Significance
To regulate the growth and size of organs, cells can use information from their neighbors to modify intracellular mediators of cell proliferation. The intracellular Hippo pathway is a widely utilized nexus for growth control in animals, but its regulation by extracellular signals is not fully understood. We here identify a pathway that regulates organ size in Drosophila, triggered by the transmembrane receptor, the giant protocadherin Fat. We show that the Fat-regulated SH3 domain adaptor protein Dlish binds to and reduces the stability of the growth suppressor Expanded, a known regulator of the Hippo pathway. The destabilization of Expanded by Dlish works in parallel to a previously established pathway in which Dlish increases levels of the growth-stimulating protein Dachs.
Keywords: Dlish, Expanded, Fat, Hippo signaling, Slimb
Abstract
The Drosophila protocadherin Fat controls organ size through the Hippo pathway, but the biochemical links to the Hippo pathway components are still poorly defined. We previously identified Dlish, an SH3 domain protein that physically interacts with Fat and the type XX myosin Dachs, and showed that Fat’s regulation of Dlish levels and activity helps limit Dachs-mediated inhibition of Hippo pathway activity. We here characterize a parallel growth control pathway downstream of Fat and Dlish. Using immunoprecipitation and mass spectrometry to search for Dlish partners, we find that Dlish binds the FERM domain growth repressor Expanded (Ex); Dlish SH3 domains directly bind sites in the Ex C terminus. We further show that, in vivo, Dlish reduces the subapical accumulation of Ex, and that loss of Dlish blocks the destabilization of Ex caused by loss of Fat. Moreover, Dlish can bind the F-box E3 ubiquitin ligase Slimb and promote Slimb-mediated ubiquitination of Expanded in vitro. Both the in vitro and in vivo effects of Dlish on Ex require Slimb, strongly suggesting that Dlish destabilizes Ex by helping recruit Slimb-containing E3 ubiquitin ligase complexes to Ex.
The intracellular domain (ICD) of the giant Drosophila protocadherin Fat reduces cell proliferation in imaginal disc tissues by regulating the Hippo pathway, an effect potentiated by heterophilic binding between Fat and the protocadherin Dachsous (Ds) (1, 2) (Fig. 1B). The Fat ICD increases the activity of NDR family kinase Warts, the Drosophila homolog of vertebrate LATS and the final effector kinase in the Hippo pathway, and decreases the activity of the Warts target Yorkie (Yki), the Drosophila homolog of the vertebrate YAP and TAZ transcriptional coactivators. Active Warts phosphorylates and inhibits Yki by increasing the binding between Yki and its cytoplasmic tethers. In the absence of Fat, Warts activity is reduced, increasing the proportion of Yki that enters the nucleus with its TEAD-family cofactor Scalloped (Sd), driving transcription and overgrowth of imaginal disc epithelia.
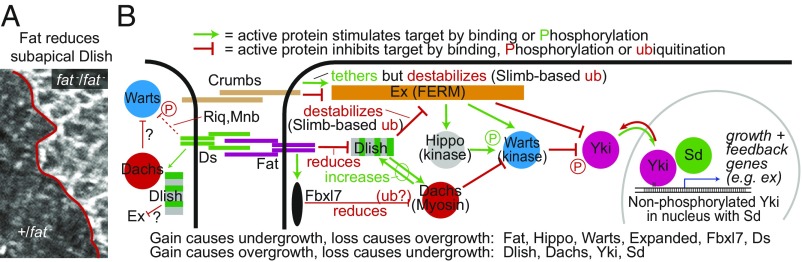
Fig. 1.
(A) Subapical Dlish is increased in wing disc epithelium cells homozygous for fat null mutation (7, 8). (Magnification: 2,400×.) (B) Model of inputs from the Fat, Ds, and Crumbs ICDs into the Hippo growth control pathway; based on this work and others (1, 2, 33). Studies differ on whether Fbxl7 regulates Dachs via ubiquitination (17, 18). Mnb, minibrain; Riq, riquiqui; ub, ubiquitination.
While Fat is one of the best-studied transmembrane regulators of the Hippo pathway, the biochemical pathways that link Fat’s ICD to changes in Warts and Yki activity have not been fully elucidated. The portions of Fat’s ICD that suppress growth lack obvious catalytic or protein-binding motifs and, until recently, binding partners (3–6). However, recent work indicates that Fat’s ICD binds to the cytoplasmic SH3-domain protein Dlish (also known as Vamana), reducing Dlish levels and activity, and thereby regulating a Dlish-binding partner, the atypical type XX myosin Dachs (7, 8) (Fig. 1). Fat, Dlish, Dachs, and Warts are all concentrated at the subapical cell cortex of disc epithelial cells near their adherens junctions, although some subapical Warts is also concentrated at nonjunction sites (9), and for both Dlish and Dachs this depends on the formation of a Dlish-Dachs complex (7, 8). When Fat is lost, the subapical levels of Dlish and Dachs greatly increase, an effect specific to the Fat branch of the Hippo pathway (7, 8, 10) (Fig. 1A). The increased Dachs binds and inhibits Warts by altering its conformation and reducing its levels, thereby inducing Yki-mediated overgrowth (11–14).
