Abstract
Drug resistance is a major obstacle in cancer treatment: A case in point is the failure of cancer patients to respond to tyrosine kinase inhibitors (TKI) of EGFR, a receptor that is highly expressed in many cancers. Identification of the targets and delineation of mechanisms of drug resistance remain major challenges. Traditional pharmacological assays of drug resistance measure the response of tumor cells using cell proliferation or cell death as readouts. These assays performed using traditional plastic tissue culture plates (2D) do not translate to in vivo realities. Here, we describe a genetic screen based on phenotypic changes that can be completed over a period of 1-1 months using functional endpoints in physiologically relevant 3D culture models. This phenotype-based assay could lead to the discovery of previously unknown therapeutic targets and could explain the source of the resistance and relapse. As a proof of principle, we performed our 3D culture assay with a small cDNA library in that yielded five unknown intermediates in EGFR and PI3K signaling pathways. Here, we describe the screening method and the characterization of one of the five molecules, but this approach could be easily expanded for a high-throughput screening to identify or evaluate many more unknown intermediates in oncogenic signaling pathways.
Keywords: 3D Culture, EGFR-TKI, Phenotypic screen, Drug resistance, Extracellular matrix, HMT3522, cDNA overexpression library
Background
Despite significant progress in therapeutic strategies, almost 30% of breast cancer patients develop chemotherapy resistance following initial treatment leading to relapse ( Brewster et al., 2008 ; Holohan et al., 2013 ). Genes or pathways that are responsible for chemoresistance have been identified through traditional screening approaches including gain-of-function and loss-of-function tools (Grimm, 2004; Arnoldo et al., 2014 ). More recently, a high-throughput screening technique of CRISPR/Cas9-based system has been used to identify molecular targets and their related pathways (Hu and Zhang, 2016; Luo, 2016). Nevertheless, most assays fail to provide a mechanism of resistance and alternative possibilities of treatment.
In a tissue, dynamic and reciprocal signaling between cells and their local microenvironment is required for maintenance of tissue architecture and homeostasis (Bissell and Hines, 2011; Furuta et al., 2018 ). In particular, biochemical and biophysical cues from the local microenvironment are recognized by cell surface receptors such as growth factor receptors and integrins that initiate a signaling cascade transduced into the nucleus to change gene expression ( Bissell et al., 1999 ; Ghajar and Bissell, 2008). In addition to mutations that drive gene expression involved in drug resistance, the microenvironment plays a significant role in multidrug resistance ( Weaver et al., 2002 ; Correia and Bissell, 2012). Therefore, a physiologically relevant assay system–one that permits screening while recapitulating the microenvironment in vivo–is critical for identifying new targets for breast cancer therapy and for elucidating mechanisms underlying the tissue specificity of drug resistance.
HMT-3522 progression series of human mammary epithelial cells was derived from a reduction mammoplasty of a woman with benign fibrocystic breast disease ( Petersen et al., 1990 ; Briand et al., 1996 ). The epithelial tissue was cultured in a chemically defined culture medium and the phenotypically normal S1 cells were established ( Briand et al., 1987 ). After over 100 passages of S1 cells in epidermal growth factor-free medium, growth factor-independent and tumorigenic T4-2 cells were obtained ( Briand et al., 1996 ). Non-malignant S1 cells in 3D culture of Laminin-rich extracellular matrix (lrECM) recapitulate an organized polar unit of breast structure referred to as acinus, in culture we refer to them as mammary organoids ( Petersen et al., 1992 ; Weaver et al., 1995 ; Lee et al., 2007 ). In the same medium and culture conditions, malignant T4-2 cells form disorganized colonies in 3D culture and tumors in mice. We have shown that malignant T4-2 cells in the presence of lrECM become quiescent and can be reverted to a structure partially resembling non-malignant counterparts, despite still containing the cancer genome (Figure 1). This “phenotypic reversion” is achieved by adjusting extracellular and intracellular signaling pathways, such as inhibitors of receptor tyrosine kinase or the humanized blocking antibody hampering oncogenic signaling which otherwise leads cells to lose architecture and grow out of control ( Weaver et al., 1997 ; Wang et al., 1998 ; Weaver and Bissell, 1999; Muschler et al., 2002 ; Wang et al., 2002 ; Kenny and Bissell, 2007; Itoh et al., 2007 ; Beliveau et al., 2010 ; Onodera et al., 2014 ). Other breast cancer cell lines, such as metastatic MDA-MB-231, can be reverted by treatment with a combination of inhibitors of integrin beta1, PI3K, or MAPK ( Wang et al., 2002 ).
