Abstract
Hepatitis B virus (HBV) is a unique, tiny, partially double-stranded, reverse-transcribing DNA virus with proteins encoded by multiple overlapping reading frames. The substitution rate is surprisingly high for a DNA virus, but lower than that of other reverse transcribing organisms. More than 260 million people worldwide have chronic HBV infection, which causes 0.8 million deaths a year. Because of the high burden of disease, international health agencies have set the goal of eliminating HBV infection by 2030. Nonetheless, the intriguing HBV genome has not been well characterized. We summarize data on the HBV genome structure and replication cycle, explain and quantify diversity within and among infected individuals, and discuss advances that can be offered by application of next-generation sequencing technology. In-depth HBV genome analyses could increase our understanding of disease pathogenesis and allow us to better predict patient outcomes, optimize treatment, and develop new therapeutics.
Keywords: Hepatitis B Virus, Genotype, Diversity, Evolution
Abbreviations used in the paper: cccDNA, covalently closed circular DNA; dsDNA, double-stranded DNA; HBsAg, hepatitis B surface antigen; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HIV, human immunodeficiency virus; NGS, next-generation sequencing; ORF, open reading frame; P, reverse transcriptase polymerase; RC-DNA, relaxed circular DNA; RT, reverse transcriptase; S, surface

Anna L. McNaughton

Valentina D’Arienzo

M. Azim Ansari

Sheila F. Lumley

Margaret Littlejohn

Peter Revill

Jane A. McKeating

Philippa C. Matthews
Hepatitis B virus (HBV) was first identified in the 1960s by Baruch Blumberg, who went on to win the Nobel prize for this discovery.1, 2 The virus is a leading cause of liver disease worldwide: an estimated 250–260 million individuals are chronically infected, and approximately one third of the world’s population has serologic evidence of exposure.3 HBV is a global public health problem with endemic levels of infection in Southeast Asia and Africa, where prevalence rates are at least 8% in many populations.4, 5 However, HBV is under-represented in terms of resource allocation, political advocacy, and research.6
Chronic HBV infection leads to liver inflammation, with long-term risks of cirrhosis and hepatocellular carcinoma (HCC).7, 8 In contrast to the decrease in mortality from human immunodeficiency virus (HIV), tuberculosis, and malaria, HBV-associated mortality is increasing.9 The United Nations Sustainable Development Goals set the challenge of eliminating HBV infection as a public health threat by 2030.10, 11 However, substantial barriers to elimination include gaps in vaccine coverage, long periods between vaccination and its effects on population prevalence,12 and lack of a cure. Other challenges include the virus’s resistance to drugs (and to a lesser extent vaccines),13, 14 HIV coinfection, stigma, poverty, lack of education, and limited access to diagnostic tests.6 HBV infection is treated with interferon and nucleos(t)ide analogue reverse transcriptase (RT) inhibitors—primarily tenofovir or entecavir—which can limit liver damage by suppressing viral replication.15 However, interferon therapy is associated with unpleasant side effects and cures only a small percentage of patients. Nucleos(t)ide analogue RT inhibitors decrease viremia but have no consistent effect on clearance. Therefore, rebound viremia after cessation is common. There is a great need to cure HBV infection if we are to achieve elimination targets; curative therapy for HBV is an important goal for individual patients and the international public health agenda.16
Curing HBV infection requires a detailed and robust understanding of the genetic sequence, structure, and diversity of HBV. Scientific investment is required to develop panels of diverse infectious clones that replicate in cell lines and in animals, to support drug resistance-screening programs.17 Detailed insights into immune control and clearance can be gained from identifying sites of immune selection pressure in the virus genome.18 This approach has helped identify immune correlates of HIV control over the past decade.18, 19 Increasing our understanding of virus genetics can improve management of patients—in stratification, selection of therapy, identification of drug- and vaccine-resistant strains, and development of new approaches to monitoring.20
HBV sequence data largely consist of consensus sequences of individual viral genes derived by Sanger sequencing. However, next-generation sequencing (NGS) platforms are rapidly becoming more accessible and affordable, in addition to new bioinformatic approaches to handle the resulting datasets.21, 22, 23 In addition to enabling whole-genome sequencing, NGS offers a powerful method for detection of minor variants relevant to the identification of drug resistance,24, 25, 26 studies of quasispecies dynamics,27 and characterization of complex viral populations.28 Together with improved curation and publication of clinical metadata, these accurate, full-length, ultra-deep HBV sequence data provide increasing opportunities for developing new insights into HBV evolution, diversity, pathogenesis, immune control, and treatment outcomes.
To provide a solid foundation for interpretation of new sequence datasets, we assimilate available data on HBV genome structure, function, and diversity and summarize gains made using NGS platforms.
Taxonomy
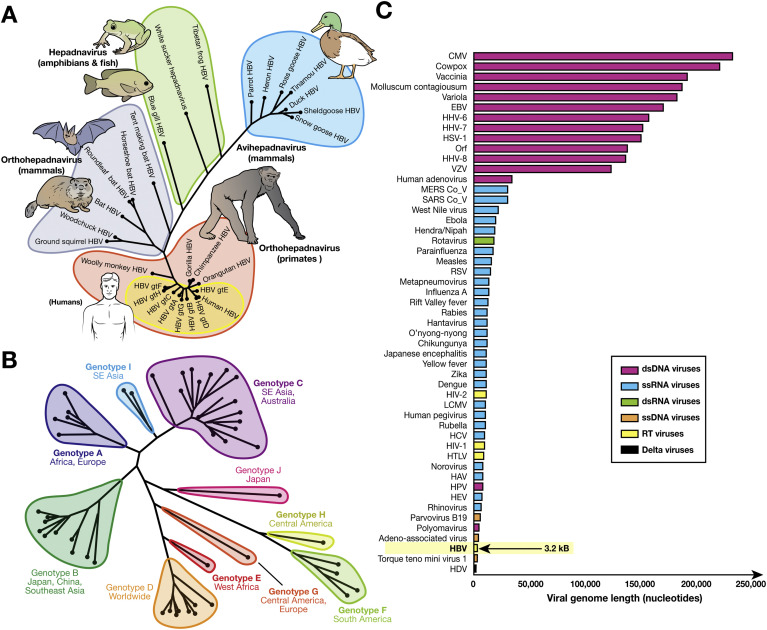
HBV is the prototype virus of the Hepadnaviridae family—small spherical viruses with icosahedral symmetry that combine a partial double-stranded (ds) DNA genome and virus-encoded RT. Within the Baltimore virus classification system, which classifies viruses based on their genomic composition and replication cycles,29 the Hepadnaviridae are classified as group VII (sometimes referred to as pararetroviruses)—they are the only animal viruses of this group. Until recently, the family was divided into 2 genera: the Orthohepadnavirus species (which infect mammals, including primates and bats) and the Avihepadnavirus species (which infect birds). However, the recent discovery of putative hepadnaviruses that infect fish30 and amphibians31 indicates that the viral family might be larger than initially believed (Figure 1 A).32, 33 Based on sequence diversity, HBV is divided into 9 genotypes and 1 putative genotype (Figure 1 B). Hepadnaviruses have some of the smallest known viral genomes, ranging from 3.0 to 3.3 kb; the HBV genome is approximately 3.2 kb34 (Figure 1 C).
Figure 1.
Relationships between HBV and other hepadnaviruses, genotype diversity, and genome size. (A) Phylogenetic tree of the relation among avian, mammalian, and other hepadnaviruses. Hepadnavirus reference sequences for avian (NC_005950.1, NC_001344.1, NC_016561.1, NC_005890.1, NC_001486.1, NC_035210.1, NC_005888.1), mammalian (NC_003977.2, NC_028129.1, NC_024445.1, NC_024444.1, NC_024443.1, NC_020881.1, NC_004107.1, NC_001484.1), and other (NC_027922.1, NC_030446.1, NC_030445.1) species were downloaded from Genbank.32 This dataset was further supplemented with hepadnavirus isolates from chimpanzees, orangutans, and gorillas (AF193863, FJ798097, FJ798098) and some widely cited HBV genotype strains (X02763, D00330, AY123041, V01460, X75657, X69798, AF160501, AY090454). (B) Midpoint-rooted maximum likelihood phylogenetic tree generated using MEGA733 with bootstrap replicates of 1000 used, indicating relations between HBV genotypes and subtypes and their typical geographic distribution. Widely used reference sequences for genotypes A–D and F are included. For genotypes with a single subtype, the reference sequences were used to generate the tree. The sequences used to generate the tree were genotype A: KP234050.1, HE974376.1, KP234052.1, AY934764.1, KP234053.1, GQ331048.1; genotype B: D50521.1, AB073825.1, GQ924628.1, AB073826.1, AB219427.1, DQ463792.1, AP011091.1, AP011093.1, GQ205440.1, GQ358146.1; genotype C: KM999990.1, KY629637.1, AB554019.1, AB554018.1, AB644281.1, AB644283.1, AB644286.1, AB644287.1, KP017272.1, KU695741.1, KF873519.1, KM999992.1, KM999993.1, AP011107.1, KP017269.1, AP011108.1; genotype D: AB104711.1, HQ700511.1, KP090181.1, FJ692533.2, DQ315780.1, KF170740.1, KP322600.1, FJ904406.1; genotype F: AF223963.1, AY311369.1, AY311370.1, AB166850.1; genotype I: AB562462.1, FJ023671.1; genotype J: AB486012.1; and HBVdb genotype reference sequences for genotypes A–H, respectively: X02763, D00331, AY123041.1, V01460.1, X75657.1, X69798.1, AF160501, AY090454. (C) Relative genome sizes of viruses pathogenic to humans including HBV (3.2 kB; arrow). Genomes were obtained from https://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=10239 and sorted by nucleotide length. For each virus type, a representative genome was selected. Metadata, including accession numbers for each organism, can be found at 10.6084/m9.figshare.6080402. CMV, cytomegalovirus; dsRNA, double-stranded RNA; EBV, Epstein-Barr virus; gt, genotype; HAV, hepatitis A virus; HCV, hepatitis C virus; HDV, hepatitis D virus; HEV, hepatitis E virus; HHV, human herpesvirus; HPV, human papillomavirus; HSV, herpes simplex virus; HTLV, human T-lymphotropic virus; LCMV, lymphocytic choriomeningitis virus; MERS, Middle East Respiratory Syndrome; SARS, Severe Acute Respiratory Syndrome; ssDNA, single-stranded DNA; ssRNA, single-stranded RNA; VZV, varicella zoster virus.
