Abstract
On the basis of TP53 mutations and standardized uptake values (SUVs) from 18F-fluorodeoxyglucose positron emission tomography (18F-FDG-PET), we sought to enhance our knowledge of the biology underlying low progesterone receptor (PR) expression in estrogen receptor (ER)-positive/human epidermal growth factor receptor-2 (HER2)-negative tumors. This study included 272 patients surgically treated for ER-positive, HER2-negative breast cancer and who had undergone TP53 gene sequencing. Of these, 229 patients also underwent 18F-FDG PET or PET/CT. Mutational analysis of exons 5–9 of the TP53 gene was conducted using PCR amplification and direct sequencing. The SUVs were measured using 18F-FDG-PET scan images. Twenty-eight (10.3%) tumors had a somatic TP53 mutation. The TP53 mutation rate was significantly higher in low-PR tumors than in high-PR tumors (17.1% vs 7.9%, P = 0.039). Low-PR tumors had significantly higher median SUVs than high-PR tumors (P = 0.046). The multivariable analysis revealed that SUV and age remained independent variables associated with low PR expression. An adverse impact of low PR expression on recurrence-free survival was observed in the multivariable Cox regression hazard model. We provide clinical evidence that genetic alteration of the TP53 gene and dysregulated glucose metabolism partly involve low PR expression in ER-positive and HER2-negative breast cancer.
Keywords: progesterone receptor, TP53 mutation, SUV, breast cancer
Background
In estrogen receptor (ER)-positive breast cancer, progesterone receptor (PR) expression is generally considered a marker of an intact estrogen-responsive pathway (Horwitz & McGuire 1975). In addition, patients with ER-positive/PR-negative breast cancers have a worse prognosis than that of patients with PR-positive tumors (Clark et al. 1983). Recently, two studies suggested that low or negative PR expression can be used to identify ‘luminal B-like’ cancer, which has a poor outcome and is distinguished from ‘luminal A-like’ cancer in ER-positive/human epidermal growth factor receptor-2 (HER2)-negative cancer (Cancello et al. 2013, Prat et al. 2013).
Therefore, many investigators have tried to dissect tumor biology regarding the loss of PR in ER-positive cancer. Previous studies using human tumor samples suggested several lines of evidence that a higher rate of TP53 mutation (Olivier et al. 2006), DNA copy-number gain or increasing PI3K/mTOR gene signature (Creighton et al. 2009), and activation of the growth factor signal pathway (Arpino et al. 2005) could be associated with loss of PR expression in ER-positive cancer. However, these studies included both ER-positive/HER2-positive and -negative tumors; thus, previous findings on the loss of PR expression might be affected by HER2 overexpression. Molecular studies based on next-generation sequencing showed that HER2-enriched tumors more frequently harbor TP53 mutation than luminal A or luminal B tumors (Desmedt et al. 2012, Bertheau et al. 2013). Furthermore, standard of care for ER-positive/HER2-positive patients is anti-HER2 therapy plus chemotherapy followed by endocrine therapy, differing from that for ER-positive/HER2-negative patients (Goldhirsch et al. 2013).
Our group previously reported the prognostic influence of the standardized uptake value (SUV) on 18F-fluorodeoxyglucose positron emission tomography (18F-FDG-PET), which represents glucose uptake, in ER-positive breast cancer (Ahn et al. 2014a ,b ). In addition, we found a positive correlation between the SUV and the 21-gene recurrence score (RS) in ER-positive/HER2-negative disease (Ahn et al. 2017).
In this study, we first questioned whether the rate of TP53 mutation was higher in low-PR tumors than that in high-PR tumors among ER-positive/HER2-negative breast cancer. Second, we compared SUVs according to PR status in these tumors. This study sought to enhance our understanding of the biology underlying low PR expression within ER-positive/HER2-negative tumors.
Patients and methods
Patients
This study included 272 patients surgically treated for ER-positive, HER2-negative breast cancer and who had undergone TP53 gene sequencing between March 2007 and December 2015 at Gangnam Severance Hospital, Yonsei University College of Medicine. Patients who were treated with neoadjuvant chemotherapy and those who were diagnosed with recurrent or metachronous breast cancer were excluded from this study. Patients with ductal carcinoma in situ were also excluded. Of the 272 patients, 229 also underwent 18F-FDG PET or PET/computed tomography (CT) as part of routine preoperative staging.