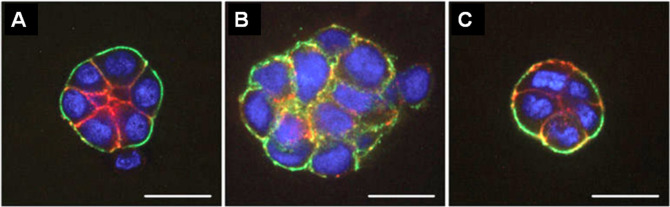
Figure 1. Fluorescence microscopy images of HMT 3522 progression series.
A. Non-malignant S1 cells organized into acini structure resembling alveoli in the mammary gland in vivo. B. Malignant T4-2 cells formed large disorganized colonies. C. “Phenotypically reverted” malignant T4-2 cell showed basal polarity and growth arrest by modulation of signal pathways. All cells were grown in 3D culture of lrECM, and stained for α6 integrin (green), β-catenin (red), and nuclei (blue). Adapted from Figure 3 in Lee et al. (2007) .
Using the three conditions described above, non-malignant, malignant and reverted malignant cells in our 3D cell culture models, we demonstrated that the efficacy of the chemotherapeutic response was strictly dependent on cell and tissue polarity and tissue architecture ( Weaver et al., 2002 ). By combining the ability to “phenotypically” revert the malignant cells with a gain-of-function genetic screen, we developed a phenotype-based functional genetic screen that allowed the identification of novel genes associated with EGFR -TKI resistance ( Lee et al., 2012 ).
In this protocol, viral cDNA library generated from malignant breast epithelial cells is transduced into the same malignant cells under 2D (tissue culture on plastic) followed by subsequent transfer to treatment with EGFR-TKI (AG1478) or PI3K inhibitor (LY294002), on 3D lrECM culture in the presence of antibiotics for selection of integrated genes. The 3D cultures are maintained for 5-7 days to allow the malignant T4-2 cells to undergo phenotypic reversion in the presence of either inhibitor. Then colonies that fail to revert and continuously grow are isolated from the cultures and subjected to sequencing for identifying the integrated cDNA that conferred resistance to EGFR-TKI or PI3K inhibitor (Figure 2).
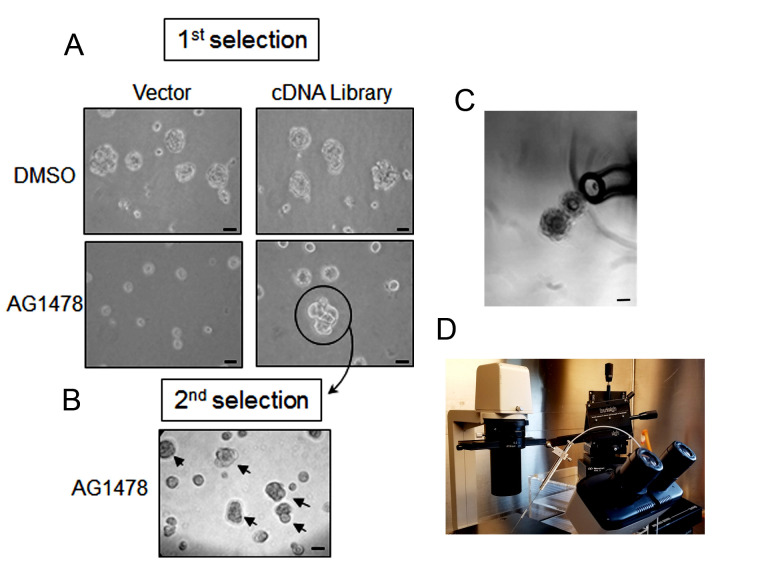
Figure 2. Workflow for phenotype-based genetic screen.
(1-2) Amphotropic retroviral cDNA library was generated and transduced into HMT3522 malignant T4-2 cells. (3) The cDNA expressing T4-2 cells were plated on 3D lrECM culture and treated with EGFR-TKI AG1478. Colonies that fail to revert (circled in blue) were isolated from the culture using a flame-drawn glass pipette under a phase-contrast microscope in a Biosafety cabinet. (4) Each isolated colony was transferred to one well of a 48-well tissue culture plate and subsequently expanded to a larger multi-well plate until the cell number reached one million cells. (5) Cells were re-plated on 3D lrECM culture and treated with EGFR-TKI AG1478, and the isolation procedure was repeated as described in (3). (6) Genomic DNA was isolated from selected clones grown in 2D and cDNA inserts were recovered by PCR using the flanking sequences of retroviral vector used for cloning. (7) The amplified PCR products were analyzed for sequencing to determine the identity of inserted cDNA.
