Abstract
Malonyl-CoA is a central metabolite in fatty acid biochemistry. It is the rate-determining intermediate in fatty acid synthesis but is also an allosteric inhibitor of the rate-setting step in mitochondrial long-chain fatty acid oxidation. While these canonical cytoplasmic roles of malonyl-CoA have been well described, malonyl-CoA can also be generated within the mitochondrial matrix by an alternative pathway: the ATP-dependent ligation of malonate to Coenzyme A by the malonyl-CoA synthetase ACSF3. Malonate, a competitive inhibitor of succinate dehydrogenase of the TCA cycle, is a potent inhibitor of mitochondrial respiration. A major role for ACSF3 is to provide a metabolic pathway for the clearance of malonate by the generation of malonyl-CoA, which can then be decarboxylated to acetyl-CoA by malonyl-CoA decarboxylase. Additionally, ACSF3-derived malonyl-CoA can be used to malonylate lysine residues on proteins within the matrix of mitochondria, possibly adding another regulatory layer to post-translational control of mitochondrial metabolism. The discovery of ACSF3-mediated generation of malonyl-CoA defines a new mitochondrial metabolic pathway and raises new questions about how the metabolic fates of this multifunctional metabolite intersect with mitochondrial metabolism.
Keywords: Malonyl-CoA, synthetase, mitochondria, fatty acid, malonate, malonic acid, succinate dehydrogenase, antimetabolite
1. Introduction.
Malonyl-coenzyme A (malonyl-CoA) is positioned at a central regulatory node in mammalian metabolism to coordinate the synthesis and oxidation of fatty acids, which are spatially- and temporally-segregated processes. Malonyl-CoA is generated by the biotin- and ATP-dependent carboxylation of acetyl-CoA by the highly regulated acetyl-CoA carboxylase (ACC) in the cytoplasm and on the cytoplasmic-surface of the mitochondrial outer membrane (Foster, 2012; Kerner et al., 2014; Ruderman et al., 2003; Saggerson, 2008; Wolfgang and Lane, 2006). Malonyl-CoA can then be used by fatty acid synthase (FASN) to generate long-chain fatty acids, or be used for chain-elongation of fatty acids (Kerner et al., 2014). Therefore, malonyl-CoA represents the rate-determining and committed metabolite in de novo fatty acid synthesis. Concomitantly, malonyl-CoA acts as an allosteric inhibitor of carnitine palmitoyltransferase 1 (CPT1), the rate-setting step in the mitochondrial β-oxidation of long-chain fatty acids (Zammit et al., 1997). Therefore, malonyl-CoA, by participating in fatty acid synthesis and inhibiting mitochondrial long-chain fatty acid oxidation, is the metabolite that mediates the basic metabolic logic whereby fatty acid synthesis and oxidation do not occur simultaneously. This regulatory role extends beyond eukaryotes, as many bacteria utilize malonyl-CoA not only as a fatty acid biosynthetic intermediate but also as a transcriptional regulator. The highly conserved bacterial fatty acid and phospholipid regulator, FapR, acts as a transcriptional repressor to inhibit the expression of genes responsible for the synthesis of saturated fatty acids (Schujman et al., 2003). Malonyl-CoA binding to FapR signals the abundance of substrate for fatty acid biosynthesis, and a conformational shift in FapR promotes its release from the operon to permit transcription (Schujman et al., 2006). In these ways, malonyl-CoA represents an important metabolic and regulatory node in fatty acid metabolism.
The synthesis of malonyl-CoA has garnered extensive investigation. Acetyl-CoA carboxylase (ACC) is a well characterized enzyme that generates most of the cellular malonyl-CoA and is highly sensitive to cellular energy charge. ACC is perhaps the best-characterized canonical target of AMP-activated protein kinase (AMPK) as AMPK phosphorylates and inhibits ACC under low energy status to suppress energydemanding fatty acid synthesis (Hardie et al., 2012; Munday and Hemingway, 1999). ACC is encoded by two separate genes in mammals, ACC1 and ACC2 that exhibit unique subcellular and tissue distributions. ACC1 is a large enzyme that forms extensive filaments when activated by one of its allosteric activators, citrate (Hunkeler et al., 2018; Meredith and Lane, 1978). ACC2 does not form filaments but is instead anchored to the outer mitochondrial membrane facing the cytoplasm and is enriched in tissues that perform a high degree of fatty acid oxidation such as skeletal muscle and heart (Abu-Elheiga et al., 2000; Ha et al., 1996). While the acetyl-CoA used by ACC to generate malonyl-CoA is localized in the cytoplasm, most of the acetyl-CoA is derived from the ATP-citrate lyase-dependent breakdown of citrate that has been transported to the cytoplasm from mitochondria. This enables carbohydrates like glucose and fructose to generate abundant substrates for fatty acid synthesis and intimately, but indirectly, involves mitochondria.
ACC-derived malonyl-CoA can then be used to extend fatty acids by two-carbon units by FASN or fatty acid elongases, which are both localized in the cytoplasm. Both acetyl-CoA and malonyl-CoA are membrane-impermeable and almost all of the canonical enzymatic machinery for the generation and use of malonyl-CoA that has been described is localized exclusively to the cytoplasm. The main purpose of malonyl-CoA is to drive de novo fatty acid synthesis, and pharmacological inhibition of FASN was sufficient to increase cellular malonyl-CoA concentrations and was cytotoxic to cultured breast cancer cells (Pizer et al., 2000; Wu et al., 2014). This increase in malonyl-CoA was ACC-dependent, and pharmacological inhibition of both ACC and FASN reduced the cytotoxicity of FASN inhibition alone (Pizer et al., 2000). In a similar manner, pharmacological activation of AMPK, particularly in combination with other inhibitors of metabolism or signal transduction, may be a useful strategy to treat a variety of cancers (Candido et al., 2018). Although the effects of malonyl-CoA concentrations in the context of disease are incompletely known, there are physiologically relevant roles for malonyl-CoA beyond its use as a lipogenic substrate and allosteric inhibitor of mitochondrial fatty acid oxidation.