The staging was performed according to the 7th edition of the American Joint Committee on Cancer system (Edge & Compton 2010). The Elston-Ellis modification of the Scarff-Bloom-Richardson grading system was used for histologic grading. Adjuvant systemic therapy and/or radiotherapy were administered according to the standard guidelines based on patient age, primary tumor characteristics and axillary lymph node status. Endocrine therapy was administered to all patients. The follow-up protocol included planned regular visits every 6 months; missed appointments were followed-up by telephone calls to minimize the number of patients lost to follow-up and to improve the accuracy of the survival data. The final update to the clinical database was made in March 2018. Our study was approved by the Institutional Review Board of Gangnam Severance Hospital, Yonsei University, Seoul, Korea, in accordance with the good clinical practice guidelines under the Declaration of Helsinki.
Definition of low PR expression and immunohistochemical (IHC) study
We defined low PR expression based on the modified Allred system (Harvey et al. 1999): low, Allred score 0–4; and high, Allred score 5–8. ER expression was also evaluated in the same manner. All tumors included in this study were ER positive (Allred scores ≥3). HER2 status was evaluated according to the 2013. American Society of Clinical Oncology/College of American Pathologists (ASCO-CAP) guidelines (Wolff et al. 2013) and was negative for all cases. The antibodies used for the IHC study were described previously. Ki67 expression was measured by an experienced pathologist and reported as a percentage score (range 0–100%) of positive tumor cells.
TP53 Sanger sequencing
Mutational analysis of exons 5–9 of the TP53 gene was conducted using PCR amplification and direct sequencing (Kim et al. 2014). The primers designed to amplify the exons and flanking introns of the TP53 gene were described previously (Kim et al. 2014). Briefly, PCR was performed using an Accu-PowerTM Premix (Bioneer, Daejeon, Korea) under the following amplification conditions: 94°C for 4 min followed by 50 cycles of 94°C for 1 min, 60°C for 30 s and 72°C for 30 s and final extension at 72°C for 15 min. Purified PCR products obtained using a QIAquick Gel Extraction kit (Qiagen) were used for sequencing with a Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems). The thermal cycler conditions were as follows: 96°C for 5 min followed by 24 cycles of 96°C for 10 s, 50°C for 5 s and 60°C for 4 min and final extension at 72°C for 5 min. The sequences were analyzed using an ABI 3500Dx system (Applied Biosystems). The TP53 sequences were compared to the GenBank database sequence (accession number NC_000017.9) (Cancer Genome Atlas Network 2012). Both forward and reverse strands were sequenced to confirm the full sequence length (bp) and nucleotide alterations (Kim et al. 2013).
18F-FDG PET or PET/CT method
The procedure for 18F-FDG PET or PET/CT was as previously reported (Ahn et al. 2014b , Lee et al. 2016). The SUV was calculated by measuring the 18F-FDG uptake by the primary tumor in the region of interest, as follows: SUV = (maximal radioactivity concentration in the region of interest)/(injected dose/patient’s weight (kg)).
Statistical analysis
Continuous variables were compared using Mann–Whitney U test. Discrete variables were compared using χ 2 or Fisher’s exact tests. The SUV was incorporated into the analyses as a continuous variable. Kolmogorov–Smirnov tests were used to assess the normal distribution of the SUV. To identify predictive factors for low PR expression, the binary logistic regression analysis was performed using all variables. The variables with P value <0.05 were included in the full multivariable model, and the stepwise backward Wald method was used to arrive at the final model. No adjustments were made for multiple statistical testing.
The recurrence-free survival time (RFS) was measured from the date of the first curative surgery to the date of the first tumor recurrence, including loco-regional recurrence, distant metastasis or death. The Kaplan–Meier method was used to estimate the RFS, and the estimated survival curves were compared using the log-rank test. The Cox’s regression-hazard model was used for univariable and multivariable survival analyses. Significant variables in univariable analysis are included in the multivariable model. Variables that were significant at the 0.05 level were then selected to build a predictive multivariable Cox regression survival model. A backwards selection method (P = .05 for inclusion and P = .05 for exclusion, wald method) was used to identify significant prognostic factors for the RFS. In addition, a predictive ability of multivariable was measured using Harrell c-statistics (Harrell et al. 1996) and the concordance index (c-index) was calculated to measure the concordance for time-to-event data, in which increasing values between 0.5 and 1.0 indicated improved prediction. PASW Statistics for Windows, version 18.0 (SPSS, Inc.) and the R software (version R-3.3.3; https://www.r-project.org) were used to perform these analyses. Statistical significance was defined as P values <0.05 or 95% CIs.