Using this technique, we identified five candidates (Table 2) and further characterized one of the genes that conferred EGFR-TKI resistance in breast cancer cells when overexpressed. This gene was identified as FAM83A, one of the eight members of a previously unidentified oncogene family. We observed that FAM83A expression was increased when cells were treated with tyrosine kinase inhibitor, lapatinib; the cells that survived became resistant to the treatment. FAM83A-associated molecular mechanism of drug resistance serves as an example of the utility of this screening method and provides a basis for predictive diagnosis and possible molecular targets for chemotherapy.
Table 2. Candidate genes that recovered from the screen.
| Gene Name | Accession Number | Cellular Functions | Resistance |
|---|---|---|---|
| FAM83A | NM207006 | Activation of RAS/MAPK pathway | EGFR Inhibitor |
| MGRBP | NM018270 | Chromatin modification | PI3K Inhibitor |
| ZFPL1 | NM006782 | Vesicle-mediated transport | PI3K Inhibitor |
| PTBP1 | NM002819 | RNA-binding protein | PI3K Inhibitor |
| Rab32 | NM006834 | Rab GTPase | PI3K Inhibitor |
This assay can be applied for identifying novel therapeutic targets by modulation of gene expression such as systemic gain-of-function genetic screens with a cDNA library or loss-of-function genetic screens with shRNA (short hairpin RNA) or sgRNA (single guide RNA) libraries. Additionally, this assay is also a useful tool for evaluation of drug response and efficacy of pharmacological candidates in different cancer cell lines allowing us to predict therapeutic outcomes.
Materials and Reagents
-
Construction of cDNA library
Retroviral pESY-Neo vector (used for the construction of cDNA library is a modified version of the pEYK3.1 vector [a kind gift from the laboratory of Dr. George Daley at the Whitehead Institute])
DNeasy® tissue kit (QIAGEN, catalog number: 69504)
Oligotex poly (A)+ RNA purification column (QIAGEN, catalog number: 70042)
SuperScript® plasmid system with Gateway® technology for cDNA synthesis and cloning (Invitrogen Life Technologies, catalog number: 18248013)
Tris-Borate-EDTA (TBE) buffer (Amresco, 10x liquid concentrate, catalog number: 0658-4l)
Ultrapure agarose (Invitrogen, catalog number: 16500-100)
XL-1 Gold Ultracompetent cells (Stratagene, catalog number: 200315)
QIAquick gel extraction kit (QIAGEN, catalog number: 28704)
Plasmid Mega/Giga purification kit (QIAGEN, catalog number: 12781)
-
Retrovirus production and titration
Phoenix–amphotropic retrovirus producing cells (Phoenix-AMPHO ATCC® CRL-3213TM)
Dulbecco’s modified eagle medium (DMEM, high glucose) (Gibco, Life Technologies, catalog number: 11965092)
Fetal bovine serum (Gibco, catalog number: 16000044)
200 mM L-Glutamine (Gibco, catalog number: 25030081)
LipofectamineTM 2000 (Invitrogen, catalog number: 16680190)
0.45 μM SFCA membrane syringe filter (Corning Incorporated, catalog number: 443220)
NIH3T3 mouse embryo fibroblast cells (ATCC, catalog number: CRL-1658)
-
Transduction of viral cDNA library
Retroviral cDNA library supernatant
Polybrene (Hexadimethrine bromide, Sigma-Aldrich, catalog number: H9268)
-
Cell culture
Pipette tips and sterile serological pipettes
100-mm tissue culture-treated Petri-dish (BD Biosciences, catalog number: 353003)
Tissue culture treated multiple well plates, 48-well (BD Bioscience, catalog number: 353078)
Falcon tubes, polypropylene, 50-ml (BD Biosciences, catalog number: 352095)
Falcon tubes, polypropylene, 15-ml (BD Biosciences, catalog number: 352096)
-
Finely flame-drawn Pasteur pipettes
Fine Pasteur pipettes were made from commercial Pasteur pipettes (VWR, catalog number: 14673-043). The tip of pipette was heated several seconds on the flame of Bunsen burner, rapidly pulled out of flame using forceps, and cut the tip of pipette to obtain a suitable diameter.
-
(Optional) HamiltonTM Micro-syringe
Caution: Flame-drawn pipettes are very sharp. Handle with care. The pipette should be autoclaved and dried before use.
