SUMMARY
Human oligodendrocyte progenitor cells (hOPCs) persist into adulthood as an abundant precursor population capable of division and differentiation. The transcriptional mechanisms that regulate hOPC homeostasis remain poorly defined. Herein, we identify paired related homeobox protein 1 (PRRX1) in primary PDGFαR+ hOPCs. We show that enforced PRRX1 expression results in reversible G1/0 arrest. While both PRRX1 splice variants reduce hOPC proliferation, only PRRX1a abrogates migration. hOPC engraftment into hypomyelinated shiverer/rag2 mouse brain is severely impaired by PRRX1a, characterized by reduced cell proliferation and migration. PRRX1 induces a gene expression signature characteristic of stem cell quiescence. Both IFN-γ and BMP signaling upregulate PRRX1 and induce quiescence. PRRX1 knockdown modulates IFN-γ-induced quiescence. In mouse brain, PRRX1 mRNA was detected in non-dividing OPCs and is upregulated in OPCs following demyelination. Together, these data identify PRRX1 as a regulator of quiescence in hOPCs and as a potential regulator of pathological quiescence.
Graphical Abstract
In Brief
Wang et al. investigate the role of transcription factor PRRX1 in human oligodendrocyte progenitor cells. PRRX1 induces reversible cell-cycle arrest, resulting in a quiescent-like state that prevents colonization and myelination of hypomyelinated shiverer mice. PRRX1 expression was regulated by interferon-γ and BMP and required for interferon-induced quiescence.
INTRODUCTION
Unlike other transient amplifying cells, oligodendrocyte progenitor cells (OPCs) persist throughout adulthood and remain a mitotic progenitor pool capable of generating new oligodendrocytes (Rivers et al., 2008; Dimou et al., 2008). Timely differentiation of these progenitors is necessary for efficient remyelination (Franklin, 2002) and motor skill learning (McKenzie et al., 2014; Marques et al., 2016). In addition to their role as a source of new oligodendrocytes, it is apparent that the function of adult OPCs is vital for normal brain function (Birey et al., 2015). OPC density is tightly regulated in vivo, such that loss of a single OPC, by death or differentiation, is replaced by mitosis and migration (Hughes et al., 2013). However, the molecular mechanisms that regulate OPC homeostasis in adult brain are poorly understood.
Although OPCs represent the most abundant progenitor in the adult CNS, most have been considered quiescent (Dawson et al., 2003). Fate-mapping studies indicate that likely all adult OPCs undergo cell division and that OPC cell-cycle length dramatically increases with age, from days to several months in aged animals (Psachoulia et al., 2009; Young et al., 2013). Thus, adult OPCs can enter a reversibly quiescent state for extended periods in adult brain but eventually reenter the cell cycle.
Cellular quiescence is characterized by reversible cell-cycle exit and downregulation of genes important for DNA replication, cell-cycle progression, metabolism, and protein synthesis (Rumman et al., 2015). Quiescence is essential for maintaining cell populations in reserve for tissue regeneration and repair. For example, loss of quiescence in muscle stem cells depletes the stem cell pool, leading to failed regeneration (Shea et al., 2010; Mourikis et al., 2012). Quiescent OPCs are observed in chronic demyelinated lesions in multiple sclerosis (MS) (Wolswijk, 1998). Thus, although OPCs are present in most chronic MS lesions, their number and/or density may be lower than is necessary for efficient repair (for review, see Dietz et al., 2016). As such, an additional explanation for failed remyelination could be due to pathological quiescence.
We have identified several transcription factors that were up-regulated in human OPCs (hOPCs) relative to neural progenitor cells (NPCs) during lineage specification (Wang et al., 2014). Among these, paired related homeobox protein 1 (PRRX1) was implicated in adult murine NPC self-renewal (Shimozaki et al., 2013). In this study, we examined the role of PRRX1 in the regulation of hOPC function in vitro and following transplantation into hypomyelinated shiverer/rag2 brain. PRRX1 overexpression led to profound and reversible arrest of the cell cycle, resulting in reduced engraftment and myelination in shiverer mice. We determined that PRRX1 induced a conserved gene signature involved in establishing cellular quiescence. PRRX1 was upregulated in response to known inducers of quiescence and was necessary for cell-cycle arrest.
RESULTS
PRRX1 Suppresses hOPC Proliferation and Migration In vitro
Because the two PRRX1 spliced variants encode structurally distinct isoforms (Reichert et al., 2013), we sought to better define the PRRX1 expression profile. Using CD140a/CD133 fluorescence-activated cell sorting (FACS) (Wang et al., 2013), both PRRX1a and PRRX1b mRNAs were enriched in platelet-derived growth factor (PDGF) αR+ OPCs compared to CD133+ NPCs (Figure 1A). In parallel experiments, hOPCs were induced to differentiate as immature oligodendrocytes for up to 4 days in vitro (Pol et al., 2017). We found that both PRRX1a and PRRX1b mRNA were downregulated as hOPCs underwent oligodendrocytic differentiation (Figure 1B). A similar pattern was found in mouse OPCs, with downregulation occurring in differentiated oligodendrocytes (Zhang et al., 2014).
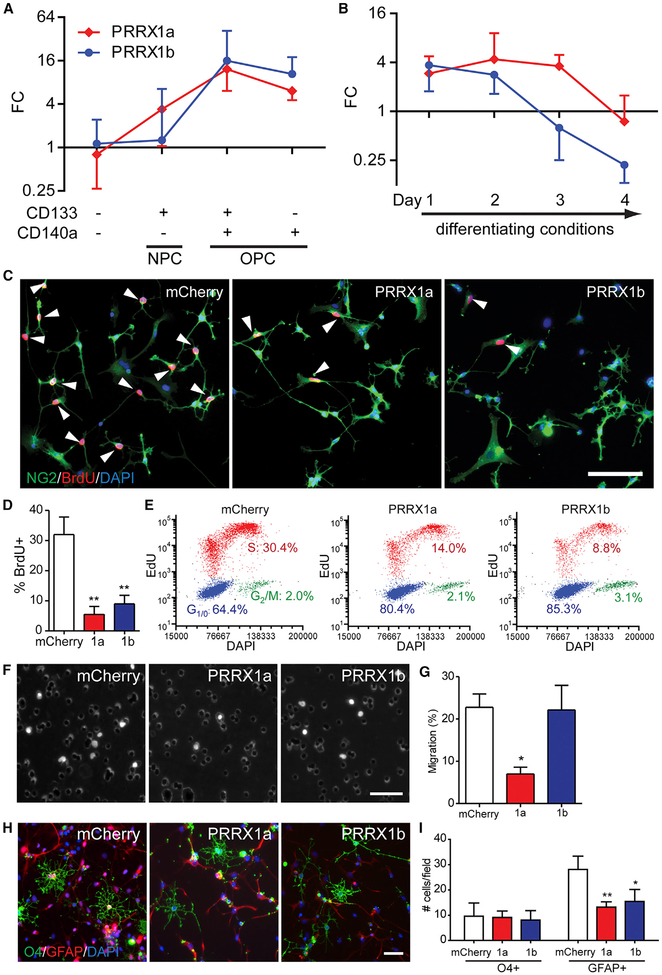
Figure 1. PRRX1a/b Are Expressed by hOPCs and Differentially Regulate Proliferation, Migration, and Differentiation.
(A) Human NPCs (CD133+CD140a−), early OPCs (CD133+CD140a+), and late OPCs (CD133−CD140a+) were isolated from fetal 18–22 weeks gestational age brain by FACS (n = 3 individual human samples).
(B) PDGFαR+ hOPCs were isolated and underwent oligodendrocyte differentiation in the absence of mitogens for up to 4 days (n = 4 human samples). qPCR was performed on RNA extracted immediately post-sort or after 1–4 days in culture. Mean ± SEM fold change (FC) shown relative to fetal dissociate (CD133−CD140a−) after GAPDH normalization.
(C–E) Fetal PDGFαR+ hOPCs infected with mCherry (control) or PRRX1 LV were maintained in SFM with platelet-derived growth factor (PDGF)-AA for 4 days.
(C) 24-hr BrdU incorporation was assessed in NG2+ OPCs (arrowheads indicate BrdU+ cells).
(D) Quantification of BrdU percentage in NG2+ hOPCs (n = 4 fetal samples, **p < 0.01 versus mCherry, one-way repeated-measures ANOVA, Dunnett’s post-test).
(E) Flow cytometry of S-phase entry (red, 24-hr EdU incorporation) and G1/0 and G2/M phases (blue and green, respectively).
(F) LV-infected hOPC migration seeded on transwell membranes. Migrant DAPI+ cells (100 ng/mL) were imaged.
(G) Percentage of migrating cells was assessed (n = 5 fetal samples, *p < 0.05 versus mCherry, one-way repeated-measures ANOVA, Dunnett’s post-test).
(H) LV-infected hOPCs were allowed to differentiate for 2 days following mitogen withdrawal in the presence of 40 ng/mL T3. Cultures were immunostained with an immature oligodendrocyte marker (O4, green) and an astrocyte marker (GFAP, red).
(I) Average number of oligodendrocyte and astrocytes in each field was quantified (n = 4 fetal samples, *p < 0.05, **p < 0.01 versus mCherry, one-way repeated-measures ANOVA, Dunnett’s multiple comparisons post-test).
For bar charts, mean ± SEM is shown. Scale: 50 μm.
In human NPCs, PRRX1 overexpression did not potentiate oligodendrocyte progenitor and oligodendrocyte generation, suggesting that it may have a role other than induction of OPC fate per se (Wang et al., 2014). As such, we investigated whether PRRX1a and PRRX1b might differentially regulate hOPC specification, migration, proliferation, and differentiation. hOPCs were infected with lentivirus (LV) encoding PRRX1a, PRRX1b, or mCherry as control. After 4 days in serum-free medium (SFM) containing PDGF AA, the percentage of proliferating (bromodeoxyuridine [BrdU]+NG2+) OPCs was significantly reduced following PRRX1a and PRRX1b overexpression compared to mCherry control cells (5.5% ± 2.6% and 9.0% ± 2.8%, respectively, versus 32.0% ± 5.8%; n = 4, p < 0.01, one-way repeated-measures ANOVA, Dunnett’s post-test) (Figures 1C and 1D). PRRX1 substantially increased the percentage of cells in G1/0, indicating that PRRX1 overexpression resulted in cell-cycle arrest (Figure 1E).
Next, because insufficient migration of OPCs may contribute to impaired remyelination (Boyd et al., 2013), we determined whether PRRX1 influenced migration. We measured migration in transwell-based assays following PRRX1a- or PRRX1b-expressing LV infection. hOPC migration was differentially regulated by PRRX1 variants (Figures 1F and 1G). Only 7.0% ± 1.6% of PRRX1a cells migrated toward PDGF-AA (100 ng/mL), whereas 22.8% ± 3.1% of control, mCherry-infected, and 22.2% ± 5.8% of PRRX1b-infected hOPCs migrated (n = 5 per group, p < 0.05, one-way repeated-measures ANOVA, Dunnett’s post-test).
We next asked whether PRRX1a/b would influence hOPC fate when cultured in differentiating conditions. PRRX1 did not influence the total number of O4+ oligodendrocytes; however, it reduced the GFAP+ astrocyte number. Specifically, the average number of O4+ cells per field did not differ (mCherry, 9.6 ± 5.2; PRRX1a, 9.1 ± 2.6; PRRX1b, 8.1 ± 3.7); however, there was a significant reduction in the GFAP+ cell absolute number with PRRX1a/b overexpression (13.2 ± 2.1 and 15.5 ± 4.7, respectively, versus 28.1 ± 5.3; n = 4, p < 0.01, one-way repeated-measures ANOVA, Dunnett’s post-test) (Figures 1H and 1I). While the absolute number of astrocytes was reduced, the proportion of GFAP+ astrocytes was unaffected. The proportion of O4+ oligodendrocytes was increased by PRRX1a/b (22.7 ± 2.9 and 19.8 ± 2.8, respectively, versus 13.3 ± 4.0; n = 4, p < 0.05, Dunnett’s post-test). Consistent with a primary effect on proliferation, the total number of cells was substantially reduced with PRRX1 overexpression (live cells/field: mCherry, 72.4 ± 22.6; PRRX1a, 43.6 ± 11.5; PRRX1b, 38.0 ± 12.2; p = 0.06, one-way repeated-measures ANOVA). This decrease was not due to increased apoptosis, because equivalent rates of cleaved caspase-3 were observed (5.9% ± 0.3% and 4.1% ± 0.5%, control and PRRX1a, respectively). To exclude other forms of cell death, we assessed cell viability and death at 48 hr. PRRX1a LV had no effect on cellular morphology or live:dead ratio compared to mCherry (mCherry: 1.3 ± 0.1, PRRX1a: 1.4 ± 0.1; ratio of calcein: DAPI; n = 3 fetal samples, p > 0.05, t test).
