Abstract
Schizophrenia is a neurodevelopmental disorder with the typical age of onset of overt symptoms and deficits occurring during late adolescence or early adulthood, coinciding with the final maturation of the cortical network involving the prefrontal cortex. These observations have led to the hypothesis that disturbances of the developmental events that take place in the prefrontal cortex during this period, specifically the remodeling of synaptic connectivities between pyramidal neurons, may contribute to the onset of illness. In this context, we investigated the gene expression changes of pyramidal neurons in the human prefrontal cortex during normal periadolescent development in order to gain insight into the possible molecular mechanisms involved in synaptic remodeling of pyramidal neuronal circuitry. Our data suggest that genes associated with the ubiquitination system, which has been implicated in the biology of synaptic plasticity, may play a major role. Among these genes, UBE3B, which encodes the ubiquitin ligase E3, was found to undergo periadolescent increase and was validated at the protein level to be upregulated during periadolescent development. Furthermore, we found that the density of UBE3B-immunoreactive pyramidal neurons was decreased in schizophrenia subjects, consistent with the result of a previous study of decreased UBE3B mRNA expression in pyramidal neurons in this illness. Altogether these findings point to the novel hypothesis that this specific ligase may play a role in the developmental pathogenesis of schizophrenia onset by possibly altering the synaptic remodeling process.
Keywords: Ubiquitin ligase, synaptic pruning, adolescence, prefrontal cortex, schizophrenia
Introduction
The human prefrontal cortex mediates the highest order of cognitive functions. It undergoes a protracted course of development through childhood and adolescence before achieving final maturation during early adulthood. This maturational process is in large part defined by the extensive pruning of synaptic connectivities between pyramidal neurons (Goldman-Rakic et al., 1997; Huttenlocher, 2002). Pyramidal neurons in the cerebral cortex exhibit layer-specific connectional patterns, providing neural circuit structures that support distinct aspects of higher cortical functions. For instance, dendritic spines on pyramidal neurons in layer 3 are targeted by both local and long-range glutamatergic projections in a highly reciprocal fashion (Levitt et al., 1993; Pucak et al., 1996). Synchronized activities of pyramidal neuronal networks through these connections, especially in the gamma frequency band (i.e. 30-100 Hz), are critical for the integrity of higher cortical functions (Buzsaki and Draguhn, 2004; Uhlhaas et al., 2008). During adolescence, prefrontal cortical circuitry becomes increasingly adept in engaging in gamma band oscillation, which coincides with the maturation of prefrontal cortex-orchestrated functions such as working memory and executive function (Fuster, 2008; Uhlhaas et al., 2009; Uhlhaas and Singer, 2011).
Pyramidal neurons in the prefrontal cortex, especially those in layer 3, have long been implicated in the pathophysiology of schizophrenia (Glantz and Lewis, 2000; Glausier and Lewis, 2013; Sweet et al., 2009; Sweet et al., 2003). Because the onset of schizophrenia typically occurs during late adolescence and early adulthood, understanding the molecular mechanisms that regulate the maturation of these pyramidal neurons during this period would provide insight into the pathophysiological mechanisms of disease onset. Hence, in this study, we examined the changes in the gene expression profile of layer 3 pyramidal neurons of the human prefrontal cortex during normal adolescent development. We identified gene networks and pathways that were developmentally differentially regulated, many of which are associated with cytoskeleton integrity, regulation of translation initiation, elongation and termination, ubiquitination and proteolysis. One of the most highly differentially regulated genes during adolescent development that has also previously been found to be differentially expressed in the cerebral cortex in subjects with schizophrenia in one of our earlier studies is the ubiquitin ligase-encoding gene UBE3B (Pietersen et al., 2014a). Furthermore, we found that, at the protein level, the number of cells that expressed UBE3B gradually increased during periadolescent development. In addition, the density of cells, including pyramidal cells, that expressed UBE3B was found to be decreased in schizophrenia subjects. Altogether, these findings converge upon the novel hypothesis that aberrant ubiquitination as a result of decreased UBE3B expression may contribute to the developmental pathophysiology of schizophrenia onset.
Materials and Methods
Postmortem human brain tissue
Fresh-frozen tissue blocks containing Brodmann's area 9 of the prefrontal cortex were obtained from the National Institute of Child and Human Development (NICHD) Brain and Tissue Bank (Table 1). In addition, tissue blocks containing the same prefrontal region from 15 schizophrenia and 15 healthy control subjects, matched for age, sex and postmortem interval (PMI), were obtained from the Harvard Brain Tissue Resource Center (Supplementary Table S1). Postmortem human brain collection procedures have been approved by the Partners Human Research Committee. Written informed consent for use of each of the brains for research has been obtained by the respective legal next-of-kin. The diagnosis of schizophrenia was made by two Board-certified psychiatrists by reviewing medical records and an extensive family questionnaire that included medical, psychiatric, family and social histories. These brains were also examined by a Board-certified neuropathologist to rule out any neurological conditions. None of the subjects had any history of active substance use disorders at the time of death, as corroborated by toxicology results.
Table 1. Subjects and demographic information.