HMT3522 T4-2 cells (or any epithelial cancer cell line that can be ‘reverted’ to a phenotypically normal phenotype)
Dulbecco’s modified Eagle’s medium (DMEM/F12, Thermo Fisher Scientific, catalog number: 12400-024)
Insulin (250 ng/ml, Sigma-Aldrich, catalog number: I6634)
Transferrin (10 μg/ml, Sigma-Aldrich, catalog number: T2252)
Sodium Selenite (2.6 ng/ml, BD Bioscience, catalog number: 354201)
Estradiol (10-10 M, Sigma-Aldrich, catalog number: E2758)
Hydrocortisone (1.4 x 10-6 M, Sigma-Aldrich, catalog number: H0888)
Prolactin (5 μg/ml, Sigma-Aldrich, catalog number: L6520)
0.25% (wt/vol) Trypsin (Thermo Fisher Scientific, catalog number: 25200-056)
Soybean trypsin inhibitor (10 mg/ml, Sigma-Aldrich, catalog number: T6522)
Na2HPO4 (Sigma-Aldrich, catalog number: S9390)
KH2PO4 (J.T. Baker Inc., catalog number: 3246-01)
NaCl (Sigma-Aldrich, catalog number: S5886)
KCl (Sigma-Aldrich, catalog number: P5405)
H14 Complete medium (without Epidermal Growth Factor) (see Recipes)
1x PBS (see Recipes)
-
Phenotypic reversion assay on 3D lrECM culture
LDEV-free growth factor reduced basement membrane matrix (MatrigelTM, Corning, catalog number: 354230; CultrexTM, Trevigen, catalog number: 3433-001-01)
G418 Sulfate (Thermo Fisher Scientific, catalog number: 10131027) to select stably transduced cells
Signaling pathway inhibitors (EGFR-TKI AG1478, EMD Millipore, catalog number: 658652; LY294002, Biomol, catalog number: L7751; Lapatinib or Gefitinib were kind gifts from Catherine Park at UCSF, but they are also commercially available at Sigma-Aldrich)
-
Validation of selected clones
Dispase II (neutral protease, grade II) (Sigma-Aldrich, catalog number: 4942078001)
DNeasy® tissue kit (QIAGEN, catalog number: 69504)
Primers to flanking sequences in the cloning vector
LA Taq DNA Polymerase (A high fidelity Polymerase Chain Reaction (PCR) enzyme for long range) (Takara Bio, catalog number: RR002)
pGEM®-T Easy cloning vector (Promega, catalog number: A1360)
Equipment
Pipettes
Water-Jacketed CO2 incubator (Thermo Scientific Forma, model: FormaTM Series II 3120)
Centrifuge (Eppendorf, model: 5804)
Inverted phase microscope (Nikon, model: Eclipse TS100)
BioSafety cabinet
Gene Pulse® electroporation cuvettes (Bio-Rad, catalog number: 1652089)
Electroporation system (Bio-Rad Gene Pulser® II electroporation system, catalog numbers: 1652105-1652110)
FluorchemTM 8900 gel imaging system (Alpha Innotech, model: 8900)
PCR thermocycler
(Optional) Micromanipulator (Burleigh, model: PCS5000)
Procedure
-
Construction of cDNA library
Construction of retroviral cDNA library used for this protocol is performed as previously described ( Lee et al., 2012 ).
Extract total RNA from HMT3522 T4-2 cells.
Apply the total RNA to Oligotex poly (A)+ RNA purification column and purify mRNA according to the manufacturer’s protocol (QIAGEN).
Synthesize double-stranded cDNA using SuperScript® plasmid system with Gateway® Technology for cDNA Synthesis and Cloning by the manufacturer’s instruction (Invitrogen life technologies).
Run cDNA on a 0.7% agarose gel at 30 W in 0.5x TBE buffer for the size-fractionation.
Excise DNA segments covering a size range of 0.5 to 5 kb and purify the fraction with agarose gel extraction kit according to the manufacturer’s manual (QIAGEN).
Perform ligation reaction by mixing the purified cDNA in a 10 μl reaction with the linearized retroviral pESY-Neo vector and incubate for 16 h at 4 °C.
Electroporate 2 μl ligation mixture into XL-1 Gold Ultracompetent cells in a 0.1 cm electroporation cuvette using the Bio-Rad Gene Pulse Controller® at 100 ohms and 25 μF.
Plate the transformed bacteria on 150 mm LB plate, containing 50 μg/ml of kanamycin and incubate them for 16-18 h at 37 °C.
Determine the complexity of the library by counting the number of transformed E. coli colonies in the plates using an auto-count program of FluorchemTM imaging system (Alpha Innotech).
Gently scrape the colonies from the plates and isolate the plasmid DNA using the plasmid Mega purification kit (QIAGEN).
Note: We calculated the titer (CFU/ml) of our library by multiplying the number of colonies on the plates by the volume of pooled library (ml). The resulting retroviral cDNA library consisted of a complexity of approximately 5 x 104 CFU/ml and an average insert size of 1.2 kb.
-
Retrovirus production and titration
Plate Phoenix amphotropic retrovirus producing cells at a density of 1.5 x 106 cells in DMEM medium containing 10% (vol/vol) FBS and 2 mM Glutamine on 100-mm tissue culture-treated Petri-dish prior to transfection.
Transfect with 10 μg of cDNA plasmid using Lipofectamine 2000 according to the manufacturer’s manual (Invitrogen).
