Abstract
Objectives
To explore the molecular epidemiology of rare deafness genes in Taiwanese sensorineural hearing impairment (SNHI) patients with cochlear implantation (CI) by performing massive parallel sequencing (MPS) and correlating genetic factors and CI outcomes.
Methods
We enrolled 41 Taiwanese non-syndromic deafness patients with CI that lacked known mutations in common deafness genes. All probands were screened by a targeted exon amplification method that used massively parallel sequencing to screen a customized panel that included 40 relatively rare non-syndromic deafness genes.
Results
Thirteen candidate variants in nine relatively rare deafness genes (MYO15A, TMC1, MYH14, MYO3A, ACTG1, COL11A2, DSPP, GRHL2, and WFS1) were identified in 24.4% (10/41) of the non-syndromic deafness probands with CI. According to the ACMG Standards and Guidelines, five variants in MYO15A and ACTG1 were classified as likely pathogenic variants. Two of three multi-generational pedigrees exhibiting deafness were analyzed for the segregation of the disorder with the possible disease-causing variants. Patients with variants detected in most of the identified variant-bearing genes showed relatively good CI outcomes.
Conclusions
We successfully identified candidate variants in partially deaf Taiwanese probands who lacked the known mutations in common deafness genes. Comparing the progress of hearing rehabilitation in CI patients with their apparent causative variants and the expression profiles of their altered genes allowed us to speculate on how alterations in specific gene sets may influence outcomes in hearing rehabilitation after CI.
Introduction
Sensorineural hearing impairment (SNHI) is a common clinical disorder that severely to profoundly affects at least 1 in 1000 children of developed countries [1]. Cochlear implantation (CI) is currently regarded as the standard treatment for severe to profound SNHI in children. CI has well-documented benefits for spoken language, reading skills, and cognitive development [2], but the outcomes after CI can vary among individuals. Age at implantation [3, 4], residual hearing [5], the presence of inner ear malformations [6], the presence of cochlear nerve deficiency [7], parent-child interactions [2], and socioeconomic status [2] have all been shown to affect the outcomes.
Genetic factors contribute to SNHI in more than 50% of these patients [8]. To date, more than 100 genes and loci have been associated with deafness, and approximately 50 genes have been shown to cause non-syndromic hereditary hearing impairment (The Hereditary Hearing Loss Homepage, http://hereditaryhearingloss.org/) [9]. Due to limitations in mutation detection methodologies, most of the existing studies have focused on the three most common deafness genes, GJB2 (or Cx26), SLC26A4 (or PDS), and MT-RNR1 [10], in the context of epidemiological studies or examining the correlation between CI outcomes and genotypes [11–14]. Only 1/3 of SNHI patients and 1/4 patients with CI can be identified as having known mutations in common deafness genes [10, 14]. The development of massively parallel sequencing (MPS), also known as next-generation sequencing, has allowed researchers and clinicians to more easily address such extraordinarily heterogenetic disorders [15–20]. The powerful strategy of simultaneously obtaining high-throughput reads from multiple targeted genes in numerous samples has reduced the sequencing cost and turnaround time of genetic examination. In recent years, MPS technology has been widely used to examine the prevalence of relatively rare deafness genes and the correlations between various genotypes and the outcomes of CI rehabilitation [15, 19–24].
The auditory performance in patients with CIs can be predicted according to the pathologies that are associated with mutations in different genes [14, 20–22, 24, 25]. When the function of the mutated gene is confined to the intra-cochlear etiology, good post-CI auditory performance is possible [14]. Conversely, poor CI performance can be predicted when the causative mutation occurs in a gene that is expressed in the spiral ganglion neurons (SGNs), brainstem auditory nuclei, or hair cell synapses. Genetic information about predicted good or poor CI outcomes could allow clinicians to counsel patients on whether to undergo an operation and/or decide on a rehabilitation program [24, 26]. Therefore, it is beneficial to identify the causative gene mutation prior to CI intervention. A greater knowledge of the genetic backgrounds of deafness patients with CI will enable clinicians to offer a more precise, genetically based prediction of CI outcome.
In this study, we sought to unveil the prevalence rate of rare deafness-associated variants in 41 Taiwanese SNHI patients with CIs, using MPS technology. We also examined the relationship between the identified variants and the outcomes of CI.
Materials and methods
Subject recruitment
A total of 41 probands who had bilateral hearing loss and lacked the known mutations in common deafness genes, including GJB2, SLCA26A4 and m.1555A>G in MT-RNR1 [27], were recruited from Chang Gung Memorial Hospital (CGMH). All participants had non-syndromic hearing loss without any other organic abnormality and had undergone unilateral CI at CGMH. The subjects included three probands from multiplex families and 38 simplex probands. Their average residual hearing before implantation was 103.3±11.4 dBHL (decibels Hearing Level). The subjects included 18 males (43.9%) and 23 females (56.1%). The average age at which they received CI was 6.6 years old (range, 0.9 years to 33.3 years).
The study protocol and written informed consent form were approved by the Chang-Gung Memorial Hospital Ethics Committee for Human Studies. Signed informed consent forms were obtained from all participants or their guardians before we began the testing procedures.
