Significance
Toxins exploit numerous pathways of their host cells to gain cellular entry and promote intoxication. Therefore, studying the action of toxins allows us to better understand basic mechanisms in cell biology. In this study, we found that ZDHHC5, an enzyme that adds a lipid posttranslational modification to cysteines of proteins, is responsible for allowing anthrax toxin to enter cells. This enzyme acts on proprotein convertases that are needed to cleave these toxins to their active forms. ZDHHC5 does not affect the enzymatic activity of these proteases, but allows them to encounter the toxin by favoring their partitioning in microdomains on the cell surface, domains where the toxin has previously been shown to preferentially reside.
Keywords: anthrax toxin, ZDHHC5, S-palmitoylation, proprotein convertases
Abstract
The protein acyl transferase ZDHHC5 was recently proposed to regulate trafficking in the endocytic pathway. Therefore, we explored the function of this enzyme in controlling the action of bacterial toxins. We found that ZDHHC5 activity is required for two very different toxins: the anthrax lethal toxin and the pore-forming toxin aerolysin. Both of these toxins have precursor forms, the protoxins, which can use the proprotein convertases Furin and PC7 for activation. We show that ZDHHC5 indeed affects the processing of the protoxins to their active forms. We found that Furin and PC7 can both be S-palmitoylated and are substrates of ZDHHC5. The impact of ZDHHC5 on Furin/PC7-mediated anthrax toxin cleavage is dual, having an indirect and a direct component. First, ZDHHC5 affects the homeostasis and trafficking of a subset of cellular proteins, including Furin and PC7, presumably by affecting the endocytic/recycling pathway. Second, while not inhibiting the protease activity per se, ZDHHC5-mediated Furin/PC7 palmitoylation is required for the cleavage of the anthrax toxin. Finally, we show that palmitoylation of Furin and PC7 promotes their association with plasma membrane microdomains. Both the receptor-bound toxin and the convertases are of very low abundance at the cell surface. Their encounter is unlikely on reasonable time scales. This work indicates that palmitoylation drives their encounter in specific domains, allowing processing and thereby intoxication of the cell.
The endocytic system in eukaryotic cells is responsible for a variety of functions, including nutrient uptake, receptor down-regulation, and membrane protein trafficking and homeostasis, as well as the delivery of a plethora of bacterial toxins. Recently, proteomic screens have revealed that the palmitoyl transferase ZDHHC5 may be a regulator of endosomal trafficking. It was identified in the cell surface and endosome-enriched fractions of avian epithelial cells (1), among tight-junction regulators in Madin-Darby canine kidney epithelial cells (2), and cell surface and endosomal cargoes in breast cancer cells (3). Furthermore, a recent small-interfering RNA (siRNA) screen implicated ZDHHC5 as an important trafficking regulator of endosome-to-Golgi retrieval (4), and multiple studies suggest either ZDHHC5 or its substrates change localization in neurons, thereby affecting their activity (5–7). These previous studies made a solid case for the involvement of ZDHHC5 in endosomes and endosomal trafficking, but how and why it plays a role there, especially in nonneuronal cells, is still elusive.
ZDHHC5 belongs to the ZDHHC S-palmitoyltransferase family. These enzymes add medium-chain fatty acids, such as palmitate (C16), to cytoplasmic cysteines of substrate proteins in a specific manner (8–10). Most of the 23 ZDHHC enzymes present in humans localize to the early biosynthetic pathway: that is, the endoplasmic reticulum and the Golgi (9). ZDHHC5 is among the rarer enzymes of the family that localize to the endosomal system. This allows it to modify a unique set of substrates of which it could potentially modify the trafficking, turnover rate, and function (11).
Given its proposed role in endocytosis, we investigated whether ZDHHC5 is involved in the mode of action of bacterial toxins. We mainly focused on the anthrax toxin, the major virulence factor of Bacillus anthracis. Anthrax toxin consists of three components: protective antigen (PA), which is the subunit that mediates specific interaction with target cells; and lethal factor (LF) and edema factor (EF), the two enzymatic subunits. The toxin enters cells using one of two receptors: CMG2 and TEM8 (12). As PA binds to CMG2 or TEM8, it becomes cleaved by the PC family of proteases, which includes Furin (13–15). PA can then oligomerize, allowing LF or EF to bind, and the whole complex, driven by the receptor, is internalized. In the acidic environment of endosomes, the PA oligomer undergoes a pH-dependent conformational change and forms a transmembrane pore that allows translocation of LF or EF across the endosomal membrane, ultimately allowing them to reach the cytosol to exert their actions (16). Anthrax toxin entry thus heavily relies on endosomal function and regulation. In this work, we found that intoxication by anthrax exquisitely depends on ZDHHC5 and reveals an unexpected role of ZDHHC5 in the process. This study once more highlights the extreme precision with which host–pathogen interactions have been optimized.
Results
ZDHHC5 Affects the Stability of Palmitoylated and Cell Surface Proteins.
Recent findings have indicated that ZDHHC5 is involved in endosomal trafficking, potentially affecting the trafficking of membrane proteins to lysosomes and thereby their turnover rate. Moreover, palmitoylation of proteins can strongly affect their half-life, either positively (17, 18) or negatively (19). These observations prompted us to first analyze the effect of ZDHHC5 on overal protein flux in the cell. To do this, we engineered retinal pigmental epithelial 1 (RPE-1) cells lacking ZDHHC5 using CRISPR/Cas9 (SI Appendix, Fig. S1A) and used stable isotope labeling with amino acids in cell culture (SILAC) to quantitatively measure the difference in protein amounts in this condition compared with control cells. Analysis of total cell extracts (TCE) revealed differential protein expression between control and ΔZDHHC5 cells (SI Appendix, Fig. S1B). Using B-significance, a statistical method that considers mass spectrometry signal intensity, we found that 3.1% of proteins statistically varied between the two cell types in the forward (control cells labeled with heavy isotopes and ΔZDHHC5 cells labeled with light isotopes) or reverse (vice versa) reactions. The agreement between the forward and reverse reaction was decent with an R2 of 0.57.
Given that ZDHHC5 is a palmitoyl transferase, we next queried this TCE dataset for potentially palmitoylated proteins—that is, proteins found in palmitoyl-proteomes or validated as palmitoylated—using the SwissPalm 2.0 palmitoylation database (https://swisspalm.org/) (20). Palmitoylated proteins were significantly more abundant in control than in ΔZDHHC5 cells, suggesting that in the absence of ZDHHC5, these proteins had a reduced half-life (SI Appendix, Fig. S1D). We also queried the Cell Surface Protein Atlas (CSPA) (21) and found that cell surface proteins were also statistically more abundant in control than in ΔZDHHC5 cells (SI Appendix, Fig. S1E). The identified surface proteins in this query were, however, different from the identified palmitoylated proteins in the SwissPalm query (SI Appendix, Fig. S1F), suggesting that at least some cell surface proteins were affected by ZDHHC5 not due to their direct palmitoylation by the enzyme, but through an indirect effect, possibly influencing the endocytic pathway.
To compare the surface proteome of control and ΔZDHHC5 cells more directly, we performed surface biotinylation, isolation, and mass spectrometry analysis of these biotinylated proteins. We found that 0.9% of proteins were B-significant in control vs. ΔZDHHC5 cells in at least the forward or reverse reactions (SI Appendix, Fig. S1B). Of these, many—but not all—were already differentially expressed when comparing the total cell proteomes. In the pull-down dataset, only 11% of the proteins were found in the CSPA, but of the ones that were B-significant between WT and ΔZDHHC5 cells, 29% were in the CSPA, supporting a role of ZDHHC5 in regulating cell surface abundance of a set of proteins.
Finally, we performed a pathway analysis on the proteins that were B-significant in either the TCE or surface proteomes in control vs. ΔZDHHC5 cells. The most enriched pathways, of proteins that were more stable in control rather than ΔZDHHC5 cells, were membrane trafficking paths, in particular related to the endocytic route including autophagy (SI Appendix, Fig. S1G). Altogether, these findings further support a role for ZDHHC5 in endocytosis and recycling.
ZDHHC5 Controls Anthrax Toxin Action.
