Abstract
Cell-type-specific gene expression is physiologically modulated by the binding of transcription factors to genomic enhancer sequences, to which chromatin modifiers such as histone deacetylases (HDACs) are recruited. Drugs that inhibit HDACs are in clinical use but lack specificity. HDAC3 is a stoichiometric component of nuclear receptor co-repressor complexes whose enzymatic activity depends on this interaction. HDAC3 is required for many aspects of mammalian development and physiology, for example, for controlling metabolism and circadian rhythms. In this Review, we discuss the mechanisms by which HDAC3 regulates cell type-specific enhancers, the structure of HDAC3 and its function as part of nuclear receptor co-repressors, its enzymatic activity and its post-translational modifications. We then discuss the plethora of tissue-specific physiological functions of HDAC3.
The precise regulation of gene expression is essential for the regulation of mammalian development and physiology to ensure proper growth, function, homeostasis and adaptation to changing conditions. Gene regulation is primarily mediated by sequence-specific transcription factors, which localize at cis-regulatory enhancer elements1–3. Binding of lineage-determining transcription factors facilitates chromatin opening and binding of signal-dependent transcription factors, including nuclear receptors4–10. This leads to the intricate assembly of transcriptional complexes that regulate gene expression, in part through the recruitment of transcription co-regulators that further modulate chromatin structure, including through the reversible acetylation of histone proteins by histone acetyltransferases (HATs) and histone deacetylases (HDACs)11,12.
HAT activity is an intrinsic property of transcription co-activators, which promote gene expression through the transfer of an acetyl group from acetyl-CoA to the ε-amine of several lysine residues on the amino-terminal tails of histones13–16. HDACs antagonize the functions of HATs through the deacetylation of histone tails and thus repress gene expression. Numerous studies have demonstrated that precise control of these opposing enzymatic functions is crucial for development and physiology17.
The mammalian genome encodes 11 HDAC isoforms that make up the HDAC superfamily, which exert both specific and sometimes overlapping functions in the nucleus and cytosol. The first HDAC to be isolated and characterized was HDAC1 (REFS18–21), and the gene encoding it is the founding member of the 11 highly conserved mammalian genes making up the HDAC family22. Those sharing the highest sequence homology to HDAC1 are referred to as class I HDACs, which include HDAC1, HDAC2, HDAC3 and HDAC8. The class II HDACs include HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10. Class I and class II HDACs all contain a conserved deacetylase domain and require zinc for enzymatic function19, although catalytic activity has been difficult to demonstrate for class IIa HDACs (HDAC4, HDAC5, HDAC7 and HDAC9)17,23. The single class IV HDAC is HDAC11, which demonstrates sequence conservation with class I and class II HDACs within the deacetylase domain. The structurally distinct, nicotinamide adenine dinucleotide-dependent class III HDACs (also known as sirtuins) do not share sequence similarity to class I and class II HDACs and have recently been reviewed elsewhere24–26, as have HDACs in general17,19,27.
In this Review, we comprehensively discuss the unique molecular and physiological functions of HDAC3. HDAC inhibitors are increasingly used in medicine, yet the drugs currently used affect the catalytic activity of multiple class I HDACs, including HDAC3 (REF28). We therefore focus on the fundamental differences between HDAC3 and other class I HDACs and on the varied, tissue-specific functions of HDAC3.
The structure and function of HDAC3
The recognition that HDAC3 is unique in comparison with other class I HDACs stems from the discovery in 2000 that HDAC3 is the predominant HDAC associated with nuclear receptor co-repressor 1 (NCoR1 or NCoR) and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT; also known as NCoR2), which are both nuclear receptor co-repressors29–31. Studies of these complexes provided valuable insights into the enzymatic activity of HDAC3.
The enzymatic activity of HDAC3 is dependent on its interaction with NCoR or SMRT.
Nuclear receptors function as signal-dependent transcription factors that integrate and deliver developmental, hormonal, environmental and nutrient cues to the genome, thereby acting as genetic switches of gene transcription32–35. In the classical model of nuclear receptor function, ligand binding elicits an allosteric change in the structure of the nuclear receptor, which facilitates differential recruitment of co-activators or co-repressors36,37 Co-activators with HAT activity bind to ligand-bound nuclear receptors, whereas co-repressors bind ligand-free nuclear receptors to directly mediate gene repression or less commonly to indirectly mediate gene activation (FIG. 1a). In the absence of activating ligands, nuclear receptors interact on chromatin with NCoR and SMRT, which dampen gene expression36,38,39.
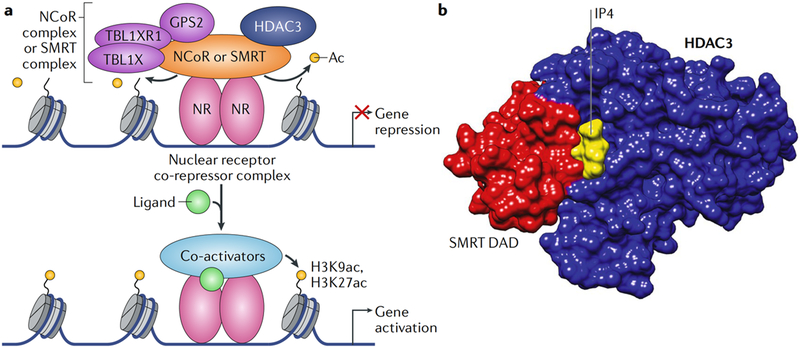
Fig. 1 |. HDAC3 is a core component of nuclear receptor co-repressor complexes that modulate nuclear receptor-mediated transcription.
a | Histone deacetylase 3 (HDAC3)-containing nuclear receptor co-repressors complexes bind to ligand-free nuclear receptors and repress transcription, partly by deacetylating histones. Ligand binding by the nuclear receptors dismisses the co-repressor complex and recruits co-activators that promote gene transcription, partly through histone acetylation. b | Crystal structure modelling (Protein Data Bank (PDB) identifier (ID): 4A69) of the HDAC3 interaction with the deacetylase-activating domain (DAD) of silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) and inositol tetraphosphate (IP4) is shown49. The interaction with the SMRT DAD is required for the enzymatic activity of HDAC3 (REFS31,47,54). Ac, acetyl group; GPS2, G protein pathway suppressor 2; H3K27ac, acetylated histone H3 lysine 27; H3K9ac, acetylated histone H3 lysine 9; NCoR, nuclear receptor co-repressor 1; NR, nuclear receptor; TBL1X, transducin β-Like 1, X-Linked; TBL1XR1, TBL1-reLated protein 1.
HDAC3 is a stoichiometric component of both the NCoR29,30 and the SMRT31 co-repressor complexes. Core NCoR and SMRT complexes contain the WD40 repeat-containing proteins TBL1X and TBL1XR131,40, which can recognize histones and recruit the ubiquitylation machinery and the 19S proteasome. The NCoR and SMRT complexes also include GPS2 (REF.41), whose molecular function within these complexes remains unclear. Together with HDAC3, these molecules represent the core stoichiometric components of the NCoR and SMRT complexes. Importantly, although earlier studies had demonstrated in vitro interactions between NCoR or SMRT and other class I HDACs, including HDAC1 (REFs42–44), HDAC3 was the only class I HDAC found in endogenous NCoR and SMRT complexes. Class II HDACs also interact with NCoR and SMRT in vitro and when overexpressed in mammalian cells42,45, but they have not been identified in endogenous NCoR or SMRT complexes.
HDAC3 is not only a stoichiometric component of the NCoR and SMRT complexes; its catalytic function requires physical interaction with a conserved domain of the NCoR and SMRT proteins, which is known as the deacetylase-activating domain (DAD)41,46–49 (FIG. 1 b). The DAD includes a SANT motif that is 57 amino acids in length, which is found in many chromatin remodellers and transcription regulators50–52; in both NCoR and SMRT, the SANT motif is flanked by a unique amino terminus of 36 amino acids in length that is required for interaction with and activation of HDAC3 by NCoR and SMRT53. The crystal structure of HDAC3 in complex with the SMRT DAD revealed extensive protein-protein interactions at the surface of the amino terminus of HDAC3 (REF49). The crystal structure also unexpectedly revealed the presence of inositol tetraphosphate (Ins(1,4,5,6)P4 or IP4), which serves as an ‘intermolecular glue’ by forming hydrogen bonds and salt bridges54 between the two proteins (FIG. 1 b). The atomic-level resolution of the DAD-HDAC3 structure also explained why a conserved tyrosine residue in SMRT (Y470) or NCoR (Y478) is crucial for the activating interaction with HDAC3 (REFS53,55,56). Interestingly, in the absence of NCoR or SMRT interactions, HDAC3 is unstable57 and sequestered into a cytosolic TCP1 ring complex58, which is a chaperone that facilitates folding of the HDAC3 polypeptide in the cytoplasm and is released upon presentation to NCoR or SMRT in the nucleus, allowing for the formation of an active HDAC3 enzyme complex in a process requiring ATP47,59.
Non-enzymatic functions of HDAC3.
The potent deacetylase activity of HDAC3 can be abolished by point mutations in the active site of the enzyme (Y298F), by mutation of two catalytically important histidine residues (H134A-H135A; known as the HAHA mutant) or by mutation of a lysine at the interface of HDAC3 with IP4 and DAD (K25A)59. Remarkably, these mutant proteins are still able to partially rescue the effect of Hdac3 deletion in mouse liver59. Furthermore, although loss of HDAC3 is lethal owing to gastrulation defects57,58,60,61, mice with mutations in the DAD of both NCoR and SMRT are born in expected Mendelian ratios, despite lacking detectable HDAC3 enzymatic activity55,62. These observations strongly suggest that HDAC3 has important non-enzymatic functions. This is consistent with the recent observation that nearly 10% of mammalian enzymes have active site-inactivating mutations yet remain widely conserved, implying selective pressure for non-catalytic functions56. The non-enzymatic mechanisms of HDAC3 function remain to be identified, but must be taken into careful consideration when evaluating HDAC3 function in different tissues, and have important implications for the development of drugs targeting the enzymatic activity of HDAC3.
Post-translational modification of HDAC3.
