Abstract
Purpose:
Quantitative parameter maps, as opposed to qualitative grayscale images, may represent the future of diagnostic MRI. A new quantitative MRI method is introduced here that requires a single 3D acquisition, allowing good spatial coverage to be achieved in relatively short scan times.
Methods:
A multi-pathway multi-echo (MPME) sequence was developed, and at least three pathways with two echo times were needed to generate T1, T2, T2*, B1+ and B0 maps. The method required the central k-space region to be sampled twice, with the same sequence but with two very different nominal flip angle settings. Consequently, scan time was only slightly longer than that of a single scan. MPME data were reconstructed into parameter maps, for phantom as well as brain acquisitions, in five healthy volunteers at 3T. Spatial resolution, matrix size and FOV were 1.2×1.0×1.2mm3, 160×192×160 and 19.2×19.2×19.2cm3 (whole brain), acquired in 11.5 min with minimal acceleration. Validation was performed against T1, T2 and T2* maps calculated from gradient-echo and spin-echo data.
Results:
In Bland-Altman plots, bias and limits of agreement for T1 and T2 results in vivo and in phantom were: −2.9/±125.5ms (T1 in vivo), −4.8/±20.8ms (T2 in vivo), −1.5/±18.1ms (T1 in phantom), and −5.3/±7.4ms (T2 in phantom), for ROIs including given brain structures or phantom compartments. Due to relatively-high noise levels, the current implementation of the approach may prove more useful for ROI-based as opposed to pixel-based interpretation.
Conclusions:
We proposed a novel approach to quantitatively map MR parameters based on an MPME acquisition.
Keywords: Multi-pathway imaging, quantitative MRI, relaxometry
Introduction
MR images are not typically quantitative in nature, in the sense that voxel values are associated with arbitrary grayscales rather than clear physical quantities. While contrast can be manipulated to great effect to produce clinically-useful images, one rarely obtains maps of the actual physical parameters that determine contrast. As imaging strategies and acceleration techniques continue to evolve, routine MR images may become more quantitative in nature, with values that have actual units and physical meaning. Existing quantitative imaging methods include the use of a multi-echo spin-echo (or gradient-echo) sequence to measure T2 (or T2*) (1,2). Alternately, the signal equation in phase-cycled SSFP and ‘dual echo in the steady state’ (DESS) (3) involves T2 along with several other parameters, and strategies can be developed to generate T2 maps from these signals (4–8). The ‘Gradient-Echo Sampling of FID and Echo’ (GESFIDE) method (9) jointly measures T2 and T2* by acquiring both free induction decay (FID) as well as spin echo signal. We adapted a GESFIDE-like strategy to a multi-echo DESS sequence (10), which is faster than the spin-echo based method originally used for GESFIDE. T1, on the other hand, can be measured by sampling the recovery that follows an inversion pulse (11), for example using a Look-Locker sequence (12), or fitting data acquired with varied TR and/or flip angle settings (13–15). Some methods can capture both T1 and T2 jointly, for example DESPOT (16,17) and the ‘Triple echo in the steady state’ (TESS) method (18). More recently, MR fingerprinting (19–21) has received much attention and has been employed in a variety of contexts.
The proposed method relies on the use of a Multi-Pathway Multi-Echo (MPME) sequence. Multi-echo imaging is commonplace on commercial scanners, and multi-pathway imaging is sometimes performed with DESS (3), TESS (18) and in thermometry (22). But the only prior MPME work appears to be our own two-pathway multi-echo approach for T2, T2* and B0 mapping (10). The present three-pathway multi-echo pulse sequence can be seen equally well as adding a third pathway to our two-pathway prior work from (10), or adding multi-echo capability to TESS (18). We show here that a single 3D MPME acquisition, supplemented with a quickly-acquired central k-space region using the same sequence but a very different flip angle setting, suffices to map T2, T2*, T1, B0, B1+ and M0.
One weakness common to many prior approaches comes from the assumption that the flip angle is known. While MRI protocols include a nominal value for the flip angle, and that prescan procedures attempt to scale the B1+ field appropriately, flip angles in MRI vary spatially and are always at best only approximately equal to the nominal user-input value, especially at higher field strengths. A reliable estimate of flip angle is often necessary to achieve a reliable measurement of T1. The B1+ field could be measured separately, for example with the Bloch-Siegert method (23,24), at the cost of extra scan time. A major strength of the present approach is that it does not assume the flip angle to be known and naturally generates spatially-varying 3D flip angle maps. On the other hand, a major weakness of the approach is that noise gets amplified from one reconstruction step to the next; for this reason, in its current implementation at least, the approach may be more appropriate for ROI-based as opposed to pixel-based interpretation.
The method was tested in simulations, phantom scans and five healthy volunteer brain exams. Validation was performed against T2 and T1 maps calculated from multiple spin-echo acquisitions.
Methods
Multi-Pathway Multi-Echo (MPME) pulse sequence
Steady-state sequences do not have RF spoiling; for this reason, they may generate many different signal pathways (25,26). These sequences can be balanced or unbalanced, depending on whether their gradient waveforms have zero total area or not, respectively. The total gradient area determines the k-space separation between pathways, which means that for balanced sequences this separation is zero and all pathways overlap near k-space center. Such multi-layer overlap creates good SNR, but also leads to well-known ‘dark-band artifacts’ wherever pathway signals interact destructively. Balanced steady-state sequences are known as bSSFP (Balanced Steady-State Free Precession) or by their vendor-specific names such as True-FISP (Fast Imaging with Steady-state Precession), FIESTA (Fast Imaging Employing Steady-state Acquisition) or bFFE (balanced Fast Field Echo), for example.
On the other hand, in the case of unbalanced steady-state sequences, most pathway signals are displaced to faraway locations in k-space, beyond the sampled region. These sequences typically sample only one pathway, the one that remained near k-space center, the so-called 0th or FISP pathway. The DESS (Dual Echo in the Steady State) sequence (3) is an exception, as it samples both the 0th and –1st pathway (also called PSIF, for inverted FISP). The TESS sequence (18) goes one step further as it samples the +1st pathway as well.
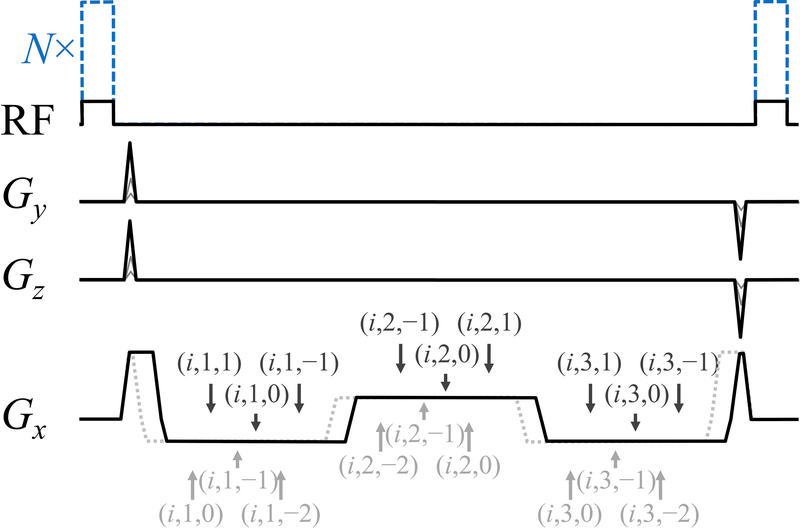
Our MPME pulse sequence is depicted in Fig. 1. Only the Gx gradient waveform is unbalanced, i.e., the Gy and Gz waveforms have zero total area. As a consequence, pathway signals were separated by a kx increment Dkx = −γ × AGx, where γ is the gyromagnetic ratio and . The MPME sequence was designed such that each readout window captured three different pathways: the +1st, 0th and −1st pathway (black Gx waveform in Fig. 1), or the 0th, −1st and −2nd pathway (dashed gray Gx waveform). The sequence is depicted in Fig. 1 for NTE = 3 readout windows, of alternating polarity.