Massively parallel sequencing
Genomic DNA (gDNA) was extracted from peripheral blood using a QIAamp DNA blood mini kit (Qiagen, Taiwan). DNA libraries were generated by the targeted exon amplification method. An Ion AmpliseqTM Custom Panel of 40 relatively rare and non-syndromic deafness genes (14 autosomal dominant, 19 autosomal recessive, 6 autosomal dominant/recessive, and 1 X-linked; S1 Table) were selected for our customized panel (Applied Biosystems, Life Technologies, Carlsbad, CA) by Ion AmpliseqTM Designer (Version 3.6, https://www.ampliseq.com/browse.action). This customized panel comprised 1319 amplicons with 5 bp exon padding, and covered 95.6% of the exonic regions. The whole panel size was 256.6 Kb. The 40 genes screened in this panel were selected from our survey, and the examined amplicons included all of the relevant gene variants identified in studies on populations from China, Korea, and Japan [17, 25, 28]. The gene set included MYO15A, TMC1, PCDH15, CDH23, MYO7A, ESRRB, MARVELD2, TECTA, WHRN, MYO6, POU3F4, CDH23, COL11A2, WFS1, EYA4, STRC, TMPRSS3, WHRN, ACTG1, DFNA5, and CRYM, in which mutations have been commonly found in hearing loss patients of Eastern Asia. For library preparation, we used an Ion Ampliseq Library Kit 2.0 (Applied Biosystems, Life Technologies) according to the manufacturer’s instructions. The purified amplicon libraries were assessed for their concentrations and size distributions using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). Each library was diluted to 10 pM and subjected to clonal amplification using an Ion OneTouchTM 2 System and an Ion OneTouch 200 Template Kit v2 (Life Technologies). The obtained products were sequenced using an Ion Torrent Personal Genome Machine (PGM) system with an Ion PGMTM 200 Sequencing Kit and an Ion 318TM Chip or 316 TM Chip (Life Technologies). The average depth of coverage for the target region obtained for the 41 samples was 425.2-fold, and 96.2% of the targeted regions were read by at least 20× coverage.
Data analysis
The raw sequence reads were processed using the Ion Torrent SuiteTM Software and aligned to a reference human genome sequence (Feb. 2009, GRCh37/hg19) with a Torrent Mapping Alignment Program optimized to the Ion TorrentTM data. The variant calling process was conducted using the Torrent Variant Caller plug-in software (all from Life Technologies).
Filtering criteria
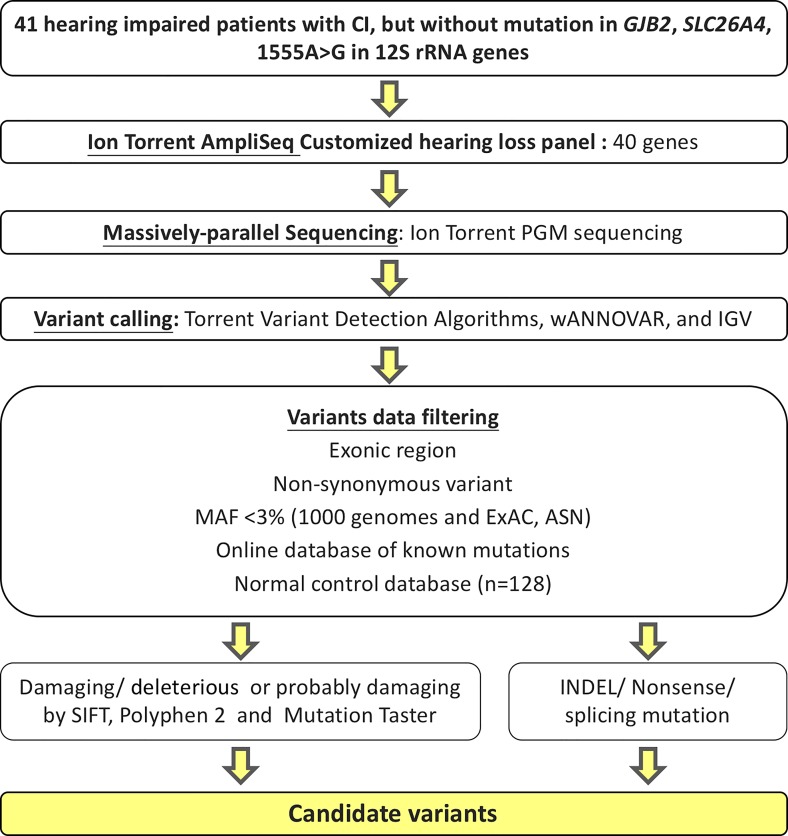
Variants called by the Torrent Variant Caller were annotated through wANNOVAR, a web-based application for gene and amino acid annotation and functional evaluation [29, 30]. The following criteria were used to select variants: a non-synonymous variant in an exonic region; an allele frequency below 3% in the Asian population, as indicated by the 1000 Genomes Project [31] and Exome Aggregation Consortium (ExAC) [32], with a variant frequency of 30–70% for heterozygous variants and 85–100% for homozygous variants; allele coverage exceeding 30X; and variant allele coverage exceeding 15X. We also included variants that could lead to the synthesis of immature proteins (e.g., nonsense, INDEL, and splicing site variants) and missense variants that were nonsynonymous and annotated as “deleterious”, “damaging”, or “possibly damaging” by SIFT [33], Polyphen 2 [34], or Mutation taster [35] (Fig 1). All of the selected variants were further screened in 128 ethnically matched normal hearing controls (see Acknowledgements). Finally, bi-allelic variants selected for autosomal recessive genes were retained. The filtered variants were confirmed by Sanger sequencing and then taken as candidate variants.
Fig 1. Pipeline used to identify candidate variants.
Forty-one patients with CI who lacked the known mutations in common deafness-associated genes were subjected to comprehensive genetic analysis by using an Ion Torrent PGM sequencer to target 40 relatively rare deafness genes. Variants were called using plug-in Torrent variant detection algorithms, annotated through wANNOVAR, and initially confirm using the integrative Genome viewer. Annotated variants were filtered by various criteria, including: being located in an exonic region; being non-synonymous; having an allele frequency < 3% in the 1000 Genomes Project; having a variant frequency of 30–70% for heterozygotes or >85% for homozygotes; and being absent from online databases and 128 ethnically matched normal hearing controls. SIFT, Polyphen 2, and Mutation taster were used to predict the functions of the identified variants; we first filtered for missense variants, and then directly identified indels, splicing site variants, and nonsense variants.