All mammalian HDACs contain putative sites of post-translational modification, including phosphorylation sites that may alter HDAC3 activity, stability or protein complex assembly63, similar to many nuclear receptor co-regulators64. Casein kinase 2 phosphorylates HDAC3 on Ser424 (REF65), a site that is not conserved in other class I HDACs, and alters the deacetylase activity of HDAC3 in the NCoR and SMRT complexes66. Biochemical co-purification studies also identified the protein serine/threonine phosphatase 4 complex to be associated with the HDAC3 complex66. The signalling mediators upstream of casein kinase 2, the functions of Ser424 phosphorylation in vivo and other functional post-translational modifications of HDAC3 in vivo remain to be identified.
Tissue-specific functions of HDAC3
Cell type-specific deletion of the Hdac3 gene is required to understand its physiological roles and the early embryonic lethality of whole-body deletion of Hdac3. The development of genetically engineered transgenic mouse lines expressing Cre recombinase (Cre) under the precise control of tissue-specific promoters has facilitated studies by generating Hdac3 deletions in an array of tissues and organs. Next, we discuss the consequences of Hdac3 deletion in different mouse tissues.
HDAC3 controls lipid metabolism and circadian histone deacetylation in the liver.
Hepatocyte-specific deletion of Hdac3 in a mouse line engineered to express Cre under the control of the albumin promoter (albumin-Cre) revealed considerable changes within liver hepatocytes in vivo in lipid, cholesterol and carbohydrate metabolism67, which were consistent with early cellular studies68. Deletion of Hdac3 in adult hepatocytes using adeno-associated virus (AAV) serotype 8 expressing Cre under control of the TBG promoter (AAV8-TBG-Cre) leads to massive hepatosteatosis owing to increased lipogenesis and decreased fatty acid oxidation69,70. Despite their hepatosteatosis, mice lacking HDAC3 in the liver are not insulin resistant but rather demonstrate improved glucose tolerance and insulin sensitivity, as intermediary metabolites are shunted away from gluconeogenesis towards de novo lipogenesis, where new lipids are safely stored by derepression of lipid storage pathways69, which likely prevents lipotoxicity and insulin resistance.
Genome-wide chromatin immunoprecipitation followed by sequencing (ChlP-seq) analysis of hepatic HDAC3-bound enhancers revealed a subset of enhancers exhibiting circadian HDAC3 recruitment70. This is mediated through interaction of the NCoR-HDAC3 complex with the circadian nuclear receptor REV-ERBα (also known as NR1D1), which has higher affinity for NCoR than for SMRT71,72. The rhythmic recruitment of HDAC3 leads to a circadian rhythm of histone acetylation specifically at REV-ERBα binding sites in the liver, which are enriched for lipogenic genes. Consistent with this, REV-ERBα-deficient mice exhibit hepatosteatosis73. Mechanistically, the circadian rhythm of genomic histone deacetylation mediated by HDAC3 at REV-ERBα-regulated enhancers opposes chromatin looping between enhancers and promoters and gene activation through eviction of the acetylation reader bromodomain-containing protein 4 and the looping factor mediator complex subunit 1 (REF74).
REV-ERBα-dependent HDAC3 recruitment to enhancers in the liver is mediated by two mechanisms. First, during the day, direct binding of REV-ERBα to the DNA motifs RevDR2 and ROR response element (RORE) mediates the recruitment of HDAC3 to repress circadian clock genes70,73; during the night, the same enhancers are bound by RAR-related orphan receptor (ROR) with its co-activators, thereby imparting clock gene expression (FIG. 2a). Second, at metabolic genes, HDAC3 is recruited through tethering of REV-ERBα to the liver-specific transcription factor hepatocyte nuclear factor 6 (HNF6)75. During the night, when REV-ERBα is not expressed, HDAC3 is released from HNF6 sites, allowing for co-activator recruitment and metabolic gene activation in anticipation of feeding (Fig. 2b).
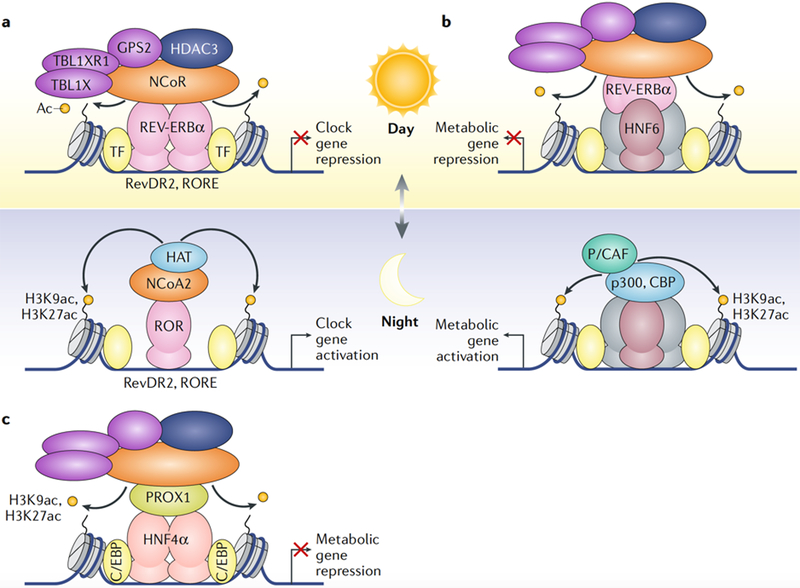
Fig. 2 |. HDAC3 suppresses liver metabolism and circadian clock genes through distinct enhancer complexes.
a | The circadian nuclear receptor REV-ERBα is maximally expressed in mice at Zeitgeber time (ZT) 10, during the light period, and is nearly absent 12 hours later in the dark period (at ZT 22). REV-ERBα recruits histone deacetylase 3 (HDAC3) through the nuclear receptor co-repressor 1 (NCoR) complex to its canonical DNA-binding motifs RevDR2 and ROR response element (RORE) to repress core circadian clock genes70,73. When REV-ERBα is not expressed, the nuclear receptor RAR-related orphan receptor (ROR) binds these motifs instead and recruits nuclear receptor co-activator 2 (NCoA2) and histone acetyltransferases (HATs) to activate gene transcription. In the depiction, ZT 0–12 is the light period and ZT 12–24 is the dark period. b | REV-ERBα tethered to hepatocyte nuclear factor 6 (HNF6) recruits HDAC3 through binding to the NCoR complex to mediate circadian repression of metabolic genes75. In the absence of REV-ERBα during the dark period, the HATs p300, CREB-binding protein (CBP) and p300/CBP-associated factor (P/CAF; also known as KAT2B) acetylate nearby histones and promote metabolic gene expression. c | Prospero homeobox protein 1 (PROX1) forms a distinct co-repressor module at enhancers bound by HNF4α independently of REV-ERBα76. PROX1 and HDAC3 are co-recruited to HNF4α-bound enhancers enriched with different C/EBP transcription factors. Ac, acetyl group; GPS2, G protein pathway suppressor 2; H3K27ac, acetylated histone H3 lysine 27; H3K9ac, acetylated histone H3 Lysine 9; TBL1X, transducin β-Like 1, X-Linked; TBL1XR1, TBL1-reLated protein 1; TF, transcription factor.
Livers lacking HDAC3 are considerably more fatty than those lacking REV-ERBα69,70, suggesting that other transcription factors are involved in the regulation of liver genes by HDAC3. Analysis of the nuclear interactome of HDAC3 in mouse livers identified a novel HDAC3 co-repressor module containing pros-pero homeobox protein 1 (PROX1), which is recruited to chromatin by the nuclear receptor HNF4α independently of REV-ERBα76 (Fig. 2c). It remains unclear how PROX1 mechanistically influences the HDAC3-containing NCoR and SMRT complexes. Other transcription factors, including nuclear receptors, are also likely to contribute to the overall function of HDAC3 in the liver76.
HDAC3 primes the thermogenic capacity of brown fat for survival in cold conditions.
In brown adipose tissue (BAT), HDAC3 is required for nonshivering thermogenesis and for the maintenance of core body temperature; thus, mice lacking HDAC3 in BAT fail to survive acute exposure to very low temperatures77. In BAT, uncoupling protein 1 (UCP1) generates heat by dissipating energy through the uncoupling of mitochondrial respiration and subsequent increase in metabolic oxidation rates78. Mice with BAT-specific deletion of Hdac3 using Ucp1-Cre or adiponectin-Cre exhibit almost no expression of UCP1, as well as transcriptional downregulation of mitochondrial and oxidative phosphorylation genes required for nonshivering thermogenesis and cold survival77 (Fig. 3a). Bioinformatics interrogation of global run-on sequencing (GRO-seq) data in BAT to map functional enhancers through the identification of enhancer RNAs revealed thousands of sites where enhancer activity was downregulated upon loss of HDAC3 binding. Additional bioinformatics integration of ChlP-seq data sets uncovered a novel mechanism of HDAC3 co-activation at enhancers bound by the thermogenic nuclear receptor oestrogen-related receptor-α (ERRα)77,79–81.
