. Olfr78 is expressed in the renal arterioles and the juxtaglomerular apparatus. In the juxtaglomerular cells, Olfr78 mediates renin secretion in response to short-chain fatty acids. Deletion of the Olfr gene in Olfr78−/− knockout mice resulted in lower BP associated with lower levels of plasma renin which was demonstrated to be due to modulation of short-chain fatty acid-mediated renin release from the juxtaglomerular cells. Gpr41 is expressed in smooth muscle cells of small resistant blood vessels. Gpr41 contributes to the hypotensive effect of the short-chain fatty acid propionate. Furthermore, a relationship was demonstrated between these two receptors, whereby Olfr78 functions to raise BP and thereby antagonizes the contribution of Gpr41 to hypotensive effects of propionate (285). These studies provided some basic insights into the relationship between gut microbiota and previously unknown mechanisms leading to hypertension.
Soon thereafter, two studies were reported on the influence of microbiota on the development of inherited hypertension in rats. The first study was conducted to test the hypothesis that the extent of hypertension of the Dahl S rat can be determined by an alteration in its gut microbiotal composition (245). Gut microbiotal composition of the Dahl S rat was altered by transplantation of the relatively normotensive Dahl R rat microbiota. As a result of the transplantation, a significant increase in BP of the Dahl S rat was documented. The reciprocal transfer of S rat microbiota to the R rats did not alter the BP of the R rats. 16S RNA analysis of the microbiotal compositions combined with plasma short-chain fatty acid profiles revealed that microbiotal compositions were indeed altered and that the short-chain fatty acids acetate and heptanoate were higher in the S rats transplanted with microbiota from the R rats compared with the S rats given microbiota from the S rats (245). These data suggest that inherited levels of elevated BP can be further augmented by the type of gut microbiota. The second reported study is on the SHR rat (394). The gut microbiota of the hypertensive SHR was decreased in microbial richness, evenness, and diversity compared with the normotensive WKY rats (394). Administration of an antibiotic, minocycline, lowered BP of the SHR. The effect of this particular antibiotic is interesting because it crosses the blood-brain barrier and has neuronal effects on lowering BP. The microbiotal influence of minocycline was demonstrated through alterations in the 16S RNA composition of microbiota post minocycline treatment that was associated with a decrease in ANG II-induced BP as well as a reduction in the Firmicutes to Bacteriodetes (F/B) ratio and an increase in plasma butyrate, a short-chain fatty acid known to induce vasodilation. Cross-transplantation of microbiota from WKY to SHR and vice versa will be interesting, but has not yet been reported. Collectively, these data point to the importance of microbiota and their genomes, referred to as the microbiome in the pathogenesis of hypertension. The historical relevance of these studies is summarized in an editorial by Honor who draws attention to studies conducted in the late 1970s and early 1980s, which have suggested a role for the gut microbiome in the development of hypertension in rats and man (147). Early work conducted in humans and experimental rats (146, 148, 149) suggests that an increase in BP through the action of steroids was prevented by administration of antibiotics (146, 148). Studies in rats were conducted in Sprague-Dawley (SD) rats rendered hypertensive with corticosterone or by administration of ACTH. Hypertension was prevented by prior treatment of the rats with neomycin and to a lesser extent with vancomycin (146, 148). Neomycin also slowed the development of hypertension in the SHRSP rat (148). Since modern methods to detect microbiota through 16S sequencing was not available at that time, gut flora was examined with conventional techniques such as diluting of fecal samples that were then spread on agar plates enriched to selectively support growth of organisms. The findings were insufficiently detailed to assign links with steroid metabolism; however, the effect of neomycin on BP was repeated in the Florey Institute, Melbourne, Australia with rats given ACTH and corticosterone (159) and in the rat model of one clip, one kidney (CSK) hypertension (94). Three decades later, the two reports on the Dahl S rat and the SHR and their links to microbiota as a factor influencing development of hypertension (245, 394) has generated much excitement in the field as is evident from a number of review articles (4, 172, 242, 246, 263, 296, 324, 407) and a working group report from the National Heart, Lung, and Blood Institute of the National Institutes of Health, USA (270) related to the importance of salt, immunity, the microbiome and hypertension, or kidney disease.
Thus the microbiome now belongs to the long list of factors beyond inheritance of the host genome that influence the etiology of hypertension (FIGURE 9). Admittedly the discoveries thus far, albeit associations, are in need of further studies for cause-effect relationships between the host genome and the microbiome to impact a change in BP. This could be challenging because unlike the single host genome, the microbiome consists of a group of genomes, which means that these groups of genomes will have collective transcriptomes that are influenced by the host genome and vice versa. However, the microbiome cannot explain inherited difference in blood pressure in human or animal studies. It serves as a complicating factor that obscures the influence of genetics on blood pressure regulation. Environmental factors such as, for example, dietary proteins, which are known to alter blood pressure, at least in the Dahl S model (61, 239), may do so via the microbiome or by altering the meta-transcriptome, epigenome, or the metabolome. Contemplation of these many possibilities is impossible unless one considers these host-microbiotal interactions in the context of a “holobiont,” which is defined as an ecosystem consisting of a macro-organism (the host) with a group of microorganisms (microbiota). Galla et al. (99) provide a perspective leading to this concept along with an update on the status of research on microbiota in hypertension.
XIII. LOOKING AHEAD
In conclusion, research into the inherited susceptibility factors for BP, which perhaps began with the ambitious view of finding a handful of causal variants with major effects on BP, which could be used as targets for better clinical management of hypertension, has not proven to be the case. Instead, a small number of genes have been identified through positional mapping approaches, and a deeper understanding of additional mechanisms as being operational at the genomic level has emerged. These mechanisms are depicted in FIGURE 10 as epistasis, epigenetics, chromosome conformations, and interactions with genomes of microbiota, all of which impact molecular pathways converging to causally influence BP regulation.
FIGURE 10.
Current views on mechanisms by which DNA impacts blood pressure regulation. The genome of the host as well as the genome of microbiota, called microbiome, are depicted as known genomic factors influencing BP. The genes researched and found to be linked or associated with the inheritance of hypertension in rat models and/or in humans are listed on the right hand side of the diagram. Please see note added in proof for Rffl-Inc1 as an additional noncoding locus validated recently as a new BP QTL.
For the future of research on the genetics of hypertension, the following are a few considerations and anticipatory remarks.
A. Missing Heritability
Despite the identification of numerous SNPs associated with hypertension and BP traits, the proportion of phenotypic variance that is explained by all of these loci together is <2.5%. This phenomenon has been described as the problem of “missing heritability” and is not restricted to BP traits (230). For instance, a classic complex trait such as height, which has a very large heritability estimate from family studies (~80%), has <10% of the phenotypic variance explained from the SNPs identified using very large sample sizes (>180,000 individuals). A different way of estimating heritability using SNP data of unrelated individuals is the GCTA approach introduced by Yang et al. (h2SNP) (393). This is based on estimating the heritability from unrelated individuals using common SNPs with the assumption that heritability estimates in unrelated individuals is only attributable to the common SNPs, while the estimation in related individuals is attributed to the entire genome. Applying this method to systolic BP has shown that h2SNP was ~24%, which is ~50% of the heritability estimates from other twin studies, and ~80% of the same study heritability estimate (h2 = 30%). Furthermore, the number of independent variants with similar effect size to those reported in the ICBP study was estimated to be 116 (95% CI: 57–174), which can collectively explain around 2.2% of the phenotypic variance for BP phenotypes, compared with only 0.9% explained by the 29 SNPs identified by ICBP (160). These findings indicate that a large proportion of the heritability of BP is “hidden” rather than “missing” because of a large number of common variants, each of which has too small an effect to be detected at the stringent genome-wide significance level using current sample sizes.
The search for missing heritability is one of the primary goals for research on hypertension and other complex conditions. Among the hypotheses contemplated to explain “missing heritability” of complex traits are overestimates of heritability, genomic regions that are unexplored, genetic variants that remain untested, rare genetic variants that are yet unknown or not associated, gene interactions or epistasis (230, 231), and more recently the underrecognized role of the host genomic interactions with the microbiome. Thus there is no question that the quest for genetic analysis of hypertension thus far has opened up more avenues to pursue. Appropriate experimental designs have to be employed that allow for investigations to be focused on causal relationships rather than mere associations. Determining the full extent to which each of these additional avenues contributes to the genotype-BP relationship is expected to improve our understanding of the biological systems that underpin variation conferring susceptibility to the development of hypertension as well as increase the accuracy of individual risk prediction.
B. Emerging Insights From Other “Omics”: Metabolomics
Other omics technologies such as metabolomics are potentially powerful tools to identify molecular pathways. They can capture both intrinsic and extrinsic factors, and their dynamic nature makes them ideal for measuring physiological response to external stimuli or the development of pathogenic processes. Metabolomics, which is the systematic study of small molecule metabolites, may have an important role in defining candidate systems/pathways in the pathogenesis of hypertension. Furthermore, metabolomic markers are closer to the phenotype of interest in contrast to the genotype which is static and unchanged throughout life. Metabolomic profiling of over 3,000 adult twins identified a putative novel pathway for BP regulation involving a dicarboxylic acid (hexadecanedioate) with a causal role supported by in vivo studies in rats (248). The role of hexadecanedioate in a vascular mechanism for hypertension is supported by evidence from a study of pulmonary hypertension, indicating a disruption of β-oxidation and an increase of ω-oxidation in this condition and pointing to a putative role in elevating pressure in both the systemic and the pulmonary circulations (405). The strongest genetic association seen with hexadecanedioate maps to SLCO1B1, an association previously reported in a metabolome-wide genetic study in caucasians (350). Targeted metabolomics profiling in the European Prospective Investigation Into Cancer and Nutrition (EPIC)-Potsdam study showed higher concentrations of serine, glycine, and acyl-alkyl-phosphatidylcholines C42:4 and C44:3 tended to be associated with higher and diacyl-phosphatidylcholines C38:4 and C38:3 with lower predicted 10-yr hypertension-free survival (70). Other metabolite associations with incident hypertension and BP come from two US studies which found 4-hydroxyhippurate, a metabolic sex steroids pattern and 2 diacylglycerols 16:0/22:5 and 16:0/22:6 to be associated with BP and incident hypertension (205, 406). Finally, Menni et al. (249) showed 12 metabolites to be strongly associated with pulse wave velocity with uridine, phenylacetylglutamine, and serine appearing to strongly correlate with PWV in women.
C. Networks of Relationships and Pathways Influencing the Inheritance of Hypertension
Beyond the factors listed above, investigations into the genotype-phenotype relationship of BP regulation is expected to follow the genetic investigations conducted in lower order organisms such as yeast and Drosophila. FIGURE 11 (154) is provided as an example to illustrate the point that quantitative traits studied extensively in Drosophila have revealed pervasive networks of epistatic interactions that recapitulate known as well as candidate genetic networks affecting complex traits (154). It is tempting to speculate that similar extensive networks of pathways govern the genotype-BP causality relationship. Others have also proposed similar regulatory networks operating within specific genomic contexts that interact with environmental factors such as dietary salt levels in the context of epigenomic contributions to salt-sensitive hypertension (216). Furthermore, a treelike paradigm has been proposed for understanding such pathway networks underlying the development of salt-sensitive hypertension (216). On one hand, reductionistic approaches of modeling single genes in suitable experimental designs for assessing causal effects on BP are expected to progress further, but on the other hand, it is also important to test the molecular network hypothesis on a genome-wide scale to fully assess the genetic contributions to the development of hypertension. Technological advances in machine-learning and other computational approaches will perhaps be required to test the molecular network hypothesis in the context of BP regulation. Mackay and Moore (230, 231) point out that “the most important short-term goal is to develop, evaluate and employ statistical and computational methods that embrace, rather than ignore, the complexity of the genotype to phenotype map.” They predict as quoted below: “artificial intelligence is poised to have a big impact on the genetic analysis of complex traits by generating interesting and unexpected models of genotype to phenotype relationships” (230, 231).
FIGURE 11.
Networks of epistatic interactions. Interaction networks are depicted for starvation resistance (A) and chill coma recovery (B). Nodes depict genes, and edges significant interactions. Red nodes are genes containing significant SNPs from the Flyland analysis. Blue nodes are genes containing significant SNPs from DGRP analysis. [From Huang et al. (123), with permission from Proceedings of the National Academy of Sciences USA.]
D. Chromosome Conformations
Another emerging hypothesis for contributions from the genome to complex traits is that alterations in nuclear organization of DNA resulting in higher order structures such as folds of DNA within chromatin confer differential susceptibility. Fueled primarily by advances in chromosome conformation capture assay (3C) and sequencing-based chromosomal contact mapping (Hi-C, 5C and 4C-seq) that is applicable on a genome-wide scale (62, 217, 315, 325, 364), it is now technically feasible to detect such spatial organization of nuclear DNA. However, there is little known about how such folds affect the ways in which cells access, read, and interpret genetic information pertinent to BP regulation.
XIV. CONCLUDING REMARKS
To conclude, advances in genomics have accelerated over the last decade leading to an unparalleled leap in our understanding of the genetic architecture of BP and hypertension. While the technological and analytic aspects of genomics have been very successful in discovering DNA sequence variants associated with BP and hypertension, the functional and biological significance of the vast number of these variants in the human genome are unknown. In the near future, integrated analyses of whole genome sequences along with other “-omics” is anticipated to mature and lead to better insights, which can then be accommodated into the decision-making tree of health care for every individual. The challenging task will be to identify variants or biomarkers or pathways that are disease-causing or disease-modifying or treatment-stratifying, and to develop strategies to prevent or treat hypertension while keeping the ethical and social implications constantly aligned with technical and clinical limitations of the diagnostic/therapeutic used.
NOTE ADDED IN PROOF
Since this article was accepted, a new study has been published (36a), wherein a precise 19 bp indel polymorphism was positionally cloned as quantitative trait nucleotides (QTNs) for blood pressure and short QT intervals. This study is a follow-up of the work presented in Table 2 on rat chromosome 10, study “o”. The BP QTL was previously located within <45 kb of the rat genome (121). ln the new study, using CRISPR/Cas9 technology, both targeted disruption and targeted knock-in rescue approaches were applied to validate the 19 bp of the DNA sequence variation between S and Lew rats as QTNs. The 19 bp sequence was discovered to be part of the genomic sequence which is transcribed into a long noncoding RNA called Rffl-lnc1. Secondary structural alterations of Rffl-lnc1 were noted as a result of the 19 bp indel polymorphism. This study is not only the first to define genomic variation at the highest level of precision for mapping inherited elements that control a complex trait among mammalian models, but also the first to identify the inheritance of a variation within a long noncoding RNA as a causal factor for BP regulation.
GRANTS
S. Padmanabhan is funded by Medical Research Council Grant MR/M016560/1, The AIM-HY Study, and British Heart Foundation Grants PG/12/85/29925 and CS/16/1/31878. B. Joe acknowledges continued grant support from National Heart, Lung, and Blood Institute Grants HL020176 and HL112641.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
Our thanks to Saroj Chakraborty, Graduate Research Assistant, and Dr. Sivarajan Kumarasamy, University of Toledo College of Medicine, for tabulating data presented in Table 1 and Figure 2B, respectively; Alisha Aman, Research Assistant, Institute of Cardiovascular and Medical Sciences, University of Glasgow, for assistance with Figures 4–8; and Prof. John Paul Rapp for critical reading and a contribution to the section on epistasis in rat models of hypertension.
Address for reprint requests and other correspondence: B. Joe, Dept. of Physiology and Pharmacology, Center for Hypertension and Personalized Medicine, University of Toledo College of Medicine and Life Sciences, 237 Block Health Science Bldg., 3000 Arlington Ave., Toledo, OH 43614 (e-mail: bina.joe@utoledo.edu).
