Significance
We have correlated signaling behaviors seen with biosensors (conformational changes of Rac1) with the nanoscale resolution of electron cryotomography. The workflow and approaches we describe bridge five orders of magnitude (tens of microns to nanometers) to identify, in the crowded environment within eukaryotic cells, distinct nanoarchitectures induced locally by Rac1 as it mediates cell motility, mechanosensing, and invasion. Our studies reveal a dynamic switching mechanism that generates a nanoscaffold system that can either propagate to highly complex actin filament architectures mandated by biological processes or completely dissipate. The workflow implemented here offers a powerful analytical tool not only for deciphering the function of Rho GTPase molecular switches but for the dynamics of many other macromolecular machines as well.
Keywords: cellular cryotomography, biosensors, Rho GTPase molecular signaling, actin cytoskeleton, correlative imaging
Abstract
Rac1 activation is at the core of signaling pathways regulating polarized cell migration. So far, it has not been possible to directly explore the structural changes triggered by Rac1 activation at the molecular level. Here, through a multiscale imaging workflow that combines biosensor imaging of Rac1 dynamics with electron cryotomography, we identified, within the crowded environment of eukaryotic cells, a unique nanoscale architecture of a flexible, signal-dependent actin structure. In cell regions with high Rac1 activity, we found a structural regime that spans from the ventral membrane up to a height of ∼60 nm above that membrane, composed of directionally unaligned, densely packed actin filaments, most shorter than 150 nm. This unique Rac1-induced morphology is markedly different from the dendritic network architecture in which relatively short filaments emanate from existing, longer actin filaments. These Rac1-mediated scaffold assemblies are devoid of large macromolecules such as ribosomes or other filament types, which are abundant at the periphery and within the remainder of the imaged volumes. Cessation of Rac1 activity induces a complete and rapid structural transition, leading to the absence of detectable remnants of such structures within 150 s, providing direct structural evidence for rapid actin filament network turnover induced by GTPase signaling events. It is tempting to speculate that this highly dynamical nanoscaffold system is sensitive to local spatial cues, thus serving to support the formation of more complex actin filament architectures—such as those mandated by epithelial−mesenchymal transition, for example—or resetting the region by completely dissipating.
Rho GTPase proteins are molecular switches that control important signal transduction pathways in cells (1). They are in an “activated” conformation, capable of interacting with downstream effectors, when other proteins such as guanine exchange factors catalyze binding to GTP. Hydrolysis of the GTP leads to adoption of an inactive conformation. Since their initial description by Hall and coworkers (2–4) in the early 1990s, biochemical, cell biological, and genetic approaches have identified various effectors through which Rho GTPase family members trigger actin polymerization, filament turnover, filament bundling, and more, leading to the formation of functionally distinct cytoskeletal actin networks (5–9). Specific actin-based morphologies are correlated with the localization and activity of specific Rho GTPase family members. For example, RhoA is thought to be associated with actin stress fibers (2), Rac1 is thought to be associated with ruffles and broader protrusions (4), and filopodia are thought to be closely associated with Cdc42 (3, 10).
We and others have investigated the role of Rho family GTPases, using fluorescent biosensors to report on their activation states in living cells, correlating these states with the actin cytoskeleton morphology and motility phenotypes using light microscopy (11–17). These studies show that GTPases are activated at the front of migrating cells and regulate one another. The activity of Rac1, the subject of this study, fluctuates on a seconds and submicron scale to mediate cell motility, mechanosensing, and invasion (1, 13, 18). Although Rac1 activation is known to drive cell protrusion by inducing local actin rearrangements, information about these rearrangements at the molecular level remains elusive. However, accurately determining the structural properties of the cell cytoskeleton as a function of Rho GTPase activation state is crucial for developing testable hypotheses regarding the precise mechanisms by which Rho-family molecular switches regulate human health.