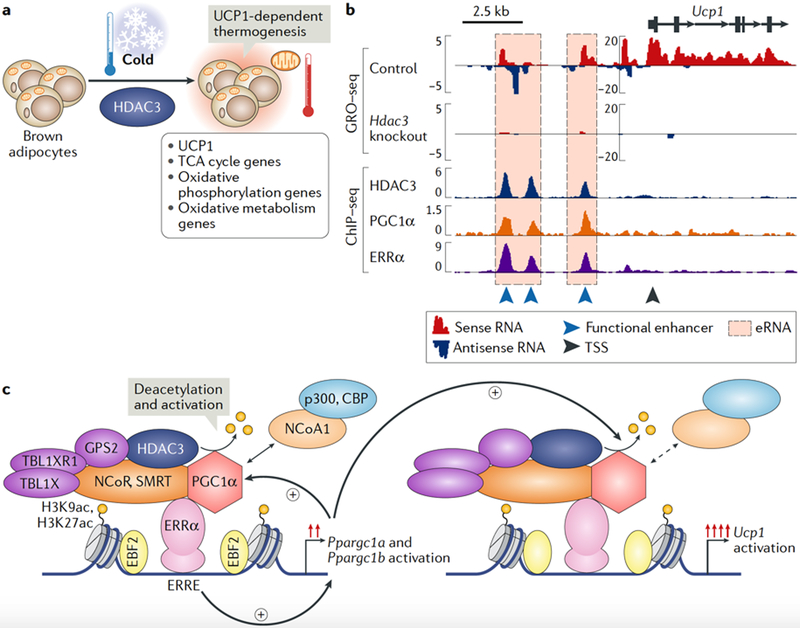
Fig. 3 |. HDAC3 primes thermogenic gene transcription in brown adipose tissue.
a | In brown adipose tissue (BAT), histone deacetylase 3 (HDAC3) primes the expression of Ucp1 (which encodes mitochondrial brown fat uncoupling protein 1), tricarboxylic acid (TCA) cycle genes, oxidative phosphorylation genes and oxidative metabolism genes to facilitate rapid UCP1-dependent thermogenesis upon exposure to acute cold77. b | A genomic view of the Ucp1 locus is shown, highlighting the co-activator activity of HDAC3 at oestrogen-related receptor-α (ERRα)-bound enhancers in BAT. Global run-on sequencing (GRO-seq) data demonstrate strong enhancer RNA (eRNA) transcription at Ucp1 enhancers and transcription of the Ucp1 gene in control BAT. Upon loss of HDAC3, eRNA transcription and expression of the Ucp1 gene are lost at enhancers bound by HDAC3, ERRα and peroxisome proliferator-activated receptor-γ co-activator 1α (PGC1α) (another ERRα co-activator)77. The GRO-seq y axis represents reads per kilobase per million; the chromatin immunoprecipitation followed by sequencing (ChlP-seq) y axis represents reads per million. c | In BAT, HDAC3 activates PGC1α by deacetyLating it77,157,158. HDAC3 and ERRα bind to enhancers of the Ppargcla and Ppargclb genes to promote their basal transcription levels. Increased expression of PGC1α facilitates an autoreguLatory loop maintained by HDAC3, ERRα and PGC1α, which drives the transcription of Ucp1 and oxidative phosphorylation genes to ensure thermogenic aptitude and survival upon exposure to a cold environment77. Notably, enhancers bound by HDAC3, ERRα and PGC1α are also marked by the brown fat lineage factor early B cell factor 2 (EBF2)159,160. CBP, CREB-binding protein; ERRE, ERR response element; GPS2, G protein pathway suppressor 2; H3K27ac, acetylated histone H3 Lysine 27; H3K9ac, acetylated histone H3 Lysine 9; NCoA1, nuclear receptor co-activator l; NCoR, nuclear receptor co-repressor l; SMRT, silencing mediator of retinoic acid and thyroid hormone receptor; TBL1X, transducin β-Like l, X-Linked; TBL1XRl, TBL1-reLated protein l; TSS, transcription start site.
As exemplified by the Ucp1 locus, HDAC3 colocalization with ERRα on chromatin is required for enhancer activation and Ucp1 transcription (FIG. 3b). These Ucp1 enhancers are also bound by peroxisome proliferator-activated receptor-γ co-activator 1α (PGC1α)82· which is essential for ERRα-dependent transcription77,83. PGC1α is repressed by acetylation and derepressed by HDAC3-mediated deacetylation (FIG. 3c). Furthermore, HDAC3, PGC1α and ERRα co-bind enhancers of the Ppargc1a and Ppargc1b genes, thereby initiating a positive transcriptional feedback loop that supports the expression of Ppargc1a, Ppargc1b, Ucp1 and oxidative phosphorylation genes that are essential for thermogenesis (FIG. 3c). Two subsequent studies demonstrated that ERRα84 and its closely related family member ERRγ85 are required for establishing the basal thermogenic and oxidative capacity of BAT; a tissue-specific deletion of these factors causes a phenotype highly similar to the HDAC3 BAT-specific knockout84,85. Additionally, the BAT transcription factor PR domain-containing protein 16 (PRDM16)86 interacts with HDAC3 and NCoR87, suggesting the existence of additional interactions among several important regulators of BAT thermogenesis. Notably, in the white adipose tissue of aged mice exhibiting impaired induction of brown-like ‘beige’ or ‘brite’ adipocytes88, HDAC3 may modulate futile metabolic cycles89. Collectively, these studies demonstrate that HDAC3 is a major regulator of BAT-mediated thermogenesis and energy homeostasis.
HDAC3 influences cardiac development and cardio-myocyte metabolism.
The prevalence of congenital heart defects is nearly 1% of live births90. Cardiac progenitor cells (CPCs) making up the first heart field (FHF) and second heart field (SHF) are controlled by distinct gene networks that facilitate the development of the major cell types of the adult heart91. Depletion of HDAC3 in CPCs in vivo initiates precocious differentiation of cardiomyocytes, indicating the existence of a HDAC3-dependent coordination of CPC specification92–94. In embryonic stem cell models, HDAC3 was shown to tether cardiac lineage genes located within lamina-associated domains (LADs) to the nuclear lamina to silence the genes through the formation of facultative heterochromatin94 (FIG. 4a). HDAC3 tethering of LADs to the nuclear periphery is independent of its catalytic function and mediated through physical interactions with LAD-associated protein complexes90,94–96. During differentiation, the HDAC3-tethered cardiac LADs are released from the nuclear periphery to dissipate the heterochromatin configuration and establish a chromatin configuration that is permissive for the expression of cardiomyocyte genes97. How HDAC3-tethered LADs are released from the nuclear lamina during development is unknown94.
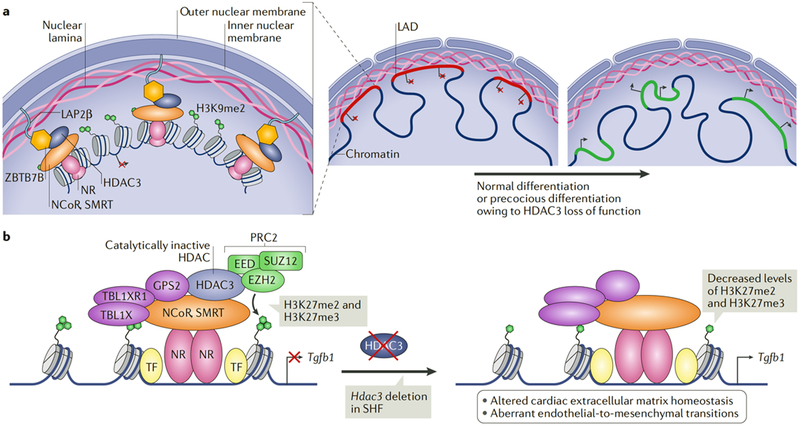
Fig. 4 |. HDAC3 influences cardiac development through deacetylase-independent mechanisms.
a | Histone deacetylase 3 (HDAC3) prevents the precocious differentiation of cardiac progenitor cells and premature expression of cardiomyocyte genes92–94. HDAC3 tethers cardiac lineage genes located in lamina-associated domains (LADs) to the nuclear periphery for silencing. HDAC3 bound to the nuclear receptor co-repressor 1 (NCoR) and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) complexes on chromatin interacts with the lamina proteins zinc-finger and BTB domain-containing protein 7B (ZBTB7B) and lamina-associated polypeptide 2, isoform-β (LAP2β) to tether LADs to the nuclear lamina, where they become facultative heterochromatin and enriched in the repressive histone H3 lysine 9 dimethylation (H3K9me2) modification90–94. The release of LADs from the nuclear periphery alters their chromatin configuration and facilitates the expression of cardiomyocyte genes94,97. b | In the second heart field (SHF), the non-enzymatic activity of HDAC3 represses transforming growth factor-β (TGFβ) signalling by recruiting the H3K27 methyltransferase Polycomb repressive complex 2 (PRC2) to the Tgfb1 gene. EED, embryonic ectoderm development protein; EZH2, enhancer of zeste homologue 2; GPS2, G protein pathway suppressor 2; H3K27me3, histone H3 lysine 27 trimethylation; NR, nuclear receptor; TF, transcription factor; SUZ12, suppressor of zeste 12 protein homologue; TBL1X, transducin β-like 1, X-linked; TBL1XR1, TBL1-related protein 1.
Deletion of Hdac3 in the FHF using Nkx2–5-Cre results in lethality between embryonic day 11.5 (E11.5) and birth owing to severe underdevelopment of ventricular walls and to ventricular septal defects92. During FHF cardiogenesis, the transcription factor TBX5 promotes HDAC3 recruitment with nuclear receptors at select enhancers to repress cardiac lineage genes. Notably, a mutation in human TBX5 (G125R) causes Holt-Oram syndrome, which is characterized by skeletal abnormalities and cardiac malformations, and may affect HDAC3 protein interactions and recruitment to chromatin92.
Progenitor cells of the SHF give rise to structures that are involved in nearly two-thirds of all human congenital cardiac defects, including those seen in tetralogy of Fallot and DiGeorge syndrome98. Deletion of Hdac3 in the SHF using Isl1-Cre and Mef2c-anterior heart field (AHF)-Cre activated transforming growth factor-β (TGFβ) signalling pathways and caused aberrant secretion of extracellular matrix proteins by myofibroblasts, which precipitated a connective tissue disorder and resulted in perinatal lethality owing to severe aortic, outflow tract, valve, ventricle and septal defects93. The phenotype of loss of HDAC3-mediated silencing of TGFβ signalling resembles the pathogenic connective tissue disruption and alteration of TGFβ signalling seen in Marfan syndrome and Loeys-Dietz syndrome93,99,100. In the SHF, HDAC3 silences TGFβl through a deacetylase-independent function by acting as a protein scaffold for the recruitment of components of the histone H3 lysine 27 (H3K27) methyltransferase Polycomb repressive complex 2 (PRC2) (FIG. 4b). Deletion of Hdac3 in neural crest cells (NCCs) using Wnt1-Cre or Pax3-Cre also causes perinatal cardiovascular abnormalities through impairment of cell-autonomous differentiation into the smooth muscle of the heart owing to altered Notch signalling101.