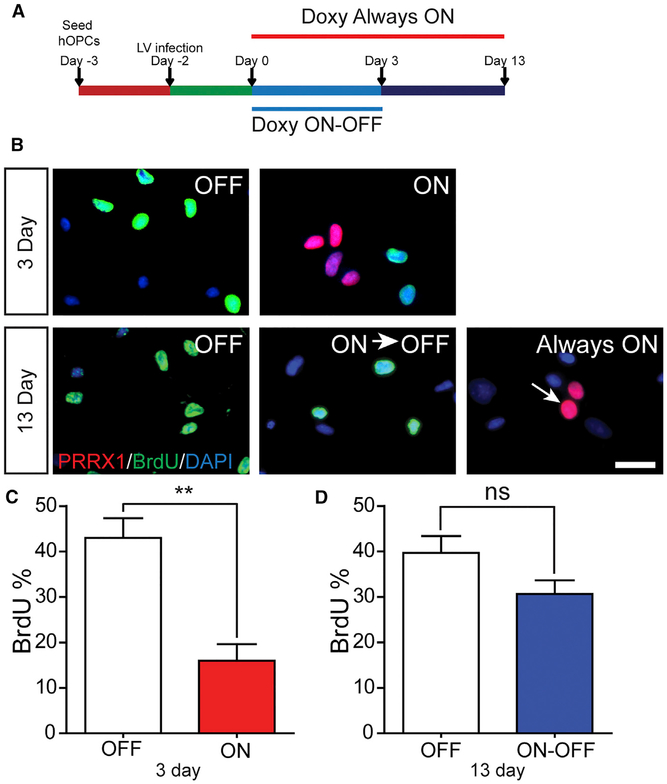
A key aspect of quiescence versus senescence is the reversible nature of cell-cycle arrest. To determine whether transient PRRX1a-FLAG expression induced reversible cell-cycle arrest, we used a doxycycline-inducible LV (Figure 2). Doxycycline-induced PRRX1a significantly reduced hOPC proliferation (control, 43% ± 4%, versus doxycycline-induced PRRX1, 16% ± 4%; n = 4 fetal samples, p < 0.01, t test). Among FLAG+ cells at 3 days, only 7.8% ± 2% were BrdU+, indicating a profound effect in PRRX1-expressing hOPCs. If doxycycline was maintained for 13 days, we did not observe BrdU+FLAG+ cells, indicating that PRRX1 was sufficient to arrest proliferation. To determine whether hOPCs could reenter the cell cycle following PRRX1 downregulation, doxycycline was removed in matched cultures at day 3 (ON-OFF) (Figure 2A) and cell-cycle reentry was determined. In ON-OFF cultures, PRRX1a-FLAG expression was no longer detected, and the proportion of proliferating hOPCs had returned to control levels (OFF, 39.7% ± 3.7%, versus ON-OFF, 30.7% ± 2.9%; n = 3, p > 0.05), representing a significant increase compared to day 3 (ON, p < 0.05). As such, these data indicate that PRRX1a can induce reversible hOPC cell-cycle arrest.
Figure 2. Transient PRRX1 Expression Results in Reversible Cell-Cycle Arrest.
(A) hOPCs were infected with doxycycline-inducible FUW-PRRX1a-FLAG LV. Matched cultures were assigned to OFF (no doxycycline), ON-OFF (doxycycline for first 3 days), or Always ON (doxycycline for 13 days).
(B) hOPC proliferation was assessed by BrdU incorporation (green, 24-hr terminal pulse) at both 3 days and 13 days. PRRX1a-FLAG expression was detected by immunostaining (red).
(C and D) Quantification of BrdU at day 3 (C) and day 13 (D) (n = 3–4 fetal samples).
**p < 0.01, paired t test. For bar charts, mean ± SEM is shown. Scale: 50 μm.
PRRX1a Overexpression Inhibits Migration of Engrafted hOPCs
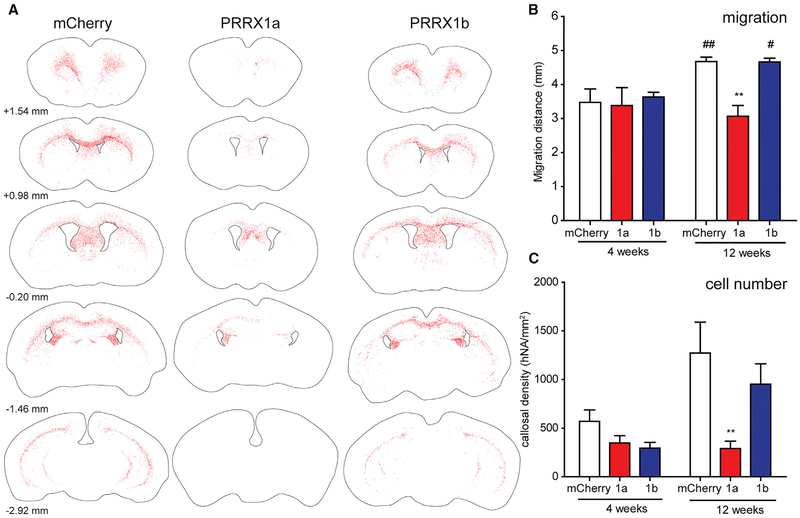
To determine whether PRRX1 overexpression would induce quiescence in hOPCs in vivo, we transplanted LV-infected hOPCs into hypomyelinated and immunocompromised shiverer/rag2 mice. PRRX1a/b overexpression was confirmed in engrafted cells (Figure S1). At 12 weeks, human nuclear antigen-positive (hNA+) cells were largely confined to white matter (n = 4–9 per group). PRRX1a overexpression significantly impaired the rostrocaudal and lateral distribution of hOPCs compared to mCherry transplants (Figure 3A). We found that PRRX1a-infected hOPCs were distributed over a smaller region of shiverer/rag2 forebrain (mCherry, 4.7 ± 0.1 mm; PRRX1a, 3.1 ± 0.3 mm; p < 0.01) (Figure 3B). As observed in vitro, PRRX1b did not affect migration after engraftment at 12 weeks. PRRX1a overexpression resulted in a substantial reduction in density and overall number of hNA+ cells at 12 weeks. The callosal density of hNA+ cells were reduced by >4-fold relative to mCherry-infected hOPCs (290.8 ± 75.9 versus 1,272.4 ± 316.8 cells/mm2; p < 0.01, two-way ANOVA, Bonferroni’s post-test) (Figure 3C). While the density of PRRX1b-infected cells was reduced by >25%, this was not statistically significant (952.6 ± 208.3 cells/mm2). Immunoreactivity for cleaved caspase-3 was similarly rare among the groups at 12 weeks (data not shown), suggesting that PRRX1 overexpression did not increase cell death.
Figure 3. PRRX1 Reduces Engraftment by hOPCs In vivo.
hOPCs infected with PRRX1a, PRRX1b, or mCherry LV were transplanted bilaterally into the corpus callosum of postnatal day 2–3 shiverer/rag2 mice. Mice were sacrificed 4 or 12 weeks following implantation.
(A) Total rostrocaudal dissemination of human nuclear antigen-positive cells (red dots) was assessed and plotted relative to bregma (according to Paxinos and Franklin, 2004).
(B and C) Average rostrocaudal migration distance (B, in millimeters) and hNA+ cell density in the engrafted corpus callosum (C, in cells per square millimeter) of human (hNA) cells were quantified at 4 and 12 weeks after implantation.
Mean ± SEM, n = 4–9 per group; **p < 0.01 versus mCherry, #p < 0.05, ##p < 0.01 versus 4 weeks, two-way ANOVA, Bonferroni’s post-test.
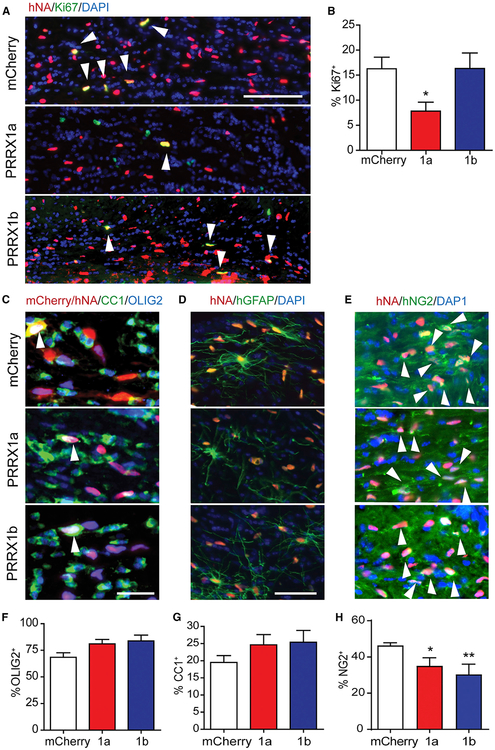
We hypothesized that the reduced cell density would be the result of impaired proliferation. Next, we determined the proportion of Ki67+ cells among hNA+ cells (Figures 4A and 4B). Because hOPC density is negatively correlated with proliferation (r = −0.836; p < 0.01) (Figure S2) (Windrem et al., 2008), proliferation was calculated from engrafted regions with equivalent density. PRRX1a overexpression significantly reduced hOPC proliferation compared to mCherry (7.8% ± 1.8% versus 16.3% ± 2.3%; n = 5 per group, p < 0.05, one-way ANOVA, Dunnett’s post-test). In contrast, PRRX1b overexpression did not significantly impair proliferation (16.4% ± 3.1%).
Figure 4. PRRX1 Alters Proliferation, but Not Differentiation, of hOPCs In vivo.
The fate and rate of hOPC differentiation was determined at 12 weeks post-implantation.
(A) Human nuclear antigen-positive (hNA+) cells (red) and Ki67 (green) in corpus callosum (white arrowheads indicate double-labeled cells).
(B) Percentage of Ki67+ among transplanted hNA+ was determined as a measure of proliferation (n = 5–9 per group; *p < 0.05 versus mCherry, one-way ANOVA, Dunnett’s post-test).
(C–H) Serial sections were stained for oligodendrocyte lineage-expressed OLIG2 (blue) and CC1, a marker of post-mitotic oligodendrocytes (green) (arrowheads indicate triple-labeled cells) (C). The extent of OLIG2+ lineage commitment (F), and the rate of human CC1+ oligodendrocyte differentiation among hNA+OLIG2+ cells was determined (G) (mean ± SEM, n = 4–9 per group). (D) Human astrocyte differentiation by transplanted cells was similar between groups (human-specific GFAP antibody). (E) hOPC fate was determined using human-specific NG2 antibody (hNG2) and quantified (H) (*p < 0.05 and **p < 0.01 versus mCherry, one-way ANOVA, Dunnett’s post-test).
Scale: 100 μm (A), 25 μm (C), and 50 μm (D).
Progressive Migration and Colonization of Shiverer White Matter Is Inhibited by PRRX1a
To determine whether the reduction in cell density and migration at 12 weeks was due to reduced rates of migration and colonization, we euthanized a second cohort of transplanted animals at 4 weeks. Migration and cell engraftment of mCherry hOPCs were substantially reduced compared with the rates at 12 weeks (Figures 3B and 3C). In PRRX1a-transplanted animals, the distribution of hOPCs at 4 weeks was similar to that at 12 weeks. This indicated that PRRX1a-overexpressing cells did not undergo progressive expansion and engraftment.
We hypothesized that PRRX1a-mediated arrest of shiverer colonization was due to induced quiescence and that active hOPCs would divide more rapidly and outcompete quiescent PRRX1a+ hOPCs when transplanted into the same microenvironment. To test this hypothesis, we infected separate hOPC cultures isolated from the same donor with either mCherry or PRRX1a-FLAG LV. Cells were mixed (1:1 ratio) immediately before unilateral implantation (n = 5) (Figure S3A). At 4 weeks, we observed hNA+ cells clustered at the implantation site and migrating hNA+ cells across the corpus callosum (Figure S3B). At the site of injection, >99% of hNA+ cells colabeled with mCherry. A few cells lacked detectable mCherry or PRRX1-FLAG-expression and likely represented uninfected hOPCs or possible silenced LV. On the contralateral side, all hNA+ cells expressed mCherry and morphologically resembled migrating bipolar precursors. We did not observe FLAG immunoreactive PRRX1a+ hOPCs in either site across five animals. In contrast, 11 animals were engrafted with hOPCs isolated from five human samples, with each successfully yielding PRRX1a-FLAG+ cells upon sectioning at 4 and 12 weeks post-transplant. The complete absence of PRRX1a-FLAG+ cells following mixed transplantation using matched donors indicates that the mCherry-infected hOPCs outcompeted PRRX1a+ hOPCs in vivo, possibly by competition for limited trophic support.