Previous evidence suggested that the increased Dlish of fat mutants stimulates growth only by increasing subapical Dachs, rather than through any direct effect on Warts or Yki (7, 8). Unlike Dachs, Dlish does not bind Warts. And while Dlish is necessary and sufficient for the localization and activity of wild-type Dachs, the overgrowth induced by a membrane-targeted Dachs construct is not reduced by the loss of Dlish; concentrating Dachs at the membrane is sufficient to bypass Dlish (7). Instead, Dlish provides a physical link between Dachs and the Fat ICD, along with the DHHC palmitoyltransferase Approximated (App), which can bind Fat and Dachs and palmitoylate Dlish (7, 8, 15, 16). Loss of either Dlish or App disrupts Fat’s ability to regulate Dachs levels and Yki activity. In the simplest view, the Fat ICD binds and inhibits Dlish and App, reducing their ability to regulate Dachs; when Fat is lost, App and palmitoylated Dlish are free to bind Dachs and concentrate it near the subapical cell membrane where it inhibits Warts (2). Dachs is also needed to concentrate the Dlish/Dachs complex in the cortex (7, 8), likely by binding to the actin cytoskeleton or other scaffolds; thus, the reduction of Dachs by the Fat-tethered ubiquitin ligase Fbxl7 (17, 18) provides a parallel route for regulating Dlish and Dachs activity (Fig. 1B). The Ds ICD also binds and recruits Dachs and Dlish (Fig. 1B), but it is uncertain how strongly the endogenous Ds ICD affects the Hippo pathway (2).
While Dachs plays a major role in growth control through its ability to inhibit Warts, we show below that this is not the sole mediator of Dlish activity. Rather, Dlish also regulates a parallel pathway mediated by the growth-inhibiting FERM domain protein Expanded (Ex) (19). Ex regulates the Hippo pathway at multiple levels that are distinct from the Dachs-mediated alterations in Warts conformation (Fig. 1B). Ex binds Hippo, Warts, and the pathway modulators Merlin and Kibra and stimulates the phosphorylation and activity of Warts (20–25); Ex can also bypass Warts by binding and inhibiting Yki (26–28).
Indeed, Fat was originally linked to the Hippo pathway, not through Dachs, but through Ex: loss of Fat decreases Ex levels in the subapical cell cortex (29–32). The Ex decrease is particularly striking as it is at odds with the increased Yki-driven ex transcription caused by loss of Fat, indicating that the effect is posttranscriptional; the effect is also specific for the Fat branch of the Hippo pathway, as increasing Yki activity through other branches increases both ex transcription and Ex protein levels as part of a negative feedback loop (21, 29, 31).
We will show here that mimicking the effects of Fat loss by increasing Dlish levels has a growth-inducing activity that is independent of the Dachs myosin, and thus Dachs-mediated inhibition of Warts. We will show that two of the three SH3 domains of Dlish bind directly to multiple sites in Ex, that Dlish decreases Ex protein levels in wing imaginal discs independently of ex transcription, and that without Dlish the loss of Fat no longer reduces Ex protein levels. Previous studies showed that Ex levels are reduced by ubiquitination mediated by the Ex-binding F-box E3 ubiquitin ligase Slimb and the Skp-Cullin-F-box (SCF) complex, a process stimulated by the Ex-binding transmembrane protein Crumbs (33, 34) (Fig. 1B). We will confirm and extend our previous finding that Dlish binds to Slimb (7) and show that Dlish stimulates the Slimb-dependent ubiquitination of Ex in vitro.
Results
Dlish Can Stimulate Overgrowth Without Dachs.
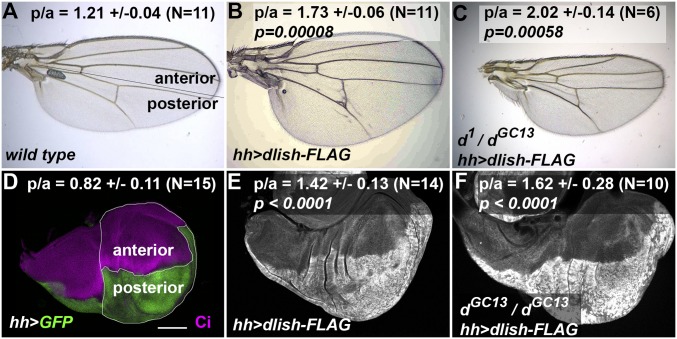
Our previous studies left open the possibility that Dlish might affect growth independently of Dachs by affecting other components of the Hippo pathway. We therefore tested the effects of driving UAS-dlish-FLAG expression in dachs mutants. Overexpression of dlish in wild-type flies increases Dachs accumulation and causes overgrowth in adult wings and imaginal discs (7, 8). Surprisingly, posterior, hh-gal4-driven overexpression of UAS-dlish-FLAG caused similar overgrowth in hypomorphic d1/dGC13 adult wings (Fig. 2 A–C), and in the prospective blade and hinge regions of null dGC13/dGC13 wing imaginal discs (Fig. 2 D–F). In fact, the increased ratio of posterior to anterior areas was as great as that caused by posterior dlish overexpression in wild-type discs. Thus, overexpressed Dlish has an overgrowth-inducing activity that is independent of Dachs.
Fig. 2.