| Group | Case ID | Age | Sex | PMI | RNA purity 260/280 | RIN | Amount of RNA after amplification (μg) | Percent present |
|---|---|---|---|---|---|---|---|---|
| Pre- and Early Adolescent | PFC002 | 2 | M | 16 | 2.66 | 7.2 | 36.21 | 38 |
| PFC003 | 4 | F | 15 | 2.39 | 7.8 | 35.63 | 25 | |
| PFC004 | 8 | M | 16 | 2.43 | 6.8 | 29.79 | 28 | |
| PFC005 | 12 | M | 15 | 2.4 | 7.0 | 32.02 | 26 | |
| PFC006 | 12 | M | 15 | 2.51 | 6.2 | 41.88 | 31 | |
| PFC007 | 13 | M | 11 | 2.74 | 6.0 | 27.60 | 25 | |
| PFC008 | 15 | M | 9 | 2.54 | 5.9 | 19.49 | 22 | |
| Late Adolescent | PFC009 | 16 | M | 16 | 2.35 | 7.1 | 43.56 | 32 |
| PFC010 | 16 | F | 13 | 2.38 | 5.8 | 19.20 | 32 | |
| PFC011 | 17 | M | 15 | 2.47 | 7.9 | 19.19 | 33 | |
| PFC012 | 18 | M | 17 | 2.32 | 6.5 | 28.14 | 34 | |
| PFC013 | 20 | M | 5 | 2.31 | 6.9 | 24.26 | 34 | |
| PFC014 | 22 | M | 13 | 2.55 | 5.8 | 41.40 | 29 |
Laser capture microdissection
A detailed methodology for tissue preparation, laser capture microdissection and RNA processing has been described in detail elsewhere (Pietersen et al., 2011; Pietersen et al., 2009a; Pietersen et al., 2009b). Briefly, sections of 8 μm were cut on a cryostat, mounted on slides and stored at -80 °C until use. Pyramidal neurons were stained with the Histogene™ quick staining kit (Applied Biosystems, Carlsbad, CA) and identified based on pyramidal morphology and clearly identifiable proximal apical and basal dendrites (Pietersen et al., 2014a). In addition, pyramidal neurons that were in close proximity to any nonpyramidal cells were excluded in order to avoid contamination. Pyramidal neurons in layer 3 were removed using the Arcturus XT™ system (Applied Biosystems, Carlsbad, CA). Approximately 500 neurons per subject were captured onto a CapSure HS™ LCM cap (Applied Biosystems, Carlsbad, CA) for microarray profiling.
Affymetrix platform-based microarray gene expression profiling
RNA processing
RNA isolation was performed using the Picopure™ RNA Isolation kit (Life Technologies, Grand Island, NY) with a DNase step (Qiagen, Valencia, CA). This typically resulted in approximately 1-25 ng of total RNA (Pietersen et al., 2011; Pietersen et al., 2009a). The extracted RNA underwent two rounds of linear amplification using the RiboAmp® kit (Life Technologies, Grand Island, NY). A dilution of the resulting products (approximately 250 ng/μl) was used to determine the distribution of transcript lengths with the Experion StdSens Labchip (Bio-Rad Laboratories, Hercules, CA). The concentration and purity of these samples were determined by absorbance measurements at the optical density of A260 and A280, using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA) (Table 1).
Microarray
The TURBO Biotin labeling™ kit (end-labeling; Life Technologies, NY) was used to label the aRNA obtained from amplified samples (∼15 μg). Gene expression profiling was performed using the Affymetrix Human X3P GeneChip®, which possesses an extreme 3′ bias in its probe design and hence is particularly suitable for samples that are prone to RNA degradation, such as postmortem human brain tissue. This chip has also been shown to be superior to the more commonly used Affymetrix human U133 plus 2.0 chip in terms of data reproducibility (Caretti et al., 2008). The hybridization and scanning procedures were performed at the Partners HealthCare Center for Personalized Genetic Medicine, Cambridge, MA.
Each array was scanned twice and the Affymetrix Microarray Suite 5.1 software averaged the two images to compute an intensity value for each probe cell within each probe set. For the quality control step, we employed the Partek® software's built-in function (Partek, St. Louis, MO). We then normalized the data with Partek's standard normalization method. Principal component analysis revealed the segregation of the data into two groups (Figure 1A), corresponding to the two age groups shown in Table 1. An Analysis of Covariance (ANCOVA) was performed with batch effect (scan date) as a covariate (Simunovic et al., 2009). Differentially expressed genes were then visualized by performing unsupervised hierarchical clustering (Figure 1B). By adjusting the stringency of the filtering criteria, i.e. fold-change (FC) and false discovery rate (FDR)-adjusted p-value, a representative gene list was determined which was then used for pathway analyses. Pathway analyses were performed with two web-based algorithms, Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA) and MetaCore (GeneGo, New York, NY), to map the differentially expressed genes onto biological functions and canonical pathways20, 21(20, 21). With Ingenuity, the significance for each of the identified pathways was determined via a Fisher's exact test, whereas GeneGo Metacore makes use of their algorithm for hypergeometric distribution, identifying pathways overrepresented with significant genes. Furthermore, literature mining was performed to elucidate which of these pathways or gene families might be particularly pertinent for pyramidal neuronal functions and dendritic/synaptic architecture and plasticity.
Figure 1.
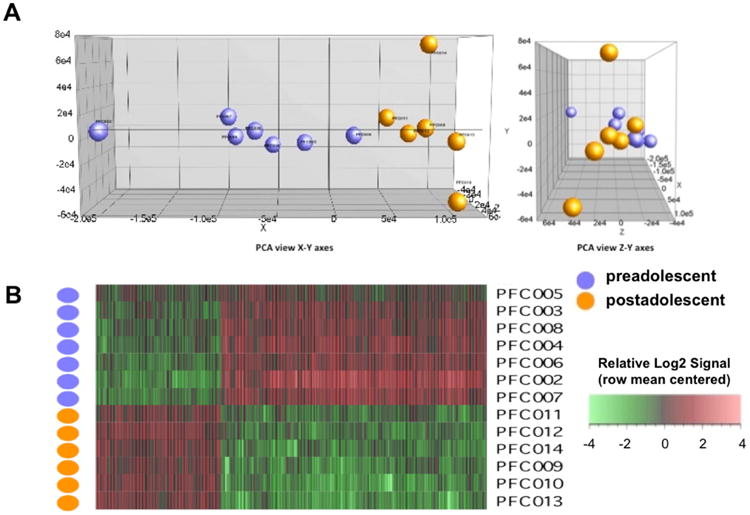
A. Principal component analysis reveals the segregation of the data into two age groups. Note that the purple and orange circles correspond to cases PFC2-8 and PFC9-14 shown in Table 1, respectively. B. Differentially expressed genes visualized by unsupervised hierarchical clustering based on the stringency criteria of FC=1.5 and p=0.05.
