Abstract
Background:
There is limited knowledge of the genetic alterations in acral melanoma metastases at different anatomic sites. Here, we characterized the genetic abnormalities of metastases in a 51-year-old man with stage IIIC heel melanoma who developed concomitant brain and cutaneous metastases in spite of multiple treatment modalities.
Methods:
Melanoma cells were isolated following palliative resection of the patient’s cortical tumor and biopsy of cutaneous thigh metastasis. Mutational analysis using polymerase chain reaction amplification and BLAST, as well as exome sequencing (160Mb coverage) was performed on the tumors, cell lines generated thereof and normal lymph nodes.
Results:
All specimens had neuroblastoma RAS viral oncogene homolog Q61K mutations. There was a 40-fold higher somatic mutation frequency in the brain metastasis compared to the cutaneous metastasis. The former showed truncations of DNA mismatch repair genes (MLH1 and MSH2), and non-canonical BRAF (v-raf murine sarcoma viral oncogene homolog B1), PIK3CA and NF-1 mutations not observed in the extracranial lesion. Genomic profiling of each cell line was concordant with the respective original tumor tissue.
Conclusions:
We present the mutational differences between brain and cutaneous acral melanoma metastases in a patient with concomitant lesions. Further genetic and functional studies are needed to understand the biology of metastatic disease appearing at different sites.
Keywords: acral melanoma, brain metastases, cutaneous metastases, mismatch repair genes, NRAS
Melanoma is one of the most aggressive malignancies and has one of the highest mutation rates of any cancer type.1,2 Significant strides have been made in melanoma therapeutics that are resonating throughout oncology,3,4 and the field’s knowledge of melanoma pathogenesis continues to advance with genomic and functional studies.5 However, the mechanism of metastasis and what accounts for the predilection for certain organ systems such as the central nervous system (CNS) is still poorly understood.
About 10% of patients with melanoma will develop brain metastases,6 thus accounting for 5–20% of all cancer patients with intracranial metastatic disease7 – a disproportionately high number given melanoma’s prevalence relative to other cancer types.8,9 Prognosis is particularly poor with median overall survival as low as 2.3 months in individuals with poor performance status.10,11 Historically, many patients with brain metastases were excluded from clinical trials owing to limited survival. Current treatment options include palliative surgical resection, stereotactic radiosurgery and whole-brain radiotherapy; these approaches have had limited success, even when combined with other modalities.9 Recently, disparate mutational patterns in brain metastases vs. primary tumors have been shown in other cancer types.12 However, there has been limited exploration of this phenomenon in melanoma.
Acral melanomas are a distinct subtype of melanoma. The majority occurs on the soles, palms or nonvolar sites, but can also be subungal.13 They are the most common type of malignant melanoma in Asians and higher phototype individuals yet account for <5% of all melanomas. The genetic underpinnings of acral melanomas are thought to be distinct from other melanoma subtypes and notably not driven by ultraviolet (UV)-induced mutations.14,15 The critical genes in acral melanomas are still being delineated;15–17 for example, KIT amplifications are more common whereas v-raf murine sarcoma viral oncogene homolog B1 (BRAF) or NRAS mutations are present but less common in comparison to superficial spreading or nodular melanomas.14,17,18
Here, we report the clinical presentation of a patient with metastatic acral melanoma and compare the genomic landscape of the intracranial vs. extracranial metastases. The intersection of these less studied phenotypes – acral melanoma and brain metastasis – presents a unique opportunity to further our understanding of melanoma pathogenesis.
Materials and methods
Human melanoma tumor samples
Tissue samples were procured from surgical specimens and full clinical data were obtained with approval from the Institutional Review Board. The operating surgeon allocated tissue samples at the end of the operation from regions most likely to harbor viable tumor without interfering with diagnosis or clinical staging.