Fig. 1.
: A three-pathway three-echo version of our MPME sequence is depicted here. Using a (i, j, k) notation to represent the jth echo from the kth pathway during the ith scan, the timing of all pathway and echo acquisitions is displayed with arrows, for two acquisition scenarios: [1,0,−1] (solid black line in Gx channel) and [0,−1,−2] (dotted gray line). In the RF channel, black lines indicate the smaller flip angle acquisition, i=1, while dashed blue lines indicate the larger flip angle acquisition, i=2. Only three of these magnetization states are needed for the proposed processing to be carried out, which explains why either a [0,−1,−2] or a [1,0,−1] scheme can be chosen.
The Gx waveform as drawn in Fig. 1 consisted of 5 separate lobes, whose relative areas must be precisely set for the desired pathways to be obtained. In units of −AGx, the 5 lobes had an area of +1.5:−3:+3:−3:+0.5 for the +1st, 0th and −1st pathways to be sampled, see black Gx waveform in Fig. 1. Alternately, to sample the 0th, −1st and −2nd pathways, areas of +0.5:−3:+3:−3:+1.5 were employed instead, see dashed gray Gx waveform in Fig. 1. A notation (i, j, k) is employed here to represent signal from the kth pathway (ranging from −2 to 1), jth echo (from 1 to NTE) and ith scan. Scan number i=1 was acquired using a relatively low flip angle setting (black RF waveform in Fig. 1) while scan i=2 was obtained with a much larger flip angle setting (dashed blue RF waveform in Fig. 1). The two scans were performed either serially, or in TR-interleaved fashion, as further detailed below. In our favored implementation only a small central region in the ky-kz plane was acquired for the i=2 scan. In such case, with one scan much shorter than the other, the two scans had to be performed in serial fashion (as opposed to TR-interleaved).
Reconstruction step 1: Evaluating B0
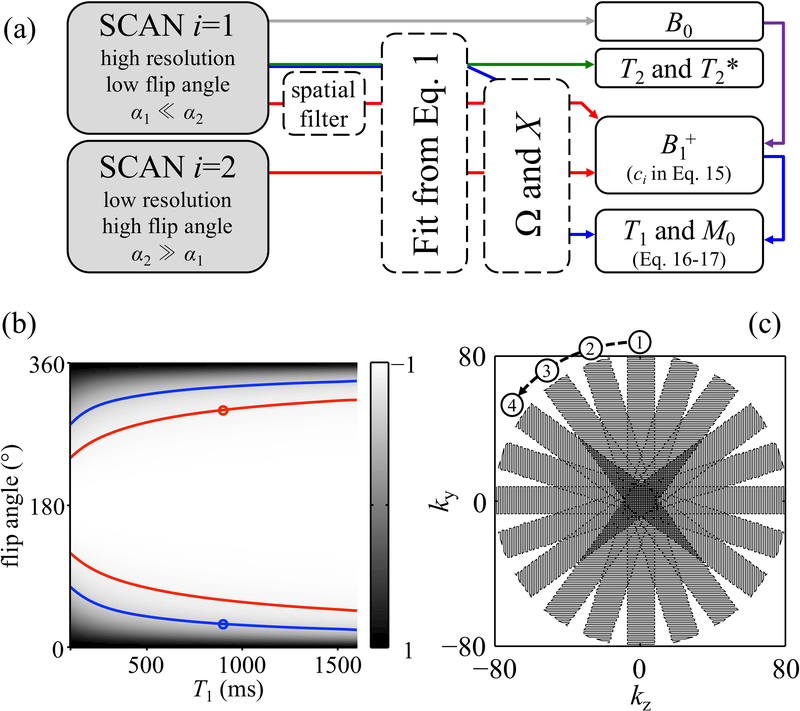
Reconstruction was performed in Matlab, on a 64-bit linux machine (Intel i7–4820K 3.7GHz × 4, 64GB of memory), and called functions from the freely-downloadable fast imaging library available from the National Center for Image Guided Therapy (27). Data acquired with an MPME sequence were converted into B0 maps in a fairly standard manner (10), i.e., by analyzing the phase evolution from one echo to the next. More specifically, both positive and negative pathway signals experience the same off-resonance effects, and as such, all pathway signals could jointly contribute toward evaluating B0. These different signals were combined through a weighted linear regression, in a manner similar to Equation 11 of Ref. (10). Because the scan i=2 had low spatial resolution in most results obtained here, and as depicted with a gray line in the flowchart from Fig. 2a, this operation was performed based on the i=1 scan only.
Fig. 2.
: a) The main reconstruction steps are shown here, in a color-coded manner, for our preferred implementation. Gray line: The complex multi-echo data from scan i=1 is employed toward evaluating B0. Green line: R2 and R2′, or equivalently T2 and T2*, were mapped by fitting the data from the full-resolution i=1 scan to Eq. 1. Red lines: Both datasets, with high and with low nominal flip angle values, were employed toward evaluating B1+, i.e., the flip angle, whose cosine appears as ci in Eq. 15. The i=1 higher spatial resolution data were filtered to match those of the i=2 scan, and both datasets were used toward solving for ci in Eq. 15. B0 was further employed in the process, as indicated with the purples line, to account for off-resonance effects (see Eq. 8). Blue lines: Using B1+ as evaluated above, the higher resolution i=1 data were employed to map T1 and M0, through Eq. 16 and 17, respectively. In (b), the mixing factor, X, is mapped here for a number of different combinations of flip angle and T1 values. For a given voxel, one MPME acquisition allows one value of the mixing factor to be calculated, which is not sufficient to pinpoint both α1 and T1 (e.g., see blue lines). However, if a 2nd scan is performed with a quantifiably-different flip angle setting, for example α2 = 10×α1, a second mixing factor value is obtained (e.g., see red lines), and T1 as well as α1 can be evaluated (see blue circle). The sampling strategy in the ky-kz plane, for in vivo scans, is shown in (c). The PROPELLER-like scheme, with oversampling near k-space center, provides extra robustness to motion. The covered region was circular in shape, with a 10-fold oversampled center and a periphery under-sampled by 40%. Overall, the number of phase encodes and corresponding scan time were adjusted to match those of a Cartesian scheme.
Reconstruction step 2: Evaluating T2 and T2*
Consider one voxel, from coil-combined images, with signal Si,j,k. These signals depended on TEi,j,k, the echo time at which they were sampled:
| [1] |
where R2 and R2′ represent irreversible and reversible decay, respectively, such that T2 = 1/R2 and T2* = 1/(R2+R2′). The meaning of will be clarified when defining F states below, but at this point it can simply be viewed as a real-valued constant for a given scan i and pathway k. Because signals from negative pathways prove similar to a spin-echo on its way to formation, reversible decay is in the process of being corrected and as such, R2′ appeared with a minus sign in Eq. 1 for cases k < 0. In contrast, for positive pathways, both R2 and R2′ combine with a same sign thus giving rise to T2* decay, as seen in Eq. 1 for cases k ≥ 0.