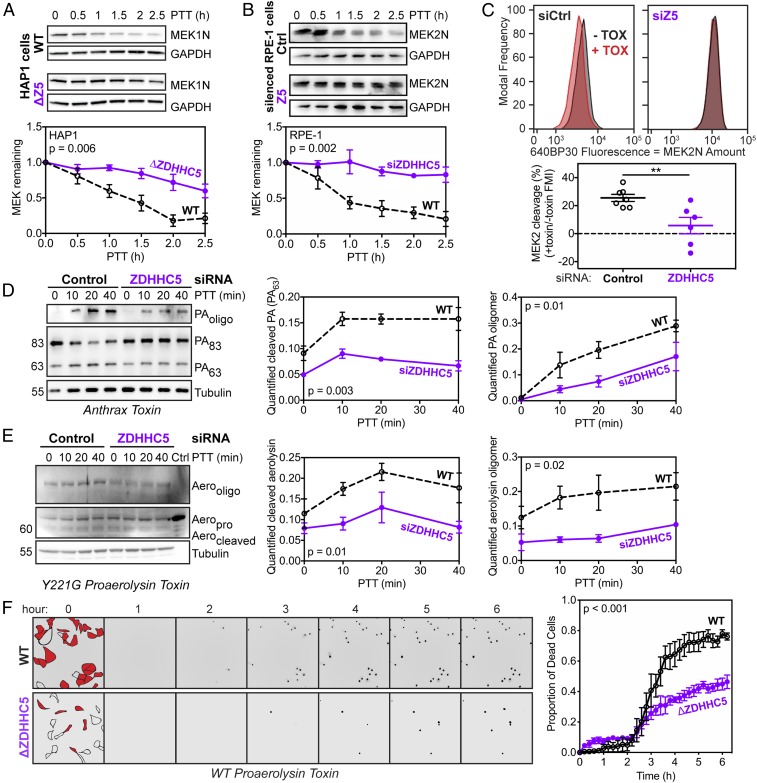
The involvement of ZDHHC5 in cell surface proteome homeostasis and endocytosis led us to investigate the importance of ZDHHC5 in the entry of anthrax toxin. To know whether any part of the endosomal system could be affected, we first monitored the action of the anthrax lethal toxin, which is cleavage of MAPKKs by the LF subunit in the cytosol. More specifically, we followed MEK1 or MEK2 using antibodies against their N termini, which are cleaved by LF. A time course of MAPKK processing after toxin treatment showed that ZDHHC5 knockdown or knockout led to a strong inhibition of toxin action in multiple cell types (Fig. 1 A and B and SI Appendix, Fig. S2A). For a more quantitative assay, we used analytical flow cytometry to monitor MEK2-N cleavage in RPE-1 cells. We chose a 1.5-h toxin treatment because this led to a significant and robust shift in fluorescence. The toxin-induced shift was essentially abolished upon ZDHHC5 silencing (Fig. 1C). Having established that ZDHHC5 was involved in toxin entry, we sought to pinpoint the spatiotemporal properties of its involvement.
Fig. 1.
ZDHHC5 is required for efficient toxin entry. Anthrax toxin (500 ng/mL PA and 50 ng/mL LF) was applied to Control- (Ctrl) or ZDHHC5- (Z5) lacking HAP1 (A) or RPE-1 (B) cells for 1 h at 4 °C and then cells were shifted to 37 °C for 0–2.5 h posttoxin treatment (PTT), after which cells were lysed and MEK level was probed by Western blot. Statistics: unpaired two-tailed t test on AUCs with n = 4 (HAP1) or 6–8 (RPE-1). (C) RPE-1 cells were silenced with Control or ZDHHC5 and then exposed to anthrax toxin (+TOX; black) or not (−TOX; red) as before. After 1.5 h, cells were detached, fixed, permeabilized with Triton X-100, stained with MEK2 antibody and a fluorescent secondary antibody, and analyzed using flow cytometry. Statistics: unpaired two-tailed t test (**P < 0.01). Anthrax toxin (500 ng/mL PA and 50 ng/mL LF) (D) or the Y221G variant of proaerolysin (500 ng/mL) (E) was applied to Control- or ZDHHC5-silenced RPE-1 cells for 1 h or 30 min, respectively, at 4 °C, and then cells were shifted to 37 °C for 0–40 min, after which cells were lysed and toxins were probed by Western blot. Statistics: unpaired two-tailed t test on AUC with n = 4 (anthrax) and n = 3 (proaerolysin). (F) Representative fields in live-cell imaging of WT or ΔZDHHC5 cells treated with WT proaerolysin toxin. Cell outlines in the fields are shown at time 0 with cells in red as the ones that die at the end of the time course (more than 6 h). Each time-point shows the channel in which a NucGreen Dead dye marks the nuclei of dead cells. Statistics: representative graph; unpaired two-tailed t test on AUCs with n = 3. (Magnification: 20×.)
The first events in the mode of action of anthrax toxin can readily be monitored by Western blot analysis. The receptor binding subunit, PA, binds to the CMG2 or TEM8 receptor. PA is initially an 83-kDa protein that requires proteolytic cleavage of its N-terminal domain, leading to the oligomerization-competent PA63 form. This cleavage is mediated at the cell surface by proprotein convertases (PCs), such as Furin (14). At first, the PA oligomer is SDS sensitive but, upon endocytosis and arrival in sorting endosomes, the low pH leads to a conformational change in the complex that triggers membrane insertion and renders the complex SDS-resistant, and therefore visible by SDS/PAGE and Western blotting. A time-course analysis revealed that ZDHHC5 knockdown led to a reduced cleavage of PA83 into PA63 and a concomitant decrease in the appearance of the SDS-resistant oligomer (Fig. 1D).
The requirement for PC-mediated cleavage is not unique to anthrax PA; it is shared by other bacterial toxins. We therefore investigated whether ZDHHC5 could also affect proteolytic activation of another toxin. We chose the pore-forming aerolysin, which is produced by Aeromonas hydrophila as a protoxin, proaerolysin, which requires C-terminal cleavage to undergo heptamerization and membrane insertion (22). Cellular conversion of proaerolysin into aerolysin and the subsequent formation of the SDS-resistant aerolysin heptamer were drastically reduced in RPE-1 cells lacking ZDHHC5 compared with control cells (Fig. 1E). Because aerolysin, as opposed to anthrax lethal toxin, leads to cell death, we monitored cell viability upon exposure of control and ΔZDHHC5 cells to proaerolysin using the NucGreen Dead dye in live-cell microscopy over 6 h (Fig. 1F). ΔZDHHC5 cells were significantly less sensitive to the protoxin than control cells. Note that during purification a minor amount of proaerolysin undergoes processing, which could contribute to the observed activity. ZDHHC5 does not influence the sensitivity of cells to precleaved aerolysin (SI Appendix, Fig. S3E). The observation that two very different toxins, a pore-forming toxin and an enzymatically active toxin, both rely on ZDHHC5 to fully exert their activity strongly points toward a general role of ZDHHC5 in PC-mediated toxin activation.
ZDHHC5 Knockdown Resembles Furin and PC7 Knockdown.
We next examined which PCs mediate PA processing in RPE-1 cells using toxin-induced MEK2 cleavage as a readout. Silencing of PC2 and PC7 had no effect on toxin entry (SI Appendix, Fig. S2 B and C), possibly due to the very low expression of these enzymes in RPE-1 cells (SI Appendix, Fig. S2D). Furin and PC7 knockdown, individually or together, in contrast, drastically slowed down MEK2 cleavage, whether monitored by Western blot (SI Appendix, Fig. S2 B and C) or by flow cytometry (SI Appendix, Fig. S2I). Their expression and silencing were evident at both mRNA and protein level (SI Appendix, Fig. S2 E–H). Knockdown of ZDHHC5, therefore, had very similar effects as knockdown of Furin and PC7.
To confirm that ZDHHC5 mainly affected the initial PA cleavage step, we treated the cells with a precleaved PA (PA63). When PA63 was added to cells in combination with LF, MEK2 cleavage in control cells could no longer be distinguished from that in cells silenced for ZDHHC5 or Furin/PC7 expression (SI Appendix, Fig. S3A). The same held true for ΔZDHHC5 HAP1 compared with control HAP1 cells treated with precleaved PA63 (SI Appendix, Fig. S3B). When we looked at kinetics of PA oligomer formation, while clear differences were observed between control cells and cells depleted of ZDHHC5 or Furin/PC7 when treated with the PA83 proform (SI Appendix, Fig. S3C), kinetics were similar to each other in all cells when adding the precleaved PA63 (SI Appendix, Fig. S3D). Thus, ZDHHC5, just like Furin and PC7, acts on the cleavage of PA, an essential step for the toxin to enter cells.
Furin and PC7 Are ZDHHC5 Substrates.