Cell line isolation
Solid tumor samples were minced mechanically in culture media [Dulbecco’s Modified Eagle Medium (Hyclone, Thermo Scientific, Waltham, MA, USA), 10% fetal bovine serum (Gemini Bioproducts, West Sacramento, CA, USA) and penicillin/streptomycin (Gibco, Life Technologies, Grand Island, NY, USA)] on 10-cm culture plates (Corning Inc., Corning, NY, USA) and left overnight in standard culture conditions (37°C, humidified atmosphere, 5% CO2). The liquid media in which the procured tissue was originally placed was spun down (1500rpm) to isolate the detached cells in solution, and the pelleted cells were resuspended in fresh culture media and propagated in culture flasks (Corning Inc.). The minced tumor samples were removed from the 10-cm culture dishes on Day 2 and mechanically forced through 100μM nylon mesh filters (Fisher Scientific, Pittsburgh, PA, USA). The cells and tissue clumps were centrifuged, resuspended in fresh culture media and propagated in culture flasks. Cells were propagated by changing culture media every 3–4days and passaging cells in 1:3 to 1:6 ratio using 0.05% trypsin (Hyclone, Thermo Scientific) when the plates became 50–80% confluent.
BRAF/NRAS mutational analysis
Genomic DNA was isolated from the clonal cell lines (Wizard genomic DNA purification kit, Promega, Madison, WI). Mutational analysis for BRAF and NRAS mutations was performed using polymerase chain reaction (PCR) amplification of exon 15 for BRAF and exons 2 and 3 for NRAS. Braf exon 15 primer sequences were 5′-AAG CAT CTC ACC TCA TAA CAC AT-3′ and 5′-CCT TCT AGT AAC TCA GCA GCA TCT CA-3′. Nras exon 2 primer sequences were 5′-CTA CTC CAG AAG TGT GAG GCC GA-3′ and 5′-TGA ACT CAA CAC TGA GTT TGC A-3′. Nras exon 3 primer sequences were 5′-TGG AGG GAC AAA CCA GAT AGG CAG-3′ and 5′-ACA ACC TAA AAC CAA CTC TTC CCA-3′. DFCI sequencing core performed the sequence analysis of the PCR-amplified fragments and the results were compared using BLAST.
Exome Plus library construction and sequencing
Exome Plus capture was performed using the Exome Plus bait (Agilent Technologies, Santa Clara, CA, USA) expanded with a set of baits designed by the Broad Institute Genomics Platform in order to cover ∼160Mb of human target sequence. The in-solution hybrid selection process established at the Broad Institute was performed. In brief, genomic DNA was sheared, end repaired, ligated with barcoded Illumina sequencing adapters (Illumina, San Diego, CA, USA), amplified, size selected and subjected to in-solution hybrid capture using the Exome Plus bait set.19,20 Resulting Exome Plus Illumina sequencing libraries were then PCR quantified, pooled and sequenced with 76 base paired-end reads using HiSeq 2000 or 2500 sequencers (Illumina). Output from Illumina software was processed by the Broad Institute Picard data-processing pipeline to yield BAM files containing aligned reads to the reference genome hg19. Cross-contamination between samples from other individuals was monitored with the ContEst algorithm.21 Somatic single nucleotide variants in targeted exons were identified with the MuTect algorithm,22 and small insertions or deletions with Indelocator (http://www.broadinstitute.org/cancer/cga/indelocator). Alterations were annotated using Oncotator (http://www.broadinstitute.org/cancer/cga/oncotator) and manually reviewed in the Integrated Genomics Viewer (IGV).23 Copy number aberrations were quantified using ReCapSeg (http://gatkforums.broadinstitute.org/discussion/5640/recapseg-overview) and reported for each gene as the segmented normalized log2-transformed copy ratio between each tumor sample and a panel of normal samples.
Depth of sequencing was assessed by determining mean target coverage (the average number of reads at each base pair throughout the entire target DNA sequence), and the percentage of bases that have coverage of at least 20 reads (20×) All sequenced samples were included in the downstream analyses.