References
- 1.Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, Blankenhorn EP, Blizard DA, Bolivar V, Brockmann GA, Buck KJ, Bureau JF, Casley WL, Chesler EJ, Cheverud JM, Churchill GA, Cook M, Crabbe JC, Crusio WE, Darvasi A, de Haan G, Dermant P, Doerge RW, Elliot RW, Farber CR, Flaherty L, Flint J, Gershenfeld H, Gibson JP, Gu J, Gu W, Himmelbauer H, Hitzemann R, Hsu HC, Hunter K, Iraqi FF, Jansen RC, Johnson TE, Jones BC, Kempermann G, Lammert F, Lu L, Manly KF, Matthews DB, Medrano JF, Mehrabian M, Mittlemann G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Mountz JD, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Paigen B, Palmer AA, Peirce JL, Pomp D, Rosemann M, Rosen GD, Schalkwyk LC, Seltzer Z, Settle S, Shimomura K, Shou S, Sikela JM, Siracusa LD, Spearow JL, Teuscher C, Threadgill DW, Toth LA, Toye AA, Vadasz C, Van Zant G, Wakeland E, Williams RW, Zhang HG, Zou F; The Complex Trait Consortium . The nature and identification of quantitative trait loci: a community’s view. Nat Rev Genet : 911–916, 2003. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abumrad NA, Goldberg IJ. CD36 actions in the heart: lipids, calcium, inflammation, repair and more? Biochim Biophys Acta : 1442–1449, 2016. doi: 10.1016/j.bbalip.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St Lezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet : 76–83, 1999. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 4.Al Khodor S, Shatat IF. Gut microbiome and kidney disease: a bidirectional relationship. Pediatr Nephrol : 921–931, 2017. doi: 10.1007/s00467-016-3392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alemayehu A, Breen L, Krenova D, Printz MP. Reciprocal rat chromosome 2 congenic strains reveal contrasting blood pressure and heart rate QTL. Physiol Genomics : 199–210, 2002. doi: 10.1152/physiolgenomics.00065.2002. [DOI] [PubMed] [Google Scholar]
- 6.Allbutt TC. Diseases of the Arteries, Including Angina Pectoris. London: Macmillan, 1915. [Google Scholar]
- 7.Aneas I, Rodrigues MV, Pauletti BA, Silva GJ, Carmona R, Cardoso L, Kwitek AE, Jacob HJ, Soler JM, Krieger JE. Congenic strains provide evidence that four mapped loci in chromosomes 2, 4, and 16 influence hypertension in the SHR. Physiol Genomics : 52–57, 2009. doi: 10.1152/physiolgenomics.90299.2008. [DOI] [PubMed] [Google Scholar]
- 8.Annest JL, Sing CF, Biron P, Mongeau JG. Familial aggregation of blood pressure and weight in adoptive families. I. Comparisons of blood pressure and weight statistics among families with adopted, natural, or both natural and adopted children. Am J Epidemiol : 479–491, 1979. doi: 10.1093/oxfordjournals.aje.a112829. [DOI] [PubMed] [Google Scholar]
- 9.Annest JL, Sing CF, Biron P, Mongeau JG. Familial aggregation of blood pressure and weight in adoptive families. II. Estimation of the relative contributions of genetic and common environmental factors to blood pressure correlations between family members. Am J Epidemiol : 492–503, 1979. doi: 10.1093/oxfordjournals.aje.a112830. [DOI] [PubMed] [Google Scholar]
- 10.Ariyarajah A, Palijan A, Dutil J, Prithiviraj K, Deng Y, Deng AY. Dissecting quantitative trait loci into opposite blood pressure effects on Dahl rat chromosome 8 by congenic strains. J Hypertens : 1495–1502, 2004. doi: 10.1097/01.hjh.0000133720.94075.6f. [DOI] [PubMed] [Google Scholar]
- 11.Arora P, Wu C, Khan AM, Bloch DB, Davis-Dusenbery BN, Ghorbani A, Spagnolli E, Martinez A, Ryan A, Tainsh LT, Kim S, Rong J, Huan T, Freedman JE, Levy D, Miller KK, Hata A, Del Monte F, Vandenwijngaert S, Swinnen M, Janssens S, Holmes TM, Buys ES, Bloch KD, Newton-Cheh C, Wang TJ. Atrial natriuretic peptide is negatively regulated by microRNA-425. J Clin Invest : 3378–3382, 2013. doi: 10.1172/JCI67383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr M, MacKenzie SM, Wilkinson DM, Holloway CD, Friel EC, Miller S, MacDonald T, Fraser R, Connell JM, Davies E. Functional effects of genetic variants in the 11beta-hydroxylase (CYP11B1) gene. Clin Endocrinol (Oxf) : 816–825, 2006. doi: 10.1111/j.1365-2265.2006.02673.x. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Ishay D, Saliternik R, Welner A. Separation of two strains of rats with inbred dissimilar sensitivity to Doca-salt hypertension. Experientia : 1321–1322, 1972. doi: 10.1007/BF01965321. [DOI] [PubMed] [Google Scholar]
- 14.Bianchi G, Ferrari P, Staessen JA. Adducin polymorphism: detection and impact on hypertension and related disorders. Hypertension : 331–340, 2005. doi: 10.1161/01.HYP.0000156497.39375.37. [DOI] [PubMed] [Google Scholar]
- 15.Bianchi G, Fox U, Imbasciati E. The development of a new strain of spontaneously hypertensive rats. Life Sci : 339–347, 1974. doi: 10.1016/0024-3205(74)90064-2. [DOI] [PubMed] [Google Scholar]
- 16.Bianchi G, Tripodi G, Casari G, Salardi S, Barber BR, Garcia R, Leoni P, Torielli L, Cusi D, Ferrandi M. Two point mutations within the adducin genes are involved in blood pressure variation. Proc Natl Acad Sci USA : 3999–4003, 1994. doi: 10.1073/pnas.91.9.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilusic M, Bataillard A, Tschannen MR, Gao L, Barreto NE, Vincent M, Wang T, Jacob HJ, Sassard J, Kwitek AE. Mapping the genetic determinants of hypertension, metabolic diseases, and related phenotypes in the lyon hypertensive rat. Hypertension : 695–701, 2004. doi: 10.1161/01.HYP.0000144542.57306.5e. [DOI] [PubMed] [Google Scholar]
- 18.Bilusić M, Moreno C, Barreto NE, Tschannen MR, Harris EL, Porteous WK, Thompson CM, Grigor MR, Weder A, Boerwinkle E, Hunt SC, Curb JD, Jacob HJ, Kwitek AE. Genetically hypertensive Brown Norway congenic rat strains suggest intermediate traits underlying genetic hypertension. Croat Med J : 586–599, 2008. doi: 10.3325/cmj.2008.5.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biron P, Mongeau JG, Bertrand D. Blood pressure values in 1116 French-Canadian children. Can Med Assoc J : 432, 1976. [PMC free article] [PubMed] [Google Scholar]
- 20.Biron P, Mongeau JG, Bertrand D. Familial aggregation of blood pressure in 558 adopted children. Can Med Assoc J : 773–774, 1976. [PMC free article] [PubMed] [Google Scholar]
- 21.Böger CA, Heid IM. Chronic kidney disease: novel insights from genome-wide association studies. Kidney Blood Press Res : 225–234, 2011. doi: 10.1159/000326901. [DOI] [PubMed] [Google Scholar]
- 22.Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, Lebel M, Gordon RD, Semmekrot BA, Poujol A, Välimäki MJ, De Ferrari ME, Sanjad SA, Gutkin M, Karet FE, Tucci JR, Stockigt JR, Keppler-Noreuil KM, Porter CC, Anand SK, Whiteford ML, Davis ID, Dewar SB, Bettinelli A, Fadrowski JJ, Belsha CW, Hunley TE, Nelson RD, Trachtman H, Cole TR, Pinsk M, Bockenhauer D, Shenoy M, Vaidyanathan P, Foreman JW, Rasoulpour M, Thameem F, Al-Shahrouri HZ, Radhakrishnan J, Gharavi AG, Goilav B, Lifton RP. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature : 98–102, 2012. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bright R. Tabular view of the morbid appearances in 100 cases connected with albuminous urine. Guys Hosp Rep : 132–152, 1836. [Google Scholar]
- 24.Brown DM, Provoost AP, Daly MJ, Lander ES, Jacob HJ. Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nat Genet : 44–51, 1996. doi: 10.1038/ng0196-44. [DOI] [PubMed] [Google Scholar]
- 25.Brown MJ. Ins and outs of aldosterone-producing adenomas of the adrenal: from channelopathy to common curable cause of hypertension. Hypertension : 24–26, 2014. doi: 10.1161/HYPERTENSIONAHA.113.01833. [DOI] [PubMed] [Google Scholar]
- 26.Browning BL, Browning SR. Haplotypic analysis of Wellcome Trust Case Control Consortium data. Hum Genet : 273–280, 2008. doi: 10.1007/s00439-008-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burt VL, Whelton P, Roccella EJ, Brown C, Cutler JA, Higgins M, Horan MJ, Labarthe D. Prevalence of hypertension in the US adult population. Results from the Third National Health and Nutrition Examination Survey, 1988-1991. Hypertension : 305–313, 1995. doi: 10.1161/01.HYP.25.3.305. [DOI] [PubMed] [Google Scholar]
- 28.Cerame BI, New MI. Hormonal hypertension in children: 11beta-hydroxylase deficiency and apparent mineralocorticoid excess. J Pediatr Endocrinol Metab : 1537–1547, 2000. doi: 10.1515/JPEM.2000.13.9.1537. [DOI] [PubMed] [Google Scholar]
- 29.Charron S, Duong C, Ménard A, Roy J, Eliopoulos V, Lambert R, Deng AY. Epistasis, not numbers, regulates functions of clustered Dahl rat quantitative trait loci applicable to human hypertension. Hypertension : 1300–1308, 2005. doi: 10.1161/01.HYP.0000192024.72367.c3. [DOI] [PubMed] [Google Scholar]
- 30.Charron S, Lambert R, Eliopoulos V, Duong C, Ménard A, Roy J, Deng AY. A loss of genome buffering capacity of Dahl salt-sensitive model to modulate blood pressure as a cause of hypertension. Hum Mol Genet : 3877–3884, 2005. doi: 10.1093/hmg/ddi412. [DOI] [PubMed] [Google Scholar]
- 31.Chauvet C, Charron S, Ménard A, Xiao C, Roy J, Deng AY. Submegabase resolution of epistatically interacting quantitative trait loci for blood pressure applicable for essential hypertension. J Hypertens : 893–901, 2008. doi: 10.1097/HJH.0b013e3282f85ded. [DOI] [PubMed] [Google Scholar]
- 32.Chauvet C, Crespo K, Ménard A, Roy J, Deng AY. Modularization and epistatic hierarchy determine homeostatic actions of multiple blood pressure quantitative trait loci. Hum Mol Genet : 4451–4459, 2013. doi: 10.1093/hmg/ddt294. [DOI] [PubMed] [Google Scholar]
- 33.Chauvet C, Ménard A, Tremblay J, Xiao C, Shi Y, L’Heureux N, Cardin S, Tardif JC, Nattel S, Deng AY. Cardiac pathways distinguish two epistatic modules enacting BP quantitative trait loci and candidate gene analysis. Hypertens Res : 631–637, 2009. doi: 10.1038/hr.2009.70. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Gu D, Jaquish CE, Chen CS, Rao DC, Liu D, Hixson JE, Hamm LL, Gu CC, Whelton PK, He J; GenSalt Collaborative Research Group . Association between blood pressure responses to the cold pressor test and dietary sodium intervention in a Chinese population. Arch Intern Med : 1740–1746, 2008. doi: 10.1001/archinte.168.16.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M, Chen X, Guo Y, Shi R, Zhang G. Brain natriuretic peptide rs198388 polymorphism and essential hypertension in Hunan Han people. Zhong Nan Da Xue Xue Bao Yi Xue Ban : 1207–1213, 2010. doi: 10.3969/j.issn.1672-7347.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Chen YP, Tsai CW, Shen CY, Day CH, Yeh YL, Chen RJ, Ho TJ, Padma VV, Kuo WW, Huang CY. Palmitic acid interferes with energy metabolism balance by adversely switching the SIRT1-CD36-fatty acid pathway to the PKC zeta-GLUT4-glucose pathway in cardiomyoblasts. J Nutr Biochem : 137–149, 2016. doi: 10.1016/j.jnutbio.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 36a.Cheng X, Waghulde H, Mell B, Morgan EE, Pruett-Miller SM, Joe B. Positional cloning of quantitative trait nucleotides for blood pressure and cardiac QT-interval by targeted CRISPR/Cas9 editing of a novel long non-coding RNA. PLoS Genet : e1006961, 2017. doi: 10.1371/journa1.pgen.1006961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cicila GT, Choi C, Dene H, Lee SJ, Rapp JP. Two blood pressure/cardiac mass quantitative trait loci on chromosome 3 in Dahl rats. Mamm Genome : 112–116, 1999. doi: 10.1007/s003359900954. [DOI] [PubMed] [Google Scholar]
- 38.Cicila GT, Dukhanina OI, Kurtz TW, Walder R, Garrett MR, Dene H, Rapp JP. Blood pressure and survival of a chromosome 7 congenic strain bred from Dahl rats. Mamm Genome : 896–902, 1997. doi: 10.1007/s003359900607. [DOI] [PubMed] [Google Scholar]
- 39.Cicila GT, Morgan EE, Lee SJ, Farms P, Yerga-Woolwine S, Toland EJ, Ramdath RS, Gopalakrishnan K, Bohman K, Nestor-Kalinoski AL, Khuder SA, Joe B. Epistatic genetic determinants of blood pressure and mortality in a salt-sensitive hypertension model. Hypertension : 725–732, 2009. doi: 10.1161/HYPERTENSIONAHA.108.126649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cicila GT, Rapp JP, Bloch KD, Kurtz TW, Pravenec M, Kren V, Hong CC, Quertermous T, Ng SC. Cosegregation of the endothelin-3 locus with blood pressure and relative heart weight in inbred Dahl rats. J Hypertens : 643–651, 1994. doi: 10.1097/00004872-199406000-00004. [DOI] [PubMed] [Google Scholar]
- 41.Cicila GT, Rapp JP, Wang JM, St Lezin E, Ng SC, Kurtz TW. Linkage of 11 beta-hydroxylase mutations with altered steroid biosynthesis and blood pressure in the Dahl rat. Nat Genet : 346–353, 1993. doi: 10.1038/ng0493-346. [DOI] [PubMed] [Google Scholar]
- 42.Clark JS, Jeffs B, Davidson AO, Lee WK, Anderson NH, Bihoreau MT, Brosnan MJ, Devlin AM, Kelman AW, Lindpaintner K, Dominiczak AF. Quantitative trait loci in genetically hypertensive rats. Possible sex specificity. Hypertension : 898–906, 1996. doi: 10.1161/01.HYP.28.5.898. [DOI] [PubMed] [Google Scholar]
- 43.Coburn CT, Hajri T, Ibrahimi A, Abumrad NA. Role of CD36 in membrane transport and utilization of long-chain fatty acids by different tissues. J Mol Neurosci : 117–122, 2001. doi: 10.1385/JMN:16:2-3:117. [DOI] [PubMed] [Google Scholar]
- 44.Cook HW. Additional notes on blood pressure in life insurance prognosis. The Medical Examiner and Practitioner : 335–336, 1906. [Google Scholar]
- 45.Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet : 829–840, 2006. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- 46.Cowley AW Jr, Moreno C, Jacob HJ, Peterson CB, Stingo FC, Ahn KW, Liu P, Vannucci M, Laud PW, Reddy P, Lazar J, Evans L, Yang C, Kurth T, Liang M. Characterization of biological pathways associated with a 1.37 Mbp genomic region protective of hypertension in Dahl S rats. Physiol Genomics : 398–410, 2014. doi: 10.1152/physiolgenomics.00179.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowley AW Jr, Nadeau JH, Baccarelli A, Berecek K, Fornage M, Gibbons GH, Harrison DG, Liang M, Nathanielsz PW, O’Connor DT, Ordovas J, Peng W, Soares MB, Szyf M, Tolunay HE, Wood KC, Zhao K, Galis ZS. Report of the National Heart, Lung, and Blood Institute Working Group on epigenetics and hypertension. Hypertension : 899–905, 2012. doi: 10.1161/HYPERTENSIONAHA.111.190116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cowley AW Jr, Roman RJ, Kaldunski ML, Dumas P, Dickhout JG, Greene AS, Jacob HJ. Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension : 456–461, 2001. doi: 10.1161/01.HYP.37.2.456. [DOI] [PubMed] [Google Scholar]
- 49.Cowley AW Jr, Stoll M, Greene AS, Kaldunski ML, Roman RJ, Tonellato PJ, Schork NJ, Dumas P, Jacob HJ. Genetically defined risk of salt sensitivity in an intercross of Brown Norway and Dahl S rats. Physiol Genomics : 107–115, 2000. [DOI] [PubMed] [Google Scholar]
- 50.Cowley AW Jr, Yang C, Kumar V, Lazar J, Jacob H, Geurts AM, Liu P, Dayton A, Kurth T, Liang M. Pappa2 is linked to salt-sensitive hypertension in Dahl S rats. Physiol Genomics : 62–72, 2016. doi: 10.1152/physiolgenomics.00097.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crespo K, Chauvet C, Blain M, Ménard A, Roy J, Deng AY. Normotension in Lewis and Dahl salt-resistant rats is governed by different genes. J Hypertens : 460–465, 2011. doi: 10.1097/HJH.0b013e328341f1cc. [DOI] [PubMed] [Google Scholar]
- 52.Crespo K, Ménard A, Deng AY. Hypertension Suppression, Not a Cumulative Thrust of Quantitative Trait Loci, Predisposes Blood Pressure Homeostasis to Normotension. Circ Cardiovasc Genet : 610–617, 2015. doi: 10.1161/CIRCGENETICS.114.000965. [DOI] [PubMed] [Google Scholar]
- 52a.Crespo K, Menard A, Deng AY. Retinoblastoma-associated protein 140 as a candidate for a novel etiological gene to hypertension. Clin Exp Hypertens : 533–540, 2016. doi: 10.3109/10641963.2016.1163373. [DOI] [PubMed] [Google Scholar]
- 53.Cui ZH, Ikeda K, Kawakami K, Gonda T, Masuda J, Nabika T. Exaggerated response to cold stress in a congenic strain for the quantitative trait locus for blood pressure. J Hypertens : 2103–2109, 2004. doi: 10.1097/00004872-200411000-00011. [DOI] [PubMed] [Google Scholar]
- 54.Cui ZH, Ikeda K, Kawakami K, Gonda T, Nabika T, Masuda J. Exaggerated response to restraint stress in rats congenic for the chromosome 1 blood pressure quantitative trait locus. Clin Exp Pharmacol Physiol : 464–469, 2003. doi: 10.1046/j.1440-1681.2003.03860.x. [DOI] [PubMed] [Google Scholar]
- 55.Dahl LK. Possible role of salt intake in the development of essential hypertension. 1960. Int J Epidemiol : 967–972, 2005. doi: 10.1093/ije/dyh317. [DOI] [PubMed] [Google Scholar]
- 56.Dahl LK, Heine M, Tassinari L. Effects of chronia excess salt ingestion. Evidence that genetic factors play an important role in susceptibility to experimental hypertension. J Exp Med : 1173–1190, 1962. doi: 10.1084/jem.115.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dahl LK, Heine M, Tassinari L. Role of genetic factors in susceptibility to experimental hypertension due to chronic excess salt ingestion. Nature : 480–482, 1962. doi: 10.1038/194480b0. [DOI] [PubMed] [Google Scholar]
- 58.Dale BL, Madhur MS. Linking inflammation and hypertension via LNK/SH2B3. Curr Opin Nephrol Hypertens : 87–93, 2016. doi: 10.1097/MNH.0000000000000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davidson AO, Schork N, Jaques BC, Kelman AW, Sutcliffe RG, Reid JL, Dominiczak AF. Blood pressure in genetically hypertensive rats. Influence of the Y chromosome. Hypertension : 452–459, 1995. doi: 10.1161/01.HYP.26.3.452. [DOI] [PubMed] [Google Scholar]