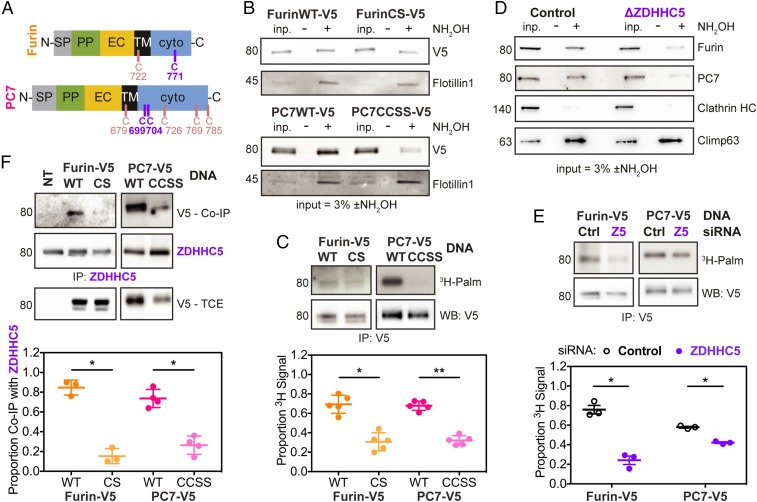
Given that the requirement of ZDHHC5 for toxin entry is similar to that of Furin/PC7, we investigated whether Furin and PC7 might actually be ZDHHC5 substrates. PC7 has previously been reported to undergo palmitoylation (23), but not Furin. Both Furin and PC7 are single-spanning type I membrane proteins with cysteines in their cytosolic domain as potential sites of palmitoylation (Fig. 2A). By labeling cells with a clickable analog of palmitic acid (17-octadecynoic acid) and performing downstream click chemistry with a fluorescent dye, we found that both Furin and PC7 show a palmitoylation signal above background levels (SI Appendix, Fig. S4A). PC2 was used as a negative control because it has no cytosolic domain and thus cannot be S-palmitoylated. The level of palmitoylation was more pronounced for PC7 than Furin, explaining why Furin palmitoylation might have been overlooked previously. We generated a double cysteine to serine mutant for PC7, corresponding to the two reported palmitoylation sites—Cys-699 and Cys-704 (24)—and a single cysteine mutant for Furin in which the unique cytosolic cysteine was mutated to serine (C771S). By acyl resin-assisted capture (Acyl-RAC), a method that captures acylated proteins, palmitoylation-deficient mutants of both proteases showed results consistent with decreased palmitoylation (Fig. 2B). The involvement of these cysteines was confirmed by 3H-palmitate labeling (Fig. 2C; see SI Appendix, Fig. S4C for controls), although a detectable signal remained for the Furin mutant, despite the absence of cytosolic cysteine. Even mutating the transmembrane cysteine, in addition to the cytosolic cysteine, did not lead to a further decrease of the signal. It is therefore still unclear what the residual signal represents. Taken together, these experiments show that Furin and PC7 can undergo palmitoylation.
Fig. 2.
Furin and PC7 are both palmitoylated, primarily by ZDHHC5. (A) Diagrams of Furin and PC7 with the transmembrane and cytosolic cysteines shown. Abbreviations: EC, extracellular; PP, propeptide; SP, signal peptide; TM, transmembrane; with “N-” and “-C” referring to the termini. The main palmitoylated cysteines are in purple (C771 for Furin, FurinCS; and C699/C704 for PC7, PC7CCSS), while the rest are shown in pink. (B) Representative blot of Acyl-RAC performed on RPE-1 cells transfected with V5-tagged Furin or PC7 WT or mutants. Hydroxylamine (NH2OH) was used to cleave off acyl chains before capturing their resultant free cysteines on beads. (C) RPE-1 cells were transfected as in B, then labeled for 2 h with 3H-palmitic acid, after which cells were lysed and immunoprecipitated. Statistics: ratio paired two-tailed t test on the original data. (D) Representative blot of Acyl-RAC performed as in B on WT and ΔZDHHC5 HAP1 cells. Endogenous Furin and PC7 are shown. (E) RPE-1 cells were silenced for Control or ZDHHC5 and then transfected with V5-tagged Furin or PC7. They were treated with 3H-palmitic acid as in C. Statistics: ratio paired two-tailed t test on the original data. (F) RPE-1 cells were transfected with V5-tagged WT and mutant Furin and PC7 and then lysed and immunoprecipitated (IP) with anti-ZDHHC5 antibody. Both ZDHHC5 (direct) and V5 Co-IP blots are shown. Statistics: ratio paired two-tailed t test on the original data. *P < 0.05, **P < 0.01.
We next tested whether Furin and PC7 are palmitoylated by ZDHHC5. Using Acyl-RAC on cells depleted of ZDHHC5, either ΔZDHHC5 HAP1 cells (Fig. 2D) or ZDHHC5-silenced RPE-1 cells (SI Appendix, Fig. S4B), we saw a decrease in endogenous Furin and PC7 captured following hydroxylamine treatment when compared to control cells. In addition, 3H-palmitate incorporation into Furin and PC7 was lower for ZDHHC5-silenced than for control RPE-1 cells (Fig. 2E). Finally, immunoprecipitation of ZDHHC5 led to the pull-down of Furin and PC7, an interaction that was diminished for their palmitoylation-deficient mutants (Fig. 2F), as observed but not understood for other ZDHHC enzyme/substrate pairs.
A previous study reported that the anthrax toxin receptors, CMG2 and TEM8, can be palmitoylated themselves (25). We tested whether they too could be ZDHHC5 targets. However, incorporation of 3H-palmitate into CMG2 (SI Appendix, Fig. S4D) or TEM8 (SI Appendix, Fig. S4E) was not affected by ZDHHC5 silencing or knockout.
Taken together, the present results indicate that Furin and PC7 can be palmitoylated, primarily on C771, and C669 and C704, respectively. Both are targets of ZDHHC5, in contrast to the anthrax receptors.
ZDHHC5 Affects PC Activity at the Plasma Membrane and in Endosomes.
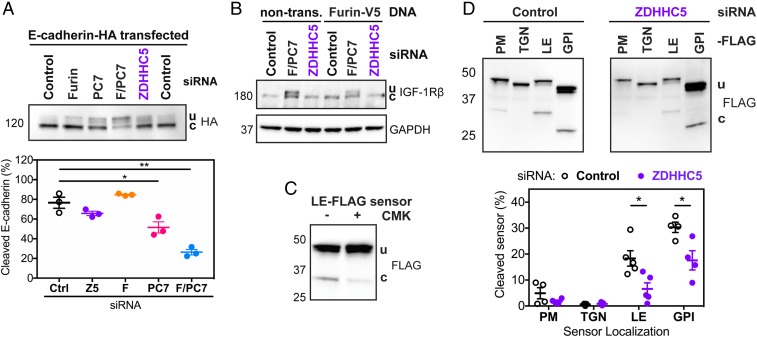
The above observations that ZDHHC5 is a palmitoyltransferase for Furin and PC7 and that ZDHHC5 is involved in cleavage of anthrax PA and aerolysin raised the possibility that ZDHHC5 might have a more general effect on PC substrate processing. To test this, we chose two well-known Furin and/or PC7 substrates: E-cadherin (26) and IGF-1R (27, 28). While we did confirm that E-cadherin (Fig. 3A) and IGF-1R (Fig. 3B) are indeed Furin and PC7 substrates, we saw no effect of ZDHHC5 silencing on their processing. Thus, the effect of ZDHHC5 on Furin/PC7-mediated toxin cleavage cannot be generalized to all PC substrates.
Fig. 3.
While ZDHHC5 does not affect processing of other Furin/PC7 substrates, it does affect cleavage in endosomes and lipid rafts. (A) HA-tagged E-cadherin was transfected into RPE-1 cells that were silenced for Control, Furin (F here), PC7, F/PC7, or ZDHHC5. Western blots using HA antibody show both uncleaved (u) and cleaved (c) versions of E-cadherin. Statistics: unpaired two-tailed t test. (B) Representative blot of IGF-1Rβ levels in cells silenced for F/PC7 or ZDHHC5, with or without transfection of Furin-V5. Both uncleaved (u) and cleaved (c) IGF-1Rβ are labeled. (C) The FLAG-tagged late endosome (LE-FLAG) sensor was transfected into RPE-1 cells and the PC-specific inhibitor chloromethyl ketone (CMK) was added for 6 h at 50 μM, after which cells were lysed and subjected to SDS/PAGE, transferred, and probed with an anti-FLAG antibody. (D) PC FLAG-tagged sensors targeting different subcellular compartments (PM for plasma membrane, TGN for trans-Golgi network, and GPI for glycosylphosphatidylinositol-anchored proteins primarily found in lipid rafts) were transfected in cells either silenced for Control or ZDHHC5. Cells were lysed and the levels of the cleaved sensors were measured using Western blots with the anti-FLAG antibody. Statistics: unpaired two-tailed t test. *P < 0.05, **P < 0.01.
Bacterial toxins undergo cleavage at the cell surface, while E-cadherin and IGF-1R were reported to undergo cleavage in the Golgi apparatus (29, 30). This raised the possibility that ZDHHC5 might affect Furin/PC7 in a subcellular localization-dependent manner. We took advantage of a recently published library of PC biosensors (31), which are sensitive to cleavage by any members of the proprotein convertase family, as demonstrated by their inhibition by chloromethyl ketone (Fig. 3C). Each sensor was designed to localize to a specific cellular site and report on the local action of PCs. Upon transfection into RPE-1 cells, we observed cleavage of the sensors at the cell surface, in late endosomes and in glycosylphosphatidylinositol (GPI)-rich domains, but not in the trans-Golgi network (Fig. 3D). Upon ZDHHC5-silencing, we saw significant decreases in biosensor-cleavage in late endosomes and in GPI-rich domains (Fig. 3D). We also observed a decrease in cleavage of the plasma membrane biosensor; however, the cleavage of this PC sensor was low even under control conditions. Therefore, the measured decrease in cleavage might not have reached statistical significance over four experiments. The analysis of the sensors indicates that ZDHHC5 affects the trafficking and activity of PCs through the endocytic pathway and likely at the plasma membrane, which may both involve GPI-rich domains.