Results
Clinical history
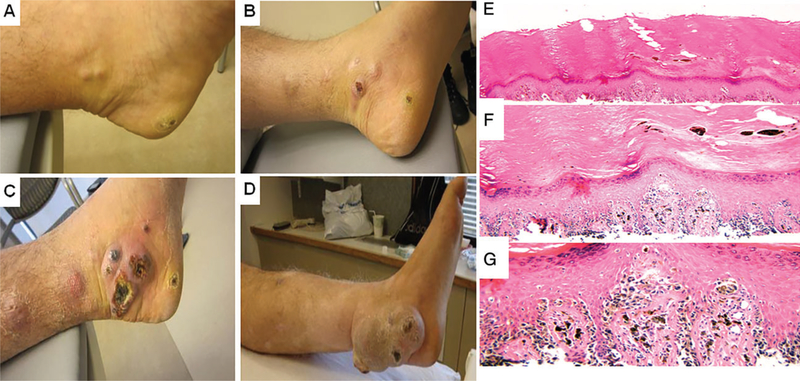
A 51-year-old phototype III man with a history of extensive sun exposure in his youth presented to his primary care physician with a rapidly enlarging 4-cm left inguinal mass (see Fig. 1 for timeline of clinical events). Examination revealed a dark brown pigmented lesion on the left plantar heel, as well, which the patient reported had first appeared 3–4years prior. A superficial shave biopsy of the heel lesion showed an acral melanoma that was at least in situ (Fig. 2A, E–G). Surgical exploration of the left groin revealed a necrotic, enlarged lymph node which was excised. Immunohistochemical analysis revealed positive MART-1 and HMB-45 staining, and focal cyclin D1 positivity (data not shown). DNA testing of the thigh metastasis showed an NRAS Q61K mutation and BRAF wild type at codon 600 (Table 1).
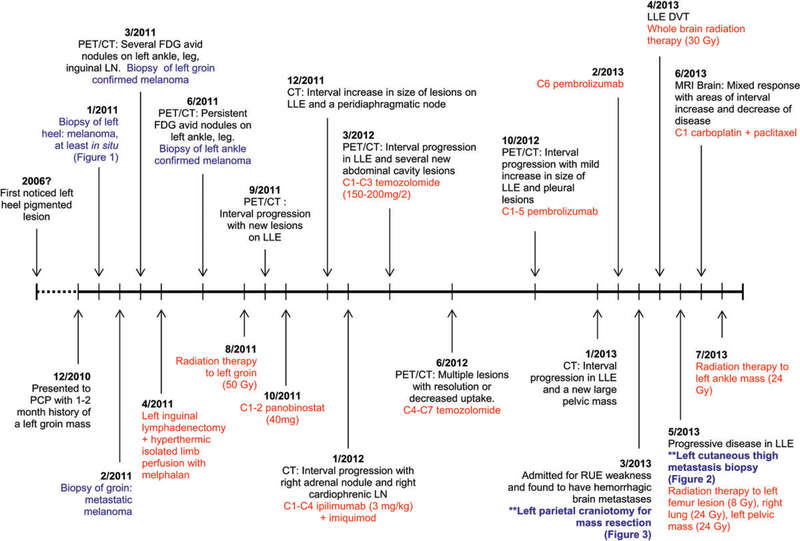
Fig. 1.
Timeline of clinical course, pathological specimen (highlighted in blue) and treatments (highlighted in red). PCP, primary care physician; PET/CT, positron emission tomography/computed tomography; FDG, fluorodeoxyglucose; LN, lymph node; Gy, gray; LLE, left lower extremity; C, cycle; RUE, right upper extremity.
Fig. 2.
Primary acral melanoma. A–D) Clinical photos of the lesion. A) s/p biopsy of heel primary melanoma (March 2011), (B) positron emission tomography shows interval increase in lesions in left lower extremity (LLE) (December 2011), (C) interval progression on LLE while on ipilimumab (March 2012) and (D) persistent lesion s/p radiation therapy (July 2013). E–G) Histology of the lesion. E) Lower power view of superficial shave biopsy of acral melanoma, (F) medium power view shows atypical melanocytes confluent along the basal layer and (G) high power view exhibits pagetoid spread, and the severe atypia of the melanocytes. There is minimal dermis in the specimen; however, no definitive invasion is seen.
Table 1.
Genetic mutations
| Cutaneous thigh metastasis tumor | Cutaneous thigh metastasis cell line | Brain metastasis tumor | Brain metastasis cell line | |
|---|---|---|---|---|
| Coverage | 48.02× | 67.72× | 60.36× | 46.53× |
| Mutation rate (mutations/Mb) | 3.36 | 5.21 | 186.91 | 215.36 |
| BRAF | p.A736T | p.A736T | ||
| NRAS | p.Q61K | p.Q61K | p.Q61K | p.Q61K |
| PIK3CA | p.E35K | p.E35K | ||
| TP53 | ||||
| ARID2 | ||||
| MLH1 | p.A589V | |||
| MLH3 | p.G1250D | p.G1250D | ||
| MSH2 | p.Q413* | p.Q413* | ||
| MSH3 | p.E1081K | p.E1081K | ||
| NF1 | p.R1870_splice | p.R1870_splice |
BRAF, v-raf murine sarcoma viral oncogene homolog B1; NRAS, neuroblastoma RAS viral oncogene homolog
premature stop codon.