- 60.De Miguel C, Das S, Lund H, Mattson DL. T lymphocytes mediate hypertension and kidney damage in Dahl salt-sensitive rats. Am J Physiol Regul Integr Comp Physiol : R1136–R1142, 2010. doi: 10.1152/ajpregu.00298.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Miguel C, Lund H, Mattson DL. High dietary protein exacerbates hypertension and renal damage in Dahl SS rats by increasing infiltrating immune cells in the kidney. Hypertension : 269–274, 2011. doi: 10.1161/HYPERTENSIONAHA.110.154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet : 390–403, 2013. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng AY. Genetic basis of polygenic hypertension. Hum Mol Genet , R2: R195–R202, 2007. doi: 10.1093/hmg/ddm126. [DOI] [PubMed] [Google Scholar]
- 64.Deng AY, Chauvet C, Ménard A. Alterations in Fibronectin Type III Domain Containing 1 Protein Gene Are Associated With Hypertension. PLoS One : e0151399, 2016. doi: 10.1371/journal.pone.0151399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng AY, Dene H, Pravenec M, Rapp JP. Genetic mapping of two new blood pressure quantitative trait loci in the rat by genotyping endothelin system genes. J Clin Invest : 2701–2709, 1994. doi: 10.1172/JCI117284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng AY, Dene H, Rapp JP. Congenic strains for the blood pressure quantitative trait locus on rat chromosome 2. Hypertension : 199–202, 1997. doi: 10.1161/01.HYP.30.2.199. [DOI] [PubMed] [Google Scholar]
- 67.Deng AY, Ménard A, Xiao C, Roy J. Sexual dimorphism on hypertension of quantitative trait loci entrapped in Dahl congenic rats. Clin Exp Hypertens : 511–519, 2008. doi: 10.1080/10641960802251933. [DOI] [PubMed] [Google Scholar]
- 68.Deng AY, Rapp JP. Locus for the inducible, but not a constitutive, nitric oxide synthase cosegregates with blood pressure in the Dahl salt-sensitive rat. J Clin Invest : 2170–2177, 1995. doi: 10.1172/JCI117906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deng Y, Rapp JP. Cosegregation of blood pressure with angiotensin converting enzyme and atrial natriuretic peptide receptor genes using Dahl salt-sensitive rats. Nat Genet : 267–272, 1992. doi: 10.1038/ng0792-267. [DOI] [PubMed] [Google Scholar]
- 70.Dietrich S, Floegel A, Weikert C, Pischon T, Boeing H, Drogan D. Identification of Serum Metabolites Associated With Incident Hypertension in the European Prospective Investigation Into Cancer and Nutrition-Potsdam Study. Hypertension : 471–477, 2016. doi: 10.1161/HYPERTENSIONAHA.116.07292. [DOI] [PubMed] [Google Scholar]
- 71.Dublin LI, Lotka AJ. Length of Life: A Study of the Life Table. New York: Ronald Press, 1936. [Google Scholar]
- 72.Dublin LI, Lotka AJ, Spiegelman M. . Length of Life: A Study of the Life Table. New York: Ronald Press, 1949. [Google Scholar]
- 73.Dukhanina OI, Dene H, Deng AY, Choi CR, Hoebee B, Rapp JP. Linkage map and congenic strains to localize blood pressure QTL on rat chromosome 10. Mamm Genome : 229–235, 1997. doi: 10.1007/s003359900399. [DOI] [PubMed] [Google Scholar]
- 74.Duong C, Charron S, Deng Y, Xiao C, Ménard A, Roy J, Deng AY. Individual QTLs controlling quantitative variation in blood pressure inherited in a Mendelian mode. Heredity (Edinb) : 165–171, 2007. doi: 10.1038/sj.hdy.6800920. [DOI] [PubMed] [Google Scholar]
- 75.Dupont J, Dupont JC, Froment A, Milon H, Vincent M. Selection of three strains of rats with spontaneously different levels of blood pressure. Biomedicine : 36–41, 1973. [PubMed] [Google Scholar]
- 76.Dutil J, Deng AY. Further chromosomal mapping of a blood pressure QTL in Dahl rats on chromosome 2 using congenic strains. Physiol Genomics : 3–9, 2001. [DOI] [PubMed] [Google Scholar]
- 77.Dutil J, Eliopoulos V, Tremblay J, Hamet P, Charron S, Deng AY. Multiple quantitative trait loci for blood pressure interacting epistatically and additively on Dahl rat chromosome 2. Hypertension : 557–564, 2005. doi: 10.1161/01.HYP.0000158841.71658.5e. [DOI] [PubMed] [Google Scholar]
- 78.Dzau VJ, Jacob HJ, Lindpainter K, Ganten D, Lander ES. Genetic mapping in hypertension. J Vasc Surg : 930–931, 1992. doi: 10.1016/0741-5214(92)90757-Y. [DOI] [PubMed] [Google Scholar]
- 79.Eaves IA, Wicker LS, Ghandour G, Lyons PA, Peterson LB, Todd JA, Glynne RJ. Combining mouse congenic strains and microarray gene expression analyses to study a complex trait: the NOD model of type 1 diabetes. Genome Res : 232–243, 2002. doi: 10.1101/gr.214102. Article published online before print in January 2002. [DOI] [PubMed] [Google Scholar]
- 80.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet : 779–797, 2013. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Efendiev R, Krmar RT, Ogimoto G, Zwiller J, Tripodi G, Katz AI, Bianchi G, Pedemonte CH, Bertorello AM. Hypertension-linked mutation in the adducin alpha-subunit leads to higher AP2-mu2 phosphorylation and impaired Na+,K+-ATPase trafficking in response to GPCR signals and intracellular sodium. Circ Res : 1100–1108, 2004. doi: 10.1161/01.RES.0000149570.20845.89. [DOI] [PubMed] [Google Scholar]
- 82.Ehret GB, O’Connor AA, Weder A, Cooper RS, Chakravarti A. Follow-up of a major linkage peak on chromosome 1 reveals suggestive QTLs associated with essential hypertension: GenNet study. Eur J Hum Genet : 1650–1657, 2009. doi: 10.1038/ejhg.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eliopoulos V, Dutil J, Deng Y, Grondin M, Deng AY. Severe hypertension caused by alleles from normotensive Lewis for a quantitative trait locus on chromosome 2. Physiol Genomics : 70–75, 2005. doi: 10.1152/physiolgenomics.00019.2005. [DOI] [PubMed] [Google Scholar]
- 84.Ely DL, Turner ME. Hypertension in the spontaneously hypertensive rat is linked to the Y chromosome. Hypertension : 277–281, 1990. doi: 10.1161/01.HYP.16.3.277. [DOI] [PubMed] [Google Scholar]
- 85.Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, Grzybowski M, Lombard JH, Staruschenko A, Moreno C, Jacob HJ, Geurts AM. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc Natl Acad Sci USA : 12817–12822, 2014. doi: 10.1073/pnas.1410745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Esunge PM. From blood pressure to hypertension: the history of research. J R Soc Med : 621, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feinleib M, Garrison RJ, Fabsitz R, Christian JC, Hrubec Z, Borhani NO, Kannel WB, Rosenman R, Schwartz JT, Wagner JO. The NHLBI twin study of cardiovascular disease risk factors: methodology and summary of results. Am J Epidemiol : 284–295, 1977. doi: 10.1093/oxfordjournals.aje.a112464. [DOI] [PubMed] [Google Scholar]
- 88.Felts JH. Stephen Hales and the measurement of blood pressure. N C Med J : 602–603, 1977. [PubMed] [Google Scholar]
- 89.Feng D, Yang C, Geurts AM, Kurth T, Liang M, Lazar J, Mattson DL, O’Connor PM, Cowley AW Jr. Increased expression of NAD(P)H oxidase subunit p67(phox) in the renal medulla contributes to excess oxidative stress and salt-sensitive hypertension. Cell Metab : 201–208, 2012. doi: 10.1016/j.cmet.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ferrandi M, Cusi D, Molinari I, Del Vecchio L, Barlassina C, Rastaldi MP, Schena FP, Macciardi F, Marcantoni C, Roccatello D, Peters LL, Armelloni S, Min L, Giardino L, Mattinzoli D, Camisasca C, Palazzo F, Manunta P, Ferrari P, Bianchi G. Alpha- and beta-adducin polymorphisms affect podocyte proteins and proteinuria in rodents and decline of renal function in human IgA nephropathy. J Mol Med (Berl) : 203–217, 2010. doi: 10.1007/s00109-009-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fishbein L. Pheochromocytoma and Paraganglioma: Genetics, Diagnosis, and Treatment. Hematol Oncol Clin North Am : 135–150, 2016. doi: 10.1016/j.hoc.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 92.Flister MJ, Tsaih SW, O’Meara CC, Endres B, Hoffman MJ, Geurts AM, Dwinell MR, Lazar J, Jacob HJ, Moreno C. Identifying multiple causative genes at a single GWAS locus. Genome Res : 1996–2002, 2013. doi: 10.1101/gr.160283.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, Liu K, Morrison AC, Ganesh S, Kutlar A, Ramachandran VS, Polak JF, Fabsitz RR, Dries DL, Farlow DN, Redline S, Adeyemo A, Hirschorn JN, Sun YV, Wyatt SB, Penman AD, Palmas W, Rotter JI, Townsend RR, Doumatey AP, Tayo BO, Mosley TH Jr, Lyon HN, Kang SJ, Rotimi CN, Cooper RS, Franceschini N, Curb JD, Martin LW, Eaton CB, Kardia SL, Taylor HA, Caulfield MJ, Ehret GB, Johnson T, Chakravarti A, Zhu X, Levy D; International Consortium for Blood Pressure Genome-wide Association Studies (ICBP-GWAS) . Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet : 2273–2284, 2011. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fraillon D, Wynne KN, Funder JW. Further studies on neomycin and experimental hypertension. Clin Exp Pharmacol Physiol : 339–342, 1984. doi: 10.1111/j.1440-1681.1984.tb00274.x. [DOI] [PubMed] [Google Scholar]
- 95.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, Tayo BO, Sun YV, Gottesman O, Adeyemo A, Johnson AD, Young JH, Rice K, Duan Q, Chen F, Li Y, Tang H, Fornage M, Keene KL, Andrews JS, Smith JA, Faul JD, Guangfa Z, Guo W, Liu Y, Murray SS, Musani SK, Srinivasan S, Velez Edwards DR, Wang H, Becker LC, Bovet P, Bochud M, Broeckel U, Burnier M, Carty C, Chasman DI, Ehret G, Chen WM, Chen G, Chen W, Ding J, Dreisbach AW, Evans MK, Guo X, Garcia ME, Jensen R, Keller MF, Lettre G, Lotay V, Martin LW, Moore JH, Morrison AC, Mosley TH, Ogunniyi A, Palmas W, Papanicolaou G, Penman A, Polak JF, Ridker PM, Salako B, Singleton AB, Shriner D, Taylor KD, Vasan R, Wiggins K, Williams SM, Yanek LR, Zhao W, Zonderman AB, Becker DM, Berenson G, Boerwinkle E, Bottinger E, Cushman M, Eaton C, Nyberg F, Heiss G, Hirschhron JN, Howard VJ, Karczewsk KJ, Lanktree MB, Liu K, Liu Y, Loos R, Margolis K, Snyder M, Psaty BM, Schork NJ, Weir DR, Rotimi CN, Sale MM, Harris T, Kardia SL, Hunt SC, Arnett D, Redline S, Cooper RS, Risch NJ, Rao DC, Rotter JI, Chakravarti A, Reiner AP, Levy D, Keating BJ, Zhu X, Go MJ, Kim YJ, Lee J-Y, Jeon J-P, Kim SS, Han B-G, Cho YS, Sim X, Tay WT, Ong RTH, Seielstad M, Liu JJ, Aung T, Wong TY, Teo YY, Tai ES, Chen C-H, Chang L, Chen Y-T, Wu J-Y, Kelly TN, Gu D, Hixson JE, Sung YJ, He J, Tabara Y, Kokubo Y, Miki T, Iwai N, Kato N, Takeuchi F, Katsuya T, Nabika T, Sugiyama T, Zhang Y, Huang W, Zhang X, Zhou X, Jin L, Zhu D; Asian Genetic Epidemiology Network Consortium . Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet : 545–554, 2013. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Frantz S, Clemitson JR, Bihoreau MT, Gauguier D, Samani NJ. Genetic dissection of region around the Sa gene on rat chromosome 1: evidence for multiple loci affecting blood pressure. Hypertension : 216–221, 2001. doi: 10.1161/01.HYP.38.2.216. [DOI] [PubMed] [Google Scholar]
- 97.Friso S, Carvajal CA, Fardella CE, Olivieri O. Epigenetics and arterial hypertension: the challenge of emerging evidence. Transl Res : 154–165, 2015. doi: 10.1016/j.trsl.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 98.Fung MM, Salem RM, Lipkowitz MS, Bhatnagar V, Pandey B, Schork NJ, O’Connor DT; AASK Study Investigators . Methylenetetrahydrofolate reductase (MTHFR) polymorphism A1298C (Glu429Ala) predicts decline in renal function over time in the African-American Study of Kidney Disease and Hypertension (AASK) Trial and Veterans Affairs Hypertension Cohort (VAHC). Nephrol Dial Transplant : 197–205, 2012. doi: 10.1093/ndt/gfr257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Galla S, Chakraborty S, Mell B, Vijay-Kumar M, Joe B. Microbiotal-host interactions and hypertension. Physiology (Bethesda) : 224–233, 2017. doi: 10.1152/physiol.00003.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garrett MR, Dene H, Rapp JP. Time-course genetic analysis of albuminuria in Dahl salt-sensitive rats on low-salt diet. J Am Soc Nephrol : 1175–1187, 2003. doi: 10.1097/01.ASN.0000060572.13794.58. [DOI] [PubMed] [Google Scholar]
- 101.Garrett MR, Dene H, Walder R, Zhang QY, Cicila GT, Assadnia S, Deng AY, Rapp JP. Genome scan and congenic strains for blood pressure QTL using Dahl salt-sensitive rats. Genome Res : 711–723, 1998. doi: 10.1101/gr.8.7.711. [DOI] [PubMed] [Google Scholar]
- 102.Garrett MR, Joe B, Dene H, Rapp JP. Identification of blood pressure quantitative trait loci that differentiate two hypertensive strains. J Hypertens : 2399–2406, 2002. doi: 10.1097/00004872-200212000-00019. [DOI] [PubMed] [Google Scholar]
- 103.Garrett MR, Joe B, Yerga-Woolwine S. Genetic linkage of urinary albumin excretion in Dahl salt-sensitive rats: influence of dietary salt and confirmation using congenic strains. Physiol Genomics : 39–49, 2006. doi: 10.1152/physiolgenomics.00150.2005. [DOI] [PubMed] [Google Scholar]
- 104.Garrett MR, Meng H, Rapp JP, Joe B. Locating a blood pressure quantitative trait locus within 117 kb on the rat genome: substitution mapping and renal expression analysis. Hypertension : 451–459, 2005. doi: 10.1161/01.HYP.0000154678.64340.7f. [DOI] [PubMed] [Google Scholar]
- 105.Garrett MR, Rapp JP. Defining the blood pressure QTL on chromosome 7 in Dahl rats by a 177-kb congenic segment containing Cyp11b1. Mamm Genome : 268–273, 2003. doi: 10.1007/s00335-002-2245-9. [DOI] [PubMed] [Google Scholar]
- 106.Garrett MR, Rapp JP. Multiple blood pressure QTL on rat Chromosome 2 defined by congenic Dahl rats. Mamm Genome : 41–44, 2002. doi: 10.1007/s00335-001-2114-y. [DOI] [PubMed] [Google Scholar]
- 107.Garrett MR, Rapp JP. Two closely linked interactive blood pressure QTL on rat chromosome 5 defined using congenic Dahl rats. Physiol Genomics : 81–86, 2002. doi: 10.1152/physiolgenomics.00080.2001. [DOI] [PubMed] [Google Scholar]
- 108.Garrett MR, Saad Y, Dene H, Rapp JP. Blood pressure QTL that differentiate Dahl salt-sensitive and spontaneously hypertensive rats. Physiol Genomics : 33–38, 2000. [DOI] [PubMed] [Google Scholar]
- 109.Garrett MR, Zhang X, Dukhanina OI, Deng AY, Rapp JP. Two linked blood pressure quantitative trait loci on chromosome 10 defined by dahl rat congenic strains. Hypertension : 779–785, 2001. doi: 10.1161/hy1001.091503. [DOI] [PubMed] [Google Scholar]
- 110.Garrod AE. About Alkaptonuria. Med Chir Trans : 69–78, 1902. [PMC free article] [PubMed] [Google Scholar]
- 111.Garrod AE. Inborn Errors of Metabolism. London: H. Frowde and Hodder & Stoughton, 1923. [Google Scholar]
- 112.Garrod AE. The incidence of alkaptonuria: a study in chemical individuality. 1902. Mol Med : 274–282, 1996. [PMC free article] [PubMed] [Google Scholar]
- 113.Garrod AE. The incidence of alkaptonuria: a study in chemical individuality. 1902 [classical article]. Yale J Biol Med : 221–231, 2002. [PMC free article] [PubMed] [Google Scholar]
- 114.Garrod AE. The Lancet. The incidence of alkaptonuria: a study in chemical individuality. Nutr Rev : 81–83, 1975. doi: 10.1111/j.1753-4887.1975.tb06025.x. [DOI] [PubMed] [Google Scholar]
- 115.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science : 433, 2009. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera, Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Worley KC, Cooney AJ, D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Albà M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Venter JC, Payseur BA, Bourque G, López-Otín C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F; Rat Genome Sequencing Project Consortium . Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature : 493–521, 2004. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 117.Glatz JF, Nabben M, Heather LC, Bonen A, Luiken JJ. Regulation of the subcellular trafficking of CD36, a major determinant of cardiac fatty acid utilization. Biochim Biophys Acta : 1461–1471, 2016. doi: 10.1016/j.bbalip.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 118.Gopalakrishnan K, Kumarasamy S, Abdul-Majeed S, Kalinoski AL, Morgan EE, Gohara AF, Nauli SM, Filipiak WE, Saunders TL, Joe B. Targeted disruption of Adamts16 gene in a rat genetic model of hypertension. Proc Natl Acad Sci USA : 20555–20559, 2012. doi: 10.1073/pnas.1211290109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gopalakrishnan K, Kumarasamy S, Mell B, Joe B. Genome-wide identification of long noncoding RNAs in rat models of cardiovascular and renal disease. Hypertension : 200–210, 2015. doi: 10.1161/HYPERTENSIONAHA.114.04498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gopalakrishnan K, Kumarasamy S, Yan Y, Liu J, Kalinoski A, Kothandapani A, Farms P, Joe B. Increased Expression of Rififylin in A < 330 Kb Congenic Strain is Linked to Impaired Endosomal Recycling in Proximal Tubules. Front Genet : 138, 2012. doi: 10.3389/fgene.2012.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gopalakrishnan K, Morgan EE, Yerga-Woolwine S, Farms P, Kumarasamy S, Kalinoski A, Liu X, Wu J, Liu L, Joe B. Augmented rififylin is a risk factor linked to aberrant cardiomyocyte function, short-QT interval and hypertension. Hypertension : 764–771, 2011. doi: 10.1161/HYPERTENSIONAHA.110.165803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gopalakrishnan K, Saikumar J, Peters CG, Kumarasamy S, Farms P, Yerga-Woolwine S, Toland EJ, Schnackel W, Giovannucci DR, Joe B. Defining a rat blood pressure quantitative trait locus to a <81.8 kb congenic segment: comprehensive sequencing and renal transcriptome analysis. Physiol Genomics : 153–161, 2010. doi: 10.1152/physiolgenomics.00122.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Graham LA, Padmanabhan S, Fraser NJ, Kumar S, Bates JM, Raffi HS, Welsh P, Beattie W, Hao S, Leh S, Hultstrom M, Ferreri NR, Dominiczak AF, Graham D, McBride MW. Validation of uromodulin as a candidate gene for human essential hypertension. Hypertension : 551–558, 2014. doi: 10.1161/HYPERTENSIONAHA.113.01423. [DOI] [PubMed] [Google Scholar]