Results
Baseline characteristics
Two hundred seventy-two patients with ER-positive, HER2-negative tumors were included in the analyses. Two hundred two (74.3%) and seventy (25.7%) patients had a high and low PR expression, respectively, while 241 (88.6%) and 31 (11.4%) patients had high and low ER expression, respectively. The baseline characteristics were compared according to PR expression (Table 1). The median age of the low PR group was significantly younger than that of the high PR group (P < 0.001). There were no differences in anatomical staging, whereas histologic grade tended to be higher in low-PR tumors (P = 0.064). Also, the proportion of tumors with ER expression differed significantly according to PR expression (P = 0.019), while the proportion of tumors with high Ki67 expression did not differ by PR expression.
Table 1.
Baseline characteristics.
| High PR (N = 202) | Low PR (N = 70) | P-Valuea | |
|---|---|---|---|
| Median age, years (range) | 57 (31–91) | 49 (26–81) | <0.001b |
| T stage | 0.139 | ||
| T1 | 113 (55.9) | 32 (45.7) | |
| T2 | 89 (44.1) | 38 (54.3) | |
| N stage | 0.194c | ||
| N0 | 124 (61.4) | 51 (72.9) | |
| N1 | 66 (32.7) | 14 (20.0) | |
| N2 | 8 (4.0) | 4 (5.7) | |
| N3 | 4 (2.0) | 1 (1.4) | |
| Stage | 0.751 | ||
| I | 79 (39.1) | 24 (34.3) | |
| II | 108 (53.5) | 41 (58.6) | |
| III | 15 (7.4) | 5 (7.1) | |
| Histologic graded | 0.064 | ||
| I | 60 (30.3) | 15 (22.4) | |
| II | 109 (55.1) | 34 (50.7) | |
| III | 29 (14.6) | 18 (26.9) | |
| Estrogen receptore | 0.019 | ||
| High | 184 (91.1) | 57 (81.4) | |
| Low | 18 (8.9) | 13 (18.6) | |
| Ki67 | 0.980 | ||
| ≥20% | 43 (21.3) | 15 (21.4) | |
| <20% | 159 (78.7) | 55 (78.6) |
aChi-square test except; bMann–Whitney U test; cFisher exact test; dmissing values; ehigh, Allred score 5–8; low, Allred score 0–4.
High TP53 mutation rates in low-PR tumors
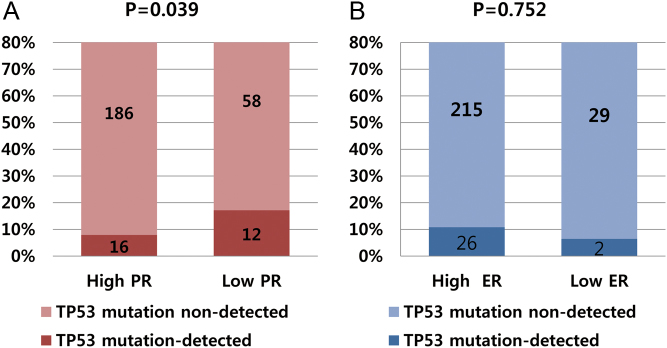
Tumors from 28 patients (10.3%) had a somatic TP53 mutation. The diverse locations of mutations within the TP53 coding sequence are shown in Supplementary data 1 (see section on supplementary data given at the end of this article). Among six ‘hotspot’ residues (R175, G245, R248, R249, R273 and R282) (Cho et al. 1994), our patients had two R248 and two R273 missense mutations. Missense mutations were most common (19 of 28; 67.9%), followed non-sense mutations (4 of 28), splicing mutations (3 of 28) and frame-shift mutations (2 of 28) (Supplementary data 1). The TP53 mutation rate was significantly higher in low-PR tumors than that in high-PR tumors (17.1% vs 7.9%, P = 0.039; Fig. 1A). However, the TP53 mutation rate was not statistically different according to ER expression (6.5% in low-ER-tumors vs 10.8% in high-ER tumors, P = 0.752; Fig. 1B).