PRRX1 Does Not Substantially Promote Myelination by hOPCs
A possible explanation for reduced migration and engraftment following PRRX1 overexpression was precocious differentiation. At 12 weeks, we found no difference in the proportion of OLIG2+ cells among engrafted human cells (mCherry, 66.5% ± 4.2%; PRRX1a, 81.0% ± 4.2%; PRRX1b, 83.4% ± 7.0%) (Figures 4C and 4D). We did not observe differences in the proportion of CC1+ oligodendrocytes among OLIG2+hNA+ cells (24.6% ± 3.0% and 25.4% ± 3.4% for PRRX1a/b, respectively, versus 19.5% ± 2.0%; n = 4–8 per group) (Figure 4F). We observed a significant decrease in the proportion of hNG2+ hOPCs following PRRX1 overexpression (35.0% ± 2.6% and 30.3% ± 3.3% for PRRX1a/b, respectively, versus 46.3% ± 0.9%) (Figures 4D and 4H). There was no difference in human GFAP+ astrocyte differentiation between groups (Figure 4D). In all three groups, we observed a similar distribution of human astrocytes throughout the corpus callosum, similar to previous observations (Sim et al., 2011). Altogether, these observations suggest that PRRX1 did not induce differentiation but instead acted to decrease the available pool of progenitors as a consequence of reduced cell-cycle entry and migration and thereby prevented effective engraftment.
PRRX1 Induces a Quiescent-like Gene Expression Signature
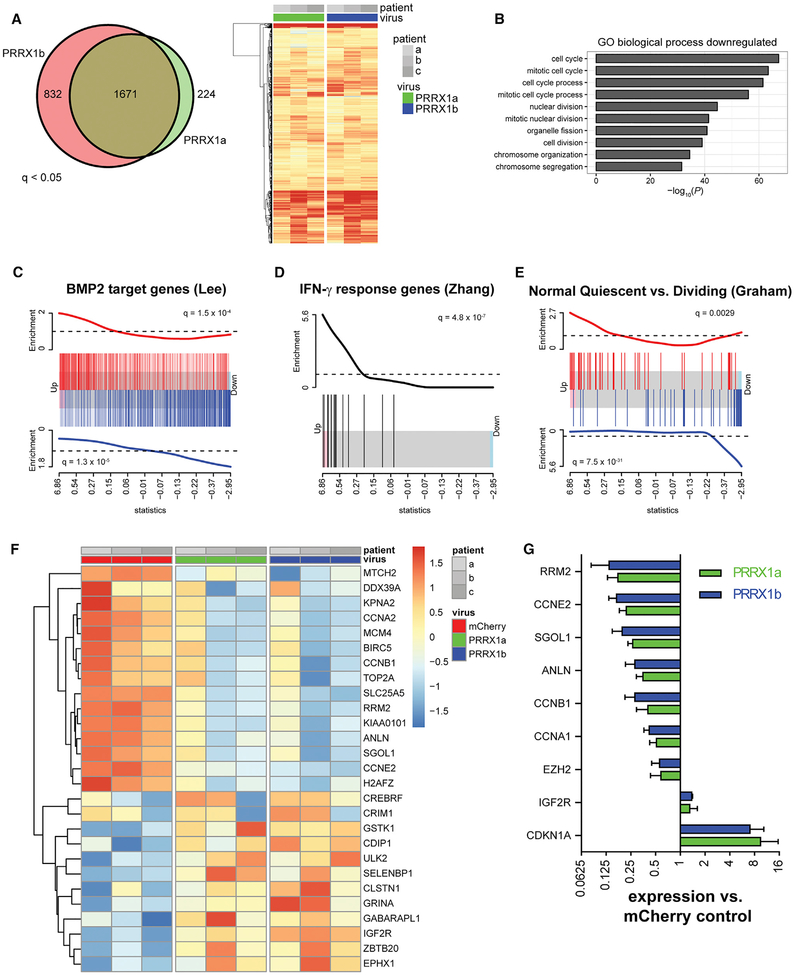
We next investigated the transcriptional mechanisms by which PRRX1 regulated hOPC proliferation and migration. We performed RNA sequencing (RNA-seq) analysis 2 days following infection with either PRRX1a/b or mCherry LV (n = 3, patient samples). Differential gene expression analysis identified more than 1,500 genes upregulated by PRRX1 compared to mCherry (q < 0.05) (Figure 5A). We found profound and highly significant downregulation of cell-cycle-related genes (cell-cycle term gene ontology [GO]: 0007049, p = 5.4 × 10−68). Almost all GO terms identified involved the regulation of cell division, proliferation, DNA replication, and cell cycle, indicating that the most profound effect of PRRX1 was to repress the cell cycle.
Figure 5. PRRX1 Overexpression Induces a Gene Signature Associated with Quiescence.
RNA-seq analysis was performed 48 hr after infection with PRRX1 or mCherry LV (n = 3 individual patient samples).
(A) Differential gene expression analysis identified >1,500 genes upregulated compared to mCherry hOPCs (q < 0.05). Venn diagram shows the high extent of overlap between genes upregulated between PRRX1a and PRRX1b. Likewise, the heatmap of all upregulated genes reflects strong correlation between PRRX1a and PRRX1b transcriptional responses (expression relative to matched mCherry-infected hOPCs; increasing intensity of red indicates upregulation).
(B) Gene ontology-based over-representation analysis revealed that cell-cycle-related biological process terms were significantly downregulated in response to PRRX1 overexpression.
(C–E) Gene set enrichment analysis showed coordinated regulation of transcripts associated with BMP2 (C), IFN-γ (D), and quiescence (E). Barcode plots are shown, and the x axis is the rank order of hOPC genes by the F-test statistic in response to PRRX1a expression. Red bars indicate the position of upregulated genes, and blue bars indicate the downregulated genes in each gene set. The red and blue traces above and below show relative enrichment of up- and downregulated genes, respectively (q value is shown).
(F) Heatmap of significantly regulated genes associated with the stem cell quiescence signature (Cheung and Rando, 2013). Scale indicates expression relative to median level across RNA samples.
(G) qPCR of cell-cycle genes following PRRX1 infection (mean ± SEM, n = 3).
Most genes were similarly regulated by PRRX1a and PRRX1b overexpression (Figure 5A). To investigate splice-specific PRRX1-dependent expression, we used the PRRX1 transcript level as an additional covariate. We found that PRRX1a and PRRX1b induced the differential regulation of 177 genes (PRRX1a versus PRRX1b, q < 0.05). A direct comparison of the PRRX1a/b transcriptomes revealed increased expression of 24 genes associated with migration following PRRX1b overexpression (ARC, MMP9, etc.), while only 10 genes were downregulated (Figure S4A). PRRX1b upregulated a significant number of genes associated with cell migration and locomotion (GO: 004001, p = 4.21 × 10−11). This is consistent with PRRX1b overexpression leading to a more promigratory phenotype in vitro and following transplantation versus PRRX1a. Among PRRX1a-specific migration-associated transcripts, the laminin receptor integrin β4 (ITGB4) was differentially upregulated by PRRX1a relative to PRRX1b and mCherry, while seizure protein 6-like (SEZ6L) was downregulated (Figure S4C). Integrin α6β4 is not typically expressed by OPCs but can mediate stable adhesion with basement membranes, while cell surface protein SEZ6L is an OPC-expressed transcript that is downregulated during differentiation (Pol et al., 2017; Zhang et al., 2014).
Next, we performed gene set enrichment analysis (GSEA) using the MSigDB C2 database of 5,000 gene sets (Subramanian et al., 2005). We noted that the profile of PRRX1a+ cells resembled that of the response to BMP2 signaling (q = 1.5 × 10−4 and 1.3 × 10−5 for up- and downregulated targets, respectively) (Lee et al., 2007) (Figure 5B). Furthermore, we observed that the PRRX1 gene signature was significantly enriched for genes responsive to interferon stimulation (ZHANG_INTERFERON_RESPONSE, q = 4.8 × 10−7, Zhang et al., 2005; DER_IFN_GAMMA _RESPONSE_UP, q = 4.1 × 10−6, Der et al., 1998; and SANA_RESPONSE_TO_IFNG_UP, q = 8.1 × 107, Sana et al., 2005) (Figure 5C). As such, these data suggested that PRRX1 overexpression has a similar mode of action to BMP and interferon signaling in hOPCs.
We also found common inductive and repressive targets between PRRX1a and transcription factor FOXO3 (q = 1.4 × 10−7) (Delpuech et al., 2007). FOXO3 is upregulated in quiescent stem cells (Cheung and Rando, 2013) and is required for adult NPC maintenance (Paik et al., 2009; Renault et al., 2009). Among the top 20 most significantly enriched signatures was the regulation of quiescence-associated transcripts in CD34+ hematopoietic stem cells in quiescent versus dividing phenotypes (q = 7.5 × 10−31) (Graham et al., 2007) (Figure 5D). Finally by GSEA, we identified significant enrichment of a consensus quiescent stem cell gene expression signature following PRRX1a overexpression (Figures 5E and 5F) (Cheung and Rando, 2013).
Among quiescence genes, we noted the downregulation of cell-cycle components. We confirmed the differential regulation of these by qRT-PCR (Figure 5G) (n = 3 fetal samples). Altogether, the gene expression establishes that PRRX1a overexpression induced quiescence in hOPC and suggested that PRRX1a may be regulated by BMP or interferon-gamma (IFN-γ) treatment.
Cytokine-Induced Quiescence Upregulates PRRX1 Expression in hOPC
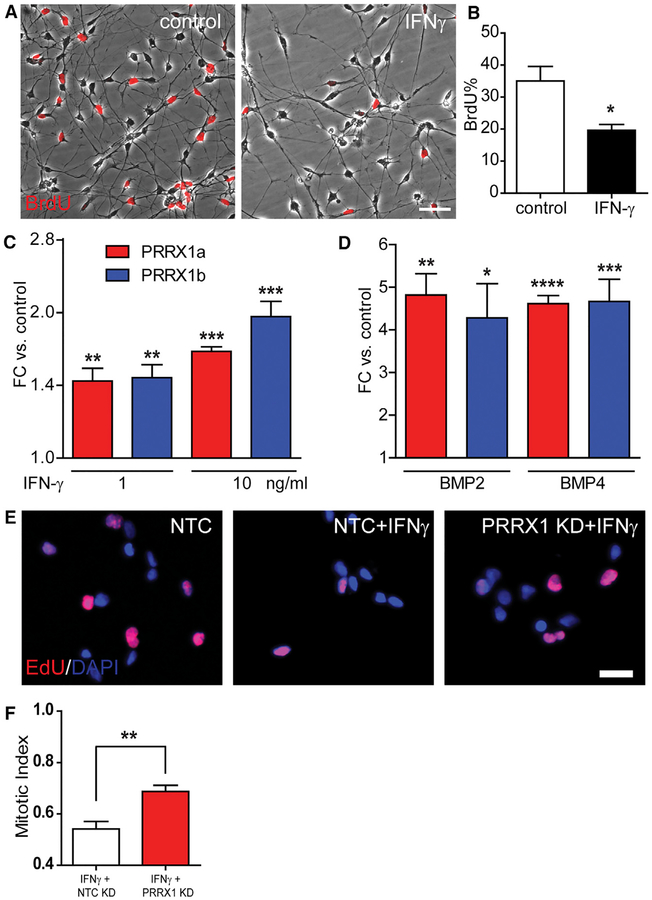
Both IFN-γ and BMP signaling are involved in the regulation of progenitor quiescence (Tanner et al., 2011; Mira et al., 2010). As in rodent OPCs, IFN-γ treatment did not induce hOPC apoptosis at a low dose (cleaved caspase-3, up to 10 ng/mL). IFN-γ significantly reduced BrdU incorporation (20% ± 1.8% versus 35% ± 4.6%; n = 3 per group, p < 0.05 by paired Student’s t test) (Figures 6A and 6B). Flow cytometry-based cell-cycle analysis revealed a 61% ± 2% reduction in S-phase entry following 24-hr treatment with IFN-γ (n = 2 fetal samples) (Figure S5).
Figure 6. IFN-γ and BMP Induce Endogenous PRRX1 Expression.
(A) Phase contrast images of hOPCs overlaid with BrdU immunofluorescence in control and after IFN-γ treatment for 7 days (10 ng/mL).
(B) Quantification of proliferating hOPC after IFN-γ treatment.
(C and D) Cytokine treatment significantly increased PRRX1a and PRRX1b mRNA following exposure to IFN-γ (C) or 50 ng/mL BMP (D) (n = 3–4 fetal samples each; *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA).
(E) To determine whether PRRX1 upregulation was necessary for IFN-γ-induced quiescence, fetal hOPCs were transfected with NTC (non-targeting control) or PRRX1 siRNA. The effect of PRRX1 knockdown on IFN-γ-induced hOPC quiescence was assessed by EdU (8-hr pulse).
(F) Mitotic index of IFN-γ-treated cells was calculated relative to the untreated matched hOPCs (mean ± SEM, n = 4 fetal samples; **p < 0.01, paired t test). Scale: 100 μm (A) and 50 μm (E).