ZDHHC5 Controls Presence and Function of Furin/PC7 at the Cell Surface.
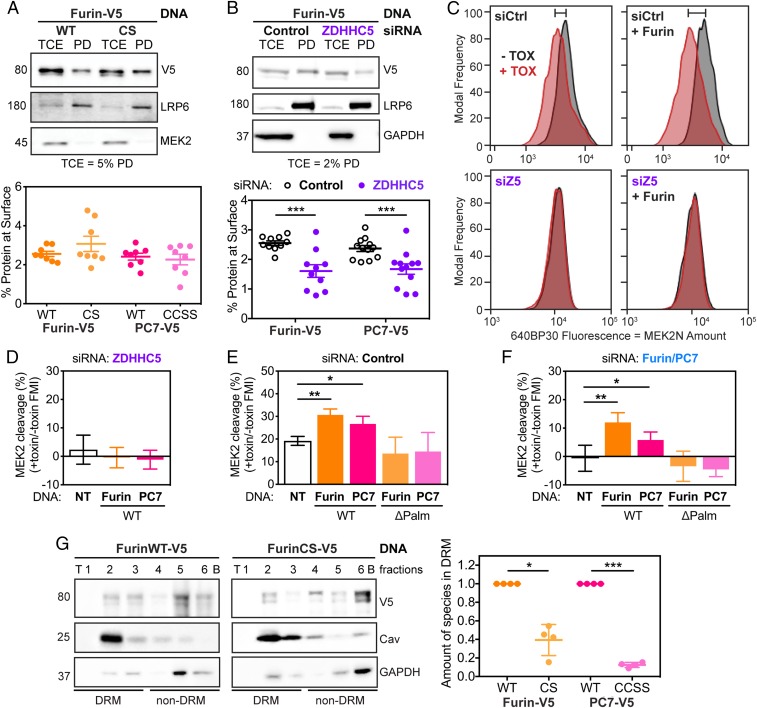
We next sought to understand why Furin/PC7-mediated toxin cleavage is impaired in the absence of ZDHHC5. It has previously been shown that the palmitoylation-deficient mutant of PC7 reaches the cell surface normally (32). Using surface biotinylation on ectopically expressed enzymes, we could confirm this observation and extend it to Furin (Fig. 4A and SI Appendix, Fig. S5A).
Fig. 4.
Palmitoylation-deficient mutants of Furin and PC7 are not active in cleaving anthrax toxin because they do not preferentially reside in microdomains. (A) RPE-1 cells were transfected with WT or palmitoylation-deficient mutants of Furin (shown) or PC7 (in SI Appendix, Fig. S5A). The cells were biotinylated for 30 min and then quenched, washed, and lysed. The surface fraction was pulled down using streptavidin beads. SDS/PAGE was performed on TCE and pull-down (PD) fractions with the percent of TCE shown. Statistics: unpaired two-tailed t test. (B) As in A, RPE-1 cells were transfected with V5-tagged Furin (shown) or PC7 (shown in SI Appendix, Fig. S5B) after being silenced for ZDHHC5 or not (Control). Then, the surface proteins were labeled with biotin and isolated. Statistics: unpaired two-tailed t test. (C) Analytical flow cytometry was performed as described in Fig. 1 but with Furin overexpression (using an antibody against V5 to select for transfected cells) or not in cells silenced for Control (Ctrl) or ZDHHC5 (Z5). The shift in MEK cleavage between no toxin (−TOX, black) and toxin (+TOX, red) is noted by the bar on top of the graph. Quantification of recomplementation flow cytometry peaks as in C with overexpression of both Furin and PC7, WT or palmitoylation-deficient mutants (ΔPalm), in ZDHHC5- (D), Control- (E), and F/PC7- (F) silenced cells. Statistics: paired two-tailed t test on the original data. (G) OptiPrep ultracentrifugation gradients were used to probe presence of Furin, WT or CS mutant in DRMs. Representative blot with DRM control (caveolin 1) and non-DRM control (GAPDH) for Furin (shown) or PC7 (shown in SI Appendix, Fig. S5G). Statistics: ratio paired two-tailed t test on the original data. *P < 0.05, **P < 0.01, and ***P < 0.001.
We also investigated the effect of ZDHHC5 expression on the surface abundance of Furin and PC7. Upon ZDHHC5-silencing, we saw a substantial decrease in the amounts of both proteases by surface biotinylation (Fig. 4B and SI Appendix, Fig. S5B). To be more quantitative, we turned to flow cytometry, relying on antibodies recognizing Furin or PC7 at the surface in nonpermeabilized cells (SI Appendix, Fig. S5D). With this method, in cells silenced for ZDHHC5 compared with control cells, we also observed a significant decrease in the surface amount of Furin and PC7 (SI Appendix, Fig. S5E). The effect of ZDHHC5 on surface expression is specific to only a subset of surface proteins because the transferrin and LRP6 receptors were unaffected by ZDHHC5 silencing (SI Appendix, Fig. S5C). This observation is consistent with our mass spectrometry experiments showing that some, but not all, proteins are differentially regulated at the surface upon ZDHHC5 knockout (SI Appendix, Fig. S1).
The observed lower surface abundance of Furin and PC7 can be due to reduced delivery or increased endocytosis. To test the latter, we used flow cytometry and found that Furin endocytosis was more rapid than that of PC7, but that entry of both enzymes was independent of ZDHHC5 expression (SI Appendix, Fig. S5F). Thus, the lower Furin and PC7 surface expression upon ZDHHC5 silencing must be due to reduced delivery to the plasma membrane. Furthermore, the fact that transport of cysteine mutants was unaltered, while transport of the WT protein upon ZDHHC5 silencing was reduced, suggests that ZDHHC5 affects transport, but not through direct palmitoylation of Furin and PC7.
We used to our advantage the fact that the palmitoylation-deficient mutants were properly delivered to the plasma membrane to test whether palmitoylation has an effect on toxin cleavage. As a read out, we applied the quantitative analytical flow cytometry MEK2 cleavage assay. Overexpression of Furin or PC7 in control cells led to an increase in MEK2 cleavage, but this increase was not observed in ZDHHC5-silenced cells (Fig. 4 C–E). Consistent with these observations, overexpression of the palmitoylation-deficient mutants of either enzyme also did not lead to an increase in MEK2 cleavage (Fig. 4E). These mutants equally failed to restore cleavage activity when expressed in cells silenced for endogenous Furin/PC7, whereas expression of the WT enzymes did (Fig. 4F). Thus, the action of anthrax toxin is exquisitely dependent on the presence of palmitoylated Furin/PC7 at the cell surface.
Palmitoylated Furin/PC7 Preferentially Reside in Microdomains.
The combined observations that palmitoylation of Furin/PC7 is not required for plasma membrane targeting nor for the enzymatic activity per se, but is required for PA cleavage to occur pointed toward a role of palmitoylation in Furin/PC7 localization at the cell surface. Interestingly, both aerolysin and anthrax PA have been found to localize to specific plasma membrane domains, in particular using association with detergent-resistant membranes (DRMs) as a read out (33). For aerolysin, this localization is due to its binding to GPI-anchored proteins and facilitates oligomerization (34). For anthrax toxin, it was found that PA63, but not PA83 nor the uncleavable mutant of PA83, preferentially associated with DRMs (35). When drugs, such as β-methylcyclodextrane (MCD) or filipin, which disrupt microdomains, were applied to cells, association with DRMs was impaired (35). Careful analysis of these published data reveal that following filipin treatment of cells, PA83 actually remained largely uncleaved, a feature that was not picked up at that time (35). In the experiments using MCD, the precleaved PA63 was added to the cells, thus the cleavage step was not analyzed (35). The presence of PA63 in DRMs was then interpreted as “moving into microdomains after cleavage.” The present study offers a different interpretation, which is that PA63 is observed in microdomains because that is where cleavage by convertases occurs.
Consistent with this, we found that WT Furin (Fig. 4G) or PC7 (SI Appendix, Fig. S5G) associate with detergent-resistant domains. This association was consistently decreased with the palmitoylation-deficient mutants. Palmitoylation-dependent association with DRMs has been observed for a variety of proteins (36).
Taking these data together, we find that ZDHHC5 mediates trafficking to the cell surface and palmitoylation of Furin and PC7, ensuring the presence of these enzymes in specific microdomains. Toxins, such as anthrax PA or proaerolysin, also concentrate in microdomains, increasing the probability of an encounter with their surface-processing enzymes, allowing their cleavage and intoxication.