Genome Structure
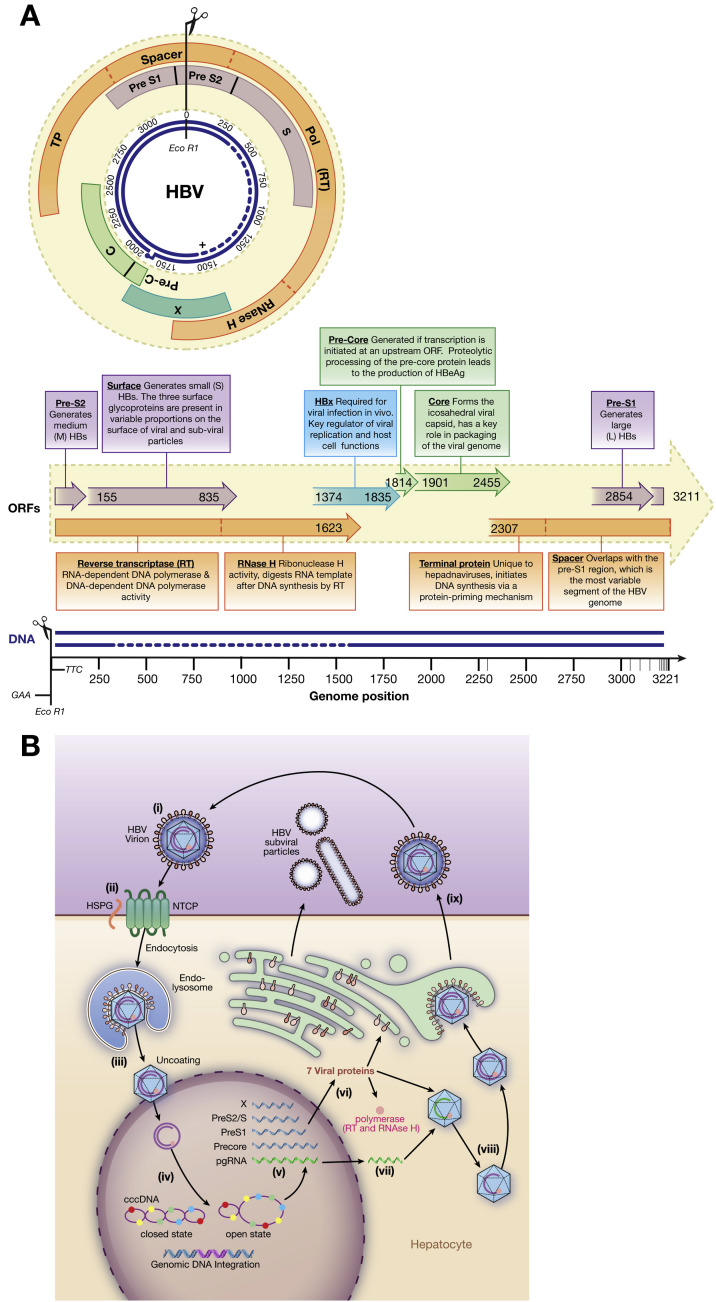
The circular partially double stranded HBV genome encodes 4 genes— polymerase (P), surface (S; pre-S1 and pre-S2), precore/core (C) and the X protein, encoded by discrete open reading frames (ORFs; Figure 2 A).35, 36, 37, 38 HBV produces 5 viral RNA transcripts of varying lengths, which are translated into 7 distinct proteins (Figure 2 A and B).
Figure 2.
Annotated HBV genome and replication cycle. (A) The 4 overlapping ORFs and the 7 products encoded. Gene products are indicated by text boxes, with start and end positions derived using X02763.1 as a reference strain. The major functional domains of the P gene product are indicated (dotted lines). Large HBs consist of pre-S1, pre-S2, and S; medium HBs consist of pre-S2 and S; and small HBs consist of S only. The overlap of >1000 nucleotides between the P and S genes is the largest gene overlap of any known animal virus.35 The near-complete negative DNA strand and partially complete positive DNA strands (dotted line indicates approximated missing region) also are shown, in addition to the position of EcoR1. The 5′ end of the complete negative-sense DNA strand is covalently bound to the viral RT. The complementary positive-sense DNA strand is partially complete, covering approximately two thirds of the viral genome.36 The 5′ end of the incomplete strand is defined by a short oligo-ribonucleotide region; the 3′ end varies within and among hosts. (B) Replication cycle (adapted from Liang, special issue). (i) Infective HBV virions in serum, often referred to as Dane particles (diameter, 42 nm). The capsid structure has icosahedral symmetry: T = 4 (31 nm; 90% of population) and T = 3 (28 nm; 10% of population).37, 38 (ii) The virus enters hepatocytes by HSPG (low-affinity binding) and solute carrier family 10 member 1 (SLC10A1; also called sodium taurocholate co-transporting polypeptide NTCP; high-affinity binding). (iii) The molecular processes of un-coating and nuclear import are unclear but likely require cell proteins. (iv) Viral DNA enters the nucleus as RC-DNA. (v) Viral DNA is reconfigured as cccDNA within the nucleus by the cell’s DNA repair factors; this stable structure occurs in association with host histones that mediate DNA packaging. (vi) The open cccDNA structure is a template for host RNA polymerase II. (vii) DNA is transcribed to pre-genomic RNA intermediates in the nucleus, creating 4 mRNAs (blue): a 3.5-kb transcript encoding precore RNA (full-length pre-genomic RNA also shown in green); 2.4- and 2.1-kb mRNA transcripts for pre-S and S, respectively; and a 0.7-kb mRNA encoding the X protein. The RNA is transported to the cytoplasm, where it is translated to 7 viral proteins (short, medium, and long S proteins, core, e antigen, polymerase, and X protein). (viii) HBV RT produces a negative-strand DNA from pre-genomic RNA. The RNA template is degraded by RNase H, and then synthesis of the positive-strand DNA is initiated. HBV DNA is repackaged in relaxed form with other proteins inside the host cell. (ix) New virions and viral proteins are released into the blood. Excess HBsAg forms small noninfectious, subviral particles (∼20 nm diameter), and long filaments161; free HBeAg and capsids also are secreted. C, core; HBeAg, hepatitis B e antigen; HBx, hepatitis B X protein; HSPG, heparan sulfate proteoglycan; NCTP, Na+-taurocholate co-transporting polypeptide pol, polymerase; TP, terminal protein.
This basic genomic organization is common to all hepadnaviruses, although the X gene is absent from most Avihepadnavirus species (with the exception of a vestigial X gene in duck hepadnaviruses).39 The compact nature of hepadnavirus genomes, which have multiple overlapping reading frames, results in approximately two thirds of nucleotides encoding more than 1 functional element.40 This genome structure encompasses virus genes and regulatory regions and restricts redundancy within coding regions (Figure 2 A). One specific example is the N-terminal region of the precore protein, which is highly conserved among Orthohepadnavirus species, likely owing to constraints from the overlapping encapsidating signal (epsilon) sequence.41 The negative-sense genomic DNA strand (complementary to the mRNA transcript) is the complete strand—it is held in a circular conformation by an overlap at the 5′ end of the genome (ranging from 50 bp in Avihepadnavirus species to 240 bp in Orthohepadnavirus species).
Partially double stranded relaxed circular (RC-DNA) in HBV virions is converted into covalently closed circular DNA (cccDNA) inside the hepatocyte nucleus by the viral polymerase filling in the partially single-stranded region of the genome (Figure 2 B). Biogenesis of cccDNA, including the exact mechanism of DNA repair of the partially single-stranded DNA region of the RC-DNA, is not fully understood. It is likely that cell enzymes such as tyrosyl-DNA phosphodiesterase 2 contribute to cccDNA formation through cleavage of the HBV P from RC-DNA.42, 43 The viral cccDNA is extremely stable and persists in the nucleus as a viral minichromosome44 for the lifespan of the cell, providing the transcriptional template for all RNA species that are translated into viral proteins (Figure 2 B).
In addition to persisting as a minichromosome in the form of cccDNA, hepadnavirus DNA also integrates into the host genome.45 In woodchucks, integration usually occurs within Myc proto-oncogenes, eventually causing HCC in almost all infected animals.45 In humans, HBV integration can occur in different sites within the genome, and the consequences are less clear, although chronic HBV infection is associated with liver cancer.46 After integration of HBV DNA into the host genome, only the S gene typically remains under the control of its native promoter,45 leaving these integrated genomes as a source of HBV surface antigen (HBsAg) production.47
During infection, infectious viral particles containing HBV genomes are secreted from infected hepatocytes, in addition to smaller subviral particles and long tubular filamentous particles. These particles are empty shells formed from the HBsAg—they lack a capsid and virus genome and are therefore noninfectious.48 The particles typically outnumber infectious virions by as much as 100,000-fold48 and are believed to be involved in immune evasion by binding neutralizing antibodies49 and potentially promoting T-cell anergy.50 Similar particles have been documented in the woodchuck HBV model,51 indicating a common role in Orthohepadnavirus infections.
Virus Genotypes and Reference Sequences
Nine different HBV genotypes (A–I) have been defined by >8% divergence at the nucleotide level; a 10th putative genotype (J) was characterized after isolation from 1 individual.52, 53 The HBV genotypes are further divided into at least 35 subtypes by >4% divergence, with wide variation observed in the numbers of subtypes described per genotype (Table 1 ).20, 54, 55, 56
Table 1.