Similar to rodent NPCs, we found that BMP treatment reduced the proportion of 5-Ethynyl-2’-deoxyuridine (EdU)+ hOPCs by 27% ± 4% (Figure S5). Because BMP induces astrocyte differentiation from hOPCs (Sim et al., 2011), we further investigated hOPC fate and OLIG2+ hOPC proliferation (Figure S6). Although BMP treatment increased the proportion of GFAP+ cells, the fraction of OLIG2+ cells remained greater than 75% at 24 hr (89% ± 4.1% to 78% ± 3.3%, n = 2 fetal samples). Consistent with induced hOPC quiescence, BMP treatment significantly reduced the fraction of dividing OLIG2-expressing EdU+ cells (n = 3 fetal samples) (Figure S6), thus indicating that both IFN-γ and BMP can inhibit cell-cycle entry in hOPCs.
Given the coregulation of PRRX1 and both BMP2 and IFN-γ target genes, we next asked whether PRRX1 was regulated by BMP or IFN-γ. IFN-γ increased PRRX1a and PRRX1b mRNA (1.66- ± 0.03-fold and 1.96- ± 0.15-fold, respectively, at 10 ng/mL IFN-γ relative to untreated control; n = 3 per group, p < 0.005, one-way ANOVA, Dunnett’s post-test) (Figure 6C). Likewise, 50 ng/mL BMP2 and BMP4 upregulated both isoforms by >4-fold. Specifically, BMP2 increased PRRX1a and PRRX1b levels 4.82 ± 0.50 and 4.28 ± 0.81, respectively (n = 3 per group, p < 0.05, paired Student’s t test). BMP4 increased PRRX1a and PRRX1b levels 4.61 ± 0.19 and 4.67 ± 0.52, respectively, compared to controls (n = 4 per group, p < 0.001, paired Student’s t test) (Figure 6D). These data indicate that endogenous PRRX1 mRNA was upregulated by cytokines capable of inducing hOPC quiescence.
PRRX1 Is Necessary for IFN-γ-Induced Quiescence in hOPCs
To determine whether PRRX1 mRNA upregulation was necessary for cytokine-induced quiescence, we transfected hOPCs with PRRX1 or non-targeting control (NTC) small interfering RNA (siRNA). The effect of PRRX1 knockdown (KD) on IFN-γ-induced quiescence was assessed by analysis of S-phase entry (8-hr EdU pulse). PRRX1 KD in the absence of IFN-γ did not significantly influence hOPC proliferation (0.90 ± 0.08 mitotic index, defined as the percentage of EdU positive relative to NTC control, n = 3 fetal samples). As expected, IFN-γ reduced the mitotic index of hOPCs transfected with NTC siRNA by 46% (0.54 ± 0.03, n = 4 fetal samples) (Figures 6E and 6F). PRRX1 KD significantly increased the mitotic index of IFN-γ-treated cells, increasing by 21% (0.69 ± 0.02, paired t test, p < 0.001). The effect of PRRX1 siRNAi on IFN-γ-induced cell-cycle arrest was verified using two separate PRRX1 siRNAs, which similarly increased proliferation following cytokine treatment (0.67 ± 0.06 and 0.64 ± 0.01, respectively, n = 3).
To better understand the mechanisms by which PRRX1 regulates hOPCs, we performed RNA-seq analysis following siRNA and/or IFN-γ treatment (Figure S7). RNA-seq confirmed PRRX1 KD (77% KD, q = 7.3 × 10−9). 646 and 796 genes were significantly regulated by PRRX1 KD in the presence of PDGFAA and neurotrophin-3 (NT-3) alone and following IFN-γ treatment, respectively (q < 0.05). In actively dividing hOPCs, PRRX1 KD resulted in upregulation of cell proliferation genes (GO: 0008283, topGO analysis; q = 4 × 10−7) and locomotion genes (GO: 0040011, q = 8.0 × 10−5), consistent with the observed repressive effects of PRRX1 (Figure S7). Basal PRRX1 expression was necessary for expression of cholesterol biosynthesis genes (HORTON_SREBF_TARGETS, q = 0.0059), and PRRX1 KD resulted in small but significant downregulation of oligodendrocyte lineage genes (SOX10 and OLIG2) and differentiation genes (MBP and PLP1) (Figure S7). Importantly, PRRX1 KD in the presence of IFN-γ resulted in downregulation of genes associated with the IFN-γ-response (SANA_RESPONSE_TO_IFNG_UP, q = 1.9 × 10−5) (Figure S7) and establishes a role for PRRX1 in meditating the IFN-γ-dependent responses.
PRRX1 Is Expressed by Ki67-Negative OPCs following Demyelination and Induced by IFN-γ
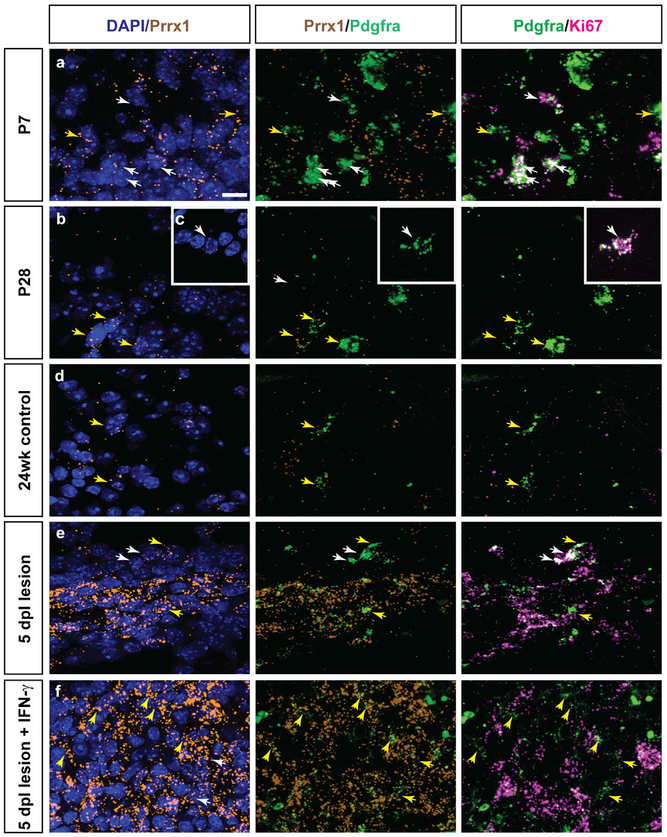
To further investigate PRRX1 expression in vivo, we performed semiquantitative fluorescence in situ hybridization. As expected, we observed high levels of Prrx1 mRNA in the adult mouse subventricular zone (Shimozaki et al., 2013). At post-natal day 7, we found that many cells in mouse white matter expressed low levels of Prrx1 (Figure 7A). In the corpus callosum, dividing OPCs defined as Pdgfra+Ki67+ cells (white arrows) were largely Prrx1 negative. In contrast, a small subpopulation of Pdgfra+ OPCs expressed Prrx1, with most of these cells negative for Ki67 (Figure 7A, yellow arrows). While Prrx1 expression in non-OPCs decreased significantly with age, the pattern of Prrx1 expression in OPCs became more pronounced after the peak of oligodendrocyte generation. In postnatal day 28 corpus callosum and in aged adult spinal cord at 24 weeks, Prrx1 expression was largely restricted to Pdgfra+Ki67−OPCs (Figures 7B and 7D). Dividing Ki67+ OPCs were negative or Prrx1low (Figure 7C). Next, we examined the expression of Prrx1 mRNA following demyelination induced by lysolecithin injection into the spinal cord of 24-week-old adult mice. We examined the expression of Prrx1 at 5 days post-lesion (dpl), because OPCs at this stage have been recruited from surrounding white matter and are actively dividing in and around the lesion (Sim et al., 2002) (data not shown). Prrx1 was substantially upregulated within the lesion and was expressed by multiple cell types, including Pdgfra+ OPCs (Figure 7E). Prrx1 expression was predictive of the Ki67 status of OPCs, with Prrx1lowKi67+ dividing OPCs found predominantly at the lesion edge and Prrx1highKi67−quiescent OPCs in the lesion center. Finally, to determine whether IFN-γ could upregulate Prrx1 expression in vivo, we coinjected IFN-γ into demyelinating lesions (300 U). At 5 dpl, Prrx1 was strongly enhanced throughout the lesion and surrounding tissue (Figure 7F). OPC division was inhibited by IFN-γ, with almost all Pdgfra+ cells within the lesion exhibiting a Prrx1highKi67−phenotype, suggesting that IFN-γ-mediated signaling could induce Prrx1 and induce OPC quiescence.
Figure 7. Prrx1 mRNA Expression in Mouse OPCs and following Demyelination.
PRRX1 expression was analyzed in mouse brain and spinal cord during normal development and following demyelination.
(A–C) Triple-labeled fluorescence in situ hybridization (FISH) of postnatal day 7 (A) and day 28 (B and C) in mouse corpus callosum: Prrx1 mRNA (orange), Pdgfra mRNA (green), and Ki67 mRNA (pink). White arrows denote Prrx1low or Prrx1-negative Pdgfra+ OPCs that coexpressed Ki67, a marker of proliferating cells. Yellow arrows denote Prrx1high Pdgfra+ OPCs. These cells were typically Ki67 negative.
(D) 24-week adult mouse spinal cord ventral white matter.
(E) 24-week adult mouse spinal cords were lesioned via direct injection of lysolecithin.
(F) Coinjection of lysolecithin and IFN-γ into ventral white matter. Animals were sacrificed at 5 days post-lesion (dpl) (n = 3 per group).
Scale: 20 μm.
DISCUSSION
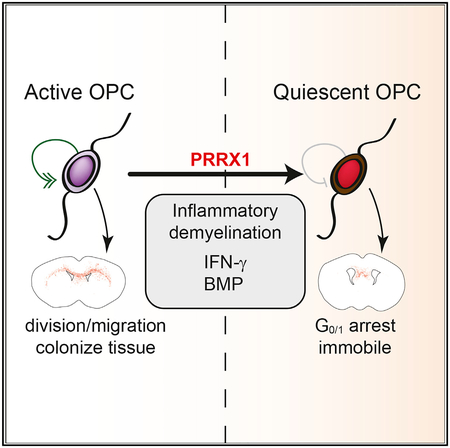
Adult OPCs are considered quiescent due to their slow rate of proliferation (Psachoulia et al., 2009) and reduced expression of cell-cycle genes (Belachew et al., 2002; Lin et al., 2009). Here, we show that quiescence can be induced in fetal hOPCs by PRRX1 overexpression and that induction of quiescence by cytokine treatment depends on PRRX1 upregulation. Prrx1 mRNA expression in mouse was associated with non-dividing OPCs and was induced following demyelination. These data provide insight into the regulation of cell-cycle dynamics and quiescence in hOPCs that may help uncover the mechanisms important for their homeostasis and activation following injury.
The principal effect of PRRX1 on hOPCs was the induction of a reversible cell-cycle arrest. We observed a dominant effect of PRRX1a both in culture and following transplantation. The expression data demonstrate that both PRRX1 isoforms are upregulated in OPCs compared to NPCs and subsequently downregulated during oligodendrocyte differentiation. Although forced expression of PRRX1 induced cell-cycle arrest, it did not prevent the progression of cells to mature oligodendrocytes, because comparable numbers of oligodendrocytes were observed in vitro and in vivo, respectively. Previously, PRRX1 expression in human and rodent NPCs induced some features of OPC fate (Wang et al., 2014; Shimozaki et al., 2013). Similar results were reported in a study by Mira et al. (2010), in which a substantial proportion of quiescent (Ki67−) NPCs were found to express oligodendroglial and astrocytic differentiation markers. This is further evidence that quiescence in OPCs is distinct from processes regulating terminal differentiation, which is not an obligatory consequence of cell-cycle exit (Levine et al., 2001). Given that PDGFαR+ hOPCs represent a mixture of different developmental stages or cell-cycle status (Dietz et al., 2016), it is also possible that PRRX1 regulates quiescence in a subset of OPCs. Only a small proportion of fetal hOPCs expressed high levels of PRRX1 by single-cell RNA-seq, and only a sub-population of OPCs express PRRX1 during mouse development, which suggests that this transcript may be differentially activated by the local environment or niche.
PRRX1 induced profound repression of cell-cycle-associated transcripts. Downregulation of CDK2/cyclin E (CCNE1) corresponds with their essential role in OPC proliferation (Belachew et al., 2002). Likewise, PRRX1 expression was associated with highly significant upregulation of cell-cycle inhibitors p21CIP1 (CDKN1A) and p15INK4B (CDKN2B). Surprisingly, we noted significant downregulation of other inhibitors, such as p57KIP1 (CDKN1B) and p16INK4A (CDKN2A). While p21 and p57 are often coordinately regulated in vitro, they are known to have distinct and non-redundant roles in OPCs (Zezula et al., 2001). The upregulation of p21 is consistent with quiescence, as observed in NPCs (Kippin et al., 2005), and downregulation of p57 may represent a resetting of the oligodendrocyte timer (Durand et al., 1998), thereby providing a stable source of progenitor cells capable of future expansion and differentiation. PRRX1 is a known inducer of epithelial-mesenchymal transition (EMT) in chick (Ocaña et al., 2012). PRRX1 does not induce expression of known mesodermal transcription factors in hOPCs. Likewise, although PRRX1 can induce SOX9 expression (Reichert et al., 2013), we did not observe any effect in hOPCs. The pattern of PRRX1 target genes appears to be tissue and cell type specific.