qRT-PCR validation
For validation of microarray data, cDNA was reverse transcribed from aRNA (200 ng input) using the High Capacity RNA-to-cDNA kit (Applied Biosystems, Carlsbad, CA). TaqMan®-based qRT-PCR (Applied Biosystems, Carlsbad, CA) was subsequently performed on selected genes within signaling pathways identified by pathway analysis as differentially expressed during development and randomly chosen genes, as previously described (Mauney et al., 2015; Pietersen et al., 2014a; Pietersen et al., 2014b). Primers were designed to span exon-intron boundaries to avoid amplification of any contaminating genomic DNA. Samples were normalized with respect to the housekeeping gene, hypoxanthine guanine phosphoribosyltransferase (HPRT), which has been shown to produce reliable results in human brain tissue, as its expression does not appear to differ in disease states (Radonic et al., 2004). Negative controls (negative reverse transcription and no template controls) were included to detect any contamination of the samples, such as genomic (g)DNA. Samples were run in duplicate. The average of the duplicates was taken as input for quantification using the 2̂-ΔCt method (Livak and Schmittgen, 2001) or the relative expression software tool (REST) for group-wise comparisons of expression ratios (Pfaffl et al., 2002). A Spearman Rho's correlation analysis was then performed on the fold-changes determined by microarray and qRT-PCR. A correlation of >0.8 with a significance of p<0.05 between qRT-PCR and microarray expression changes was considered validation of the microarray result (Morey et al., 2006).
UBE3B immunohistochemistry
Tissue processing
Frozen tissue blocks were sectioned at 20 μm, mounted on gelatin-subbed slides, and post-fixed in 4% paraformaldehyde for 20 minutes at room temperature. Sections were then incubated in endogenous enzyme block (1% H2O2, 10% MeOH) for 15 minutes and, additionally, blocked using 2% bovine serum albumin (BSA) in 10 % goat serum (Life Technologies, 16210-072, Grand Island, NY) at room temperature for 1 hour, followed by incubation in an anti-UBE3B (1:100, Novus Biologicals, NBP1-92559, Littleton, CO) antibody produced in rabbit (Sigma-Aldrich, St. Louis, MO) at 4°C overnight. The specificity of this antibody has been validated by protein array, Western blot and antigen preabsorption. Sections were then incubated at room temperature in biotinylated anti-rabbit IgG antibody produced in goat (1:500, Vector Labs, Burlingame, CA), followed by a 2-hour incubation in horseradish peroxidase-conjugated streptavidin (1:5,000, Zymed, San Francisco, CA) made in 0.1 Mol/L of phosphate buffer (PB) at room temperature, and finally in diaminobenzidine/peroxidase reaction (0.02% diaminobenzidine, 0.08% nickel-sulphate, 0.006% hydrogen peroxide in 1 Mol/L PB) before they were dehydrated and coverslipped.
Quantification
The number of all UBE3B-immunoreactive cells and pyramidal cells that were UBE3B-immunoreactive within three 500 μm × 500 mm squares that were 500 μm apart from one another placed within layer 3 of the prefrontal cortex were quantified by the same investigator (EAK) in a blind fashion using a StereoInvestigator system (MBG Bioscience, Williston, VT). Cell densities were compared between subjects with schizophrenia and normal control subjects using unpaired t-test. We also used correlation analysis to evaluate any potential effect of each of the numerical covariates (i.e. age, PMI, antipsychotic exposure in terms of chlorpromazine equivalent) on the cell density measures. The possible effect of sex on each of the density measures was evaluated by comparing cell densities between the two sexes within each of the two subject groups.
Results
Affymetrix-based microarray gene expression profiling
The efficiency of microarray hybridization appeared to be adequate in terms of probe intensity and percentage of present calls, and these parameters were highly comparable between the two age groups, with average (± SD) probe intensity being 81.8 ± 10.5 and 70.7 ± 6.2, and percent present calls 27.9 ± 5.3 and 32.3 ± 1.9. Overall, these percentages of present calls are lower than what have been reported in previously published microarray studies performed on RNA extracted from homogenized cortical gray matter, which contains a much greater number of RNA species in significantly larger quantities. Our data, however, are comparable in magnitude to what have been reported in recent microarray studies of laser-dissected hippocampal subfields and laser-captured homogeneous neuronal populations in humans (Benes et al., 2008; Pietersen et al., 2014a; Pietersen et al., 2014b; Simunovic et al., 2009) and to what has been described in previous microarray studies based on laser-captured cells from clinical samples or other specimens of single cells (Luzzi et al., 2001; Mahadevappa and Warrington, 1999).
Based on the stringency criteria of FC of 1.5 and FDR-adjusted p-values of 0.05, we identified 460 genes as differentially expressed during adolescent development (Supplementary Table S2). Hierarchical clustering revealed that these 460 genes appeared to be well segregated according to age group (Figure 1B). Some of the most highly differentially expressed genes are listed in Table 2; Of note, all of these genes were found to be upregulated during adolescent development. The canonical pathways and gene networks that were found to be the most significantly differentially regulated included pathways related to cytoskeletal remodeling, neurite outgrowth, regulation of translation initiation, elongation and termination, ubiquitination and proteolysis (Tables 3 and 4). The specific differentially expressed genes associated with these pathways are shown in Supplementary Tables S3-13.