Discussion
During infection, bacteria and viruses are dependent on a plethora of host mechanisms to promote infection and spreading. This is why host–pathogen interaction studies have significantly contributed to mammalian cell biology. The anthrax toxin is a textbook hijacker of the endocytic pathway (12). Its cellular entry requires binding to specific receptors, which themselves need to undergo sequential posttranslational modifications to ensure uptake not only of PA but also the enzymatic subunits LF and EF. These receptor modifications include palmitoylation, phosphorylation, and ubiquitination (25, 37). Once removed from the cell surface, the toxin makes use of the multivesicular body biogenesis pathway to make its way into the lumen of intraluminal vesicles (38). This allows both long-term action of the toxin as well as delivery of LF to distant cells through the exosomal pathway (39).
The present work once more highlights how anthrax toxin exquisitely depends on very specific features of its target cells. For a long time, PA has been known to rely on the cleavage at the cell surface by members of the proprotein convertase family, such as Furin and PC7. When discovered, this finding was surprising because the vast majority of these convertases have been reported to reside inside the cell. Thus, PA cleavage was proposed to depend on a minute population of Furin/PC7 in transit at the cell surface, as it is undergoing recycling back to the Golgi apparatus (14). We here confirm that a minor population of Furin or PC7 reaches the cell surface. The requirement for proprotein convertase cleavage at the host cell surface was subsequently extended to other toxins and viruses (22, 29, 40). We now find that for PA cleavage to occur, not only must Furin/PC7 be localized at the plasma membrane, but these transmembrane proteases must be S-palmitoylated by ZDHHC5 in their cytosolic domains.
ZDHHC5 is one of the more studied palmitoyl transferases, and possibly one the most abundant ZDHHCs (7). ZDHHC5 appears to play an important role in the brain because mice lacking the enzyme have altered learning and memory abilities (41). Reported substrates include: stress-regulated exon of voltage-activated potassium channel, STREX BK (42); postsynaptic density protein 95, PSD-95 or DLG4 (6, 41); somatostatin receptor 5, SSTR5 (43); flotillin-1/2, FLOT1/2 (44); glutamate receptor interacting protein 1, GRIP1 (7); sodium/calcium exchange 1, NCX1 (45, 46); phospholemman, PLM (47, 48); δ-catenin (5); enhancer of zeste homolog 2, EZH2 (49); and sphingosine-1-phosphate receptor 1, S1PR1 (50). While ZDHHC5 mediated-palmitoylation may affect substrate trafficking (6, 7), it may also indirectly affect these substrates by influencing endosomal transport and function (1, 3, 4). In our work, we find evidence of both those roles of ZDHHC5, especially in assisting proteins to reach the cell surface. These proteins seem to be enriched in endocytic and autophagic pathways, which is also consistent with the observation that ZDHHC5 is involved in massive endocytosis, or MEND (47). One interesting ZDHHC5-regulated protein that arose in our analysis was SQSTM1, which interacts with adaptors on endosomes to control their movement in the perinuclear area (51). After the canonical four transmembrane domains that contain the active site, ZDHHC5 has a long, mostly unfolded, cytoplasmic domain (500 amino acids) proposed to mediate substrate interaction (7, 41, 48, 52). This cytoplasmic tail contains two LC3-interacting region domains (53), consistent with a role in autophagy and SQSTM1 connection. Interaction with substrates has also been found to dependent on phosphorylation of the substrate (54). Because Furin has been shown to be phosphorylated (55, 56) and PC7 has one proposed phosphorylation site (57) in its cytoplasmic tail, such a regulatory mechanism could also operate for ZDHHC5-mediated modification of PCs.
In this work, we found that ZDHHC5 affects the targeting of Furin and PC7 to the plasma membrane. This trafficking alteration is likely due to a general effect of ZDHHC5 on trafficking in the Golgi–plasma membrane–endosomes circuit, because palmitoylation-deficient Furin and PC7 were not altered in their transport to the surface. These PC mutants were, however, severely altered in their ability to process PA. The absence of the palmitate modification of these proteases does not affect the cleavage capacity per se because ZDHHC5 silencing did not prevent processing of the known Furin/PC7 targets E-cadherin and IGF-1R (27, 29). Palmitoylation has been reported to target a variety of substrates to specific membrane subdomains (10, 58), possibly lipid rafts (36). We also find that palmitoylation of Furin/PC7 allows their partitioning into specific membrane domains, consistent with our observation that the GPI-domain–specific PC biosensor is sensitive to ZDHHC5 silencing. Interestingly, we have previously observed that PA63 also associates with DRMs, in contrast to uncleaved PA83, which associated with detergent-soluble membranes (35). Our current observations that palmitoylation of Furin/PC7 allows microdomain association and is necessary for efficient protein cleavage led us to reinterpret the earlier data to posit that the cleavage of PA83 actually occurs in these microdomains, leading to the detection only of PA63. This association would also allow the concentration of PA63 in these domains, favoring the oligomerization step, as observed for many pore-forming toxins (35). Therefore, ZDHHC5-mediated palmitoylation of Furin/PC7 leads to the association of these enzymes with plasma membrane subdomains, favoring the encounter between the low-abundance toxin and the equally low-abundant enzyme.
Being a proprotein convertase, Furin is necessary for the maturation of a variety of host proteins but also bacterial and viral proteins (59). Therefore, several studies have focused on the identification of Furin-inhibitors to prevent cellular intoxication or viral infection (60). These inhibitors are, however, likely to have severe side effects due to the inhibition of host cell target processing, such as for E-cadherin. Our study suggests a possibly more specific alternative to block pathogenic agents that depend on cell surface processing by Furin or PC7, which is ZDHHC5.
Materials and Methods
Cell Lines.
RPE-1 (CVCL_4388), HeLa (CVCL_0030), and haploid HAP1 (CVCL_Y019) cells were used in this study. RPE-1 and HAP1 cells were grown in DMEM GlutaMAX (Gibco 31966) supplemented with 10% FBS (Pan Biotech) and 2 mM antibiotics (P/S: penicillin and streptomycin). HeLa cells were maintained in MEM (Sigma-Aldrich M4655) supplemented with 10% FBS, 2 mM l-glutamine (Gibco 25030081), and 1× MEM Non-Essential Amino Acid Solution (Gibco 11140035) and 2 mM antibiotics (P/S). HAP1 cells were obtained from the Horizon Discovery Group and the ΔZDHHC5 HAP1 cells contained a 10-bp deletion in exon 2. RPE-1 ZDHHC5 knockout (ΔZDHHC5) cells were constructed using CRISPR/Cas9 with guide RNA 5′-AAGGATACGTGACAGCCGTG-3′. Transfected cells were singly sorted into 96-well plates and knockout clones were detected by Western blot. The resultant clone used throughout the study (#16) contains a homozygous 1-bp insertion in exon 5 that leads to premature termination 35 bases downstream (c.549_550insG/p.Ala184GlyfsX35).
Gene Silencing and Overexpression.
Genes were silenced using siRNAs obtained from Qiagen targeting (all 5′ to 3′): ZDHHC5: ACCACCATTGCCAGACTACAA, PC2: AAGGTTATGGTCAATCCCAAA, Furin: 1-CCCGAGGATGACGGCAAGACA and 8-TTCCCTGTCCCTCTAAAGCAA, PC7: CAGCAAGTACGGATTCATCAA, PC7: 1-TAGCTATGACCTCAACTCTAA and 8-CAGGAGCGCATCTCAATGGAA, and viral glycoprotein VSV-G (as negative control): ATTGAACAAACGAAACAAGGA. For Furin and PC7, two different siRNAs were used because one was outside the coding sequence for recomplementation purposes. Silencing was performed for 72–96 h using Lipofectamine RNAiMAX (Thermo Fisher Scientific 13778150) or INTERFERrin (Polyplus 409-10) following the manufacturer’s protocol. Silencing efficiency was checked via Western blot and qPCR.
Plasmids were constructed using Gateway cloning (Thermo Fisher Scientific) for PC2, PC7, and E-cadherin or TOPO cloning (Thermo Fisher Scientific) for Furin. PC-sensor plasmids were a gift from the Constam laboratory, Ecole Polytechnique Federale de Lausanne, Lausanne, Switzerland. Point mutations to change or insert residues were performed using QuikChange XL site-directed mutagenesis kit (Agilent Technologies) or Q5 Site-directed mutagenesis kit (New England Biolabs). Proteins were expressed in RPE-1 cells for 24–48 h using FuGENE 6 HD Transfection Reagent (Promega E2691) following the manufacturer’s protocol.
Antibodies.