HBV Subtypes and Genotype Features
| Genotype | Subtypes | Geographic distribution | Genome length (bp) | Distinguishing features of HBV sequencea |
|---|---|---|---|---|
| A | A1–A4 | Africa, Europe | 3221 | 6-bp insertion in core gene; G1896Ab mutations rare; BCP mutationsc common |
| B | B1–B5 | Japan, China, Southeast Asia | 3215 | B1 and B5 are pure strains, whereas B2, B3, and B4 are recombinants with genotype C in the core region |
| C | C1–C16 | Southeast Asia, Australia | 3215 | BCP mutationsb common |
| D | D1–D7 | Worldwide, Middle East, West Africa | 3182 | 33-bp deletion in pre-S1 |
| E | No subtypes described | West Africa | 3212 | 3-bp deletion in pre-S1 |
| F | F1–F4 | North and South America | 3215 | G1896Ab mutations rare |
| G | No subtypes described | Central America and Europe | 3248 | 36-bp insertion in core and 3-bp deletion in pre-S1; insertion results in a high level of core expression; stop codons at positions 2 and 28 (G1896Ab) of the precore protein render it unable to express HBeAg; often found in coinfection with other genotypes that express HBeAg |
| H | No subtypes described | Central America | 3215 | G1896Ab mutations rare |
| I | I1–I2 (putatived) | Southeast Asia | 3215 | Evolved as a recombinant of genotypes A, C, and G56 |
| J (putative) | No subtypes described | Japan | 3182 | Single isolate identified in elderly Japanese patient with HCC; highly divergent from other human HBV strains; likely a genotype C–gibbon Orthohepadnavirus recombinant53 |
NOTE. Further details about genotypes and subtypes can be found in Rajoriya et al20 and Tong and Revill.55
BCP, basal core promoter; HBeAg, hepatitis B e antigen.
Insertions and deletions relative to 3215-bp genome length.
G1896A mutation introduces a premature stop codon in the precore, resulting in loss of HBeAg expression.
Basal core promoter mutations at A1762T and G1764A result in decreased HBeAg expression.
Few sequences of genotype I have been characterized, although the genetic distance between isolates suggests there might be 2 subtypes.56
There are substantial differences among genotypes in geographic distribution, transmission mode, and clinical outcomes, including emergence of drug resistance and response to therapy20, 57 (Table 1). However, the data are incomplete—particularly from low- and middle-income countries.6, 58 Furthermore, it is difficult to associate differences in disease progression and outcome with HBV genetic sequences vs population behavior, coinfections, exposures to drugs and hepatotoxins, and human genetic factors.5 Most studies have focused on small numbers of individuals in relatively restricted areas.57 Prospective high-resolution genome-wide association studies of large numbers of patients are required to determine how interactions between human and virus genomes affect outcomes.
Sequence data indicate wide variation in the numbers of subtypes within each genotype, ranging from genotype C, with 16 distinct subtypes,52 to genotypes E, G, H, and J, each of which consists of a single subtype (Figure 1 B).55, 56 Molecular clock analysis has indicated that genotype C is likely to be the oldest genotype59—the large number of subtypes is in keeping with its protracted endemic association with human populations.60, 61 However, it has been a challenge to study the evolution of HBV, because the lack of temporal structure has confounded molecular clock analyses.62 Genotypes F and H, which have a smaller number of subtypes but are highly divergent from other genotypes, might have higher rates of substitution.63 Genotype F has higher inter-subtype diversity than other genotypes,64 which could be due to the geographic range of the populations it infects—from native Alaskan to Latin American populations.65, 66, 67
Genotypes B and C have been associated with higher rates of vertical transmission than other genotypes.68 Genotypes A1,69 C,68 and F have been associated with earlier progression to HCC (particularly in Alaskan natives).66 Routine genotype analysis of HBV in infected individuals in clinical practice has not been recommended by US, Asia-Pacific, or European clinical guidelines,70, 71, 72 largely because results do not affect treatment decisions. However, more recent European and US guidelines recognize that genotype variations are associated with responses to therapy with pegylated interferon alfa. This treatment is not recommended for patients who are negative for the HBV e antigen (HBeAg) and infected with genotype D or E, and different stopping points are proposed for patients infected with genotypes A–D who have not responded to therapy.15, 71 As new therapies are developed, and genotype and subtype data become more widely available, we will develop a better understanding of the effects of genotype on treatment outcome.
Reference Genomes
There are few robust molecular biology and comparative bioinformatic studies of diverse HBV strains; most published sequences are from HBV genotypes B and C, which together account for >60% of published full-length genomes (data downloaded August 2017). Universally accepted reference sequence(s) and numbering of amino acid residues provide an important foundation for unifying research efforts. With this in mind, we used previously published HBV protein alignments73 as a point of reference for pinpointing the sites of immune epitopes within the HBV genome.74
Consistent numbering of the HBV genome is a challenge because of genotype-specific differences in genome length and the circular genome. A unified system would be valuable, similar to that proposed for HCV,75 in which numbering is based on a reference strain, and a consistent approach has been proposed for documentation of insertions and deletions. Conventional HBV numbering, based on molecular cloning of the genome, typically uses X02763 (genotype A) or NC_003977.2 (genotype D) as a reference strain and defines the genome origin at an EcoRI restriction site (GAA/TCC, with nucleotide 1 starting at T), which is embedded within the overlapping P and S genes.76 The presence of this restriction site is hypothetical in many HBV isolates,77 and this numbering convention is not always followed. Sequence data must be examined and realigned to ensure consistent numbering.77
There are several central databases of HBV sequences, including HBVdb77 and HEPseq (http://www.hepseq.org/Public/Web_Front/main.php), and recent studies have reported reference sequences for subtypes of genotypes A78 and C.79 However, there is no unified set of reference sequences of HBV genotypes and subtypes. This differs from HCV and HIV, for which there are large sequence databases and consistently used reference sequences and supporting resources.80, 81, 82 As the HBV field works progressively toward developing unbiased methods for whole-genome sequencing, the numbers of sequences deposited into such databases is likely to increase considerably. A curated alignment of validated genotype and subtype reference sequences would be a valuable resource for researchers, ensuring that comparative analyses are conducted based on a consistent approach.
Recombination
Inter-genotype HBV recombination has been reported in situations that range from individual case reports to recombinants that have reached fixation and meet the criteria for classification as separate genotypes (genotype I) or subtypes (B2–B4). Breakpoints for the recombinants are not randomly distributed throughout the genotype. Based on the EcoRI numbering convention, breakages tend to occur at sites within nucleotides 1700–2000 and 2100–2300,83 possibly because of the decreased between-genotype diversity observed in these regions. In addition to the genotype I recombinant, examples include B and C recombinants in parts of mainland East Asia that are now defined as sub-genotypes B2,22 , 84 B3,85 and B485; C and D recombinants reported from Tibet83 and China84, 86; D and E recombinants reported from different parts of Africa87, 88; A, C, and G recombinants reported in 2 patients in China—although this is based on sequencing only a 1-kb stretch of the genome89; and A and G recombinant sequences identified in a patient with A2 infection90 and in several patients in Canada.91
Intra-genotype HBV recombinants also have been described; genotypes B, C, and E have an increased frequency of intra-genotype recombinant strains compared with other genotypes.64 For genotypes B and C, this likely reflects their long association with humans.59 Well-defined subtype reference strains could be useful to identify recombination events, to explain current distribution and diversity of viral variants, and to predict future evolutionary directions of the epidemic.92
Approaches to Deep Sequencing
Deep sequencing analyses can increase our understanding of HBV diversity and evolution, control by the immune response, resistance to treatment, and disparities in clinical outcomes. After the success of second-generation short-read sequencing by synthesis approaches (such as the Roche 454 platform [Hoffmann-La Roche, Basel Switzerland] and Illumina [San Diego, CA]), third-generation long-read sequencing technology is advancing (sequencing approaches summarised in a supplementary table on-line at: https://doi.org/10.6084/m9.figshare.7106288). Oxford Nanopore Technologies (Oxford Science Park, UK) provides a radically different approach by generating sequence data in real time directly from samples. It produces complete genomic haplotypes, albeit with some constraints because of the high rate of errors in base-calling algorithms.
NGS studies have the potential to increase our understanding of viral diversity. For example, these studies have detected minor variant populations at low levels24, 25 and associated quasispecies diversity with treatment outcome and HBeAg status.27 There are several factors that have hampered our understanding of the nature and effects of intra-host diversity of HBV. Few studies have used whole-genome sequencing analyses, and sequence output can be biased by the need for prior DNA amplification (especially when viral loads are low) and by representation of the RC-DNA reservoir rather than cccDNA sequences. Studies of HCV have found that diversity in different regions of the genome can indicate contrasting biological processes.93, 94
In regions of the world where HBV is endemic and mixed infections are common, it can be a challenge to differentiate between coinfection and true recombination using current sequencing approaches.92 Several NGS platforms, including Illumina and Roche 454, rely on short reads and amplicon-based approaches, respectively. Therefore, full-genome reconstruction of individual quasispecies can be difficult; inference when multiple genotypes are detected can be unclear.28 The development of new long-read sequencing technologies such as those from Oxford Nanopore Technologies and Pacific Biosciences (Menlo Park, CA) will enable more accurate haplotype reconstruction and increase the specificity with which recombinant strains can be distinguished from mixed infection.95
Virus Evolution and Diversity
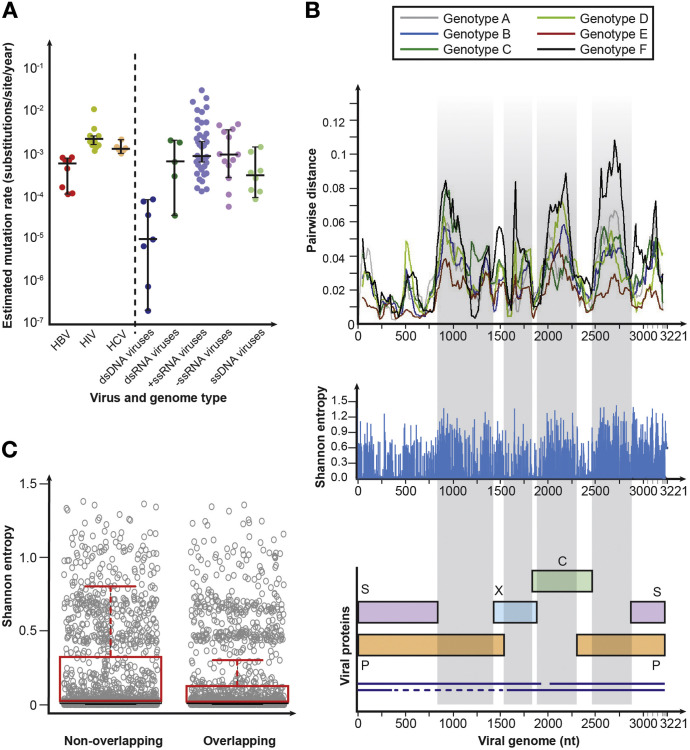
The unique combination of a DNA genome coupled with multiple overlapping ORFs, an RT step, and a stable cccDNA reservoir leads to a complex and unique replicative process. On the one hand, there is evidence that HBV has a relatively low mutation rate (0.0005 substitutions per site per year) compared with other RT viruses—for example, its rate of mutation is 5-fold less than HIV (0.003 substitutions per site per year)96, 97, 98 (Figure 3 A).77, 99, 100, 101 On the other hand, HBV is more diverse than other dsDNA viruses (Figure 3 A) with a level of variation and rate of evolution that is more comparable to an RNA virus than a DNA virus.102 This unusual replication cycle and genomic structure make it difficult to estimate a genome-wide rate of virus evolution.