Figure 1.
TP53 mutation rates according to PR and ER status. (A) The TP53 mutation rate was significantly higher in low-PR tumors than that in high-PR tumors (17.1% vs 7.9%, P = 0.039). (B) The rates were not statistically different according to ER expression (10.8% in high-ER tumors vs 6.5% in low ER-tumors, P = 0.752).
SUV in relation to PR expression
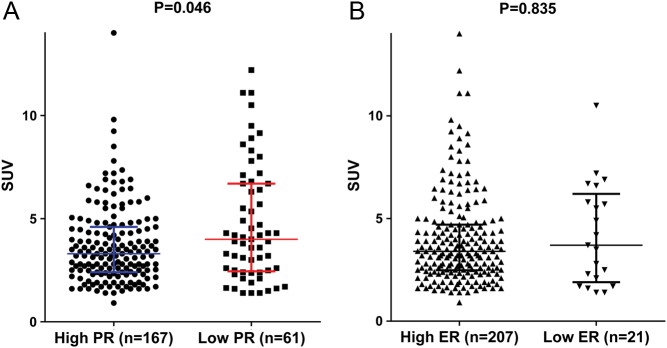
To investigate the pathologic characteristics of tumors in terms of glucose uptake, we measured SUV in patients undergoing preoperative 18f-FDG-PET. We obtained SUVs from 229 patients (84.2%). We compared the SUV distributions according to PR expression because the SUVs were not normally distributed (P < 0.001, Kolmogorov–Smirnov test). The SUV distributions differed significantly according to PR expression (P = 0.046, Mann–Whitney U test), indicating that low-PR tumors had a significantly higher median SUV than that of high-PR tumors (Fig. 2A). However, the median SUVs were not statistically different according to ER expression (P = 0.835, Fig. 2B).
Figure 2.
Distribution of SUVs according to PR and ER status. (A) Low-PR tumors had a significantly higher median SUV than that of high-PR tumors (P = 0.046, Mann–Whitney U test). The median SUVs for low- and high-PR tumors were 4 (1.4–12.2) and 3.3 (0.91–14), respectively. (B) The median SUVs were not statistically different according to ER expression (P = 0.835, Mann–Whitney U test). The median SUVs for low- and high-ER tumors were 3.7 (1.4–10.5) and 3.4 (0.91–14.0), respectively.
SUV is predictive of low PR expression
We identified the factors associated with low-PR expression using binary logistic regression analysis. The variables with P < 0.05 in univariate analysis included age, TP53 mutation, tumor grade and SUV (Table 2). Multivariable analysis revealed that SUV and age remained independent variables associated with low PR expression (Table 2). Continuous SUV had an odds ratio (OR = 1.220; 95% confidence interval (CI) = 1.068–1.394) for predicting low-PR tumors in multivariable analysis.
Table 2.
Binary logistic regression analysis to identify predictive factors for low PR expression.
| Variables | Univariable | Multivariable | ||
|---|---|---|---|---|
| P-Value | OR (95% CI) | P-Value | OR (95% CI) | |
| Age (per 1 year) | 0.001 | 1.044 (1.018–1.071) | 0.003 | 1.043 (1.014–1.073) |
| TP53 mutation | 0.033 | 2.405 (1.076–5.377) | ||
| Grade | ||||
| I | Ref | |||
| II | 0.526 | 1.248 (0.629–2.474) | ||
| III | 0.029 | 2.483 (1.098–5.615) | ||
| SUV (per 1 unit) | 0.003 | 1.218 (1.070–1.387) | 0.003 | 1.220 (1.068–1.394) |
| T stage | ||||
| T1 | Ref | |||
| T2 | 0.141 | 1.508 (0.873–2.603) | ||
| N stage | ||||
| N0 | Ref | |||
| N1 | 0.050 | 0.516 (0.266–1.000) | ||
| N2 | 0.758 | 1.216 (0.350–4.217) | ||
| N3 | 0.660 | 0.608 (0.066–5.571) | ||
| Stage | ||||
| I | Ref | |||
| II | 0.453 | 1.250 (0.699–2.235) | ||
| III | 0.870 | 1.097 (0.361–3.331) | ||
| ER expressiona | ||||
| High | Ref | |||
| Low | 0.021 | 2.351 (1.135–4.868) | ||
| Ki67 | ||||
| ≥20% | Ref | |||
| <20% | 0.98 | 0.992 (0.511–1.924) | ||
aHigh, Allred score 5–8; low, Allred score 0–4.