Evaluation of auditory and speech performance
To evaluate auditory performance in CI patients, we used the Categories of Auditory Performance (CAP) scale [36] and the Speech Intelligibility Rating (SIR) scale [37]. The CAP, which evaluates the auditory receptive ability, places an individual into one of eight categories ranging from “no awareness of environmental sound” (CAP score = 0) to “uses the telephone” (CAP score = 7). The SIR, which evaluates spontaneous speech intelligibility, places an individual into one of five categories ranging from “unintelligible speech” (SIR score = 1) to “speech intelligible to all listeners” (SIR score = 5). The CAP and SIR scales have been confirmed as reliable instruments for measuring CI outcomes [36, 37]. We referred to the literature to identify appropriate criteria [24], and classified a poor CI outcome as corresponding to CAP ≤ 5 and SIR ≤ 3.
Result
Identification of candidate variants
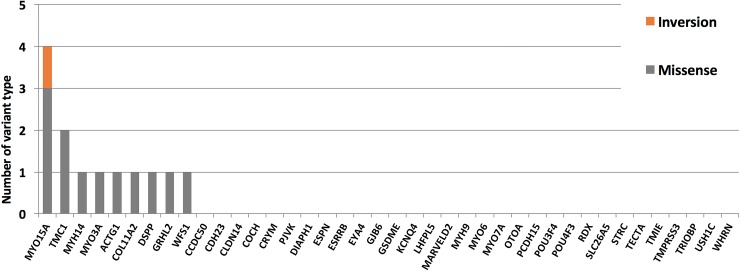
We identified a total of 13 previously unidentified candidate variants in nine deafness genes, as found in 10 of the 41 CI patients (24.4%) (Fig 1 and Table 1). The allele frequency of each of the 13 candidate variants was very low in the east Asian populations captured in the 1000 Genomes Project and ExAC, and these variants were all absent from our 128 normal Taiwanese controls. The 13 candidate variants included 12 missense variants in nine cases that were predicted to have damaging or disease-causing effects by causing amino acid changes, as assessed by protein-impact predictors, and one inversion variant found in one case (Table 1 and Fig 2). We identified four heterozygous variants responsible for autosomal dominant SNHI (ADSNHI) in four probands (DE3864, DE3335, DE3395, and DE4467). One homozygous variant and two compound heterozygous variants were consistent with recessive inheritance in two probands (DE3241 and DE4372). However, DE3386 harbored three candidate variants in genes whose mutations had previously been associated with dominant or recessive inheritance, making it impossible for us to conclusively determine how any one of the variants impacted the patient’s phenotype. Three probands (DE3221, DE4377, and DE4702) were identified as having three heterozygous variants in two genes, COL11A2 and TMC1; as mutations in these genes had previously been associated with autosomal dominant and autosomal recessive inheritance patterns [38], we were unable to confirm the pathogenicity of either variant. The gene most frequently associated with a putatively pathogenic variant was MYO15A (4 variants), followed by TMC1 (2), with the remaining genes each having a single putatively pathogenic variant (MYH14, MYO3A, ACTG1, COL11A2, DSPP, GRHL2, and WFS1 (Fig 2). Beyond the variants specified above, five of the identified variants could be classified as likely pathogenic variants, and the remaining variants were of uncertain significance according to the AMA and ACMG guidelines [39].
Table 1. Thirteen different variants were identified in 10 patients.
| Case | Gene | Variant type | Inheritance | Nucleotide change | Protein change | Genotype | SIFT | Polyphen2 | Mutation Taster | 1000 Genomes ASN |
ExAC ASN |
Normal control (allele fre, %) | Pathogenicity (Deafness variation databasea) |
AMA and ACMG guidelines |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DE2864 | WFS1 | Missense | AD | NM_006005.3:c.173C>T | NP_005996.2:p.(Ala58Val) | Het | T | PD | N | 0 | 0.0046 | 0 | Pathogenic (Wolfram syndrome) | Uncertain significance |
| DE3221 | COL11A2 | Missense | AR/AD | NM_080680.2:c.191G>A | NP_542411.2:p.(Arg64Gln) | Het | T | PD | D | 0.001 | 0.0011 | 0 | VUS | Uncertain significance |
| DE3241 | MYO3A | Missense | AR | NM_017433.4:c.1256T>C | NP_059129.3:p.(Ile419Thr) | Hom | D | B | D | 0 | 0.0023 | 0 | VUS | Uncertain significance |
| DE3335 | DSPP | Inversion | AD | NM_014208.3:c.3021_3022inv | NP_055023.2:p.(Asp1008Asn) | Het | T | PD | P | 0 | 0 | 0 | - | Uncertain significance |
| DE3386 | MYH14 | Missense | AD | NM_024729.3:c.2080C>T | NP_079005.3:p.(Arg694Cys) | Het | D | D | D | 0 | 0 | 0 | - | Likely pathogenic |
| MYO15A | Missense | AR | NM_016239.3:c.4457G>T | NP_057323.3:p.(Gly1486Val) | Het | D | D | D | 0 | 0 | 0 | - | Likely pathogenic | |
| MYO15A | Missense | AR | NM_016239.3:c.4101C>A | NP_057323.3:p.(Asn1367Lys) | Het | D | D | D | 0 | 0 | 0 | - | Likely pathogenic | |
| DE3395 | ACTG1 | Missense | AD | NM_001199954.2:c.830C>T | NP_001186883.1:(p.Thr277Ile) | Het | D | D | D | 0 | 0 | 0 | - | Likely pathogenic |
| DE4372 | MYO15A | Missense | AR | NM_016239.3:c.5443C>A | NP_057323.3:p.(Gln1815Lys) | Het | D | D | D | 0 | 0 | 0 | - | Uncertain significance |
| MYO15A | Missense | AR | NM_016239.3:c.5977C>T | NP_057323.3:p.(Arg1993Trp) | Het | D | D | D | 0 | 0 | 0 | VUS | Likely pathogenic | |
| DE4377 | TMC1 | Missense | AR/AD | NM_138691.2:c.1632T>G | NP_619636.2:p.(Phe544Leu) | Het | T | PD | D | 0 | 0 | 0 | VUS | Uncertain significance |
| DE4467 | GRHL2 | Missense | AD | NM_024915.3:c.193G>A | NP_079191.2:p.(Gly65Ser) | Het | T | PD | D | 0 | 0 | 0 | - | Uncertain significance |
| DE4702 | TMC1 | Missense | AR/AD | NM_138691.2:c.1777T>C | NP_619636.2:p.(Phe593Leu) | Het | D | B | D | 0 | 0 | 0 | - | Uncertain significance |
a Deafness variation database (http://deafnessvariationdatabase.org/)
B: Benign; T: Tolerated; D: Damaging/deleterious; PD: Probably damaging; NA: Not available; N: Polymorphism
VUS: Variant of uncertain significance
AD: Autosomal dominant; AR: Autosomal recessive
Fig 2. Candidate variants detected in nine genes of 41 patients.