Primary antibodies used in this study that are commercially available include: rabbit anti-MEK2 (Santa Cruz Biotechnology sc-523, AB_2281672), goat antiprotective antigen from Bacillus anthracis (List Biological Laboratories #771B), mouse anti-V5 (Thermo Fisher Scientific R960-25, AB_2556564), rabbit or goat anti-Furin (Thermo Fisher Scientific PA1-062, AB_2105077; R&D Systems AF1503), rabbit anti-PC7 (Cell Signaling Technology D4I5G #19346), rabbit anti-ZDHHC5 (Sigma-Aldrich HPA014670, AB_2257442), mouse anti-GAPDH (Sigma-Aldrich G8795, AB_1078991), mouse anti–α-tubulin (Sigma-Aldrich T5168, AB_477579), mouse antitransferrin receptor (Thermo Fisher Scientific 13–6800, AB_2533029), rabbit anticlathrin heavy chain (Santa Cruz Biotechnology sc-9069, AB_2083184), rabbit anti-Climp63 (Bethyl Laboratories A302-257A, AB_1731083), rat anti–HA-HRP (Roche Diagnostics 12013819001, AB_390917), mouse anti–IGF-1Rβ (Santa Cruz Biotechnology F-1 sc-390130), mouse anti-FLAG M2 (Sigma-Aldrich F1804, AB_262044), rabbit anti-LRP6 (Cell Signaling Technology C5C7 #2560, AB_2139329), rabbit anticaveolin 1 (Santa Cruz Biotechnology sc-894, AB_2072042), rabbit anti-CMG2 (Proteintech Group 16723–1-AP, AB_2056741), and goat anti-TEM8 (Sigma-Aldrich SAB2501028, AB_10611834). Primary antibodies that were homemade include: anti-MEK1 raised in rabbit (35), antiaerolysin raised in chicken (33), and anti-LF raised in rabbit (35). Secondary antibodies for Western blotting include anti–mouse-HRP (GE Healthcare NA931, AB_772210), anti–rabbit-HRP (GE Healthcare NA934, AB_772206), anti–goat-HRP (Sigma-Aldrich A5420, AB_258242), and anti–chicken-HRP (Sigma A-9046, AB_248432). For immunoprecipitation, mouse anti-V5 agarose affinity gel (Sigma-Aldrich A7345; AB_10062721) was used. For flow cytometry, fluorescence secondary antibodies used include goat anti-mouse Alexa Fluor 488 (Thermo Fisher Scientific A11029; AB_2534088), donkey anti-rabbit Alexa Fluor 568 (Thermo Fisher Scientific A10042; AB_2534017), and donkey anti-rabbit Alex Flour 647 (Thermo Fisher Scientific A31573; AB_2536183).
SDS/PAGE and Western Blots.
Cells were normally lysed in immunoprecipitation buffer [IPB: 0.5% Nonidet P-40, 500 mM Tris pH 7.4, 20 mM EDTA, 10 mM NaF, 2 mM benzamidin, and a Roche mini-protease inhibitor mixture tablet (PI tab; Sigma-Aldrich 11836153001)] and spun at 5,000 × g for 3 min to eliminate the DNA. Exceptionally, cells transfected with the PC-sensors were lysed in PBS supplemented with 1% Triton X-100, 1 mM EDTA, and 1 PI tab, and spun at 21,000 × g for 10 min. To inhibit PC-sensors, cells were incubated with decanoyl-RVKR-chloromethyl ketone (Enzo Life Sciences ALX-260-022) before lysis. DNA-free lysates were quantified using Bicinchoninic acid (BCA) protein assay (Interchim Uptima 40840A) and denatured by addition of SDS sample buffer with β-mercaptoethanol and incubation for 5–10 min at 95 °C. Samples were migrated on precast Novex 4–20% or 4–12% polyacrylamide gels (Thermo Fisher Scientific), then transferred to Novex nitrocellulose membranes (Thermo Fisher Scientific) using iBlot 2. Blocking and antibody steps were performed using 5% milk in PBST (PBS with 0.5% Tween-20). Primary antibody steps were incubated overnight at 4 °C, while the membranes were incubated with secondary antibodies for 1 h at room temperature, both with gentle shaking. Three to five washes of PBST were performed before developing using the Super Signal West Dura solutions (Thermo Fisher Scientific) and the Fusion Solo chemiluminescence imaging system.
Quantitative Real-Time PCR and RNA-Sequencing.
For quantitative real-time PCR and RNA-sequencing, see SI Appendix.
Toxin Purification.
For toxin purification, see SI Appendix.
Toxin Entry Assays.
Cells at 80–90% confluence were washed two times with minimal media [Glasgow minimal essential media (Sigma-Aldrich G6148) buffered with 10 mM Hepes] at 4 °C and then incubated with toxin (anthrax toxin: 500 ng/mL PA and 50 ng/mL LF; aerolysin Y221G, which does not kill cells, 500 ng/mL) for 30 min to 1 h at 4 °C. For cleaved toxin, PA was incubated for 10 min with 100 μg/mL trypsin then stopped with 1 mg/mL of trypsin inhibitor for 1 min, and added to minimal media as for the uncleaved toxin. After the toxin incubation, cells were washed three times with minimal media and moved to 37 °C. The cells were lysed at the appropriate times shown in the experiments (0–2.5 h for experiments with anthrax toxin showing MEK cleavage; 0–40 min for experiments showing PA or aerolysin cleavage). Samples were run on gels as described above with tubulin or GAPDH as loading controls. For comparison purposes, quantified values were normalized to time 0 (longer MEK experiments) or within each experiment (shorter PA or aerolysin experiments). For all representative Western blots, MEK1 or MEK2 has a molecular mass of 45 kDa and GAPDH has a molecular mass of 37 kDa. The molecular weight of the SDS-resistant PAoligo or aerooligo is not defined as it is above the top marker in SDS/PAGE.
Live-Cell Cytotoxicity Imaging.
Complete DMEM GlutaMax media supplemented with aerolysin toxin (either proaerolysin or precleaved aerolysin) at 25 ng/mL and 1 drop per milliliter of NuncGreen Dead 488 ReadyProbes Reagent (Thermo Fisher Scientific R37109) was added to RPE-1 WT or ΔZDHHC5 cells at 30–50% confluence in PerkinElmer CellCarrier Ultra 96-well plates. Plates were put directly in the PerkinElmer Operetta CLS in which brightfield and green fluorescent (excitation BP460-490 and emission BP500-550) channels were initialized at the right focal plane using the 20× Plan Apochromat objective. Five to six fields were imaged per well once every 12–15 min over 5–6 h. CellProfiler (61) and Knime were used to quantify and export the data for each field, well, and time-point. For the statistics, the cells in the fields for each well were added together and wells for the same condition were averaged over their replicates.
SILAC Cell Surface Protein Pull-Down Coupled to Mass Spectrometry.
For SILAC cell surface protein pull-down coupled to mass spectrometry, see SI Appendix.
S-Palmitoylation Methods.
Acyl-RAC and 3H-labeled palmitate incorporation assays were performed as previously described with a few minor modifications (19). Briefly, for Acyl-RAC, cells were lysed in buffer A (25 mM Hepes, 25 mM NaCl, 1 mM EDTA, 1 PI tab) supplemented with 1.5% Triton X-100. Disulfide bonds were broken during a 30-min incubation with 10 mM TCEP. To block free SH groups, the sample was diluted in two volumes of blocking buffer (100 mM Hepes, 1 mM EDTA, 2.5% SDS, and 1 PI tab) with 1.5% S-methyl methanethiosulfonate (MMTS) and incubated for 4 h at 40 °C. Subsequently, proteins were acetone precipitated, the pellet was thoroughly washed (five times) in 70% acetone, and resuspended in binding buffer (100 mM Hepes, 1 mM EDTA, 1% SDS, and 1 PI tab). To cleave the thioester bonds in S-acylated cysteines and capture the corresponding proteins, samples were treated (0.5 M final concentration hydroxylamine in 1 M Tris) or not (1 M Tris) and incubated with 10% (wt/vol) Thiopropyl Sepharose 6B beads (Sigma GE17-0420-01) overnight with rotation. The beads were washed with binding buffer and the proteins were eluted by addition of SDS sample buffer with β-mercaptoethanol and incubation for 5–10 min at 95 °C. Finally, samples were run on SDS/PAGE and analyzed by Western blotting. Flotillin-1, Caveolin 1, or Climp63 were used as positive controls, while Clathrin heavy chain (HC) was used as a negative control.
For 3H-palmitate labeling, cells were starved for 1 h in minimal media and then incubated for 2 h in minimal media supplemented with 200 µCi/mL 3H palmitic acid [9,10-3H(N)] (American Radiolabeled Chemicals ART 0129). Cells were then lysed as usual and lysates were immunoprecipitated with antibodies against the protein of interest. After immunoprecipitation, proteins were eluted off the beads as described above. Samples were divided for Western blotting (one fourth) and gel fixation (three fourths). Gels were exposed to fixing solution (25% isopropanol and 10% acetic acid in H2O), followed by a 30-min incubation with signal enhancer Amplify NAMP100 (Amersham) and gel drying. Dried gels were exposed to Hyperfilm MP (Amersham) film for 10 d to 6 wk. In some cases, 1 M hydroxylamine (pH 7.4) was added and samples were incubated at room temperature for 5 min before loading on SDS/PAGE. Western blots were done in parallel to show the efficiency of the immunoprecipitation.