ER, estrogen receptor; SUV, standardized uptake.
Survival analysis
At a median follow-up of 50 months (3–134), 18 patients experienced tumor recurrence, including 17 distant metastases and one loco-regional recurrence. Among the 18 patients with recurrence, three had both metastases and loco-regional recurrences. During the follow-up period, one patient died from breast cancer, and one patient died from a non-cancerous cause. The adjuvant treatments, presented online, were not statistically different according to PR expression (Supplementary data 2).
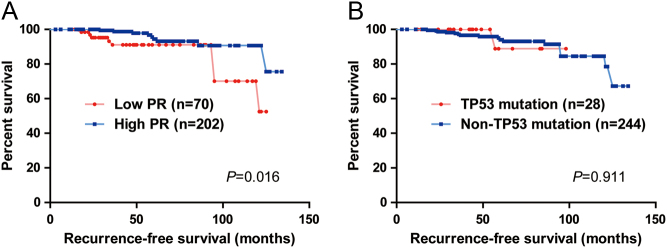
The Kaplan–Meier plots for RFS differed significantly according to PR expression (P = 0.016; Fig. 3A). The RFS was longer in the groups with high-PR tumors; however, a significant difference of the RFS according to the presence of TP53 mutation was not observed (P = 0.911; Fig. 3B).
Figure 3.
Kaplan–Meier plots of recurrence-free survival (RFS). (A) The RFS differed significantly according to PR expression (P = 0.016, log-rank test). (B) However, the RFS did not differ significantly according to the presence of TP53 mutation (P = 0.911, log-rank test).
In the univariable analyses using the Cox regression model, T stage, N stage, stage, SUV and PR expression were significant (Table 3). In the multivariable model, stage and PR expression were significant prognostic factors for the RFS (Table 3). Then, we compared a predictive ability of the multivariable model with or without SUV using the Harrell c-statistics. For the model 1 (without SUV), the Harrell c-index was 0.713. For the model 2 (with SUV), the Harrell c-index was 0.687, indicating that SUV does not add a predictive power to the model 1 that includes stage and PR expression.
Table 3.
A cox-regression hazard model for recurrence-free survival.
| Variables | Univariable | Multivariable | ||||
|---|---|---|---|---|---|---|
| P-Value | HR (95% CI) | Model 1 (backward) | Model 2 (backward + SUV) | |||
| P-Value | HR (95% CI) | P-Value | HR (95% CI) | |||
| Age | 0.740 | 1.008 (0.963–1.054) | ||||
| TTP53 mutation | 0.912 | 0.891 (0.116–6.848) | ||||
| SUV | 0.046 | 1.231 (1.004–1.509) | 0.577 | 1.071 (0.842–1.362) | ||
| Grade | ||||||
| I | 0.482 | Ref | ||||
| II | 0.381 | 1.528 (0.468–4.987) | ||||
| III | 0.046 | 1.970 (0.432–8.985) | ||||
| T stage | ||||||
| T1 | Ref | |||||
| T2 | 0.004 | 5.235 (1.674–16.372) | ||||
| N stage | ||||||
| N0 | Ref | |||||
| N1 | 0.376 | 0.562 (0.157–2.010) | ||||
| N2 | 0.700 | 1.497 (0.192–11.709) | ||||
| N3 | 0.000 | 18.300 (3.873–86.470) | ||||
| Stage | ||||||
| I | Ref | Ref | Ref | |||
| II | 0.080 | 3.190 (0.872–11.661) | 0.079 | 3.208 (0.872–11.795) | 0.245 | 2.237 (0.576–8.684) |
| III | 0.007 | 8.031 (1.791–36.015) | 0.007 | 7.980 (1.776–35.852) | 0.061 | 7.164 (0.911–56.332) |
| ER expression | ||||||
| High | Ref | |||||
| Low | 0.473 | 0.580 (0.131–2.572) | ||||
| Ki67 | ||||||
| ≥20% | Ref | |||||
| <20% | 0.175 | 0.506 (0.189–1.355) | ||||
| PR expression | 0.024 | 0.067 | ||||
| High | Ref | Ref | Ref | |||
| Low | 0.023 | 2.955 (1.165–7.499) | 2.953 (1.152–7.567) | 2.793 (0.932–8.367) | ||
| Harrell c-index | 0.718 | 0.687 | ||||
ER, estrogen receptor; PR, progesterone receptor; SUV, standardized uptake value.