Tissues with limited expression of FASN, such as mammalian muscle, generate cytoplasmic malonyl-CoA via ACC to regulate fatty acid oxidation (Funai et al., 2013; Pender et al., 2006). It has been proposed that ACC2 is anchored on the mitochondrial outer membrane so that its product, malonyl-CoA is juxtaposed to its allosteric target, Cpt1b (Abu-Elheiga et al., 2001). However, empirical evidence for the physiological significance for this juxtaposition on the regulation of fatty acid oxidation is lacking (Hoehn et al., 2010; Olson et al., 2010). Muscle regulates malonyl-CoA concentrations primarily by its decarboxylation via malonyl-CoA decarboxylase, MLYCD, rather than its use as a substrate for FASN (Rodriguez et al., 2014; Saha et al., 2000). Curiously, MLYCD contains putative peroxisomal and mitochondrial targeting sequences and can be readily found in mitochondria (FitzPatrick et al., 1999; Kerner and Hoppel, 2002; Laurent et al., 2013; Sambandam et al., 2004). Its localization to the cytoplasm is likely due to a weak initial Kozak sequence allowing transcription to begin downstream of the putative mitochondrial targeting signal (Voilley et al., 1999). The mitochondrial localization of MLYCD suggests that its substrate, malonyl-CoA, may be present within that organelle. How malonyl-CoA, which is membrane-impermeable, can be generated in or gain access to the mitochondrial matrix has been a longstanding mystery. This has been at least partly resolved by the discovery of a eukaryotic mitochondrial malonyl-CoA synthetase, ACSF3, localized to the mitochondrial matrix (Chen et al., 2011).
2. ACSF3, a mitochondrial malonyl-CoA synthetase.
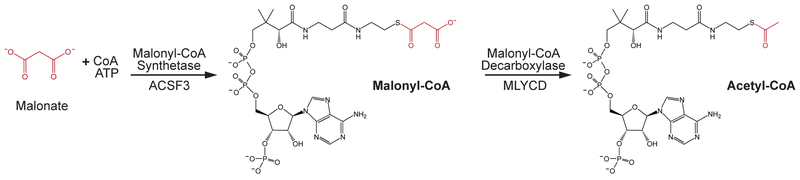
Acyl-CoA synthetase (ACS) enzymes are essential for activating fatty acids for further metabolic processes by catalyzing the ATP-dependent ligation of CoA onto a diverse array of fatty acids. Members of the ACS enzyme family are broadly distributed across tissues and organelles and act upon a wide range of structurally diverse fatty acids (Ellis et al., 2015). In addition to activating fatty acids, there is evidence that ACS isoforms can also direct fatty acids into specific metabolic fates (Ellis et al., 2010; Ellis et al., 2011). ACSF3 was originally described as an orphan member of the ACS family of enzymes because it was part of a structurally unique and enigmatic branch of ACS enzymes and its preferred fatty acid substrate was not known (Watkins et al., 2007). The Arabidopsis ACSF3 ortholog, Acyl Activating Enzyme 13, was described as a eukaryotic malonyl-CoA synthetase essential for plant growth and viability, especially in the presence of exogenous malonate (Chen et al., 2011; Guan and Nikolau, 2016). Like other acyl-CoA synthetases, ACSF3 ligates Coenzyme A to its co-substrate, malonate, in an ATP-dependent manner (Figure 1). Human ACSF3 localizes to the mitochondrial matrix to produce malonyl-CoA from malonate within that organelle (Witkowski et al., 2011). Mitochondrial malonate is an endogenous dicarboxylic acid that is a classic competitive inhibitor of succinate dehydrogenase, a component of the tricarboxylic acid (TCA) cycle and Complex II of the electron transport chain (Quastel and Wooldridge, 1928). As such, malonate is often used as a metabolic toxin to destroy striatal neurons in models of Parkinson’s disease (Beal et al., 1993; Moy et al., 2000; Zeevalk et al., 1997) or to inhibit mitochondrial respiration (Salabei et al., 2014). Since malonate is an effective inhibitor of mitochondrial respiration, a robust detoxifying enzyme would be important to eliminate this endogenous antimetabolite and prevent it from disrupting normal metabolism. In this way, ACSF3 may participate in a metabolite proofreading system to clear endogenous malonate and prevent inhibition of mitochondrial respiration (Van Schaftingen et al., 2013).
Figure 1.