- 123a.Grondin M, Eliopoulos V, Lambert R, Deng Y, Ariyarajah A, Moujahidine M, DutiI J, Charron S, Deng AY. Complete and overlapping congenics proving the existence of a quantitative trait locus for blood pressure on Dahl rat chromosome 17. Physiol Genomics : 112–116, 2005. [DOI] [PubMed] [Google Scholar]
- 124.Guerrera D, Shah J, Vasileva E, Sluysmans S, Méan I, Jond L, Poser I, Mann M, Hyman AA, Citi S. PLEKHA7 Recruits PDZD11 to Adherens Junctions to Stabilize Nectins. J Biol Chem : 11016–11029, 2016. doi: 10.1074/jbc.M115.712935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hamilton M, Pickering GW, Roberts JA, Sowry GS. The aetiology of essential hypertension. 4. The role of inheritance. Clin Sci : 273–304, 1954. [PubMed] [Google Scholar]
- 126.Hamilton M, Pickering GW, Roberts JA, Sowry GS. The aetiology of essential hypertension. I. The arterial pressure in the general population. Clin Sci : 11–35, 1954. [PubMed] [Google Scholar]
- 127.Hamilton M, Pickering GW, Roberts JA, Sowry GS. The aetiology of essential hypertension. II. Scores for arterial blood pressures adjusted for differences in age and sex. Clin Sci : 37–49, 1954. [PubMed] [Google Scholar]
- 128.Hanlon P, Lorenz WA, Shao Z, Harper JM, Galecki AT, Miller RA, Burke DT. Three-locus and four-locus QTL interactions influence mouse insulin-like growth factor-I. Physiol Genomics : 46–54, 2006. doi: 10.1152/physiolgenomics.00247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harris EL, Grigor MR, Thompson CM. Cosegregation of the Tnfalpha locus with cardiovascular phenotypes in the F2 generation of a New Zealand genetically hypertensive and Brown Norway cross. Clin Exp Pharmacol Physiol : 204–207, 1998. doi: 10.1111/j.1440-1681.1998.t01-17-.x. [DOI] [PubMed] [Google Scholar]
- 130.Harris EL, Phelan EL, Thompson CM, Millar JA, Grigor MR. Heart mass and blood pressure have separate genetic determinants in the New Zealand genetically hypertensive (GH) rat. J Hypertens : 397–404, 1995. doi: 10.1097/00004872-199504000-00004. [DOI] [PubMed] [Google Scholar]
- 131.Havlik RJ, Garrison RJ, Feinleib M, Kannel WB, Castelli WP, McNamara PM. Blood pressure aggregation in families. Am J Epidemiol : 304–312, 1979. doi: 10.1093/oxfordjournals.aje.a112815. [DOI] [PubMed] [Google Scholar]
- 132.Heller J, Hellerová S, Dobesová Z, Kunes J, Zicha J. The Prague Hypertensive Rat: a new model of genetic hypertension. Clin Exp Hypertens : 807–818, 1993. doi: 10.3109/10641969309041643. [DOI] [PubMed] [Google Scholar]
- 133.Hennekens CH, Jesse MJ, Klein BE, Gourley JE, Blumenthal S. Aggregation of blood pressure in infants and their siblings. Am J Epidemiol : 457–463, 1976. doi: 10.1093/oxfordjournals.aje.a112247. [DOI] [PubMed] [Google Scholar]
- 134.Hernandez AL, Kitz A, Wu C, Lowther DE, Rodriguez DM, Vudattu N, Deng S, Herold KC, Kuchroo VK, Kleinewietfeld M, Hafler DA. Sodium chloride inhibits the suppressive function of FOXP3+ regulatory T cells. J Clin Invest : 4212–4222, 2015. doi: 10.1172/JCI81151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Herrera VL, Lopez LV, Ruiz-Opazo N. Alpha1 Na,K-ATPase and Na,K,2Cl-cotransporte/D3mit3 loci interact to increase susceptibility to salt-sensitive hypertension in Dahl S(HSD) rats. Mol Med : 125–134, 2001. [PMC free article] [PubMed] [Google Scholar]
- 136.Herrera VL, Pasion KA, Moran AM, Ruiz-Opazo N. Dahl (S x R) congenic strain analysis confirms and defines a chromosome 5 female-specific blood pressure quantitative trait locus to <7 Mbp. PLoS One : e42214, 2012. doi: 10.1371/journal.pone.0042214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Herrera VL, Pasion KA, Tan GA, Moran AM, Ruiz-Opazo N. Sex-specific effects on spatial learning and memory, and sex-independent effects on blood pressure of a <3.3 Mbp rat chromosome 2 QTL region in Dahl salt-sensitive rats. PLoS One : e67673, 2013. doi: 10.1371/journal.pone.0067673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Herrera VL, Pasion KA, Tan GA, Ruiz-Opazo N. Dahl (S × R) rat congenic strain analysis confirms and defines a chromosome 17 spatial navigation quantitative trait locus to <10 Mbp. PLoS One : e58280, 2013. doi: 10.1371/journal.pone.0058280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Herrera VL, Tsikoudakis A, Ponce LR, Matsubara Y, Ruiz-Opazo N. Sex-specific QTLs and interacting loci underlie salt-sensitive hypertension and target organ complications in Dahl S/jrHS hypertensive rats. Physiol Genomics : 172–179, 2006. doi: 10.1152/physiolgenomics.00285.2005. [DOI] [PubMed] [Google Scholar]
- 140.Herrera VL, Xie HX, Lopez LV, Schork NJ, Ruiz-Opazo N. The alpha1 Na,K-ATPase gene is a susceptibility hypertension gene in the Dahl salt-sensitiveHSD rat. J Clin Invest : 1102–1111, 1998. doi: 10.1172/JCI3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Heslop BF, Phelan EL. The GH and AS hypertensive rat strains. Lab Anim : 41–46, 1973. doi: 10.1258/002367773781005932. [DOI] [PubMed] [Google Scholar]
- 142.Hilbert P, Lindpaintner K, Beckmann JS, Serikawa T, Soubrier F, Dubay C, Cartwright P, De Gouyon B, Julier C, Takahasi S, et al. Chromosomal mapping of two genetic loci associated with blood-pressure regulation in hereditary hypertensive rats. Nature : 521–529, 1991. doi: 10.1038/353521a0. [DOI] [PubMed] [Google Scholar]
- 143.Hirano K, Kuwasako T, Nakagawa-Toyama Y, Janabi M, Yamashita S, Matsuzawa Y. Pathophysiology of human genetic CD36 deficiency. Trends Cardiovasc Med : 136–141, 2003. doi: 10.1016/S1050-1738(03)00026-4. [DOI] [PubMed] [Google Scholar]
- 144.Ho JE, Levy D, Rose L, Johnson AD, Ridker PM, Chasman DI. Discovery and replication of novel blood pressure genetic loci in the Women’s Genome Health Study. J Hypertens : 62–69, 2011. doi: 10.1097/HJH.0b013e3283406927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hong KW, Jin HS, Lim JE, Kim S, Go MJ, Oh B. Recapitulation of two genomewide association studies on blood pressure and essential hypertension in the Korean population. J Hum Genet : 336–341, 2010. doi: 10.1038/jhg.2010.31. [DOI] [PubMed] [Google Scholar]
- 146.Honour J. The possible involvement of intestinal bacteria in steroidal hypertension. Endocrinology : 285–287, 1982. doi: 10.1210/endo-110-1-285. [DOI] [PubMed] [Google Scholar]
- 147.Honour JW. Historical perspective: gut dysbiosis and hypertension. Physiol Genomics : 443–446, 2015. doi: 10.1152/physiolgenomics.00063.2015. [DOI] [PubMed] [Google Scholar]
- 148.Honour JW, Borriello SP, Ganten U, Honour P. Antibiotics attenuate experimental hypertension in rats. J Endocrinol : 347–350, 1985. doi: 10.1677/joe.0.1050347. [DOI] [PubMed] [Google Scholar]
- 149.Honour JW, Tourniaire J, Biglieri EG, Shackleton CH. Urinary steroid excretion in 17 alpha-hydroxylase deficiency. J Steroid Biochem : 495–505, 1978. doi: 10.1016/0022-4731(78)90115-2. [DOI] [PubMed] [Google Scholar]
- 150.Huan T, Esko T, Peters MJ, Pilling LC, Schramm K, Schurmann C, Chen BH, Liu C, Joehanes R, Johnson AD, Yao C, Ying SX, Courchesne P, Milani L, Raghavachari N, Wang R, Liu P, Reinmaa E, Dehghan A, Hofman A, Uitterlinden AG, Hernandez DG, Bandinelli S, Singleton A, Melzer D, Metspalu A, Carstensen M, Grallert H, Herder C, Meitinger T, Peters A, Roden M, Waldenberger M, Dörr M, Felix SB, Zeller T, Vasan R, O’Donnell CJ, Munson PJ, Yang X, Prokisch H, Völker U, van Meurs JB, Ferrucci L, Levy D; International Consortium for Blood Pressure GWAS (ICBP) . A meta-analysis of gene expression signatures of blood pressure and hypertension. PLoS Genet : e1005035, 2015. doi: 10.1371/journal.pgen.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huan T, Meng Q, Saleh MA, Norlander AE, Joehanes R, Zhu J, Chen BH, Zhang B, Johnson AD, Ying S, Courchesne P, Raghavachari N, Wang R, Liu P, O’Donnell CJ, Vasan R, Munson PJ, Madhur MS, Harrison DG, Yang X, Levy D. Integrative network analysis reveals molecular mechanisms of blood pressure regulation. Mol Syst Biol : 799, 2015. doi: 10.15252/msb.20145399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huang BS, Ahmad M, Deng AY, Leenen FH. Neuronal responsiveness to central Na+ in 2 congenic strains of Dahl salt-sensitive rats. Hypertension : 1315–1320, 2007. doi: 10.1161/HYPERTENSIONAHA.106.086363. [DOI] [PubMed] [Google Scholar]
- 153.Huang BS, Van Vliet BN, Leenen FH. Increases in CSF [Na+] precede the increases in blood pressure in Dahl S rats and SHR on a high-salt diet. Am J Physiol Heart Circ Physiol : H1160–H1166, 2004. doi: 10.1152/ajpheart.00126.2004. [DOI] [PubMed] [Google Scholar]
- 154.Huang W, Richards S, Carbone MA, Zhu D, Anholt RR, Ayroles JF, Duncan L, Jordan KW, Lawrence F, Magwire MM, Warner CB, Blankenburg K, Han Y, Javaid M, Jayaseelan J, Jhangiani SN, Muzny D, Ongeri F, Perales L, Wu YQ, Zhang Y, Zou X, Stone EA, Gibbs RA, Mackay TF. Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci USA : 15553–15559, 2012. doi: 10.1073/pnas.1213423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hübner N, Lee YA, Lindpaintner K, Ganten D, Kreutz R. Congenic substitution mapping excludes Sa as a candidate gene locus for a blood pressure quantitative trait locus on rat chromosome 1. Hypertension : 643–648, 1999. doi: 10.1161/01.HYP.34.4.643. [DOI] [PubMed] [Google Scholar]
- 156.Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Müller A, Cook SA, Kurtz TW, Whittaker J, Pravenec M, Aitman TJ. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet : 243–253, 2005. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 157.Huggins GS, Bacani CJ, Boltax J, Aikawa R, Leiden JM. Friend of GATA 2 physically interacts with chicken ovalbumin upstream promoter-TF2 (COUP-TF2) and COUP-TF3 and represses COUP-TF2-dependent activation of the atrial natriuretic factor promoter. J Biol Chem : 28029–28036, 2001. doi: 10.1074/jbc.M103577200. [DOI] [PubMed] [Google Scholar]
- 158.Hunt SC, Stephenson SH, Hopkins PN, Williams RR. Predictors of an increased risk of future hypertension in Utah. A screening analysis. Hypertension : 969–976, 1991. doi: 10.1161/01.HYP.17.6.969. [DOI] [PubMed] [Google Scholar]
- 159.Ingram BW, Funder JW. Neomycin and experimental hypertension. Clin Exp Pharmacol Physiol : 331–333, 1982. doi: 10.1111/j.1440-1681.1982.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 160.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sõber S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjögren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimäki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kähönen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Köttgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grässler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stančáková A, Raffel LJ, Yao J, Kathiresan S, O’Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikäinen LP, Soininen P, Tukiainen T, Würtz P, Ong RT, Dörr M, Kroemer HK, Völker U, Völzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Järvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T; International Consortium for Blood Pressure Genome-Wide Association Studies; CARDIoGRAM consortium; CKDGen Consortium; KidneyGen Consortium; EchoGen consortium; CHARGE-HF consortium . Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature : 103–109, 2011. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Iwai N, Inagami T. Quantitative analysis of renin gene expression in extrarenal tissues by polymerase chain reaction method. J Hypertens : 717–724, 1992. doi: 10.1097/00004872-199208000-00004. [DOI] [PubMed] [Google Scholar]
- 162.Jacob HJ, Lazar J, Dwinell MR, Moreno C, Geurts AM. Gene targeting in the rat: advances and opportunities. Trends Genet : 510–518, 2010. doi: 10.1016/j.tig.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jacob HJ, Lindpaintner K, Lincoln SE, Kusumi K, Bunker RK, Mao YP, Ganten D, Dzau VJ, Lander ES. Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell : 213–224, 1991. doi: 10.1016/0092-8674(91)90584-L. [DOI] [PubMed] [Google Scholar]
- 164.Jaworski RL, Jirout M, Closson S, Breen L, Flodman PL, Spence MA, Kren V, Krenova D, Pravenec M, Printz MP. Heart rate and blood pressure quantitative trait loci for the airpuff startle reaction. Hypertension : 348–352, 2002. doi: 10.1161/hy0202.103419. [DOI] [PubMed] [Google Scholar]
- 165.Jiang S, Hsu YH, Xu X, Xing H, Chen C, Niu T, Zhang Y, Peng S, Xu X. The C677T polymorphism of the methylenetetrahydrofolate reductase gene is associated with the level of decrease on diastolic blood pressure in essential hypertension patients treated by angiotensin-converting enzyme inhibitor. Thromb Res : 361–369, 2004. doi: 10.1016/j.thromres.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 166.Joe B, Garrett M.. Substitution Mapping using congenic strains to detect genes controlling blood pressure In: Cardiovascular Genomics, edited by Raizada MK, Paton JFR, Kasparov S, Katovich MJ. New York: Humana, 2005, p. 41–58. doi: 10.1385/1-59259-883-8:041 [DOI] [Google Scholar]
- 167.Joe B, Garrett MR. Genetic analysis of inherited hypertension in the rat In: Genetics of Hypertension, edited by Dominiczak AF, Connell JMC. Netherlands: Elsevier, 2007, p. 177–200. [Google Scholar]
- 168.Joe B, Garrett MR, Dene H, Rapp JP. Substitution mapping of a blood pressure quantitative trait locus to a 2.73 Mb region on rat chromosome 1. J Hypertens : 2077–2084, 2003. doi: 10.1097/00004872-200311000-00017. [DOI] [PubMed] [Google Scholar]
- 169.Joe B, Letwin NE, Garrett MR, Dhindaw S, Frank B, Sultana R, Verratti K, Rapp JP, Lee NH. Transcriptional profiling with a blood pressure QTL interval-specific oligonucleotide array. Physiol Genomics : 318–326, 2005. doi: 10.1152/physiolgenomics.00164.2004. [DOI] [PubMed] [Google Scholar]
- 170.Joe B, Saad Y, Dhindaw S, Lee NH, Frank BC, Achinike OH, Luu TV, Gopalakrishnan K, Toland EJ, Farms P, Yerga-Woolwine S, Manickavasagam E, Rapp JP, Garrett MR, Coe D, Apte SS, Rankinen T, Pérusse L, Ehret GB, Ganesh SK, Cooper RS, O’Connor A, Rice T, Weder AB, Chakravarti A, Rao DC, Bouchard C. Positional identification of variants of Adamts16 linked to inherited hypertension. Hum Mol Genet : 2825–2838, 2009. doi: 10.1093/hmg/ddp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170a.Johnson MD, He L, Herman D, Wakimoto H, Wallace CA, Zidek V, Mlejnek P, Musilova A, Simakova M, Vorlicek J, Kren V, Viklicky O, Qi NR, Wang J, Seidman CE, Seidman J, Kurtz TW, Aitman TJ, Pravenec M. Dissection of chromosome 18 blood pressure and salt-sensitivity quantitative trait loci in the spontaneously hypertensive rat. Hypertension : 639–645, 2009. doi: 10.1161/HYPERTENSIONAHA.108.126664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, Morris RW, Tzoulaki I, O’Brien ET, Poulter NR, Sever P, Shields DC, Thom S, Wannamethee SG, Whincup PH, Brown MJ, Connell JM, Dobson RJ, Howard PJ, Mein CA, Onipinla A, Shaw-Hawkins S, Zhang Y, Davey Smith G, Day IN, Lawlor DA, Goodall AH, Fowkes FG, Abecasis GR, Elliott P, Gateva V, Braund PS, Burton PR, Nelson CP, Tobin MD, van der Harst P, Glorioso N, Neuvrith H, Salvi E, Staessen JA, Stucchi A, Devos N, Jeunemaitre X, Plouin PF, Tichet J, Juhanson P, Org E, Putku M, Sõber S, Veldre G, Viigimaa M, Levinsson A, Rosengren A, Thelle DS, Hastie CE, Hedner T, Lee WK, Melander O, Wahlstrand B, Hardy R, Wong A, Cooper JA, Palmen J, Chen L, Stewart AF, Wells GA, Westra HJ, Wolfs MG, Clarke R, Franzosi MG, Goel A, Hamsten A, Lathrop M, Peden JF, Seedorf U, Watkins H, Ouwehand WH, Sambrook J, Stephens J, Casas JP, Drenos F, Holmes MV, Kivimaki M, Shah S, Shah T, Talmud PJ, Whittaker J, Wallace C, Delles C, Laan M, Kuh D, Humphries SE, Nyberg F, Cusi D, Roberts R, Newton-Cheh C, Franke L, Stanton AV, Dominiczak AF, Farrall M, Hingorani AD, Samani NJ, Caulfield MJ, Munroe PB; Cardiogenics Consortium; Global BPgen Consortium . Blood pressure loci identified with a gene-centric array. Am J Hum Genet : 688–700, 2011. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jose PA, Raj D. Gut microbiota in hypertension. Curr Opin Nephrol Hypertens : 403–409, 2015. doi: 10.1097/MNH.0000000000000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kalousová M, Muravská A, Zima T. Pregnancy-associated plasma protein A (PAPP-A) and preeclampsia. Adv Clin Chem : 169–209, 2014. doi: 10.1016/B978-0-12-800094-6.00005-4. [DOI] [PubMed] [Google Scholar]
- 174.Kantachuvesiri S, Haley CS, Fleming S, Kurian K, Whitworth CE, Wenham P, Kotelevtsev Y, Mullins JJ. Genetic mapping of modifier loci affecting malignant hypertension in TGRmRen2 rats. Kidney Int : 414–420, 1999. doi: 10.1046/j.1523-1755.1999.00571.x. [DOI] [PubMed] [Google Scholar]
- 175.Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Köhl J, Wahl L, Kuperman D, Germer S, Aud D, Peltz G, Wills-Karp M. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol : 221–226, 2000. doi: 10.1038/79759. [DOI] [PubMed] [Google Scholar]
- 176.Kato N, Hyne G, Bihoreau MT, Gauguier D, Lathrop GM, Rapp JP. Complete genome searches for quantitative trait loci controlling blood pressure and related traits in four segregating populations derived from Dahl hypertensive rats. Mamm Genome : 259–265, 1999. doi: 10.1007/s003359900983. [DOI] [PubMed] [Google Scholar]
- 177.Kato N, Mashimo T, Nabika T, Cui ZH, Ikeda K, Yamori Y. Genome-wide searches for blood pressure quantitative trait loci in the stroke-prone spontaneously hypertensive rat of a Japanese colony. J Hypertens : 295–303, 2003. doi: 10.1097/00004872-200302000-00020. [DOI] [PubMed] [Google Scholar]