As shown in our prior work (10), Eq. 1 can be solved for R2 and R2′ (or equivalently, T2 and T2*), for example by taking the natural logarithm of Eq. 1 and solving the resulting system of linear equations. In (10), a single scan and only two pathways were needed to map R2 and R2′. The third pathway obtained here can be included in the solution to Eq. 1, for improved SNR. Information from different pathways and/or scans should be combined in an SNR-optimum manner, using the signal strength |Si,j,k| as a weight. As in Step 1 above, and as shown with a green line in the flowchart from Fig. 2a, T2 and T2* maps were evaluated here based on the i=1 data alone because the i=2 scan typically had lower spatial resolution.
Step 3: Calculating the magnetization states F and Z
The derivation below employs a notation for magnetization states that is very similar to that originally introduced by Hennig (25,28). Fk and Zk represent the transverse and longitudinal magnetization for pathway k, respectively. The superscripts −, + and ⇒ are further used here to distinguish between magnetization states just before, just after and a time TR after an RF pulse, respectively.
The signal immediately after an idealized zero-length RF pulse would be , as obtained when setting TE to zero in Eq. 1. Similarly, Eq. 1 also allows to be evaluated, by setting TE equal to TR and replacing both R2 and R2′ by their values as found in Step 1, above. The phase of these complex-valued F states is not known at this point, but as explained later this will not cause any problem here. At steady-state, the magnetization states just before an RF pulse (i.e., those with a ‘−’ superscript) are closely related to those a time TR after the RF pulse (i.e., those with a ‘⇒’ superscript); however, as explained further below, the details of this relation depend on whether the scans i=1 and i=2 were obtained in a sequential vs. an interleaved manner, hence the need to consider both − and ⇒ superscripts at this point.
Unlike transverse F states, longitudinal Z states cannot readily be sampled by a pulse sequence, at least not without interrupting the steady-state. The longitudinal states and are linked through T1:
| [2] |
The 0th and non-zero pathways behave differently with respect to T1, as the latter represents dephased magnetization states decaying with T1 while the k = 0 pathway represents a magnetization vector recovering with T1. As described in (28), all Z states had Hermitian symmetry with respect to k:
| [3] |
where Ο is a placeholder for ‘+’, ‘−’ or ‘⇒’ superscripts, and the notation represents the complex conjugate of Z. Still from Ref. (28), the Zi,k and Fi,k states just before and after an RF pulse were related through:
| [4] |
where αi is the flip angle for ith scan. As in Eq. 2, Eq. 4 had different form for the 0th pathway because it represents a refocused vector rather than a dephased distribution of vectors. Equation 15 of Ref. (28) provided another relationship between Zi,k and Fi,k states, before and after an RF pulse:
| [5] |
Adding the +k and −k versions of Eq. 5, and employing the symmetry from Eq. 3, one obtains:
| [6] |
Finding both and for the |k| > 0 case from Eq. 4, and relating them through Eq. 3, an equation is obtained that involves only F states:
| [7] |
Step 4: Evaluating the flip angles, or more precisely, their cosine value
As shown with red lines in Fig. 2a, data from both i=1 and i=2 scans were employed toward mapping B1+, i.e., the flip angle map. A spatial filter was applied to the i=1 data, to bring spatial resolution down to the level of the i=2 data. As a result, reconstructed flip angle maps had low spatial resolution, which should not be a problem as B1+ is expected to vary smoothly in space.
The main difference between scans i=1 and i=2, besides spatial resolution, is their nominal flip angles, and , and also possibly their TR settings, TR1 and TR2. The ‘true’ flip angle values differ from their corresponding nominal values, and , for two main reasons: First, the strength of the RF field, B1+, varies spatially and essentially ‘scales’ the non-selective RF waveforms depicted in Fig. 1. If such spatially-variable scaling were the only issue, the ratio of nominal flip angles, , would still be correct. However, as described in more detail below, off-resonance effects can further affect the flip angle values, in a non-linear fashion.
The flip angles α1 and α2 are defined here as the ‘true’ flip angles associated with nominal flip angles and whenever/wherever the offset frequency, Δf, is null, leading to . More generally, for cases where Δf is non-zero, a nutation function v(α,Δf) is introduced below to account for off resonance effects on the cosine of these flip angles:
| [8] |
where v(αi,0) = cos(αi) wherever Δf = 0. In practice, a one-time application of a Bloch equation simulation was used to generate a two-dimensional v(α1, Δf) matrix, specific to the shape of the RF waveform being employed (see Fig. 1). In the flowchart from Fig. 2a, the fact that the cosines c1 and c2 had a dependency on Δf is captured using a purple arrow.
From Eq. 2, because T1 must be the same for both scans, one obtains:
| [9] |
A factor Ωi,k is defined whose effect is that of a scaling factor, for a given scan i and pathway k:
| [10] |
Furthermore, a most-useful quantity Xi,k, named here ‘mixing factor’, is defined as:
| [11] |
The mixing factor captures how +k and –k transverse magnetization states get intermixed when an RF pulse is played out. Please note that the factors Ωi,k and Xi,k as defined above involve only transverse magnetization states F, and as such can be calculated from acquired data, through Eq. 1. Equations 4 and 11 were combined along with the trigonometric identity cos(α) = 2×cos2(α/2) – 1:
| [12] |
Replacing in Eq. 6 with the expression from Eq. 12 one can obtain:
| [13] |
In a sequential acquisition, whereby the same flip angle is used in consecutive TRs, the steady-state condition is simply:
| [14] |
Replacing all of the longitudinal Z states in Eq. 9 with their corresponding expressions from Eqs 12–14, one obtains with some effort:
| [15] |
Replacing c2 in Eq. 15 with its expression from Eq. 8, one obtains an equation of a single unknown, α1, which can readily be solved numerically. Reconstructed maps of α1 had the same low spatial resolution as the acquired i=2 data, and were then further fitted to a polynomial to help suppress noise.
Please note that acquired MRI data actually enter Eq. 15 only though the form of the mixing factor Xi,k. Using a setting of k=−1 in Eq. 10 and 11, Xi,k and Ωi,k then involve only , and , all of which can be calculated from the −2nd, −1st and 0th pathway acquisition (gray dashed Gx waveform in Fig. 1). Using a k=1 setting instead, Xi,k and Ωi,k involve , and , all of which can be calculated from the −1st, 0th and +1st pathway acquisition (black Gx waveform in Fig. 1).
Because Eq. 15 involves only the cosine of the flip angles, and that cos(θ) = cos(n×2π+θ) = cos(n×2π−θ) where n is an integer, an infinite number of degeneracies can be expected on flip angle values. The normal mode of operation for the proposed method is to use a smaller α1 << 180o, and a much larger α2 ⊰ [180o 360o], in which case the complex value of is about 180° out of phase with that of . If and were found to be in phase instead, it would be assumed that α2 got so large as to reach/exceed 360o, and lies in the [360o 540o] interval instead. Accordingly, using the relative phase of and as a guide, the flip angles can be evaluated without degeneracy as long as α1 remains non-zero, positive and smaller than 180o, and that α2 remains within the [180o 540o] interval.