For nonradioactive metabolic labeling followed by click chemistry, please see SI Appendix.
Coimmunoprecipitation.
Coimmunoprecipitations were performed as described previously (19). Briefly, RPE-1 cells were lysed normally (as described above). A tenth of the lysate was taken as TCE control and the rest was added to washed Protein G-coupled Sepharose beads (Sigma-Aldrich GE Healthcare 17-0618-01) for preclearing (30 min). The precleared lysate was added to freshly washed Protein G beads containing anti-ZDHHC5 antibody for an overnight incubation at 4 °C. Proteins were eluted off the beads and divided for Western blots for direct immunoprecipitation (one eighth) and coimmunoprecipitation (seven eighths). Normalization of quantified coimmunoprecipitation amounts considered both V5 expression level and ZDHHC5 immunoprecipitation efficiency.
Cell Surface Protein Pull-Down.
Surface biotinylation assays were performed as described previously (62). Briefly, cells were shifted to ice and incubated for 30 min with cold biotinylation solution (Thermo Fisher Scientific EZ-link Sulfo-NHS 21327) diluted in PBS (0.17-mg/mL final concentration), then quenched three times with cold 100 mM NH4Cl. Cells were lysed as normally and about 5% of the DNA-cleared lysate was taken as a TCE control. The rest of the lysate was incubated with prewashed streptavidin-coupled beads (Sigma S1638) overnight at 4 °C. Finally, beads were washed and proteins were eluted from beads as above. LRP6 was used as a control for the surface fraction while GAPDH or MEK2 were used as intracellular controls.
Analytical Flow Cytometry.
Cells were treated as described above for the anthrax toxin entry assays, except that instead of lysing the cells, at 1.5-h posttoxin treatment, they were harvested by trypsinization. Cells were washed two times with FACS buffer (PBS/EDTA with 1% FBS), and then once again in PBS. The cells were incubated with 1/1,000 diluted CellTrace violet (Thermo Fisher Scientific L34955) in PBS to stain for dead cells. Cells were subsequently washed in FACS buffer and fixed using 3% paraformaldehyde (PFA) for 20 min at room temperature. Afterward, cells were washed twice more in FACS buffer and permeabilized with 0.1% Triton X-100 for 5 min. Exceptionally, for distinguishing surface fraction from internalized fractions, cells were permeabilized with 0.1% saponin instead of Triton X-100. Cells were then incubated with primary (1 h, room temperature) and secondary antibodies (30 min, room temperature) in FACS buffer with 0.1% Triton X-100. Analytical flow cytometry was performed using an LSRII (Becton Dickinson) instrument and results were analyzed using FlowJo 10.0. For the toxin entry experiments, quantification of the fluorescence median intensity (FMI) of peaks was used to quantify MEK amount, and normalization of +toxin divided by –toxin was done. For overexpression experiments, signal in transfected and nontransfected controls were analyzed.
DRM Fractionation.
DRMs were prepared from RPE-1 cells using OptiPrep (Alere Technologies #1114542) gradients as previously published (35). Briefly, cells were lysed on ice in TNE buffer (25 mM TrisHCl, pH 7.4; 150 mM NaCl; 5 mM EDTA) supplemented with 1% Triton X-100 and applied to the bottom of a gradient consisting of three layers: 100% OptiPrep, 50% OptiPrep and 50% TNE, and 100% TNE. After a 2-h run at 55,000 rpm in a Thermo Fisher Scientific S55-S rotor, six equal fractions were carefully collected from the top. Fractions were precipitated with final concentrations of 2% sodium deoxycholate and 6% trichoroacetic acid and run on SDS/PAGE. Caveolin 1 was used as a control for DRMs while GAPDH and Climp63 were used as controls for non-DRM fractions.
Quantification, Statistics, Figures.
Western blot and film quantifications were done using Fiji (63). Figures were generated and statistics were performed as described in the figure legends using GraphPad Prism or R statistical computing environment (64). Throughout the study, indicated significance asterisks are as follows: *P < 0.05, **P < 0.01, and ***P < 0.001, while unmarked comparisons with control are not significant. All error bars are SEMs. For all areas under curve (AUC) t testing compared with the control, specific P values are reported on each graph. The pathway analysis figure was made using the ClueGo application of Cytoscape v3.7 (65).
Supplementary Material
Acknowledgments
We thank Thomas Jenny for his excellent technical assistance throughout the project; Caroline Desmurget for her assistance in some preliminary experiments; Dr. Laurence Abrami for the CMG2 and TEM8 data; Sylvia Ho for the purification of the proaerolysin toxin; Drs. Pierpaolo Ginefra and Daniel Constam for sharing their proprotein convertase biosensors; and Drs. Maria Eugenia Zaballa and Laurence Abrami for critical reading of this manuscript. Flow cytometry analyses were performed in the Ecole Polytechnique Fédérale de Lausanne Flow Cytometry Core Facility, proteomic analyses in the Proteomics Core Facility, microscopy in the Bioimaging and Optics Facility, and RNA sequencing in the Gene Expression Core Facility. This work received funding from the Swiss National Science Foundation, the Swiss National Centre of Competence in Research Chemical Biology, and the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013/ERC Grant Agreement 340260-PalmERa, to F.G.v.d.G.). O.A.S. was a recipient of a Marie Skłodowska Curie Ecole Polytechnique Fédérale de Lausanne Cofund Postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812588116/-/DCSupplemental.
References
- 1.Zhang L, et al. Proteomic analysis of surface and endosomal membrane proteins from the avian LMH epithelial cell line. J Proteome Res. 2011;10:3973–3982. doi: 10.1021/pr200179r. [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson K, et al. Proteomic analysis of proteins surrounding occludin and claudin-4 reveals their proximity to signaling and trafficking networks. PLoS One. 2015;10:e0117074. doi: 10.1371/journal.pone.0117074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz-Vera J, et al. A proteomic approach to identify endosomal cargoes controlling cancer invasiveness. J Cell Sci. 2017;130:697–711. doi: 10.1242/jcs.190835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breusegem SY, Seaman MNJ. Genome-wide RNAi screen reveals a role for multipass membrane proteins in endosome-to-Golgi retrieval. Cell Rep. 2014;9:1931–1945. doi: 10.1016/j.celrep.2014.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brigidi GS, et al. Palmitoylation of δ-catenin by DHHC5 mediates activity-induced synapse plasticity. Nat Neurosci. 2014;17:522–532. doi: 10.1038/nn.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brigidi GS, Santyr B, Shimell J, Jovellar B, Bamji SX. Activity-regulated trafficking of the palmitoyl-acyl transferase DHHC5. Nat Commun. 2015;6:8200. doi: 10.1038/ncomms9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas GM, Hayashi T, Chiu S-L, Chen C-M, Huganir RL. Palmitoylation by DHHC5/8 targets GRIP1 to dendritic endosomes to regulate AMPA-R trafficking. Neuron. 2012;73:482–496. doi: 10.1016/j.neuron.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain LH, Shipston MJ. The physiology of protein S-acylation. Physiol Rev. 2015;95:341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemonidis K, et al. Substrate selectivity in the zDHHC family of S-acyltransferases. Biochem Soc Trans. 2017;45:751–758. doi: 10.1042/BST20160309. [DOI] [PubMed] [Google Scholar]
- 10.Zaballa M-E, van der Goot FG. The molecular era of protein S-acylation: Spotlight on structure, mechanisms, and dynamics. Crit Rev Biochem Mol Biol. 2018;53:420–451. doi: 10.1080/10409238.2018.1488804. [DOI] [PubMed] [Google Scholar]
- 11.Blaskovic S, Blanc M, van der Goot FG. What does S-palmitoylation do to membrane proteins? FEBS J. 2013;280:2766–2774. doi: 10.1111/febs.12263. [DOI] [PubMed] [Google Scholar]
- 12.Friebe S, van der Goot FG, Bürgi J. The ins and outs of anthrax toxin. Toxins (Basel) 2016;8:E69. doi: 10.3390/toxins8030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon VM, Klimpel KR, Arora N, Henderson MA, Leppla SH. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect Immun. 1995;63:82–87. doi: 10.1128/iai.63.1.82-87.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klimpel KR, Molloy SS, Thomas G, Leppla SH. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc Natl Acad Sci USA. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remacle AG, et al. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J Biol Chem. 2008;283:20897–20906. doi: 10.1074/jbc.M803762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krantz BA, Finkelstein A, Collier RJ. Protein translocation through the anthrax toxin transmembrane pore is driven by a proton gradient. J Mol Biol. 2006;355:968–979. doi: 10.1016/j.jmb.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 17.Dallavilla T, et al. Model-driven understanding of palmitoylation dynamics: Regulated acylation of the endoplasmic reticulum chaperone calnexin. PLOS Comput Biol. 2016;12:e1004774. doi: 10.1371/journal.pcbi.1004774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakkaraju AK, et al. Palmitoylated calnexin is a key component of the ribosome-translocon complex. EMBO J. 2012;31:1823–1835. doi: 10.1038/emboj.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrami L, et al. Identification and dynamics of the human ZDHHC16-ZDHHC6 palmitoylation cascade. eLife. 2017;6:e27826. doi: 10.7554/eLife.27826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanc M, et al. SwissPalm: Protein palmitoylation database. F1000 Res. 2015;4:261. doi: 10.12688/f1000research.6464.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bausch-Fluck D, et al. A mass spectrometric-derived cell surface protein atlas. PLoS One. 2015;10:e0121314. doi: 10.1371/journal.pone.0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrami L, et al. The pore-forming toxin proaerolysin is activated by furin. J Biol Chem. 1998;273:32656–32661. doi: 10.1074/jbc.273.49.32656. [DOI] [PubMed] [Google Scholar]