Each color bar indicates a different variant type, as indicated.
Co-segregation analysis for multiplex probands
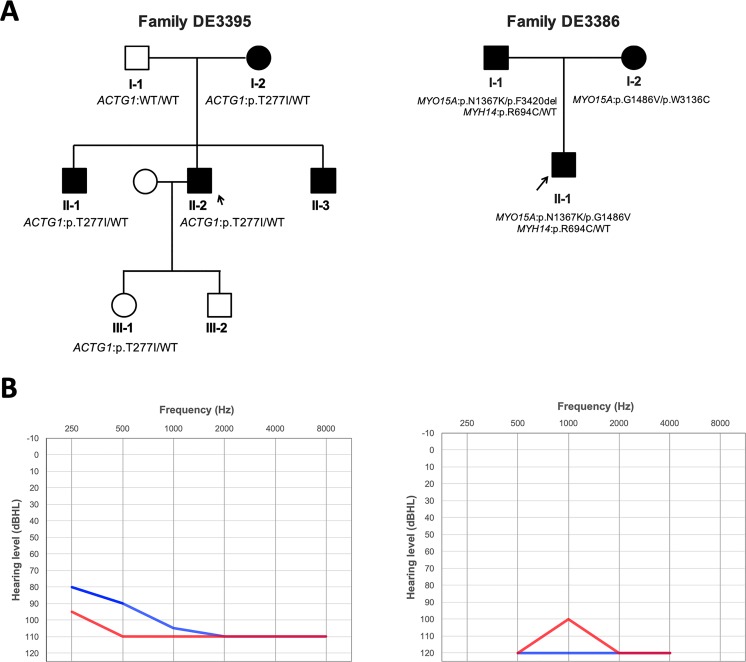
We identified four possible candidate variants in two of three multiplex probands, and performed segregation analysis using limited affected or unaffected familial members in Family DE3386 and Family DE3395 (Fig 3A and Fig 3B).
Fig 3. Families DE3395 and DE3386.
(A) Probands of each family are indicated by arrows. (B) Audiograms of DE3395 (left) and DE3386 (right). Both recipients had bilateral symmetric flat-type audiograms of profound severity. Hearing levels of the right ear and left ear are marked with red and blue lines, respectively.
DE3395 is a late-onset hearing loss patient who was found to have p.(Thr277Ile) (c.830C>T) in ACTG1 (Table 1). Among the family members of DE3395, his mother and two brothers also experienced a progressive deafness phenotype beginning in the first or second decade of life (as seen in the proband). Our direct sequencing identified the p.(Thr277Ile) variant in ACTG1 in two additional affected members of this family (I-2 and II-1) and the proband’s daughter (III-1), who had a normal hearing ability at the age of 8 years. The final affected individual (II-3) did not consent to testing, so the genetic result was unavailable.
Three candidate variants, compound heterozygous variants (p.(Gly1486Val) and p.(Asn1367Lys)) in MYO15A and heterozygous variant (p.Arg694Cys) in MYH14, were identified in individual DE3386 (Table 1). Sanger sequencing revealed that p.(Arg694Cys) and p.(Asn1367Lys) were co-inherited from his father (I-1) and p.(Gly1486Val) came from his mother (I-2). Because both of his parents are also SNHI patients, we further applied MPS to reveal his parents’ variant profiles to know the family’s inherited pattern and discovered that his mother (I-2) also had p.(Trp3136Cys) in MYO15A while his father (I-1) had p.(Phe3420del) in MYO15A (Table 2). Notably, p.(Phe3420del) in MYO15A has been reported in hearing loss patients from the Chinese Han population [17].