Our transcriptional analyses implicated PRRX1 as a target gene for both BMP- and IFN-γ-induced quiescence. While prolonged BMP treatment of rodent and hOPCs induces astrocyte differentiation (Mabie et al., 1997; Sim et al., 2006, 2011), brief BMP exposure induces a transient state before astrocytic commitment that can revert OPCs to a multipotent stem cell-like state characterized by SOX2 induction, chromatin remodeling, and increased neurogenic capacity (Kondo and Raff, 2000, 2004). Our data suggest that during the induction of this transient state, PRRX1 expression is upregulated, along with SOX2. BMP4 treatment of NPCs is known to induce reversible quiescence (Mira et al., 2010) and has been shown to upregulate PRRX1 mRNA (Martynoga et al., 2013). In that study, NFIX was identified as a key regulator of NPC quiescence. Therefore, it is intriguing that chromatin immunoprecipitation sequencing (ChIP-seq) revealed NFIX is bound directly to the promoter of PRRX1, suggesting that PRRX1 may partly contribute to BMP-induced quiescence in NPCs. This parallels with observations in other systems in which PRRX1 expression is induced by BMP (Ocaña et al., 2012; Salazar et al., 2016).
However, IFN-γ-induced PRRX1 upregulation appears to be relatively OPC specific. PRRX1 upregulation was not observed following IFN-γ treatment in several other peripheral cell types (Johnson-Huang et al., 2012; Calderon et al., 2008; Banno et al., 2003; Tassiulas et al., 2004), including microglia (Rock et al., 2005). IFN-γ induces quiescence in rodent glia (Agresti et al., 1996; Tanner et al., 2011), and at high doses, it causes cytotoxicity (Vartanian et al., 1995; Baerwald and Popko, 1998; Horiuchi et al., 2006; Wang et al., 2010). Excessive IFN-γ levels are known to induce severe hypomyelination in CNS development (Corbin et al., 1996; LaFerla et al., 2000) and demyelination (Horwitz et al., 1997). Enforced IFN-γ expression in adult brain impaired remyelination due to either reduced OPC proliferation or differentiation (Lin et al., 2006). In MS lesions, IFN-γ mRNA is associated with perivascular inflammatory cuffs (Woodroofe and Cuzner, 1993), which is likely in apposition with migrating OPCs, which use brain microvasculature to migrate in developing brain (Tsai et al., 2016). There is a clear pathogenic role of IFN-γ in MS (Popko et al., 1997). For example, IFN-γ treatment resulted in a severe exacerbation of MS (Panitch et al., 1987). IFN-γ levels are elevated before exacerbations and remain elevated in progressive MS, suggesting a potential pathogenic role for chronic low levels of IFN-γ (Beck et al., 1988). In this study, we found that Prrx1 mRNA was upregulated in OPCs following demyelination and further induced by IFN-γ treatment. While the expression of PRRX1 has not been assessed in MS lesions, the induction of pathological quiescence by cytokine-mediated PRRX1 upregulation may prevent effective OPC recruitment in MS lesions by impairing proliferation and migration away from blood vessels.
We found that both PRRX1 variants impaired proliferation in vitro but that only PRRX1a was sufficient to reduce migration. PRRX1a influenced hOPC engraftment, resulting in a profound failure of cell expansion and migration in vivo. PRRX1a/b-specific transcripts were identified, and a significant over-representation of locomotion-associated genes were specifically upregulated by PRRX1b relative to PRRX1a. This is consistent with the observed negative effect of PRRX1a over-expression on migration in vitro and following transplantation. Furthermore, this suggests that PRRX1b may relieve the inhibitory effects of quiescence on cell motility by directly driving migration. In addition, increased expression of integrin α6β4 by PRRX1a may prevent detachment from endothelial basement membrane and thereby reduce OPC migration along microvasculature (Tsai et al., 2016). Previous studies in pancreatic cancer have indicated differential effects of PRRX1a/b, with variant-specific effects on proliferation, migration, and metastasis (Takano et al., 2016; Reichert et al., 2013). These differences were attributed to isotype-specific cofactor interactions. While identical in DNA binding homeobox domain, mouse PRRX1a (equivalent to human PRRX1b) contains an OAR domain with unknown function (Norris and Kern, 2001). PRRX1 interacts with SOX2, and its activity is greatly enhanced by SOX2 coexpression in NPCs (Shimozaki et al., 2013). The splice-specific mechanisms remain to be determined.
Following shiverer transplantation, hOPCs outcompete their host counterparts such that by 1 year, most OPCs were derived from donor human cells (Windrem et al., 2014). Herein, hOPC migration approximated to at least 0.2 mm/week. The 2.5- to 3-fold expansion in cell density between 8 and 12 weeks was consistent with previous studies (Sim et al., 2011; Windrem et al., 2008). In contrast, the abject failure of PRRX1a+ hOPCs to engraft following mixed transplantation with mCherry+ cells indicates that PRRX1 provides a strong competitive disadvantage to engrafted cells. Because PRRX1a+ cells were present at 12 weeks when transplanted alone, this result suggests that PRRX1+ hOPCs were either unable to compete for limiting levels of trophic factors immediately following transplantation or, after initial engraftment, that control OPCs outcompeted PRRX1+ cells for trophic support and subsequently died. This is reminiscent of previous rat transplantation studies, which suggested competition between endogenous and transplanted OPCs and that a niche could only be created by depleting the host progenitor pool (Franklin et al., 1996). Among several possible trophic factors, PDGF-AA clearly regulates OPC density (Woodruff et al., 2004), and as such, PRRX1-induced changes in signaling competence might contribute to the failure of these cells to compete with control hOPCs.
Numerous studies have demonstrated that OPCs in adult human and rodent brain are relatively quiescent with a prolonged cell cycle (Belachew et al., 2002; Dawson et al., 2003; Dowling et al., 1997; Geha et al., 2010; Morris et al., 1994; Psachoulia et al., 2009; Rivers et al., 2008; Simon et al., 2011; Young et al., 2013). Cell-cycle quiescence preserves the longevity of stem cell pools to prevent excessive proliferation and exhaustion of the proliferative capacity of adult stem and progenitor cells. The linkage of progenitor exhaustion and aging has been well established in other systems (López-Otín et al., 2013). OPC exhaustion was proposed as a mechanism for failure of remyelination in a subset of chronic demyelinating MS lesions (Lucchinetti et al., 1996), and OPC recruitment is substantially delayed in aged animals following demyelination (Sim et al., 2002). Yet OPC exhaustion does not appear to be relevant in several animal modules of OPC depletion following either irradiation (Chari and Blakemore, 2002) or focal toxic injury (Penderis et al., 2003). Likewise, several studies have identified OPCs in MS lesions, albeit at densities lower than expected for successful repair (reviewed in Dietz et al., 2016). Thus, it remains unclear whether OPC exhaustion contributes to MS progression and chronic disease.
It is possible that therapies aimed at accelerating the activation of resident OPCs from a state of pathological quiescence would greatly benefit recovery in patients with MS or other demyelinated lesions. Activation of adult OPC proliferation by upregulation of the transcription factor SOX2 is required for efficient remyelination (Zhao et al., 2015). Because failure to remyelinate has been attributed to impaired activation of OPC proliferation (Wolswijk, 1998; Morris et al., 1994; Dowling et al., 1997), this process may also occur in chronic MS lesions. However, further studies are needed to determine whether OPCs in and around chronic lesions are quiescent and whether it is the inability to overcome this pathological quiescence that impedes remyelination and repair. The results presented here suggest that PRRX1 may be an important target for such studies and those examining the regulation and maintenance of oligodendrocyte progenitors in adult brain.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fraser Sim (fjsim@buffalo.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and housing conditions
All experiments were performed according to protocols approved by the University at Buffalo’s Institutional Animal Care and Use Committee. Animals were maintained in specific pathogen free facility with continuous access to food and water. Health of mice were monitored regularly by veterinarian. Mice used in this study were 1–24 weeks old for development and demyelination experiments while for transplantation mice were less than 1 week. For all experimental groups involving cell transplantation, animals could not be sexed at postnatal day 1–3 and were assigned randomly to groups receiving LV-infected hOPCs. After weaning recipient mice were sexed and found to be in a ratio of 1:1.3 male:female. For other experiments examining expression of Prrx1 mRNA, both males and females were used (5:7 male:female). Mice were maintained on diurnal 12 hr light/dark cycle.
Human CD140a/PDGFαR cell culture
Fetal brain tissue samples (17–22 weeks gestational age) were obtained from patients who consented to tissue use under protocols approved by the University at Buffalo Research Subjects Institutional Review Board. Forebrain tissue was minced and dissociated using papain and DNase as previously described (Conway et al., 2012). Magnetic sorting of CD140a positive cells was performed as described (Pol et al., 2013).
Cell Lines
HEK293T cells were obtained from American Type Culture Collection (ATCC; CRL-11268). HEK293T-17 cells were grown in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen) and penicillin/streptomycin (Invitrogen) under optimum growth conditions (37°C, 5% CO2).
METHOD DETAILS
Quantitative PCR analyses
RNA was extracted from freshly sorted human cells and after 1–4 d or indicated treatment length in culture using a total RNA isolation kit (Omega Bio-Tek). cDNA was synthesized using SuperScript III reverse transcriptase (ThermoFisher Scientific). Human-specific primers designed using NCBI Primer-BLAST or Primer Express (v1; Applied Biosystems) (Table S1)(Wang et al., 2014). GAPDH was used as a control gene. Samples were run in duplicate and gene expression was calculated by ΔΔCt analysis.
Lentivirus cloning and packaging
Complete coding sequences for a flag-tagged PRRX1a and PRRX1b with additional restriction enzyme sites (SpeI and XhoI) were obtained by PCR using the following primer sequences: PRRX1b, 5′-AAATCTAGACCCATGACCTCCAGCTACGGGCACG-3′ (forward), 5′-AAACTCGAGCCTCAGTTGACTGTTGGCACCTGG-3′ (reverse); PRRX1a-flag, 5′-TTTACTAGTGACCATGACCTCCA GCTA-3′ (forward), 5′-AAACTCGAGTTACTTGTCGTCATCGTCTTTGTAGTCGAATCCGTTATGAAGCCC-3′ (reverse). PCR products were then cloned into a lentiviral backbone in place of the mCherry sequence (excised with SpeI and XhoI) (derived from pTRIP-EF1a, in Sevin et al., 2006) (gift of Abdel Benraiss, University of Rochester).
Lentiviruses were prepared as described previously (Sim et al., 2006). Briefly, following triple transfection of HEK293T cells with pTRIP and packaging plasmids pLP/VSVG (ThermoFisher Scientific) and psPAX2 (AddGene), viral supernatant was collected at 48 and 72 h. Titration of virus was performed by flow cytometry for mCherry-expressing virus and using real-time quantitative PCR for the WPRE sequence (Geraerts et al., 2006). Human primary cells were infected at 1 MOI for 24 h followed by complete media replacement unless stated otherwise.
Lentivirus infection of hOPCs
Human CD140a+ OPCs were plated at 5 × 104/ml on poly-L-ornithine- and laminin-coated tissue-culture plates and cultured for 48 h in serum-free medium (SFM) supplemented with 20 ng/ml PDGF-AA and 5 ng/ml NT-3 (PeproTech). Cells were then infected with the above-described lentiviruses at 1 MOI and maintained for at least 24 h prior to subsequent experiments.
Proliferation assay
Human CD140a+ OPCs were infected with lentivirus as described above and cultured for 48 h in SFM supplemented with 20 ng/ml PDGF-AA and 5 ng/ml NT-3. BrdU (30 μM, Sigma) was then added, and cells were fixed 24 h later. Cells were immunostained for BrdU (1:1000; AbD Serotec) and the percentage of BrdU+ DAPI-stained nuclei was obtained from 5–6 fields for each fetal sample.
Migration assay
Human CD140a+ OPCs (5 × 104/ml) were cultured on laminin-coated membrane inserts with 8-μm diameter pores (Corning) and infected for 24 h with lentiviruses. Cells were maintained in medium containing 20 ng/ml PDGF-AA and 5 ng/ml NT-3 for 2 d. PDGF was then removed for 24 h, and subsequently reintroduced to the bottom chamber only at 100 ng/ml. After 16 h, cells were fixed with a 4% paraformaldehyde solution and DAPI-stained nuclei were imaged (five fields per membrane) before and after cells were scraped from the top surface of the insert membrane. Migration was calculated as the number of remaining (migrated) nuclei divided by the total number of nuclei.