Here, we present a correlative spatiotemporal multiscale workflow that identifies quantifiable direct coupling between Rac1 activation states and its functional output, spatially distinct cytoskeletal nanoarchitectures within eukaryotic cells. In contrast to previous uses of correlative light and electron microscopy technologies, which target scale integration for localizing the entities between the two imaging modalities (19–21), here we expanded this technology to directly tie high-resolution structural information to spatiotemporal dynamics of cellular events at spatially defined regions of the cell. We used live-cell imaging of a FRET-based Rac1 biosensor to produce spatiotemporal maps of the activation dynamics of Rac1 at the cell periphery. We then employed new-generation transmission electron microscopes, direct electron imaging devices, and electron cryotomography (cryo-ET) to image the same region after the last frame captured by live-cell imaging was recorded. Cryo-ET technology is the only imaging modality that enables the analysis of biological samples in three dimensions and in their hydrated state at increasingly higher resolutions (22–24). For selected purified samples, cryo-ET combined with subtomogram averaging is now capable of reaching subnanometer resolution (25, 26), and even near-atomic resolution has now been attained (27). For motifs extracted from tomograms of mammalian cells, the resolution has so far been restricted to 2 nm to 4 nm. The actin filament structures within these cellular volumes are amenable to tracing of individual filaments (28, 29), allowing quantitative analysis of filament arrangements (30–33). With the advent of direct detector imaging technology, robust cryo-ET hardware, and high-throughput image acquisition schemes, it is now more feasible to obtain larger amounts of cryo-ET data from various regions and various cells, paving the way for statistical scrutiny of the tomographic analyses (30).
For this correlative light−cryo-ET study, we applied segmentation and template matching technology (34) to quantitatively define the structural organization and content of the cell regions at distinct Rac1 activation states. Alignment between the live-cell imaging and cryo-ET to match Rac1 activity with structural features was achieved within 100 nm (35). Using this workflow, we provided quantitative nanoscale description of a scaffold nanoarchitecture that directly correlates with a defined signaling event. We observed large amounts of ribosomes, various other macromolecular machines, all three types of cytoskeletal filaments, and vesicles in the 3D volumetric reconstructions (tomograms) of regions that do not have high Rac1 activity, by biosensor imaging. Regions with high Rac1 activity contain exclusively actin filament structures composed of densely packed, short actin filaments lacking mutual alignment or branches. These unique actin filament structures were devoid of other discernible macromolecular assemblies such as ribosomes or other types of filaments. Furthermore, 150 s from cessation of high Rac1 activity, no remnants of these actin assemblies were present, and the region shows the same organization as regions with sustained low Rac1 activity.
The correlative spatiotemporal multiscale workflow described here offers a powerful analytical tool not only for deciphering the function of Rho GTPase molecular switches but also for the dynamics of many other macromolecular machines. The ability to assign spatial details and associated numerical values for the interior of a cell at the nano scale and to correlate them with signaling events is of particular importance for biophysical modeling.
Results
To investigate the spatiotemporal coordination between Rac1 activation and the nanoorganization within cell edges undergoing protrusions and retractions, Mouse Embryo Fibroblasts (MEFs) stably expressing the Rac1 biosensor [Rac1 fluorescent activation reporter (FLARE), type DC1g] (18) were plated on fibronectin-coated substrates suitable for both high-resolution cryo-ET and FRET live-cell imaging modalities. Live-cell imaging of Rac1 biosensors generated short movies (5 min) recording the history of both Rac1 activity and edge dynamics (Fig. 1 and Movies S1 and S2). Time-lapse image sequences show fluctuations in Rac1 GTPase activity at the periphery of protruding and retracting cells. Peak activity was observed within 2 μm to 4 μm of the edge, as was seen in our previous reports (13, 18).
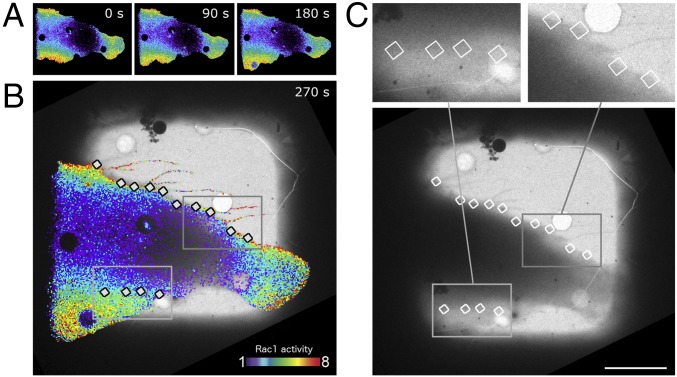
Fig. 1.
Correlating spatiotemporal activity of Rac1 GTPases with the underlying nanoscale morphology of MEF cells. (A) Biosensors showing Rac1 activation at different times during live-cell imaging. (The width of each time-point image is 89 μm.) The pseudocolor scale shows the increase in ratio relative to the lowest 5% of ratio values in the cell. Note protrusion at lower left. Activity dynamics depicted as stills here are shown in Movies S1 and S2. (B) Close-up still at 270 s overlaid with cryo-EM image used for correlation between the imaging modalities. The cell was rapidly cryofixed for data collection at the 270-s time point. Regions used for cryo-ET are indicated as white boxes. (Scale bar, 20 μm.) (C) (Lower) The overview image used in B for the biosensor overlay. This image was taken after cryofreezing in the electron cryomicroscope just before cryo-ET data collection. (The width of each box is 26 μm.) (Upper) Enlargements of the areas marked by gray boxes in B and in Lower. (Scale bar, 20 μm.)