Table 2. The twenty most highly differentially regulated genes during periadolescent development.
| Gene name | Fold-Change |
|---|---|
| ZNF7 (zinc finger protein 7) | 18.69 |
| UBE3B (ubiquitin protein ligase E3B) | 12.20 |
| STMN1 (stathmin 1) | 6.50 |
| CC2D1A (coiled-coil and C2 domain containing 1A) | 6.49 |
| C1orf227 (chromosome 1 open reading frame 227) | 6.44 |
| KLK3 (kallikrein-related peptidase 3) | 6.21 |
| MLL2 (myeloid/lymphoid or mixed-lineage leukemia 2) | 6.00 |
| ZNF37A///ZNF37BP (zinc finger protein 37A/// zinc finger protein 37B, pseudogene) | 5.93 |
| ATF7IP (activating transcription factor 7 interacting protein) | 5.92 |
| ALB (albumin) | 5.79 |
| ATOH (Atonal Homolog 8 (drosophila)) | 5.53 |
| KIAA1257///LOC100132731 | 5.46 |
| CARD11 (caspase recruitment domain family, member 11) | 5.30 |
| SP3 (Sp3 transcription factor) | 5.01 |
| RUNX3 (runt-related transcription factor 3) | 4.91 |
| CIAO1 (cytosolic iron-sulfur protein assembly 1) | 4.80 |
| C8orf56 (chromosome 8 open reading frame 56) | 4.79 |
| FUS (fused in sarcoma) | 4.79 |
| S100A1 (s100 calcium binding protein A1) | 4.62 |
| DHX9 (DEAH (Asp-Glu-Ala-His) boxpolypeptide 9 | 4.58 |
Table 3. Gene networks differentially regulated during periadolescent development.
| Gene Networks | p-value | Number of Genes |
|---|---|---|
| Translation_Translation initiation | 3.302E-12 | 25 |
| Translation_Elongation-Termination | 1.640E-05 | 19 |
| Transcription_Nuclear receptors transcriptional regulation | 7.201E-03 | 12 |
| Cytoskeleton_Actin filaments | 8.250E-03 | 11 |
| Development_Neurogenesis_Synaptogenesis | 2.441E-02 | 10 |
| Development_Neurogenesis_Axonal guidance | 4.817E-02 | 11 |
| Cell cycle_G1-S | 7.542E-02 | 8 |
| Cell cycle_S phase | 1.103E-01 | 7 |
| DNA damage_Checkpoint | 1.206E-01 | 6 |
| Cytoskeleton_Regulation of cytoskeleton rearrangement | 1.244E-01 | 8 |
| Cardiac development_Role of NADPH oxidase and ROS | 1.566E-01 | 6 |
| Cell adhesion_Amyloid proteins | 1.603E-01 | 8 |
| Proteolysis_Ubiquitin-proteasomal proteolysis | 1.648E-01 | 7 |
Table 4. Pathways differentially regulated during periadolescent development.
| Pathways | p-value | Number of Genes |
|---|---|---|
| Cytoskeleton remodeling_RalB regulation pathway | 8.785E-04 | 3 |
| Cytoskeleton remodeling_Cytoskeleton remodeling | 8.944E-04 | 7 |
| Proteolysis_Putative SUMO-1 pathway | 9.031E-04 | 4 |
| Cytoskeleton remodeling_TGF, WNT and cytoskeletal remodeling | 1.469E-03 | 7 |
| Development_MAG-dependent inhibition of neurite outgrowth | 2.286E-03 | 4 |
| Transcription_Androgen Receptor nuclear signaling | 4.697E-03 | 4 |
| Proteolysis_Role of Parkin in the Ubiquitin-Proteasomal Pathway | 5.502E-03 | 3 |
| Development_S1P2 and S1P3 receptors in cell proliferation and differentiation | 6.914E-03 | 3 |
| Translation_Regulation of translation initiation | 7.693E-03 | 3 |
| Neurophysiological process_Synaptic vesicle fusion and recycling in nerve terminals | 7.865E-03 | 4 |
| Translation_Regulation of EIF4F activity | 8.409E-03 | 4 |
| Cytoskeleton remodeling_RalA regulation pathway | 1.033E-02 | 3 |
| Cell cycle_Role of APC in cell cycle regulation | 1.235E-02 | 3 |
| Oxidative stress_Role of Sirtuin1 and PGC1-alpha in activation of antioxidant defense system | 1.291E-02 | 4 |
| Neurophysiological process_Activity-dependent synaptic AMPA receptor removal | 1.443E-02 | 4 |
| Translation _Regulation of EIF2 activity | 2.107E-02 | 3 |
| Immune response_Th1 and Th2 cell differentiation | 2.253E-02 | 3 |
| Cytoskeleton remodeling_Role of PKA in cytoskeleton reorganisation | 2.253E-02 | 3 |
Finally, we found that the FC of two highly differentially expressed genes and three randomly selected differentially expressed genes, as determined by microarray, were highly significantly correlated with FC determined by qRT-PCR (R=0.86, p=0.001; Figure 2).
Figure 2.
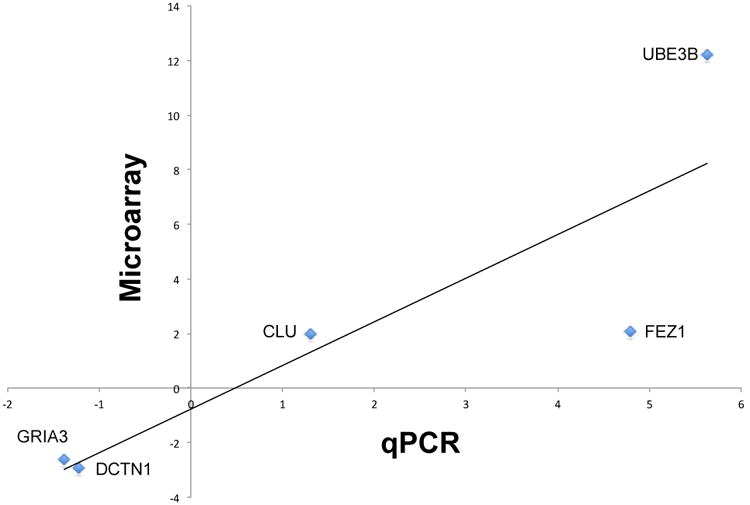
Expression levels of two highly differentially expressed genes and three randomly selected genes as determined by microarray and qRT-PCR were significantly correlated (R=0.86, p=0.001).