Figure 3.
HBV diversity. (A) Relation between genome type and substitution rate. Estimates of evolutionary rate (substitutions per nucleotide per year) were taken from Sanjuán99 and were calculated using Bayesian molecular clock approaches. For the different genome types, median rates of evolution were 9.32 × 10−6 (interquartile range [IQR], 7.00 × 10−7–7.20 × 10−5) for dsDNA, 6.36 × 10−4 (IQR, 1.60 × 10−4–1.88 × 10−3) for dsRNA, 1.10 × 10−3 (IQR, 4.52 × 10−4–2.69 × 10−3) for +ssRNA, 9.17 × 10−4 (IQR, 3.55 × 10−4–3.40 × 10−3) for −ssRNA, and 2.08 × 10−4 (IQR, 1.36 × 10−4–5.65 × 10−4) for ssDNA. (B) Distribution of diversity along the HBV genome. Full-length HBV genome sequences were obtained from HBVdb77 in August 2017 (n = 5383). Sequences were aligned using MAFFT (https://mafft.cbrc.jp/alignment/server/).100 Sequences for each genotype were randomly shuffled using a function within SSE 1.3101 and 250 sequences of each genotype were randomly selected for analysis to normalize the number of sequences of each genotype analyzed. Only 225 sequences were available for genotype F; genotypes G, H, I, and J were excluded from the analysis because there were insufficient numbers of sequences available for comparison with other genotypes. Within-genotype pairwise nucleotide distances were calculated for genotypes A–F using SSE 1.3 using a window size of 150 bp and increments of 20 bp. The greatest variability (typically >5% sequence divergence) is observed in regions where there are no overlapping ORFs. Entropy at each nucleotide within the dataset was calculated using SSE 1.3. (C) Comparison of Shannon entropy at each site of overlapping and nonoverlapping regions of the HBV genome. Genotypes were analyzed individually and regions of the genome were divided into overlapping and nonoverlapping regions using an annotated genome (https://hbvdb.ibcp.fr/HBVdb/HBVdbGenome). Mean Shannon entropy in overlapping regions is significantly lower at 0.16 (95% confidence interval, 0.14–0.17) than in nonoverlapping regions (0.20; 95% confidence interval, 0.18–0.21; P < .0001 by Mann-Whitney U-test). C, core; dsRNA, double-stranded RNA; HCV, hepatitis C virus; ssDNA, single-stranded DNA; ssRNA, single-stranded RNA.
There is considerable variation in rates of HBV evolution.62, 103 Faster rates of evolution have been observed in individuals with chronic infection, over specific time periods, or in analyses of families.103 Greater diversity has been observed in HBeAg-negative infection.97 The long-term rate of HBV evolution is lower than rates reported from short-term studies.62, 63 For example, sequences of HBV isolated from 2 sets of 400-year-old mummified remains from Korea and Italy62, 104 had minimal genetic divergence from modern HBV sequences.
Overlapping ORFs can offer a fitness advantage to viruses with high rates of mutation, because substitutions in these regions have higher odds of producing detrimental effects.105, 106 In addition, there is evidence that many substitutions that occur in the viral genome during the course of chronic infection might not generate variation, but are reversions back to the genotype consensus.107 Therefore, most substitutions in the HBV genome are not maintained over the long term.
The rate of substitution in overlapping regions of the HBV genome is 40% lower than in nonoverlapping regions63 (Figure 3 B), and there is a significant difference in entropy between these regions (Figure 3 C). Overlapping regulatory elements and encoded RNA secondary structures required for replication with the ORFs provide further constraint to nucleotide substitution in the HBV genome. For example, diversity within genotypes in the nonoverlapping region within the X gene is decreased relative to the other nonoverlapping regions (Figure 3 B), most likely a result of overlap with the basal core promoter region, a regulatory element of the genome that controls expression of precore mRNA and pre-genomic RNA.
Many HBV genotypes have an unexpectedly high level of diversity at the start of the S gene, although this region overlaps with the P gene. Intriguingly, this divergent region of the P–S overlap (often referred to as the spacer region in Pol) has a pattern of codon use that is distinct from the 3′ two thirds of the overlap.35 It has been proposed that the P sequence in the 5′ region of the P–S overlap evolved independently,108 and that mutations and deletions in this region do not greatly affect the function of the encoded polymerase.109, 110 This region corresponds to a hydrophilic region under strong immune pressure on the overlapping S gene, indicating that the spacer region of P allows conformational adaptability under selective pressures.111 Analysis of HBV sequences from a family transmission network found the precore and middle region of the S gene to be hotspots for sequence diversity compared with the relative conservation of these regions between genotypes.112 Opposing effects on HBV genetic diversity are presented in Table 2 .43, 63, 64, 74, 102, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125
Table 2.
Determinants of Diversity and Conservation Within HBV
| HBV attribute | Factors associated with sequence conservation | Factors associated with sequence diversity |
|---|---|---|
| Genome structure and sequence | Overlapping ORFs and regulatory regions in viral genome (Figures 2A and 3B) impose constraint on viral plasticity102 because nonsynonymous mutations have to be accommodated within 2 different proteins to be viable; this makes most mutations disadvantageous,113 an example of constrained evolution.114 This is highlighted by greater diversity in nonoverlapping regions (Figure 3B and C). Examples include the highly conserved epsilon sequence, which overlaps the unique N-terminus of the precore gene and A–T nucleotide substitution at position 1858; it constrains selection of G1896A precore start codon mutation on the opposing strand.115 |
Redundancy within the third codon position in regions where ORFs overlap allows selection of mutations (Figure 3C). Host immune responses select escape mutations (eg, within or flanking T-cell epitopes).74 Exposure to antiviral therapy selects drug resistance mutations.116 Selection of G1896A precore stop codon and BCP mutations.117 |
| Persistence and transmission | Superior transmission potential of wild-type variants.118 Transmission bottlenecks limit diversity at onset of new infections.119, 120 |
Long duration of infection can generate diverse quasispecies populations within hosts.119, 120, 121 |
| Replication cycle | Stable reservoir of cccDNA with long half-life (Figure 2Bv); estimated cccDNA half-life 33–57 d in duck hepadnaviruses.43 In humans, average cccDNA half-life has been estimated at 9.2 mo but differed markedly in HBeAg-positive (8.6 mo) and HBeAg-negative (26.2 mo) individuals.122 Studies are needed to determine half-life in humans at different stages of disease progression. | Error-prone viral RT enzyme with high substitution rate when transcribing pgRNA into RC-DNA (Figure 2Bvii). HBV is produced at high rate of replication.123 |
| Genotypes | Lowest level of diversity is observed in genotype E (Figures 1B and 3B), where there is only a single reported subtype. | Increased diversity is a feature of some specific genotypes; genotype F diverges considerably from other genotypes124 and shows a high level of inter-subtype diversity,64, 125 (Figures 1B and 3B). Adaption to a genetically diverse population at some point in the evolutionary history could explain the increased substitution rates.63 |
BCP, basal core promoter; HBeAg, hepatitis B e antigen.
As with determination of genotype, baseline testing for drug resistance is rarely performed. The prevalence of mutations in HBV that cause resistance to treatment varies worldwide, from <2% in the United States and Canada126 to >20% in some African cohorts,116 but more data are needed. In areas where many individuals also have HIV infection, many patients have been exposed to antiretroviral therapy, including the nucleos(t)ide analogues lamivudine and tenofovir. Prior exposure to antiretroviral therapy could increase the risk for mutations in HBV that mediate resistance to treatment—particularly to lamivudine, which has a low barrier to resistance. Although strains of HBV that are resistant to tenofovir have been described, they are unusual and their effects on patient outcome are not clear.116 Therefore, tenofovir is often recommended after nonresponse to alternative therapies.15, 127
Within-Patient Diversity
Although some regions of the HBV genome are highly conserved, there are few data on intrapatient diversity. Simultaneous, competing evolutionary pressures can create different subpopulations of HBV within patients (quasispecies) and at the population level. These can produce a more diverse RC-DNA population and a less diverse and stable cccDNA population, with different sequence polymorphisms potentially archived in the cccDNA pool128 (Figure 2 Bv).