- 178.Kato N, Nabika T, Liang YQ, Mashimo T, Inomata H, Watanabe T, Yanai K, Yamori Y, Yazaki Y, Sasazuki T. Isolation of a chromosome 1 region affecting blood pressure and vascular disease traits in the stroke-prone rat model. Hypertension : 1191–1197, 2003. doi: 10.1161/01.HYP.0000103161.27190.67. [DOI] [PubMed] [Google Scholar]
- 179.Kato N, Sugiyama T, Morita H, Nabika T, Kurihara H, Yamori Y, Yazaki Y. Genetic analysis of the atrial natriuretic peptide gene in essential hypertension. Clin Sci (Lond) : 251–258, 2000. doi: 10.1042/cs0980251. [DOI] [PubMed] [Google Scholar]
- 180.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K, Katsuya T, Yokota M, Kim YJ, Ong RT, Nabika T, Gu D, Chang LC, Kokubo Y, Huang W, Ohnaka K, Yamori Y, Nakashima E, Jaquish CE, Lee JY, Seielstad M, Isono M, Hixson JE, Chen YT, Miki T, Zhou X, Sugiyama T, Jeon JP, Liu JJ, Takayanagi R, Kim SS, Aung T, Sung YJ, Zhang X, Wong TY, Han BG, Kobayashi S, Ogihara T, Zhu D, Iwai N, Wu JY, Teo YY, Tai ES, Cho YS, He J. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet : 531–538, 2011. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Katsuya T, Higaki J, Miki T, Kohara K, Yagisawa H, Tanase H, Mikami H, Serikawa T, Nojima H, Ogihara T. Hypotensive effect associated with a phospholipase C-delta 1 gene mutation in the spontaneously hypertensive rat. Biochem Biophys Res Commun : 1359–1366, 1992. doi: 10.1016/0006-291X(92)90452-Q. [DOI] [PubMed] [Google Scholar]
- 182.Kennedy DJ, Kashyap SR. Pathogenic role of scavenger receptor CD36 in the metabolic syndrome and diabetes. Metab Syndr Relat Disord : 239–245, 2011. doi: 10.1089/met.2011.0003. [DOI] [PubMed] [Google Scholar]
- 183.Kim M, Platt MJ, Shibasaki T, Quaggin SE, Backx PH, Seino S, Simpson JA, Drucker DJ. GLP-1 receptor activation and Epac2 link atrial natriuretic peptide secretion to control of blood pressure. Nat Med : 567–575, 2013. doi: 10.1038/nm.3128. [DOI] [PubMed] [Google Scholar]
- 184.Klimes I, Weston K, Kovacs P, Gasperikova D, Jezova D, Kvetnansky R, Thompson JR, Sebokova E, Samani NJ. Mapping of genetic loci predisposing to hypertriglyceridaemia in the hereditary hypertriglyceridaemic rat: analysis of genetic association with related traits of the insulin resistance syndrome. Diabetologia : 352–358, 2003. doi: 10.1007/s00125-003-1035-6. [DOI] [PubMed] [Google Scholar]
- 185.Klöting I, Kovács P, van den Brandt J. Quantitative trait loci for body weight, blood pressure, blood glucose, and serum lipids: linkage analysis with wild rats (Rattus norvegicus). Biochem Biophys Res Commun : 1126–1133, 2001. doi: 10.1006/bbrc.2001.5091. [DOI] [PubMed] [Google Scholar]
- 185a.Klöting I, Voigt B, Kovács P.. Metabolic features of newly established congenic diabetes-prone BB. SHR rat strains. Life Sci : 973–979, 1998. [DOI] [PubMed] [Google Scholar]
- 186.Koffler M. Ambulatory treatment of hypertension with the Kempner rice diet. Harefuah : 45–47, 1955. [PubMed] [Google Scholar]
- 187.Koh-Tan HH, McBride MW, McClure JD, Beattie E, Young B, Dominiczak AF, Graham D. Interaction between chromosome 2 and 3 regulates pulse pressure in the stroke-prone spontaneously hypertensive rat. Hypertension : 33–40, 2013. doi: 10.1161/HYPERTENSIONAHA.111.00814. [DOI] [PubMed] [Google Scholar]
- 188.Koonen DP, Febbraio M, Bonnet S, Nagendran J, Young ME, Michelakis ED, Dyck JR. CD36 expression contributes to age-induced cardiomyopathy in mice. Circulation : 2139–2147, 2007. doi: 10.1161/CIRCULATIONAHA.107.712901. [DOI] [PubMed] [Google Scholar]
- 189.Korner PI. Essential Hypertension and Its Causes: Neural and Non-neural Mechanisms. Oxford, UK: Oxford Univ. Press, 2007. [Google Scholar]
- 190.Kotchen TA. Historical trends and milestones in hypertension research: a model of the process of translational research. Hypertension : 522–538, 2011. doi: 10.1161/HYPERTENSIONAHA.111.177766. [DOI] [PubMed] [Google Scholar]
- 191.Kotchen TA, Cowley AW Jr, Liang M. Ushering Hypertension Into a New Era of Precision Medicine. JAMA : 343–344, 2016. doi: 10.1001/jama.2015.18359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kotchen TA, Kotchen JM, Grim CE, George V, Kaldunski ML, Cowley AW, Hamet P, Chelius TH. Genetic determinants of hypertension: identification of candidate phenotypes. Hypertension : 7–13, 2000. doi: 10.1161/01.HYP.36.1.7. [DOI] [PubMed] [Google Scholar]
- 193.Kourtidis A, Anastasiadis PZ. Bringing together cell-to-cell adhesion and miRNA biology in cancer research. Future Oncol : 1211–1214, 2016. doi: 10.2217/fon-2016-0012. [DOI] [PubMed] [Google Scholar]
- 194.Kourtidis A, Anastasiadis PZ. PLEKHA7 defines an apical junctional complex with cytoskeletal associations and miRNA-mediated growth implications. Cell Cycle : 498–505, 2016. doi: 10.1080/15384101.2016.1141840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kourtidis A, Ngok SP, Pulimeno P, Feathers RW, Carpio LR, Baker TR, Carr JM, Yan IK, Borges S, Perez EA, Storz P, Copland JA, Patel T, Thompson EA, Citi S, Anastasiadis PZ. Distinct E-cadherin-based complexes regulate cell behaviour through miRNA processing or Src and p120 catenin activity. Nat Cell Biol : 1145–1157, 2015. doi: 10.1038/ncb3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kovács P, Voigt B, Klöting I. Alleles of the spontaneously hypertensive rat decrease blood pressure at loci on chromosomes 4 and 13. Biochem Biophys Res Commun : 586–589, 1997. doi: 10.1006/bbrc.1997.7342. [DOI] [PubMed] [Google Scholar]
- 197.Kovács P, Voigt B, Klöting I. Novel quantitative trait loci for blood pressure and related traits on rat chromosomes 1, 10, and 18. Biochem Biophys Res Commun : 343–348, 1997. doi: 10.1006/bbrc.1997.6782. [DOI] [PubMed] [Google Scholar]
- 198.Kren V, Pravenec M, Lu S, Krenova D, Wang JM, Wang N, Merriouns T, Wong A, St Lezin E, Lau D, Szpirer C, Szpirer J, Kurtz TW. Genetic isolation of a region of chromosome 8 that exerts major effects on blood pressure and cardiac mass in the spontaneously hypertensive rat. J Clin Invest : 577–581, 1997. doi: 10.1172/JCI119198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198a.Kren V, Qi N, Krenova D, Zidek V, Sladká M, Jáchymová M, Míková B, Horky K, Bonne A, Van Lith HA, Van Zutphen BF, Lau YF, Pravenec M, St Lezin E. Y-chromosome transfer induces changes in blood pressure and blood lipids in SHR. Hypertension : 1147–1152, 2001. [DOI] [PubMed] [Google Scholar]
- 199.Kreutz R, Hübner N, James MR, Bihoreau MT, Gauguier D, Lathrop GM, Ganten D, Lindpaintner K. Dissection of a quantitative trait locus for genetic hypertension on rat chromosome 10. Proc Natl Acad Sci USA : 8778–8782, 1995. doi: 10.1073/pnas.92.19.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kreutz R, Struk B, Rubattu S, Hübner N, Szpirer J, Szpirer C, Ganten D, Lindpaintner K. Role of the alpha-, beta-, and gamma-subunits of epithelial sodium channel in a model of polygenic hypertension. Hypertension : 131–136, 1997. doi: 10.1161/01.HYP.29.1.131. [DOI] [PubMed] [Google Scholar]
- 201.Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics : 237–244, 2012. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res : 1639–1645, 2009. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ku DD, Guo L, Dai J, Acuff CG, Steinhelper ME. Coronary vascular and endothelial reactivity changes in transgenic mice overexpressing atrial natriuretic factor. Am J Physiol Heart Circ Physiol : H2368–H2376, 1996. [DOI] [PubMed] [Google Scholar]
- 204.Kuijpers MH, Gruys E. Spontaneous hypertension and hypertensive renal disease in the fawn-hooded rat. Br J Exp Pathol : 181–190, 1984. [PMC free article] [PubMed] [Google Scholar]
- 205.Kulkarni H, Meikle PJ, Mamtani M, Weir JM, Barlow CK, Jowett JB, Bellis C, Dyer TD, Johnson MP, Rainwater DL, Almasy L, Mahaney MC, Comuzzie AG, Blangero J, Curran JE. Plasma lipidomic profile signature of hypertension in Mexican American families: specific role of diacylglycerols. Hypertension : 621–626, 2013. doi: 10.1161/HYPERTENSIONAHA.113.01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kumarasamy S, Waghulde H, Gopalakrishnan K, Mell B, Morgan E, Joe B. Mutation within the hinge region of the transcription factor Nr2f2 attenuates salt-sensitive hypertension. Nat Commun : 6252, 2015. doi: 10.1038/ncomms7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kupper N, Willemsen G, Riese H, Posthuma D, Boomsma DI, de Geus EJ. Heritability of daytime ambulatory blood pressure in an extended twin design. Hypertension : 80–85, 2005. doi: 10.1161/01.HYP.0000149952.84391.54. [DOI] [PubMed] [Google Scholar]
- 208.Kurtz TW, Simonet L, Kabra PM, Wolfe S, Chan L, Hjelle BL. Cosegregation of the renin allele of the spontaneously hypertensive rat with an increase in blood pressure. J Clin Invest : 1328–1332, 1990. doi: 10.1172/JCI114572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kuwasako T, Hirano K, Sakai N, Ishigami M, Hiraoka H, Yakub MJ, Yamauchi-Takihara K, Yamashita S, Matsuzawa Y. Lipoprotein abnormalities in human genetic CD36 deficiency associated with insulin resistance and abnormal fatty acid metabolism. Diabetes Care : 1647–1648, 2003. doi: 10.2337/diacare.26.5.1647-a. [DOI] [PubMed] [Google Scholar]
- 210.Lauzier B, Merlen C, Vaillant F, McDuff J, Bouchard B, Beguin PC, Dolinsky VW, Foisy S, Villeneuve LR, Labarthe F, Dyck JR, Allen BG, Charron G, Des Rosiers C. Post-translational modifications, a key process in CD36 function: lessons from the spontaneously hypertensive rat heart. J Mol Cell Cardiol : 99–108, 2011. doi: 10.1016/j.yjmcc.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 211.Lee SJ, Liu J, Westcott AM, Vieth JA, DeRaedt SJ, Yang S, Joe B, Cicila GT. Substitution mapping in Dahl rats identifies two distinct blood pressure quantitative trait loci within 1.12- and 1.25-mb intervals on chromosome 3. Genetics : 2203–2213, 2006. doi: 10.1534/genetics.106.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lee YH, Rosner B, Gould JB, Lowe EW, Kass EH. Familial aggregation of blood pressures of newborn infants and their mother. Pediatrics : 722–729, 1976. [PubMed] [Google Scholar]
- 213.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Köttgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genome-wide association study of blood pressure and hypertension. Nat Genet : 677–687, 2009. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet , Suppl 1: S3, 2007. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lewis O. Stephen Hales and the measurement of blood pressure. J Hum Hypertens : 865–871, 1994. [PubMed] [Google Scholar]
- 216.Liang M, Cowley AW Jr, Mattson DL, Kotchen TA, Liu Y. Epigenomics of hypertension. Semin Nephrol : 392–399, 2013. doi: 10.1016/j.semnephrol.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science : 289–293, 2009. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, Ulick S, Lalouel JM. A chimaeric 11 beta-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature : 262–265, 1992. doi: 10.1038/355262a0. [DOI] [PubMed] [Google Scholar]
- 219.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell : 545–556, 2001. doi: 10.1016/S0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 220.Lin Y, Lai X, Chen B, Xu Y, Huang B, Chen Z, Zhu S, Yao J, Jiang Q, Huang H, Wen J, Chen G. Genetic variations in CYP17A1, CACNB2 and PLEKHA7 are associated with blood pressure and/or hypertension in She ethnic minority of China. Atherosclerosis : 709–714, 2011. doi: 10.1016/j.atherosclerosis.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 221.Liu C, Li H, Qi Q, Lu L, Gan W, Loos RJ, Lin X. Common variants in or near FGF5, CYP17A1 and MTHFR genes are associated with blood pressure and hypertension in Chinese Hans. J Hypertens : 70–75, 2011. doi: 10.1097/HJH.0b013e32833f60ab. [DOI] [PubMed] [Google Scholar]
- 222.Liu X, Meng F, Yang P. Association study of CD36 single nucleotide polymorphisms with essential hypertension in the Northeastern Han Chinese. Gene : 410–415, 2013. doi: 10.1016/j.gene.2013.05.077. [DOI] [PubMed] [Google Scholar]
- 223.Liu Y, Liu P, Yang C, Cowley AW Jr, Liang M. Base-resolution maps of 5-methylcytosine and 5-hydroxymethylcytosine in Dahl S rats: effect of salt and genomic sequence. Hypertension : 827–838, 2014. doi: 10.1161/HYPERTENSIONAHA.113.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension : 974–982, 2010. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lodwick D, Kaiser MA, Harris J, Cumin F, Vincent M, Samani NJ. Analysis of the role of angiotensinogen in spontaneous hypertension. Hypertension : 1245–1251, 1995. doi: 10.1161/01.HYP.25.6.1245. [DOI] [PubMed] [Google Scholar]
- 226.Longini IM Jr, Higgins MW, Hinton PC, Moll PP, Keller JB. Environmental and genetic sources of familial aggregation of blood pressure in Tecumseh, Michigan. Am J Epidemiol : 131–144, 1984. doi: 10.1093/oxfordjournals.aje.a113862. [DOI] [PubMed] [Google Scholar]
- 227.Lu X, Wang L, Lin X, Huang J, Charles Gu C, He M, Shen H, He J, Zhu J, Li H, Hixson JE, Wu T, Dai J, Lu L, Shen C, Chen S, He L, Mo Z, Hao Y, Mo X, Yang X, Li J, Cao J, Chen J, Fan Z, Li Y, Zhao L, Li H, Lu F, Yao C, Yu L, Xu L, Mu J, Wu X, Deng Y, Hu D, Zhang W, Ji X, Guo D, Guo Z, Zhou Z, Yang Z, Wang R, Yang J, Zhou X, Yan W, Sun N, Gao P, Gu D. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet : 865–874, 2015. doi: 10.1093/hmg/ddu478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Luft FC. Twins in cardiovascular genetic research. Hypertension : 350–356, 2001. doi: 10.1161/01.HYP.37.2.350. [DOI] [PubMed] [Google Scholar]
- 229.Mackay TF. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet : 22–33, 2014. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Mackay TF, Moore JH. Erratum to: Why epistasis is important for tackling complex human disease genetics. Genome Med : 85, 2015. doi: 10.1186/s13073-015-0205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Mackay TF, Moore JH. Why epistasis is important for tackling complex human disease genetics. Genome Med : 124, 2014. doi: 10.1186/gm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Manunta P, Cusi D, Barlassina C, Righetti M, Lanzani C, D’Amico M, Buzzi L, Citterio L, Stella P, Rivera R, Bianchi G. Alpha-adducin polymorphisms and renal sodium handling in essential hypertensive patients. Kidney Int : 1471–1478, 1998. doi: 10.1046/j.1523-1755.1998.00931.x. [DOI] [PubMed] [Google Scholar]
- 233.Manunta P, Ferrandi M, Cusi D, Ferrari P, Staessen J, Bianchi G. Personalized Therapy of Hypertension: the Past and the Future. Curr Hypertens Rep : 24, 2016. doi: 10.1007/s11906-016-0632-y. [DOI] [PubMed] [Google Scholar]
- 234.Manunta P, Lavery G, Lanzani C, Braund PS, Simonini M, Bodycote C, Zagato L, Delli Carpini S, Tantardini C, Brioni E, Bianchi G, Samani NJ. Physiological interaction between alpha-adducin and WNK1-NEDD4L pathways on sodium-related blood pressure regulation. Hypertension : 366–372, 2008. doi: 10.1161/HYPERTENSIONAHA.108.113977. [DOI] [PubMed] [Google Scholar]
- 235.Manunta P, Maillard M, Tantardini C, Simonini M, Lanzani C, Citterio L, Stella P, Casamassima N, Burnier M, Hamlyn JM, Bianchi G. Relationships among endogenous ouabain, alpha-adducin polymorphisms and renal sodium handling in primary hypertension. J Hypertens : 914–920, 2008. doi: 10.1097/HJH.0b013e3282f5315f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Markel’ AL. [Genetic model of stress-induced arterial hypertension]. Izv Akad Nauk SSSR Biol : 466–469, 1985. [PubMed] [Google Scholar]
- 237.Mashimo T, Nabika T, Matsumoto C, Tamada T, Ueno K, Sawamura M, Ikeda K, Kato N, Nara Y, Yamori Y. Aging and salt-loading modulate blood pressure QTLs in rats. Am J Hypertens : 1098–1104, 1999. doi: 10.1016/S0895-7061(99)00084-9. [DOI] [PubMed] [Google Scholar]
- 238.Matsukawa N, Nonaka Y, Higaki J, Nagano M, Mikami H, Ogihara T, Okamoto M. Dahl’s salt-resistant normotensive rat has mutations in cytochrome P450(11 beta), but the salt-sensitive hypertensive rat does not. J Biol Chem : 9117–9121, 1993. [PubMed] [Google Scholar]
- 239.Mattson DL, Kunert MP, Kaldunski ML, Greene AS, Roman RJ, Jacob HJ, Cowley AW Jr. Influence of diet and genetics on hypertension and renal disease in Dahl salt-sensitive rats. Physiol Genomics : 194–203, 2004. doi: 10.1152/physiolgenomics.00151.2003. [DOI] [PubMed] [Google Scholar]
- 240.Mattson DL, Meister CJ, Marcelle ML. Dietary protein source determines the degree of hypertension and renal disease in the Dahl salt-sensitive rat. Hypertension : 736–741, 2005. doi: 10.1161/01.HYP.0000153318.74544.cc. [DOI] [PubMed] [Google Scholar]
- 241.McBride MW, Carr FJ, Graham D, Anderson NH, Clark JS, Lee WK, Charchar FJ, Brosnan MJ, Dominiczak AF. Microarray analysis of rat chromosome 2 congenic strains. Hypertension : 847–853, 2003. doi: 10.1161/01.HYP.0000047103.07205.03. [DOI] [PubMed] [Google Scholar]
- 242.Mehta G, Mookerjee RP. Breaking bad: the two sides of gut microbiota in portal hypertension. Liver Int : 1295–1297, 2014. doi: 10.1111/liv.12598. [DOI] [PubMed] [Google Scholar]
- 243.Mei H, Rice TK, Gu D, Hixson JE, Jaquish CE, Zhao Q, Chen JC, Cao J, Li J, Kelly TN, Rao DC, He J. Genetic correlation of blood pressure responses to dietary sodium and potassium intervention and cold pressor test in Chinese population. J Hum Hypertens : 500–508, 2011. doi: 10.1038/jhh.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Mell B, Abdul-Majeed S, Kumarasamy S, Waghulde H, Pillai R, Nie Y, Joe B. Multiple blood pressure loci with opposing blood pressure effects on rat chromosome 1 in a homologous region linked to hypertension on human chromosome 15. Hypertens Res : 61–67, 2015. doi: 10.1038/hr.2014.134. [DOI] [PubMed] [Google Scholar]