In vitro differentiation
Human CD140a+ OPCs were maintained in culture medium containing 20 ng/ml PDGF-AA and 5 ng/ml NT-3 for 2 d after lentivirus infection. Then, PDGF-AA was removed and 40 ng/ml T3 (Sigma) was added to promote OPC differentiation for 2 d. Cells were live-stained with O4 primary antibody (1:25; ATCC), then fixed for subsequent secondary and anti-GFAP (1:500; Sigma) immunocyto-chemistry. Six random fields were imaged with a 20 × objective for each condition, and at least 200 total cells were counted. The density of O4+ and GFAP+ cells are expressed as number of cells per field. Apoptosis post-infection was assessed by staining parallel cultures with anti-cleaved caspase-3 (1:400; Cell Signaling Technology). In addition, overall viability was measured in matched cultures of hOPCs 48 hr post-infection, by live staining with calcein (3μg/mL; Millipore) and DAPI (1μg/mL; Sigma) for 30 min at 37°C. Cultures were then imaged for calcein and DAPI fluorescence with a 10x objective with >20 fields analyzed per condition (n = 3 fetal human samples).
Transient PRRX1a expression
The PRRX1 coding sequence (CDS) was cloned into FUW-tetO-lox-NKX2.2 plasmid, as described previously (Wang et al., 2014). Human CD140a+ OPCs were coinfected with FUW-M2rtTA (Addgene; 20342) and FUW-PRRX1a LV, at 0.5 and 0.15 multiplicity of infection, respectively. Cells were maintained in SFM as described above and treated with doxycycline 48 hr post-infection (0.19 μM, VWR International). Media and doxycycline were replaced every other day. 0.19 μM doxycycline treatment had no effect on hOPC growth. Cultures were given a 24-hr terminal pulse of 5-bromo-2′-deoxyuridine (BrdU, 10 mM) prior to fixation for assessment of cell proliferation.
Flow cytometry cell cycle analysis
Human CD140a+ OPCs were infected with lentivirus as described above and cultured for 48 h in SFM supplemented with 20 ng/ml PDGF-AA and 5 ng/ml NT-3. EdU (50 μM) was then added, and cells were collected 24 h later and processed using the Click-iT EdU Flow Cytometry Assay Kit (ThermoFisher Scientific) according to the manufacturer’s protocol. DNA was stained with 1 μg/ml DAPI and cells were analyzed on a Fortessa SORP flow cytometer (BD Biosciences) and using FCS Express software (version 4; De Novo Software).
Transplantation into shiverer/rag2 mice
All transplantation experiments were performed using shiverer/rag2 mice (a gift of Dr. Steven A. Goldman, University of Rochester)(Windrem et al., 2008) according to protocols approved by the University at Buffalo Institutional Animal Care and Use Committee. A total of nine fetal brain samples at 17–20 weeks gestational age were used for transplantation experiments. CD140a+ hOPCs were cultured for up to 1 week in SFM containing PDGF-AA (20 ng/ml) and NT-3 (5 ng/ml) and frozen using ProFreeze (Lonza) prior to surgery. Prior to injection, cells were thawed, infected with lentiviruses, and cultured for 24–48 h before resuspending in HBSS (Corning) at 1 × 105/μl. To better account for human sample differences, cells from human tissue donors were often combined prior to infection and subsequent transplantation. Injections were performed as previously described (Sim et al., 2011). Briefly, postnatal day 2–3 pups were anesthetized by hypothermia and bilateral injections of 5 × 104 cells were made with pulled glass pipettes inserted directly through the skull into the corpus callosum (~2.5 mm posterior to bregma) to a depth of 1.1 mm. Animals were sacrificed after 4 or 12 weeks by transcardial perfusion with HBSS followed by a 4% formaldehyde solution.
Immunohistochemistry
Immunohistochemistry was performed on coronal sections (16 μm) of mouse forebrain, as previously described (Sim et al., 2011). Human cells were identified with mouse anti-human nuclei antibodies (hNA; 1:400; Millipore), and co-labeled with markers for oligodendrocytes (CC1, 1:50; OLIG2, 1:750; Millipore), astrocytes (GFAP, SMI21; 1:1000; Covance) and OPCs (hNG2, 9.2.27; 1:200; Millipore). Overexpression was detected using anti-PRRX1 (1:300; OriGene) and anti-FLAG (1:200; Sigma) antibodies. Proliferating cells were detected using antibodies against Ki67 (1:25; BD Biosciences) or BrdU (1:2000; AbD Serotec). Alexa Fluor (488, 594, and 647) secondary antibodies (ThermoFisher Scientific) were used at 1:500 dilution. Sections were imaged with a 20 × objective using an inverted fluorescence microscope (Olympus IX51). The quantification of hNA+ and double-positive cells was performed by counting cells in midline and lateral regions of the corpus callosum; images from at least 9 fields containing >200 cells were collected per animal. To assess the migration of implanted cells, sections spanning from bregma +3.9 to −3.4 mm (Paxinos and Franklin, 2004) were sampled every 160 μm and immunostained for hNA. The rostrocaudal migration distance was calculated as the distance between the first and last sections containing hNA+ cells.
RNA sequencing
Human fetal CD140a+ OPCs were maintained in proliferative conditions with SFM supplemented with 20 ng/ml PDGF-AA and 5 ng/ml NT-3 after lentivirus infection with mCherry (control) or PRRX1a overexpression or PRRX1b overexpression, for 2 days before mRNA extraction. RNA extraction and first stand synthesis was performed as described previously (Conway et al., 2012). RNA sequencing was performed at the UB sequencing core on an Illumina HiSeq2500 using 100 cycle paired-end sequencing. Sequences were aligned to the UCSC Hg19 mouse genome using tophat (v2.1.1) and counts per gene determined using htseq (v0.6.1). R/Bioconductor was used for subsequent analysis. Following loading of read counts using DESeq2, fragments per kilobase per million mapped fragments (FPKM) were calculated. hOPCs isolated from three separate fetal samples between 20- and 21-week gestation age were used. edgeR was used for differential expression analysis of read counts (Robinson et al., 2010). Genes with low counts (less than one count-per-million) in at least three libraries were removed from the analysis, and TMM normalized to account for differences between libraries (Robinson et al., 2010). A quasi-likelihood negative bionomial generalized log-linear model was fitted and empirical Bayes F-test performed to identify differentially expressed transcripts in PRRX1-expressing hOPCs. This was preferred as it provides a robust method given the small number of biological replicates (n = 3, fetal samples). p values were controlled for multiple testing by determining the false discovery rate (FDR). Pathway analysis was performed in edgeR using gene ontology (GO) enrichment analysis, KEGG pathway enrichment analysis, and gene set enrichment analysis of Broad C2 gene sets (v5) using camera. Heatmaps were plotted using on expression values following variance stabilizing transformation of normalized count data (DESeq2).
Treatment with IFN-γ and BMPs
hOPCs were then grown in SFM supplemented with 20ng/ml PDGF-AA and 5ng/ml of NT-3 prior to and during the treatment with IFN-γ or BMP (Peprotech). For mRNA-extraction, hOPCs were treated with IFN-γ for 18 hr. For BMP2/BMP4, hOPCs were treated for 40 hr before mRNA extraction. For assessment of proliferation following cytokine treatment, hOPCs were treated with IFN-γ (0–100 ng/ml) and added at each media change. The culture was given an 18-hr terminal BrdU pulse prior to fixation and assessment of mitotic index. For assessment of cell fate, cultures were treated with cytokines for 24 hr prior to fixation. OLIG2 (Millipore) and GFAP (Covance) were stained as described above.10 random fields were imaged at 20X, and at least 500 cells were counted in each condition.
siRNA-mediated PRRX1 knockdown
Stealth RNAi molecules were obtained from ThermoFisher Scientific, three targeting human PRRX1 (HSS143371, HSS143372, HSS143373), and a non-targeting control RNAi (NTC #12935112). hOPCs were seeded at 80% confluence onto 48-well plates. 24 hr later, combined or individual siRNAs (total concentration 100 nM) were transfected using Lipofectamine RNAiMAX transfection reagent (ThermoFisher Scientific). The three individual PRRX1 siRNA reduced mRNA expression by 65 ± 3, 45 ± 5 and 76 ± 3% respectively, while the pooled siRNA reduced expressed 73 ± 7% (n = 2, human samples). hOPCs were treated with IFN-γ 10 ng/ml at 8 hr post transfection and cells fixed 24hrs later. Proliferation was assessed by measurement of EdU incorporation (8-hour terminal pulse, 10 μM EdU) using the Click-iT Plus Edu Alexa imaging kit (ThermoFisher Scientific). Experiments were performed on hOPCs from individual human fetal samples, and the mitotic index was calculated relative to cells transfected with NTC in the absence of IFN-γ. Identical experiments performed in 6-well plates were performed with subsequent RNA extraction and RNA-seq as described above (n = 2, human samples).
Fluorescent in situ hybridization (FISH)
Triple label FISH was performed with probes targeting Prrx1 (GenBank: NM_011127.2), Pdgfra (GenBank: NM_011058.2), Ki67 (GenBank: NM_001081117.2) mRNA using RNAscope Fluorescence V2 assay (Advanced Cell Diagnostics) according to the manufacturer’s instructions. Postnatal day 7 (p7), day 28, and 24-week-old mice were euthanized and perfused with 0.9% saline followed by 4% paraformaldehyde. Dissected mice brains and spinal cords were cryopreserved in sucrose gradient (10, 20, 30%) for 24hr each and then snap frozen in OCT prior to sectioning at 16 μm thickness. Fixed sections were baked at 60°C for 1 hr, washed with ethanol, followed by tissue pretreatment, probe hybridization following the RNAscope fluorescence multiplex assay and sections were counterstained with DAPI to visualize nuclei. Positive stained cells were identified as punctate dots present in nucleus and cytoplasm. Confocal Z stack images (every 0.2 μm) within the corpus callosum were taken (Zeiss LSM 510 Meta), and Z stacked images generated.
Animals and spinal cord surgery
All experiments were performed according to protocols approved by the University at Buffalo’s Institutional Animal Care and Use Committee. Focal demyelination of adult (24 week) mouse spinal cord was induced as previously described (Zhao et al., 2006; Well-iver et al., 2018). Briefly, animals were anesthetized under isoflurane, and 0.5 μL 1% lysolecithin (Lα-lysophosphatidylcholine, Sigma) was directly injected into the dorsal and ventral funiculus of the spinal cord between two thoracolumbar vertebrae. For experiments involving IFN-γ, was diluted in 0.9% saline, mixed with 1% lysolecithin to a final concentration of 300U per injection and demyelination induced by injection of the mixture into the spinal cord. Animals were sacrificed at 5 days post-lesion (dpl) by transcardial perfusion of saline followed by 4% paraformaldehyde under deep anesthesia. The spinal column was extracted, post-fixed for 30 min in 4% paraformaldehyde, the spinal cord isolated and processed as described above.