The source of malonate in mammalian systems has not been well worked out, but it likely comes at least in part from the non-enzymatic hydrolysis of cytoplasmic malonyl-CoA. CoA thioesters are highly susceptible to non-enzymatic hydrolysis, and malonyl-CoA concentrations correlate with conditions of high de novo fatty acid synthesis (Bandyopadhyay et al., 2006; Zhao et al., 2009). Malonyl-CoA concentrations are maintained lower than other short-chain acyl-CoAs in vivo (Sadhukhan et al., 2016), and this may be related to the potency of malonate as an inhibitor of mitochondrial respiration. In addition to non-enzymatic hydrolysis of malonyl-CoA, there could be a role for enzymatic hydrolysis by the acyl-CoA thioesterase family of enzymes (Tillander et al., 2017). Another possible source of malonate could be the unintended production from an enzymatic reaction for which malonate precursors are not the primary substrates. For example, propionyl-CoA carboxylase in the mitochondrial matrix can carboxylate acetyl-CoA to malonyl-CoA; however, this reaction occurs at a rate 100 times slower than propionyl-CoA carboxylation (Hegre et al., 1959). There is also evidence that, in plants and in pig heart, malonate could be generated from the decarboxylation of oxaloacetate (de Vellis et al., 1963; Vennesland and Evans, 1944). Additionally, some bacterial species can generate malonate from pyrimidines (Hayaishi and Kornberg, 1952). A pathway of malonate generation from beta-alanine and malonate semialdehyde has also been proposed in humans, but the enzymatic requirements for these metabolic conversions have not been fully established (Scholem and Brown, 1983). Malonate may also be formed from the oxidation of malondialdehyde, an indicator of oxidative stress (Chen et al., 2011). While the sources of cellular malonate are incompletely defined, nevertheless, a major role for ACSF3 is to activate the toxic, endogenous antimetabolite malonate into malonyl-CoA that can be decarboxylated to acetyl-CoA and therefore fully oxidized within the TCA cycle (Figure 1). Next, the metabolic consequences of impaired malonate and malonyl-CoA metabolism in human disease are discussed.
3. Human genetics of malonate and malonyl-CoA metabolism
Inborn errors of MLYCD result in a combined malonic and methylmalonic aciduria (FitzPatrick et al., 1999; Gao et al., 1999; Wightman et al., 2003). The severe malonic and methylmalonic aciduria characteristic of MLYCD deficiency is accompanied by developmental delay, seizure disorders, hypoglycemia, and cardiomyopathy. Recently, patients that presented with combined malonic and methylmalonic aciduria that did not exhibit mutations in MLYCD were found to have nonsynonymous mutations in the ACSF3 gene (Alfares et al., 2011; Sloan et al., 2011). The similarities in the phenotypes of MLYCD and ACSF3 deficiencies suggest that they exist in the same biochemical pathway. Additionally, they are genetically linked in mammals suggesting their coinheritance may be evolutionarily advantageous. This presents a metabolic rationale for why MLYCD is localized within mitochondria—toxic malonate may be metabolized within mitochondria through the subsequent activities of ACSF3 and MLYCD. Together, the mitochondrial malonyl-CoA synthetase and decarboxylase perform a critical metabolic editing function, allowing highly oxidative tissues to continue to respire unabated by malonate inhibition. Importantly, malonate is a more potent inhibitor of succinate dehydrogenase than the other acidemias that accompany these metabolic deficiencies (Kolker et al., 2003). In addition, ACS enzymes are feedback inhibited by their product, therefore, the loss of MLYCD and subsequent increase in malonyl-CoA would be predicted to inhibit ACSF3 activity and overwhelm its ability to detoxify malonate. Therefore, the coordinated metabolism of malonate by these two enzymes is critical for malonate metabolism.
While the evidence from human genetics presented above is rather convincing that mutations in ACSF3 cause a devastating inborn error of metabolism, more recent evidence has called this into question. Due to the serious nature of malonic and methylmalonic aciduria, some regions have begun screening newborns for these metabolites. The screening has discovered patients with malonic and methylmalonic aciduria that have mutations in ACSF3 with no apparent adverse phenotype (Levtova et al., 2018). These mutations are not null for ACSF3; therefore, it is not clear how much residual ACSF3 activity exists in these patients given that the adverse actions of malonic and methylmalonic aciduria did not reach a clinically significant threshold. Additionally, while a full phenotype of ACSF3 knockout mice has not been described, a preliminary report suggests they have neurological impairments (Epping et al., 2015). Clearly, more work is needed to elucidate the clinical significance of ACSF3 mutations.
One function of ACSF3 that has been proposed was that it generated malonyl-CoA in the matrix of the mitochondria to enable mitochondrial type II fatty acid synthesis (Guan and Nikolau, 2016; Witkowski et al., 2011). Mitochondrial fatty acid synthesis is to be contrasted with type I fatty acid synthesis that exists in the cytoplasm of mammalian cells. Type II fatty acid synthesis is a more ancient form of lipogenesis that occurs exclusively in the mitochondrial matrix to generate octanoyl-ACP (acyl-carrier protein), which is a substrate for the synthesis of lipoic acid, a required cofactor for 2-oxoacid dehydrogenases such as pyruvate dehydrogenase and α-ketoglutarate dehydrogenase (Hiltunen et al., 2010; Hiltunen et al., 2009; Solmonson and DeBerardinis, 2018). Others have shown that ACSF3-mediated generation of mitochondrial malonyl-CoA contributed to lipoic acid synthesis by shRNA knockdown in cells in vitro (Witkowski et al., 2011). Our laboratory has generated human cells with a null mutation in ACSF3 by CRISPR/Cas9 genome engineering that recapitulated many of the biochemical features of ACSF3 deficiency and could not metabolize malonate (Bowman et al., 2017). These ACSF3-deficient cells, however, did not exhibit any defects in protein lipoylation. Cells cannot scavenge lipoic acid from cell culture media so this suggests that ACSF3 is not required for mitochondrial type II de novo fatty acid synthesis (Habarou et al., 2017; Mayr et al., 2014). Another putative source of mitochondrial malonyl-CoA for type II fatty acid synthesis is the carboxylation of acetyl-CoA to malonyl-CoA by propionyl-CoA carboxylase in the mitochondrial matrix; however, this reaction occurs at a very low rate compared to propionyl-CoA carboxylation (Hegre et al., 1959). The loss of lipoic acid synthesis or other components of mitochondrial type II de novo fatty acid synthesis results in a devastating phenotype and almost total loss of mitochondrial respiration (Smith et al., 2012; Yi and Maeda, 2005). Given that ACSF3 deficiency does not phenocopy defects in lipoic acid synthesis, the genetics and biochemistry do not support a requirement for ACSF3 in mitochondrial type II de novo fatty acid synthesis. Interestingly, some plants such as petunia flowers do utilize a malonyl-CoA synthetase for lipid and pigment biosynthesis, including the anthocyanins that give them their vibrant colors (Chen et al., 2017). While ACSF3-derived malonyl-CoA contributes to biosynthetic routes in certain organisms, human cells are able to catabolize mitochondrial malonyl-CoA. Another fate of mitochondrial malonyl-CoA is described next and depends on the chemical energy that is used by ACSF3 to generate the malonyl-CoA thioester bond.