In Fig. 2b, the mixing factor X was simulated over a range of T1 and flip angle settings (with T2=100 ms, TR=30 ms) to provide a visual representation of how the proposed method works. One MPME scan allows the mixing factor X to be calculated through Eq. 11, but a single X value would not suffice to pinpoint both α and T1 (e.g., see blue lines in Fig. 2b). But two scans along with a known relation between them, for example α2 = 10×α1, does allow a solution to be found (e.g., see ‘○’ marks in Fig. 2b).
Step 5: Evaluating T1 and M0
As shown with blue lines in Fig. 2a, Ω1,k and X1,k were evaluated, through Eq. 10–11, based on full-resolution i=1 data. Ω1,k and X1,k, along with c1 as found above, were then employed toward calculating T1 and M0. More specifically, combining Eq. 2 with Eqs 12–14, one obtains:
| [16] |
Using the k=0 case from Eqs 2 and 4, along with Eqs 6, 14 and 16, as well as the fact that represents refocused magnetization and can be assumed to be real-valued without loss of generality, one obtains:
| [17] |
where the identity was employed. Given that M0 must be positive the ambivalence in sign was resolved by applying an absolute value operator. M0 is found through Eq. 17 using known F states, along with ci and T1 values as found above. It is worth noting that M0 is unavoidably weighed by the receive-coil sensitivity, and thus is not a truly quantitative proton density map, and for this reason will be referred to as C×M0 below. Equations 14 through 17, above, apply to cases where the low and high flip angle scans are performed in a sequential manner; in contrast, equations A1 through A5 in the Appendix apply to cases where these two scans would be performed in an interleaved manner instead.
On performing all of the processing above using real (non-complex) F states
The F and Z magnetization states used throughout the derivations above were complex numbers. A great simplification came from realizing that real numbers could be used instead. With the MPME sequence from Fig. 1, the longitudinal magnetization was always flipped to the x or ‘real’ axis in the transverse plane, and in an ideal case all signals Si,j,k should have been real. In reality, variations in B0 and B1 fields made the measured signals complex in nature. But as confirmed through simulations, such variations affected only the phase of F states and not their magnitude. In other words, by ignoring the phase of F states one merely ignores undesired B0 and B1 frequency and phase variations unrelated to the derivations above.
Of course, phase information was valuable, and was employed in usual ways toward frequency mapping. But idealized real-valued F states free of phase errors were obtained here by taking the magnitude of complex ones, and adjusting polarity as follows:
| [18] |
where Ο is a placeholder for ‘+’, ‘−’ or ‘⇒’ superscripts. Even in cases where RF chopping was used Eq. 18 was still valid: while RF chopping might swap αi for −αi, it would not affect ci in the equations above, as cos(αi) = cos(−αi). Using Eq. 18 and performing all steps above on real rather than complex numbers led to a vastly simplified and more robust implementation.
Simulated acquisition with the proposed MPME sequence
Signals from the MPME sequence in Fig. 1 were simulated as follows. Isochromats were rotated to represent RF pulses, frequency offsets and unbalanced gradients, while T2 and T1 recovery were applied. Several different frequency offsets were simulated and combined with Lorentzian weighting to simulate the effect of R2′. Five hundred TR periods were simulated to reach steady-state before signals were ‘sampled’. To validate the simulation program, simulated MPME signals were compared to acquired ones. Simulations were performed using the nominal flip angle value, as well as ±10% the nominal value, as the nominal flip angle value cannot be expected to be accurate. A number of experiments were performed using an fBIRN phantom (29), with flip angle = 15°, 20°, and 30°; TR = 25 ms, 35 ms, and 45 ms, and with either [0,−1,−2] or [1,0,−1] pathway acquisitions (see Fig. 1). Signal strength was averaged spatially over the phantom and compared with simulated signals with Bland-Altman plots. Once validated, the simulated version of the MPME pulse sequence helped provide insights about the overall method, as shown in Results.
Parameter mapping from noiseless simulated data – looking for degeneracies
The same reconstruction code was used to convert either simulated or acquired MPME signals into basic MR parameters, namely T2, T2*, T1, B0, B1+ and M0. In a first step, noiseless simulations were performed to help confirm that the proposed algorithm seems effectively free of degeneracies or local minima. Simulations were made while modifying the MR parameters over a range of values: T1 from 300 to 1800 ms (in 100 ms steps), T2 from 30 to 120 ms (in 10 ms steps), T2*= 0.8×T2, and Δf from −100Hz to +100Hz (in 25 Hz steps). The simulated acquisition parameters were α1 = 15o, α2 = 22×α1, TR1 = TR2 = 25 ms, and NTE = 3 echoes uniformly distributed across TR. MR parameters were computed based on the simulated MPME signals, and then compared to the known ‘true’ values.
Parameter mapping from noisy simulated data – parameter optimization
Simulations with added noise were performed to help optimize the imaging parameters. Simulations were made over a range of TR1 from 15 to 50 ms, TR2 also from 15 ms to 50 ms, α1 from 10o to 30o, α2 from 310o to 350o, using NTE = TR/8ms. MR parameters typical of brain imaging were selected, T1/T2/T2* = 1500/70/60 ms (30–32), with Δf = 0. Each pathway and echo signal, in turn, was disrupted by a small perturbation equal to +0.1% and then −0.1% of M0. A setting was sought for the acquisition parameters TR1, TR2, α1 and α2 that would minimize T1 errors, as caused by the perturbations. After the acquisition parameters were set, Gaussian noise (SNR = 30) was added to the simulated signals and noise propagation, from one parameter-mapping step to the next, was evaluated.
Phantom data
Scans were performed on a Siemens Trio 3T system with a product 8-channel knee coil (Invivo Corporation, Gainesville, FL, USA) and the pulse sequences from Fig. 1, with hard RF pulses (see Fig. 1). A sequential acquisition was performed using a [1,0,−1] pathway scheme with parameters from Table 1. Data from the [1,0,−1] scheme were processed using a k=1 setting in Eqs 10–11. A multi-compartment gel phantom was built specifically for these experiments, using gelatin (Knox, E.D. Smith Foods, ON, Canada) doped with a gadolinium contrast agent (Magnevist, Bayer HealthCare LLC, NJ, USA). The concentration of contrast agent in the gel was varied from one compartment to the next.
Table 1:
Acquisition parameters: MPME scans for phantoms and healthy volunteers
| Number of TE |
Pathway spacing time (ms) |
TR (ms) |
Flip angle (°) |
BW/pixel (Hz) |
Voxel size (mm3) |
FOV size (cm3) |
Scan time (min) |
|
|---|---|---|---|---|---|---|---|---|
| Gelatin phantom | 3 | 1.86 | 25 | 15/330 | 539 | 1×0.5×1 | 16×16×16 | 11:24 |
| Subjects A-D Subject E |
3 same |
2 same |
25 same |
15/330 same |
501 same |
1.2×1×1.2 same |
19.2×19.2×19.2 21.1×19.2×21.1 |
11:26 13:39 |
In our favored implementation, only the central k-space region was sampled for the i=2 scan, which nearly halved scan time as compared to fully sampling both scans. More specifically, only 25% of the ky and of the kz axis were sampled for the i=2 scan, a 16× reduction compared to full sampling. Having one scan that requires 16× less TR intervals than the other is not compatible with a TR-interleaved acquisition of the two, and for this reason the sequential acquisition associated with Eqs 15–17 was preferred here; the TR-interleaved scheme is presented in the Appendix. The [1,0,−1] scheme was selected here, although the [0,−1,−2] scheme appeared to be mostly equivalent in terms of quality of calculated parameter maps.