Prenatal cardiac-specific deletion of Hdac3 using Myhca-Cre resulted in lethality by 16 weeks of age. Mice with loss of HDAC3 in cardiomyocytes developed severe hypertrophic cardiomyopathy and interstitial fibrosis of the left ventricle and septum, which were caused by pathological ventricular dysfunction owing to severe mitochondrial dysfunction, reduced activity of the electron transport chain and elevated levels of fasting myocardial triglycerides61. Postnatal deletion of Hdac3 using Mck-Cre in cardiac and skeletal muscles resulted in a milder phenotype of ventricular and septal thickening, enlarged atria and elevated levels of fasting myocardial triglycerides associated with repression of cardiac oxidative phosphorylation and fatty acid metabolism pathways102. Challenging these mice with a high-fat diet caused severe hypertrophic cardiomyopathy and lethal heart failure owing to impaired fatty acid oxidation and myocardial energy starvation102, suggesting that HDAC3 modulates cardiomyocytes to respond to dynamic environmental and nutrient conditions. The cardiomyopathies of both mouse models are reminiscent of those of mice lacking peroxisome proliferator-activated receptors or ERRs103, or their co-activators PGC1α and PGC1β104. The complex consequences of Hdac3 deletion in the heart clearly require further characterization of the specific functions of HDAC3 in development and in specific cell populations.
HDAC3 regulates neuronal cell fate and function.
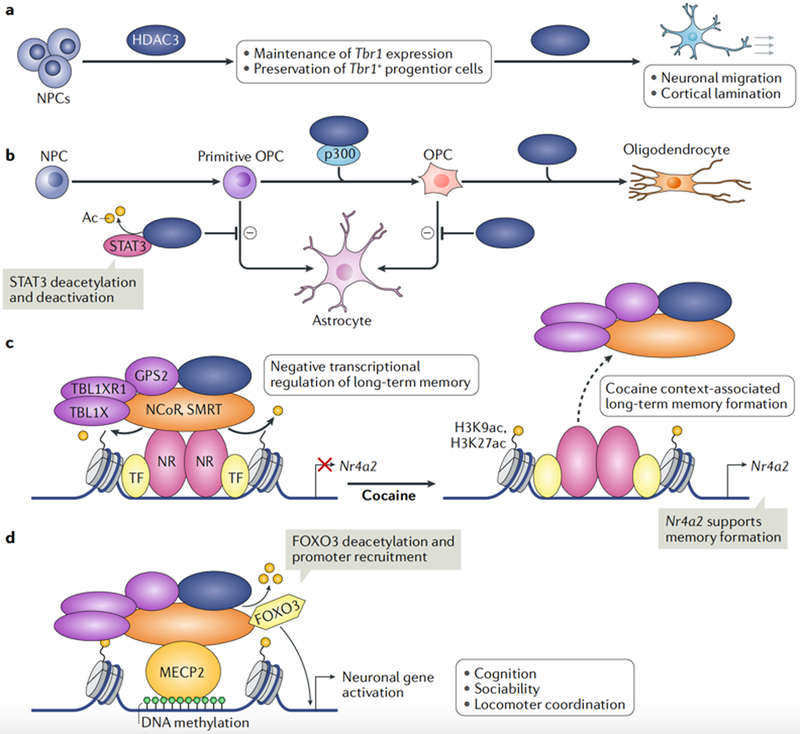
HDAC3 also has an important role in neurodevelopment. Loss of HDAC3 in neural progenitor cells (NPCs) of the central nervous system leads to death within 16 hours of birth, which is attributed to impaired neuronal migration and cortical lamination, possibly owing to loss of HDAC3-dependent expression of the transcription factor T-box brain protein 1 (REF105) (FIG. 5a). HDAC3 in NPCs also controls glial cell fate, as mice lacking HDAC3 in NPCs exhibit increased numbers of astrocytes with a concomitant decrease in oligodendrocytes105 (FIG. 5b). Indeed, deletion of Hdac3 in primitive oligodendrocyte progenitor cells (OPCs) leads to a significant increase in the number of astrocytes relative to oligodendrocytes, resulting in pathological tremors and optic nerve impairment owing to myelination defects106. Neurodevelopmental cell fate mapping in mice demonstrated that HDAC3 guides OPCs towards the oligodendrocyte fate and away from the astrocyte fate106. ChlP-seq of HDAC3 in combination with global gene expression profiling in OPCs revealed a mechanism whereby HDAC3 deacetylates and inactivates the transcription factor signal transducer and activator of transcription 3 (STAT3) to inhibit astrogliogenesis; HDAC3 also interacts with the HAT p300 to activate enhancers and genes that control oligodendrocyte formation presumably through deacetylation of non-histone proteins106 (FIG. 5b). Recent studies also revealed that HDAC3 represses pro-myelination expression programmes that include the HIPPO signalling effector TEAD4107.
Fig. 5 |. HDAC3 controls brain development, glial cell fate and the formation of long-term memory.
a | Histone deacetylase 3 (HDAC3) controls brain development through maintenance of T-box brain 1 (Tbr1) gene expression or preservation of the Tbr1+ progenitor cell population. Loss of HDAC3 leads to impaired differentiation and migration of neural progenitor cells (NPCs), resulting in severe brain cytoarchitectural defects105. b | HDAC3 controls glial cell fate. HDAC3 inhibits intermediate primitive oligodendrocyte progenitor cells (OPCs) and OPCs from undergoing astrogliogenesis, thereby promoting the formation of oligodendrocytes. HDAC3 inhibition of astroglial differentiation occurs through deacetylation and inhibition of signal transducer and activator of transcription 3 (STAT3), and by interaction with the histone acetyltransferase p300 to activate oligodendrocyte gene enhancers105,106. Loss of HDAC3 decreases oligodendrocyte numbers and increases astrocyte numbers105,106. c | In the hippocampus, HDAC3 represses nuclear receptor subfamily 4, group A, member 2 (Nr4a2) gene, which is important for the formation of long-term memory and object recognition108. HDAC3 also regulates the Nr4a2 gene in the nucleus accumbens, where HDAC3 binding at enhancers is attenuated by cocaine use, which may facilitate long-term memories associated with drug abuse and reinforce future use109. d | HDAC3 is recruited to specific sites of methylated DNA by the nuclear receptor co-repressor 1 (NCoR) and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) complexes through their interaction with methyl-CpG-binding protein 2 (MECP2), which is implicated in Rett syndrome110,111. There, HDAC3 deacetylates and activates the transcription factor forkhead box protein O3 (FOXO3) to promote neuronal gene expression112. Ac, acetyl group; GPS2, G protein pathway suppressor 2; H3K27ac, acetylated histone H3 lysine 27; H3K9ac, acetylated histone H3 lysine 9; NR, nuclear receptor; TBL1X, transducin β-like 1, X-linked; TBL1XR1, TBL1-related protein 1; TF, transcription factor.
Thy1-Cre-mediated and Camk2a-Cre-mediated knockout of Hdac3 in the postnatal forebrain and cerebellum led to lethality by 6 weeks of age. These mice had developed severe locomotor deficits, progressive ataxia and paralysis secondary to the loss of Purkinje neurons and substantial cerebellum abnormalities105. As HDAC3 is the most highly expressed class I HDAC in the adult brain, there is great interest in its influence on the transcriptional regulation of learning and memory. Loss of HDAC3 within the adult CA1 hippocampal region through bilateral intracranial injection of AAV-Cre revealed that HDAC3 is an important inhibitor of the formation of long-term memory through repression of the gene nuclear receptor subfamily 4, group A, member 2 (Nr4a2)108 (FIG. 5c). Mice with the Y478A mutation in the NCoR DAD phenocopied the hippocampal deletion of Hdac3 and thus established the existence of a HDAC3 catalytic-dependent mechanism that restricts the formation of long-term memory. Similarly, cocaine use decreases HDAC3 occupancy at the Nr4a2 enhancer and promotes Nr4a2 expression in the nucleus accumbens109. Thus, cocaine-induced Nr4a2 expression may enhance memory formation and promote future drug abuse (Fig. 5c).
HDAC3 has recently been associated with the neurological disorder Rett syndrome. A missense mutation (R306C) in methyl-CpG-binding protein 2 (MECP2) impairs the recruitment of the NCoR-HDAC3 complex to MECP2-bound methylated DNA to repress gene transcription110,111. Deletion of Hdac3 using Camk2-Cre in forebrain excitatory neurons causes a phenotype similar to that of the Mecp2 knockout mouse model of Rett syndrome, including cognitive deficits and impaired sociability and locomotor coordination112. HDAC3 cistrome and transcriptome analyses in vivo showed that HDAC3 is a positive transcriptional regulator at a subset of neuronal genes; it is recruited to these genes by MECP2 and deacetylates the neuronal transcription factor FOXO3 (Fig. 5d). Notably, NPCs derived from human induced pluripotent stem cells harbouring the Rett syndrome MECP2-R306C mutation and corrected by CRISPR-Cas gene editing mechanistically re-capitulated and validated the mouse models demonstrating HDAC3 working as a positive transcriptional regulator at enhancers with FOXO3 (REP112).
HDAC3 coordinates lung development.