4. Lysine Malonylation.
The high-energy thioester bond of an acyl-CoA is ideal for acyltransferase reactions to facilitate anabolic or catabolic metabolism. It also enables their transacylation to proteins facilitated both by enzymatic transfer and non-enzymatic addition under high substrate concentrations. Acetylation of proteins is the most well characterized post-translational protein acylation having empirically established roles in regulating enzyme activity, transcription factor regulation, and affecting histone-mediated transcriptional activity. Although acetylation is the most widely studied, essentially all acyl-CoAs can have their acyl group transferred to a protein substrate including the short-chain acyl-CoAs derived from dicarboxylic acids: malonyl-CoA, succinyl-CoA, and glutaryl-CoA (Azevedo and Saiardi, 2016; Hirschey and Zhao, 2015; Lin et al., 2012). These CoA esters of dicarboxylic acids have been shown to robustly acylate lysine residues (Kulkarni et al., 2017; Pougovkina et al., 2014; Wagner et al., 2017). While a dedicated enzyme responsible for malonylating proteins has not been identified, there is a report of carnitine palmitoyltransferase 1a, Cpt1a, exhibiting moonlighting activities as a succinyltransferase (Kurmi et al., 2018). While the addition of acyl moieties to lysine residues occurs via an unidentified acyltransferase or via non-enzymatic addition, Sirt5 has been shown to be an NAD-dependent lysine demalonylase and desuccinylase that is localized mainly to the mitochondrial matrix (Du et al., 2011; Peng et al., 2011). A knockout mouse model of Sirt5 is associated with a 2-fold increase in fasting blood ammonia with modest suppression of Cps1 activity which is required for mitochondrial urea cycle function (Nakagawa et al., 2009). It is not clear if the small change in Cps1 activity in this model impacts the urea cycle, increases fasting blood ammonia directly, or results in broader mitochondrial defects, but Sirt5 deficiency did not result in further overt metabolic consequences (Yu et al., 2013). Recent studies suggest that Sirt5 regulates heart function and is protective in models of cardiac ischemia-reperfusion injury and pressure overload (Boylston et al., 2015; Hershberger et al., 2017; Sadhukhan et al., 2016). Proteomic investigations have demonstrated that the loss of Sirt5 drives the up-regulation of succinylated, malonylated, and glutarylated proteins in multiple cellular compartments (Nishida et al., 2015; Park et al., 2013; Rardin et al., 2013; Tan et al., 2014). Reciprocally, the overexpression of Sirt5 down-regulates succinylated and malonylated proteins, but mice that globally and robustly overexpress Sirt5 exhibit a fairly mild phenotype (Bentley et al., 2018). Although examples of lysine malonylation and succinylation of specific target proteins have been highlighted as the parsimonious explanation for the observed phenotypes of Sirt5 knockout mice, a broader explanation of the biological function and regulatory potential of these modifications has not yet been demonstrated.
Mouse models of obesity and diabetes that exhibit enhanced de novo lipogenesis, such as the ob/ob mouse, show an increase in protein lysine malonylation (Bowman et al., 2017; Du et al., 2015). Additionally, fasting, a time of suppressed de novo lipogenesis, correlates with a decrease in malonylation (Nishida et al., 2015). These data suggest that protein malonylation and fatty acid synthesis are linked and that cytoplasmic malonyl-CoA is likely an endogenous source of malonate. Consistent with our model of malonate metabolism, the loss of MLYCD generates a dramatic increase in mitochondrial malonylated proteins due to an increase in malonyl-CoA concentration (Colak et al., 2015). Our laboratory has shown that a loss of ACSF3 results in the inability of cells to malonylate mitochondrial proteins due to a loss of mitochondrial malonyl-CoA without affecting succinylation (Bowman et al., 2017). In a setting of ACSF3 loss-of-function, there are metabolic effects that are independent of malonate-mediated inhibition of SDH, suggesting that malonylation can have independent effects on mitochondrial function.
Eukaryotes express an abundant number of protein acyltranferses within the cytoplasm and nucleus. However, it is not apparent that mitochondria express enzymes capable of lysine acylation or at least evidence for their existence has not been strong. Alternatively, it has been argued that mitochondrial lysine acylation occurs via non-enzymatic addition (Weinert et al., 2015). This mode of post-translational modification does not preclude an important role in regulation, as this would likely be a way to evolve post-translational functionality. However, it may suggest that for many of the sites identified as modified by acetylation, succinylation, malonylation, etc., there may not be direct regulatory functions. Rather, removal of the acyl modification may be required to retain efficient activity or protein-protein interactions. The function of deacetylases and demalonylases may be less akin to a novel regulatory function but may be a mechanism of protein quality control (Wagner and Hirschey, 2014). None-the-less, a better understanding of the roles and mechanism of mitochondrial protein acylation is needed.