Posterior, hh-gal4-induced overexpression of Dlish causes Dachs-independent overgrowth. (A–C) Adult wings and ratios of posterior to anterior area (p/a) ±SD. (A) Wild type, with line showing subdivision into anterior and posterior (hh-gal4-expressing) regions. (B) hh-gal4 UAS-dlish-FLAG. (C) hh-gal4 > dlish-FLAG in hypomorphic d1/dGC13 background. The p/a ratio is significantly higher in both compared with wild type using a two-tailed Mann–Whitney U test. (Magnification: 50×.) (D–F) Example late third instar imaginal wing discs and average ratios of p/a area ±SD in wing pouch and hinge regions from N discs. (D) hh-gal4 UAS-GFP; posterior marked with GFP (green) and anterior with anti-Ci (magenta); lines show regions measured. (E and F) hh-gal4 UAS-dlish-FLAG (posterior marked with anti-FLAG) in wild type (E) and dGC13/dGC13 (F) wing discs. The p/a ratio is significantly higher in both compared with hh-gal4 UAS-GFP using either a two-tailed Mann–Whitney U test or two-tailed t test. (Scale bar [D–F]: 100 μm.)
Dlish Directly Binds Ex at Multiple Sites.
We next looked for protein-binding partners that could explain the Dachs-independent activity of Dlish. Since we have not detected binding between Dlish and the Hippo pathway candidates Hpo, Warts, Mats, or Yki (SI Appendix, Fig. S1A), we took a more open-ended approach, expressing Dlish-FLAG in S2R+ cells and identifying binding partners by coimmunoprecipitation (co-IP) and liquid chromatography-tandem mass spectrometry (LC-MS/MS). We filtered the raw results (Dataset S1) to remove contaminants isolated by FLAG IP and LC-MS/MS from control cells lacking Dlish-FLAG and those commonly seen in Drosophila IP + mass spectrometry analyses (https://reprint-apms.org) (35). Among the highest-ranked targets (SI Appendix, Table S1), Ex was the only protein known to be directly involved in the Hippo pathway.
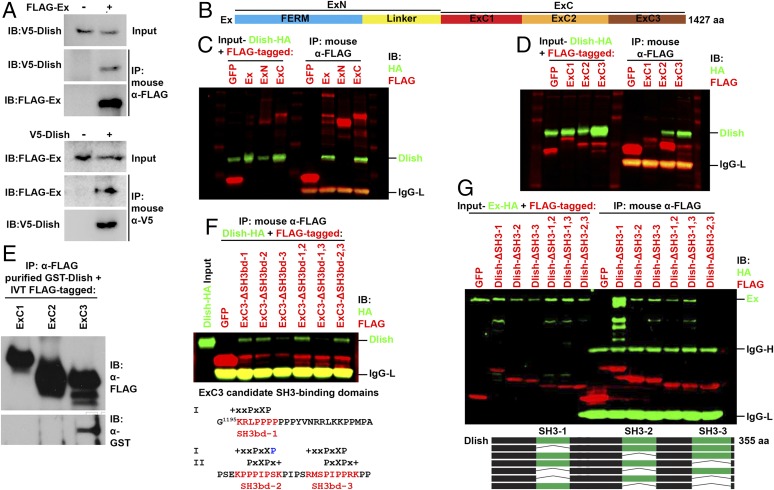
We confirmed strong binding between Dlish and Ex in S2R+ cells by reciprocal co-IP (Fig. 3A and SI Appendix, Fig. S1B). The N-terminal half of Ex (ExN) contains a FERM domain and a linker sequence, while its C-terminal half (ExC) contains several regions whose sequences conform to the consensus for SH3 binding (19), suggestive since Dlish binds other partners via its three SH3 domains. A recent structure-function study arbitrarily subdivided ExC into three regions: ExC1, ExC2, and ExC3 (34) (Fig. 3B). When coexpressed in S2R+ cells, Dlish co-IPs with ExC but not Ex-N (Fig. 3C), and with both ExC2 and ExC3 but not ExC1 (Fig. 3D).
Fig. 3.
Characterization of the binding between Dlish and Ex. (A) Reciprocal co-IP between V5-tagged Dlish and FLAG-tagged Ex made in S2R+ cells. (B) Domain organization of Ex. (C) HA-tagged Dlish co-IPs with full-length Ex-FLAG and ExC-FLAG (aa 654–1427) but not ExN-FLAG (aa 1–653) in S2R+ cells. (D) HA-tagged Dlish co-IPs with ExC2-FLAG (aa 912–1164) and ExC3-FLAG (aa 1168–1427) but not ExC1-FLAG. (aa 654–911). (E) GST-purified Dlish co-IPs with in vitro translated (IVT) FLAG-ExC3 but not FLAG-ExC1 or FLAG-ExC2. (F) Co-IP of HA-tagged Dlish is lost when candidate SH3-binding domains (SH3bd) 1 and 3 are both removed from ExC3-FLAG. Matches of the candidate domains to class I and II SH3-binding domains are shown below; + = basic, X = non-G hydrophobic, x = any, blue = mismatch. (G) Co-IP of HA-tagged Ex is lost when SH3 domains 2 and 3 are both removed from Dlish-FLAG.
The binding to ExC2 and ExC3 is direct. While purified GST-Dlish only co-IPs with in vitro translated ExC3-FLAG (Fig. 3E), it pulls down His-ExC2 and His-ExC3 purified from bacteria (see below, Fig. 6C). Dlish bound to multiple sites within ExC3. ExC3 contains three regions that match the consensus for canonical class I and/or class II SH3-binding domains (36); co-IP of Dlish-FLAG by ExC3 was blocked only by the simultaneous removal of the first and third candidate SH3-binding domains (Fig. 3F). Conversely, two of the three SH3 domains in Dlish can bind Ex. Co-IP of Ex-HA was only blocked when both the second and third SH3 domains of Dlish were removed (Fig. 3G). Similarly, Ex-FLAG from S2R+ cells was pulled down by GST-Dlish-SH3-2 or GST-Dlish-SH3-3, but not GST-Dlish-SH3-1 (SI Appendix, Fig. S1C).