Two months after his initial presentation, the patient was found to have subcutaneous nodules on his left foot and ankle. Whole-body positron emission tomography/computed tomography (CT) confirmed these as fluorodeoxyglucose (FDG) avid sites as well as identifying FDG-avid lymph nodes in the left femoral basin. There was no evidence of intracranial metastases on magnetic resonance imaging (MRI) at that time. The patient underwent left inguinal lymphadenectomy and left leg hyperthermic isolated limb perfusion with melphalan. Pathology revealed 4 of the 13 positive lymph nodes with extracapsular extension into adjacent soft tissue and lymphatics consistent with stage IIIC (N3M0) disease. Given the high risk for locoregional recurrence, he underwent 4 weeks of adjuvant radiation therapy (total 5000Gy) 4 months after lymphadenectomy. The following month he had progression of existing in-transit lesions as well as new biopsy-proven melanoma more proximally on the thigh. Six months after lymphadenectomy, he was started on a clinical trial of panobinostat (LBH-589), a histone deacetylase (HDAC) inhibitor with microphthalmia-associated transcription factor inhibitory activity for two cycles, and then switched to topical imiquimod therapy and ipilimumab for a total of four cycles for progression of left lower extremity disease (Figs. 2B and3) and new right cardiophrenic angle peri-diaphragmatic lymphadenopathy.
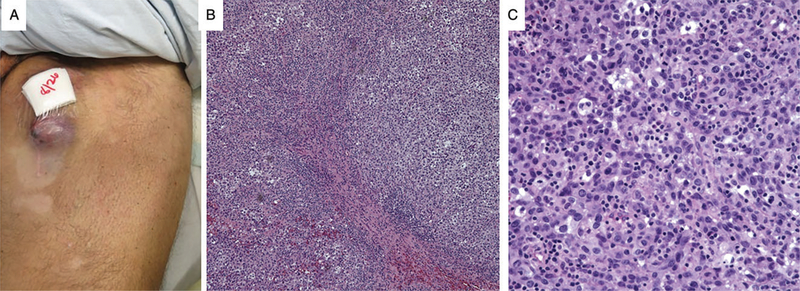
Fig. 3.
Left cutaneous thigh metastasis. A) Gross photograph of left cutaneous thigh metastasis, (B) low and C) high power view of the H&E from the cutaneous thigh metastasis shows sheets of atypical melanocytes with nuclear pleomorphism and several mitoses.
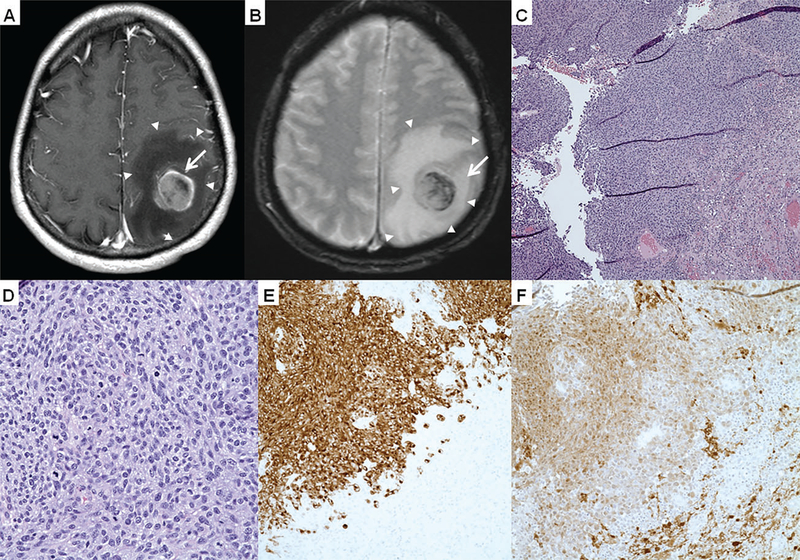
In the ensuing 15 months, he developed widespread metastatic lesions of the right adrenal gland, kidney, anterior abdominal wall and cardiophrenic angle, internal mammary, external iliac, bilateral inguinal lymph nodes and in the left lower extremity (Fig. 2C) which progressed in spite of temozolomide followed by the programmed cell death 1 (PD-1) inhibitor, pembrolizumab. After cycle 6 of pembrolizumab (27 months after initial presentation), the patient was admitted to the hospital with right upper extremity paresis. CT scan revealed a new 30-mm enhancing hemorrhagic lesion in the left motor cortex and a second 7-mm lesion in the left cerebellar hemisphere also seen on MRI (Fig. 4A,B) He underwent left parietal craniotomy and palliative resection of brain lesions with pathology confirming metastatic melanoma (Fig. 4C,D) with positive Mart-1(Fig. 4E), S100 (Fig. 4F) and HMB-45 immunohistochemistry. Cells were isolated and cultured from excess tumor. One month later, he began whole-brain radiotherapy for 10 fractions in the setting of new intracranial lesions. During this time, he had progression of his left lower extremity and pelvic lesions (Fig. 2D) for which he received additional radiotherapy. As part of the treatment protocol, an incisional biopsy was performed of a left thigh in-transit lesion prior to palliative radiotherapy. As was performed for the intracranial metastasis, melanoma cells were isolated and cell lines established from excess thigh tumor tissue. Following one cycle of carboplatin AUC2 plus paclitaxel, he experienced progressive clinical deterioration with worsening pulmonary metastases and recurrent malignant pleural effusion requiring placement of an indwelling pleural catheter, as well as a lower extremity venous thromboembolism requiring inferior vena cava filter.