To determine the 3D organization and molecular content of these regions at high resolution and defined Rac1 activation states, cell movement had to be halted while maintaining the structural integrity of the site. This was achieved via rapid fixation at the final time point of FRET imaging. Next, samples were vitrified to prevent structural collapse or shrinkage associated with dehydration. We employed cryo-ET to image these cells while hydration of the samples was maintained.
For high-precision capture of regions designated to be imaged at high resolution, we need to span five orders of magnitude: FRET and light microscopy (LM) (tens of microns) to cryo-ET (nanometers and angstroms). We used an intermediate imaging step, cryo-light imaging aided by a cryo-fluorescence microscope set up at liquid nitrogen temperatures (CorrSight) to identify, under cryogenic conditions, the cells and regions of interest (SI Appendix, Figs. S1 and S2). Using a marker-free alignment method (35) and including the cryo-fluorescence and LM-phase images of the cells, we identified the biosensor-loaded cells and the respective regions within 100-nm accuracy (SI Appendix, Fig. S1). Guided by the correlated last frame of the FRET live-cell imaging movies of Rac1 biosensors, regions along the cell periphery were then selected for cryo-ET data collection acquisition with the Titan Krios cryomicroscope (Fig. 1 and SI Appendix, Fig. S2). We acquired a total of 171 tomograms along the peripheries of 25 cells. Thirty of these tomograms were fully correlated with biosensor data and cell dynamics. The cryo-ET acquisition window width was between 1 μm and 2 μm (Fig. 1 and SI Appendix, Fig. S1, XY dimensions) resulting in pixel sizes between 0.25 and 0.5 nm. The nanometer-scale morphology of the cell peripheries was then determined using 3D reconstruction (36), segmentation (37, 38), and automated feature detection technologies (34). The high accuracy with which we aligned biosensor images and the extracted features of 3D volumes allowed us to derive a meaningful correlation of morphological features with Rac1 activation states.
The imaged regions of the MEF cell peripheries were, on average, 156.0 ± 57.4 nm thick. The extracellular matrix in these regions contributed an additional 50.2 ± 17.2 nm thickness and showed a smooth appearance without any discernable internal features. In about 20% of the tomograms, dendritic actin networks of up to about 0.5-μm width were discernable near the cell periphery (SI Appendix, Fig. S3A). The main characteristic of these dendritic networks was the presence of actin branch junctions, where shorter actin filaments emanated from the sides of longer filaments at angles of ∼70° to 80°, suggesting that these were mediated by Arp2/3 complexes (39). There were also many bundles of long parallel actin filaments running along the cell periphery or passing through the volume away (>2 µm) from the cell periphery, as well as loosely arranged long actin filaments (SI Appendix, Fig. S3 B–D). In addition to these actin filament structures, we observed clustered and unclustered ribosomes, various types of vesicles, endoplasmic reticulum, intermediate filaments, microtubules, and an occasional mitochondrion interspaced in the dendritic network. Generally, these nonactin features only started appearing at a distance of 30.9 ± 9.3 nm from the ventral membrane [in Z (perpendicular to the substrate)] and were present up to the dorsal membrane. Between these features and the ventral membrane, long, loosely aligned actin filaments, consistent with a cortical actin filament layer, tended to be visible.
To explore directly whether Rac1 activation produces localized actin behavior in an intact cell, we first defined the subvolumes within the overall volumes of the cell periphery that were correlated with elevated biosensor activity (Fig. 2, SI Appendix, Fig. S4, and Movie S3). Regions from the ventral membrane up to a height of about 60 nm above that membrane were densely packed with short actin filaments, most shorter than 150 nm (Fig. 2). These filaments were poorly aligned with respect to each other (angular dispersion 18.16° ± 2.03°; angular dispersion for random orientations is ∼26°) and did not emanate from existing filaments. These volumes are devoid of other large-scale macromolecular features such as ribosomes, intermediate filaments, or long, parallel actin filaments.