In vivo brain data
Five healthy volunteers (4 males and 1 female, age: 27.2±4.1 years old) were imaged following informed consent using an IRB-approved protocol, with same scanner and a product 12-channel head coil. In early results, motion artifacts were seen emanating from ventricles and CSF-containing regions. For this reason, a more motion-robust coverage of the ky-kz plane was developed for in vivo data compared to phantom data, loosely inspired from the ‘periodically rotated overlapping parallel lines with enhanced reconstruction’ (PROPELLER) method (33,34). As shown in Fig. 2c, the central ky-kz region was oversampled using overlapping ‘blades’ to improve motion robustness, assuming irregular events with respect to k-space acquisition were averaged out. The phase-encoding scheme was designed such that all k-space samples fell on Cartesian coordinate. Ten blades were used to cover the k-space, while the width was reduced by 40% to match the overall scan time of a Cartesian scan: As an example of a 160×160 acquisition matrix, the ky-kz pattern from Fig. 2c had 25664 phase encodes, leading to 0.25% increase in scan time as compared with the Cartesian scheme. K-space density correction was applied prior to a conjugate gradient-based parallel imaging reconstruction method (35,36). The readout direction was oriented in the superior-inferior direction, and the FOV was set large enough along the left-right and anterior-posterior directions to encompass the entire cross section of the head.
As explained in the text above Eq. 8, one would expect the relationship to hold wherever the offset frequency, Δf, is null. However, for in vivo scans, one further scaling factor had to be introduced:
| [19] |
This factor β was determined through one single fit involving all available in vivo data.
Validation
In addition to our MPME scans, a series of gradient-echo and spin-echo images were acquired for multiple TE settings, for a single centrally-located 2D axial slice. Moreover, several inversion-recovery spin-echo (IR-SE) images were obtained for the same slice while varying the inversion time. Parameters for the validation scans are summarized in Table 2.
Table 2:
Acquisition parameters for validation scans
| Sequence: | TE (ms) | TR (ms) | TI (ms) | Flip angle (°) | BW/pixel (Hz) | Voxel size (mm3) | |
|---|---|---|---|---|---|---|---|
| Gelatin phantom | 2D IR-SE | 10 | 5000 | 50, 100, 200, 400, 800, 1600, 3200 | 180–90-180 | 183 | 1×1×3 |
| 2D SE | 20, 30, 60, 120, 200 |
1500 | N/A | 90–180 | 183 | 1×1×3 | |
| in vivo | 2D IR-SE | 10 | 2500 | 50, 300, 800, 1600, 2400 | 180–90-180 | 473 | 1.2×1.2×4 |
| 2D SE | 25, 50, 90, 150 | 800 | N/A | 90–180 | 501 | 1.2×1.2×4 | |
| 2D GRE | 16, 24, 32, 40, 48, 56, 64, 72 | 800 | N/A | 60 | 501 | 1.2×1.2×2 |
Log-linear fitting allowed T2* and T2 maps to be calculated from the gradient-echo and spin-echo data, respectively, while T1 maps were generated using a non-linear least-square fit of the IR-SE data. These independently-obtained T2*, T2, and T1 maps were used as reference standards toward validating the present parameter-mapping method. The reference T1 maps were further employed to help fit for β in Eq. 19: a value for β was sought that minimized T1 errors for all five volunteers jointly. The same β value was then employed in the reconstruction of all MPME in vivo datasets.
Because they were generated in a standard manner, B0 maps were not further validated. As for C×M0 and B1+, the value of all MR parameters were so intertwined in the equations above that not all of them may require independent testing; although not explicitly validated here, obtaining correct values for T2*, T2, and T1 would strongly suggest that C×M0 and B1+ values may be correct as well, given how tightly these parameters are interconnected.
Results
Simulated results
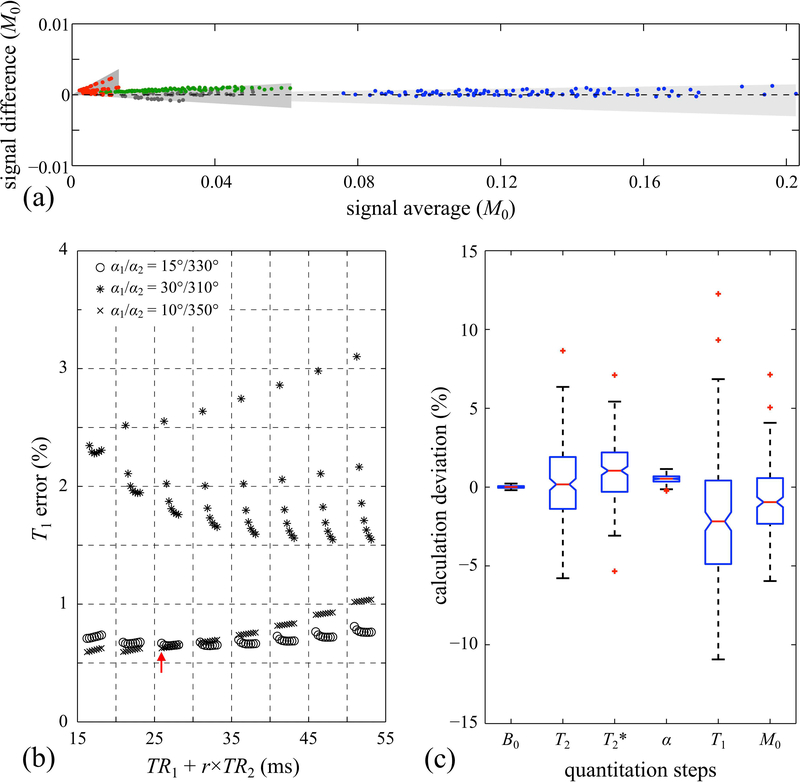
The Bland-Altman plot in Fig. 3a compares simulated and acquired signal strengths, for signal pathways ranging from −2 to +1. One single global fit/scaling was performed to make M0 in the acquired data match the (arbitrary) value for M0 employed in the MPME simulation code. The good agreement obtained in Fig. 3a was interpreted as a validation of the MPME simulation code.
Fig. 3.
: (a) Signals acquired and spatially-averaged over an fBIRN gel phantom were compared in a Bland-Altman manner with corresponding MPME simulations. Overall, the good agreement between simulated and sampled MPME data was interpreted as validation of the simulation. Red, gray, blue, and green circles represent data from the −2nd, −1st, 0th, and 1st pathways, respectively. Gray shaded areas were drawn to show the effect of varying the simulated flip angle from −1% to +1% of the nominal values. In (b), simulated MPME with added noise were converted into T1 estimates, for a number of different TR1, TR2, α1 and α2 settings. A solution was sought that would lead to relatively small T1 errors and relatively short scan times. The red arrow points to the practical solution selected here for in vivo scans. The parameter r represents the ratio in matrix size between scans i=2 and i=1, which is 1/16 here, such that (TR1+r×TR2) as plotted on the horizontal axis is proportional to total scan time. (c) Deviations from the ground truth at each quantitation step were simulated and are displayed in a box-and-whisker plot, showing how noise propagates from one processing step to the next. Red ‘+’ markers, whiskers and boxes indicate, respectively, outliers, min/max values, and the 1st and 3rd quartiles.