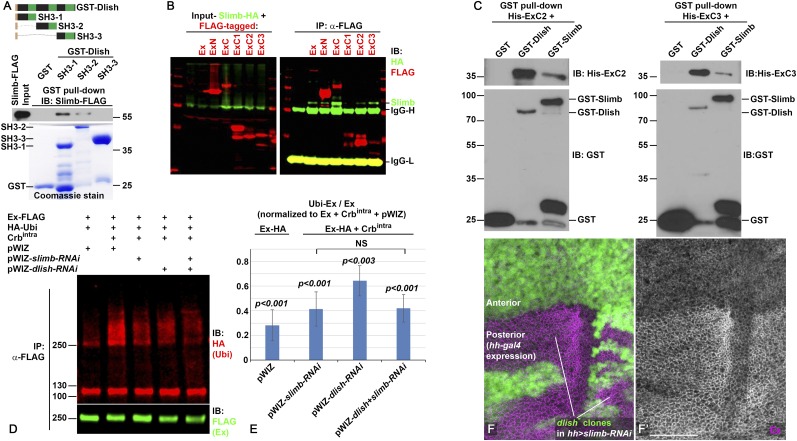
Fig. 6.
Dlish binds Slimb and regulates Ex ubiquitination. (A) Slimb-FLAG from S2R+ cells is pulled down by GST-Dlish-SH3-1 and GST-Dlish-SH3-2 but not by GST-Dlish-SH3-3. (B) FLAG-tagged Ex, ExN, ExC, ExC2, and ExC3 co-IP with Slimb-HA in S2R+ cells. (C) His-ExC2 and His-ExC3 purified from Escherichia coli BL21(DE3) are pulled down by GST-Dlish and GST-Slimb. (D and E) RNAi-mediated depletion of Dlish or Slimb in S2R+ cells reduces Crbintra-stimulated ubiquitination of Ex. (D) Example Western IB of ubiquitination assay; Ubi, ubiqutin. (E) Quantification of five ubiquitination assays. Ratios of ubiquitinated Ex (Ubi-Ex) to total Ex on each IB were normalized to the control Ex-FLAG + Crbintra condition (=1.0). Changes were compared with the control using paired two-tailed t tests; NS, not significant. Error bars show ±SD. (F) The increased subapical Ex (magenta, gray in F′) in the posterior of hh-gal4 UAS-slimb-RNAi wing discs is not affected by homozygous dlish4506 clones (loss of green). (Scale bar: 50 μm.)
Ex also stabilized Dlish in S2R+ cells in a manner largely consistent with the binding interactions shown above: Dlish stabilization was strongest with coexpression of Ex, Ex-C, and Ex-C3 (SI Appendix, Fig. S2 A–D). Since Dlish and Ex are both concentrated in the subapical cell cortex of imaginal disc cells (7, 8, 19), we further tested whether Ex might tether or stabilize a subset of the cell’s Dlish in this region. Driving expression of UAS-ex-GFP in the posterior of wing discs with hh-gal4 weakly increased posterior Dlish in wing discs, a difference not observed in wild-type discs (SI Appendix, Fig. S2 E–G). However, clonal loss of Ex did not obviously reduce Dlish levels (SI Appendix, Fig. S2H), suggesting that, at endogenous levels, binding between Dlish and Ex has a role beyond the stabilization of Dlish.
Dlish Reduces Ex Levels in Vivo.
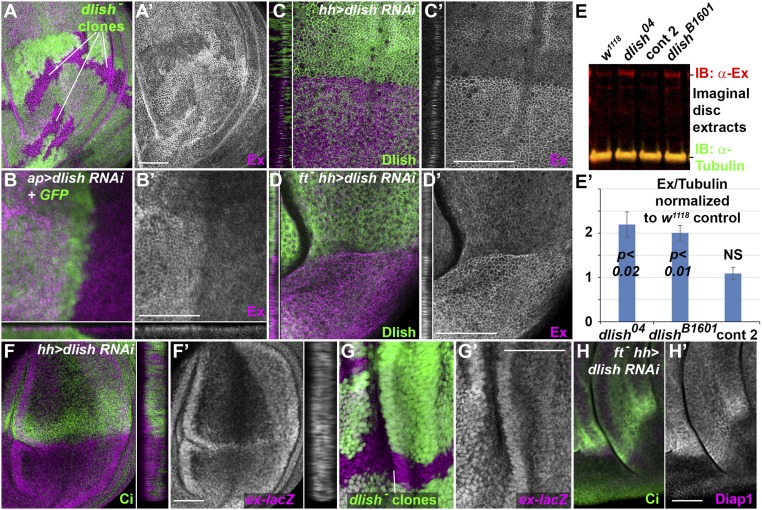
We next tested whether the Ex in imaginal discs responded to changes in Dlish and found that eliminating or reducing Dlish increased anti-Ex staining in the subapical cell cortex. For instance, when clones of homozygous dlish─/dlish─ cells were generated in dlish+/dlish− wing discs using FRT/FLPase-mediated mitotic recombination, Ex levels were higher in the dlish−/dlish− cells; this was especially obvious in the wing pouch region of the wing discs (Fig. 4A). Subapical Ex was also increased by dorsal, ap-gal4-driven and posterior, hh-gal4-driven expression of dlish RNAi (Fig. 4 B and C).