Table 2. Variants identified in the parents of DE3386 (DE4451 and DE4452).
| Case | Gene | Variant type | Inheritance | Nucleotide change | Protein change | Genotype | SIFT | Polyphen2 | Mutation Taster | 1000 Genomes ASN |
ExAC ASN |
Normal control (allele fre, %) | Pathogenicity (Deafness variation databasea) |
AMA and ACMG guidelines |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DE4451 | MYO15A | Missense | AR | NM_016239.3:c.9408G>C | NP_057323.3:p.(Trp3136Cys) | Het | D | D | D | 0 | 0.0002 | 0 | - | Likely pathogenic |
| MYO15A | Missense | AR | NM_016239.3:c.4457G>T | NP_057323.3:p.(Gly1486Val) | Het | D | D | D | 0 | 0 | 0 | - | Likely pathogenic | |
| DE4452 | MYO15A | Deletion | AR | NM_016239.3:c.10258_10260del | NP_057323.3:p.(Phe3420del) | Het | . | . | . | 0 | 0 | 0 | Pathogenic | Likely pathogenic |
| MYO15A | Missense | AR | NM_016239.3:c.4101C>A | NP_057323.3:p.(Asn1367Lys) | Het | D | D | D | 0 | 0 | 0 | - | Likely pathogenic | |
| MYH14 | Missense | AD | NM_024729.3:c.2080C>T | NP_079005.3:p.(Arg694Cys) | Het | D | D | D | 0 | 0 | 0 | - | Likely pathogenic |
a Deafness variation database
B: Benign; T: Tolerated; D: Damaging/deleterious; PD: Probably damaging; NA: Not available; N: Polymorphism
VUS: Variant of uncertain significance
AD: Autosomal dominant; AR: Autosomal recessive
Correlation of genotype and CI outcomes in patients with candidate variants
The CI outcomes of the 10 patients who harbored the identified candidate variants are listed in Table 3. All patients had received CI at least 4.9 years prior to enrolling in our study. Among them, patients DE3221 and DE4702 had variants in COL11A2 and TMC1, respectively, and were classified as having poor outcomes. Conversely, patient DE4377, who also harbored a variant in TMC1, showed a good CI outcome. The remaining seven patients had variants in MYO15A, MYO3A, MYH14, ACTG1, WFS1, DSPP, and GRHL2, and tended to have good outcomes (CAP and SIR scores of more than 5 and 3, respectively).
Table 3. CI outcomes of the 10 patients with the identified candidate variants.
| Sample ID | Sex | Genotype | Age at implantation, yr | Preoperative hearing, dBHL | Duration of rehabilitation, yr | CAP | SIR |
|---|---|---|---|---|---|---|---|
| DE2864 | F | WFS1 p.(Ala58Val)/ WT | 5.6 | 100 | 6.4 | 5 | 4 |
| DE3221 | M | COL11A2 p.(Arg64Gln)/ WT | 1.7 | 120 | 6.5 | 4 | 2 |
| DE3241 | M | MYO3A p.(Ile419Thr)/ WT | 25.1 | N/A | 12 | 5 | 4 |
| DE3335 | M | DSPP p.(Asp1008Asn) / WT | 4 | 120 | 4.9 | 6 | 4 |
| DE3386 | F | MYH14 p.(Arg694Cys)/ WT | 1.5 | 106.6 | 5.7 | 5 | 4 |
| MYO15A p.(Gly1486Val)/ p.(Asn1367Lys) | |||||||
| DE3395 | F | ACTG1 p.(Thr277Ile)/ WT | 26.3 | 108.3 | 6 | 6 | 5 |
| DE4372 | M | MYO15A p.(Gln1815Lys)/ p.(Arg1993Trp) | 2.3 | 116.6 | 12.1 | 6 | 5 |
| DE4377 | F | TMC1 p.(Phe544Leu)/ WT | 9.6 | 101.6 | 14.6 | 6 | 5 |
| DE4467 | F | GRHL2 p.(Gly65Ser)/ WT | 4.2 | 90 | 14 | 6 | 5 |
| DE4702 | M | TMC1 p.(Phe593Leu)/ WT | 5.5 | 100 | 13 | 4 | 3 |
CAP: Categories of Auditory Performance
SIR: Speech Intelligibility Rating
Discussion
In the present study, we sought to clarify the genetic characteristics of non-syndromic deafness patients without the known mutations in common deafness-related genes. Using MPS, we successfully identified candidate variants in 24.4% of these patients (10/41) by targeting 40 relatively less common and non-syndromic hearing loss genes. We also examined the segregation of the potential disease-causing variants in the families of two multiplex probands. Among the patients harboring the identified candidate variants, eight showed good CI outcomes, while two showed poor outcomes.
Several studies conducted in Eastern Asia have detected deafness genes at different rates in various targeted cohorts. For example, Wu et al. found that approximately 20.6% (37/180) of Taiwanese children with CI exhibited mutations in four common deafness-related genes: GJB2, SLC26A4, the mitochondrial 12S rRNA gene, and OTOF [14]. A phenotype-driven candidate gene approach of screening 204 reported hearing loss-related genes followed by targeted resequencing was used to find causative variants in 54.8% of sporadic severe-to-profound hearing loss patients in a Korean population [28]. In another study, MPS was used to genetically screen 60% and 36% of patients with prelingual onset hearing loss and postlingual onset hearing loss, respectively, in a Japanese population [25]. Another study used a comprehensive deafness gene panel (including 50 non-syndromic and 7 non-syndromic/syndromic deafness genes) to screen 125 deaf probands without common mutations in GJB2, SLC26A4, or MT-RNR1, and found potentially causative mutations in 26.4% (33/125) of the patients [17]. Collectively, these studies showed that different targeted genes and objects were selected at different detection rates in Asian populations, and that genetic causes play important roles in different classes of deafness. However, two limitations should be noted with respect to our study: (1) we examined a relatively small population, and (2) our utilized genetic panel did not include all of the reported non-syndromic deafness genes. Thus, some relevant variations may exist in untargeted regions (e.g., introns or regulatory elements) of screened genes and/or untargeted genes. That said, our results could facilitate the design or updating of genetic screening panels that include both common and rare deafness-related variants. Future studies could seek to increase the sample size of non-syndromic hearing loss patients and/or seek to design a new sequencing panel for clinical genomic diagnosis.