Discussion
In this study, tumors with low PR expression more frequently had TP53 mutations and elevated glucose uptake compared to those in tumors with high PR expression, providing a biological rationale for why low PR expression could be a biomarker to distinguish aggressive tumors from ER-positive/HER2-negative tumors, similar to high Ki67 level or poor histologic grade. Our findings are in line with those of previous studies indicating that the loss of PR expression correlates with aggressive tumor characteristics (Arpino et al. 2005, Olivier et al. 2006) and with a gene signature of the PI3K/Akt/mTOR pathway (Creighton et al. 2009).
An earlier study of 1794 breast cancer patients showed that TP53 mutations were more frequent in PR-negative tumors than those in PR-positive tumors (Olivier et al. 2006) but did not identify HER2 status, remaining unclear whether the TP53 mutation rate is higher in PR-lacking tumors within ER-positive/HER2-negative tumors. We observed that the rate was significantly higher in low-PR tumors within luminal/HER2-negative tumors, suggesting that frequent TP53 pathway defects could characterize ER-positive/PR-low tumors even without HER2 amplification.
A comprehensive genomic study using whole genome analyses showed that TP53 mutations contribute to endocrine failure in luminal breast cancer (Ellis et al. 2012). A bioinformatics analysis integrating gene copy number aberrations, gene expression profiles and mutations indicated that TP53 mutations play an integral role in determining luminal B status and resistance to endocrine therapy (Ellis et al. 2012). Furthermore, ~30% of luminal B tumors harbored TP53 mutant in TCGA data (Cancer Genome Atlas Network 2012), suggesting that a considerable number of luminal B tumors have an impaired TP53 pathway. Our observations of TP53 mutations and low PR expression consolidate a current consensus that low PR expression can be used to identify luminal B-like tumors (Goldhirsch et al. 2013, Coates et al. 2015) and provide a rationale that low-PR tumors have a worse prognosis than high-PR tumors even though they are all ER-positive/HER2-negative tumors.
In addition to TP53 mutations, we also characterized the low-PR tumors using the SUV as an objective numerical value indicative of glucose uptake within tumors. Based on a study elucidating the tumor biology of increasing SUV such as MYC-overexpression (Palaskas et al. 2011) or centromere protein F-expression, breast cancer with high SUV levels could indicate aggressive tumors. Moreover, our previous study showed that high SUV could be a poor prognostic factor in luminal breast cancer, also concordant with another study (Aogi et al. 2015), which similarly evaluated the prognosis of luminal breast cancer according to SUV. In another study of luminal/HER2-negative tumors, we found that SUV is correlated with the 21-gene recurrence score and inversely correlated with PR mRNA expression (Ahn et al. 2017). Our previous finding on the inverse correlation between SUV and PR mRNA expression is concordant with the findings of this study that low-PR tumors had a higher SUV. Several lines of evidence suggest that growth factor signaling is frequently activated in PR-lacking tumors (Cui et al. 2005), supporting our findings that enhanced growth factor pathway expression may contribute to increased glycolysis (Vander Heiden et al. 2009). Further studies on the relationship between the loss of PR and increased glycolytic activity in this subset of breast cancer are warranted.
Our major limitation is the potential argument against the optimal PR expression threshold. Although Prat et al. suggested a cut-off of 20% (Prat et al. 2013) based on the relationship between mRNA levels of the PR gene and IHC examination, it is still hard to clearly distinguish low and high PR tumors using IHC examination alone. Expert panels largely agreed that low PR expression can be utilized as a determinant for luminal B-like tumors (Goldhirsch et al. 2013, Coates et al. 2015), but recommended its combination with other pathologic markers such as Ki67 and grade (Coates et al. 2015). More research is needed to determine the optimal PR expression cut-off.