5. Summary.
Malonyl-CoA lies at a critical metabolic mode in cellular metabolism. It has been shown to play important roles in cytoplasmic fatty acid biochemistry as all of the canonical enzymatic machinery for its production and use is located there. However, the discovery of a malonyl-CoA generating enzyme within the matrix of mitochondria opens new avenues for discovery of the roles and requirements of this important metabolite in an unexpected cellular location. ACSF3 is a mitochondrial enzyme that generates malonyl-CoA from the toxic antimetabolite malonate. Therefore, ACSF3 performs a critical metabolic editing function to enable highly metabolically active cells to continue to respire. Although it is likely that malonate is endogenously generated, the route of synthesis and how it is regulated needs to be defined empirically. A better understanding of malonate generation and the effects of mitochondrial protein malonylation could also be used to devise rational nutritional and/or pharmacological interventions for malonic acidurias.
Acknowledgements
We acknowledge the support of the National Institutes of Health grant R01NS072241 to M.J.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References.
- Abu-Elheiga L, Brinkley WR, Zhong L, Chirala SS, Woldegiorgis G, Wakil SJ, 2000. The subcellular localization of acetyl-CoA carboxylase 2. Proc. Natl. Acad. Sci. U. S. A 97(4), 1444–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ, 2001. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science 291(5513), 2613–2616. [DOI] [PubMed] [Google Scholar]
- Alfares A, Nunez LD, Al-Thihli K, Mitchell J, Melancon S, Anastasio N, Ha KC, Majewski J, Rosenblatt DS, Braverman N, 2011. Combined malonic and methylmalonic aciduria: exome sequencing reveals mutations in the ACSF3 gene in patients with a non-classic phenotype. J. Med. Genet 48(9), 602–605. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Saiardi A, 2016. Why always lysine? The ongoing tale of one of the most modified amino acids. Adv Biol Regul 60, 144–150. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay GK, Yu JG, Ofrecio J, Olefsky JM, 2006. Increased malonyl-CoA levels in muscle from obese and type 2 diabetic subjects lead to decreased fatty acid oxidation and increased lipogenesis; thiazolidinedione treatment reverses these defects. Diabetes 55(8), 2277–2285. [DOI] [PubMed] [Google Scholar]
- Beal MF, Brouillet E, Jenkins B, Henshaw R, Rosen B, Hyman BT, 1993. Age-dependent striatal excitotoxic lesions produced by the endogenous mitochondrial inhibitor malonate. J. Neurochem 61(3), 1147–1150. [DOI] [PubMed] [Google Scholar]
- Bentley NL, Fiveash CE, Osborne B, Quek LE, Ogura M, Inagaki N, Cooney GJ, Polly P, Montgomery MK, Turner N, 2018. Protein hypoacylation induced by Sirt5 overexpression has minimal metabolic effect in mice. Biochem. Biophys. Res. Commun [DOI] [PubMed] [Google Scholar]
- Bowman CE, Rodriguez S, Selen Alpergin ES, Acoba MG, Zhao L, Hartung T, Claypool SM, Watkins PA, Wolfgang MJ, 2017. The Mammalian Malonyl-CoA Synthetase ACSF3 Is Required for Mitochondrial Protein Malonylation and Metabolic Efficiency. Cell Chem Biol 24(6), 673–684 e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylston JA, Sun J, Chen Y, Gucek M, Sack MN, Murphy E, 2015. Characterization of the cardiac succinylome and its role in ischemia-reperfusion injury. J. Mol. Cell. Cardiol 88, 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido S, Abrams SL, Steelman L, Lertpiriyapong K, Martelli AM, Cocco L, Ratti S, Follo MY, Murata RM, Rosalen PL, Lombardi P, Montalto G, Cervello M, Gizak A, Rakus D, Suh PG, Libra M, McCubrey JA, 2018. Metformin influences drug sensitivity in pancreatic cancer cells. Adv Biol Regul 68, 13–30. [DOI] [PubMed] [Google Scholar]
- Chen G, Liu H, Wei Q, Zhao H, Liu J, Yu Y, 2017. The acyl-activating enzyme PhAAE13 is an alternative enzymatic source of precursors for anthocyanin biosynthesis in petunia flowers. J Exp Bot 68(3), 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kim HU, Weng H, Browse J, 2011. Malonyl-CoA synthetase, encoded by ACYL ACTIVATING ENZYME13, is essential for growth and development of Arabidopsis. Plant Cell 23(6), 2247–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak G, Pougovkina O, Dai L, Tan M, Te Brinke H, Huang H, Cheng Z, Park J, Wan X, Liu X, Yue WW, Wanders RJ, Locasale JW, Lombard DB, de Boer VC, Zhao Y, 2015. Proteomic and Biochemical Studies of Lysine Malonylation Suggest Its Malonic Aciduria-associated Regulatory Role in Mitochondrial Function and Fatty Acid Oxidation. Mol. Cell. Proteomics 14(11), 3056–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vellis J, Shannon LM, Lew JY, 1963. Malonic Acid Biosynthesis in Bush Bean Roots. I. Evidence for Oxaloacetate as Immediate Precursor. Plant Physiol. 38(6), 686–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H, 2011. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334(6057), 806–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Cai T, Li T, Xue P, Zhou B, He X, Wei P, Liu P, Yang F, Wei T, 2015. Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol. Cell. Proteomics 14(1), 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JM, Bowman CE, Wolfgang MJ, 2015. Metabolic and tissue-specific regulation of acyl-CoA metabolism. PLoS One 10(3), e0116587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JM, Li LO, Wu PC, Koves TR, Ilkayeva O, Stevens RD, Watkins SM, Muoio DM, Coleman RA, 2010. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell metabolism 12(1), 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JM, Mentock SM, Depetrillo MA, Koves TR, Sen S, Watkins SM, Muoio DM, Cline GW, Taegtmeyer H, Shulman GI, Willis MS, Coleman RA, 2011. Mouse cardiac acyl coenzyme a synthetase 1 deficiency impairs Fatty Acid oxidation and induces cardiac hypertrophy. Molecular and cellular biology 31(6), 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epping MW, Wang CX, Manoli I, Fraser JL, Gomez-Rodriguez J, Elliot G, Sloan JL, Hoffman V, Schwartzberg PL, Venditti CP, 2015. A knock-out mouse model of CMAMMA (Acsf3 deficiency) displays neurological phenotype and methylmalonic acidemia, The 65th Annual Meeting of the American Society of Human Genetics Baltimore, MD, USA. [Google Scholar]
- FitzPatrick DR, Hill A, Tolmie JL, Thorburn DR, Christodoulou J, 1999. The molecular basis of malonyl-CoA decarboxylase deficiency. Am. J. Hum. Genet 65(2), 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DW, 2012. Malonyl-CoA: the regulator of fatty acid synthesis and oxidation. J Clin Invest 122(6), 1958–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funai K, Song H, Yin L, Lodhi IJ, Wei X, Yoshino J, Coleman T, Semenkovich CF, 2013. Muscle lipogenesis balances insulin sensitivity and strength through calcium signaling. J. Clin. Invest 123(3), 1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Waber L, Bennett MJ, Gibson KM, Cohen JC, 1999. Cloning and mutational analysis of human malonyl-coenzyme A decarboxylase. J. Lipid Res 40(1), 178–182. [PubMed] [Google Scholar]
- Guan X, Nikolau BJ, 2016. AAE13 encodes a dual-localized malonyl-CoA synthetase that is crucial for mitochondrial fatty acid biosynthesis. Plant J. 85(5), 581–593. [DOI] [PubMed] [Google Scholar]
- Ha J, Lee JK, Kim KS, Witters LA, Kim KH, 1996. Cloning of human acetyl-CoA carboxylase-beta and its unique features. Proc. Natl. Acad. Sci. U. S. A 93(21), 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habarou F, Hamel Y, Haack TB, Feichtinger RG, Lebigot E, Marquardt I, Busiah K, Laroche C, Madrange M, Grisel C, Pontoizeau C, Eisermann M, Boutron A, Chretien D, Chadefaux-Vekemans B, Barouki R, Bole-Feysot C, Nitschke P, Goudin N, Boddaert N, Nemazanyy I, Delahodde A, Kolker S, Rodenburg RJ, Korenke GC, Meitinger T, Strom TM, Prokisch H, Rotig A, Ottolenghi C, Mayr JA, de Lonlay P, 2017. Biallelic Mutations in LIPT2 Cause a Mitochondrial Lipoylation Defect Associated with Severe Neonatal Encephalopathy. Am J Hum Genet 101(2), 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA, 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Reviews Molecular Cell Biology 13, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayaishi O, Kornberg A, 1952. Metabolism of cytosine, thymine, uracil, and barbituric acid by bacterial enzymes. J. Biol. Chem 197(2), 717–732. [PubMed] [Google Scholar]
- Hegre CS, Halenz DR, Lane MD, 1959. The Enzymatic Carboxylation of Butyryl Coenzyme-A. Journal of the American Chemical Society 81(24), 6526–6527. [Google Scholar]
- Hershberger KA, Abraham DM, Martin AS, Mao L, Liu J, Gu H, Locasale JW, Hirschey MD, 2017. Sirtuin 5 is required for mouse survival in response to cardiac pressure overload. 292(48), 19767–19781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen JK, Autio KJ, Schonauer MS, Kursu VA, Dieckmann CL, Kastaniotis AJ, 2010. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta 1797(6–7), 1195–1202. [DOI] [PubMed] [Google Scholar]
- Hiltunen JK, Schonauer MS, Autio KJ, Mittelmeier TM, Kastaniotis AJ, Dieckmann CL, 2009. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 284(14), 9011–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Zhao Y, 2015. Metabolic regulation by lysine malonylation, succinylation and glutarylation. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn KL, Turner N, Swarbrick MM, Wilks D, Preston E, Phua Y, Joshi H, Furler SM, Larance M, Hegarty BD, Leslie SJ, Pickford R, Hoy AJ, Kraegen EW, James DE, Cooney GJ, 2010. Acute or chronic upregulation of mitochondrial fatty acid oxidation has no net effect on whole-body energy expenditure or adiposity. Cell metabolism 11(1), 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkeler M, Hagmann A, Stuttfeld E, Chami M, Guri Y, Stahlberg H, Maier T, 2018. Structural basis for regulation of human acetyl-CoA carboxylase. Nature 558(7710), 470–474. [DOI] [PubMed] [Google Scholar]
- Kerner J, Hoppel CL, 2002. Radiochemical malonyl-CoA decarboxylase assay: activity and subcellular distribution in heart and skeletal muscle. Anal. Biochem 306(2), 283–289. [DOI] [PubMed] [Google Scholar]
- Kerner J, Minkler PE, Lesnefsky EJ, Hoppel CL, 2014. Fatty acid chain elongation in palmitate-perfused working rat heart: mitochondrial acetyl-CoA is the source of two-carbon units for chain elongation. The Journal of biological chemistry 289(14), 10223–10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker S, Schwab M, Horster F, Sauer S, Hinz A, Wolf NI, Mayatepek E, Hoffmann GF, Smeitink JA, Okun JG, 2003. Methylmalonic acid, a biochemical hallmark of methylmalonic acidurias but no inhibitor of mitochondrial respiratory chain. J. Biol. Chem 278(48), 47388–47393. [DOI] [PubMed] [Google Scholar]