Statistical analysis
Statistical analyses were performed using Prism software (GraphPad Software). In vitro experiments were performed with at least three independent fetal samples, and differences among overexpression groups were compared using one-way repeated-measures ANOVA with Dunnett’s multiple comparisons post-test. Differences among the groups for in vivo experiments were compared using one-way or two-way ANOVA with Bonferroni’s or Dunnett’s post-test where appropriate. Statistical significance was considered at p < 0.05. All data are reported as mean ± SEM.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-BrdU antibody (1:2000) | Bio-Rad / AbD Serotec | Cat# MCA2060; RRID:AB_323427 |
| 04 primary antibody (1:25) | Dr. James Goldman (Columbia University) | N/A |
| Monoclonal Anti-Glial Fibrillary Acidic Protein (GFAP) antibody (G-A-5; 1:800) | Sigma-Aldrich | Cat# G3893; RRID:AB_477010 |
| Cleaved Caspase-3 (Asp175) Antibody (1:400) | Cell Signaling Technology | Cat# 9661; RRID:AB_2341188 |
| Human Nuclei (hNA) antibody (1:100) | Millipore Sigma | Millipore Cat# MAB1281; RRID:AB_94090 |
| Anti-APC (Ab-7) Mouse mAb (CC-1) antibody (1:50) | Millipore Sigma | Cat# OP8O; RRID:AB_2057371 |
| Anti-Olig-2 Antibody (1:750) | Millipore Sigma | Millipore Cat# AB9610; RRID:AB_570666 |
| Mouse Anti-GFAP Monoclonal Antibody (SMI21; 1:1000) | BioLegend (Covance) | Cat# SMI-21 R-100; RRID:AB_509978 |
| Anti-Chondroitin Sulfate Proteoglycan Antibody (9.2.27; 1:200) | Millipore | Cat# MAB2029; RRID:AB_94509 |
| PRRX1 mouse monoclonal antibody;clone 1C2 (1:300) | OriGene | Cat# TA803112; RRID:AB_2626674 |
| Rabbit Anti-FLAG antibody (1:200) | Sigma-Aldrich | Cat# F7425; RRID:AB_439687 |
| Mouse Ki-67 antibody (1:25) | BD Biosciences | Cat# 550609; RRID:AB_393778 |
| Bacterial and Virus Strains | ||
| FUW-PRRX1a LV | This Paper | N/A |
| pTRIP-EF1a LV | Sevin et al., 2006 | N/A |
| PRRXIa overexpression LV | This Paper | N/A |
| PRRX1 b overexpression LV | Wang et al., 2014 | N/A |
| FUW-PRRX1a LV | This Paper | N/A |
| Biological Samples | ||
| Fetal brain tissue samples (17–22 week gestational age) | N/A | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Papain | Worthington Biochemical | Cat# LS003126 |
| DNase | Sigma-Aldrich | Cat# D4263–5VL |
| poly-L-ornithine | Sigma-Aldrich | Cat# P3655–100MG |
| Laminin, Mouse | ThermoFisher Scientific | Cat# 23017–015 |
| Recombinant Human PDGF-AA, | PeproTech | Cat#100–13A |
| Recombinant Human NT-3, | PeproTech | Cat#450–03 |
| 3,3′,5-Triiodo-L-thyronine sodium salt | Sigma-Aldrich | Cat# T5516–1 MG |
| BrdU | Sigma-Aldrich | Cat# B5002–1G |
| EdU | VWR | Cat# TCE1057–50MG |
| DAPI | Sigma-Aldrich | Cat# D8417–5MG |
| Paraformaldehyde | VWR International | Cat# JTS898–7 |
| Calcein | VWR International | Cat# 80050–600 |
| Doxycycline hyclate | VWR International | Cat# AAJ60579–14 |
| Recombinant Human FGF-basic | Peprotech | Cat# 100–18 |
| ProFreeze | VWR International | Cat# 12002–076 |
| HBSS | VWR International | Cat# 45000–458 |
| Recombinant Human IFN-gamma | Peprotech | Cat# 300–02 |
| Recombinant Murine IFN-γ | Peprotech | Cat# 315–05 |
| Recombinant Human BMP-2 (CHO cell derived) | Peprotech | Cat# 120–02C |
| Recombinant Human BMP-4 (HeLa cell derived) | Peprotech | Cat# 120–05 |
| OCT | VWR International | Cat# 25608–930 |
| Lipofectamine RNAiMAX transfection reagent | ThermoFisher Scientific | Cat# 13778030 |
| Lysolecithin, L-a-Lysophosphatidylcholine from egg yolk | Sigma-Aldrich | Cat# L4129–100MG |
| Critical Commercial Assays | ||
| Total RNA isolation kit | Omega Bio-Tek | Cat# 101319–242 |
| SuperScript III Reverse transcriptase | ThermoFisher Scientific | Cat# 18080–051 |
| Click-iT® EdU Alexa Fluor® 647 Flow Cytometry Assay Kit | ThermoFisher Scientific | Cat# C-10424 |
| RNAscope Multiplex Fluorscent Reagent Kit V2 | Advanced Cell Diagnostics | Cat# 323100 |
| TSA® Plus Cyanine 3 kit | Perkin Elmer | Cat# NEL744E001KT |
| TSA® Plus Cyanine 5 kit | Perkin Elmer | Cat# NEL745E001 KT |
| TSA Plus Fluorescein kit | Perkin Elmer | Cat# NEL741E001 KT |
| Deposited Data | ||
| RNA-seq | NCBI GEO | GSE122563 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | Cat# CRL-3216, RRID:CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse:shiverer/rag2 mice | Windrem et al., 2008 | N/A |
| BALB/c inbred mice | Envigo | Cat# 047 |
| Oligonucleotides | ||
| FITC BLOCK-iT, Fluorescent oligo | ThermoFisher Scientific | Cat# 2013 |
| Stealth RNAi PRRX1 Human 1 (HSS143371, HSS143372, HSS143373) | ThermoFisher Scientific | Cat# 1299003 |
| Stealth RNAi siRNA Negative Control Med GC Duplex | ThermoFisher Scientific | Cat# 12935112 |
| RNAscope® Probe - Mm-Pdgfra-C2 | Advanced Cell Diagnostics | Cat# 480661-C2 |
| RNAscope Probe - Mm-Prrx1 | Advanced Cell Diagnostics | Cat# 485231 |
| RNAscope Probe - Mm-Mki67-C3 | Advanced Cell Diagnostics | Cat# 416771-C3 |
| Recombinant DNA | ||
| Plasmid: FUW-M2rtTA | Addgene | Cat# 20342 |
| ViraPower Lentiviral Packaging Mix | ThermoFisher Scientific | Cat# K497500 |
| Plasmid: FUW-tetO-lox-NKX2.2 | Wang et al., 2014 | N/A |
| Plasmid: psPAX2 | Addgene | Cat# 12260 |
| Software and Algorithms | ||
| Fiji/I mageJ | National Institutes of Health (NIH) | SCR_003070 |
| Prism software | GraphPad Software | SCR_002798 |
| AutoQuant | Media cybernetics | http://www.mediacy.com/autoquantx3 |
| tophat (v2.1.1) | Johns Hopkins CCB | https://ccb.jhu.edu/software/tophat/index.shtml |
| htseq (vO.6.1) | https://doi.org/10.1093/bioinformatics/btu638 | https://htseq.readthedocs.io/en/release_0.10.0/ |
| R/Bioconductor | http://www.bioconductor.org/ | |
| DESeq2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| edgeR | Bioconductor | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| FCS Express software (version 4) | De Novo Software | N/A |
| Other | ||
| 24-well transwell inserts, 6.5mm, 0.8μm | Laboratory Product Sales (LPS), Inc | Cat# 3464 |
Highlights.
Transcription factor PRRX1 is expressed by quiescent human OPCs (hOPCs)
PRRX1a induces hOPC cell-cycle arrest and inhibits migration in vitro and in vivo
PRRX1 prevents efficient engraftment and tissue colonization by transplanted OPCs
PRRX1 is upregulated by IFN-γ and BMP and is required for IFN-induced quiescence
ACKNOWLEDGMENTS
This work was supported by an NINDS grant (R01NS104021), NCATS (UL1TR001412), National Multiple Sclerosis Society (RG 5505-A-2 and RG 5110A1/1), the Kalec Multiple Sclerosis Foundation, the Change MS Foundation, the Skarlow Memorial Trust, and the Empire State Stem Cell Fund through a New York State Department of Health contract (C028108). J.P. received additional support from NIGMS (R25GM09545902) and NCATS (UL1TR001412-S1). The single-cell RNA-seq was carried out by Genomics Shared Resources at the Roswell Park Cancer Institute, which is supported by grants from the NIH/National Cancer Institute (P30 CA016056, RPCI Cancer Center support grant). We acknowledge the assistance of the Confocal Microscope and Flow Cytometry Facility in the School of Medicine and Biomedical Sciences, University at Buffalo. We thank Dr. Brad Poulos of the Human Fetal Tissue Repository at the Albert Einstein College of Medicine and Advanced Bioscience Resources for assistance with tissue acquisition. We thank Beverly DiCorso, Shangru Lyu, Lili Lin, Joseph E. Noble, and Nicole Wawrzyniak for technical assistance.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and one table and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.068.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Agresti C, D’Urso D, and Levi G (1996). Reversible inhibitory effects of interferon-gamma and tumour necrosis factor-alpha on oligodendroglial line-age cell proliferation and differentiation in vitro. Eur. J. Neurosci 8, 1106–1116. [DOI] [PubMed] [Google Scholar]
- Baerwald KD, and Popko B (1998). Developing and mature oligodendrocytes respond differently to the immune cytokine interferon-gamma. J. Neurosci. Res 52, 230–239. [DOI] [PubMed] [Google Scholar]
- Banno T, Adachi M, Mukkamala L, and Blumenberg M (2003). Unique keratinocyte-specific effects of interferon-gamma that protect skin from viruses, identified using transcriptional profiling. Antivir. Ther. (Lond.) 8, 541–554. [PubMed] [Google Scholar]
- Beck J, Rondot P, Catinot L, Falcoff E, Kirchner H, and Wietzerbin J (1988). Increased production of interferon gamma and tumor necrosis factor precedes clinical manifestation in multiple sclerosis: do cytokines trigger off exacerbations? Acta Neurol. Scand 78, 318–323. [DOI] [PubMed] [Google Scholar]
- Belachew S, Aguirre AA, Wang H, Vautier F, Yuan X, Anderson S, Kirby M, and Gallo V (2002). Cyclin-dependent kinase-2 controls oligodendrocyte progenitor cell cycle progression and is downregulated in adult oligodendrocyte progenitors. J. Neurosci 22, 8553–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F, Kloc M, Chavali M, Hussein I, Wilson M, Christoffel DJ, Chen T, Frohman MA, Robinson JK, Russo SJ, et al. (2015). Genetic and stress-induced loss of NG2 glia triggers emergence of depressive-like behaviors through reduced secretion of FGF2. Neuron 88, 941–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A, Zhang H, and Williams A (2013). Insufficient OPC migration into demyelinated lesions is a cause of poor remyelination in MS and mouse models. Acta Neuropathol. 125, 841–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon B, Suri A, Pan XO, Mills JC, and Unanue ER (2008). IFN-gamma-dependent regulatory circuits in immune inflammation highlighted in diabetes. J. Immunol 181, 6964–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari DM, and Blakemore WF (2002). Efficient recolonisation of progenitor-depleted areas of the CNS by adult oligodendrocyte progenitor cells. Glia 37, 307–313. [PubMed] [Google Scholar]
- Cheung TH, and Rando TA (2013). Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway GD, O’Bara MA, Vedia BH, Pol SU, and Sim FJ (2012). His-tone deacetylase activity is required for human oligodendrocyte progenitor differentiation. Glia 60, 1944–1953. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Kelly D, Rath EM, Baerwald KD, Suzuki K, and Popko B (1996). Targeted CNS expression of interferon-gamma in transgenic mice leads to hypomyelination, reactive gliosis, and abnormal cerebellar development. Mol. Cell. Neurosci 7, 354–370. [DOI] [PubMed] [Google Scholar]
- Dawson MR, Polito A, Levine JM, and Reynolds R (2003). NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol. Cell. Neurosci 24, 476–488. [DOI] [PubMed] [Google Scholar]
- Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, Downward J, and Schulze A (2007). Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol. Cell. Biol 27, 4917–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BR, and Silverman RH (1998). Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligo-nucleotide arrays. Proc. Natl. Acad. Sci. USA 95, 15623–15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KC, Polanco JJ, Pol SU, and Sim FJ (2016). Targeting human oligodendrocyte progenitors for myelin repair. Exp. Neurol 283 (Pt B), 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimou L, Simon C, Kirchhoff F, Takebayashi H, and Götz M (2008). Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. J. Neurosci 28, 10434–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling P, Husar W, Menonna J, Donnenfeld H, Cook S, and Sidhu M (1997). Cell death and birth in multiple sclerosis brain. J. Neurol. Sci 149, 1–11. [DOI] [PubMed] [Google Scholar]
- Durand B, Fero ML, Roberts JM, and Raff MC (1998). p27Kip1 alters the response of cells to mitogen and is part of a cell-intrinsic timer that arrests the cell cycle and initiates differentiation. Curr. Biol 8, 431–440. [DOI] [PubMed] [Google Scholar]
- Franklin RJM (2002). Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci 3, 705–714. [DOI] [PubMed] [Google Scholar]
- Franklin RJ, Bayley SA, and Blakemore WF (1996). Transplanted CG4 cells (an oligodendrocyte progenitor cell line) survive, migrate, and contribute to repair of areas of demyelination in X-irradiated and damaged spinal cord but not in normal spinal cord. Exp. Neurol 137, 263–276. [DOI] [PubMed] [Google Scholar]
- Geha S, Pallud J, Junier MP, Devaux B, Leonard N, Chassoux F, Chneiweiss H, Daumas-Duport C, and Varlet P (2010). NG2+/Olig2+ cells are the major cycle-related cell population of the adult human normal brain. Brain Pathol. 20, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraerts M, Willems S, Baekelandt V, Debyser Z, and Gijsbers R (2006). Comparison of lentiviral vector titration methods. BMC Biotechnol 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SM, Vass JK, Holyoake TL, and Graham GJ (2007). Transcriptional analysis of quiescent and proliferating CD34+ human hemopoietic cells from normal and chronic myeloid leukemia sources. Stem Cells 25, 3111–3120. [DOI] [PubMed] [Google Scholar]
- Horiuchi M, Itoh A, Pleasure D, and Itoh T (2006). MEK-ERK signaling is involved in interferon-gamma-induced death of oligodendroglial progenitor cells. J. Biol. Chem 281, 20095–20106. [DOI] [PubMed] [Google Scholar]