To be stably fixed in the virus population, mutant genomes egressed in virions must effectively compete with circulating wild-type viruses to infect hepatocytes and generate cccDNA. This unique population structure maximizes the potential pool of mutants, enabling advantageous virus adaptation within each patient and still eliminating viruses with deleterious mutations. Some less-fit RC-DNA genomes might persist by bypassing egression and being recycled directly back into the nucleus to replenish the viral cccDNA population. However, this model was based on observations from the duck hepadnavirus129 and has not been clearly documented in human HBV infection.130
Certain HBV polymorphisms and deletions have been associated with specific clinical outcomes, such as cirrhosis and HCC. Examples include diversity in the pre-S region, which has been correlated with progression from chronic HBV infection to HCC.131 However, deletions in the pre-S region have been associated with HCC in patients infected with HBV genotypes B and C (in particular, pre-S deletions of nt 2977–3013 in HBV genotype C).132, 133 Large deletions that result from splicing of the HBV pre-genomic RNA are associated with advanced liver disease, including cirrhosis134, 135 and HCC.136 Mutations in the basal core promoter (A1762T and G1764A), detected by pyrosequencing, have been associated with increased risk of disease progression to cirrhosis or HCC in some populations137—mostly in patients with HBV genotype B or C infection—independent of viral load.138, 139 Likewise, viral diversity is likely to affect response to treatment, although this relation is not clearly defined. A large study correlated a higher level of virus diversity (particularly in the basal core promoter and precore regions) with a lower probability of HBsAg loss.117 Other studies have associated HBV heterogeneity with positive effects of treatment.140, 141
A wide range of measures can be used to assess the diversity in HBV in infected individuals. Broad estimates of virus diversity have been made using pairwise and entropy-based measures,142 detection of minor variant viral populations with specific polymorphisms (often associated with drug resistance),26, 28, 143 and detection of mixed-genotype or subtype infections.28, 92 It is important to increase our understanding of quasispecies dynamics if we are to better understand how selection and fixation of polymorphisms affect patient outcomes, including virus resistance to drugs and vaccines, the antivirus immune response, and development of chronic liver disease. Diverse virus populations could arise through immune selective pressure; there is a balance between the benefit of immune-escape mutations and the deleterious effects of mutations on HBV fitness or replicative capacity.
A study of mother–child pairs demonstrated a relatively tight bottleneck at transmission, with limited virus diversity in infected children compared with their mothers—suggesting only a proportion of HBV strains in the mother are transferred to the child.119, 144 In mothers with HBV and HIV coinfection, minor HBV variants may be established as the dominant virus in their infants. Mutations in HBsAg were frequently observed in these strains,145 indicating that HIV infection opens the HBV transmission bottleneck. Analyses of intrahepatic quasispecies demonstrated an association between intrahepatic diversity (focused within T-cell epitopes) and off-treatment control, indicating a role for immune-mediated selection pressure in control of viremia.119 Similarly, increased diversity of quasispecies has been associated with effective therapy,141, 146 although this observation has not been consistent.117
For other blood-borne virus infections, intrapatient virus diversity has been associated with strong suppression by treatment93, 147 or conversely with poor patient outcome.94 However, studies of factors that affect the diversity of HBV are confounded by factors such as genotype and subtype, small heterologous cohorts, variations in sequencing methods, and examination of different areas of the genome. Therefore, it is a challenge to uncover true associations. Studies also are confounded by the geographic distribution of genotypes and the ethnicity of affected individuals.
Future Directions
Chronic HBV infection is a fundamental global public health challenge for the 21st century. There is not enough unbiased generation and interpretation of sequence data or attempts to unify such data with relevant resources (such as genome annotation, reference sequences, and robust linked clinical data). The development of unbiased and meta-genomic pipelines, alongside carefully collated host metadata, has begun to affect management of patients with infectious diseases.148, 149, 150 Although deep sequencing approaches have not been robustly applied to HBV, there are several situations in which NGS data could be of substantial value, such as in development of diagnostic tools, selection of treatment, analyses of transmission, and studies of HBV pathogenesis.
In virus diagnostics, NGS could be used to identify known or novel viruses or to exclude infectious etiology of clinical syndromes.151, 152 Previously unrecognized HBV coinfection was detected using a meta-genomic approach in a cohort of patients with acute liver failure,151 and new splice variants were identified using Pacific Biosciences technology.153 At the same time, NGS might be used to identify existing and new drug-resistant mutations and study their dynamics.117, 154, 155, 156, 157
Strategies are in development to bring genome sequence analysis to the clinical virology laboratory.158 For example, pre-S deletion patterns, combined with quantitative NGS data and machine learning methods, might be used to identify patients at risk for liver disease progression.133, 137 NGS also might be used to characterize the vertical transmission bottleneck and identify and track outbreaks in a range of settings.159, 160, 161
Challenges remain in the widespread application of NGS platforms, including the need to deplete host reads, which could require enrichment and amplification steps (particularly in detecting viruses at low copy numbers). Systems and reagents are expensive, and interpretation of NGS data requires considerable bioinformatic support and adaptation for different genomic configurations. For HBV, this means refining methods for a circular and partial dsDNA genome. However, the development of portable, real-time, third-generation sequencing platforms, such as the Nanopore MinION (Oxford Nanopore Technologies),158 have made the prospect of deep sequencing as a point of care test increasingly feasible. Relatively short and simple sample preparation protocols, minimal setup requirements (a laptop computer), and low costs relative to convention benchtop sequencers make the technology particularly appealing for resource-limited settings. Although the error rate of Nanopore has been too high for robust application to studies of pathogen diversity, rapid improvements are being made to laboratory and bioinformatic protocols.162
Substantial gaps remain in our understanding of the relationship between HBV genome structure, replication cycle, diversity, transmission, and clinical outcomes. Recent sequencing advances offer an enormous opportunity to generate datasets that can help to address some of these questions. The generation of standardized reference genomes of all HBV genotypes and subtypes to enable robust and consistent collation and analysis is required to develop insights into current and future epidemiology, to inform better clinical assessment and prognostication, to improve deployment of current antiviral drugs and vaccines, and to drive discovery of new therapeutic agents.
Acknowledgments
Author contributions: Jane A. McKeating and Philippa C. Matthews conceived, designed, and supervised the project. Anna L. McNaughton and Philippa C. Matthews undertook a literature review and wrote the main manuscript draft. M. Azim Ansari, Anna L. McNaughton, Margaret Littlejohn and Peter Revill researched viral evolution and diversity. Sheila F. Lumley, Anna L. McNaughton, and Philippa C. Matthews researched next generation sequencing. Valentina D’Arienzo and Jane A. McKeating researched the genomic structure, lifecycle, and molecular biology of HBV. M. Azim Ansari, Peter Revill, Margaret Littlejohn, and Anna L. McNaughton researched the evolutionary biology and phylogenetics. M. Azim Ansari researched recombination. Sheila F. Lumley and Philippa C. Matthews researched clinical microbiology (diagnostics, treatment, and public health). For the figures and tables, Anna L. McNaughton and M. Azim Ansari contributed information on phylogenetic trees; Anna L. McNaughton and Philippa C. Matthews contributed information on viral genome size in context; Anna L. McNaughton, Valentina D’Arienzo, Sheila F. Lumley, and Jane A. McKeating contributed information on genome structure; Anna L. McNaughton, Valentina D’Arienzo, Jane A. McKeating, Peter Revill, and Philippa C. Matthews contributed information on the replication cycle; and Anna L. McNaughton and M. Azim Ansari contributed information on HBV diversity. All authors contributed editorial input to refine the manuscript and figures.
Footnotes
Conflicts of interest Peter Revill has received research funding from Gilead Sciences. Philippa C. Matthews has a consultancy role in Immunocore and is on the Editorial Board of BMC Infectious Diseases.
Funding Philippa C. Matthews is funded by a Wellcome Trust Intermediate Fellowship (grant 110110). Peter Revill’s research program is funded in part by the NHMRC (APP1159305). Jane A. McKeating is funded by the EU 2020 Research and Innovation Programme (grant agreement 667273 Hep-CAR consortia) and the Wellcome Trust (IA 200838/Z/16/Z).
References
- 1.London W.T., Sutnick A.I., Blumberg B.S. Australia antigen and acute viral hepatitis. Ann Intern Med. 1969;70:55–59. doi: 10.7326/0003-4819-70-1-55. [DOI] [PubMed] [Google Scholar]
- 2.Blumberg B.S., Alter H.J., Visnich S. A “new” antigen in leukemia sera. JAMA. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization Preventing perinatal hepatitis B virus transmission : a guide for introducing and strengthening hepatitis B birth dose vaccination. http://apps.who.int/iris/bitstream/10665/208278/1/ Available at: Published 2015.