Another limitation is noted in the survival analyses. In multivariable model 1, the poor prognostic impact of low PR expression was observed but disappeared in multivariable model 2. Also, the negative impact of high SUV on survival suggested by previous studies was not found in model 2. This might be due to the small number of recurrences and missing SUV data. Much longer follow-up is required to identify the influence of PR status and SUV in those patients because luminal/HER2-negative patients generally have a very favorable outcome. In addition, a method of selective hot-spot sequencing could be regarded as a limitation because it does not discover any kinds of genetic change of whole exome. Also, high SUV in low PR-tumors cannot be excluded as a mere reflection of poorly differentiated carcinoma because low-PR tumors tend to have high histologic grade.
Despite these limitations, our study characterized low-PR tumors in terms of altered TP53 pathway and glucose metabolism. It is worth exploring low-PR tumors in viewpoints of these biologic pathways. Moreover, these characteristics of low-PR tumors partly explain why low-PR tumors have a worse outcome among luminal/HER2-negative breast cancer that generally shows a favorable course.
Conclusions
We provide evidence that genetic alteration of the TP53 gene and dysregulated glucose metabolism partly involve low PR expression in ER-positive and HER2-negative breast cancer.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This research was supported by the Basic Science Research Program through the NRF, funded by the Ministry of Science, ICT, & Future Planning (NRF-2015R1C1A1A02037104 and NRF-2016R1D1A1A09917675); a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1520120); and a new faculty research seed money grant from Yonsei University College of Medicine (2016-32-0025).
Availability of data and materials
The datasets generated and analyzed during the current study are available from the corresponding author on request.
Ethics approval and consent to participate
The study protocol was reviewed and approved by the Institutional Review Boards of Gangnam Severance Hospital, Yonsei University, Seoul, Korea, and adhered to the tenets of the Declaration of Helsinki. Owing to the retrospective approach of this study, the need for informed consent was waived by the ethics committees.
References
- Ahn SG, Lee M, Jeon TJ, Han K, Lee HM, Lee SA, Ryu YH, Son EJ, Jeong J. 2014a. [18F]-fluorodeoxyglucose positron emission tomography can contribute to discriminate patients with poor prognosis in hormone receptor-positive breast cancer. PLoS ONE 9 e105905 ( 10.1371/journal.pone.0105905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SG, Park JT, Lee HM, Lee HW, Jeon TJ, Han K, Lee SA, Dong SM, Ryu YH, Son EJ, et al 2014b. Standardized uptake value of (1)(8)F-fluorodeoxyglucose positron emission tomography for prediction of tumor recurrence in breast cancer beyond tumor burden. Breast Cancer Research 16 502 ( 10.1186/s13058-014-0502-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SG, Lee JH, Lee HW, Jeon TJ, Ryu YH, Kim KM, Sohn J, Yun M, Lee SA, Jeong J, et al 2017. Comparison of standardized uptake value of 18F-FDG-PET-CT with 21-gene recurrence score in estrogen receptor-positive, HER2-negative breast cancer. PLoS ONE 12 e0175048 ( 10.1371/journal.pone.0175048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aogi K, Kadoya T, Sugawara Y, Kiyoto S, Shigematsu H, Masumoto N, Okada M. 2015. Utility of (18)F FDG-PET/CT for predicting prognosis of luminal-type breast cancer. Breast Cancer Research and Treatment 150 209–217. ( 10.1007/s10549-015-3303-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpino G, Weiss H, Lee AV, Schiff R, De Placido S, Osborne CK, Elledge RM. 2005. Estrogen receptor-positive, progesterone receptor-negative breast cancer: association with growth factor receptor expression and tamoxifen resistance. Journal of the National Cancer Institute 97 1254–1261. ( 10.1093/jnci/dji249) [DOI] [PubMed] [Google Scholar]
- Bertheau P, Lehmann-Che J, Varna M, Dumay A, Poirot B, Porcher R, Turpin E, Plassa LF, de Roquancourt A, Bourstyn E, et al 2013. p53 in breast cancer subtypes and new insights into response to chemotherapy. Breast 22 (Supplement 2) S27–S29. ( 10.1016/j.breast.2013.07.005) [DOI] [PubMed] [Google Scholar]