Fig. 4.
Loss of Dlish increases Ex in wing imaginal discs. (A) Increased subapical Ex (magenta, grey in A′) in dlish4506 homozygous clones (loss of green). (B–D) Increased subapical Ex (magenta, grey in B′–D′) after driving UAS-dlish-RNAi and UAS-GFP dorsally with ap-gal4 (B), or posteriorly with hh-gal4 in wild type (C) or ftfd homozygotes (D), marked by loss of Dlish (green). Cross-sections show subapical regions. (E) Example Western IB (E) and quantification from multiple IBs (E′) of Ex levels in extracts from w1118 control, phenotypically wild-type control 2 (Bloomington 51324) and dlish mutant discs. Quantifications were normalized to w1118 lanes from the same IB, and average changes (three extracts for each) compared using paired two-tailed t tests; NS, not significant; error bars = ±SD. (F and G) ex-lacZ expression (anti-βgal, magenta, gray in F′ and G′) is not altered by posterior, hh-gal4-driven UAS-dlish-RNAi (F, posterior marked by absence of green Ci; detail shows cross-section through disc) or by dlish04 mutant clones (G, marked by loss of green). (H) Expression of the Yki target Diap1 (magenta, grey in H′) is decreased by posterior, hh-gal4-driven UAS-dlish-RNAi in ftfd homozygote. (Scale bars: 50 μm.)
Loss of Dlish increased not only subapical Ex, but also total Ex levels in imaginal discs. We measured Ex in Western immunoblots (IBs) from wing imaginal disc extracts, normalizing Ex levels to Tubulin levels to control for changes in cell number (Fig. 4E). Compared with the levels from two independent sets of control discs, Ex levels from two different dlish mutant lines were roughly twofold higher (Fig. 4E′).
The Ex-suppressing activity of Dlish is posttranscriptional, as Dlish had opposite effects on Ex protein levels and reporters of ex transcription. As noted in the Introduction, ex transcription in imaginal discs is increased during Yki-induced overgrowth as part of a negative feedback loop. In a wild-type background, loss of dlish causes very weak undergrowth and reduced Yki activity, and thus levels of the ex-lacZ reporter of ex transcription are unchanged (Fig. 4 F and G) or very slightly reduced (8), showing Dlish had opposite effects on ex transcription and Ex protein. In a fat mutant background, loss of dlish reverses the overgrowth and increased Yki activity normally caused by loss of fat (7, 8), as shown by the reduced expression of the Yki activity reporter Diap1 (Fig. 4H). Nonetheless, subapical Ex levels were increased by posterior, hh-gal4-driven expression of UAS-dlish-RNAi in a homozygous fat mutant wing disc (Fig. 4D).
Dlish overexpression also has disparate effects on ex transcription and Ex protein accumulation. Posterior, hh-gal4-driven expression of UAS-dlish-FLAG causes overgrowth and increased Yki activity (7, 8), and so increases the level of ex transcription as indicated by the ex-lacZ reporter (SI Appendix, Fig. S3A). Nonetheless, Ex protein levels are largely unaltered (SI Appendix, Fig. S3 B and C). In contrast, posterior overexpression of a membrane-tethered form of Dachs (Dachs-CAAX), although causing overgrowth similar to that caused by UAS-dlish-FLAG (7), increased posterior Ex (SI Appendix, Fig. S3 D and E).
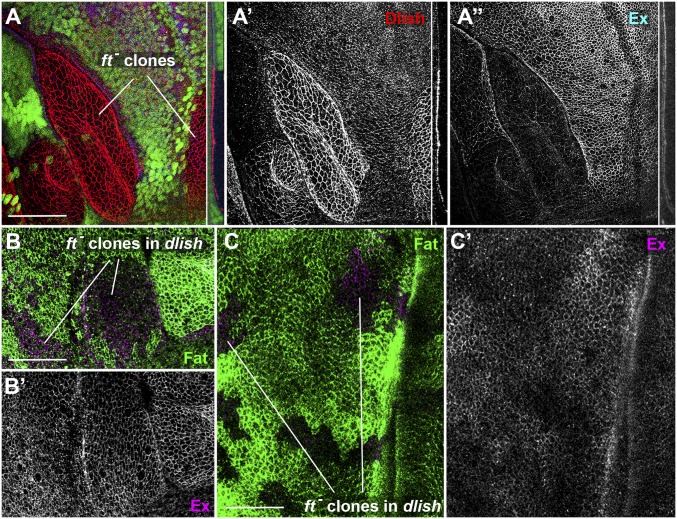
The disparate effects of Dlish on ex transcription and Ex protein are thus reminiscent of those caused by loss of Fat, which increases Yki activity and thus ex transcription, but often decreases subapical Ex protein in wing discs (Fig. 5A) (29–32). Loss of Fat causes a strong accumulation of subapical Dlish (7, 8) (Fig. 5A), and we hypothesize that the increased Dlish may help destabilize Ex protein. Consistent with this hypothesis, we found that Ex was not obviously decreased in fat clones when they were generated in dlish mutant wing discs, and in some cells Ex increased slightly (Fig. 5 B and C).
Fig. 5.