Fig. 4.
Intracranial metastasis. Magnetic resonance imaging in (A) axial T1 and (B) T2-weighted imaging showed an enhancing 3-cm lesion (arrows) with areas of hemorrhage. The mass involved the motor cortex with marked, surrounding vasogenic edema (arrowheads) and local mass effect manifested as sulcal effacement. The histopathology shown on H&E with a (C) low power image, (D) high power image and positive immmunostains (E) Mart-1 and (F) S100 exhibits diffuse melanoma.
Approximately 32 months after initial presentation, the patient was admitted to the neurologic intensive care unit after presenting with weakness, headache, nausea, vomiting, somnolence, waxing and waning level of consciousness and intermittent right leg numbness. Head CT revealed hemorrhage of the brain metastases. After discussion with the patient and his family, the decision was made to focus on comfort measures. He was discharged to home hospice and expired.
Genomic analysis
The mutation rate displayed by the cutaneous metastasis and corresponding cell line (Table 1) was within the range found previously in other cutaneous melanoma samples.1,2 In contrast, the brain metastasis and its associated cell line displayed a much higher mutation rate (Table 1), with over 12,000 mutations called. Given this high mutation rate, analysis for mutations in DNA repair genes was undertaken. Notably, an early stop codon mutation was identified in the DNA mismatch repair gene MHS2 in these samples (Table 1) suggesting reduced MSH2 activity in these cells. In addition, missense mutations were identified in MLH1, MLH3 and MSH3 genes in both the brain tumor and cell line samples. In contrast, no mismatch repair gene mutations were identified in the thigh tumor sample or cell line (Table 1). Analysis of copy number data did not show clear copy number loss affecting any of the genes in question.
Analysis of the mutation signatures revealed no UV or microsatellite instability (MSI) signatures in these samples. Interestingly, however, a mutational pattern similar to that caused by alkylating agents was found in both the brain metastasis and its corresponding cell line24 (signature 11).
A summary of alterations found in key melanoma oncogenes and tumor suppressor genes is shown in Table 1. All cells regardless of site of metastasis displayed a canonical activating NRAS Q61K mutation. In addition to the NRAS mutation, the metastatic brain tumor and cell line samples from this patient also carried BRAF A736T and PIK3CA E35K mutations which were not seen in the thigh metastasis. The brain metastasis also harbored a mutation within a NF1 splice site.
Discussion
Here, we report a 51-year-old man who initially presented with a stage IIIC lower extremity NRAS mutant acral melanoma and developed progressive disease marked by in-transit, ipsilateral inguinal lymph node, pelvic, intra-abdominal, thoracic and intracranial metastases despite surgical debulking, radiotherapy, HDAC inhibitor, cytotoxic T lymphocyte antigen-4 inhibitor, alkylating agent, PD-1 inhibitor and platinum-based chemotherapy. We present genomic analysis of his tumors from the intracranial and thigh metastases and additionally contribute genomically annotated cell lines generated from these specimens.