- Horwitz MS, Evans CF, Mcgavern DB, Rodriguez M, and Oldstone MBA (1997). Primary demyelination in transgenic mice expressing interferon-gamma. Nat. Med 3, 1037–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes EG, Kang SH, Fukaya M, and Bergles DE (2013). Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci 16, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Huang LM, Suárez-Fariñas M, Pierson KC, Fuentes-Duculan J, Cueto I, Lentini T, Sullivan-Whalen M, Gilleaudeau P, Krueger JG, Haider AS, and Lowes MA (2012). A single intradermal injection of IFN-γ induces an inflammatory state in both non-lesional psoriatic and healthy skin. J. Invest. Dermatol 132, 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippin TE, Martens DJ, and van der Kooy D (2005). p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 19, 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, and Raff M (2000). Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science 289, 1754–1757. [DOI] [PubMed] [Google Scholar]
- Kondo T, and Raff M (2004). Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 18, 2963–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFerla FM, Sugarman MC, Lane TE, and Leissring MA (2000). Regional hypomyelination and dysplasia in transgenic mice with astrocyte-directed expression of interferon-gamma. J. Mol. Neurosci 15, 45–59. [DOI] [PubMed] [Google Scholar]
- Lee KY, Jeong JW, Wang J, Ma L, Martin JF, Tsai SY, Lydon JP, and DeMayo FJ (2007). Bmp2 is critical for the murine uterine decidual response. Mol. Cell. Biol 27, 5468–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JM, Reynolds R, and Fawcett JW (2001). The oligodendrocyte precursor cell in health and disease. Trends Neurosci. 24, 39–47. [DOI] [PubMed] [Google Scholar]
- Lin W, Kemper A, Dupree JL, Harding HP, Ron D, and Popko B (2006). Interferon-gamma inhibits central nervous system remyelination through a process modulated by endoplasmic reticulum stress. Brain 129, 1306–1318. [DOI] [PubMed] [Google Scholar]
- Lin G, Mela A, Guilfoyle EM, and Goldman JE (2009). Neonatal and adult O4(+) oligodendrocyte lineage cells display different growth factor responses and different gene expression patterns. J. Neurosci. Res 87, 3390–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks of aging. Cell 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchinetti CF, Brück W, Rodriguez M, and Lassmann H (1996). Distinct patterns of multiple sclerosis pathology indicates heterogeneity on pathogenesis. Brain Pathol. 6, 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabie PC, Mehler MF, Marmur R, Papavasiliou A, Song Q, and Kessler JA (1997). Bone morphogenetic proteins induce astroglial differentiation of oligodendroglial-astroglial progenitor cells. J. Neurosci 17, 4112–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Mateo JL, Zhou B, Andersen J, Achimastou A, Urbán N, van den Berg D, Georgopoulou D, Hadjur S, Wittbrodt J, et al. (2013). Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 27, 1769–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li H, de Faria JP, Emery B, Tohyama K, and Richardson WD (2014). Motor skill learning requires active central myelination. Science 346, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortigüela R, Marqués-Torrejón MA, Nakashima K, et al. (2010). Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89. [DOI] [PubMed] [Google Scholar]
- Morris CS, Esiri MM, Sprinkle TJ, and Gregson N (1994). Oligodendrocyte reactions and cell proliferation markers in human demyelinating diseases. Neuropathol. Appl. Neurobiol 20, 272–281. [DOI] [PubMed] [Google Scholar]
- Mourikis P, Sambasivan R, Castel D, Rocheteau P, Bizzarro V, and Tajbakhsh S (2012). A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 30, 243–252. [DOI] [PubMed] [Google Scholar]
- Norris RA, and Kern MJ (2001). The identification of Prx1 transcription regulatory domains provides a mechanism for unequal compensation by the Prx1 and Prx2 loci. J. Biol. Chem 276, 26829–26837. [DOI] [PubMed] [Google Scholar]
- Ocaña OH, Córcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, and Nieto MA (2012). Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724. [DOI] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, et al. (2009). FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5, 540–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panitch HS, Hirsch RL, Haley AS, and Johnson KP (1987). Exacerbations of multiple sclerosis in patients treated with gamma interferon. Lancet 1, 893–895. [DOI] [PubMed] [Google Scholar]
- Paxinos G, and Franklin KB (2004). The Mouse Brain in Stereotaxic Coordinates (Elsevier Academic Press; ). [Google Scholar]
- Penderis J, Shields SA, and Franklin RJ (2003). Impaired remyelination and depletion of oligodendrocyte progenitors does not occur following repeated episodes of focal demyelination in the rat central nervous system. Brain 126, 1382–1391. [DOI] [PubMed] [Google Scholar]
- Pol SU, Lang JK, O’Bara MA, Cimato TR, McCallion AS, and Sim FJ (2013). Sox10-MCS5 enhancer dynamically tracks human oligodendrocyte progenitor fate. Exp. Neurol 247, 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol SU, Polanco JJ, Seidman RA, O’Bara MA, Shayya HJ, Dietz KC, and Sim FJ (2017). Network-based genomic analysis of human oligodendrocyte progenitor differentiation. Stem Cell Reports 9, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popko B, Corbin JG, Baerwald KD, Dupree J, and Garcia AM (1997). The effects of interferon-gamma on the central nervous system. Mol. Neurobiol 14, 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psachoulia K, Jamen F, Young KM, and Richardson WD (2009). Cell cycle dynamics of NG2 cells in the postnatal and ageing brain. Neuron Glia Biol. 5, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M, Takano S, von Burstin J, Kim SB, Lee JS, Ihida-Stansbury K, Hahn C, Heeg S, Schneider G, Rhim AD, et al. (2013). The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev. 27, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. (2009). FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, and Richardson WD (2008). PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat. Neurosci 11, 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock RB, Hu S, Deshpande A, Munir S, May BJ, Baker CA, Peterson PK, and Kapur V (2005). Transcriptional response of human microglial cells to interferon-gamma. Genes Immun. 6, 712–719. [DOI] [PubMed] [Google Scholar]
- Rumman M, Dhawan J, and Kassem M (2015). Concise review: quiescence in adult stem cells: biological significance and relevance to tissue regeneration. Stem Cells 33, 2903–2912. [DOI] [PubMed] [Google Scholar]
- Salazar VS, Gamer LW, and Rosen V (2016). BMP signalling in skeletal development, disease and repair. Nat. Rev. Endocrinol 12, 203–221. [DOI] [PubMed] [Google Scholar]
- Sana TR, Janatpour MJ, Sathe M, McEvoy LM, and McClanahan TK (2005). Microarray analysis of primary endothelial cells challenged with different inflammatory and immune cytokines. Cytokine 29, 256–269. [DOI] [PubMed] [Google Scholar]
- Sevin C, Benraiss A, Van Dam D, Bonnin D, Nagels G, Verot L, Lauren-deau I, Vidaud M, Gieselmann V, Vanier M, et al. (2006). Intracerebral adeno-associated virus-mediated gene transfer in rapidly progressive forms of metachromatic leukodystrophy. Hum. Mol. Genet 15, 53–64. [DOI] [PubMed] [Google Scholar]
- Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, and Brack AS (2010). Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell 6, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozaki K, Clemenson GD Jr., and Gage FH (2013). Paired related homeobox protein 1 is a regulator of stemness in adult neural stem/progenitor cells. J. Neurosci 33, 4066–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim FJ, Zhao C, Penderis J, and Franklin RJ (2002). The age-related decrease in CNS remyelination efficiency is attributable to an impairment of both oligodendrocyte progenitor recruitment and differentiation. J. Neurosci 22, 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim FJ, Lang JK, Waldau B, Roy NS, Schwartz TE, Pilcher WH, Chandross KJ, Natesan S, Merrill JE, and Goldman SA (2006). Complementary patterns of gene expression by human oligodendrocyte progenitors and their environment predict determinants of progenitor maintenance and differentiation. Ann. Neurol 59, 763–779. [DOI] [PubMed] [Google Scholar]
- Sim FJ, McClain CR, Schanz SJ, Protack TL, Windrem MS, and Goldman SA (2011). CD140a identifies a population of highly myelinogenic, migration-competent and efficiently engrafting human oligodendrocyte progenitor cells. Nat. Biotechnol 29, 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Götz M, and Dimou L (2011). Progenitors in the adult cerebral cortex: cell cycle properties and regulation by physiological stimuli and injury. Glia 59, 869–881. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano S, Reichert M, Bakir B, Das KK, Nishida T, Miyazaki M, Heeg S, Collins MA, Marchand B, Hicks PD, et al. (2016). Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev. 30, 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner DC, Cherry JD, and Mayer-Pröschel M (2011). Oligodendrocyte progenitors reversibly exit the cell cycle and give rise to astrocytes in response to interferon-g. J. Neurosci 31, 6235–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassiulas I, Hu X, Ho H, Kashyap Y, Paik P, Hu Y, Lowell CA, and Ivashkiv LB (2004). Amplification of IFN-alpha-induced STAT1 activation and inflammatory function by Syk and ITAM-containing adaptors. Nat. Immunol 5, 1181–1189. [DOI] [PubMed] [Google Scholar]
- Tsai HH, Niu J, Munji R, Davalos D, Chang J, Zhang H, Tien AC, Kuo CJ, Chan JR, Daneman R, and Fancy SP (2016). Oligodendrocyte precursors migrate along vasculature in the developing nervous system. Science 351, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian T, Li Y, Zhao M, and Stefansson K (1995). Interferon-gamma-induced oligodendrocyte cell death: implications for the pathogenesis of multiple sclerosis. Mol. Med 1, 732–743. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ren Z, Tao D, Tilwalli S, Goswami R, and Balabanov R (2010). STAT1/IRF-1 signaling pathway mediates the injurious effect of interferon-gamma on oligodendrocyte progenitor cells. Glia 58, 195–208. [DOI] [PubMed] [Google Scholar]
- Wang J, O’Bara MA, Pol SU, and Sim FJ (2013). CD133/CD140a-based isolation of distinct human multipotent neural progenitor cells and oligodendrocyte progenitor cells. Stem Cells Dev. 22, 2121–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pol SU, Haberman AK, Wang C, O’Bara MA, and Sim FJ (2014). Transcription factor induction of human oligodendrocyte progenitor fate and differentiation. Proc. Natl. Acad. Sci. USA 111, E2885–E2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welliver RR, Polanco JJ, Seidman RA, Sinha AK, O’Bara MA, Khaku ZM, Santiago González DA, Nishiyama A, Wess J, Feltri ML, et al. (2018). Muscarinic receptor M3R signaling prevents efficient remyelination by human and mouse oligodendrocyte progenitor cells. J. Neurosci 38, 6921–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windrem MS, Schanz SJ, Guo M, Tian GF, Washco V, Stanwood N, Rasband M, Roy NS, Nedergaard M, Havton LA, et al. (2008). Neonatal chimerization with human glial progenitor cells can both remyelinate and rescue the otherwise lethally hypomyelinated shiverer mouse. Cell Stem Cell 2, 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windrem MS, Schanz SJ, Morrow C, Munir J, Chandler-Militello D, Wang S, and Goldman SA (2014). A competitive advantage by neonatally engrafted human glial progenitors yields mice whose brains are chimeric for human glia. J. Neurosci 34, 16153–16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolswijk G (1998). Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J. Neurosci 18, 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodroofe MN, and Cuzner ML (1993). Cytokine mRNA expression in inflammatory multiple sclerosis lesions: detection by non-radioactive in situ hybridization. Cytokine 5, 583–588. [DOI] [PubMed] [Google Scholar]
- Woodruff RH, Fruttiger M, Richardson WD, and Franklin RJ (2004). Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination. Mol. Cell. Neurosci 25, 252–262. [DOI] [PubMed] [Google Scholar]
- Young KM, Psachoulia K, Tripathi RB, Dunn SJ, Cossell L, Attwell D, Tohyama K, and Richardson WD (2013). Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron 77, 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zezula J, Casaccia-Bonnefil P, Ezhevsky SA, Osterhout DJ, Levine JM, Dowdy SF, Chao MV, and Koff A (2001). p21cip1 is required for the differentiation of oligodendrocytes independently of cell cycle withdrawal. EMBO Rep. 2, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara H, Heller-mann G, Behera S, Singam R, Lockey RF, and Mohapatra SS (2005). Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med 11, 56–62. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Li WW, and Franklin RJ (2006). Differences in the early inflammatory responses to toxin-induced demyelination are associated with the age-related decline in CNS remyelination. Neurobiol. Aging 27, 1298–1307. [DOI] [PubMed] [Google Scholar]
- Zhao C, Ma D, Zawadzka M, Fancy SPJ, Elis-Williams L, Bouvier G, Stockley JH, de Castro GM, Wang B, Jacobs S, et al. (2015). Sox2 sustains recruitment of oligodendrocyte progenitor cells following CNS demyelination and primes them for differentiation during remyelination. J. Neurosci 35, 11482–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.