- Cancello G, Maisonneuve P, Rotmensz N, Viale G, Mastropasqua MG, Pruneri G, Montagna E, Iorfida M, Mazza M, Balduzzi A, et al 2013. Progesterone receptor loss identifies luminal B breast cancer subgroups at higher risk of relapse. Annals of Oncology 24 661–668. ( 10.1093/annonc/mds430) [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2012. Comprehensive molecular portraits of human breast tumours. Nature 490 61–70. ( 10.1038/nature11412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. 1994. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265 346–355. ( 10.1126/science.8023157) [DOI] [PubMed] [Google Scholar]
- Clark GM, McGuire WL, Hubay CA, Pearson OH, Marshall JS. 1983. Progesterone receptors as a prognostic factor in Stage II breast cancer. New England Journal of Medicine 309 1343–1347. ( 10.1056/NEJM198312013092240) [DOI] [PubMed] [Google Scholar]
- Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Thurlimann B, Senn HJ. 2015. Tailoring therapies – improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Annals of Oncology 26 1533–1546. ( 10.1093/annonc/mdv221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Kent Osborne C, van de Vijver MJ, Foekens JA, Klijn JG, Horlings HM, Nuyten D, Wang Y, Zhang Y, Chamness GC, et al 2009. Molecular profiles of progesterone receptor loss in human breast tumors. Breast Cancer Research and Treatment 114 287–299. ( 10.1007/s10549-008-0017-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. 2005. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. Journal of Clinical Oncology 23 7721–7735. ( 10.1200/jco.2005.09.004) [DOI] [PubMed] [Google Scholar]
- Desmedt C, Voet T, Sotiriou C, Campbell PJ. 2012. Next-generation sequencing in breast cancer: first take home messages. Current Opinion in Oncology 24 597–604. ( 10.1097/CCO.0b013e328359554e) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge SB, Compton CC. 2010. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of Surgical Oncology 17 1471–1474. ( 10.1245/s10434-010-0985-4) [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al 2012. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486 353–360. ( 10.1038/nature11143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Senn HJ. 2013. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Annals of Oncology 24 2206–2223. ( 10.1093/annonc/mdt303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE, Jr, Lee KL, Mark DB. 1996. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in Medicine 15 361–387. () [DOI] [PubMed] [Google Scholar]
- Harvey JM, Clark GM, Osborne CK, Allred DC. 1999. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. Journal of Clinical Oncology 17 1474–1481. ( 10.1200/jco.1999.17.5.1474) [DOI] [PubMed] [Google Scholar]
- Horwitz KB, McGuire WL. 1975. Predicting response to endocrine therapy in human breast cancer: a hypothesis. Science 189 726–727. ( 10.1126/science.168640) [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim J, Lee HD, Jeong J, Lee W, Lee KA. 2013. Spectrum of EGFR gene copy number changes and KRAS gene mutation status in Korean triple negative breast cancer patients. PLoS ONE 8 e79014 ( 10.1371/journal.pone.0079014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HW, Lee HM, Hwang SH, Ahn SG, Lee KA, Jeong J. 2014. Patterns and biologic features of p53 mutation types in korean breast cancer patients. Journal of Breast Cancer 17 1–7. ( 10.4048/jbc.2014.17.1.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Lee HM, Choi SE, Yoo H, Ahn SG, Lee MK, Jeong J, Jung WH. 2016. The prognostic impact of early change in 18F-FDG PET SUV after neoadjuvant chemotherapy in patients with locally advanced breast cancer. Journal of Nuclear Medicine 57 1183–1188. ( 10.2967/jnumed.115.166322) [DOI] [PubMed] [Google Scholar]
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, et al 2006. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clinical Cancer Research 12 1157–1167. ( 10.1158/1078-0432.Ccr-05-1029) [DOI] [PubMed] [Google Scholar]
- Palaskas N, Larson SM, Schultz N, Komisopoulou E, Wong J, Rohle D, Campos C, Yannuzzi N, Osborne JR, Linkov I, et al 2011. 18F-fluorodeoxy-glucose positron emission tomography marks MYC-overexpressing human basal-like breast cancers. Cancer Research 71 5164–5174. ( 10.1158/0008-5472.can-10-4633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Cheang MC, Martin M, Parker JS, Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen TO, et al 2013. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. Journal of Clinical Oncology 31 203–209. ( 10.1200/jco.2012.43.4134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324 1029–1033. ( 10.1126/science.1160809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, et al 2013. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Journal of Clinical Oncology 31 3997–4013. ( 10.1200/jco.2013.50.9984) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on request.



 This work is licensed under a
This work is licensed under a