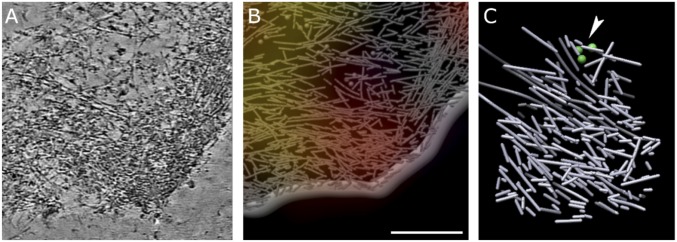
Fig. 2.
The macromolecular organization of cytoskeleton assemblies in regions of high Rac1 activity. (A) A 20-nm-thick slice through tomogram of region with high Rac1 activity. Cell edge is facing the lower right corner. (B) A 3D rendering of traced actin filaments, cell membrane, and ribosomes (spheres) in the region shown in A. The Rac1 activity map is overlaid; red corresponds to high Rac1 activity. (Scale bar, A and B, 300 nm.) (C) Actin filaments extracted from the red region in B. In regions with high Rac1 activity, we consistently observe densely packed, short actin filaments, most of them shorter than 150 nm, with random orientation. Only a very small number of longer filaments and very few ribosomes (green spheres, near arrowhead) are present. Successive slices through cryo-ET reconstruction of region with high Rac1 activity showing short, actin filaments with little mutual alignment can be seen in Movie S3.
Many of the regions with high Rac1 activity were less than 0.5 μm from the cell periphery. In these regions, the height of the actin structure coincided with cell thickness, and there were no additional features present. For regions with high Rac1 activity that were in somewhat thicker regions of the cell, these actin structures terminated at a height of about 60 nm above the ventral membrane. Beyond that height, toward the dorsal part of the cell, other features such as longer, mutually aligned actin filaments or ribosomes were present. The regions of high Rac1 activity ranged in size between 0.5 μm to a few microns diameter. The unique actin filament organization in regions with high Rac1 activity differed significantly from that in the dendritic networks. The Arp2/3-complex mediated branches characteristic for dendritic networks (40) are not present in regions with high Rac1 activity, and there is no dendritic organization apparent. Actin organization appeared the same in high Rac1 activity regions whether they were protruding or quiescent (Fig. 3A). Some actively protruding volumes with high Rac1 activity were accompanied by filopodia, which contain parallel actin bundles (Fig. 3A).
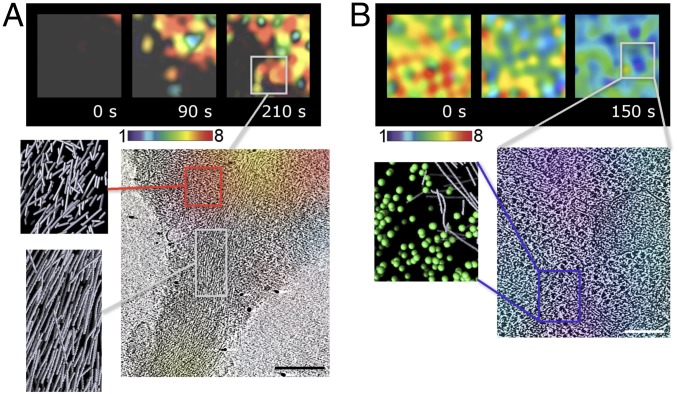
Fig. 3.
Time evolution of Rac1 activity defined at the nanoscale. (A) Molecular organization of cytoskeleton assemblies at actively protruding filopodium. (Upper) Successive images of Rac1 activity near the base of an actively extending filopodium, taken from live-cell ratio imaging of an MEF cell. (The width of each box is 3 μm.) The pseudocolor scale shows the increase in ratio relative to the lowest 5% of ratio values in the cell. (Lower) The last time frame captured by cryo-ET. Regions with high Rac1 activity (Upper, dark-gray box) show the characteristic short actin filament morphology with little directional alignment. Note the appearance of the characteristic morphology of filopodial bundle (Lower, light gray box) at close spatial proximity. (Scale bar, 300 nm.) (B) Molecular organization of cytoskeleton assemblies at site of lowering Rac1 activity. (Upper) Successive images of a region taken from live-cell ratio imaging of an MEF cell. (The width of each box is 3 μm.) The pseudocolor scale shows the increase in ratio relative to the lowest 5% of ratio values in the cell. The region turned from primarily high Rac1 activity (red) to low Rac1 activity (purple/blue) within 150 s. (Lower) The last time frame captured by cryo-ET. The region is dominated by ribosomes interspersed with long, aligned actin filament and vesicular assemblies. Short actin filaments, characteristic of regions with high Rac1 activity, are no longer present, suggesting that, in less than 150 s this morphology is replaced. (Scale bar, 150 nm.)