Our study population included the probands of two deafness-segregating families in which additional family members agreed to participate, allowing us to examine the potential clinical significance of the candidate variants by performing linkage analysis [40]. ACTG1 p.(Thr277Ile) was identified and confirmed in Family DE3395. ACTG1 encodes γ-actin, which is the predominant actin isoform in auditory hair cells, particularly those of the cuticular plate, adherens junction, and stereocilia [41, 42], and mutations in this gene have been reported to cause the autosomal dominant sensorineural hearing loss, DFNA20/26 [41, 42]. Previous reports found that the onset age of hearing loss in most cases caused by ACTG1 mutation is in the first decade or second decade of life [20, 43–46]. A similar pattern was seen in DE3395 (II-2) and his affected family members (I-2, II-1, and II-3). We found that three of the four affected family members exhibited the p.(Thr277Ile) variant, with the remaining affected individual (II-3) refusing to participate in genetic screening. The daughter of DE3395 (III-1) was found to have inherited the candidate variant; she currently has normal hearing, but her age (8 years at the time of screening) suggests that the phenotype may not have appeared yet. Thus, she has a high possibility of experiencing progressive hearing loss in the future. The family of III-1 should be counseled to have her hearing ability checked periodically. It is conceivable that patients with CIs could be expected to acquire a good outcome because ACTG1 acts in the hair cells of the cochlea.
In the proband of the second multiplex family, DE3386, we identified three candidate variants, MYH14 p.(Arg694Cys) (heterozygous variant), MYO15A p.(Gly1486Val) (compound heterozygous variant), and MYO15A p.(Asn1367Lys) (compound heterozygous variant). Mutations in MYH14, which encodes a long-chain non-muscle myosin IIC protein, were previously reported as causative for ADSNHI and DFNA4 [47–49]. MYH14 gene expression is detected in the organ of Corti and the stria vascularis in the inner ear; and Hensen cells, Claudius cells, and external sulcus cells which are surrounding the cochlea, all of which are important for the neural system that enables hearing [48]. Although the p.(Arg694Cys) variant is located in a non-functional domain, our findings suggest that additional work may be warranted to examine its potential impact. MYO15A (DFNB3) has been reported as a causative gene for ARSNHI [50]; it encodes myosin XV, which is an actin-dependent molecular motor family member that can hydrolyze ATP to enable actin filament movement [51]. The amino acids altered by the p.(Gly1486Val) and p.(Asn1367Lys) are located in the myosin motor domain and might impact the protein function in patient DE3386. Since this individual harbored three potentially relevant variants, we were unable to determine whether his deafness phenotype was due to the compound heterozygous variants in MYO15A and/or the heterozygous variant in MYH14. As the parents of DE3389 both had severe hearing loss (Fig 3A), we recruited them for additional analysis. Direct Sanger sequencing provided limited results: each parent was heterozygous for one of the variants in MYO15A, while the father additionally carried the MYH14 variant. Further MPS-based screening of the parents with our panel revealed that each had an additional variant in MYO15A: the father had p.(Phe3420del), which was reported as a pathogenic allele in a Chinese Han population [17], and the mother had p.(Trp3136Cys) in MYO15A, which is a novel candidate variant identified herein. This demonstrates that the MPS technique is a powerful tool for identifying genetic defects in hereditary hearing loss.
Several studies in CI patients have demonstrated that those with certain deafness gene mutations showed different treatment responses [14, 20, 24, 25]. For example, mutations in PCDH15 or DFNB59 tended to be associated with poor CI outcomes [24], whereas good outcomes were usually seen in patients with mutations in MYO6, ACTG1, and or MYO15A who received electric acoustic stimulation (EAS) and CI [21–23]. Here, we observed good CI performance in patients with variants in MYO15A, MYO3A, MYH14, ACTG1, WFS1, DSPP, and GRHL2. Some reports have described good performance in CI patients with mutations in MYO15A [20, 21], MYO3A [52], ACTG1 [22], and WFS1 [53]. However, our patient DE3221, who had a variant in COL11A2 (Table 3), had a poor CI outcome. Among our patients, there was a discrepancy in the CI outcomes of two patients with variants in TMC1. Although this gene is expressed in inner and outer hair cells, it does not correspond to our current hypothesis about the relationship of CI outcomes and location of mutated genes. Moreover, as we did not examine the inheritance mode(s) of the TMC1 variants in patients DE4377 and DE4702, it remains possible that one could represent a dominantly inherited pathogenic variant while the other was a recessively inherited carrier allele. We also note that environmental factors might influence the success of CI, leading to unexpected results. Nevertheless, most of the previous studies and our present results are consistent with the idea that the treatment outcome can be roughly predicted from the expression pattern of the variant protein, with a good outcome generally seen for variants in proteins expressed in portions on the cochlea, while a poor outcome seems more likely when the variant is in a protein related to sensory neurons. In the future, this general paradigm could help clinicians and patients make strategic decisions prior to beginning treatment.
In conclusion, we herein used MPS and a filtering strategy to identify candidate variants of rare deafness-related genes in SNHI patients. We also examined potential correlations between the identified variants and CI performance. Our findings support the idea that genetic examination could help predict the performance of implantation, which could assist deaf patients and clinicians in deciding whether or not pursue surgery.