- 245.Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, Haribabu B, Vijay-Kumar M, Pennathur S, Joe B. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics : 187–197, 2015. doi: 10.1152/physiolgenomics.00136.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mendelsohn AR, Larrick JW. Dietary modification of the microbiome affects risk for cardiovascular disease. Rejuvenation Res : 241–244, 2013. doi: 10.1089/rej.2013.1447. [DOI] [PubMed] [Google Scholar]
- 247.Meng H, Garrett MR, Dene H, Rapp JP. Localization of a blood pressure QTL to a 2.4-cM interval on rat chromosome 9 using congenic strains. Genomics : 210–220, 2003. doi: 10.1016/S0888-7543(03)00003-X. [DOI] [PubMed] [Google Scholar]
- 248.Menni C, Graham D, Kastenmüller G, Alharbi NH, Alsanosi SM, McBride M, Mangino M, Titcombe P, Shin SY, Psatha M, Geisendorfer T, Huber A, Peters A, Wang-Sattler R, Xu T, Brosnan MJ, Trimmer J, Reichel C, Mohney RP, Soranzo N, Edwards MH, Cooper C, Church AC, Suhre K, Gieger C, Dominiczak AF, Spector TD, Padmanabhan S, Valdes AM. Metabolomic identification of a novel pathway of blood pressure regulation involving hexadecanedioate. Hypertension : 422–429, 2015. doi: 10.1161/HYPERTENSIONAHA.115.05544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Menni C, Mangino M, Cecelja M, Psatha M, Brosnan MJ, Trimmer J, Mohney RP, Chowienczyk P, Padmanabhan S, Spector TD, Valdes AM. Metabolomic study of carotid-femoral pulse-wave velocity in women. J Hypertens : 791–796, 2015. doi: 10.1097/HJH.0000000000000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Miyaoka K, Kuwasako T, Hirano K, Nozaki S, Yamashita S, Matsuzawa Y. CD36 deficiency associated with insulin resistance. Lancet : 686–687, 2001. doi: 10.1016/S0140-6736(00)04138-6. [DOI] [PubMed] [Google Scholar]
- 251.Mo R, Omvik P, Lund-Johansen P. The Bergen blood pressure study: offspring of two hypertensive parents have significantly higher blood pressures than offspring of one hypertensive and one normotensive parent. J Hypertens : 1614–1617, 1995. [PubMed] [Google Scholar]
- 252.Mongeau JG. Heredity and blood pressure in humans: an overview. Pediatr Nephrol : 69–75, 1987. doi: 10.1007/BF00866887. [DOI] [PubMed] [Google Scholar]
- 253.Monti J, Plehm R, Schulz H, Ganten D, Kreutz R, Hübner N. Interaction between blood pressure quantitative trait loci in rats in which trait variation at chromosome 1 is conditional upon a specific allele at chromosome 10. Hum Mol Genet : 435–439, 2003. doi: 10.1093/hmg/ddg041. [DOI] [PubMed] [Google Scholar]
- 254.Monti J, Zimdahl H, Schulz H, Plehm R, Ganten D, Hübner N. The role of Wnk4 in polygenic hypertension: a candidate gene analysis on rat chromosome 10. Hypertension : 938–942, 2003. doi: 10.1161/01.HYP.0000063147.92433.7D. [DOI] [PubMed] [Google Scholar]
- 255.Moreno C, Dumas P, Kaldunski ML, Tonellato PJ, Greene AS, Roman RJ, Cheng Q, Wang Z, Jacob HJ, Cowley AW Jr. Genomic map of cardiovascular phenotypes of hypertension in female Dahl S rats. Physiol Genomics : 243–257, 2003. doi: 10.1152/physiolgenomics.00105.2003. [DOI] [PubMed] [Google Scholar]
- 256.Moreno C, Hoffman M, Stodola TJ, Didier DN, Lazar J, Geurts AM, North PE, Jacob HJ, Greene AS. Creation and characterization of a renin knockout rat. Hypertension : 614–619, 2011. doi: 10.1161/HYPERTENSIONAHA.110.163840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Moreno C, Kaldunski ML, Wang T, Roman RJ, Greene AS, Lazar J, Jacob HJ, Cowley AW Jr. Multiple blood pressure loci on rat chromosome 13 attenuate development of hypertension in the Dahl S hypertensive rat. Physiol Genomics : 228–235, 2007. doi: 10.1152/physiolgenomics.00280.2006. [DOI] [PubMed] [Google Scholar]
- 258.Moreno C, Williams JM, Lu L, Liang M, Lazar J, Jacob HJ, Cowley AW Jr, Roman RJ. Narrowing a region on rat chromosome 13 that protects against hypertension in Dahl SS-13BN congenic strains. Am J Physiol Heart Circ Physiol : H1530–H1535, 2011. doi: 10.1152/ajpheart.01026.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258a.Moujahidine M, Dutil J, Hamet P, Deng AY. Congenic mapping of a blood pressure QTL on chromosome 16 of DahI rats. Mamm Genome : 153–156, 2002. doi: 10.1007/s00335-00L-21-3. [DOI] [PubMed] [Google Scholar]
- 258b.Moujahidine M, Lambert R, Duti L J, Palijan A, Sivo Z, Ariyarajah A, Deng AY. Combining congenic coverage with gene profiling in search of candidates for blood pressure quantitative trait loci in Dahl rats. Hypertens Res : 203–212, 2004. [DOI] [PubMed] [Google Scholar]
- 259.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature : 714–719, 2010. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mutig K, Kahl T, Saritas T, Godes M, Persson P, Bates J, Raffi H, Rampoldi L, Uchida S, Hille C, Dosche C, Kumar S, Castañeda-Bueno M, Gamba G, Bachmann S. Activation of the bumetanide-sensitive Na+,K+,2Cl- cotransporter (NKCC2) is facilitated by Tamm-Horsfall protein in a chloride-sensitive manner. J Biol Chem : 30200–30210, 2011. doi: 10.1074/jbc.M111.222968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Nabika T, Ohara H, Kato N, Isomura M. The stroke-prone spontaneously hypertensive rat: still a useful model for post-GWAS genetic studies? Hypertens Res : 477–484, 2012. doi: 10.1038/hr.2012.30. [DOI] [PubMed] [Google Scholar]
- 262.Nakajima T, Wooding S, Sakagami T, Emi M, Tokunaga K, Tamiya G, Ishigami T, Umemura S, Munkhbat B, Jin F, Guan-Jun J, Hayasaka I, Ishida T, Saitou N, Pavelka K, Lalouel JM, Jorde LB, Inoue I. Natural selection and population history in the human angiotensinogen gene (AGT): 736 complete AGT sequences in chromosomes from around the world. Am J Hum Genet : 898–916, 2004. doi: 10.1086/420793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl Res : 24–37, 2017. doi: 10.1016/j.trsl.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263a.Negrín CD, McBride MW, Carswell HV, Graham D, Carr FJ, Clark JS, Jeffs B, Anderson NH, Macrae IM, Dominczak AF. Reciprocal consomic strains to evaluate y chromosome effects. Hypertension : 391–397, 2001. [DOI] [PubMed] [Google Scholar]
- 264.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Döring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvänen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dörr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Völker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Völzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB; Wellcome Trust Case Control Consortium . Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet : 666–676, 2009. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J, Morgenthaler NG, Bergmann A, Blankenberg S, Kee F, Nilsson P, Yin X, Peltonen L, Vartiainen E, Salomaa V, Hirschhorn JN, Melander O, Wang TJ. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet : 348–353, 2009. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Nguyen KD, Pihur V, Ganesh SK, Rakha A, Cooper RS, Hunt SC, Freedman BI, Coresh J, Kao WH, Morrison AC, Boerwinkle E, Ehret GB, Chakravarti A. Effects of rare and common blood pressure gene variants on essential hypertension: results from the Family Blood Pressure Program, CLUE, and Atherosclerosis Risk in Communities studies. Circ Res : 318–326, 2013. doi: 10.1161/CIRCRESAHA.112.276725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Nie Y, Kumarasamy S, Waghulde H, Cheng X, Mell B, Czernik PJ, Lecka-Czernik B, Joe B. High-resolution mapping of a novel rat blood pressure locus on chromosome 9 to a region containing the Spp2 gene and colocalization of a QTL for bone mass. Physiol Genomics : 409–419, 2016. doi: 10.1152/physiolgenomics.00004.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nie Y, Kumarasamy S, Waghulde H, Cheng X, Mell B, Czernik PJ, Lecka-Czernik B, Joe B. High-resolution mapping of a novel rat blood pressure locus on chromosome 9 to a region containing the Spp2 gene and colocalization of a QTL for bone mass. Physiol Genomics : 409–419, 2016. doi: 10.1152/physiolgenomics.00004.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Nonaka Y, Fujii T, Kagawa N, Waterman MR, Takemori H, Okamoto M. Structure/function relationship of CYP11B1 associated with Dahl’s salt-resistant rats–expression of rat CYP11B1 and CYP11B2 in Escherichia coli. Eur J Biochem : 869–878, 1998. doi: 10.1046/j.1432-1327.1998.2580869.x. [DOI] [PubMed] [Google Scholar]
- 270.Oh YS, Appel LJ, Galis ZS, Hafler DA, He J, Hernandez AL, Joe B, Karumanchi SA, Maric-Bilkan C, Mattson D, Mehta NN, Randolph G, Ryan M, Sandberg K, Titze J, Tolunay E, Toney GM, Harrison DG. National Heart, Lung, and Blood Institute Working Group Report on Salt in Human Health and Sickness: Building on the Current Scientific Evidence. Hypertension : 281–288, 2016. doi: 10.1161/HYPERTENSIONAHA.116.07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ohno Y, Tanase H, Nabika T, Otsuka K, Sasaki T, Suzawa T, Morii T, Yamori Y, Saruta T. Selective genotyping with epistasis can be utilized for a major quantitative trait locus mapping in hypertension in rats. Genetics : 785–792, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Okamoto K, Aoki K. Development of a strain of spontaneously hypertensive rats. Jpn Circ J : 282–293, 1963. doi: 10.1253/jcj.27.282. [DOI] [PubMed] [Google Scholar]
- 273.Oldham PD, Pickering G, Roberts JA, Sowry GS. The nature of essential hypertension. Lancet : 1085–1093, 1960. doi: 10.1016/S0140-6736(60)90982-X. [DOI] [PubMed] [Google Scholar]
- 274.Overgaard MT, Boldt HB, Laursen LS, Sottrup-Jensen L, Conover CA, Oxvig C. Pregnancy-associated plasma protein-A2 (PAPP-A2), a novel insulin-like growth factor-binding protein-5 proteinase. J Biol Chem : 21849–21853, 2001. doi: 10.1074/jbc.M102191200. [DOI] [PubMed] [Google Scholar]
- 275.Padmanabhan S, Caulfield M, Dominiczak AF. Genetic and molecular aspects of hypertension. Circ Res : 937–959, 2015. doi: 10.1161/CIRCRESAHA.116.303647. [DOI] [PubMed] [Google Scholar]
- 276.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, Laing S, Corso B, Navis G, Kwakernaak AJ, van der Harst P, Bochud M, Maillard M, Burnier M, Hedner T, Kjeldsen S, Wahlstrand B, Sjögren M, Fava C, Montagnana M, Danese E, Torffvit O, Hedblad B, Snieder H, Connell JM, Brown M, Samani NJ, Farrall M, Cesana G, Mancia G, Signorini S, Grassi G, Eyheramendy S, Wichmann HE, Laan M, Strachan DP, Sever P, Shields DC, Stanton A, Vollenweider P, Teumer A, Völzke H, Rettig R, Newton-Cheh C, Arora P, Zhang F, Soranzo N, Spector TD, Lucas G, Kathiresan S, Siscovick DS, Luan J, Loos RJ, Wareham NJ, Penninx BW, Nolte IM, McBride M, Miller WH, Nicklin SA, Baker AH, Graham D, McDonald RA, Pell JP, Sattar N, Welsh P, Munroe P, Caulfield MJ, Zanchetti A, Dominiczak AF; Global BPgen Consortium . Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet : e1001177, 2010. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Palijan A, Dutil J, Deng AY. Quantitative trait loci with opposing blood pressure effects demonstrating epistasis on Dahl rat chromosome 3. Physiol Genomics : 1–8, 2003. doi: 10.1152/physiolgenomics.00084.2003. [DOI] [PubMed] [Google Scholar]
- 278.Palijan A, Lambert R, Dutil J, Sivo Z, Deng AY. Comprehensive congenic coverage revealing multiple blood pressure quantitative trait loci on Dahl rat chromosome 10. Hypertension : 515–522, 2003. doi: 10.1161/01.HYP.0000090096.88509.15. [DOI] [PubMed] [Google Scholar]
- 279.Paschoud S, Jond L, Guerrera D, Citi S. PLEKHA7 modulates epithelial tight junction barrier function. Tissue Barriers : e28755, 2014. doi: 10.4161/tisb.28755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279a.Pausova Z, Sedova L, Berube J, Hamet P, Tremblay J, Dumont M, Gaudet D, Pravenec M, Kren V, Kunes J. Segment of rat chromosome 20 regulates diet-induced augmentations in adiposity, glucose intolerance, and blood pressure. Hypertension : 1047–1055, 2003. [DOI] [PubMed] [Google Scholar]
- 280.Pickering G. Hypertension. Definitions, natural histories and consequences. Am J Med : 570–583, 1972. doi: 10.1016/0002-9343(72)90049-6. [DOI] [PubMed] [Google Scholar]
- 281.Pickering G. Normotension and hypertension: the mysterious viability of the false. Am J Med : 561–563, 1978. doi: 10.1016/0002-9343(78)90839-2. [DOI] [PubMed] [Google Scholar]
- 282.Pickering GW. High Blood Pressure. New York: Grune & Stratton, 1968, p. viii. [Google Scholar]
- 283.Pickering GW. [Relation between genetic and social factors and arterial pressure]. Recenti Prog Med : 397–416, 1961. [PubMed] [Google Scholar]
- 284.Pillai R, Waghulde H, Nie Y, Gopalakrishnan K, Kumarasamy S, Farms P, Garrett MR, Atanur SS, Maratou K, Aitman TJ, Joe B. Isolation and high-throughput sequencing of two closely linked epistatic hypertension susceptibility loci with a panel of bicongenic strains. Physiol Genomics : 729–736, 2013. doi: 10.1152/physiolgenomics.00077.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA : 4410–4415, 2013. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Pravenec M, Churchill PC, Churchill MC, Viklicky O, Kazdova L, Aitman TJ, Petretto E, Hubner N, Wallace CA, Zimdahl H, Zidek V, Landa V, Dunbar J, Bidani A, Griffin K, Qi N, Maxova M, Kren V, Mlejnek P, Wang J, Kurtz TW. Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat Genet : 952–954, 2008. doi: 10.1038/ng.164. [DOI] [PubMed] [Google Scholar]
- 287.Pravenec M, Gauguier D, Schott JJ, Buard J, Kren V, Bila V, Szpirer C, Szpirer J, Wang JM, Huang H. Mapping of quantitative trait loci for blood pressure and cardiac mass in the rat by genome scanning of recombinant inbred strains. J Clin Invest : 1973–1978, 1995. doi: 10.1172/JCI118244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Pravenec M, Klír P, Kren V, Zicha J, Kunes J. An analysis of spontaneous hypertension in spontaneously hypertensive rats by means of new recombinant inbred strains. J Hypertens : 217–221, 1989. doi: 10.1097/00004872-198903000-00008. [DOI] [PubMed] [Google Scholar]
- 289.Pravenec M, Kren V, Krenová D, Zídek V, Simáková M, Musilová A, Vorlícek J, Lezin ES, Kurtz TW. Genetic isolation of quantitative trait loci for blood pressure development and renal mass on chromosome 5 in the spontaneously hypertensive rat. Physiol Res : 285–289, 2003. [PubMed] [Google Scholar]
- 290.Pravenec M, Kren V, Kunes J, Scicli AG, Carretero OA, Simonet L, Kurtz TW. Cosegregation of blood pressure with a kallikrein gene family polymorphism. Hypertension : 242–246, 1991. doi: 10.1161/01.HYP.17.2.242. [DOI] [PubMed] [Google Scholar]
- 291.Pravenec M, Kurtz TW. Genetics of Cd36 and the hypertension metabolic syndrome. Semin Nephrol : 148–153, 2002. doi: 10.1053/snep.2002.2002.30218. [DOI] [PubMed] [Google Scholar]
- 292.Pravenec M, Simonet L, Kren V, Kunes J, Levan G, Szpirer J, Szpirer C, Kurtz T. The rat renin gene: assignment to chromosome 13 and linkage to the regulation of blood pressure. Genomics : 466–472, 1991. doi: 10.1016/0888-7543(91)90412-8. [DOI] [PubMed] [Google Scholar]
- 293.Pravenec M, Zídek V, Musilová A, Vorlícek J, Kren V, St Lezin E, Kurtz TW. Genetic isolation of a blood pressure quantitative trait locus on chromosome 2 in the spontaneously hypertensive rat. J Hypertens : 1061–1064, 2001. doi: 10.1097/00004872-200106000-00010. [DOI] [PubMed] [Google Scholar]
- 294.Pravenec M, Zidek V, Simakova M, Kren V, Krenova D, Horky K, Jachymova M, Mikova B, Kazdova L, Aitman TJ, Churchill PC, Webb RC, Hingarh NH, Yang Y, Wang JM, Lezin EM, Kurtz TW. Genetics of Cd36 and the clustering of multiple cardiovascular risk factors in spontaneous hypertension. J Clin Invest : 1651–1657, 1999. doi: 10.1172/JCI6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Pulimeno P, Bauer C, Stutz J, Citi S. PLEKHA7 is an adherens junction protein with a tissue distribution and subcellular localization distinct from ZO-1 and E-cadherin. PLoS One : e12207, 2010. doi: 10.1371/journal.pone.0012207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J Am Soc Nephrol : 657–670, 2014. doi: 10.1681/ASN.2013080905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ramos A, Moisan MP, Chaouloff F, Mormède C, Mormède P. Identification of female-specific QTLs affecting an emotionality-related behavior in rats. Mol Psychiatry : 453–462, 1999. doi: 10.1038/sj.mp.4000546. [DOI] [PubMed] [Google Scholar]
- 298.Rapp JP. Genetic analysis of inherited hypertension in the rat. Physiol Rev : 135–172, 2000. [DOI] [PubMed] [Google Scholar]
- 299.Rapp JP. Theoretical model for gene-gene, gene-environment, and gene-sex interactions based on congenic-strain analysis of blood pressure in Dahl salt-sensitive rats. Physiol Genomics : 737–750, 2013. doi: 10.1152/physiolgenomics.00046.2013. [DOI] [PubMed] [Google Scholar]
- 300.Rapp JP, Dahl LK. 18-Hydrox-deoxycorticosterone secretion in experimental hypertension in rats. Circ Res , Suppl 2: 153–159, 1971. doi: 10.1161/01.RES.28.5.II-153. [DOI] [PubMed] [Google Scholar]
- 301.Rapp JP, Dahl LK. Adrenal steroidogenesis in rats bred for susceptibility and resistance to the hypertensive effect of salt. Endocrinology : 52–65, 1971. doi: 10.1210/endo-88-1-52. [DOI] [PubMed] [Google Scholar]
- 302.Rapp JP, Dahl LK. Mendelian inheritance of 18- and ll beta-steroid hydroxylase activities in the adrenals of rats genetically susceptible or resistant to hypertension. Endocrinology : 1435–1446, 1972. doi: 10.1210/endo-90-6-1435. [DOI] [PubMed] [Google Scholar]
- 303.Rapp JP, Dahl LK. Mutant forms of cytochrome P-450 controlling both 18- and 11beta-steroid hydroxylation in the rat. Biochemistry : 1235–1242, 1976. doi: 10.1021/bi00651a010. [DOI] [PubMed] [Google Scholar]