The decrease of Ex in ft mutant clones requires Dlish. (A) Homozygous ftfd clones, marked by absence of green marker, in wing imaginal disc, showing increased Dlish (red, gray in A′) and decreased Ex (blue, gray in A″). Details show subapical cross-sections. (B and C) Homozygous ftfd clones, marked by absence of green Fat, in dlishY003/dlishB1601 mutant wing discs. Ex (magenta, gray in B′ and C′) is not decreased in any clones, and is slightly increased in parts of some clones (C). (Scale bars: 50 μm.)
fat clones can also decrease subapical Ex in the retinal region of eye-antennal discs (30), but the effect of fat loss on total Ex protein levels in eye-antennal discs appears weaker than in the wing (29) (although see ref. 31 for contradictory data on wing disc levels). We found that fat mutant clones increased subapical Dlish in the disc epithelia of both wing and eye, but Dlish levels were lower in the eye than the wing in both normal and fat mutant cells (SI Appendix, Fig. S4 A and B). Accordingly, the effect of dlish clones on subapical Ex was weaker in the eye than in the wing (SI Appendix, Fig. S4 C–F).
Dlish Regulates the Ubiquitination of Ex.
One mechanism for the posttranscriptional regulation of Ex protein levels is via ubiquitination. The ICD of the transmembrane protein Crumbs can decrease Ex levels; Crumbs binds the Ex FERM domain, increasing the binding between Ex and the F-box E3 ubiquitin ligase Slimb and stimulating Ex ubiquitination via the SCF complex (33, 34, 37–39). This suggests a possible route for Dlish function, especially as we previously found that Dlish can co-IP with several members of the ubiquitination pathway, including Cullin1 and Slimb (7) (SI Appendix, Fig. S1D). We extended this data by showing that Slimb was pulled down by GST-Dlish fragments containing SH3-1 and SH3-2, but not SH3-3 (Fig. 6A); this differs from the Dlish domains required for Ex binding, making it unlikely that the Dlish-Slimb binding is mediated by Ex.
Rather, Slimb and Dlish bind to nearby sites in the Ex protein. Previous studies disagreed about whether Slimb binds only to a single phosphodegron in the linker domain of ExN (33), or to additional sites (34) nearer the Dlish-binding domains in ExC2 and ExC3 that we identified above. In our S2R+ cells, ExN and ExC can both co-IP Slimb-HA, as can ExC2 and ExC3 but not ExC1 (Fig. 6B). We confirmed binding of both Slimb and Dlish to ExC2 and ExC3 domains using GST pull-downs (Fig. 6C).
We next tested the effects of dlish and slimb, both separately and in combination, on the ubiquitination of Ex in S2R+ cells. To sensitize the assay, we drove strong Ex ubiquitination by expressing the ICD of Crumbs (Crbintra) (33), and knocked down endogenous dlish or slimb levels using RNAi. We confirmed that slimb knockdown decreased Ex ubiquitination and found a weaker but significant effect with dlish knockdown (Fig. 6 D and E). Our results further suggest that the Dlish effect depended on the presence of Slimb. The effects of dlish and slimb knockdown on Ex ubiquitination were not additive; dlish knockdown had no obvious effect in slimb knockdown cells (Fig. 6 D and E).
To confirm this result in vivo, we also generated dlish mutant clones in hh-gal4 UAS-slimb-RNAi wing discs. Ex levels were increased by the posterior knockdown of Slimb, in agreement with earlier studies (33, 34), but showed no further increase in clones lacking dlish (Fig. 6F). This suggests that Dlish regulates Ex stability through Slimb and is not acting independently on Ex ubiquitination or trafficking. Dlish does not appear to affect Slimb function in any general way. dlish null mutants (7, 8) show no signs of the increased Hedgehog and Wingless signaling seen in slimb mutants (40), and the levels of the Slimb target Cubitus interruptus (Ci) (41) were not increased by ap-gal4-driven UAS-dlish-RNAi (SI Appendix, Fig. S1E). We hypothesize that Dlish helps ubiquitinate Ex by reinforcing binding between Ex and Slimb or helping recruit other members of the SCF complex.
Discussion
The evidence to date demonstrates two parallel routes from the Fat ICD to the stimulation of the Hippo pathway and the subsequent reduction in Yki-mediated growth; both routes require the Fat-binding SH3 domain protein Dlish. In the first, Fat-mediated inhibition of Dlish levels and activity reduces the subapical localization of the Dlish-binding partner Dachs (7, 8), thereby reducing Dachs-mediated inhibition of the Warts kinase (11, 13, 14). We here elucidate a second route in which Fat maintains the subapical levels of a second growth-suppressing protein, the FERM-domain protein Ex (29–32). Our findings provide a mechanism for the maintenance of Ex by Fat.
We showed that Dlish can induce overgrowth that is independent of Dachs, and used a mass spectrometric approach to search for Dlish-binding partners that mediate this effect. That and our subsequent biochemical analyses showed that Dlish binds directly to multiple sites in the C terminus of Ex. While Ex has little effect on Dlish levels in vivo, we found that Dlish reduces subapical and total levels of Ex in vivo, and that this effect is posttranscriptional. In fat mutants the levels of subapical Dlish increase and Ex levels decrease, but blocking dlish blocks the Ex decrease in fat mutants. This last result suggests that Fat is not simply tethering Ex in the subapical cell cortex; rather, it is the increase in Dlish activity that destabilizes Ex in fat mutants.