Melanoma displays a disproportionate propensity to metastasize in the brain despite the brain often being considered as a protected site. Recent work primarily in lung, renal cell and breast carcinoma suggests that brain metastases develop oncogenic alterations which diverge from the primary tumor, and may account for treatment failures.12 In comparing this patient’s cutaneous and brain metastases with exome sequencing, we identified several differences. In particular, we identified nonsense mutations for MSH2. These alterations, which were only found in the brain metastasis, support the possibility that it is this impairment in DNA repair mechanism that led to further genomic instability and thereby allowed for additional mutations that contribute to metastasis to the CNS. Mutations have previously been reported in MLH1 and MSH2 in roughly one-third of melanoma brain metastases,25 and have been thought to cause genomic instability in melanoma.26 Other mutations leading to genomic instability have been reported, if not via MLH1 or MSH2 mutations. In comparing primary acral melanomas vs. metastases from any site, ERCC5 nonsense mutations have been speculated to contribute to the highest mutational burden,15 although no significant abnormality in ERCC5 was noted in our tumor specimens. In addition, although we found no UV or MSI signatures, we did find a mutational pattern consistent with alkylating agents in only the brain metastasis and its cell line;24 this corresponded with recent treatment with temozolomide prior to development of neurologic symptoms and discovery of intracranial spread of disease.
It is also interesting that the cutaneous thigh metastasis of an acral melanoma showed a similar number of mutations as in prior reports of cutaneous melanomas.1,16 In a study by the Marais’ group, the mutational burden of five acral melanomas were evaluated and compared to uveal, mucosal and cutaneous melanomas;15 acral melanomas were found to have a much lower mutational burden on average.
The cancer genome atlas network working group recently proposed four major categories of melanomas: BRAF mutant, NRAS mutant, NF1 mutant and triple wild type.27 In this context, our patient appears to fall into at least two categories. Both metastatic sites harbored Q61K NRAS mutations, thus a likely alteration to have been present in the primary melanoma. Approximately 15–17% of acral melanomas bear an NRAS mutation.28,29 The brain metastasis alone had additional unique PIK3CA and NF-1 mutations. The functional consequence of these non-canonical mutations is unknown; however, these could represent genetic redundancies. NF1 is a negative regulator of RAS signaling, and it is known to be affected negatively by splice-site mutations through the disruption of its mRNA sequence.30 Although the consequence of this mutation has not been tested, it is possible that NF1 function was reduced in this tumor, resulting in more robust constitutively activated, mutant NRAS signaling. In the setting of a hyper-mutated phenotype, it is perhaps not surprising to see redundancy to ensure MAP kinase pathway activation, particularly post-treatment. Moreover, NRAS and PI3K mutations may lead to redundancy in activation of the PI3K/AKT pathway. This may be particularly relevant in patients with brain metastases as the PI3K/AKT pathway has been found to be activated in particular.31 Finally, exome sequencing revealed non-canonical A736T BRAF mutation in the brain metastasis which was not observed in the thigh metastasis. Canonical activating BRAF and NRAS mutations have been considered as nearly mutually exclusive events.32 However, in the setting of the high mutation rate this generality may not apply. While the functional consequence of this mutation is unknown, it is interesting that the mutation is not in the kinase domain33 and thus could represent a mutation to alter BRAF regulatory signals.
The findings in this study must be interpreted within the context of the study design of a single patient and with a lower depth of coverage. The lower coverage was potentially the result of1 formalin fixation of tumor and normal samples prior to DNA extraction, and2 the number of sequencing rounds. Despite the low coverage, basic quality control metrics were met and analyses were performed taking this into account. While no additional systemic therapy was administered between collection of the brain and cutaneous samples, they were obtained after numerous treatment modalities which undoubtedly affected the mutational landscape.
Overall, the molecular characteristics of brain metastases remain poorly understood, and currently do not guide treatment options. This case identifies mutational differences between cutaneous and brain metastases in one patient with acral melanoma and highlights that treating brain metastases based on the genomic profile of cutaneous metastases may result in a suboptimal response of intracranial lesions. We also provide to the field genomically annotated cell lines that resemble their tumor counterparts for further functional study.