Supporting information
(XLSX)
Acknowledgments
This study was supported by research grants from the Chang-Gung Memorial Hospital (CMRPG3E0413). The frequencies of the candidate variants in the Taiwanese population were kindly provided by the Whole- Genome Research Core Laboratory of Human Diseases at Chang Gung Medical Foundation Chang Gung Memorial Hospital, Keelung. We thank Chen-Chi Wu and Yin-Hung Lin for reviewing and providing suggestions for this article. We also thank all subjects for participating in the present study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Chang-Gung Memorial Hospital Research Program CMRPG3C1403 to CMW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Morton CC, Nance WE. Newborn hearing screening—a silent revolution. The New England journal of medicine. 2006;354(20):2151–64. 10.1056/NEJMra050700 [DOI] [PubMed] [Google Scholar]
- 2.Niparko JK, Tobey EA, Thal DJ, Eisenberg LS, Wang NY, Quittner AL, et al. Spoken language development in children following cochlear implantation. JAMA. 2010;303(15):1498–506. 10.1001/jama.2010.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikolopoulos TP, O'Donoghue GM, Archbold S. Age at implantation: its importance in pediatric cochlear implantation. Laryngoscope. 1999;109(4):595–9. 10.1097/00005537-199904000-00014 [DOI] [PubMed] [Google Scholar]
- 4.Waltzman SB, Roland JT Jr. Cochlear implantation in children younger than 12 months. Pediatrics. 2005;116(4):e487–93. 10.1542/peds.2005-0282 [DOI] [PubMed] [Google Scholar]
- 5.Cullen RD, Higgins C, Buss E, Clark M, Pillsbury HC 3rd, Buchman CA. Cochlear implantation in patients with substantial residual hearing. Laryngoscope. 2004;114(12):2218–23. 10.1097/01.mlg.0000149462.88327.7f [DOI] [PubMed] [Google Scholar]
- 6.Papsin BC. Cochlear implantation in children with anomalous cochleovestibular anatomy. Laryngoscope. 2005;115(1 Pt 2 Suppl 106):1–26. [DOI] [PubMed] [Google Scholar]
- 7.Walton J, Gibson WP, Sanli H, Prelog K. Predicting cochlear implant outcomes in children with auditory neuropathy. Otol Neurotol. 2008;29(3):302–9. 10.1097/MAO.0b013e318164d0f6 [DOI] [PubMed] [Google Scholar]
- 8.Rennels M, Pickering LK. Sensorineural hearing loss in children. Lancet. 2005;365(9477):2085–6. [DOI] [PubMed] [Google Scholar]
- 9.Hilgert N, Smith RJ, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681(2–3):189–96. 10.1016/j.mrrev.2008.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu CC, Chen PJ, Chiu YH, Lu YC, Wu MC, Hsu CJ. Prospective mutation screening of three common deafness genes in a large Taiwanese Cohort with idiopathic bilateral sensorineural hearing impairment reveals a difference in the results between families from hospitals and those from rehabilitation facilities. Audiol Neurootol. 2008;13(3):172–81. 10.1159/000112425 [DOI] [PubMed] [Google Scholar]
- 11.Wu CC, Lee YC, Chen PJ, Hsu CJ. Predominance of genetic diagnosis and imaging results as predictors in determining the speech perception performance outcome after cochlear implantation in children. Arch Pediatr Adolesc Med. 2008;162(3):269–76. 10.1001/archpediatrics.2007.59 [DOI] [PubMed] [Google Scholar]
- 12.Dahl HH, Wake M, Sarant J, Poulakis Z, Siemering K, Blamey P. Language and speech perception outcomes in hearing-impaired children with and without connexin 26 mutations. Audiol Neurootol. 2003;8(5):263–8. 10.1159/000071998 [DOI] [PubMed] [Google Scholar]
- 13.Sinnathuray AR, Raut V, Awa A, Magee A, Toner JG. A review of cochlear implantation in mitochondrial sensorineural hearing loss. Otol Neurotol. 2003;24(3):418–26. [DOI] [PubMed] [Google Scholar]
- 14.Wu CC, Liu TC, Wang SH, Hsu CJ, Wu CM. Genetic characteristics in children with cochlear implants and the corresponding auditory performance. Laryngoscope. 2011;121(6):1287–93. 10.1002/lary.21751 [DOI] [PubMed] [Google Scholar]
- 15.Nishio SY, Usami S. Deafness gene variations in a 1120 nonsyndromic hearing loss cohort: molecular epidemiology and deafness mutation spectrum of patients in Japan. Ann Otol Rhinol Laryngol. 2015;124 Suppl 1:49S–60S. [DOI] [PubMed] [Google Scholar]
- 16.Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J 2nd, Scherer S, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107(49):21104–9. 10.1073/pnas.1012989107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang T, Wei X, Chai Y, Li L, Wu H. Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis. 2013;8:85 10.1186/1750-1172-8-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagawa M, Naito T, Nishio SY, Kamatani N, Usami S. Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of Japanese deafness patients. PLoS One. 2013;8(8):e71381 10.1371/journal.pone.0071381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu CC, Lin YH, Lu YC, Chen PJ, Yang WS, Hsu CJ, et al. Application of massively parallel sequencing to genetic diagnosis in multiplex families with idiopathic sensorineural hearing impairment. PLoS One. 2013;8(2):e57369 10.1371/journal.pone.0057369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyagawa M, Nishio SY, Ikeda T, Fukushima K, Usami S. Massively parallel DNA sequencing successfully identifies new causative mutations in deafness genes in patients with cochlear implantation and EAS. PLoS One. 2013;8(10):e75793 10.1371/journal.pone.0075793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyagawa M, Nishio SY, Hattori M, Moteki H, Kobayashi Y, Sato H, et al. Mutations in the MYO15A gene are a significant cause of nonsyndromic hearing loss: massively parallel DNA sequencing-based analysis. Ann Otol Rhinol Laryngol. 2015;124 Suppl 1:158S–68S. [DOI] [PubMed] [Google Scholar]
- 22.Miyagawa M, Nishio SY, Ichinose A, Iwasaki S, Murata T, Kitajiri S, et al. Mutational spectrum and clinical features of patients with ACTG1 mutations identified by massively parallel DNA sequencing. Ann Otol Rhinol Laryngol. 2015;124 Suppl 1:84S–93S. [DOI] [PubMed] [Google Scholar]