Incubate the culture for 48 h.
-
Harvest retrovirus containing medium and filter through a 0.45 μm syringe filter.
Note: The supernatant can be frozen at -80 °C for later use.
Determine the titer of harvested retrovirus as described previously in Kwon et al. (2003) .
-
Transduction of viral cDNA library
Plate growing target cells at a density of 1 x 104 cells/cm2 on 100 mm Petri-dish in H14 complete medium for 12-18 h prior to transduction at 37 °C in a humidified incubator in an atmosphere of 5% CO2.
Remove culture medium from the cells and add virus particles with Polybrene at a concentration of 8 μg/ml.
Incubate the cells with virus containing medium for 8-16 h at 37 °C in a humidified incubator in an atmosphere of 5% CO2.
Remove the virus-containing medium and add 10 ml fresh medium to the culture.
Incubate the culture for 48 h 37 °C in a humidified incubator in an atmosphere of 5% CO2.
-
Replace medium with fresh H14 complete medium containing G418 at a concentration of
250 μg/ml for selecting the cells that carry integrated cDNA into their genome.
Note: We used here neomycin resistant gene, however, puromycin resistance gene as a selection marker is preferable because the time for selection is typically shorter than neomycin or hygromycin resistance gene.
-
On-Top 3D culture assay
To study functional differentiation and alveolar morphogenesis of mammary epithelial cells in reconstituted basement membrane, several 3D cell culture techniques have been developed ( Petersen et al., 1992 ; Kaminker et al., 2005 ; Lee et al., 2007 ). In embedded 3D assay, cells are cultured within laminin-rich extracellular matrix (lrECM) gel, such as MatrigelTM. In “On-Top” 3D assay, cells are cultured on top of a layer of lrECM coat, and a 5% (vol/vol) lrECM-containing medium is dropped on top of the cells to provide the 3D laminin-rich environment. In both assays, cells form proper cell-cell and cell-ECM interaction and recapitulate physiologically and functionally similar polarized alveolar structure as in vivo. The difference between the embedded 3D assay versus the On-Top is that the embedded assay more tightly controls the morphology and the growth rate of the colonies due to the cells being encapsulated within the lrECM (MatrigelTM) at the time of plating. Thus, the embedded method provides more homogeneous physical constraints and biochemical cues to the cells. This method is most useful for morphology study. However, the embedded method takes 10 days to complete, and the colonies are difficult to remove from the gel. In this study, we employed On-Top 3D culture because: (1) The proliferation and morphological differences between nonmalignant and malignant cells can be distinguished by five days in culture. (2) Colonies can be readily isolated from the culture using finely flame-drawn drown glass pipette or micromanipulator syringe needle. This assay was ended on Day five to minimize artifacts that might have been generated by a less-strict On-Top 3D culture setting.
-
Coat 60 mm culture dish with 850 μl cold Matrigel® and incubate for 20-30 min at 37 °C.
Note: Matrigel® should be stored at -80 °C and thawed on ice overnight before use. It solidifies quickly when warmed. Therefore, it should be kept on ice at all times. It is important to incubate the Matrigel-coated plate for the suggested time in order to achieve proper gelling of Matrigel (Table 1).
-
Dislodge the cell layer to single cells with 0.25% Trypsin for 3 min at 37 °C.
Note: Complete dissociation of cells to single cells is critical, otherwise clumping will lead to false positive in 3D lrECM culture.
Count the cells, transfer 6 x 105 cells to a 15 ml conical tube and pellet them by centrifugation at 130 × g at room temperature for 5 min.
-
Re-suspend the cells to single cells in 2,500 μl H14 complete medium. This is half the suggested volume of growth medium.
Note: It is crucial that cells be dissociated thoroughly to single cells, since clumping of cells will cause false positive in 3D lrECM cultures. Add G418 at a concentration of 250 μg/ml in the growth medium to select the cells containing integrated cDNA.
Plate the cells onto the surface of gelled Matrigel and allow them to settle for 10 min at 37 °C in an incubator.
Add 250 μl Matrigel and G418 to the remaining 2250 μl cold H14 medium.
Add the Matrigel and G418-mixed H14 medium drop-wise to the plated culture.
Maintain the culture for 5-7 d and replace the growth medium with G418 every other day.
-
-
Isolation of clones from On-Top 3D culture assay and expansion of selected clones
-
Remove culture medium and rinse briefly with 5 ml sterile PBS.
Note: Warm up culture media and Dispase II in a 37 °C water bath for 10 min before use.
Incubate the culture with 1-5 ml of Dispase II solution (2 U/ml) for 30 min at 37 °C in an incubator to allow colonies to gently dissociate from Matrigel.