References
- 1.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell 2012; 150: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long GV, Stroyakovskiy D, Gogas H, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 2014; 371: 1877. [DOI] [PubMed] [Google Scholar]
- 4.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 2015; 373: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger MF, Levin JZ, Vijayendran K, et al. Integrative analysis of the melanoma transcriptome. Genome Res 2010; 20: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zakrzewski J, Geraghty LN, Rose AE, et al. Clinical variables and primary tumor characteristics predictive of the development of melanoma brain metastases and post-brain metastases survival. Cancer 2011; 117: 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schouten LJ, Rutten J, Huveneers HA, Twijnstra A. Incidence of brain metastases in a cohort of patients with carcinoma of the breast, colon, kidney, and lung and melanoma. Cancer 2002; 94: 2698. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JD, Young B. Demographics of brain metastasis. Neurosurg Clin N Am 1996; 7: 337. [PubMed] [Google Scholar]
- 9.Neagu MR, Gill CM, Batchelor TT, Brastianos PK. Genomic profiling of brain metastases: current knowledge and new frontiers. Chin Clin Oncol 2015; 4: 22. [DOI] [PubMed] [Google Scholar]
- 10.Gaspar L, Scott C, Rotman M, et al. Recursive partitioning analysis (RPA) of prognostic factors in three Radiation Therapy Oncology Group (RTOG) brain metastases trials. Int J Radiat Oncol Biol Phys 1997; 37: 745. [DOI] [PubMed] [Google Scholar]
- 11.Sperduto PW, Berkey B, Gaspar LE, et al. A new prognostic index and comparison to three other indices for patients with brain metastases: an analysis of 1,960 patients in the RTOG database. Int J Radiat Oncol Biol Phys 2008; 70: 510. [DOI] [PubMed] [Google Scholar]
- 12.Brastianos PK, Carter SL, Santagata S, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov 2015; 5: 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phan A, Touzet S, Dalle S, Ronger-Savlé S, Balme B, Thomas L. Acral lentiginous melanoma: a clinicoprognostic study of 126 cases. Br J Dermatol 2006; 155: 561. [DOI] [PubMed] [Google Scholar]
- 14.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005; 353: 2135. [DOI] [PubMed] [Google Scholar]
- 15.Furney SJ, Turajlic S, Stamp G, et al. The mutational burden of acral melanoma revealed by whole-genome sequencing and comparative analysis. Pigment Cell Melanoma Res 2014; 27: 835. [DOI] [PubMed] [Google Scholar]
- 16.Turajlic S, Furney SJ, Lambros MB, et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res 2012; 22: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furney SJ, Turajlic S, Fenwick K, et al. Genomic characterisation of acral melanoma cell lines. Pigment Cell Melanoma Res 2012; 25: 488. [DOI] [PubMed] [Google Scholar]
- 18.Lin WM, Baker AC, Beroukhim R, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res 2008; 68: 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher S, Barry A, Abreu J, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 2011; 12: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 2009; 27: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 2011; 27: 2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korabiowska M, König F, Verheggen R, et al. Altered expression and new mutations in DNA mismatch repair genes MLH1 and MSH2 in melanoma brain metastases. Anticancer Res 2004; 24: 981. [PubMed] [Google Scholar]
- 26.Garcia JJ, Kramer MJ, O’Donnell RJ, Horvai AE. Mismatch repair protein expression and microsatellite instability: a comparison of clear cell sarcoma of soft parts and metastatic melanoma. Mod Pathol 2006; 19: 950. [DOI] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015; 161: 1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zebary A, Omholt K, Vassilaki I, et al. KIT, NRAS, BRAF and PTEN mutations in a sample of Swedish patients with acral lentiginous melanoma. J Dermatol Sci 2013; 72: 284. [DOI] [PubMed] [Google Scholar]
- 29.Puig-Butillé JA, Badenas C, Ogbah Z, et al. Genetic alterations in RAS-regulated pathway in acral lentiginous melanoma. Exp Dermatol 2013; 22: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pros E, Gómez C, Martín T, Fábregas P, Serra E, Lázaro C. Nature and mRNA effect of 282 different NF1 point mutations: focus on splicing alterations. Hum Mutat 2008; 29: E173. [DOI] [PubMed] [Google Scholar]
- 31.Chen G, Chakravarti N, Aardalen K, et al. Molecular profiling of patient-matched brain and extracranial melanoma metastases implicates the PI3K pathway as a therapeutic target. Clin Cancer Res 2014; 20: 5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol 2004; 122: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell 2004; 6: 313. [DOI] [PubMed] [Google Scholar]