UBE3B expression is upregulated during adolescent development and downregulated in schizophrenia
We compared the current dataset with previously published pyramidal cell gene expression profiling data from schizophrenia subjects (Pietersen et al., 2014a) and identified genes that were differentially expressed both during adolescent development and in schizophrenia (Table 5). Presumably these may include genes that play a role in the developmental pathogenesis of the illness. We found that UBE3B was among the most highly differentially expressed genes in pyramidal neurons during normal adolescent development and it was also downregulated in schizophrenia subjects. Specifically, UBE3B was found to be upregulated by 12.20-fold (p=0.031) during adolescent development and downregulated by 1.24-fold (p=0.037) in subjects with schizophrenia (Pietersen et al., 2014a). Although very little is currently known about the functions of this specific gene, these findings raised our interests because of the known roles the ubiquitination system plays in regulating experience-dependent synaptic plasticity (Mabb and Ehlers, 2010; Tsai, 2014; Yashiro et al., 2009; Yi and Ehlers, 2005). We then used qRT-PCR to corroborate these microarray findings by showing that UBE3B was upregulated by 5.63-fold during development and downregulated by 3.36-fold in schizophrenia compared to normal control subjects. Furthermore, we validated the developmental microarray finding at the protein level. We found that the density of UBE3B-immunoreactive cells underwent postnatal developmental increase (Figure 3). We also found that the density of all UBE3B-immunoreactive cells was significantly decreased by 22.3% (p=0.026) in schizophrenia (1,483±437/mm2) compared to normal control (1,909±546/mm2) subjects (Figure 4A). More specifically and consistent with the observation of decreased UBE3B mRNA expression in pyramidal cells, we found that the density of pyramidal cells that were immunoreactive for UBE3B was significantly decreased by 48.0% (p=0.022) in schizophrenia (594±328/mm2) compared to normal control (1,136±364/mm2) subjects (Figure 4B). These findings were not affected by any of the potential confounding variables including age, PMI, and antipsychotic medications (Supplementary Table S14).
Table 5. Genes differentially expressed during adolescent development and in schizophrenia.
| Gene name | Gene symbol |
|---|---|
| active BCR-related gene | ABR |
| acyl-CoA dehydrogenase family, member 10 | ACAD10 |
| ARP2 actin-related protein 2 homolog (yeast) | ACTR2 |
| A kinase (PRKA) anchor protein 13 | AKAP13 |
| bromodomain adjacent to zinc finger domain, 2A | BAZ2A |
| chromosome 2 open reading frame 68 | C2orf68 |
| clusterin | CLU |
| Crystallin, beta B2 pseudogene 1 | CRYBB2P1 |
| diacylglycerol kinase, epsilon 64kDa | DGKE |
| dyskeratosis congenita 1, dyskerin | DKC1 |
| fasciculation and elongation protein zeta 1 (zygin I) | FEZ1 |
| fused in sarcoma | FUS |
| Immunoglobulin heavy constant alpha 1 | IGHA1 |
| large subunit GTPase 1 homolog (S. cerevisiae) | LSG1 |
| MAP kinase interacting serine/threonine kinase 1 | MKNK1 |
| myeloid/lymphoid or mixed-lineage leukemia 2 | MLL2 |
| natural cytotoxicity triggering receptor 1 | NCR1 |
| pecanex-like 2 (Drosophila) | PCNXL2 |
| phosphatidylinositol binding clathrin assembly protein | PICALM |
| proteasome (prosome, macropain) assembly chaperone 4 | PSMG4 |
| pyrroline-5-carboxylate reductase family, member 2 | PYCR2 |
| recombination signal binding protein for immunoglobulin kappa J region | RBPJ |
| ribosomal protein L22 | RPL22 |
| SEC14-like 1 (S. cerevisiae) | SEC14L1 |
| ubiquitin protein ligase E3B | UBE3B |
| zinc finger, CCHC domain containing 2 | ZCCHC2 |
| zinc finger protein 37A /// zinc finger protein 37B, pseudogene | ZNF37A /// ZNF37BP |
Figure 3.
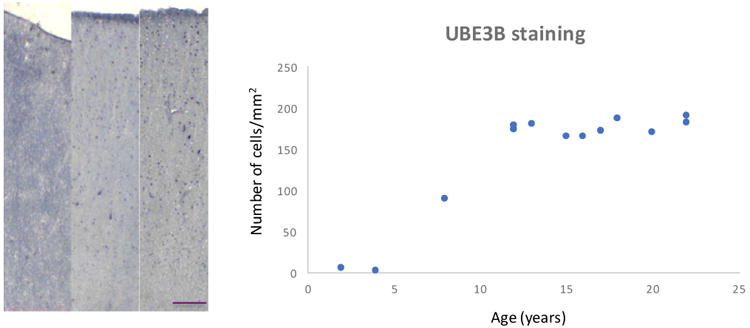
The density of UBE3B-immunoreactive cells in the prefrontal cortex progressively increases during postnatal development. Scale bar=100 μm.
Figure 4.
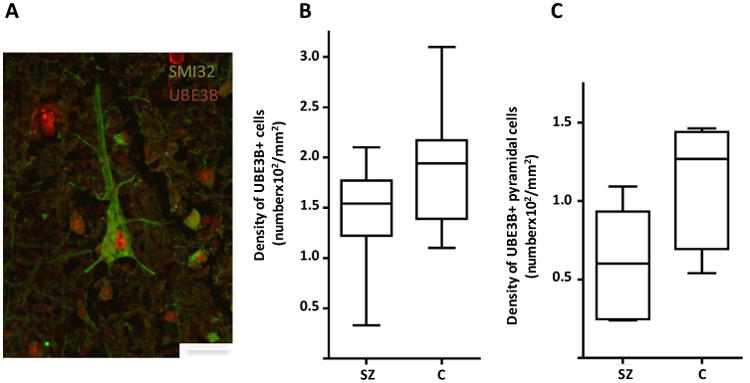
Photomicrograph showing a pyramidal neuron (labeled with SMI32) that expresses UBE3B. UBE3B is also localized to other cell types, including nonpyramidal neurons and possibly glia. Scale bar=25 μm. Densities of UBE3B-immunoreactive cells (B) and UBE3B-immunoreactive pyramidal cells (B) are significantly decreased by 22.3 % (p=0.026) and 48.0% (p=0.022), respectively, in schizophrenia (1,483±437/mm2 and 594±328/mm2, respectively) compared to normal control (1,909±546/mm2 and 1,136±364/mm2, respectively) subjects. These findings were not affected by any of the potential confounding variables including age, PMI, and antipsychotic medications (Supplementary Table S14).