- 4.Schweitzer A., Horn J., Mikolajczyk R.T. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- 5.Matthews P.C., Geretti A.M., Goulder P.J.R. Epidemiology and impact of HIV coinfection with hepatitis B and hepatitis C viruses in Sub-Saharan Africa. J Clin Virol. 2014;61:20–33. doi: 10.1016/j.jcv.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 6.O’Hara G.A., McNaughton A.L., Maponga T. Hepatitis B virus as a neglected tropical disease. PLoS Negl Trop Dis. 2017;11:e0005842. doi: 10.1371/journal.pntd.0005842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akinyemiju T., Abera S., Ahmed M. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertuccio P., Turati F., Carioli G. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67:302–309. doi: 10.1016/j.jhep.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization World Health Organization global hepatitis report 2017. http://apps.who.int/iris/bitstream/10665/255016/1/ Available at:
- 10.United Nations United Nations sustainable development goals 2012. http://www.un.org/sustainabledevelopment/sustainab Available at:
- 11.Griggs D., Stafford-Smith M., Gaffney O. Policy: sustainable development goals for people and planet. Nature. 2013;495:305–307. doi: 10.1038/495305a. [DOI] [PubMed] [Google Scholar]
- 12.McNaughton A., Lourenço J., Hattingh L. Utilising a cohort study of hepatitis B virus (HBV) vaccine-mediated immunity in South African children to model infection dynamics: can we meet global targets for elimination by 2030? Biorxiv 2017. https://www.biorxiv.org/content/early/2018/09/24/162594 Available at:
- 13.Locarnini S. The hepatitis B virus and antiviral drug resistance: causes, patterns, and mechanisms. In: Mayers D.L., editor. Antimicrobial drug resistance: mechanisms of drug resistance. Humana Press; Totowa, NJ: 2009. pp. 519–530. [Google Scholar]
- 14.Romanò L., Paladini S., Galli C. Hepatitis B vaccination. Hum Vaccin Immunother. 2015;11:53–57. doi: 10.4161/hv.34306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Association for Study of Liver EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Tseng T.C., Kao J.H. Elimination of hepatitis B: is it a mission possible? BMC Med. 2017;15:53. doi: 10.1186/s12916-017-0820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sozzi V., Walsh R., Littlejohn M. In vitro studies show that sequence variability contributes to marked variation in hepatitis B virus replication, protein expression, and function observed across genotypes. J Virol. 2016;90:10054–10064. doi: 10.1128/JVI.01293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klenerman P., McMichael A. AIDS/HIV: finding footprints among the trees. Science. 2007;315:1505–1507. doi: 10.1126/science.1140768. [DOI] [PubMed] [Google Scholar]
- 19.Kawashima Y., Pfafferott K., Frater J. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajoriya N., Combet C., Zoulim F. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualized approach? J Hepatol. 2017;67:1281–1297. doi: 10.1016/j.jhep.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Quick J., Grubaugh N.D., Pullan S.T. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Biorxiv. 2017;12:098913. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bui T.T.T., Tran T.T., Nghiem M.N. Molecular characterization of hepatitis B virus in Vietnam. BMC Infect Dis. 2017;17:601. doi: 10.1186/s12879-017-2697-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y.Y., Hsieh C.H., Chen J.H. De novo assembly of highly polymorphic metagenomic data using in situ generated reference sequences and a novel BLAST-based assembly pipeline. BMC Bioinformatics. 2017;18:223. doi: 10.1186/s12859-017-1630-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez C., Chevaliez S., Bensadoun P. Characterization of the dynamics of hepatitis B virus resistance to adefovir by ultra-deep pyrosequencing. Hepatology. 2013;58:890–901. doi: 10.1002/hep.26383. [DOI] [PubMed] [Google Scholar]
- 25.Solmone M., Vincenti D., Prosperi M.C.F. Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen. J Virol. 2009;83:1718–1726. doi: 10.1128/JVI.02011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirvani-Dastgerdi E., Winer B.Y., Celià-Terrassa T. Selection of the highly replicative and partially multidrug resistant rtS78T HBV polymerase mutation during TDF-ETV combination therapy. J Hepatol. 2017;67:246–254. doi: 10.1016/j.jhep.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homs M., Caballero A., Gregori J. Clinical application of estimating hepatitis b virus quasispecies complexity by massive sequencing: correlation between natural evolution and on-treatment evolution. PLoS One. 2014;9:e112306. doi: 10.1371/journal.pone.0112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caballero A., Gregori J., Homs M. Complex genotype mixtures analyzed by deep sequencing in two different regions of hepatitis B virus. PLoS One. 2015;10:e0144816. doi: 10.1371/journal.pone.0144816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn C.M., Iwanowicz L.R., Cornman R.S. Characterization of a novel hepadnavirus in the white sucker (Catostomus commersonii) from the Great Lakes Region of the United States. J Virol. 2015;89:11801–11811. doi: 10.1128/JVI.01278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dill J.A., Camus A.C., Leary J.H. Distinct viral lineages from fish and amphibians reveal the complex evolutionary history of hepadnaviruses. J Virol. 2016;90:7920–7933. doi: 10.1128/JVI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson D.A., Cavanaugh M., Clark K. GenBank. Nucleic Acids Res. 2013;41(D1):36–42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.International Committee on Taxonomy of Viruses ICTV Ninth Report; Family: Hepadnaviridae 2009. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/reverse-transcribing-dna-and-rna-viruses-2011/w/rt_viruses/155/hepadnaviridae Available at:
- 35.Angelo P. Different patterns of codon usage in the overlapping polymerase and surface genes of hepatitis B virus suggest a de novo origin by modular evolution. J Gen Virol. 2015;96:3577–3586. doi: 10.1099/jgv.0.000307. [DOI] [PubMed] [Google Scholar]
- 36.Kay A., Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 37.Liu C., Fan G., Wang Z. Allosteric conformational changes of human HBV core protein transform its assembly. Sci Rep. 2017;7:1404. doi: 10.1038/s41598-017-01568-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dryden K.A., Wieland S.F., Whitten-Bauer C. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol Cell. 2006;22:843–850. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 39.Lin B., Anderson D.A. A vestigial X open reading frame in duck hepatitis B virus. Intervirology. 2000;43:185–190. doi: 10.1159/000025037. [DOI] [PubMed] [Google Scholar]
- 40.Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82:693–712. doi: 10.1099/0022-1317-82-4-693. [DOI] [PubMed] [Google Scholar]
- 41.Revill P., Yuen L., Walsh R. Bioinformatic analysis of the hepadnavirus e-antigen and its precursor identifies remarkable sequence conservation in all orthohepadnaviruses peter. J Med Virol. 2010;82:105–115. doi: 10.1002/jmv.21645. [DOI] [PubMed] [Google Scholar]
- 42.Lucifora J., Salvetti A., Marniquet X. Detection of the hepatitis B virus (HBV) covalently-closed-circular DNA (cccDNA) in mice transduced with a recombinant AAV-HBV vector. Antiviral Res. 2017;145:14–19. doi: 10.1016/j.antiviral.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 44.Revill P., Locarnini S. Antiviral strategies to eliminate hepatitis B virus covalently closed circular DNA (cccDNA) Curr Opin Pharmacol. 2016;30:144–150. doi: 10.1016/j.coph.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 45.Tu T., Budzinska M., Shackel N. HBV DNA integration: molecular mechanisms and clinical implications. Viruses. 2017;9(4) doi: 10.3390/v9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Z., Jhunjhunwala S., Liu J. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593–601. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wooddell C.I., Yuen M.F., Chan H.L. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med. 2017;9(409) doi: 10.1126/scitranslmed.aan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chai N., Chang H.E., Nicolas E. Properties of subviral particles of hepatitis B virus. J Virol. 2008;82:7812–7817. doi: 10.1128/JVI.00561-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganem D. Assembly of hepadnaviral virions and subviral particles. Curr Top Microbiol Immunol. 1991;168:61–83. doi: 10.1007/978-3-642-76015-0_4. [DOI] [PubMed] [Google Scholar]
- 50.Reignat S., Webster G.J.M., Brown D. Escaping high viral load exhaustion. J Exp Med. 2002;195:1089–1101. doi: 10.1084/jem.20011723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tennant B.C., Gerin J.L. The woodchuck model of hepatitis B virus infection. ILAR J. 2001;42:89–102. doi: 10.1093/ilar.42.2.89. [DOI] [PubMed] [Google Scholar]
- 52.Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141–150. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- 53.Tatematsu K., Tanaka Y., Kurbanov F. A genetic variant of hepatitis b virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype. J. J Virol. 2009;83:10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13:14–21. doi: 10.3748/wjg.v13.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong S., Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64:S4–S16. doi: 10.1016/j.jhep.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olinger C.M., Jutavijittum P., Hübschen J.M. Possible new hepatitis B virus genotype, southeast Asia. Emerg Infect Dis. 2008;14:1777–1780. doi: 10.3201/eid1411.080437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guettouche T., Hnatyszyn H.J. Chronic hepatitis B and viral genotype: the clinical significance of determining HBV genotypes. Antiviral Ther. 2005;10:593–604. [PubMed] [Google Scholar]
- 58.Niebel M., Singer J.B., Nickbakhsh S., Gifford R.J., Thomson E.C. Hepatitis C and the absence of genomic data in low income countries; a barrier on the road to eradication? Lancet Gastroenterol Hepatol. 2017;2:700–701. doi: 10.1016/S2468-1253(17)30257-1. [DOI] [PubMed] [Google Scholar]
- 59.Paraskevis D., Magiorkinis G., Magiorkinis E. Dating the origin and dispersal of hepatitis B virus infection in humans and primates. Hepatology. 2013;57:908–916. doi: 10.1002/hep.26079. [DOI] [PubMed] [Google Scholar]
- 60.Mulyanto, Pancawardani P., Depamede S.N. Identification of four novel subgenotypes (C13-C16) and two inter-genotypic recombinants (C12/G and C13/B3) of hepatitis B virus in Papua province, Indonesia. Virus Res. 2012;163:129–140. doi: 10.1016/j.virusres.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 61.Shi W., Zhu C., Zheng W. Subgenotyping of genotype C hepatitis B virus: correcting misclassifications and identifying a novel subgenotype. PLoS One. 2012;7:e47271. doi: 10.1371/journal.pone.0047271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross Z.P., Klunk J., Fornaciari G. The paradox of HBV evolution as revealed from a 16th century mummy. PLoS Pathog. 2018;14:e1006750. doi: 10.1371/journal.ppat.1006750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paraskevis D., Angelis K., Magiorkinis G. Dating the origin of hepatitis B virus reveals higher substitution rate and adaptation on the branch leading to F/H genotypes. Mol Phylogenet Evol. 2015;93:44–54. doi: 10.1016/j.ympev.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 64.Castelhano N., Araujo N.M., Arenas M. Heterogeneous recombination among hepatitis B virus genotypes. Infect Genet Evol. 2017;54:486–490. doi: 10.1016/j.meegid.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 65.Devesa M., Loureiro C.L., Rivas Y. Subgenotype diversity of hepatitis B virus American genotype F in Amerindians from Venezuela and the general population of Colombia. J Med Virol. 2008;80:20–26. doi: 10.1002/jmv.21024. [DOI] [PubMed] [Google Scholar]
- 66.Livingston S.E., Simonetti J.P., McMahon B.J. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5–11. doi: 10.1086/509894. [DOI] [PubMed] [Google Scholar]
- 67.Mello F.C.A., Araujo O.C., Lago B.V. Phylogeography and evolutionary history of hepatitis B virus genotype F in Brazil. Virol J. 2013;10:236. doi: 10.1186/1743-422X-10-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin C.L., Kao J.H. Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med. 2015;5 doi: 10.1101/cshperspect.a021436. a021436–a021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramvis A., Kew M.C. Molecular characterization of subgenotype A1 (subgroup Aa) of hepatitis B virus. Hepatol Res. 2007;37(Suppl 1):27–32. doi: 10.1111/j.1872-034X.2007.00100.x. [DOI] [PubMed] [Google Scholar]
- 70.Sarin S.K., Kumar M., Lau G.K. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int. 2016;10:1–98. doi: 10.1007/s12072-015-9675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Terrault N.A., Lok A.S., McMahon B.J. Update on prevention, diagnosis, and treatment and of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67:1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.NICE NICE guideline: Hepatitis B (chronic): diagnosis and management. https://www.nice.org.uk/Guidance/CG165 Available at: Published June 2013. Updated October 2017.
- 73.Matthews P. Hepatitis B virus (HBV) amino acid alignments and genotype-specific consensus sequences. Figshare. https://figshare.com/articles/Hepatitis_B_Virus_HBV_amino_acid_alignments_and_genotype-specific_consensus_sequences/4040700 Available at: Published 2016.