- Kulkarni RA, Worth AJ, Zengeya TT, Shrimp JH, Garlick JM, Roberts AM, Montgomery DC, Sourbier C, Gibbs BK, Mesaros C, Tsai YC, Das S, Chan KC, Zhou M, Andresson T, Weissman AM, Linehan WM, Blair IA, Snyder NW, Meier JL, 2017. Discovering Targets of Non-enzymatic Acylation by Thioester Reactivity Profiling. Cell Chem Biol 24(2), 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmi K, Hitosugi S, Wiese EK, Boakye-Agyeman F, Gonsalves WI, Lou Z, Karnitz LM, Goetz MP, Hitosugi T, 2018. Carnitine Palmitoyltransferase 1A Has a Lysine Succinyltransferase Activity. Cell Rep 22(6), 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, Haigis MC, 2013. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell 50(5), 686–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levtova A, Waters PJ, Buhas D, Levesque S, Auray-Blais C, Clarke JTR, Laframboise R, Maranda B, Mitchell GA, Brunel-Guitton C, Braverman NE, 2018. Combined malonic and methylmalonic aciduria due to ACSF3 mutations: benign clinical course in an unselected cohort. J. Inherit. Metab. Dis [DOI] [PubMed] [Google Scholar]
- Lin H, Su X, He B, 2012. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol 7(6), 947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr JA, Feichtinger RG, Tort F, Ribes A, Sperl W, 2014. Lipoic acid biosynthesis defects. Journal of inherited metabolic disease 37(4), 553–563. [DOI] [PubMed] [Google Scholar]
- Meredith MJ, Lane MD, 1978. Acetyl-CoA carboxylase. Evidence for polymeric filament to protomer transition in the intact avian liver cell. J. Biol. Chem 253(10), 3381–3383. [PubMed] [Google Scholar]
- Moy LY, Zeevalk GD, Sonsalla PK, 2000. Role for dopamine in malonate-induced damage in vivo in striatum and in vitro in mesencephalic cultures. J. Neurochem 74(4), 1656–1665. [DOI] [PubMed] [Google Scholar]
- Munday MR, Hemingway CJ, 1999. The regulation of acetyl-CoA carboxylase--a potential target for the action of hypolipidemic agents. Adv Enzyme Regul 39, 205–234. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L, 2009. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137(3), 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW, Verdin E, 2015. SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol. Cell 59(2), 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson DP, Pulinilkunnil T, Cline GW, Shulman GI, Lowell BB, 2010. Gene knockout of Acc2 has little effect on body weight, fat mass, or food intake. Proceedings of the National Academy of Sciences of the United States of America 107(16), 7598–7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, Zhao Y, 2013. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50(6), 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pender C, Trentadue AR, Pories WJ, Dohm GL, Houmard JA, Youngren JF, 2006. Expression of genes regulating malonyl-CoA in human skeletal muscle. J. Cell. Biochem. 99(3), 860–867. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y, 2011. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10(12), M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer ES, Thupari J, Han WF, Pinn ML, Chrest FJ, Frehywot GL, Townsend CA, Kuhajda FP, 2000. Malonyl-coenzyme-A is a potential mediator of cytotoxicity induced by fatty-acid synthase inhibition in human breast cancer cells and xenografts. Cancer Res. 60(2), 213–218. [PubMed] [Google Scholar]
- Pougovkina O, Te Brinke H, Wanders RJ, Houten SM, de Boer VC, 2014. Aberrant protein acylation is a common observation in inborn errors of acyl-CoA metabolism. J. Inherit. Metab. Dis 37(5), 709–714. [DOI] [PubMed] [Google Scholar]
- Quastel JH, Wooldridge WR, 1928. Some properties of the dehydrogenating enzymes of bacteria. Biochem. J 22(3), 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens RD, Ilkayeva OR, Newgard CB, Jacobson MP, Hellerstein M, Goetzman ES, Gibson BW, Verdin E, 2013. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell metabolism 18(6), 920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S, Ellis JM, Wolfgang MJ, 2014. Chemical-genetic induction of Malonyl-CoA decarboxylase in skeletal muscle. BMC Biochem 15, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman NB, Saha AK, Kraegen EW, 2003. Minireview: malonyl CoA, AMP-activated protein kinase, and adiposity. Endocrinology 144(12), 5166–5171. [DOI] [PubMed] [Google Scholar]
- Sadhukhan S, Liu X, Ryu D, Nelson OD, Stupinski JA, Li Z, Chen W, Zhang S, Weiss RS, Locasale JW, Auwerx J, Lin H, 2016. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc. Natl. Acad. Sci. U. S. A 113(16), 4320–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggerson D, 2008. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr 28, 253–272. [DOI] [PubMed] [Google Scholar]
- Saha AK, Schwarsin AJ, Roduit R, Masse F, Kaushik V, Tornheim K, Prentki M, Ruderman NB, 2000. Activation of malonyl-CoA decarboxylase in rat skeletal muscle by contraction and the AMP-activated protein kinase activator 5-aminoimidazole-4-carboxamide-1-beta -D-ribofuranoside. J. Biol. Chem 275(32), 24279–24283. [DOI] [PubMed] [Google Scholar]
- Salabei JK, Gibb AA, Hill BG, 2014. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc 9(2), 421–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam N, Steinmetz M, Chu A, Altarejos JY, Dyck JR, Lopaschuk GD, 2004. Malonyl-CoA decarboxylase (MCD) is differentially regulated in subcellular compartments by 5'AMP-activated protein kinase (AMPK). Studies using H9c2 cells overexpressing MCD and AMPK by adenoviral gene transfer technique. Eur J Biochem 271(13), 2831–2840. [DOI] [PubMed] [Google Scholar]