- 304.Rapp JP, Dahl LK. Possible role of 18-hydroxy-deoxycorticosterone in hypertension. Nature : 338–339, 1972. doi: 10.1038/237338a0. [DOI] [PubMed] [Google Scholar]
- 305.Rapp JP, Garrett MR, Dene H, Meng H, Hoebee B, Lathrop GM. Linkage analysis and construction of a congenic strain for a blood pressure QTL on rat chromosome 9. Genomics : 191–196, 1998. doi: 10.1006/geno.1998.5394. [DOI] [PubMed] [Google Scholar]
- 306.Rapp JP, Garrett MR, Deng AY. Construction of a double congenic strain to prove an epistatic interaction on blood pressure between rat chromosomes 2 and 10. J Clin Invest : 1591–1595, 1998. doi: 10.1172/JCI2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Rapp JP, Joe B. Do epistatic modules exist in the genetic control of blood pressure in Dahl rats? A critical perspective. Physiol Genomics : 1193–1195, 2013. doi: 10.1152/physiolgenomics.00159.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Rapp JP, Joe B. Use of contiguous congenic strains in analyzing compound QTLs. Physiol Genomics : 117–120, 2012. doi: 10.1152/physiolgenomics.00136.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rapp JP, Tan SY, Margolius HS. Plasma mineralocorticoids, plasma renin, and urinary kallikrein in salt-sensitive and salt-resistant rats. Endocr Res Commun : 35–41, 1978. doi: 10.3109/07435807809073634. [DOI] [PubMed] [Google Scholar]
- 310.Rapp JP, Wang SM, Dene H. Effect of genetic background on cosegregation of renin alleles and blood pressure in Dahl rats. Am J Hypertens : 391–396, 1990. doi: 10.1093/ajh/3.5.391. [DOI] [PubMed] [Google Scholar]
- 311.Rapp JP, Wang SM, Dene H. A genetic polymorphism in the renin gene of Dahl rats cosegregates with blood pressure. Science : 542–544, 1989. doi: 10.1126/science.2563177. [DOI] [PubMed] [Google Scholar]
- 312.Redina OE, Lapteva NE, Khanina SL, Machanova NA, Dymshits GM, Markel AL. The region of rat chromosome 10 (the ngfr gene locus) is associated with blood pressure increase in response to emotional stress. Dokl Biochem Biophys : 349–351, 2001. doi: 10.1023/A:1012352512006. [DOI] [PubMed] [Google Scholar]
- 313.Redina OE, Machanova NA, Efimov VM, Markel AL. Rats with inherited stress-induced arterial hypertension (ISIAH strain) display specific quantitative trait loci for blood pressure and for body and kidney weight on chromosome 1. Clin Exp Pharmacol Physiol : 456–464, 2006. doi: 10.1111/j.1440-1681.2006.04387.x. [DOI] [PubMed] [Google Scholar]
- 314.Redina OE, Smolenskaya SE, Maslova LN, Markel AL. The genetic control of blood pressure and body composition in rats with stress-sensitive hypertension. Clin Exp Hypertens : 484–495, 2013. doi: 10.3109/10641963.2012.758274. [DOI] [PubMed] [Google Scholar]
- 315.Vietri Rudan M, Hadjur S, Sexton T. Detecting Spatial Chromatin Organization by Chromosome Conformation Capture II: Genome-Wide Profiling by Hi-C. Methods Mol Biol : 47–74, 2017. doi: 10.1007/7651_2015_261. [DOI] [PubMed] [Google Scholar]
- 316.Rudemiller N, Lund H, Jacob HJ, Geurts AM, Mattson DL; PhysGen Knockout Program . CD247 modulates blood pressure by altering T-lymphocyte infiltration in the kidney. Hypertension : 559–564, 2014. doi: 10.1161/HYPERTENSIONAHA.113.02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Rudemiller NP, Lund H, Priestley JR, Endres BT, Prokop JW, Jacob HJ, Geurts AM, Cohen EP, Mattson DL. Mutation of SH2B3 (LNK), a genome-wide association study candidate for hypertension, attenuates Dahl salt-sensitive hypertension via inflammatory modulation. Hypertension : 1111–1117, 2015. doi: 10.1161/HYPERTENSIONAHA.114.04736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Saad Y, Garrett MR, Manickavasagam E, Yerga-Woolwine S, Farms P, Radecki T, Joe B. Fine-mapping and comprehensive transcript analysis reveals nonsynonymous variants within a novel 1.17 Mb blood pressure QTL region on rat chromosome 10. Genomics : 343–353, 2007. doi: 10.1016/j.ygeno.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Saad Y, Garrett MR, Rapp JP. Multiple blood pressure QTL on rat chromosome 1 defined by Dahl rat congenic strains. Physiol Genomics : 201–214, 2001. [DOI] [PubMed] [Google Scholar]
- 320.Saad Y, Yerga-Woolwine S, Saikumar J, Farms P, Manickavasagam E, Toland EJ, Joe B. Congenic interval mapping of RNO10 reveals a complex cluster of closely-linked genetic determinants of blood pressure. Hypertension : 891–898, 2007. doi: 10.1161/HYPERTENSIONAHA.107.097105. [DOI] [PubMed] [Google Scholar]
- 321.Salvi E, Kutalik Z, Glorioso N, Benaglio P, Frau F, Kuznetsova T, Arima H, Hoggart C, Tichet J, Nikitin YP, Conti C, Seidlerova J, Tikhonoff V, Stolarz-Skrzypek K, Johnson T, Devos N, Zagato L, Guarrera S, Zaninello R, Calabria A, Stancanelli B, Troffa C, Thijs L, Rizzi F, Simonova G, Lupoli S, Argiolas G, Braga D, D’Alessio MC, Ortu MF, Ricceri F, Mercurio M, Descombes P, Marconi M, Chalmers J, Harrap S, Filipovsky J, Bochud M, Iacoviello L, Ellis J, Stanton AV, Laan M, Padmanabhan S, Dominiczak AF, Samani NJ, Melander O, Jeunemaitre X, Manunta P, Shabo A, Vineis P, Cappuccio FP, Caulfield MJ, Matullo G, Rivolta C, Munroe PB, Barlassina C, Staessen JA, Beckmann JS, Cusi D. Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension : 248–255, 2012. doi: 10.1161/HYPERTENSIONAHA.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Samani NJ, Gauguier D, Vincent M, Kaiser MA, Bihoreau MT, Lodwick D, Wallis R, Parent V, Kimber P, Rattray F, Thompson JR, Sassard J, Lathrop M. Analysis of quantitative trait loci for blood pressure on rat chromosomes 2 and 13. Age-related differences in effect. Hypertension : 1118–1122, 1996. doi: 10.1161/01.HYP.28.6.1118. [DOI] [PubMed] [Google Scholar]
- 323.Samani NJ, Lodwick D, Vincent M, Dubay C, Kaiser MA, Kelly MP, Lo M, Harris J, Sassard J, Lathrop M. A gene differentially expressed in the kidney of the spontaneously hypertensive rat cosegregates with increased blood pressure. J Clin Invest : 1099–1103, 1993. doi: 10.1172/JCI116616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Santisteban MM, Kim S, Pepine CJ, Raizada MK. Brain-Gut-Bone Marrow Axis: Implications for Hypertension and Related Therapeutics. Circ Res : 1327–1336, 2016. doi: 10.1161/CIRCRESAHA.116.307709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Sati S, Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma : 33–44, 2017. doi: 10.1007/s00412-016-0593-6. [DOI] [PubMed] [Google Scholar]
- 326.Schlager G, Chao CS. The role of dominance and epistasis in the genetic control of blood pressure in rodent models of hypertension. Clin Exp Hypertens A : 947–953, 1991. [DOI] [PubMed] [Google Scholar]
- 327.Schork NJ, Krieger JE, Trolliet MR, Franchini KG, Koike G, Krieger EM, Lander ES, Dzau VJ, Jacob HJ. A biometrical genome search in rats reveals the multigenic basis of blood pressure variation. Genome Res : 164–172, 1995. doi: 10.1101/gr.5.2.164. [DOI] [PubMed] [Google Scholar]
- 328.Schulz A, Litfin A, Kossmehl P, Kreutz R. Genetic dissection of increased urinary albumin excretion in the Munich Wistar Frömter rat. J Am Soc Nephrol : 2706–2714, 2002. doi: 10.1097/01.ASN.0000031803.55613.86. [DOI] [PubMed] [Google Scholar]
- 329.Schüpbach T, Xenarios I, Bergmann S, Kapur K. FastEpistasis: a high performance computing solution for quantitative trait epistasis. Bioinformatics : 1468–1469, 2010. doi: 10.1093/bioinformatics/btq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Schutte AE, Huisman HW, van Rooyen JM, Malan L, Malan NT, Fourie CM, Louw R, van der Westhuizen FH, Schutte R. A significant decline in IGF-I may predispose young Africans to subsequent cardiometabolic vulnerability. J Clin Endocrinol Metab : 2503–2507, 2010. doi: 10.1210/jc.2009-2329. [DOI] [PubMed] [Google Scholar]
- 331.Seidlerová J, Staessen JA, Bochud M, Nawrot T, Casamassima N, Citterio L, Kuznetsova T, Jin Y, Manunta P, Richart T, Struijker-Boudier HA, Fagard R, Filipovský J, Bianchi G. Arterial properties in relation to genetic variations in the adducin subunits in a white population. Am J Hypertens : 21–26, 2009. doi: 10.1038/ajh.2008.261. [DOI] [PubMed] [Google Scholar]
- 332.Shah J, Guerrera D, Vasileva E, Sluysmans S, Bertels E, Citi S. PLEKHA7: Cytoskeletal adaptor protein at center stage in junctional organization and signaling. Int J Biochem Cell Biol : 112–116, 2016. doi: 10.1016/j.biocel.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 333.Shear CL, Burke GL, Freedman DS, Berenson GS. Value of childhood blood pressure measurements and family history in predicting future blood pressure status: results from 8 years of follow-up in the Bogalusa Heart Study. Pediatrics : 862–869, 1986. [PubMed] [Google Scholar]
- 334.Shear CL, Freedman DS, Burke GL, Harsha DW, Berenson GS. Body fat patterning and blood pressure in children and young adults. The Bogalusa Heart Study. Hypertension : 236–244, 1987. doi: 10.1161/01.HYP.9.3.236. [DOI] [PubMed] [Google Scholar]
- 335.Shibao CA, Celedonio JE, Ramirez CE, Love-Gregory L, Arnold AC, Choi L, Okamoto LE, Gamboa A, Biaggioni I, Abumrad NN, Abumrad NA. A Common CD36 Variant Influences Endothelial Function and Response to Treatment with Phosphodiesterase 5 Inhibition. J Clin Endocrinol Metab : 2751–2758, 2016. doi: 10.1210/jc.2016-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR Jr, Ulick S, Milora RV, Findling JW, Canessa CM, Rossier BC, Lifton RP. Liddle’s syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell : 407–414, 1994. doi: 10.1016/0092-8674(94)90250-X. [DOI] [PubMed] [Google Scholar]
- 337.Shiozawa M, Provoost AP, van Dokkum RP, Majewski RR, Jacob HJ. Evidence of gene-gene interactions in the genetic susceptibility to renal impairment after unilateral nephrectomy. J Am Soc Nephrol : 2068–2078, 2000. [DOI] [PubMed] [Google Scholar]
- 338.Siegel AK, Kossmehl P, Planert M, Schulz A, Wehland M, Stoll M, Bruijn JA, de Heer E, Kreutz R. Genetic linkage of albuminuria and renal injury in Dahl salt-sensitive rats on a high-salt diet: comparison with spontaneously hypertensive rats. Physiol Genomics : 218–225, 2004. doi: 10.1152/physiolgenomics.00068.2004. [DOI] [PubMed] [Google Scholar]
- 339.Siegel AK, Planert M, Rademacher S, Mehr AP, Kossmehl P, Wehland M, Stoll M, Kreutz R. Genetic loci contribute to the progression of vascular and cardiac hypertrophy in salt-sensitive spontaneous hypertension. Arterioscler Thromb Vasc Biol : 1211–1217, 2003. doi: 10.1161/01.ATV.0000079509.20542.C9. [DOI] [PubMed] [Google Scholar]
- 340.Silva GJ, Pereira AC, Krieger EM, Krieger JE. Genetic mapping of a new heart rate QTL on chromosome 8 of spontaneously hypertensive rats. BMC Med Genet : 17, 2007. doi: 10.1186/1471-2350-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Simon DB, Karet FE, Hamdan JM, DiPietro A, Sanjad SA, Lifton RP. Bartter’s syndrome, hypokalaemic alkalosis with hypercalciuria, is caused by mutations in the Na-K-2Cl cotransporter NKCC2. Nat Genet : 183–188, 1996. doi: 10.1038/ng0696-183. [DOI] [PubMed] [Google Scholar]
- 342.Simon DB, Nelson-Williams C, Bia MJ, Ellison D, Karet FE, Molina AM, Vaara I, Iwata F, Cushner HM, Koolen M, Gainza FJ, Gitleman HJ, Lifton RP. Gitelman’s variant of Bartter’s syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat Genet : 24–30, 1996. doi: 10.1038/ng0196-24. [DOI] [PubMed] [Google Scholar]
- 343.Smirk FH, Hall WH. Inherited hypertension in rats. Nature : 727–728, 1958. doi: 10.1038/182727a0. [DOI] [PubMed] [Google Scholar]
- 344.Smolarek I, Wyszko E, Barciszewska AM, Nowak S, Gawronska I, Jablecka A, Barciszewska MZ. Global DNA methylation changes in blood of patients with essential hypertension. Med Sci Monit : CR149–CR155, 2010. [PubMed] [Google Scholar]
- 345.Soler JM, Pereira AC, Tôrres CH, Krieger JE. Gene by environment QTL mapping through multiple trait analyses in blood pressure salt-sensitivity: identification of a novel QTL in rat chromosome 5. BMC Med Genet : 47, 2006. doi: 10.1186/1471-2350-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.St Lezin E, Liu W, Wang JM, Yang Y, Qi N, Kren V, Zidek V, Kurtz TW, Pravenec M. Genetic analysis of rat chromosome 1 and the Sa gene in spontaneous hypertension. Hypertension : 225–230, 2000. doi: 10.1161/01.HYP.35.1.225. [DOI] [PubMed] [Google Scholar]
- 346a.St Lezin E, Zhang L, Yang Y, Wang JM, Wang N, Qi N, Steadman JS, Liu W, Kren V, Zidek V, Krenova D, Churchill PC, Churchill MC, Pravenec M. Effect of chromosome 19 transfer on blood pressure in the spontaneously hypertensive rat. Hypertension : 256–260, 1999. [DOI] [PubMed] [Google Scholar]
- 347.Staessen JA, Thijs L, Stolarz-Skrzypek K, Bacchieri A, Barton J, Espositi ED, de Leeuw PW, Dłużniewski M, Glorioso N, Januszewicz A, Manunta P, Milyagin V, Nikitin Y, Souček M, Lanzani C, Citterio L, Timio M, Tykarski A, Ferrari P, Valentini G, Kawecka-Jaszcz K, Bianchi G. Main results of the ouabain and adducin for Specific Intervention on Sodium in Hypertension Trial (OASIS-HT): a randomized placebo-controlled phase-2 dose-finding study of rostafuroxin. Trials : 13, 2011. doi: 10.1186/1745-6215-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Stec DE, Trolliet MR, Krieger JE, Jacob HJ, Roman RJ. Renal cytochrome P4504A activity and salt sensitivity in spontaneously hypertensive rats. Hypertension : 1329–1336, 1996. doi: 10.1161/01.HYP.27.6.1329. [DOI] [PubMed] [Google Scholar]
- 349.Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW. Dissecting the architecture of a quantitative trait locus in yeast. Nature : 326–330, 2002. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 350.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wägele B, Altmaier E, Deloukas P, Erdmann J, Grundberg E, Hammond CJ, de Angelis MH, Kastenmüller G, Köttgen A, Kronenberg F, Mangino M, Meisinger C, Meitinger T, Mewes HW, Milburn MV, Prehn C, Raffler J, Ried JS, Römisch-Margl W, Samani NJ, Small KS, Wichmann HE, Zhai G, Illig T, Spector TD, Adamski J, Soranzo N, Gieger C; CARDIoGRAM . Human metabolic individuality in biomedical and pharmaceutical research. Nature : 54–60, 2011. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Sun L, McArdle S, Chun M, Wolff DW, Pettinger WA. Cosegregation of the renin gene with an increase in mean arterial blood pressure in the F2 rats of SHR-WKY cross. Clin Exp Hypertens : 797–805, 1993. doi: 10.3109/10641969309041642. [DOI] [PubMed] [Google Scholar]
- 352.Takami S, Higaki J, Miki T, Katsuya T, Nakata Y, Rakugi H, Serikawa T, Ogihara T. Analysis and comparison of new candidate loci for hypertension between genetic hypertensive rat strains. Hypertens Res : 51–56, 1996. doi: 10.1291/hypres.19.51. [DOI] [PubMed] [Google Scholar]
- 353.Taylor MB, Ehrenreich IM. Higher-order genetic interactions and their contribution to complex traits. Trends Genet : 34–40, 2015. doi: 10.1016/j.tig.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Thiel BA, Chakravarti A, Cooper RS, Luke A, Lewis S, Lynn A, Tiwari H, Schork NJ, Weder AB. A genome-wide linkage analysis investigating the determinants of blood pressure in whites and African Americans. Am J Hypertens : 151–153, 2003. doi: 10.1016/S0895-7061(02)03246-6. [DOI] [PubMed] [Google Scholar]
- 355.Toland EJ, Saad Y, Yerga-Woolwine S, Ummel S, Farms P, Ramdath R, Frank BC, Lee NH, Joe B. Closely linked non-additive blood pressure quantitative trait loci. Mamm Genome : 209–218, 2008. doi: 10.1007/s00335-008-9093-1. [DOI] [PubMed] [Google Scholar]
- 356.Toland EJ, Yerga-Woolwine S, Farms P, Cicila GT, Saad Y, Joe B. Blood pressure and proteinuria effects of multiple quantitative trait loci on rat chromosome 9 that differentiate the spontaneously hypertensive rat from the Dahl salt-sensitive rat. J Hypertens : 2134–2141, 2008. doi: 10.1097/HJH.0b013e32830ef95c. [DOI] [PubMed] [Google Scholar]
- 357.Tomaszewski M, Debiec R, Braund PS, Nelson CP, Hardwick R, Christofidou P, Denniff M, Codd V, Rafelt S, van der Harst P, Waterworth D, Song K, Vollenweider P, Waeber G, Zukowska-Szczechowska E, Burton PR, Mooser V, Charchar FJ, Thompson JR, Tobin MD, Samani NJ. Genetic architecture of ambulatory blood pressure in the general population: insights from cardiovascular gene-centric array. Hypertension : 1069–1076, 2010. doi: 10.1161/HYPERTENSIONAHA.110.155721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Ménard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science : 808–813, 2004. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 359.Torielli L, Tivodar S, Montella RC, Iacone R, Padoani G, Tarsini P, Russo O, Sarnataro D, Strazzullo P, Ferrari P, Bianchi G, Zurzolo C. alpha-Adducin mutations increase Na/K pump activity in renal cells by affecting constitutive endocytosis: implications for tubular Na reabsorption. Am J Physiol Renal Physiol : F478–F487, 2008. doi: 10.1152/ajprenal.90226.2008. [DOI] [PubMed] [Google Scholar]
- 360.Tripodi G, Florio M, Ferrandi M, Modica R, Zimdahl H, Hubner N, Ferrari P, Bianchi G. Effect of Add1 gene transfer on blood pressure in reciprocal congenic strains of Milan rats. Biochem Biophys Res Commun : 562–568, 2004. doi: 10.1016/j.bbrc.2004.09.079. [DOI] [PubMed] [Google Scholar]
- 361.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell’Antonio G, Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L; Swiss Kidney Project on Genes in Hypertension (SKIPOGH) team . Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med : 1655–1660, 2013. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Tyler AL, Lu W, Hendrick JJ, Philip VM, Carter GW. CAPE: an R package for combined analysis of pleiotropy and epistasis. PLOS Comput Biol : e1003270, 2013. doi: 10.1371/journal.pcbi.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Ueno T, Tremblay J, Kunes J, Zicha J, Dobesova Z, Pausova Z, Deng AY, Sun YL, Jacob HJ, Hamet P. Resolving the composite trait of hypertension into its pharmacogenetic determinants by acute pharmacological modulation of blood pressure regulatory systems. J Mol Med (Berl) : 51–60, 2003. doi: 10.1007/s00109-002-0394-7. [DOI] [PubMed] [Google Scholar]