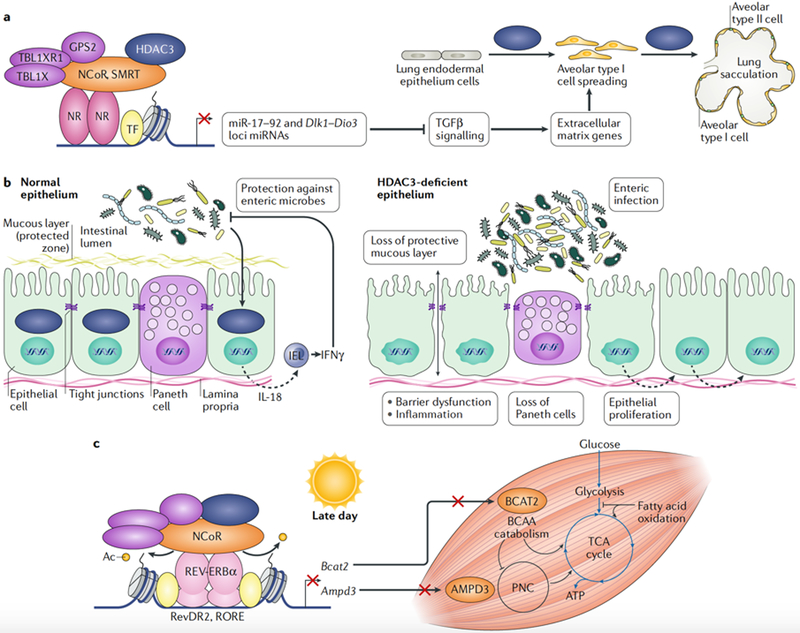
Conditional deletion of Hdac3 using sonic hedgehog (Shh-Cre) in the lung endodermal epithelium at E8.75 revealed a requirement for HDAC3 in lung sacculation and early alveologenesis, resulting in lethality between 2 and 10 days after birth113. In lung endodermal epithelium, HDAC3 is recruited to enhancers through an undetermined nuclear receptor to repress the transcription of the miR-17–92 microRNA cluster and many of the nearly 50 miRNAs transcribed from the imprinted Dlk1-Dio3 gene cluster (FIG. 6a). Expression of miR-17–92 inhibits the cell-autonomous TGFβ signalling that is required for cell flattening and spreading during sacculation of lung alveolar type I cells, thereby ensuring efficient gas exchange113. Deletion of Hdac3 in the developing lung mesenchyme by twist basic helix-loop-helix transcription factor 2 (Twist2)-Cre also resulted in complete lethality at birth as a result of respiratory failure114. Unlike Hdac3 deletion in lung endoderm, deletion in the underlying lung mesoderm causes lung hypoplasia and loss of alveolar type I cell differentiation owing to decreased WNT-β-catenin signalling from the mesenchyme to the epithelium114.
Fig. 6 |. HDAC3 regulates lung development, intestinal homeostasis, pancreatic β-cell insulin secretion and skeletal muscle metabolism.
a | Histone deacetylase 3 (HDAC3) regulates the differentiation of lung epithelial cells through repression of the Dlk1-Dio3 and miR-17–92 microRNA (miRNA) clusters. Increased expression of miRNAs represses genes in the transforming growth factor-β (TGFβ) signalling pathway, leading to defective extracellular matrix formation and failure of alveolar cell spreading. Defects in cell spreading and flattening impair lung sacculation and aίveoίogenesis113. b | In intestinal epithelial cells (IECs), HDAC3 is required for maintenance of the epithelial barrier that separates the gut lumen from the systemic circulation, ensures Paneth cell survival, controls cell proliferation and protects against enteric bacteria. HDAC3 in IECs prevents enteric infection by signalling to intraepithelial CD8+ T lymphocytes (IELs) to mount a protective interferon-γ (IFNγ) immune response116. c | Late in the day, at pre-dusk hours (Zeitgeber time (ZT) 10–12), HDAC3 and REV-ERBα repress branched-chain amino acid (BCAA) catabolism and the purine nucleotide cycle (PNC) pathway through repression of branched-chain amino acid aminotransferase, mitochondrial (BCAT2) and AMP deaminase 3 (AMPD3), respectively, to promote glucose utilization during the upcoming feeding cycle when glucose supply is abundant125. At pre-dawn hours (ZT 22–24), when REV-ERBα and HDAC3 do not bind, increased BCAT2 and AMPD3 expression promotes the utilization of protein and lipid fuels over glucose to supply metabolic intermediates to the tricarboxylic acid (TCA) cycle. Muscle utilization of protein and lipid fuels during periods of daily fasting preserves glucose supplies for the brain and other vital organs125. Ac, acetyl group; GPS2, G protein pathway suppressor 2; IL-18, interίeukin-18; NCoR, nuclear receptor co-repressor 1; NR, nuclear receptor; RORE, ROR response element; SMRT, silencing mediator of retinoic acid and thyroid hormone receptor; TBL1X, transducin β-like 1, X-linked; TBL1XR1, TBL1-reίated protein 1; TF, transcription factor.
HDAC3 governs intestinal homeostasis and host defence.
In intestinal epithelial cells (IECs), HDAC3 maintains intestinal homeostasis by coordinating intestinal lymphocyte-mediated host defences against pathogenic microorganisms, with implications for the course of inflammatory bowel disease (IBD) and bacterial enteritis115,116. Deletion of Hdac3 in IECs using Vil1-Cre or inducible Vil1-CreER resulted in the downregulation of IEC antimicrobial defence genes, the loss of Paneth cells and impairment in the function of the IEC barrier, causing increased intestinal damage and inflammation115 (FIG. 6b). Notably, the phenotype of IEC-specific Hdac3 knockout mice re-derived in germ-free conditions was largely normal, suggesting that HDAC3 is a hub of commensal-bacteria-derived signals that maintains intestinal homeostasis115. Furthermore, in IECs, HDAC3 orchestrates a host defence programme that is mediated through cytokine communication from IECs to resident CD8+ T cell lymphocytes in the intestines to protect against pathogenic gut bacteria116. Intestinal epithelial HDAC3 has also been shown to contribute to diet-induced obesity in mice117.
HDAC3 in pancreatic β-cells controls glucose-stimulated insulin secretion.
Loss of Hdac3 in insulin-producing pancreatic β-cells in the mouse insulin promoter CreERT transgenic mouse line resulted in improved glucose tolerance owing to enhanced insulin secretion without changes in insulin production or β-cell mass118. β-Cells deficient for HDAC3 demonstrate increased response to glucose levels through secretion of greater amounts of insulin into the systemic circulation. The phenotype of β-cells lacking HDAC3 is consistent with the use of a selective HDAC3 inhibitor, which improved glucose metabolism in diabetic rats119. Moreover, HDAC3 depletion alleviated progressive cytokine-mediated β-cell apoptosis, which was hypothesized to contribute to type 2 diabetes120,121. By contrast, the rat insulin II promoter Cre HDAC3 β-cell deletion mouse model exhibited decreased insulin content in the pancreas and impaired glucose-stimulated insulin secretion122, which was perhaps confounded by aberrant Cre-mediated excision in the central nervous system123,124. Future studies of HDAC3 function in the endocrine pancreas will have to target specific endocrine cell types and use more refined Cre deletion strategies in order to explain these conflicting results.
HDAC3 impacts skeletal muscle metabolism and a fuel source switch.
Deletion of Hdac3 in skeletal muscles using Mlc-Cre causes severe systemic and skeletal muscle-specific insulin resistance, impaired insulin and glucose tolerance, and diminished glucose uptake into skeletal muscles, outcomes that are independent of changes in insulin signalling. Paradoxically, deletion of Hdac3 in skeletal muscles improved exercise endurance despite these metabolic alterations125. ChIP-seq, RNA sequencing and quantitative proteomic analyses revealed that, during the daytime, skeletal muscle HDAC3 in concert with REV-ERBα represses the expression of the catabolic enzyme branched-chain amino acid aminotransferase, mitochondrial (BCAT2) and AMP deaminase 3 (AMPD3), which is the first enzyme of the purine nucleotide cycle pathway125 (FIG. 6c). In the absence of HDAC3 in skeletal muscles, elevated expression of BCAT2 and AMPD3 feeds metabolic intermediates into the tricarboxylic acid cycle to promote a fuel source switch favouring protein catabolism and lipid oxidation over glucose utilization125. Thus, HDAC3 and REV-ERBα control a gene regulatory circuit that imparts a circadian rhythm of skeletal muscle fuel source usage to anticipate muscle endurance requirements during periods of daily fasting125.
HDAC3 promotes haematopoietic cell lineages and prevents a lethal autoimmune disease.
HDAC3 orchestrates a remarkable number of processes and functions in immune cells, which influence cell homeostasis and transit through development stages and systemic immune responses. For example, Lysm-Cre-mediated depletion of HDAC3 in macrophages promoted an anti-inflammatory histone acetylation profile and gene expression pattern and enhanced interleukin-4-induced alternative macrophage activation, with a protective effect from parasitic inflammation in vivo126. Reciprocally, loss of HDAC3 has been shown to impair canonical pro-inflammatory gene expression programmes induced by lipopolysaccharides127. Highlighting the importance of HDAC3 in macrophage physiology, transplantation of anti-inflammatory HDAC3-lacking macrophages into atherosclerosis-prone low-density lipoprotein receptor (LDLR)-null mice induced the formation of more stable arterial plaque lesions with decreased lipid accumulation within foam cells and no effect on systemic lipid levels128. It remains unclear which nuclear receptor recruits HDAC3 to chromatin in macrophages; however, the lineage-determining transcription factor PU.1, which is enriched at HDAC3 binding sites, likely facilitates adjacent binding of nuclear receptors126.
Within the haematopoietic compartment, deletion of Hdac3 using Vav1-Cre produced a dysfunctional thymus with significant loss of lymphoid cells, hypocellular bone marrow and anaemia arising from DNA replication defects and genomic instability129,130. Furthermore, HDAC3 is required for stem cell proliferation, renewal and repopulation in bone marrow transplantation assays130. Deletion of Hdac3 in early B cell progenitors using Cd79a-Cre impedes B cell development owing to a substantial hindrance of distal VH-DJH immunoglobulin heavy chain recombination, which is caused by altered chromatin structure that is thought to impair the formation of long-distance chromatin interactions (chromatin looping), which, notably, requires the catalytic activity of HDAC3 (REF.131).
Deletion of Hdac3 using Lck-Cre at the early double-negative (CD4−CD8−) stage of T cell development impaired the transition to the double-positive (CD4+CD8+) stage and revealed a downregulation of a majority of genes for T cell development, function and cell cycle by an unclear mechanism132. Consistent with the phenotype of Lck-Cre-mediated loss of HDAC3 (REF132), deletion of Hdac3 at the CD4+CD8+ stage using Cd2-Cre or Cd4-Cre led to a considerable block in T cell positive selection, maturation and ultimately T cell survival133,134. The Cd2-Cre mouse model of HDAC3 deletion uniquely exhibited rectal prolapse and predisposition to IBD by 2 months of age, perhaps through altered function of peripheral regulatory T (Treg) cells133. Deletion of Hdac3 in Treg cells using Foxp3-Cre disrupted the HDAC3-dependent function of immunosuppressive Treg cells, which caused activation of conventional T cells and B cells and initiated lethal autoimmunity in animals by 4–6 weeks of age, perhaps through interactions of FOXP3 and nuclear factor-κB with HDAC3 (REFS135,136). Similarly to the Cd2-Cre mouse model, Treg cell HDAC3 was required to suppress IBD133,135.