Our evidence further indicates that Dlish helps in the ubiquitination of Ex that is mediated by the F-box E3 ubiquitin ligase Slimb. Slimb can bind and ubiquitinate Ex in vitro and reduce Ex levels in vivo, an effect stimulated by the ICD of Crumbs and mediated by the SCF complex (33, 34). We had shown previously that Dlish bound Slimb (7), and showed here that Slimb bound specific Dlish domains. We further found that Dlish increased Ex ubiquitination in vitro, and that this effect was lost when Slimb was absent. Similarly, Ex levels in vivo were no longer affected by loss of Dlish when Slimb was absent. This strongly suggests that Dlish, by binding both Ex and Slimb, acts as a cofactor that potentiates the Slimb-based ubiquitination of Ex (modeled in SI Appendix, Fig. S5).
Dlish and Slimb likely act in parallel to another E3 ubiquitin ligase, the RING domain protein POSH. POSH increases Ex ubiquitination in vitro, and while in vivo knockdown of POSH has little effect on its own, it partially reverses the loss of Ex induced by overexpression of Crbintra (42). Intriguingly, POSH, like Dlish, contains multiple SH3 domains that bind to sites in the C-terminal half of Ex (42). The domains bound appear to differ, however: Dlish binds directly to ExC2 and two SH3-binding domains in ExC3, while POSH binds only to ExC2. We have not detected binding between POSH and Dlish (SI Appendix, Fig. S6).
Dlish may regulate the stability of not only Ex, but other proteins, mostly notably the Dlish-binding partner Dachs. Total Dachs levels are strongly increased in dlish mutants (7). While this could be an indirect effect mediated by the changes in Dachs localization in cells, we note that the loss of Slimb in vivo slightly up-regulates subapical Dachs, as well as Dlish (7). Dlish also binds the F-box protein Fbxl7, which regulates the subapical accumulation of Dachs and Dlish (7, 17, 18). That said, the increased Dachs levels in dlish mutants are larger than can be accounted for by loss of Slimb or Fbxl7 function, and thus likely involve additional regulators. Dlish also strongly binds Elongin-C in S2R+ cells (SI Appendix, Fig. S1D), suggesting that Dlish might act, not only with F-box-containing SCF E3 complexes, but also on Elongin-C-Cullin-SOCS-box E3 complexes (43).
While Dlish-aided ubiquitination of Ex provides a growth control pathway independent of the Dachs-mediated inhibition of Warts, previous evidence suggests that Dachs can also regulate Ex: as in dlish mutants, in dachs mutants fat clones no longer reduce Ex levels (32). While more complex roles are possible, the simplest explanation is that Dachs is needed as a scaffold to properly localize Dlish; in dachs mutants, endogenous Dlish no longer preferentially concentrates in the subapical cell cortex where Ex is found (7, 8). One additional finding supports an indirect role for Dachs in Ex destabilization: overexpression of Dlish or Dachs is sufficient to induce Yki activity and thus ex transcription, but Dlish did not increase Ex protein while Dachs did (SI Appendix, Fig. S3). The interdependence of Dlish and Dachs makes it difficult, however, to analyze their epistatic relationship, except when overexpression of one overrides the defects in cortical localization caused by loss of the other.
The existence of an Ex-based pathway downstream of Fat and Dlish provides the opportunity for crosstalk between Fat and other Ex-regulating pathways, especially that mediated the apical transmembrane protein Crumbs, which can tether and localize Ex apically, but also stimulates Slimb-mediated ubiquitination of Ex (33, 34, 37–39) (modeled in SI Appendix, Fig. S5). We also note that Ex provides an input to the Hippo pathway that can be regulated without affecting the Fat-dependent regulation of planar cell polarity and axial patterning of Drosophila epithelia. Unlike Ex, the protocadherin binding partners Fat and Ds and both Dlish and Dachs are not only concentrated apically, they are also planar polarized in epithelial cells, and the excess and mis-polarized Dachs in fat mutants is thought to underlie some of the mutant’s defects in the polarity of cell divisions, hairs, and differentiation decisions (2, 44–46).
Experimental Procedures
Fly strains, immunostaining and microscopy, immunoprecipitation, GST pull-down, binding between in vitro translated and GST-purified proteins, and Western IBs were as previously described (7) with additions and DNA constructs listed in the SI Appendix. For mass spectrometry, Dlish-FLAG was IP’d from S2R+ cells, binding proteins were separated by SDS/PAGE, and peptide mixtures analyzed with LC-MS/MS. The ubiquitination assay was as described (17) with modifications described in the SI Appendix.
Supplementary Material
Acknowledgments
We thank H. McNeill, R. G. Fehon, N. Tapon, B. W. Lu, A. J. Zhu, the Developmental Studies Hybridoma Bank, the Bloomington Drosophila Stock Center, and Vienna Drosophila Resource Center for reagents and fly stocks used in this study. The work was supported by NIH Grants R01-NS028202 and R01-GM124377 (to S.S.B.), by the National Natural Science Foundation of China (31801190) (to X.W.), by the Fundamental Research Funds for the Central Universities (2018QC155 and 2018ZH003) (to X.W.), by the University of Wisconsin Research Committee, and by Guyer Fellowships from the University of Wisconsin’s Integrative Biology Department.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811891116/-/DCSupplemental.