In noiseless simulations, the precise value of all mapped parameters could be recovered in all simulated cases, suggesting there is no degeneracy in the solution. More specifically, the maximum deviation for T1 and T2 was 0.014% and 0.006%, respectively.
Results from noisy simulations are shown in Fig. 3b. The horizontal axis, TR1+r×TR2, where r is the ratio in k-space coverage between scan i=1 and i=2, is proportional to overall scan time. As such, a solution reasonably close to the lower left corner in Fig. 3b was sought, which would provide small T1 errors with relatively short scan time. Based on these results, a setting of TR1=25 ms, TR2=25 ms, r=1/16, =15° and =330° was selected for in vivo scans. Using these parameters and added Gaussian noise, Fig. 3c shows noise propagation through the proposed processing steps, as the different quantitative parameters get evaluated in turn.
Phantom results
Phantom results are shown in Fig. 4, for a [1,0,−1] sequential acquisition. Validation in Fig. 4c,d shows good agreement with the spin-echo reference standard. The bias and the limits of agreement for T1 and T2 measurements were: −1.5/±18.1ms and −5.3/±7.4ms, respectively. No scaling factor β (Eq. 19) was required for phantom scans, i.e., β = 1 for all phantom datasets.
Fig. 4.
: (a,b) T1 and T2 maps derived from spin-echo data were compared to those obtained with the proposed approach, for a given 2D slice in a custom-made multi-compartment gelatin phantom. (c,d) Agreement was tested using Bland-Altman plots. Black and gray dashed lines represent the bias and limits of agreement, respectively. (e-h) Quantitative maps of T2*, offset frequency, flip angle and sensitivity-weighted proton density were also reconstructed, using the proposed methodology.
In vivo brain results
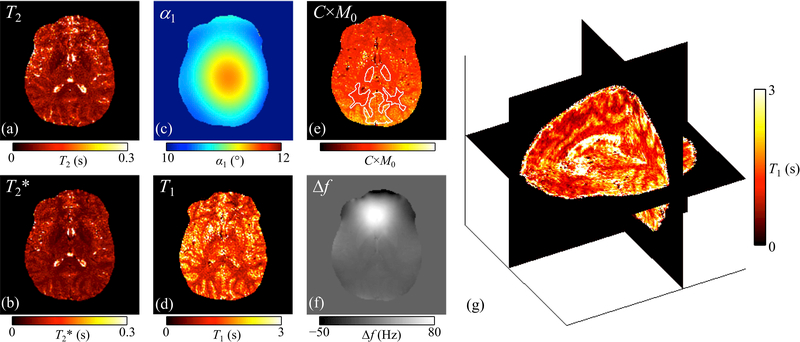
Scan parameters, as optimized through the simulation above, are listed in Table 1. One of the volunteers was imaged with a FOV 10% larger in both phase-encoding directions, due to larger head size, resulting in a slightly longer acquisition time (see Table 1). Figure 5 shows examples of individual pathway images, and reconstructed MR parameter maps are shown in Fig 6, for one given slice from one given volunteer. The display in Fig. 6g is meant to visually emphasize the 3D nature of the parameter maps reconstructed here. Overall, the noise level of the reconstructed 3D maps tended to be high, which can be considered a weakness of the approach as currently implemented.
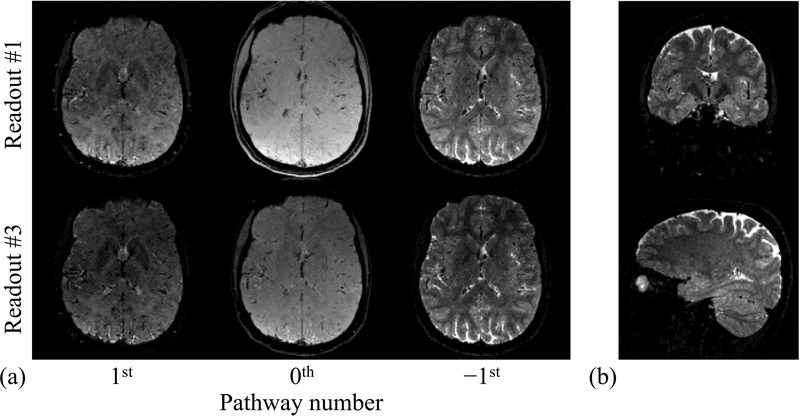
Fig. 5.
: Images are shown for three different pathways at two different echo times, for one axial slice (out of 160) and one subject (out of 5). These images provide drastically different tissue contrasts, allowing the present method to function. In (b), images that correspond to the 2nd readout (out of 3) of the −1st pathway are shown, in coronal and sagittal views.
Fig. 6.
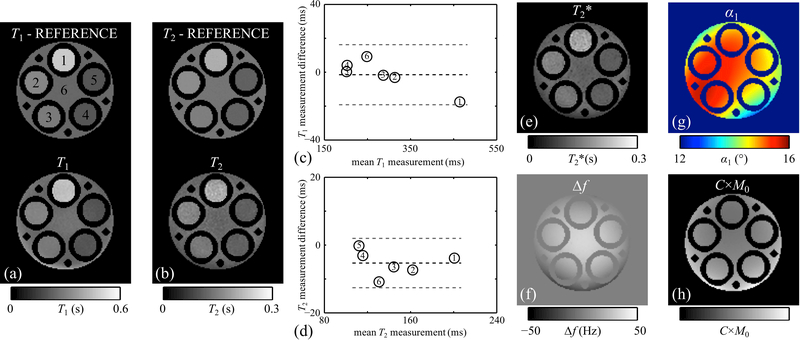
: (a-f) Reconstructed parameter maps are shown, for one given slice from one given subject (subject #1). The slice shown here was further involved in the Bland-Altman validation process in Fig. 7. More specifically, this slice was further imaged with 2D spin-echo and gradient-echo sequences, and the ROIs used for validation are shown in (e), overlaid on the C×M0 map using white contours. A 3-plane visualization of the whole-brain T1 map is shown in (g) to emphasize the 3D nature of the reconstructed results.
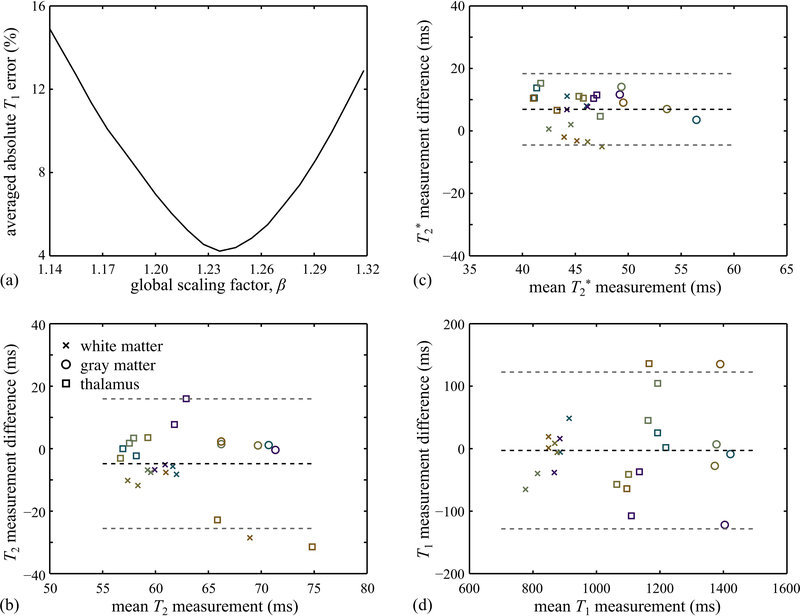
ROIs were drawn for validation purposes (e.g., white contours in Fig. 6e) in both MPME results and reference gradient echo and spin echo results, to enclose specific brain structures. While the choice = 330°/15° = 22 should arguably have led to α2/α1 = 22, the plot in Fig. 7a shows that minimized T1 errors is achieved with β = 1.24, as defined in Eq. 19. The same β was employed here for all in vivo datasets. Bland-Altman plots comparing MPME results to reference results are presented in Fig. 7b-d for T2, T2* and T1, respectively.