HDAC3 is required for the development and physiological remodelling of bones.
Following genetic studies in NCCs, mesenchymal progenitors, osteoblasts and osteoclasts, which covered many stages of skeletogenesis, HDAC3 has emerged as a regulator of bone development and growth137. Wnt-Cre-driven and Pax3-Cre-driven deletion of Hdac3 in pre-migratory NCCs, which make up the craniofacial mesenchyme, causes severe microcephaly, micrognathia, cleft palate and impaired odontogenesis, ultimately resulting in perinatal lethality101,138. Sp7-Cre-mediated deletion of Hdac3 in osteochondral progenitor cells resulted in less-severe craniofacial developmental abnormalities but significantly shortened long bones and caused severe osteopenia affecting bone strength, a phenotype that ultimately decreased lifespan139. Specific deletion of Hdac3 during limb bud development using Prrxl-Cre caused severe shortening of both the forelimbs and the hindlimbs and perinatal lethality140. Deletion of Hdac3 in early developing chondrocytes using Col2a1-Cre was embryonic lethal141.
Deletion of Hdac3 in mature chondrocytes using tamoxifen-inducible Col2a1-CreERT delayed secondary ossification, altered growth plate and long bone development and increased osteoclast activity, which caused significant reductions in bone density141. In adult osteoblasts, HDAC3 deficiency considerably increased age-related osteopenia, osteoporosis and bone fractures of relevance in human ageing142. At bone gene enhancers, HDAC3 may function in part through the transcription factors RUNX2 and zinc-finger protein 521 and yet unknown nuclear receptors, resulting in complex and temporal effects in bone and cartilage143–145. The phenotypes caused by loss of HDAC3 may in part explain the axial and cranial skeletal defects and predisposition to bone fractures observed with the use of HDAC inhibitors146–153.
Conclusions and future perspective
HDAC3 is unique among the HDAC family proteins as the major HDAC that functions as part of nuclear receptor co-repressor complexes and requires them for its catalytic activity29–31. HDAC3 is vital to life because it regulates many developmental, physiological and metabolic processes. Throughout development, HDAC3 serves to integrate complex signals from the environment and deliver them onto the genome to influence cellular migration, differentiation, fate and growth. In adult mammals, HDAC3 uniquely regulates major metabolic and energy utilization pathways and circadian, nutrient and environmental challenges, influences memory formation, limits autoimmunity and protects skeletal integrity, among other homeostatic functions.
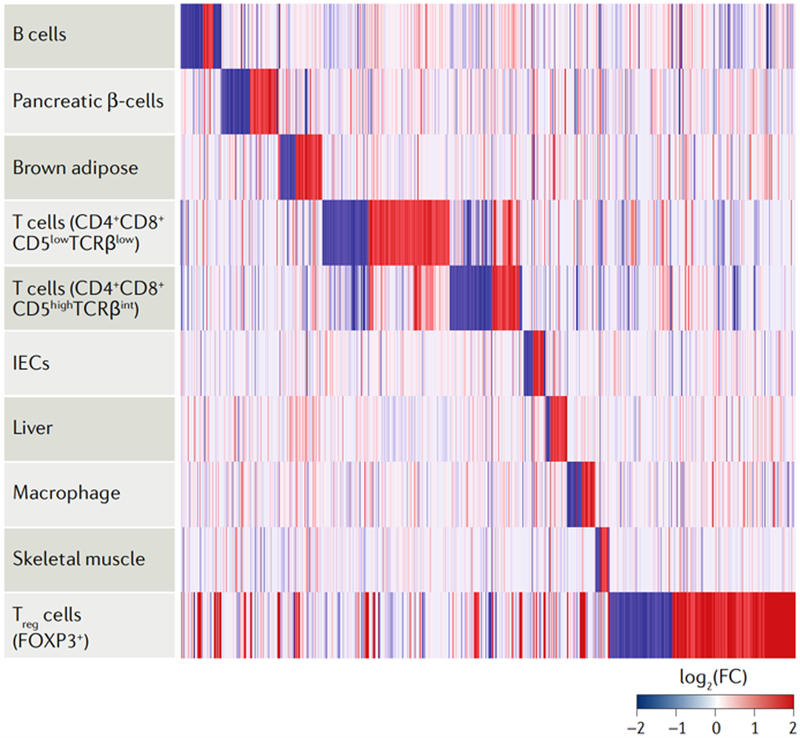
Transcriptomics analyses of mouse models with conditional HDAC3 loss of function have revealed an extraordinarily diverse set of tissue-specific HDAC3 targets. Indeed, many of the genes regulated by HDAC3 are highly tissue-specific (FIG. 7), which is consistent with cell context-dependent functions of, and tissue-specific mechanisms of, gene regulation by HDAC3, reinforcing the notion that the function and gene targets of HDAC3 cannot be predicted a priori. Although less complete, genome-wide analyses of the mechanisms of HDAC3 activity have revealed a remarkably diverse set of nuclear receptors and other transcription factors that recruit HDAC3 to chromatin, histone and non-histone targets and to participate in enzymatic and non-enzymatic functions. Thus, HDAC3 evolved as a dynamic chromatin regulator that is co-opted by many tissue-specific factors to exert biological effects, many of which remain to be discovered.
Fig. 7 |. HDAC3 regulates distinct mouse tissue-specific gene expression programmes.
A heat map illustrating the differential gene expression and hierarchical clustering of ten mouse tissues is shown, which characterize tissue-specific histone deacetylase 3 (HDAC3)-dependent gene expression programmes achieved through the numerous and complex tissue-specific regulatory mechanisms highlighted in this Review. We display all the expressed genes in each tissue as log2 of the fold change (FC) of HDAC3 knockout versus controls (false discovery rate <0.05); the rows (tissues) were sorted by hierarchical clustering. Gene expression data were obtained from publicly available Gene Expression Omnibus (GEO) data sets, normalized for cross-platform comparisons161 and log-transformed; comparisons were made using the R package limma162. The GEO data sets used for this analysis are GSE98650, GSE90531, GSE83927, GSE72917, GSE50188, GSE85929, GSE33609, GSE79696 and GSE68991. IEC, intestinal epithelial cell; FOXP3, forkhead box protein P3; TCRβ, T cell receptor-β; Treg cell, regulatory T cell.
To date, no human disease-causing mutations have been found in the HDAC3 gene, likely owing to the lethality of such germline mutations, which are expected to affect multiple tissues. Despite this, enhancers may exist with single nucleotide polymorphisms that fail to recruit HDAC3 owing to impaired binding of transcription factors or nuclear receptors10,154,155, causing pathological gene expression. Other HDAC3 ‘enhanceropathies’156 may occur as a result of mutations in HDAC3-associated proteins.
Many questions remain about the biology of HDAC3. One glaring question is whether all functions of HDAC3 require its presence in the NCoR and SMRT co-repressor complexes. Because interaction with NCoR or SMRT is required for HDAC3 activity in vivo55, it seems likely that the enzymatic functions of HDAC3 are mediated by these complexes, but this is not the case for the non-enzymatic functions of HDAC3. Indeed, although HDAC3 loss-of-function mouse models have provided great insight into the physiological roles of HDAC3, we currently have little mechanistic understanding of the non-enzymatic functions of HDAC3. This is particularly important given the pharmaceutical interest in targeting the enzymatic activity of HDAC3 (REF.150).
Acknowledgements
The authors thank members of the Lazar laboratory for discussion and comments on the manuscript. They also thank H.-W. Lim for processing ChlP-seq and GRO-seq data from GEO data sets (GSE83928 and GSE110056), H. B. Nguyen for bioinformatics analysis of gene expression (GSE98650, GSE90531, GSE83927, GSE72917, GSE50188, GSE85929, GSE33609, GSE79696 and GSE68991) and H. J. Richter for assistance with modelling the crystal structure of HDAC3. US National Institutes of Health (NIH) grant R01 DK45586 (M.A.L.) and NIH F30 DK104513 (M.J.E.) supported this work.
Glossary
- Lineage-determining transcription factors
Transcription factors that determine the differentiation fate of cells.
- Signal-dependent transcription factors
Transcription factors that associate with or bind to specific DNA sequences near lineage-determining transcription factors in response to environmental or physiological cues.
- Nuclear receptors
A superfamily of transcription factors that bind to highly specific DNA motifs in direct response to ligand binding to activate or repress gene transcription.
- Stoichiometric component
A protein whose interaction with another component of a protein complex is based on their equal molarity.
- WD40 repeat-containing proteins
Proteins containing a motif of 40 amino acids that assumes a β-propeller structure and functions in establishing protein-protein interactions, signal transduction and transcription regulation.
- Cre recombinase
(Cre). A bacteriophage P1 type I topoisomerase that catalyses DNA recombination between site-specific loxP sites.
- Adeno-associated virus
(AAV). A small, non-enveloped, replication-incompetent virus with a small singlestranded DNA genome, which is maintained extrachromosomally and used for gene delivery.
- Hepatosteatosis
Fatty liver, often owing to the pathological accumulation of triglycerides.
- Nonshivering thermogenesis
A facultative and adaptive process that protects core body temperature.
- Oxidative phosphorylation
The transfer of electrons in mitochondria between energy carriers down the electron transport chain to generate a proton gradient and produce ATP.
- Global run-on sequencing
(GRO-seq). A nuclear run-on assay coupled to next-generation sequencing to map all transcription by engaged RNA polymerases throughout the genome.
- Futile metabolic cycles
Biochemical pathways that operate simultaneously in opposing directions, often leading to the dissipation of energy.
- First heart field
(FHF). A developmental structure that gives rise to the cardiac crescent, early cardiac tube and left ventricle.
- Second heart field
(SHF). A developmental structure that gives rise to the right ventricle, atrial myocardium and cardiac outflow tract.