Discussion
In this study, we investigated the gene expression changes in pyramidal neurons from layer 3 of the prefrontal cortex during adolescent development in humans. Our findings suggest that, perhaps not surprisingly, many gene networks and pathways that are known to be associated with cytoskeletal and synaptic plasticity are differentially regulated during adolescent development (Tables 3 and 4).
The human prefrontal cortex has long been known to undergo synaptic refinement during adolescence (Huttenlocher, 2002). Studies in nonhuman primates suggest that it is the asymmetric synapses, which are localized to dendritic spines on pyramidal neurons, that are predominantly affected, whereas the density of symmetric presumably inhibitory synapses appears to be largely unchanged (Anderson et al., 1995; Bourgeois et al., 1994; Goldman-Rakic et al., 1997). Consistent with these findings, our data indicate that genes and pathways that regulate cytoskeleton and actin filaments, which provide the structural architecture for dendritic spines, appear to be developmentally regulated. In addition, we found that genes associated with translational control are also significantly differentially regulated during adolescent development. Because protein synthesis is necessary for long-term memory formation and synaptic stability (Costa-Mattioli et al., 2009), these findings shed light onto the mechanisms that mediate the functional maturation of human prefrontal cortical circuitry.
Under the assumption that genes that are differentially regulated both during adolescent development and in schizophrenia subjects may play a role in the developmental pathogenesis of schizophrenia onset, we compared the developmental dataset obtained in this study with genes that were previously found to be differentially expressed in pyramidal neurons in subjects with schizophrenia (Pietersen et al., 2014a). Among the genes that were identified, UBE3B is of particular interest as ubiquitination has been strongly implicated in the regulation of synaptic plasticity (Mabb and Ehlers, 2010; Tsai, 2014; Yashiro et al., 2009; Yi and Ehlers, 2005). Hence, developmental upregulation of UBE3B may play a role in regulating the adolescent synaptic refinement process (Bourgeois et al., 1994; Huttenlocher, 2002; Woo et al., 1997). In fact, we were able to immunohistochemically validate that, at the protein level, the number of cells that contained UBE3B protein also underwent developmental increase. This finding was obtained using a two-dimensional counting approach; the appropriateness and limitations of this especially in the context of postmortem human brain studies, as opposed to the unbiased stereological method, have been extensively discussed (Benes and Lange, 2001). In addition, we found that the density of UBE3B-expressing pyramidal neurons was also significantly decreased in schizophrenia subjects. Altogether, these findings converge upon the hypothesis that decreased expression of UBE3B could contribute to the developmental pathophysiology of schizophrenia by altering adolescent synaptic refinement of prefrontal cortical circuitry. The fact that the level of UBE3B expression appears to remain elevated after the adolescent period would suggest that it may have more to do with the process of synaptic stabilization rather than the process of synaptic pruning. A caveat regarding the observation that UBE3B expression remains elevated after adolescent development is that increased ubiquitination may be associated less with development than with the cumulative burden of protease-resistant material, but these two interpretations need not be mutually exclusive.
It should be noted that the schizophrenia pyramidal neuronal gene expression data used to be compared with the developmental data obtained in the present study were derived from the superior temporal cortex (Pietersen et al., 2014a). Nevertheless, evidence from neuroimaging studies suggests that the superior temporal cortex in schizophrenia appears to undergo similar, if not more severe, periadolescent attrition of gray matter (Kasai et al., 2003; Takahashi et al., 2009; Witthaus et al., 2009), which is generally interpreted as a manifestation of the underlying excessive pruning of synapses. Hence, if aberrant synaptic pruning contributes to the developmental pathogenesis of schizophrenia onset, it is likely that similar mechanisms may operate in both the prefrontal and superior temporal cortices. This notion is also supported by the fact that we were able to validate UBE3B expression change at the protein level in the prefrontal cortex (Figure 4) even though the comparison of gene expression profiles was made across cortical regions.
Ubiquitination is a post-translation protein modification process that plays a critical role in protein degradation. During this process, an E1 ubiquitin-activating enzyme first binds to an ubiquitin protein, followed by the transfer of ubiquitin from the E1 to the E2 site through an E2 ubiquitin-conjugating enzyme. Finally, an E3 ligase catalyzes the rate-limiting step of the transfer of ubiquitin from E2 to the target protein, “tagging” this protein for degradation via the proteasome system. This ubiquitination-proteosome protein degradation system has been implicated in the functional and structural plasticity of synapses (Mabb and Ehlers, 2010; Ryan et al., 2006; Tsai, 2014; Yi and Ehlers, 2005, 2007). Because E3 ligases regulate the rate-limiting step of the entire process, these enzymes are thought to be of particular importance. For instance, UBE3A, a paralog of UBE3B, has been found to play an important role in regulating postnatal experience-dependent synaptic plasticity (Wallace et al., 2012; Yashiro et al., 2009). Specifically, in mice lacking the maternal Ube3a allele, which in humans is the cause of Angelman syndrome, the density of dendritic spines on cortical pyramidal neurons is decreased by ∼25% and this reduction is accompanied by a significant attenuation of both long-term potentiation and long-term depression (Wallace et al., 2012; Yashiro et al., 2009). In contrast, very little is currently known about the possible functional roles of UBE3B, although this gene has been linked to autism (Chahrour et al., 2012) and the loss of function of UBE3B results in a rare autosomal recessive disorder, Kaufman oculocerebrofacial syndrome, of which microcephaly, developmental delay and intellectual retardation are some of the distinct features (Basel-Vanagaite et al., 2012; Basel-Vanagaite et al., 2014; Flex et al., 2013). Our findings that UBE3B is developmentally regulated at a time when schizophrenia typically emerges and that UBE3B-expressing pyramidal cells are decreased in subjects with schizophrenia suggest that further elucidation of the roles of UBE3B in postnatal cortical development will provide insight into the developmental pathogenetic mechanism of schizophrenia onset.