- 74.Lumley S., Noble H., Hadley M.J. Hepitopes: a live interactive database of HLA class I epitopes in hepatitis B virus. Wellcome Open Res. 2016;1:9. doi: 10.12688/wellcomeopenres.9952.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuiken C., Combet C., Bukh J. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology. 2006;44:1355–1361. doi: 10.1002/hep.21377. [DOI] [PubMed] [Google Scholar]
- 76.Ono Y., Onda H., Sasada R. The complete nucleotide sequences of the cloned hepatitis B virus DNA; subype adr and adw. Nucleic Acids Research. 1983;11:1747–1757. doi: 10.1093/nar/11.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hayer J., Jadeau F., Deléage G. HBVdb: a knowledge database for hepatitis B virus. Nucleic Acids Res. 2013;41(D1):566–570. doi: 10.1093/nar/gks1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai Q., Zhu H., Zhang Y. Hepatitis B virus genotype A: design of reference sequences for sub-genotypes. Virus Genes. 2016;52:325–333. doi: 10.1007/s11262-016-1307-0. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Z.H., Zhang L., Lu M.J. Establishment of reference sequences of hepatitis B virus genotype B and C in China. Genet Mol Res. 2015;14:16521–16534. doi: 10.4238/2015.December.9.24. [DOI] [PubMed] [Google Scholar]
- 80.Kuiken C., Yusim K., Boykin L. The Los Alamos hepatitis C sequence database. Bioinformatics. 2005;21:379–384. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- 81.Smith D.B., Bukh J., Kuiken C. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y.P., Ramirez S., Humes D. Differential sensitivity of 5′UTR-NS5A recombinants of hepatitis C virus genotypes 1-6 to protease and NS5A inhibitors. Gastroenterology. 2014;146:812–821. doi: 10.1053/j.gastro.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 83.Araujo N.M. Hepatitis B virus intergenotypic recombinants worldwide: an overview. Infect Genet Evol. 2015;36:500–510. doi: 10.1016/j.meegid.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 84.Liao H., Li X., Liu Y. Intergenotype recombinant analysis of full-length hepatitis B virus genomes from 516 Chinese patients with different illness categories. J Med Virol. 2017;89:139–145. doi: 10.1002/jmv.24609. [DOI] [PubMed] [Google Scholar]
- 85.Kuiken C., Korber B., Shafer R.W. HIV sequence databases. AIDS Rev. 2003;5:52–61. [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou B., Wang Z., Yang J. Novel evidence of HBV recombination in family cluster infections in Western China. PLoS One. 2012;7:e38241. doi: 10.1371/journal.pone.0038241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boyce C.L., Ganova-Raeva L., Archampong T.N.A. Identification and comparative analysis of hepatitis B virus genotype D/E recombinants in Africa. Virus Genes. 2017;53:538–547. doi: 10.1007/s11262-017-1469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chekaraou M.A., Brichler S., Mansour W. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J Gen Virol. 2010;91:1609–1620. doi: 10.1099/vir.0.018127-0. [DOI] [PubMed] [Google Scholar]
- 89.Su H., Liu Y., Xu Z. A novel complex A/C/G intergenotypic recombinant of hepatitis B virus isolated in Southern China. PLoS One. 2014;9:e84005. doi: 10.1371/journal.pone.0084005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adachi E., Sugiyama M., Shimizu S. Human immunodeficiency virus and hepatitis B genotype G/A2 recombinant co-infection: a case study. Springerplus. 2016;5:1502. doi: 10.1186/s40064-016-3169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Osiowy C., Gordon D., Borlang J. Hepatitis B virus genotype G epidemiology and co-infection with genotype A in Canada. J Gen Virol. 2008;89:3009–3015. doi: 10.1099/vir.0.2008/005124-0. [DOI] [PubMed] [Google Scholar]
- 92.Candotti D., Diarra B., Bisseye C. Molecular characterization of hepatitis B virus in blood donors from Burkina Faso: prevalence of quasi-subgenotype A3, genotype E, and mixed infections. J Med Virol. 2016;88:2145–2156. doi: 10.1002/jmv.24589. [DOI] [PubMed] [Google Scholar]
- 93.Fan X., Mao Q., Zhou D. High diversity of hepatitis C viral quasispecies is associated with early virological response in patients undergoing antiviral therapy. Hepatology. 2009;50:1765–1772. doi: 10.1002/hep.23290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jardim A.C., Bittar C., Matos R.P.A. Analysis of HCV quasispecies dynamic under selective pressure of combined therapy. BMC Infect Dis. 2013;13:61. doi: 10.1186/1471-2334-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jaworski E., Routh A. Parallel ClickSeq and Nanopore sequencing elucidates the rapid evolution of defective-interfering RNAs in Flock House virus. PLoS Pathog. 2017;13:e1006365. doi: 10.1371/journal.ppat.1006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pybus O.G., Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nature Rev Genet. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harrison A., Lemey P., Hurles M. Genomic analysis of hepatitis B virus reveals antigen state and genotype as sources of evolutionary rate variation. Viruses. 2011;3:83–101. doi: 10.3390/v3020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fares M.A., Holmes E.C. A revised evolutionary history of hepatitis B virus (HBV) J Mol Evol. 2002;54:807–814. doi: 10.1007/s00239-001-0084-z. [DOI] [PubMed] [Google Scholar]
- 99.Sanjuán R. From molecular genetics to phylodynamics: evolutionary relevance of mutation rates across viruses. PLoS Pathog. 2012;8:e1002685. doi: 10.1371/journal.ppat.1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform doi:10.1093/bib/bbx108. E-pub ahead of print. [DOI] [PMC free article] [PubMed]
- 101.Simmonds P. SSE: a nucleotide and amino acid sequence analysis platform. BMC Res Notes. 2012;5:50. doi: 10.1186/1756-0500-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cento V., Mirabelli C., Dimonte S. Overlapping structure of hepatitis B virus (HBV) genome and immune selection pressure are critical forces modulating HBV evolution. J Gen Virol. 2013;94:143–149. doi: 10.1099/vir.0.046524-0. [DOI] [PubMed] [Google Scholar]
- 103.Zhou Y., Holmes E.C. Bayesian estimates of the evolutionary rate and age of hepatitis B virus. J Mol Evol. 2007;65:197–205. doi: 10.1007/s00239-007-0054-1. [DOI] [PubMed] [Google Scholar]
- 104.Kahila Bar-Gal G., Kim M.J., Klein A. Tracing hepatitis B virus to the 16th century in a Korean mummy. Hepatology. 2012;56:1671–1680. doi: 10.1002/hep.25852. [DOI] [PubMed] [Google Scholar]
- 105.Krakauer D.C. Stability and evolution of overlapping genes. Evolution. 2000;54:731–739. doi: 10.1111/j.0014-3820.2000.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 106.Krakauer D.C. Evolutionary principles of genomic compression. Comments Theor Biol. 2002;7:215–236. [Google Scholar]
- 107.Tedder R.S., Bissett S.L., Myers R. The “Red Queen” dilemma—running to stay in the same place: reflections on the evolutionary vector of HBV in humans. Antiviral Ther. 2013;18:489–496. doi: 10.3851/IMP2655. [DOI] [PubMed] [Google Scholar]
- 108.Zaaijer H.L., Van Hemert F.J., Koppelman M.H. Independent evolution of overlapping polymerase and surface protein genes of hepatitis B virus. J Gen Virol. 2007;88:2137–2143. doi: 10.1099/vir.0.82906-0. [DOI] [PubMed] [Google Scholar]
- 109.Bartenschlager R., Schaller H. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. EMBO J. 1988;7:4185–4192. doi: 10.1002/j.1460-2075.1988.tb03315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Radziwill G., Tucker W., Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen P., Gan Y., Han N. Computational evolutionary analysis of the overlapped surface (S) and polymerase (P) region in hepatitis B virus indicates the spacer domain in P is crucial for survival. PLoS One. 2013;8:e60098. doi: 10.1371/journal.pone.0060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin Y., Liu C., Chien W. New insights into the evolutionary rate of hepatitis B virus at different biological scales. J Virol. 2015;89:3512–3522. doi: 10.1128/JVI.03131-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chirico N., Vianelli A., Belshaw R. Why genes overlap in viruses. Proc R Soc B Biol Sci. 2010;277:3809–3817. doi: 10.1098/rspb.2010.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mizokami M., Orito E., Ohba K. Constrained evolution with respect to gene overlap of hepatitis B virus. J Mol Evol. 1997;44(Suppl 1):S83–S90. doi: 10.1007/pl00000061. [DOI] [PubMed] [Google Scholar]
- 115.Homs M., Buti M., Quer J. Ultra-deep pyrosequencing analysis of the hepatitis B virus preCore region and main catalytic motif of the viral polymerase in the same viral genome. Nucleic Acids Rese. 2011;39:8457–8471. doi: 10.1093/nar/gkr451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mokaya J., Mcnaughton A.L., Hadley M.J. A systematic review of hepatitis B virus (HBV) drug and vaccine escape mutations in Africa: a call for urgent action. PLoS Negl Trop Dis. 2018;12:e0006629. doi: 10.1371/journal.pntd.0006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bayliss J., Yuen L., Rosenberg G. Deep sequencing shows that HBV basal core promoter and precore variants reduce the likelihood of HBsAg loss following tenofovir disoproxil fumarate therapy in HBeAg-positive chronic hepatitis B. Gut. 2016;66:2013–2023. doi: 10.1136/gutjnl-2015-309300. [DOI] [PubMed] [Google Scholar]
- 118.Lythgoe K.A., Gardner A., Pybus O.G. Short-sighted virus evolution and a germline hypothesis for chronic viral infections. Trends Microbiol. 2017;25:336–348. doi: 10.1016/j.tim.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang G., Yang J., Luo K. Quasispecies characteristics in mother-to-child transmission of hepatitis B virus by next-generation sequencing. J Infect. 2017;75:48–58. doi: 10.1016/j.jinf.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 120.Eschlimann M., Malvé B., Velay A. The variability of hepatitis B envelope is associated with HBs antigen persistence in either chronic or acute HBV genotype A infection. J Clin Virol. 2017;94:115–122. doi: 10.1016/j.jcv.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 121.Yang Z.T., Huang S.Y., Chen L. Characterization of full-length genomes of hepatitis B virus quasispecies in sera of patients at different phases of infection. J Clin Microbiol. 2015;53:2203–2214. doi: 10.1128/JCM.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boyd A., Lacombe K., Lavocat F. Decay of ccc-DNA marks persistence of intrahepatic viral DNA synthesis under tenofovir in HIV-HBV co-infected patients. J Hepatol. 2016;65:683–691. doi: 10.1016/j.jhep.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 123.Locarnini S., Zoulim F. Molecular genetics of HBV infection. Antiviral Ther. 2010;15:3–14. doi: 10.3851/IMP1619. [DOI] [PubMed] [Google Scholar]