- Nuclear lamina
A cellular structure adjacent to the inner nuclear membrane that is composed of lamin polymers and other proteins and forms a skeletal nuclear structure that interacts with nuclear scaffold proteins and chromatin.
- Facultative heterochromatin
Tightly packed, inaccessible chromatin containing inactive genes, which may become accessible and expressed in certain cell types or following certain cellular cues.
- Marfan syndrome
A connective tissue disorder caused by mutations in the fibrillin 1 gene, often presenting as tall, slender individuals with arachnodactyly, cardiac valve disease and predisposition to aortic aneurysm and dissection.
- Loeys–Dietz syndrome
A connective tissue disorder with five subtypes, each caused by unique mutations in five genes of the TGFβ signalling pathway, which cause congenital cardiac defects and predisposition to aortic aneurysm and dissection.
- CA1 hippocampal region
An area of the brain that is important for memory.
- Nucleus accumbens
A region of the brain that is involved in cognitive processing of reward, with neurons containing mostly dopamine receptors; the nucleus accumbens is thought to be the major nucleus involved in addiction.
- Paneth cells
Epithelial cells of the small intestine that secrete antimicrobial peptides and proteins, mediate host-microorganism interactions and help defend against enteric pathogens in the gut lumen.
- Regulatory T cells
(Treg cells). CD4+, CD25+ and FOXP3+ T cells that suppress the activation of the immune system to maintain tolerance to self-antigens and prevent autoimmune disease.
Footnotes
Competing interests
M.A.L. is a consultant to KDAC, a company developing his-tone deacetylase (HDAC) inhibitors, and Novartis, and serves on scientific advisory boards for Pfizer and Eli Lilly and Co.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Molecular Cell Biology thanks C. Trivedi and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Ong C-T & Corces VG Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet 12, 283–293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitz F & Furlong EEM Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet 13, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Glass CK & Natoli G Molecular control of activation and priming in macrophages. Nat. Immunol 17, 26–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll JS et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Lefterova MI et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev 22, 2941–2952 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biddie SC et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John S et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet 43, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grøntved L et al. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J 32, 1568–1583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madsen MS, Siersbæk R, Boergesen M, Nielsen R & Mandrup S Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol. Cell. Biol 34, 939–954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soccio RE et al. Genetic variation determines PPARγ function and anti-diabetic drug response in vivo. Cell 162, 33–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X-J & Seto E Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31, 449–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdin E & Ott M 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol 16, 258–264 (2015).This is an excellent review of the role of protein acetylation in biology.
- 13.Bannister AJ & Kouzarides T The CBP co-activator is a histone acetyltransferase. Nature 384, 641–643 (1996) [DOI] [PubMed] [Google Scholar]
- 14.Ogryzko VV, Schiltz RL, Russanova V, Howard BH & Nakatani Y The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH & Nakatani Y A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382, 319–324 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Spencer TE et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389, 194–198 (1997) [DOI] [PubMed] [Google Scholar]
- 17.Haberland M, Montgomery RL & Olson EN The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet 10, 32–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taunton J, Hassig CA & Schreiber SL A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Sci 272, 408–411 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Yang X-J & Seto E The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol 9, 206–218 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadosh D & Struhl K Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev 12, 797–805 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rundlett SE et al. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA 93, 14503–14508 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregoretti IV, Lee YM & Goodson HV Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol 338, 17–31 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Lahm A et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl Acad. Sci. USA 104, 17335–17340 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwer B & Verdin E Conserved metabolic regulatory functions of sirtuins. Cell. Metab 7, 104–112 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Chang H-C & Guarente L SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab 25, 138–145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalkiadaki A & Guarente L Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol 8, 287–296 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Seto E & Yoshida M Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol 6, a018713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West AC & Johnstone RW New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest 124, 30–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J 19, 4342–4350 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen YD et al. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl Acad. Sci. USA 97, 7202–7207 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guenther MG et al. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev 14, 1048–1057 (2000).References 29–31 identify the stoichiometric relationship between nuclear receptor co-repressors and HDAC3.
- 32.Everett LJ & Lazar MA Cell-specific integration of nuclear receptor function at the genome. Wiley Interdiscip. Rev. Syst. Biol. Med 5, 615–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francis GA, Fayard E, Picard F & Auwerx J Nuclear receptors and the control of metabolism. Annu. Rev. Physiol 65, 261–311 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Perissi V & Rosenfeld MG Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol 6, 542–554 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Hong SH et al. Nuclear receptors and metabolism: from feast to famine. Diabetologia 57, 860–867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glass CK & Rosenfeld MG The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14, 121–141 (2000). [PubMed] [Google Scholar]
- 37.McKenna NJ, Lanz RB & O’Malley BW Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev 20, 321–344 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Hu X & Lazar MA The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402, 93–96 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Perissi V, Jepsen K, Glass CK & Rosenfeld MG Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet 11, 109–123 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Yoon HG et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J 22, 1336–1346 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Kalkum M, Chait BT & Roeder RG The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9, 611–623 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Kao HY, Downes M, Ordentlich P & Evans RM Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev 14, 55–66 (2000). [PMC free article] [PubMed] [Google Scholar]
- 43.Heinzel T et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387, 43–48 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Nagy L et al. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89, 373–380 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Huang EY et al. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev 14, 45–54 (2000). [PMC free article] [PubMed] [Google Scholar]
- 46.Perissi V et al. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev 13, 3198–3208 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guenther MG, Barak O & Lazar MA The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol 21, 6091–6101 (2001).This paper reports that HDAC3 activity requires nuclear receptor co-repressors.
- 48.Ishizuka T & Lazar MA The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol. Endocrinol 19, 1443–1451 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Codina A et al. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proc. Natl Acad. Sci. USA 102, 6009–6014 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aasland R, Stewart AF & Gibson T The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci 21, 87–88 (1996). [PubMed] [Google Scholar]
- 51.Boyer LA et al. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10, 935–942 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Boyer LA, Latek RR & Peterson CL The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol 5, 158–163 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Yu J, Li Y, Ishizuka T, Guenther MG & Lazar MAA SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J 22, 3403–3410 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson PJ, Fairall L, Santos GM & Schwabe JWR Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481, 335–340 (2012).This paper reports the crystal structure of HDAC3 bound to a nuclear receptor DAD, identifying crucial interactions between the two proteins and a role for IP4 in maintaining them.
- 55.You S-H et al. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol 20, 182–187 (2013).This paper demonstrates that the enzymatic activity of HDAC3 requires nuclear receptor co-repressors in vivo.
- 56.Adrain C & Freeman M New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat. Rev. Mol. Cell Biol 13, 489–498 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Guo C et al. Regulated clearance of histone deacetylase 3 protects independent formation of nuclear receptor corepressor complexes. J. Biol. Chem 287, 12111–12120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guenther MG, Yu J, Kao GD, Yen T J. & Lazar, M. A. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev 16, 3130–3135 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Z et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell 52, 769–782 (2013).This paper describes non-enzymatic functions of HDAC3.
- 60.Bhaskara S et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol. Cell 30, 61–72 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montgomery RL et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J. clin. invest 118, 3588–3597 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alenghat T et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456, 997–1000 (2008).This paper is the first to demonstrate a physiological role of the NCoR1-HDAC3 complex.