- 23.van de Loo JW, et al. Biosynthesis, distinct post-translational modifications, and functional characterization of lymphoma proprotein convertase. J Biol Chem. 1997;272:27116–27123. doi: 10.1074/jbc.272.43.27116. [DOI] [PubMed] [Google Scholar]
- 24.van de Loo JW, et al. Dynamic palmitoylation of lymphoma proprotein convertase prolongs its half-life, but is not essential for trans-Golgi network localization. Biochem J. 2000;352:827–833. doi: 10.1042/0264-6021:3520827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abrami L, Leppla SH, van der Goot FG. Receptor palmitoylation and ubiquitination regulate anthrax toxin endocytosis. J Cell Biol. 2006;172:309–320. doi: 10.1083/jcb.200507067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bessonnard S, Mesnard D, Constam DB. PC7 and the related proteases Furin and Pace4 regulate E-cadherin function during blastocyst formation. J Cell Biol. 2015;210:1185–1197. doi: 10.1083/jcb.201503042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khatib AM, et al. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: Importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J Biol Chem. 2001;276:30686–30693. doi: 10.1074/jbc.M101725200. [DOI] [PubMed] [Google Scholar]
- 28.Scamuffa N, et al. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J Clin Invest. 2008;118:352–363. doi: 10.1172/JCI32040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posthaus H, et al. Proprotein cleavage of E-cadherin by furin in baculovirus over-expression system: Potential role of other convertases in mammalian cells. FEBS Lett. 1998;438:306–310. doi: 10.1016/s0014-5793(98)01330-1. [DOI] [PubMed] [Google Scholar]
- 30.Robertson DM, Zhu M, Wu Y-C. Cellular distribution of the IGF-1R in corneal epithelial cells. Exp Eye Res. 2012;94:179–186. doi: 10.1016/j.exer.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginefra P, Filippi BGH, Donovan P, Bessonnard S, Constam DB. Compartment-specific biosensors reveal a complementary subcellular distribution of bioactive furin and PC7. Cell Rep. 2018;22:2176–2189. doi: 10.1016/j.celrep.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Rousselet E, Benjannet S, Hamelin J, Canuel M, Seidah NG. The proprotein convertase PC7: Unique zymogen activation and trafficking pathways. J Biol Chem. 2011;286:2728–2738. doi: 10.1074/jbc.M110.192344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abrami L, Fivaz M, Glauser P-E, Parton RG, van der Goot FG. A pore-forming toxin interacts with a GPI-anchored protein and causes vacuolation of the endoplasmic reticulum. J Cell Biol. 1998;140:525–540. doi: 10.1083/jcb.140.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrami L, van Der Goot FG. Plasma membrane microdomains act as concentration platforms to facilitate intoxication by aerolysin. J Cell Biol. 1999;147:175–184. doi: 10.1083/jcb.147.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abrami L, Liu S, Cosson P, Leppla SH, van der Goot FG. Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J Cell Biol. 2003;160:321–328. doi: 10.1083/jcb.200211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci USA. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milne JC, Furlong D, Hanna PC, Wall JS, Collier RJ. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J Biol Chem. 1994;269:20607–20612. [PubMed] [Google Scholar]
- 38.Abrami L, Lindsay M, Parton RG, Leppla SH, van der Goot FG. Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J Cell Biol. 2004;166:645–651. doi: 10.1083/jcb.200312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abrami L, et al. Hijacking multivesicular bodies enables long-term and exosome-mediated long-distance action of anthrax toxin. Cell Rep. 2013;5:986–996. doi: 10.1016/j.celrep.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klimstra WB, Heidner HW, Johnston RE. The furin protease cleavage recognition sequence of Sindbis virus PE2 can mediate virion attachment to cell surface heparan sulfate. J Virol. 1999;73:6299–6306. doi: 10.1128/jvi.73.8.6299-6306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, et al. DHHC5 interacts with PDZ domain 3 of post-synaptic density-95 (PSD-95) protein and plays a role in learning and memory. J Biol Chem. 2010;285:13022–13031. doi: 10.1074/jbc.M109.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian L, McClafferty H, Jeffries O, Shipston MJ. Multiple palmitoyltransferases are required for palmitoylation-dependent regulation of large conductance calcium- and voltage-activated potassium channels. J Biol Chem. 2010;285:23954–23962. doi: 10.1074/jbc.M110.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kokkola T, et al. Somatostatin receptor 5 is palmitoylated by the interacting ZDHHC5 palmitoyltransferase. FEBS Lett. 2011;585:2665–2670. doi: 10.1016/j.febslet.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Martin BR, Cravatt BF, Hofmann SL. DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J Biol Chem. 2012;287:523–530. doi: 10.1074/jbc.M111.306183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin M-J, et al. Massive palmitoylation-dependent endocytosis during reoxygenation of anoxic cardiac muscle. eLife. 2013;2:e01295. doi: 10.7554/eLife.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuller W, Reilly L, Hilgemann DW. S-palmitoylation and the regulation of NCX1. Channels (Austin) 2016;10:75–77. doi: 10.1080/19336950.2015.1099329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hilgemann DW, Fine M, Linder ME, Jennings BC, Lin M-J. Massive endocytosis triggered by surface membrane palmitoylation under mitochondrial control in BHK fibroblasts. eLife. 2013;2:e01293. doi: 10.7554/eLife.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howie J, et al. Substrate recognition by the cell surface palmitoyl transferase DHHC5. Proc Natl Acad Sci USA. 2014;111:17534–17539. doi: 10.1073/pnas.1413627111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, et al. EZH2 palmitoylation mediated by ZDHHC5 in p53-mutant glioma drives malignant development and progression. Cancer Res. 2017;77:4998–5010. doi: 10.1158/0008-5472.CAN-17-1139. [DOI] [PubMed] [Google Scholar]
- 50.Badawy SMM, Okada T, Kajimoto T, Ijuin T, Nakamura S-I. DHHC5-mediated palmitoylation of S1P receptor subtype 1 determines G-protein coupling. Sci Rep. 2017;7:16552. doi: 10.1038/s41598-017-16457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jongsma MLM, et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell. 2016;166:152–166. doi: 10.1016/j.cell.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacomin A-C, Samavedam S, Promponas V, Nezis IP. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy. 2016;12:1945–1953. doi: 10.1080/15548627.2016.1207016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howie J, et al. Greasing the wheels or a spanner in the works? Regulation of the cardiac sodium pump by palmitoylation. Crit Rev Biochem Mol Biol. 2018;53:175–191. doi: 10.1080/10409238.2018.1432560. [DOI] [PubMed] [Google Scholar]
- 55.Jones BG, et al. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teuchert M, Berghöfer S, Klenk HD, Garten W. Recycling of furin from the plasma membrane. Functional importance of the cytoplasmic tail sorting signals and interaction with the AP-2 adaptor medium chain subunit. J Biol Chem. 1999;274:36781–36789. doi: 10.1074/jbc.274.51.36781. [DOI] [PubMed] [Google Scholar]
- 57.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naumenko VS, Ponimaskin E. Palmitoylation as a functional regulator of neurotransmitter receptors. Neural Plast. 2018;2018:5701348. doi: 10.1155/2018/5701348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rey FA, Lok S-M. Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell. 2018;172:1319–1334. doi: 10.1016/j.cell.2018.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shiryaev SA, et al. Targeting host cell furin proprotein convertases as a therapeutic strategy against bacterial toxins and viral pathogens. J Biol Chem. 2007;282:20847–20853. doi: 10.1074/jbc.M703847200. [DOI] [PubMed] [Google Scholar]
- 61.Kamentsky L, et al. Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perrody E, et al. Ubiquitin-dependent folding of the Wnt signaling coreceptor LRP6. eLife. 2016;5:e19083. doi: 10.7554/eLife.19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R Core Team 2013 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at www.R-project.org/. Accessed February 4, 2018.
- 65.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.