-
Isolate the colonies that have grown to large clusters and had failed to ‘revert’ to a phenotypically ‘normal’ architecture using a finely flame-drawn Pasteur pipette under a phase-contrast microscope in a BioSafety cabinet (Figures 3A and 3C).
Note: Special attention to sterile technique; it is important to wipe the microscope with 70% ethanol or other disinfectant before putting it in a BioSafety cabinet.
(Optional) HamiltonTM Micro-syringe attached Micromanipulator is an alternative option for collecting colonies (Figure 3D).
Transfer the isolated colonies to 48-well tissue culture plates and add 250 μg/ml G418-containing H14 medium for continuing culture.
To scale up, gradually expand the cells you have picked to larger plates.
-
-
Identification and validation of selected clones
Extract genomic DNA from the selected clones cultured on 35 mm tissue culture dish (approximately 5 x 105 cells) using DNeasy Tissue Kit following the manufacturer’s instruction (QIAGEN).
-
Run Polymerase Chain Reaction (PCR) to recover cDNA insert using flanking sequences in the viral vector (Figure 4).
Note: It is critical that the PCR reaction for genomic DNA should be performed using Taq DNA polymerase that is suitable for amplification of long-range DNA fragments (> 1 kb).
Ligate amplified DNA fragments from the PCR reaction into pGEM-T sub-cloning vector.
Perform DNA sequencing.
-
Analyze sequence homology to determine the identity of the integrated cDNA.
Note: Clones that do not contain in-frame cDNA insert should be excluded from the candidates.
-
Second round of screening of selected clones
Follow the steps in Procedure B-C to repeat the screening procedure for enrichment of the selected clones (Figure 3B).
Gradually and independently, expand the selected colonies on the monolayer.
Analyze the biological functions of the candidate genes.
Table 1. Suggested volume and number of cells for 3D On-Top culture .
| Diameter (mm) |
Approximate Surface Area (cm2) |
Cell Numbers for plating |
Medium Volume (μl) |
LrECM Coat (μl) | |
|---|---|---|---|---|---|
| Dish | 35 | 9 | 2 x 105 | 2,000 | 500 |
| 60 | 21 | 6 x 105 | 5,000 | 850 | |
| 100 | 55 | 1.6 x 106 | 10,000 | 2,500 |
Figure 3. Isolation of colonies that are resistant to the phenotypic reversion in 3D lrECM culture.
A. The cDNA expressing malignant T4-2 cells were plated and treated with EGFR-TKI AG1478. The culture was maintained for five days and the medium was changed to fresh with EGFR-TKI AG1478 and 250 μg/ml G418 on alternate days. Colonies that are bigger than 100 μm were selected. Selected resistant colony is circled. B. Each isolated colony was separately transferred to a multi-well plate, expanded to a sufficient number of cells, re-plated them on 3D lrECM culture with EGFR-TKI AG1478 and 250 μg/ml G418, and repeated the isolation procedure for enrichment of the isolated clone. Selected resistant colony is indicated with arrow. C. Clusters of cells were isolated by a fine-tip Pasteur pipette under a phase-contrast microscope in a Biosafety cabinet. D. HamiltonTM Micro-syringe attached Micromanipulator (Burleigh PCS-5000) is an alternative option for collecting colonies. Scale bars in A, B, C = 50 μm.
Figure 4. Fourteen representative cDNA inserts recovered by PCR using the flanking sequences of retroviral cloning vector for sequencing analysis.
Notes
In vivo, basement membrane is crucial for tissue morphogenesis and maintenance of tissue-specific structure and function. To provide physiologically relevant in vitro models, a reconstituted basement membrane can be made from an extract of gelatinous mixture secreted by the Engelbreth-Holm-Swarm (EHS) mouse sarcoma that is commercially available under the names Corning® Matrigel® matrix, and Cultrex® 3-D Culture MatrixTM. The major components of these matrices are laminin (~60%), collagen IV (~30%), and entactin (~8%), and we refer them to laminin-rich extracellular matrix (lrECM). We choose Matrigel with protein concentrations ranging from 10 to 12 mg/ml and with low levels of endotoxin (< 2 U/ml). Due to lot-to-lot variability, we usually test multiple lots before use and purchase large quantities to ensure reproducibility. Criteria for testing of Matrigel lots include morphology, cell proliferation, polarity, and levels of integrin and EGFR.
The concept of the assay includes gain-of-function of cDNA library in mammalian cells where increased expression of each gene in the library reflects a readout. A more ambitious cDNA library with a higher viral titer than the one we used here will increase the chances of identifying more candidate genes from the screen. In addition, size range of cDNA inserts between 500 bp and 4 kb is critical for achieving a full-length-enriched cDNA library. Having used a retroviral cDNA library consisting of a complexity of approximately 5 x 104 colony forming units (CFU) and an average insert size of 1.2 kb, our screen yielded 85 colonies resistant to signaling inhibitor(s), 35 colonies of which contained cDNA inserts. After excluding clones that did not express open reading frame cDNA, five candidate genes were identified (Table 2). Once clones containing genes of interest are identified, they must be validated using different cancer cell lines and/or animal models (Figure 5).