- 364.van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, Mirny LA, Dekker J, Lander ES. Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp : 1869, 2010. doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Vincent M, Samani NJ, Gauguier D, Thompson JR, Lathrop GM, Sassard J. A pharmacogenetic approach to blood pressure in Lyon hypertensive rats. A chromosome 2 locus influences the response to a calcium antagonist. J Clin Invest : 2000–2006, 1997. doi: 10.1172/JCI119731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Wade B, Abais-Battad JM, Mattson DL. Role of immune cells in salt-sensitive hypertension and renal injury. Curr Opin Nephrol Hypertens : 22–27, 2016. doi: 10.1097/MNH.0000000000000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, Bochud M, Rice KM, Henneman P, Smith AV, Ehret GB, Amin N, Larson MG, Mooser V, Hadley D, Dörr M, Bis JC, Aspelund T, Esko T, Janssens AC, Zhao JH, Heath S, Laan M, Fu J, Pistis G, Luan J, Arora P, Lucas G, Pirastu N, Pichler I, Jackson AU, Webster RJ, Zhang F, Peden JF, Schmidt H, Tanaka T, Campbell H, Igl W, Milaneschi Y, Hottenga JJ, Vitart V, Chasman DI, Trompet S, Bragg-Gresham JL, Alizadeh BZ, Chambers JC, Guo X, Lehtimäki T, Kühnel B, Lopez LM, Polašek O, Boban M, Nelson CP, Morrison AC, Pihur V, Ganesh SK, Hofman A, Kundu S, Mattace-Raso FU, Rivadeneira F, Sijbrands EJ, Uitterlinden AG, Hwang SJ, Vasan RS, Wang TJ, Bergmann S, Vollenweider P, Waeber G, Laitinen J, Pouta A, Zitting P, McArdle WL, Kroemer HK, Völker U, Völzke H, Glazer NL, Taylor KD, Harris TB, Alavere H, Haller T, Keis A, Tammesoo ML, Aulchenko Y, Barroso I, Khaw KT, Galan P, Hercberg S, Lathrop M, Eyheramendy S, Org E, Sõber S, Lu X, Nolte IM, Penninx BW, Corre T, Masciullo C, Sala C, Groop L, Voight BF, Melander O, O’Donnell CJ, Salomaa V, d’Adamo AP, Fabretto A, Faletra F, Ulivi S, Del Greco F, Facheris M, Collins FS, Bergman RN, Beilby JP, Hung J, Musk AW, Mangino M, Shin SY, Soranzo N, Watkins H, Goel A, Hamsten A, Gider P, Loitfelder M, Zeginigg M, Hernandez D, Najjar SS, Navarro P, Wild SH, Corsi AM, Singleton A, de Geus EJ, Willemsen G, Parker AN, Rose LM, Buckley B, Stott D, Orru M, Uda M, van der Klauw MM, Zhang W, Li X, Scott J, Chen YD, Burke GL, Kähönen M, Viikari J, Döring A, Meitinger T, Davies G, Starr JM, Emilsson V, Plump A, Lindeman JH, Hoen PA, König IR, Felix JF, Clarke R, Hopewell JC, Ongen H, Breteler M, Debette S, Destefano AL, Fornage M, Mitchell GF, Smith NL, Holm H, Stefansson K, Thorleifsson G, Thorsteinsdottir U, Samani NJ, Preuss M, Rudan I, Hayward C, Deary IJ, Wichmann HE, Raitakari OT, Palmas W, Kooner JS, Stolk RP, Jukema JW, Wright AF, Boomsma DI, Bandinelli S, Gyllensten UB, Wilson JF, Ferrucci L, Schmidt R, Farrall M, Spector TD, Palmer LJ, Tuomilehto J, Pfeufer A, Gasparini P, Siscovick D, Altshuler D, Loos RJ, Toniolo D, Snieder H, Gieger C, Meneton P, Wareham NJ, Oostra BA, Metspalu A, Launer L, Rettig R, Strachan DP, Beckmann JS, Witteman JC, Erdmann J, van Dijk KW, Boerwinkle E, Boehnke M, Ridker PM, Jarvelin MR, Chakravarti A, Abecasis GR, Gudnason V, Newton-Cheh C, Levy D, Munroe PB, Psaty BM, Caulfield MJ, Rao DC, Tobin MD, Elliott P, van Duijn CM; LifeLines Cohort Study; EchoGen consortium; AortaGen Consortium; CHARGE Consortium Heart Failure Working Group; KidneyGen consortium; CKDGen consortium; Cardiogenics consortium; CardioGram . Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet : 1005–1011, 2011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Wang F, Li L, Xu H, Liu Y, Yang C, Cowley AW Jr, Wang N, Liu P, Liang M. Characteristics of long non-coding RNAs in the Brown Norway rat and alterations in the Dahl salt-sensitive rat. Sci Rep : 7146, 2014. doi: 10.1038/srep07146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Wang NY, Young JH, Meoni LA, Ford DE, Erlinger TP, Klag MJ. Blood pressure change and risk of hypertension associated with parental hypertension: the Johns Hopkins Precursors Study. Arch Intern Med : 643–648, 2008. doi: 10.1001/archinte.168.6.643. [DOI] [PubMed] [Google Scholar]
- 370.Wang QZ, Gao HQ, Liang Y, Zhang J, Wang J, Qiu J. Cofilin1 is involved in hypertension-induced renal damage via the regulation of NF-κB in renal tubular epithelial cells. J Transl Med : 323, 2015. doi: 10.1186/s12967-015-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wang X, Falkner B, Zhu H, Shi H, Su S, Xu X, Sharma AK, Dong Y, Treiber F, Gutin B, Harshfield G, Snieder H. A genome-wide methylation study on essential hypertension in young African American males. PLoS One : e53938, 2013. doi: 10.1371/journal.pone.0053938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Wang X, Prins BP, Sõber S, Laan M, Snieder H. Beyond genome-wide association studies: new strategies for identifying genetic determinants of hypertension. Curr Hypertens Rep : 442–451, 2011. doi: 10.1007/s11906-011-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Wang X, Snieder H. Genome-wide association studies and beyond: what’s next in blood pressure genetics? Hypertension : 1035–1037, 2010. doi: 10.1161/HYPERTENSIONAHA.110.157214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, Ntalla I, Surendran P, Liu C, Cook JP, Kraja AT, Drenos F, Loh M, Verweij N, Marten J, Karaman I, Lepe MP, O’Reilly PF, Knight J, Snieder H, Kato N, He J, Tai ES, Said MA, Porteous D, Alver M, Poulter N, Farrall M, Gansevoort RT, Padmanabhan S, Mägi R, Stanton A, Connell J, Bakker SJ, Metspalu A, Shields DC, Thom S, Brown M, Sever P, Esko T, Hayward C, van der Harst P, Saleheen D, Chowdhury R, Chambers JC, Chasman DI, Chakravarti A, Newton-Cheh C, Lindgren CM, Levy D, Kooner JS, Keavney B, Tomaszewski M, Samani NJ, Howson JM, Tobin MD, Munroe PB, Ehret GB, Wain LV, Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O’Reilly P, Cabrera CP, Warren HR, Rose LM, Verwoert GC, Hottenga J-J, Strawbridge RJ, Esko T, Arking DE, Hwang S-J, Guo X, Kutalik Z, Trompet S, Shrine N, Teumer A, Ried JS, Bis JC, Smith AV, Amin N, Nolte IM, Lyytikäinen L-P, Mahajan A, Wareham NJ, Hofer E, Joshi PK, Kristiansson K, Traglia M, Havulinna AS, Goel A, Nalls MA, Sõber S, Vuckovic D, Luan J, M FDG, Ayers KL, Marrugat J, Ruggiero D, Lopez LM, Niiranen T, Enroth S, Jackson AU, Nelson CP, Huffman JE, Zhang W, Marten J, Gandin I, Harris SE, Zemonik T, Lu Y, Evangelou E, Shah N, de Borst MH, Mangino M, Prins BP, Campbell A, Li-Gao R, Chauhan G, Oldmeadow C, Abecasis G, Abedi M, Barbieri CM, Barnes MR, Batini C, Blake T, Boehnke M, Bottinger EP, Braund PS, Brown M, Brumat M, Campbell H, Chambers JC, Cocca M, Collins F, Connell J, Cordell HJ, Damman JJ, Davies G, de Geus EJ, de Mutsert R, Deelen J, Demirkale Y, Doney ASF, Dörr M, Farrall M, Ferreira T, Frånberg M, Gao H, Giedraitis V, Gieger C, Giulianini F, Gow AJ, Hamsten A, Harris TB, Hofman A, Holliday EG, Jarvelin M-R, Johansson Å, Johnson AD, Jousilahti P, Jula A, Kähönen M, Kathiresan S, Khaw K-T, Kolcic I, Koskinen S, Langenberg C, Larson M, Launer LJ, Lehne B, Liewald DCM, Lin L, Lind L, Mach F, Mamasoula C, Menni C, Mifsud B, Milaneschi Y, Morgan A, Morris AD, Morrison AC, Munson PJ, Nandakumar P, Nguyen QT, Nutile T, Oldehinkel AJ, Oostra BA, Org E, Padmanabhan S, Palotie A, Paré G, Pattie A, Penninx BWJH, Poulter N, Pramstaller PP, Raitakari OT, Ren M, Rice K, Ridker PM, Riese H, Ripatti S, Robino A, Rotter JI, Rudan I, Saba Y, Pierre AS, Sala CF, Sarin A-P, Schmidt R, Scott R, Seelen MA, Shields DC, Siscovick D, Sorice R, Stanton A, Stott DJ, Sundström J, Swertz M, Taylor KD, Thom S, Tzoulaki I, Tzourio C, Uitterlinden AG, Völker U, Vollenweider P, Wild S, Willemsen G, Wright AF, Yao J, Thériault S, Conen D, John A, Sever P, Debette S, Mook-Kanamori DO, Zeggini E, Spector TD, van der Harst P, Palmer CNA, Vergnaud A-C, Loos RJF, Polasek O, Starr JM, Girotto G, Hayward C, Kooner JS, Lindgren CM, Vitart V, Samani NJ, Tuomilehto J, Gyllensten U, Knekt P, Deary IJ, Ciullo M, Elosua R, Keavney BD, Hicks AA, Scott RA, Gasparini P, Laan M, Liu YM, Watkins H, Hartman CA, Salomaa V, Toniolo D, Perola M, Wilson JF, Schmidt H, Zhao JH, Lehtimäki T, van Duijn CM, Gudnason V, Psaty BM, Peters A, Rettig R, James A, Jukema JW, Strachan DP, Palmas W, Metspalu A, Ingelsson E, Boomsma DI, Franco OH, Bochud M, Newton-Cheh C, Munroe PB, Elliott P, Chasman DI, Chakravarti A, Knight J, Morris AP, Levy D, Tobin MD, Snieder H, Caulfield MJ, Ehret GB, Barnes MR, Tzoulaki I, Caulfield MJ, Elliott P; International Consortium of Blood Pressure (ICBP) 1000G Analyses; BIOS Consortium; Lifelines Cohort Study; Understanding Society Scientific group; CHD Exome+ Consortium; ExomeBP Consortium; T2D-GENES Consortium; GoT2DGenes Consortium; Cohorts for Heart and Ageing Research in Genome Epidemiology (CHARGE) BP Exome Consortium; International Genomics of Blood Pressure (iGEN-BP) Consortium; UK Biobank CardioMetabolic Consortium BP working group . Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet : 403–415, 2017. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Watkin DM, Froeb HG, Hatch FT, Gutman AB. Effects of diet in essential hypertension. II. Results with unmodified Kempner rice diet in 50 hospitalized patients. Am J Med : 441–493, 1950. doi: 10.1016/0002-9343(50)90200-2. [DOI] [PubMed] [Google Scholar]
- 376.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature : 737–738, 1953. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 377.Watson JD, Crick FH. The structure of DNA. Cold Spring Harb Symp Quant Biol : 123–131, 1953. doi: 10.1101/SQB.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- 378.Watt G. Design and interpretation of studies comparing individuals with and without a family history of high blood pressure. J Hypertens : 1–7, 1986. doi: 10.1097/00004872-198602000-00001. [DOI] [PubMed] [Google Scholar]
- 379.Weder AB. Evolution and hypertension. Hypertension : 260–265, 2007. doi: 10.1161/01.HYP.0000255165.84684.9d. [DOI] [PubMed] [Google Scholar]
- 380.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature : 661–678, 2007. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.White PC. Inherited forms of mineralocorticoid hypertension. Hypertension : 927–936, 1996. doi: 10.1161/01.HYP.28.6.927. [DOI] [PubMed] [Google Scholar]
- 382.White PC, Hautanen A, Kupari M. Aldosterone synthase (CYP11B2) polymorphisms and cardiovascular function. Endocr Res : 797–804, 1998. doi: 10.3109/07435809809032690. [DOI] [PubMed] [Google Scholar]
- 383.Williamson CR. Observations on the management of hypertension by the Kempner rice diet. N Engl J Med : 177–182, 1950. doi: 10.1056/NEJM195008032430502. [DOI] [PubMed] [Google Scholar]
- 384.Wilson FH, Disse-Nicodème S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. Human hypertension caused by mutations in WNK kinases. Science : 1107–1112, 2001. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 385.Woelfle J, Roth CL, Wunsch R, Reinehr T. Pregnancy-associated plasma protein A in obese children: relationship to markers and risk factors of atherosclerosis and members of the IGF system. Eur J Endocrinol : 613–622, 2011. doi: 10.1530/EJE-11-0423. [DOI] [PubMed] [Google Scholar]
- 386.Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, Kuchroo VK. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature : 513–517, 2013. doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Xiao B, Harada Y, Kawakami K, Nabika T. A 1.8-Mbp fragment on chromosome 1 affects sympathetic response to stress: evaluation in reciprocal congenic strains between stroke-prone spontaneously hypertensive rat and Wistar-Kyoto rat. J Hypertens : 257–265, 2011. doi: 10.1097/HJH.0b013e32834137cd. [DOI] [PubMed] [Google Scholar]
- 388.Yagil C, Hubner N, Kreutz R, Ganten D, Yagil Y. Congenic strains confirm the presence of salt-sensitivity QTLs on chromosome 1 in the Sabra rat model of hypertension. Physiol Genomics : 85–95, 2003. doi: 10.1152/physiolgenomics.00111.2002. [DOI] [PubMed] [Google Scholar]
- 389.Yagil C, Sapojnikov M, Kreutz R, Katni G, Lindpaintner K, Ganten D, Yagil Y. Salt susceptibility maps to chromosomes 1 and 17 with sex specificity in the Sabra rat model of hypertension. Hypertension : 119–124, 1998. doi: 10.1161/01.HYP.31.1.119. [DOI] [PubMed] [Google Scholar]
- 390.Yagil C, Sapojnikov M, Kreutz R, Zürcher H, Ganten D, Yagil Y. Role of chromosome X in the Sabra rat model of salt-sensitive hypertension. Hypertension : 261–265, 1999. doi: 10.1161/01.HYP.33.1.261. [DOI] [PubMed] [Google Scholar]
- 391.Yamori Y, Ooshima A, Okamoto K. Genetic factors involved in spontaneous hypertension in rats an analysis of F2 segregate generation. Jpn Circ J : 561–568, 1972. doi: 10.1253/jcj.36.561. [DOI] [PubMed] [Google Scholar]
- 392.Yan X, Baxter RC, Firth SM. Involvement of pregnancy-associated plasma protein-A2 in insulin-like growth factor (IGF) binding protein-5 proteolysis during pregnancy: a potential mechanism for increasing IGF bioavailability. J Clin Endocrinol Metab : 1412–1420, 2010. doi: 10.1210/jc.2009-2277. [DOI] [PubMed] [Google Scholar]
- 393.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet : 565–569, 2010. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, Zadeh M, Gong M, Qi Y, Zubcevic J, Sahay B, Pepine CJ, Raizada MK, Mohamadzadeh M. Gut dysbiosis is linked to hypertension. Hypertension : 1331–1340, 2015. doi: 10.1161/HYPERTENSIONAHA.115.05315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Ye P, West MJ. Cosegregation analysis of natriuretic peptide genes and blood pressure in the spontaneously hypertensive rat. Clin Exp Pharmacol Physiol : 930–936, 2003. doi: 10.1111/j.1440-1681.2003.03937.x. [DOI] [PubMed] [Google Scholar]
- 396.Young JH, Chang YP, Kim JD, Chretien JP, Klag MJ, Levine MA, Ruff CB, Wang NY, Chakravarti A. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet : e82, 2005. doi: 10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Yu H, Harrap SB, Di Nicolantonio R. Cosegregation of spontaneously hypertensive rat renin gene with elevated blood pressure in an F2 generation. J Hypertens : 1141–1147, 1998. doi: 10.1097/00004872-199816080-00010. [DOI] [PubMed] [Google Scholar]
- 398.Zagato L, Modica R, Florio M, Torielli L, Bihoreau MT, Bianchi G, Tripodi G. Genetic mapping of blood pressure quantitative trait loci in Milan hypertensive rats. Hypertension : 734–739, 2000. doi: 10.1161/01.HYP.36.5.734. [DOI] [PubMed] [Google Scholar]
- 399.Zennaro MC, Boulkroun S, Fernandes-Rosa F. An update on novel mechanisms of primary aldosteronism. J Endocrinol : R63–R77, 2015. doi: 10.1530/JOE-14-0597. [DOI] [PubMed] [Google Scholar]
- 400.Zhang L, Summers KM, West MJ. Angiotensin I converting enzyme gene cosegregates with blood pressure and heart weight in F2 progeny derived from spontaneously hypertensive and normotensive Wistar-Kyoto rats. Clin Exp Hypertens : 753–771, 1996. doi: 10.3109/10641969609081779. [DOI] [PubMed] [Google Scholar]
- 401.Zhang L, Xu D, West MJ, Summers KM. Association of the brain natriuretic peptide gene with blood pressure and heart weight in the rat. Clin Exp Pharmacol Physiol : 442–444, 1997. doi: 10.1111/j.1440-1681.1997.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 402.Zhang QY, Dene H, Deng AY, Garrett MR, Jacob HJ, Rapp JP. Interval mapping and congenic strains for a blood pressure QTL on rat chromosome 13. Mamm Genome : 636–641, 1997. doi: 10.1007/s003359900528. [DOI] [PubMed] [Google Scholar]
- 403.Zhang S, Mao G, Zhang Y, Tang G, Wen Y, Hong X, Jiang S, Yu Y, Xu X. Association between human atrial natriuretic peptide Val7Met polymorphism and baseline blood pressure, plasma trough irbesartan concentrations, and the antihypertensive efficacy of irbesartan in rural Chinese patients with essential hypertension. Clin Ther : 1774–1784, 2005. doi: 10.1016/j.clinthera.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 404.Zhang Y, Zang J, Wang B, Li B, Yao X, Zhao H, Li W. CD36 genotype associated with ischemic stroke in Chinese Han. Int J Clin Exp Med : 16149–16157, 2015. [PMC free article] [PubMed] [Google Scholar]
- 405.Zhao Y, Peng J, Lu C, Hsin M, Mura M, Wu L, Chu L, Zamel R, Machuca T, Waddell T, Liu M, Keshavjee S, Granton J, de Perrot M. Metabolomic heterogeneity of pulmonary arterial hypertension. PLoS One : e88727, 2014. doi: 10.1371/journal.pone.0088727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Zheng Y, Yu B, Alexander D, Mosley TH, Heiss G, Nettleton JA, Boerwinkle E. Metabolomics and incident hypertension among blacks: the atherosclerosis risk in communities study. Hypertension : 398–403, 2013. doi: 10.1161/HYPERTENSIONAHA.113.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Zhu Z, Xiong S, Liu D. The Gastrointestinal Tract: an Initial Organ of Metabolic Hypertension? Cell Physiol Biochem : 1681–1694, 2016. doi: 10.1159/000443107. [DOI] [PubMed] [Google Scholar]
- 408.Zhuo JL. SH2B3 (LNK) as a novel link of immune signaling, inflammation, and hypertension in Dahl salt-sensitive hypertensive rats. Hypertension : 989–990, 2015. doi: 10.1161/HYPERTENSIONAHA.115.04887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Zinner SH, Levy PS, Kass EH. Familial aggregation of blood pressure in childhood. N Engl J Med : 401–404, 1971. doi: 10.1056/NEJM197102252840801. [DOI] [PubMed] [Google Scholar]