- Scholem RD, Brown GK, 1983. Metabolism of malonic semialdehyde in man. Biochem. J 216(1), 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schujman GE, Guerin M, Buschiazzo A, Schaeffer F, Llarrull LI, Reh G, Vila AJ, Alzari PM, de Mendoza D, 2006. Structural basis of lipid biosynthesis regulation in Gram-positive bacteria. The EMBO journal 25(17), 4074–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schujman GE, Paoletti L, Grossman AD, de Mendoza D, 2003. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Developmental cell 4(5), 663–672. [DOI] [PubMed] [Google Scholar]
- Sloan JL, Johnston JJ, Manoli I, Chandler RJ, Krause C, Carrillo-Carrasco N, Chandrasekaran SD, Sysol JR, O'Brien K, Hauser NS, Sapp JC, Dorward HM, Huizing M, Barshop BA, Berry SA, James PM, Champaigne NL, de Lonlay P, Valayannopoulos V, Geschwind MD, Gavrilov DK, Nyhan WL, Biesecker LG, Venditti CP, 2011. Exome sequencing identifies ACSF3 as a cause of combined malonic and methylmalonic aciduria. Nat. Genet 43(9), 883–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Witkowski A, Moghul A, Yoshinaga Y, Nefedov M, de Jong P, Feng D, Fong L, Tu Y, Hu Y, Young SG, Pham T, Cheung C, Katzman SM, Brand MD, Quinlan CL, Fens M, Kuypers F, Misquitta S, Griffey SM, Tran S, Gharib A, Knudsen J, Hannibal-Bach HK, Wang G, Larkin S, Thweatt J, Pasta S, 2012. Compromised mitochondrial fatty acid synthesis in transgenic mice results in defective protein lipoylation and energy disequilibrium. PLoS One 7(10), e47196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solmonson A, DeBerardinis RJ, 2018. Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem 293(20), 7522–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y, 2014. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell metabolism 19(4), 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillander V, Alexson SEH, Cohen DE, 2017. Deactivating Fatty Acids: Acyl-CoA Thioesterase-Mediated Control of Lipid Metabolism. Trends Endocrinol Metab 28(7), 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaftingen E, Rzem R, Marbaix A, Collard F, Veiga-da-Cunha M, Linster CL, 2013. Metabolite proofreading, a neglected aspect of intermediary metabolism. J. Inherit. Metab. Dis 36(3), 427–434. [DOI] [PubMed] [Google Scholar]
- Vennesland B, Evans EA, 1944. The formation of malonic acid from oxalacetic acid by pig heart preparations. J. Biol. Chem 156(2), 783–784. [Google Scholar]
- Voilley N, Roduit R, Vicaretti R, Bonny C, Waeber G, Dyck JR, Lopaschuk GD, Prentki M, 1999. Cloning and expression of rat pancreatic beta-cell malonyl-CoA decarboxylase. Biochem. J 340 (Pt 1), 213–217. [PMC free article] [PubMed] [Google Scholar]
- Wagner GR, Bhatt DP, O'Connell TM, Thompson JW, Dubois LG, Backos DS, Yang H, Mitchell GA, Ilkayeva OR, Stevens RD, Grimsrud PA, Hirschey MD, 2017. A Class of Reactive Acyl-CoA Species Reveals the Non-enzymatic Origins of Protein Acylation. Cell metabolism 25(4), 823–837.e828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GR, Hirschey MD, 2014. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 54(1), 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA, Maiguel D, Jia Z, Pevsner J, 2007. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. Journal of lipid research 48(12), 2736–2750. [DOI] [PubMed] [Google Scholar]
- Weinert BT, Moustafa T, Iesmantavicius V, Zechner R, Choudhary C, 2015. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. The EMBO journal 34(21), 2620–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman PJ, Santer R, Ribes A, Dougherty F, McGill N, Thorburn DR, FitzPatrick DR, 2003. MLYCD mutation analysis: evidence for protein mistargeting as a cause of MLYCD deficiency. Hum. Mutat 22(4), 288–300. [DOI] [PubMed] [Google Scholar]
- Witkowski A, Thweatt J, Smith S, 2011. Mammalian ACSF3 protein is a malonyl-CoA synthetase that supplies the chain extender units for mitochondrial fatty acid synthesis. The Journal of biological chemistry 286(39), 33729–33736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MJ, Lane MD, 2006. The role of hypothalamic malonyl-CoA in energy homeostasis. The Journal of biological chemistry 281(49), 37265–37269. [DOI] [PubMed] [Google Scholar]
- Wu X, Qin L, Fako V, Zhang JT, 2014. Molecular mechanisms of fatty acid synthase (FASN)-mediated resistance to anti-cancer treatments. Adv Biol Regul 54, 214–221. [DOI] [PubMed] [Google Scholar]
- Yi X, Maeda N, 2005. Endogenous production of lipoic acid is essential for mouse development. Molecular and cellular biology 25(18), 8387–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS, Lin H, Schoonjans K, Auwerx J, 2013. Metabolic characterization of a Sirt5 deficient mouse model. Sci. Rep 3, 2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit VA, Fraser F, Orstorphine CG, 1997. Regulation of mitochondrial outer-membrane carnitine palmitoyltransferase (CPT I): role of membrane-topology. Adv Enzyme Regul 37, 295–317. [DOI] [PubMed] [Google Scholar]
- Zeevalk GD, Manzino L, Hoppe J, Sonsalla P, 1997. In vivo vulnerability of dopamine neurons to inhibition of energy metabolism. Eur. J. Pharmacol 320(2–3), 111–119. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Lee YJ, Kim SK, Kim HJ, Shim WS, Ahn CW, Lee HC, Cha BS, Ma ZA, 2009. Rosiglitazone and fenofibrate improve insulin sensitivity of pre-diabetic OLETF rats by reducing malonyl-CoA levels in the liver and skeletal muscle. Life Sci. 84(19–20), 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]