Fig. 7.
: (a) Absolute T1 measurement errors (%), averaged over all volunteers and all tissue types, were calculated for values of the calibration factor β (as defined in Eq. 19) ranging from 1.14 to 1.32. A clear minimum was obtained near β=1.24, and this calibration value for β was employed for all in vivo results presented here. (b-d) T2, T2* and T1 values, averaged over ROIs corresponding to different tissue types (see Fig. 6e), were obtained both for the proposed method as well as for the reference (GRE- and SE-based) scans. In the resulting Bland-Altman plots, the markers ‘×’, ‘○’ and ‘□’ represent white matter, gray matter and thalamus tissues, respectively. Five different colors were used to distinguish between the five Subjects. Black and gray dashed lines represent the bias and limits of agreement, respectively.
Discussion
A method was introduced for parameter mapping based on a MPME pulse sequence. As seen in Fig. 5, the pathway images acquired with this sequence had full brain coverage, overall good quality, diversified contrasts and good resolution (1.2×1.0×1.2 mm); as such, these images might have intrinsic diagnostic value beyond the processed maps that can be derived from them. Using the present equations, these MPME images were converted into 3D quantitative maps for T2, T2*, T1, B0, B1+ and C×M0 (i.e., M0 weighed by the receive-coil sensitivity). The main weakness of the proposed approach, as currently implemented, was noise amplification from one processing step to the next, which led to visibly-noisy parameter maps for in vivo results. For this reason, the present method might currently be restricted to ROI-based applications such as tissue characterization, as these would enable some degree of noise averaging to occur over the ROI. In future work, several approaches may be envisioned to help control noise amplification. Most notably, a regularization scheme tailored to the present equations might be devised to potentially lower noise levels to a considerable extent. Alternately, convolutional neural networks might prove well-suited to the conversion of acquired MPME images into parameter maps in an SNR-advantageous manner.
Scan time was relatively long here, about 12 min for full brain coverage. As suggested by Fig. 3b, shorter TR settings might be achievable at little to no cost in terms of T1 precision. Furthermore, readily available acceleration methods could be included at a cost in SNR; especially if noise amplification could be better controlled (paragraph above), reductions in scan time might be achieved in a simple manner with parallel imaging, for example. Other limitations included partial voluming, which was not accounted for here. In principle at least, because the proposed approach is analytical in nature, it should be possible to acquire more data and resolve partial voluming effects. Furthermore, while many interesting MR parameters were mapped others were not, such as the apparent diffusion coefficient (ADC), for example. Because gradient pulses were fairly small and/or quickly rephased here, the data did not capture diffusion effects in any practical manner. Furthermore, the need for a calibration factor β in Eq. 19 is not explained here; this calibration was performed once and for all and the same value was employed for all in vivo datasets. Other limitations included a small number of healthy subjects (five), in a single application (brain), and a small loss of resolution from the circular coverage in the ky-kz plane (see Fig. 2c) as opposed to a full, square coverage.
The use of very large flip angles in the partially-sampled i=2 scan, with nearly 360o of rotation, would suggest that high specific absorption rate (SAR) might be a problem with the proposed approach. In fact, three factors combine to make SAR a minor issue here: TR is not particularly short, hard pulses were employed, and most of the acquisition time is actually spent on the full-resolution small flip angle scan. However, as a limitation, the low-flip angle RF pulse had to be played here with longer duration and lower voltage than would otherwise have been necessary, because its duration was matched to that of the larger pulse (see Fig. 1). Please note that in the present method, a small negative flip angle could not replace a large positive one, for example −10o instead of +350o. This is because flip angles are not assumed to be correct and B1+ is actually being mapped. Imagine a location where B1+ would happen to be only 90% of its full expected value; in such location, nominal −10o and 350o pulses would turn out as −9o and 315o instead, two very different angles.
The main contribution of the present work might arguably be the set of equations derived here, which demonstrate that MPME acquisitions capture the information required for parameter mapping. The manner in which the parameter maps are generated, whether by solving small sets of equations in a serial manner as proposed here, or possibly through a regularized numerical solution, or a convolutional neural network, may evolve and improve in future work. While some of these processing strategies may prove more SNR-advantageous than others, in all cases the present equations should remain relevant, as they demonstrate that MPME scans capture the information content needed for parameter mapping. Lastly, the proposed method functions in a manner very distinct from prior quantitative imaging methods, offering a different set of strengths and weaknesses; as such, it might in time play a key role in hybrid methods combining different strategies for added performance.
Conclusion
A multi-pathway multi-echo sequence and associated processing were developed for quantitative MRI, and maps of T2, T2*, T1, B0, B1+ and C×M0 were generated.
Acknowledgements
Financial support from grants NIH R21EB019500, P41EB015898 and R01CA149342 is duly acknowledged. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Appendix
While the present method involves a lower and higher flip angle scan, these two scans can be acquired in sequential or interleaved fashion. Equations presented in the main body of the text were for the sequential case, while their equivalent for the interleaved case are presented here instead. For a TR-interleaved acquisition, whereby the flip angle flip-flops between two very different values from TR to TR, the steady-state condition is:
| [A1] |
where h(1) = 2 and h(2) = 1. Using Eq. A1 instead of Eq. 14, the TR-interleaved equivalent of Eq. 15 became:
| [A2] |
In the special case where TR1 = TR2, Eq. A2 simplifies to:
| [A3] |
Like for Eq. 15 in the sequential case, c2 was replaced by its expression from Eq. 8, and Eq. A3 could be solved for α1.