Despite the number of steps listed, we believe that a functional screen under physiologically-relevant conditions will yield important candidate genes to increase chances of finding target pathways that are responsible for chemotherapy resistance. In addition, this data will provide important insight about how these candidate genes would perform in vivo.
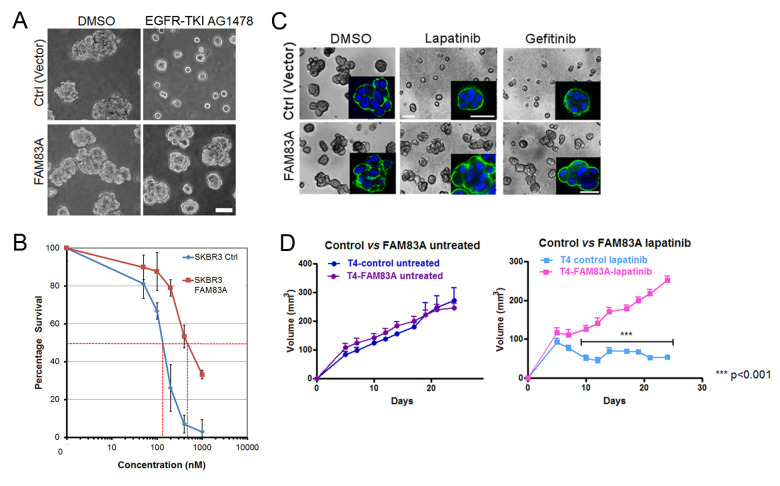
Figure 5. Validation of FAM83A conferring resistance to EGFR inhibitor.
A. Vector control or FAM83A overexpressing T4-2 cells were plated in 3D lrECM with reverting compound, EGFR-TKI AG1478. B. IC50 of lapatinib for vector control or FAM83 overexpression SKBR3 cells. C. Drug response to lapatinib or gefitinib in vector control or FAM83A overexpressing T4-2 cells in 3D lrECM culture. D. Tumor growth derived from vector or FAM83A–overexpression T4-2 cells xenografted in nude mice. (C) and (D) were adapted from Figures 3 and 4 in Lee et al. (2012) . Scale bars in A, C = 50 μm.
Recipes
-
H14 complete medium (without Epidermal Growth Factor)
DMED/F12
250 ng/ml Insulin
10 μg/ml transferrin
2.6 ng/ml sodium selenite
1.4 x 10-6 M hydrocortisone
10-10 M β-estradiol
5 μg/ml prolactin
-
1x PBS (pH 7.4)
8 mM Na2HPO4
2 mM KH2PO4
137 mM NaCl
2.7 mM KCl
Acknowledgments
We thank Drs. Richard Schwarz, Joni Mott, and Claire Robertson for critical reading of the manuscript. This work was supported by grants to Mina Bissell from the U.S. Department of Defense Innovator Award (W81XWH0810736), Breast Cancer Research Foundation (BCRF) and in part by NIH research project grant program (R01CA064786).
Competing interests
The authors declare no competing financial interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Arnoldo A., Kittanakom S., Heisler L. E., Mak A. B., Shukalyuk A. I., Torti D., Moffat J., Giaever G. and Nislow C.(2014). A genome scale overexpression screen to reveal drug activity in human cells. Genome Med 6(4): 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beliveau A., Mott J. D., Lo A., Chen E. I., Koller A. A., Yaswen P., Muschler J. and Bissell M. J.(2010). Raf-induced MMP9 disrupts tissue architecture of human breast cells in three-dimensional culture and is necessary for tumor growth in vivo . Genes Dev 24(24): 2800-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bissell M. J. and Hines W. C.(2011). Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 17: 320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bissell M. J., Weaver V. M., Lelievre S. A., Wang F., Petersen O. W. and Schmeichel K. L.(1999). Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res 59(7 Suppl): 1757-1763s; discussion 1763s-1764s. [PubMed] [Google Scholar]
- 5. Brewster A. M., Hortobagyi G. N., Broglio K. R., Kau S. W., Santa-Maria C. A., Arun B., Buzdar A. U., Booser D. J., Valero V., Bondy M. and Esteva F. J.(2008). Residual risk of breast cancer recurrence 5 years after adjuvant therapy. J Natl Cancer Inst 100(16): 1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Briand P., Petersen O. W. and Van Deurs B.(1987). A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol 23(3): 181-188. [DOI] [PubMed] [Google Scholar]
- 7. Briand P., Nielsen K. V., Madsen M. W. and Petersen O. W.(1996). Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Res 56(9): 2039-2044. [PubMed] [Google Scholar]
- 8. Correia A. L. and Bissell M. J.(2012). The tumor microenvironment is a dominant force in multidrug resistance. Drug Resist Updat 15(1-2): 39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furuta S., Ren G., Mao J. H. and Bissell M. J.(2018). Laminin signals initiate the reciprocal loop that informs breast-specific gene expression and homeostasis by activating NO, p53 and microRNAs. Elife 7: e26148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghajar C. M. and Bissell M. J.(2008). Extracellular matrix control of mammary gland morphogenesis and tumorigenesis: insights from imaging. Histochem Cell Biol 130(6): 1105-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grimm S.(2004). The art and design of genetic screens: mammalian culture cells. Nat Rev Genet 5(3): 179-189. [DOI] [PubMed] [Google Scholar]