References
- 1.Irvine KD, Harvey KF. Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harb Perspect Biol. 2015;7:a019224. doi: 10.1101/cshperspect.a019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair S, McNeill H. Big roles for Fat cadherins. Curr Opin Cell Biol. 2018;51:73–80. doi: 10.1016/j.ceb.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matakatsu H, Blair SS. Separating planar cell polarity and Hippo pathway activities of the protocadherins Fat and Dachsous. Development. 2012;139:1498–1508. doi: 10.1242/dev.070367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X, Yang CH, Simon MA. The Drosophila Cadherin Fat regulates tissue size and planar cell polarity through different domains. PLoS One. 2013;8:e62998. doi: 10.1371/journal.pone.0062998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan G, et al. Signal transduction by the Fat cytoplasmic domain. Development. 2013;140:831–842. doi: 10.1242/dev.088534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossuyt W, et al. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene. 2014;33:1218–1228. doi: 10.1038/onc.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Wang X, Matakatsu H, Fehon R, Blair SS. The novel SH3 domain protein Dlish/CG10933 mediates fat signaling in Drosophila by binding and regulating Dachs. eLife. 2016;5:e16624. doi: 10.7554/eLife.16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misra JR, Irvine KD. Vamana couples Fat signaling to the Hippo pathway. Dev Cell. 2016;39:254–266. doi: 10.1016/j.devcel.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su T, Ludwig MZ, Xu J, Fehon RG. Kibra and Merlin activate the Hippo pathway spatially distinct from and independent of Expanded. Dev Cell. 2017;40:478–490.e3. doi: 10.1016/j.devcel.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao Y, et al. Dachs: An unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development. 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- 11.Cho E, et al. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 12.Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauskolb C, Pan G, Reddy BV, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS Biol. 2011;9:e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vrabioiu AM, Struhl G. Fat/dachsous signaling promotes Drosophila wing growth by regulating the conformational state of the NDR kinase Warts. Dev Cell. 2015;35:737–749. doi: 10.1016/j.devcel.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matakatsu H, Blair SS. The DHHC palmitoyltransferase approximated regulates Fat signaling and Dachs localization and activity. Curr Biol. 2008;18:1390–1395. doi: 10.1016/j.cub.2008.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matakatsu H, Blair SS, Fehon RG. The palmitoyltransferase Approximated promotes growth via the Hippo pathway by palmitoylation of Fat. J Cell Biol. 2017;216:265–277. doi: 10.1083/jcb.201609094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosch JA, et al. The Drosophila F-box protein Fbxl7 binds to the protocadherin fat and regulates Dachs localization and Hippo signaling. eLife. 2014;3:e03383. doi: 10.7554/eLife.03383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigues-Campos M, Thompson BJ. The ubiquitin ligase FbxL7 regulates the Dachsous-Fat-Dachs system in Drosophila. Development. 2014;141:4098–4103. doi: 10.1242/dev.113498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boedigheimer M, Laughon A. Expanded: A gene involved in the control of cell proliferation in imaginal discs. Development. 1993;118:1291–1301. doi: 10.1242/dev.118.4.1291. [DOI] [PubMed] [Google Scholar]
- 20.McCartney BM, Kulikauskas RM, LaJeunesse DR, Fehon RG. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- 21.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 22.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, et al. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy BV, Irvine KD. Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development. 2011;138:5201–5212. doi: 10.1242/dev.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun S, Reddy BV, Irvine KD. Localization of Hippo signalling complexes and Warts activation in vivo. Nat Commun. 2015;6:8402. doi: 10.1038/ncomms9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badouel C, et al. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu L, et al. Ack promotes tissue growth via phosphorylation and suppression of the Hippo pathway component Expanded. Cell Discov. 2016;2:15047. doi: 10.1038/celldisc.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 30.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Willecke M, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci USA. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribeiro P, Holder M, Frith D, Snijders AP, Tapon N. Crumbs promotes expanded recognition and degradation by the SCF(Slimb/β-TrCP) ubiquitin ligase. Proc Natl Acad Sci USA. 2014;111:E1980–E1989. doi: 10.1073/pnas.1315508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, et al. SCF(Slmb) E3 ligase-mediated degradation of Expanded is inhibited by the Hippo pathway in Drosophila. Cell Res. 2015;25:93–109. doi: 10.1038/cr.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellacheruvu D, et al. The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaneko T, Li L, Li SS. The SH3 domain–A family of versatile peptide- and protein-recognition module. Front Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- 37.Chen CL, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 39.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 41.Smelkinson MG, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr Biol. 2006;16:110–116. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Ma X, Guo X, Richardson HE, Xu T, Xue L. POSH regulates Hippo signaling through ubiquitin-mediated expanded degradation. Proc Natl Acad Sci USA. 2018;115:2150–2155. doi: 10.1073/pnas.1715165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linossi EM, Nicholson SE. The SOCS box-adapting proteins for ubiquitination and proteasomal degradation. IUBMB Life. 2012;64:316–323. doi: 10.1002/iub.1011. [DOI] [PubMed] [Google Scholar]
- 44.Ayukawa T, et al. Dachsous-dependent asymmetric localization of spiny-legs determines planar cell polarity orientation in Drosophila. Cell Rep. 2014;8:610–621. doi: 10.1016/j.celrep.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Bosveld F, et al. Modulation of junction tension by tumor suppressors and proto-oncogenes regulates cell-cell contacts. Development. 2016;143:623–634. doi: 10.1242/dev.127993. [DOI] [PubMed] [Google Scholar]
- 46.Ambegaonkar AA, Irvine KD. Coordination of planar cell polarity pathways through Spiny-legs. eLife. 2015;4:e09946. doi: 10.7554/eLife.09946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.