- 63.Sengupta N & Seto E Regulation of histone deacetylase activities. J. Cell. Biochem 93, 57–67 (2004) [DOI] [PubMed] [Google Scholar]
- 64.Lonard DM & O’Malley BW Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell 27, 691–700 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Tsai S-C & Seto E Regulation of histone deacetylase 2 by protein kinase CK2. J. Biol. Chem 277, 31826–31833 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Zhang X et al. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev 19, 827–839 (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knutson SK et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J 27, 1017–1028 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Villagra A et al. Histone deacetylase 3 down-regulates cholesterol synthesis through repression of lanosterol synthase gene expression. J. Biol. Chem 282, 35457–35470 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Sun Z et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med 18, 934–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng D et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zamir I, Zhang J & Lazar MA Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev 11, 835–846 (1997). [DOI] [PubMed] [Google Scholar]
- 72.Hu X, Li Y & Lazar M Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol. Cell. Biol 21, 1747–1758 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bugge A et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev 26, 657–667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim YH et al. Rev-erbα dynamically modulates chromatin looping to control circadian gene transcription. Science 359, 1274–1277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y et al. Gene regulation. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348, 1488–1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Armour SM et al. An HDAC3-PROX1 corepressor module acts on HNF4α to control hepatic triglycerides. Nat. Commun 8, 549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emmett MJ et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 546, 544–548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Enerbäck S et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387, 90–94 (1997). [DOI] [PubMed] [Google Scholar]
- 79.Villena JA et al. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc. Natl Acad. Sci. USA 104, 1418–1423 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giguère V Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev 29, 677–696 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Luo J et al. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol. Cell. Biol 23, 7947–7956 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang JS, Ghosh S, Newman S & Salbaum JM A map of the PGC-1 α- and NT-PGC-1 α-regulated transcriptional network in brown adipose tissue. Sci. Rep 8, 7876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schreiber SN, Knutti D, Brogli K, Uhlmann T & Kralli A The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha). J. Biol. Chem 278, 9013–9018 (2003). [DOI] [PubMed] [Google Scholar]
- 84.Brown EL et al. Estrogen-related receptors mediate the adaptive response of brown adipose tissue to adrenergic stimulation. iScience 2, 221–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahmadian M et al. ERRγ preserves brown fat innate thermogenic activity. Cell Rep 22, 2849–2859 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seale P et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab 6, 38–54 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hasegawa Y et al. Repression of adipose tissue fibrosis through a PRDM16-GTF2IRD1 complex improves systemic glucose homeostasis. Cell Metab 27, 180–194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berry DC et al. Cellular aging contributes to failure of cold-induced beige adipocyte formation in old mice and humans. Cell Metab 25, 481 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Ferrari A et al. HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat. Commun 8, 93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Somech R et al. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci 118, 4017–4025 (2005). [DOI] [PubMed] [Google Scholar]
- 91.Vincent SD & Buckingham ME How to make a heart. The origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol 90, 1–41 (2010). [DOI] [PubMed] [Google Scholar]
- 92.Lewandowski SL et al. Histone deacetylase 3 modulates Tbx5 activity to regulate early cardiogenesis. Hum. Mol. Genet 23, 3801–3809 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewandowski SL, Janardhan HP & Trivedi CM Histone deacetylase 3 coordinates deacetylase-independent epigenetic silencing of transforming growth factor-β1 (TGF-β 1) to orchestrate second heart field development. J. Biol. Chem 290, 27067–27089 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Poleshko A et al. Genome-nuclear lamina interactions regulate cardiac stem cell lineage restriction. Cell 171, 573–587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Demmerle J, Koch AJ & Holaska JM The nuclear envelope protein emerin binds directly to histone deacetylase 3 (HDAC3) and activates HDAC3 activity. J. Biol. Chem 287, 22080–22088 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zullo JM et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149, 1474–1487 (2012). [DOI] [PubMed] [Google Scholar]
- 97.Jain R & Epstein JA Competent for commitment: you’ve got to have heart! Genes Dev 32, 4–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Van Der Linde D et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J. Am. Coll. Cardiol 58, 2241–2247 (2011). [DOI] [PubMed] [Google Scholar]
- 99.Loeys BL et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet 37, 275–281 (2005). [DOI] [PubMed] [Google Scholar]
- 100.Neptune ER et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat. Genet 33, 407–411 (2003). [DOI] [PubMed] [Google Scholar]
- 101.Singh N et al. Histone deacetylase 3 regulates smooth muscle differentiation in neural crest cells and development of the cardiac outflow tract. Circ. Res 109, 1240–1249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun Z et al. Diet-induced lethality due to deletion of the Hdac3 gene in heart and skeletal muscle. J. Biol. Chem 286, 33301–33309 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vega RB & Kelly DP Cardiac nuclear receptors: architects of mitochondrial structure and function. J. Clin. invest 127, 1155–1164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Finck BN & Kelly DP PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Invest 116, 615–622 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Norwood J, Franklin JM, Sharma D & D’Mello SR Histone deacetylase 3 is necessary for proper brain development. J. Biol. Chem 289, 34569–34582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang L et al. Hdac3 interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev Cell 36, 316–330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.He X et al. A histone deacetylase 3-dependent pathway delimits peripheral myelin growth and functional regeneration. Nat. Med 24, 338–351 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McQuown SC et al. HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci 31, 764–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rogge GA, Singh H, Dang R & Wood MA HDAC3 is a negative regulator of cocaine-context-associated memory formation. J. Neurosci 33, 6623–6632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ebert DH et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature 499, 341–345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lyst MJ et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci 16, 898–902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nott A et al. Histone deacetylase 3 associates with MeCP2 to regulate FOXO and social behavior. Nat. Neurosci 19, 1497–1505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y et al. HDAC3-dependent epigenetic pathway controls lung alveolar epithelial cell remodeling and spreading via miR-17–92 and TGF-β signaling regulation. Dev. Cell 36, 303–315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang X et al. Expression of histone deacetylase 3 instructs alveolar type I cell differentiation by regulating a Wnt signaling niche in the lung. Dev Biol 414, 161–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alenghat T et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature 504, 153–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Navabi N et al. Epithelial histone deacetylase 3 instructs intestinal immunity by coordinating local lymphocyte activation. Cell Rep 19, 1165–1175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Whitt J et al. Disruption of epithelial HDAC3 in intestine prevents diet-induced obesity in mice. Gastroenterology 155, 501–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Remsberg JR et al. Deletion of histone deacetylase 3 in adult beta cells improves glucose tolerance via increased insulin secretion. Mol. Metab 6, 30–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lundh M, Galbo T, Poulsen SS & Mandrup-Poulsen T Histone deacetylase 3 inhibition improves glycaemia and insulin secretion in obese diabetic rats. Diabetes Obes. Metab 17, 703–707 (2015). [DOI] [PubMed] [Google Scholar]
- 120.Chou DHC et al. Inhibition of histone deacetylase 3 protects beta cells from cytokine-induced apoptosis. Chem. Biol 19, 669–673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lundh M et al. Histone deacetylases 1 and 3 but not 2 mediate cytokine-induced beta cell apoptosis in INS-1 cells and dispersed primary islets from rats and are differentially regulated in the islets of type 1 diabetic children. Diabetologia 55, 2421–2431 (2012). [DOI] [PubMed] [Google Scholar]
- 122.Chen W-B et al. Conditional ablation of HDAC3 in islet beta cells results in glucose intolerance and enhanced susceptibility to STZ-induced diabetes. Oncotarget 7, 57485–57497 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Magnuson MA & Osipovich AB Pancreas-specific Cre driver lines and considerations for their prudent use. Cell Metab 18, 9–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Song J, Xu Y, Hu X, Choi B & Tong Q Brain expression of Cre recombinase driven by pancreas-specific promoters. Genesis 48, 628–634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hong S et al. Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat. Med 23, 223–234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mullican SE et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev 25, 2480–2488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen X et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl Acad. Sci. USA 109, E2865–E2874 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hoeksema M a et al. Targeting macrophage Histone deacetylase 3 stabilizes atherosclerotic lesions. EMBO Mol. Med 6, e201404170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bhaskara S et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 18, 436–447 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Summers AR et al. HDAC3 is essential for DNA replication in hematopoietic progenitor cells. J. Clin. invest 123, 3112–3123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stengel KR et al. Deacetylase activity of histone deacetylase 3 is required for productive VDJ recombination and B cell development. Proc. Natl Acad. Sci. USA 114, 8608–8613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stengel KR et al. Histone deacetylase 3 is required for efficient T cell development. Mol. Cell. Biol 35, 3854–3865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Philips RL et al. HDAC3 is required for the downregulation of RORγt during thymocyte positive selection. J. Immunol 197, 541–554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hsu F-C et al. Histone deacetylase 3 is required for T cell maturation. J. Immunol 195, 1578–1590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang L et al. FOXP3 + regulatory T cell development and function require histone / protein deacetylase 3. J. Clin. Invest 125, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lf Chen, Fischle, W., Verdin, E. & Greene, W. C. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 293, 1653–1657 (2001). [DOI] [PubMed] [Google Scholar]
- 137.Bradley EW, Carpio LR, van Wijnen AJ, McGee-Lawrence ME & Westendorf JJ Histone deacetylases in bone development and skeletal disorders. Physiol. Rev 95, 1359–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Singh N et al. Murine craniofacial development requires Hdac3-mediated repression of Msx gene expression. Dev Biol 377, 333–344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Razidlo DF et al. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLOS ONE 5, e11492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Feigenson M et al. Histone deacetylase 3 deletion in mesenchymal progenitor cells hinders long bone development. J. Bone Miner. Res 32, 2453–2465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carpio LR et al. Histone deacetylase 3 supports endochondral bone formation by controlling cytokine signaling and matrix remodeling. Sci. Signal 9, ra79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McGee-Lawrence ME et al. Histone deacetylase 3 is required for maintenance of bone mass during aging. Bone 52, 296–307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schroeder TM, Kahler RA, Li X & Westendorf JJ Histone deacetylase 3 interacts with Runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J. Biol. Chem 279, 41998–42007 (2004). [DOI] [PubMed] [Google Scholar]
- 144.McGee-Lawrence ME et al. Runx2 protein represses axin2 expression in osteoblasts and is required for craniosynostosis in axin2-deficient mice. J. Biol. Chem 288, 5291–5302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hesse E et al. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J. Cell Biol 191, 1271–1283 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Giavini E & Menegola E Teratogenic activity of HDAC inhibitors. Curr. Pharm. Des 20, 5438–5442 (2014). [DOI] [PubMed] [Google Scholar]
- 147.Boluk A et al. The effect of valproate on bone mineral density in adult epileptic patients. Pharmacol. Res 50, 93–97 (2004). [DOI] [PubMed] [Google Scholar]
- 148.Guo CY, Ronen GM & Atkinson SA Long-term valproate and lamotrigine treatment may be a marker for reduced growth and bone mass in children with epilepsy. Epilepsia 42, 1141–1147 (2001). [DOI] [PubMed] [Google Scholar]
- 149.Oner N et al. Bone mineral metabolism changes in epileptic children receiving valproic acid. J. Paediatr. Child Health 40, 470–473 (2004). [DOI] [PubMed] [Google Scholar]
- 150.Sato Y et al. Decreased bone mass and increased bone turnover with valproate therapy in adults with epilepsy. Neurology 57, 445–449 (2001). [DOI] [PubMed] [Google Scholar]
- 151.Lajeunie E et al. Craniosynostosis and fetal exposure to sodium valproate. J. Neurosurg 95, 778–782 (2001). [DOI] [PubMed] [Google Scholar]
- 152.Sharony R et al. Preaxial ray reduction defects as part of valproic acid embryofetopathy. Prenat. Diagn 13, 909–918 (1993). [DOI] [PubMed] [Google Scholar]
- 153.Paradis F-H & Hales BF The effects of class-specific histone deacetylase inhibitors on the development of limbs during organogenesis. Toxicol. Sci 148, 220–228 (2015). [DOI] [PubMed] [Google Scholar]
- 154.Heinz S et al. Effect of natural genetic variation on enhancer selection and function. Nature 503, 487–492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Janardhan HP et al. Hdac3 regulates lymphovenous and lymphatic valve formation. J. Clin. Invest 127, 4193–4206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smith E & Shilatifard A Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol 21, 210–219 (2014). [DOI] [PubMed] [Google Scholar]
- 157.Lerin C et al. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab 3, 429–438 (2006). [DOI] [PubMed] [Google Scholar]
- 158.Rodgers JT et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113–118 (2005). [DOI] [PubMed] [Google Scholar]
- 159.Wang W et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl Acad. Sci. USA 111, 14466–14471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shapira SN et al. EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev 31, 660–673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thompson JA, Tan J & Greene CS Cross-platform normalization of microarray and RNA-seq data for machine learning applications. PeerJ 4, e1621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ritchie ME et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]