- 124.Stuyver L., De Gendt S., Van Geyt C. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 125.Norder H., Couroucé A.-M., Magnius L.O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 126.Lok A.S., Ganova-Raeva L., Cloonan Y. Prevalence of hepatitis B antiviral drug resistance variants in North American patients with chronic hepatitis B not receiving antiviral treatment. J Viral Hepat. 2017;24:1032–1042. doi: 10.1111/jvh.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Patterson S.J., George J., Strasser S.I. Tenofovir disoproxil fumarate rescue therapy following failure of both lamivudine and adefovir dipivoxil in chronic hepatitis B. Gut. 2011;60:247–254. doi: 10.1136/gut.2010.223206. [DOI] [PubMed] [Google Scholar]
- 128.Rybicka M., Woziwodzka A., Romanowski T. Differences in sequences between HBV-relaxed circular DNA and covalently closed circular DNA. Emerg Microbes Infect. 2017;6:e55. doi: 10.1038/emi.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu T.T., Coates L., Aldrics C.E. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology. 1990;175:255–261. doi: 10.1016/0042-6822(90)90206-7. [DOI] [PubMed] [Google Scholar]
- 130.Köck J., Rösler C., Zhang J.J. Generation of covalently closed circular DNA of hepatitis B viruses via intracellular recycling is regulated in a virus specific manner. PLoS Pathog. 2010;6:e1001082. doi: 10.1371/journal.ppat.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang A.Y., Lai C.L., Huang F.Y. Deep sequencing analysis of quasispecies in the HBV pre-S region and its association with hepatocellular carcinoma. J Gastroenterol. 2017;52:1064–1074. doi: 10.1007/s00535-017-1334-1. [DOI] [PubMed] [Google Scholar]
- 132.Liu W.C., Wu I.C., Lee Y.C. Hepatocellular carcinoma-associated single-nucleotide variants and deletions identified using genome-wide high throughput analysis of HBV. J Pathol. 2017;243:176–192. doi: 10.1002/path.4938. [DOI] [PubMed] [Google Scholar]
- 133.Jia J., Liang X., Chen S. Next-generation sequencing revealed divergence in deletions of the preS region in the HBV genome between different HBV-related liver diseases. J Gen Virol. 2017;98:2748–2758. doi: 10.1099/jgv.0.000942. [DOI] [PubMed] [Google Scholar]
- 134.Soussan P., Tuveri R., Nalpas B. The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosi. J Hepatol. 2003;38:343–348. doi: 10.1016/s0168-8278(02)00422-1. [DOI] [PubMed] [Google Scholar]
- 135.Soussan P., Garreau F., Zylberberg H. In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J Clin Investig. 2000;105:55–60. doi: 10.1172/JCI8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bayliss J., Lim L., Thompson A.J. Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J Hepatol. 2013;59:1022–1028. doi: 10.1016/j.jhep.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 137.Tseng T.C., Liu C.J., Yang H.C. Higher proportion of viral basal core promoter mutant increases the risk of liver cirrhosis in hepatitis B carriers. Gut. 2015;64:292–302. doi: 10.1136/gutjnl-2014-306977. [DOI] [PubMed] [Google Scholar]
- 138.Fang Z.L., Sabin C.A., Dong B. HBV A 1762 T, G 1764 A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol. 2008;103:2254–2262. doi: 10.1111/j.1572-0241.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yin J., Xie J., Liu S. Association between the various mutations in viral core promoter region to different stages of hepatitis B, ranging of asymptomatic carrier state to hepatocellular carcinoma. Am J Gastroenterol. 2011;106:81–92. doi: 10.1038/ajg.2010.399. [DOI] [PubMed] [Google Scholar]
- 140.Chen L., Gan Q.R., Zhang D.Q. Increased intrahepatic quasispecies heterogeneity correlates with off-treatment sustained response to nucleos(t)ide analogues in e antigen-positive chronic hepatitis B patients. Clin Microbiol Infect. 2016;22:201–207. doi: 10.1016/j.cmi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 141.Han Y., Gong L., Sheng J. Prediction of virological response by pretreatment hepatitis B virus reverse transcriptase quasispecies heterogeneity: the advantage of using next-generation sequencing. Clin Microbiol Infect. 2015;21:e797. doi: 10.1016/j.cmi.2015.03.021. e1–e8. [DOI] [PubMed] [Google Scholar]
- 142.Gregori J., Perales C., Rodriguez-Frias F. Viral quasispecies complexity measures. Virology. 2016;493:227–237. doi: 10.1016/j.virol.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 143.Rodriguez-Frías F., Tabernero D., Quer J. Ultra-deep pyrosequencing detects conserved genomic sites and quantifies linkage of drug-resistant amino acid changes in the hepatitis B virus genome. PLoS One. 2012;7:e37874. doi: 10.1371/journal.pone.0037874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Du Y., Chi X., Wang C. Quantifying perinatal transmission of hepatitis B viral quasispecies by tag linkage deep sequencing. Sci Rep. 2017;7:10168. doi: 10.1038/s41598-017-10591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Khamduang W., Gaudy-Graffin C., Ngo-Giang-Huong N. Analysis of residual perinatal transmission of hepatitis B virus (HBV) and of genetic variants in human immunodeficiency virus and HBV co-infected women and their offspring. J Clin Virol. 2013;58:415–421. doi: 10.1016/j.jcv.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wong D.K., Kopaniszen M., Omagari K. Effect of hepatitis B virus reverse transcriptase variations on entecavir treatment response. J Infect Dis. 2014;210:701–707. doi: 10.1093/infdis/jiu133. [DOI] [PubMed] [Google Scholar]
- 147.Bittar C., Jardim A.C.G., Yamasaki L.H.T. Genetic diversity of NS5A protein from hepatitis C virus genotype 3a and its relationship to therapy response. BMC Infect Dis. 2010;10:36. doi: 10.1186/1471-2334-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Votintseva A.A., Bradley P., Pankhurst L. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol. 2017;55:1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Atkinson K.V., Bishop L.A., Rhodes G. Data descriptor: nasopharyngeal metagenomic deep sequencing. Scientific Data. 2017;4:170161. doi: 10.1038/sdata.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chan B.K., Wilson T., Fischer K.F. Deep sequencing to identify the causes of viral encephalitis. PLoS One. 2014;9:e93993. doi: 10.1371/journal.pone.0093993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Somasekar S., Lee D., Rule J. Viral surveillance in serum samples from patients with acute liver failure by metagenomic next-generation sequencing. Clin Infect Dis. 2017;65:1477–1485. doi: 10.1093/cid/cix596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ganova-Raeva L., Punkova L., Campo D.S. Cryptic hepatitis B and e in patients with acute hepatitis of unknown etiology. J Infect Dis. 2015;212:1962–1969. doi: 10.1093/infdis/jiv315. [DOI] [PubMed] [Google Scholar]
- 153.Betz-Stablein B.D., Töpfer A., Littlejohn M. Single-molecule sequencing reveals complex genome variation of hepatitis B virus during 15 years of chronic infection following. J Virol. 2016;90:7171–7183. doi: 10.1128/JVI.00243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jones L.R., Sede M., Manrique J.M. Hepatitis B virus resistance substitutions: long-term analysis by next-generation sequencing. Arch Virol. 2016;161:2885–2891. doi: 10.1007/s00705-016-2959-8. [DOI] [PubMed] [Google Scholar]
- 155.Wasityastuti W., Yano Y., Widasari D.I. Different variants in reverse transcriptase domain determined by ultra-deep sequencing in treatment-naïve and treated Indonesian patients infected with hepatitis B virus. Kobe J Med Sci. 2016;62:E1–E8. [PMC free article] [PubMed] [Google Scholar]
- 156.Lee Z., Nishikawa S., Gao S. Detection of hepatitis B virus (HBV) genomes and HBV drug resistant variants by deep sequencing analysis of HBV genomes in immune cell subsets of HBV mono-infected and/or human immunodeficiency virus type-1 (HIV-1) and HBV co-infected individuals. PLoS One. 2015;10:e0137568. doi: 10.1371/journal.pone.0137568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hayashi S., Murakami S., Omagari K. Characterization of novel entecavir resistance mutations. J Hepatol. 2015;63:546–553. doi: 10.1016/j.jhep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 158.Lowe C.F., Merrick L., Harrigan P.R. Implementation of next-generation sequencing for hepatitis B resistance and genotyping in a clinical microbiology laboratory. J Clin Microbiol. 2015;54:127–133. doi: 10.1128/JCM.02229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Campo D.S., Xia G.L., Dimitrova Z. Accurate genetic detection of hepatitis C virus transmissions in outbreak settings. J Infect Dis. 2016;213:957–965. doi: 10.1093/infdis/jiv542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Caro-Pérez N., Martínez-Rebollar M., Gregori J. Phylogenetic analysis of an epidemic outbreak of acute hepatitis C in HIV-infected patients by ultra-deep pyrosequencing. J Clin Virol. 2017;92:42–47. doi: 10.1016/j.jcv.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 161.Escobar-Gutiérrez A., Vazquez-Pichardo M., Cruz-Rivera M. Identification of hepatitis C virus transmission using a next-generation sequencing approach. J Clin Microbiol. 2012;50:1461–1463. doi: 10.1128/JCM.00005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.McNaughton A., Bonsall D., Cesare M.D. Rolling circle amplification and nanopore-based deep sequencing of full-length HBV genomes. J Hepatol. 2018;68(Suppl 1):S762. [Google Scholar]