- 23.Miyagawa M, Nishio SY, Kumakawa K, Usami S. Massively parallel DNA sequencing successfully identified seven families with deafness-associated MYO6 mutations: the mutational spectrum and clinical characteristics. Ann Otol Rhinol Laryngol. 2015;124 Suppl 1:148S–57S. [DOI] [PubMed] [Google Scholar]
- 24.Wu CC, Lin YH, Liu TC, Lin KN, Yang WS, Hsu CJ, et al. Identifying Children With Poor Cochlear Implantation Outcomes Using Massively Parallel Sequencing. Medicine (Baltimore). 2015;94(27):e1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyagawa M, Nishio SY, Usami S. A Comprehensive Study on the Etiology of Patients Receiving Cochlear Implantation With Special Emphasis on Genetic Epidemiology. Otol Neurotol. 2016;37(2):e126–34. 10.1097/MAO.0000000000000936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eppsteiner RW, Shearer AE, Hildebrand MS, Deluca AP, Ji H, Dunn CC, et al. Prediction of cochlear implant performance by genetic mutation: the spiral ganglion hypothesis. Hear Res. 2012;292(1–2):51–8. 10.1016/j.heares.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu CC, Lu YC, Chen PJ, Liu AY, Hwu WL, Hsu CJ. Application of SNaPshot multiplex assays for simultaneous multigene mutation screening in patients with idiopathic sensorineural hearing impairment. Laryngoscope. 2009;119(12):2411–6. 10.1002/lary.20621 [DOI] [PubMed] [Google Scholar]
- 28.Chang MY, Choi BY. Strategy for the customized mass screening of genetic sensorineural hearing loss in koreans. Korean J Audiol. 2014;18(2):45–9. 10.7874/kja.2014.18.2.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49(7):433–6. 10.1136/jmedgenet-2012-100918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10(10):1556–66. 10.1038/nprot.2015.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic acids research. 2003;31(13):3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–9. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods. 2010;7(8):575–6. 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 36.Archbold S, Lutman ME, Nikolopoulos T. Categories of auditory performance: inter-user reliability. Br J Audiol. 1998;32(1):7–12. [DOI] [PubMed] [Google Scholar]
- 37.Allen C, Nikolopoulos TP, Dyar D, O'Donoghue GM. Reliability of a rating scale for measuring speech intelligibility after pediatric cochlear implantation. Otol Neurotol. 2001;22(5):631–3. [DOI] [PubMed] [Google Scholar]
- 38.Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30(3):277–84. 10.1038/ng842 [DOI] [PubMed] [Google Scholar]
- 39.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brownstein Z, Bhonker Y, Avraham KB. High-throughput sequencing to decipher the genetic heterogeneity of deafness. Genome Biol. 2012;13(5):245 10.1186/gb-2012-13-5-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morin M, Bryan KE, Mayo-Merino F, Goodyear R, Mencia A, Modamio-Hoybjor S, et al. In vivo and in vitro effects of two novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum Mol Genet. 2009;18(16):3075–89. 10.1093/hmg/ddp249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khaitlina SY. Functional specificity of actin isoforms. Int Rev Cytol. 2001;202:35–98. [DOI] [PubMed] [Google Scholar]
- 43.Rendtorff ND, Zhu M, Fagerheim T, Antal TL, Jones M, Teslovich TM, et al. A novel missense mutation in ACTG1 causes dominant deafness in a Norwegian DFNA20/26 family, but ACTG1 mutations are not frequent among families with hereditary hearing impairment. Eur J Hum Genet. 2006;14(10):1097–105. 10.1038/sj.ejhg.5201670 [DOI] [PubMed] [Google Scholar]
- 44.Liu P, Li H, Ren X, Mao H, Zhu Q, Zhu Z, et al. Novel ACTG1 mutation causing autosomal dominant non-syndromic hearing impairment in a Chinese family. J Genet Genomics. 2008;35(9):553–8. 10.1016/S1673-8527(08)60075-2 [DOI] [PubMed] [Google Scholar]
- 45.de Heer AM, Huygen PL, Collin RW, Oostrik J, Kremer H, Cremers CW. Audiometric and vestibular features in a second Dutch DFNA20/26 family with a novel mutation in ACTG1. Ann Otol Rhinol Laryngol. 2009;118(5):382–90. 10.1177/000348940911800511 [DOI] [PubMed] [Google Scholar]
- 46.van Wijk E, Krieger E, Kemperman MH, De Leenheer EM, Huygen PL, Cremers CW, et al. A mutation in the gamma actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26). J Med Genet. 2003;40(12):879–84. 10.1136/jmg.40.12.879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lalwani AK, Goldstein JA, Kelley MJ, Luxford W, Castelein CM, Mhatre AN. Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. American journal of human genetics. 2000;67(5):1121–8. 10.1016/S0002-9297(07)62942-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4). Am J Hum Genet. 2004;74(4):770–6. 10.1086/383285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mhatre AN, Li J, Kim Y, Coling DE, Lalwani AK. Cloning and developmental expression of nonmuscle myosin IIA (Myh9) in the mammalian inner ear. Journal of neuroscience research. 2004;76(3):296–305. 10.1002/jnr.20065 [DOI] [PubMed] [Google Scholar]
- 50.Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, et al. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280(5368):1447–51. [DOI] [PubMed] [Google Scholar]
- 51.Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiology. 2005;20:239–51. 10.1152/physiol.00014.2005 [DOI] [PubMed] [Google Scholar]
- 52.Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, et al. From flies' eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci U S A. 2002;99(11):7518–23. 10.1073/pnas.102091699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hogewind BF, Pennings RJ, Hol FA, Kunst HP, Hoefsloot EH, Cruysberg JR, et al. Autosomal dominant optic neuropathy and sensorineual hearing loss associated with a novel mutation of WFS1. Mol Vis. 2010;16:26–35. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.