Supplementary Material
Acknowledgments
This study was supported by grants P50MH080272 and R01MH076060 from the National Institutes of Health.
Footnotes
Authors' Contributions: EAK, NS and CYP carried out LCM and RNA processing for microarray experiments. EAK performed qRT-PCR validation and immunohistochemistry, participated in the microarray data analysis and drafting of the manuscript. KCS participated in data analysis and preparation of the manuscript. TUWW conceived of the study, participated in its design and data interpretation, and helped draft the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson SA, Classey JD, Conde F, Lund JS, Lewis DA. Synchronous development of pyramidal neuron dendritic spines and parvalbumin-immunoreactive chandelier neuron axon terminals in layer III of monkey prefrontal cortex. Neuroscience. 1995;67(1):7–22. doi: 10.1016/0306-4522(95)00051-j. [DOI] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Dallapiccola B, Ramirez-Solis R, Segref A, Thiele H, Edwards A, Arends MJ, Miro X, White JK, Desir J, Abramowicz M, Dentici ML, Lepri F, Hofmann K, Har-Zahav A, Ryder E, Karp NA, Estabel J, Gerdin AK, Podrini C, Ingham NJ, Altmuller J, Nurnberg G, Frommolt P, Abdelhak S, Pasmanik-Chor M, Konen O, Kelley RI, Shohat M, Nurnberg P, Flint J, Steel KP, Hoppe T, Kubisch C, Adams DJ, Borck G. Deficiency for the ubiquitin ligase UBE3B in a blepharophimosis-ptosis-intellectual-disability syndrome. Am J Hum Genet. 2012;91(6):998–1010. doi: 10.1016/j.ajhg.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Yilmaz R, Tang S, Reuter MS, Rahner N, Grange DK, Mortenson M, Koty P, Feenstra H, Farwell Gonzalez KD, Sticht H, Boddaert N, Desir J, Anyane-Yeboa K, Zweier C, Reis A, Kubisch C, Jewett T, Zeng W, Borck G. Expanding the clinical and mutational spectrum of Kaufman oculocerebrofacial syndrome with biallelic UBE3B mutations. Human genetics. 2014;133(7):939–949. doi: 10.1007/s00439-014-1436-2. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lange N. Two-dimensional versus three-dimensional cell counting: a practical perspective. Trends Neurosci. 2001;24(1):11–17. doi: 10.1016/s0166-2236(00)01660-x. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Subburaju S, Walsh JP. Circuitry-based gene expression profiles in GABA cells of the trisynaptic pathway in schizophrenics versus bipolars. Proc Natl Acad Sci U S A. 2008;105(52):20935–20940. doi: 10.1073/pnas.0810153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois JP, Goldman-Rakic PS, Rakic P. Synaptogenesis in the prefrontal cortex of rhesus monkeys. Cerebral Cortex. 1994;4(1):78–96. doi: 10.1093/cercor/4.1.78. [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Draguhn A. Neuronal oscillations in cortical networks. Science. 2004;304(5679):1926–1929. doi: 10.1126/science.1099745. [DOI] [PubMed] [Google Scholar]
- Caretti E, Devarajan K, Coudry R, Ross E, Clapper ML, Cooper HS, Bellacosa A. Comparison of RNA amplification methods and chip platforms for microarray analysis of samples processed by laser capture microdissection. J Cell Biochem. 2008;103(2):556–563. doi: 10.1002/jcb.21426. [DOI] [PubMed] [Google Scholar]
- Chahrour MH, Yu TW, Lim ET, Ataman B, Coulter ME, Hill RS, Stevens CR, Schubert CR, Collaboration AAS, Greenberg ME, Gabriel SB, Walsh CA. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS genetics. 2012;8(4):e1002635. doi: 10.1371/journal.pgen.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61(1):10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flex E, Ciolfi A, Caputo V, Fodale V, Leoni C, Melis D, Bedeschi MF, Mazzanti L, Pizzuti A, Tartaglia M, Zampino G. Loss of function of the E3 ubiquitin-protein ligase UBE3B causes Kaufman oculocerebrofacial syndrome. Journal of medical genetics. 2013;50(8):493–499. doi: 10.1136/jmedgenet-2012-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster JM. The prefrontal cortex. Elsevier; New York: 2008. [Google Scholar]
- Geschwind DH, Ou J, Easterday MC, Dougherty JD, Jackson RL, Chen Z, Antoine H, Terskikh A, Weissman IL, Nelson SF, Kornblum HI. A genetic analysis of neural progenitor differentiation. Neuron. 2001;29(2):325–339. doi: 10.1016/s0896-6273(01)00209-4. [DOI] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Archives of General Psychiatry. 2000;57(1):65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS, Bourgeois JP, Rakic P. Synaptic substrate of cognitive development: Life-span analysis of synaptogenesis in the prefrontal cortex of the nonhuman primate. In: Krasnegor NA, Lyon GR, Goldman-Rakic PS, editors. Development of the prefontal cortex. Paul H. Brookes Publishing; Baltimore: 1997. pp. 27–48. [Google Scholar]
- Hanover JL, Huang ZJ, Tonegawa S, Stryker MP. Brain-derived neurotrophic factor overexpression induces precocious critical period in mouse visual cortex. Journal of Neuroscience. 1999;19(22):RC40. doi: 10.1523/JNEUROSCI.19-22-j0003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR. Neural Plasticity. Harvard University Press; Cambridge: 2002. [Google Scholar]
- Kasai K, Shenton ME, Salisbury DF, Hirayasu Y, Lee CU, Ciszewski AA, Yurgelun-Todd D, Kikinis R, Jolesz FA, McCarley RW. Progressive decrease of left superior temporal gyrus gray matter volume in patients with first-episode schizophrenia. Am J Psychiatry. 2003;160(1):156–164. doi: 10.1176/appi.ajp.160.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt JB, Lewis DA, Yoshioka T, Lund JS. Topography of pyramidal neuron intrinsic connections in macaque monkey prefrontal cortex (areas 9 and 46) Journal of Comparative Neurology. 1993;338(3):360–376. doi: 10.1002/cne.903380304. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luzzi V, Holtschlag V, Watson MA. Expression profiling of ductal carcinoma in situ by laser capture microdissection and high-density oligonucleotide arrays. Am J Pathol. 2001;158(6):2005–2010. doi: 10.1016/S0002-9440(10)64672-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevappa M, Warrington JA. A high-density probe array sample preparation method using 10- to 100-fold fewer cells. Nat Biotechnol. 1999;17(11):1134–1136. doi: 10.1038/15124. [DOI] [PubMed] [Google Scholar]