- 12. Holohan C., Van Schaeybroeck S., Longley D. B. and Johnston P. G.(2013). Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 13(10): 714-726. [DOI] [PubMed] [Google Scholar]
- 13. Hu X. and Zhang Z.(2016). Understanding the genetic mechanisms of cancer drug resistance using genomic approaches. Trends Genet 32(2): 127-137. [DOI] [PubMed] [Google Scholar]
- 14. Itoh M., Nelson C. M., Myers C. A. and Bissell M. J.(2007). Rap1 integrates tissue polarity, lumen formation, and tumorigenic potential in human breast epithelial cells. Cancer Res 67(10): 4759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kenny P. A. and Bissell M. J.(2007). Targeting TACE-dependent EGFR ligand shedding in breast cancer. J Clin Invest 117(2): 337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaminker P., Plachot C., Kim S. H., Chung P., Crippen D., Petersen O. W., Bissell M. J., Campisi J. and Lelievre S. A.(2005). Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2. J Cell Sci 6): 1321-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwon Y. J., Hung G., Anderson W. F., Peng C. A. and Yu H.(2003). Determination of infectious retrovirus concentration from colony-forming assay with quantitative analysis. J Virol 77(10): 5712-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee G. Y., Kenny P. A., Lee E. H. and Bissell M. J.(2007). Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 4(4): 359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee S. Y., Meier R., Furuta S., Lenburg M. E., Kenny P. A., Xu R. and Bissell M. J.(2012). FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest 122(9): 3211-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo J.(2016). CRISPR/Cas9: From genome engineering to cancer drug discovery. Trends Cancer 2(6): 313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muschler J., Levy D., Boudreau R., Henry M., Campbell K. and Bissell M. J.(2002). A role for dystroglycan in epithelial polarization: loss of function in breast tumor cells. Cancer Res 62(23): 7102-7109. [PubMed] [Google Scholar]
- 22. Onodera Y., Nam J. M. and Bissell M. J.(2014). Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-GlcNAc pathways. J Clin Invest 124: 367-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petersen O. W., Ronnov-Jessen L., Howlett A. R. and Bissell M. J.(1992). Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A 89(19): 9064-9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petersen O. W., van Deurs B., Nielsen K. V., Madsen M. W., Laursen I., Balslev I. and Briand P.(1990). Differential tumorigenicity of two autologous human breast carcinoma cell lines, HMT-3909S1 and HMT-3909S8, established in serum-free medium. Cancer Res 50(4): 1257-1270. [PubMed] [Google Scholar]
- 25. Wang F., Hansen R. K., Radisky D., Yoneda T., Barcellos-Hoff M. H., Petersen O. W., Turley E. A. and Bissell M. J.(2002). Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst 94(19): 1494-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang F., Weaver V. M., Petersen O. W., Larabell C. A., Dedhar S., Briand P., Lupu R. and Bissell M. J.(1998). Reciprocal interactions between β1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci U S A 95(25): 14821-14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weaver V. M. and Bissell M. J.(1999). Functional culture models to study mechanisms governing apoptosis in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia 4(2): 193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weaver V. M., Howlett A. R., Langton-Webster B., Petersen O. W. and Bissell M. J.(1995). The development of a functionally relevant cell culture model of progressive human breast cancer. Semin Cancer Biol 6(3): 175-184. [DOI] [PubMed] [Google Scholar]
- 29. Weaver V. M., Lelievre S., Lakins J. N., Chrenek M. A., Jones J. C., Giancotti F., Werb Z. and Bissell M. J.(2002). β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2(3): 205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weaver V. M., Petersen O. W., Wang F., Larabell C. A., Briand P., Damsky C. and Bissell M. J.(1997). Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies . J Cell Biol 137(1): 231-245. [DOI] [PMC free article] [PubMed] [Google Scholar]