Relationships similar to Eq. 16–17 were also obtained for the TR-interleaved case, using the steady-state definition from Eq. A1 rather than that from Eq. 14:
| [A4] |
| [A5] |
References
- 1.Poon CS, Henkelman RM. Practical T2 quantitation for clinical applications. J Magn Reson Imaging 1992;2(5):541–553. [DOI] [PubMed] [Google Scholar]
- 2.Hahn EL. Spin echoes. Phys Rev 1950;80:580–594. [Google Scholar]
- 3.Bruder H, Fischer H, Graumann R, Deimling M. A new steady-state imaging sequence for simultaneous acquisition of two MR images with clearly different contrasts. Magn Reson Med 1988;7(1):35–42. [DOI] [PubMed] [Google Scholar]
- 4.Welsch GH, Scheffler K, Mamisch TC, Hughes T, Millington S, Deimling M, Trattnig S. Rapid estimation of cartilage T2 based on double echo at steady state (DESS) with 3 Tesla. Magn Reson Med 2009;62(2):544–549. [DOI] [PubMed] [Google Scholar]
- 5.Deoni SC. Transverse relaxation time (T2) mapping in the brain with off-resonance correction using phase-cycled steady-state free precession imaging. J Magn Reson Imaging 2009;30(2):411–417. [DOI] [PubMed] [Google Scholar]
- 6.Bieri O, Scheffler K, Welsch GH, Trattnig S, Mamisch TC, Ganter C. Quantitative mapping of T2 using partial spoiling. Magn Reson Med 2011;66(2):410–418. [DOI] [PubMed] [Google Scholar]
- 7.Bieri O, Ganter C, Scheffler K. Quantitative in vivo diffusion imaging of cartilage using double echo steady-state free precession. Magn Reson Med 2012;68(3):720–729. [DOI] [PubMed] [Google Scholar]
- 8.Staroswiecki E, Granlund KL, Alley MT, Gold GE, Hargreaves BA. Simultaneous estimation of T(2) and apparent diffusion coefficient in human articular cartilage in vivo with a modified three-dimensional double echo steady state (DESS) sequence at 3 T. Magn Reson Med 2012;67(4):1086–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J, Wehrli FW. Method for image-based measurement of the reversible and irreversible contribution to the transverse-relaxation rate. Journal of magnetic resonance Series B 1996;111(1):61–69. [DOI] [PubMed] [Google Scholar]
- 10.Cheng CC, Mei CS, Duryea J, Chung HW, Chao TC, Panych LP, Madore B. Dual-pathway multi-echo sequence for simultaneous frequency and T2 mapping. J Magn Reson 2016;265:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kay I, Henkelman RM. Practical implementation and optimization of one-shot T1 imaging. Magn Reson Med 1991;22(2):414–424. [DOI] [PubMed] [Google Scholar]
- 12.Look D, Locker D. Time saving in measurement of NMR and EPR relaxation times. Rev Sci Instrum 1970;41:250–251. [Google Scholar]
- 13.Helms G, Dathe H, Dechent P. Quantitative FLASH MRI at 3T using a rational approximation of the Ernst equation. Magn Reson Med 2008;59(3):667–672. [DOI] [PubMed] [Google Scholar]
- 14.Treier R, Steingoetter A, Fried M, Schwizer W, Boesiger P. Optimized and combined T1 and B1 mapping technique for fast and accurate T1 quantification in contrast-enhanced abdominal MRI. Magn Reson Med 2007;57(3):568–576. [DOI] [PubMed] [Google Scholar]
- 15.Hurley SA, Yarnykh VL, Johnson KM, Field AS, Alexander AL, Samsonov AA. Simultaneous variable flip angle-actual flip angle imaging method for improved accuracy and precision of three-dimensional T1 and B1 measurements. Magn Reson Med 2012;68(1):54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deoni SC, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med 2003;49(3):515–526. [DOI] [PubMed] [Google Scholar]
- 17.Deoni SC, Rutt BK, Arun T, Pierpaoli C, Jones DK. Gleaning multicomponent T1 and T2 information from steady-state imaging data. Magn Reson Med 2008;60(6):1372–1387. [DOI] [PubMed] [Google Scholar]
- 18.Heule R, Ganter C, Bieri O. Triple echo steady-state (TESS) relaxometry. Magn Reson Med 2014;71(1):230–237. [DOI] [PubMed] [Google Scholar]
- 19.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature 2013;495(7440):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma D, Gulani V, Seiberlich N, Duerk JL, Griswold MA. MR Fingerprinting: Rapid Simultaneous Quantification of T1, T2, Proton Density and Off-resonance using a Spiral. Proceedings of the International Society of Magnetic Resonance in Medicine. Salt Lake City, UT, USA2013. p 18. [Google Scholar]
- 21.Ma D, Jiang Y, Chen Y, McGivney D, Mehta B, Gulani V, Griswold M. Fast 3D magnetic resonance fingerprinting for a whole-brain coverage. Magn Reson Med 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madore B, Panych LP, Mei CS, Yuan J, Chu R. Multipathway sequences for MR thermometry. Magn Reson Med 2011;66(3):658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sacolick LI, Wiesinger F, Hancu I, Vogel MW. B1 mapping by Bloch-Siegert shift. Magn Reson Med 2010;63(5):1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacolick LI, Sun L, Vogel MW, Dixon WT, Hancu I. Fast radiofrequency flip angle calibration by Bloch-Siegert shift. Magn Reson Med 2011;66(5):1333–1338. [DOI] [PubMed] [Google Scholar]
- 25.Hennig J Multiecho Imaging Sequences with Low Refocusing Flip Angles. Journal of magnetic Resonance (1969) 1988;78(3):397–407. [Google Scholar]
- 26.Mizumoto CT, Yoshitome E. Multiple echo SSFP sequences. Magn Reson Med 1991;18(1):244–250. [DOI] [PubMed] [Google Scholar]
- 27.Madore B, Hoge WS. NC-IGT Fast Imaging Library. https://ncigt.org/fast-imaging-library 2009.
- 28.Hennig J Echoes—how to generate, recognize, use or avoid them in MR-imaging sequences. Part I: Fundamental and not so fundamental properties of spin echoes. Concepts in Magnetic Resonance 1991;3(3):125–143. [Google Scholar]
- 29.Friedman L, Glover GH. Report on a multicenter fMRI quality assurance protocol. J Magn Reson Imaging 2006;23(6):827–839. [DOI] [PubMed] [Google Scholar]
- 30.Gelman N, Gorell JM, Barker PB, Savage RM, Spickler EM, Windham JP, Knight RA. MR imaging of human brain at 3.0 T: preliminary report on transverse relaxation rates and relation to estimated iron content. Radiology 1999;210(3):759–767. [DOI] [PubMed] [Google Scholar]
- 31.Ethofer T, Mader I, Seeger U, Helms G, Erb M, Grodd W, Ludolph A, Klose U. Comparison of longitudinal metabolite relaxation times in different regions of the human brain at 1.5 and 3 Tesla. Magn Reson Med 2003;50(6):1296–1301. [DOI] [PubMed] [Google Scholar]
- 32.Sedlacik J, Boelmans K, Lobel U, Holst B, Siemonsen S, Fiehler J. Reversible, irreversible and effective transverse relaxation rates in normal aging brain at 3T. Neuroimage 2014;84:1032–1041. [DOI] [PubMed] [Google Scholar]
- 33.Pipe JG. Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn Reson Med 1999;42(5):963–969. [DOI] [PubMed] [Google Scholar]
- 34.Zaitsev M, Maclaren J, Herbst M. Motion artifacts in MRI: A complex problem with many partial solutions. J Magn Reson Imaging 2015;42(4):887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoge WS, Brooks DH, Madore B, Kyriakos WE. A tour of accelerated parallel MR imaging from a linear systems perspective. Concepts in Magnetic Resonance Part A 2005;27A(1):17–37. [Google Scholar]
- 36.Hoge WS, Kilmer ME, Haker SJ, Brooks DH, Kyriakos WE. Fast regularized reconstruction of non-uniformly subsampled parallel MRI data. Proc IEEE Intl Symp on Biomedical Imaging 2006:714–717. [Google Scholar]