- Mauney SA, Pietersen CY, Sonntag KC, Woo TW. Differentiation of oligodendrocyte precursors is impaired in the prefrontal cortex in schizophrenia. Schizophr Res. 2015 doi: 10.1016/j.schres.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–193. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Lim MP, Macey L, Woo TUW, Sonntag KC. Neuronal type-specific gene expression profiling and laser-capture microdissection. Methods Mol Biol. 2011;755:327–343. doi: 10.1007/978-1-61779-163-5_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Lim MP, Woo TUW. Obtaining high quality RNA from single cell populations in human postmortem brain tissue. J Vis Exp. 2009a;(30) doi: 10.3791/1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Lim MP, Woo TU. Obtaining high quality RNA from single cell populations in human postmortem brain tissue. J Vis Exp. 2009b;(30) doi: 10.3791/1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Mauney SA, Kim SS, Lim MP, Rooney RJ, Goldstein JM, Petryshen TL, Seidman LJ, Shenton ME, McCarley RW, Sonntag KC, Woo TU. Molecular profiles of pyramidal neurons in the superior temporal cortex in schizophrenia. Journal of neurogenetics. 2014a;28(1-2):53–69. doi: 10.3109/01677063.2014.882918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Mauney SA, Kim SS, Passeri E, Lim MP, Rooney RJ, Goldstein JM, Petreyshen TL, Seidman LJ, Shenton ME, McCarley RW, Sonntag KC, Woo TU. Molecular profiles of parvalbumin-immunoreactive neurons in the superior temporal cortex in schizophrenia. Journal of neurogenetics. 2014b;28(1-2):70–85. doi: 10.3109/01677063.2013.878339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucak ML, Levitt JB, Lund JS, Lewis DA. Patterns of intrinsic and associational circuitry in monkey prefrontal cortex. Journal of Comparative Neurology. 1996;376(4):614–630. doi: 10.1002/(SICI)1096-9861(19961223)376:4<614::AID-CNE9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313(4):856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, Webster MJ, Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol Psychiatry. 2006;11(10):965–978. doi: 10.1038/sj.mp.4001875. [DOI] [PubMed] [Google Scholar]
- Simunovic F, Yi M, Wang Y, Macey L, Brown LT, Krichevsky AM, Andersen SL, Stephens RM, Benes FM, Sonntag KC. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson's disease pathology. Brain. 2009;132(Pt 7):1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet RA, Henteleff RA, Zhang W, Sampson AR, Lewis DA. Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology. 2009;34(2):374–389. doi: 10.1038/npp.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet RA, Pierri JN, Auh S, Sampson AR, Lewis DA. Reduced pyramidal cell somal volume in auditory association cortex of subjects with schizophrenia. Neuropsychopharmacology. 2003;28(3):599–609. doi: 10.1038/sj.npp.1300120. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Wood SJ, Yung AR, Soulsby B, McGorry PD, Suzuki M, Kawasaki Y, Phillips LJ, Velakoulis D, Pantelis C. Progressive gray matter reduction of the superior temporal gyrus during transition to psychosis. Arch Gen Psychiatry. 2009;66(4):366–376. doi: 10.1001/archgenpsychiatry.2009.12. [DOI] [PubMed] [Google Scholar]
- Tsai NP. Ubiquitin proteasome system-mediated degradation of synaptic proteins: An update from the postsynaptic side. Biochim Biophys Acta. 2014;1843(12):2838–2842. doi: 10.1016/j.bbamcr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Uhlhaas PJ, Haenschel C, Nikolic D, Singer W. The role of oscillations and synchrony in cortical networks and their putative relevance for the pathophysiology of schizophrenia. Schizophr Bull. 2008;34(5):927–943. doi: 10.1093/schbul/sbn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Roux F, Singer W, Haenschel C, Sireteanu R, Rodriguez E. The development of neural synchrony reflects late maturation and restructuring of functional networks in humans. Proc Natl Acad Sci U S A. 2009;106(24):9866–9871. doi: 10.1073/pnas.0900390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. The development of neural synchrony and large-scale cortical networks during adolescence: relevance for the pathophysiology of schizophrenia and neurodevelopmental hypothesis. Schizophr Bull. 2011;37(3):514–523. doi: 10.1093/schbul/sbr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron. 2012;74(5):793–800. doi: 10.1016/j.neuron.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witthaus H, Kaufmann C, Bohner G, Ozgurdal S, Gudlowski Y, Gallinat J, Ruhrmann S, Brune M, Heinz A, Klingebiel R, Juckel G. Gray matter abnormalities in subjects at ultra-high risk for schizophrenia and first-episode schizophrenic patients compared to healthy controls. Psychiatry Res. 2009;173(3):163–169. doi: 10.1016/j.pscychresns.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Woo TUW, Pucak ML, Kye CH, Matus CV, Lewis DA. Peripubertal refinement of the intrinsic and associational circuitry in monkey prefrontal cortex. Neuroscience. 1997;80(4):1149–1158. doi: 10.1016/s0306-4522(97)00059-6. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, Weinberg RJ, Ehlers MD, Philpot BD. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12(6):777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Ubiquitin and protein turnover in synapse function. Neuron. 2005;47(5):629–632. doi: 10.1016/j.neuron.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacological reviews. 2007;59(1):14–39. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