In stark contrast to the short, poorly aligned actin filaments detected in regions with high Rac1 activity, volumes with low Rac1 activity contained longer, much more well-aligned actin filaments (angular dispersion 5.90° ± 0.95°) as well as large numbers of ribosomes (Fig. 4 and Movie S4). The actin filaments in low Rac1 activity regions were about 3 to 4 times longer than those in regions with high Rac1 activity, and many of those are not fully contained in the tomograms. In fact, some of those filaments are spanning the entire 2-μm length of the tomograms, continuing in both directions beyond the edge of the tomogram. For the filaments in regions with low Rac1 activity, the packing density (occupied volume fraction) was 10 to 20%, similar to the packing density predicted by simulations of unbranched actin filament growth near the cell edge (41). In regions with high Rac1 activity, packing density was significantly higher, 20 to 25%. The maximum packing density of randomly oriented filaments varies approximately as the inverse of their aspect ratio and decreases dramatically as filaments get longer (42). The maximum packing density of 150-nm-long actin filaments calculated from this relationship (5.1/aspect ratio with average filament diameter of 8 nm) is about 27%, indicating that the packing density of actin filaments in regions with high Rac1 activity is nearly at its maximum. Despite the larger packing density, there was no detectable difference in filament curvature and thus persistence length, which is about 10 μm to 17 μm for actin filaments (43–45). This suggests that local changes in filament stiffness do not occur at these sites. However, the stiffness can be altered quite quickly by appearance of elongation in the presence of cross-linking factors (46).
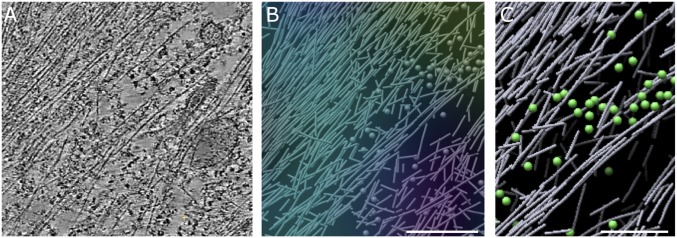
Fig. 4.
Nanometer-scale organization of cytoskeleton assemblies in regions with low Rac1 activity. (A) A 20-nm-thick slice through tomogram of region with low Rac1 activity. (B) A 3D rendering of traced actin filaments (rods) and ribosomes (spheres) in the region shown in A. The Rac1 activity map is overlaid; purple/blue corresponds to low Rac1 activity. (Scale bar, A and B, 300 nm.) (C) Enlarged region. Note the presence of longer, mutually aligned actin filaments (up to >1 µm in length) and ribosomes (green spheres). (Scale bar, 200 nm.) Successive slices through cryo-ET reconstruction of region with low Rac1 activity showing long mutually aligned actin filaments, ribosomes, intermediate filaments, and vesicular structures can be seen in Movie S4.
Various types of vesicles, endoplasmic reticulum, and occasionally intermediate filaments or microtubules were seen on top of an ∼30-nm-thick zone of cortical actin near the ventral membrane (Fig. 4 and SI Appendix, Figs. S5 and S6). The cortical layer was disrupted in regions of high Rac1 activity where only short, poorly aligned filaments were present near the ventral membrane.
One of the most prominent features of regions with low Rac1 activity was the occurrence of large amounts of ribosomes. Subtomogram averaging of 2,098 ribosomes detected in regions of low Rac1 activity (SI Appendix, Fig. S7A) yielded a resolution estimate of about 3 nm to 4 nm (SI Appendix, Fig. S7C), providing a measure for the tentative resolution of the cryo-ET reconstructions. Nearest-neighbor cluster analysis (47) revealed that about half of the ribosomes were tightly clustered (SI Appendix, Fig. S7D), containing between 4 and 16 ribosomes. With an average center-to-center distance of 37.2 nm, these clusters were somewhat more loosely arranged than those related to membrane-bound polysomes near the nuclear envelope (48) or polysomes in human glioma cells (47). While the ribosomes clusters observed here are strikingly similar to arrangements of rotary shadowed polysomes on RNA (49) as well as some of the arrangements found in glioma cells (47), there is no conclusive proof that these clusters correspond to polysomes. In addition to ribosomes, assemblies consistent with the appearance of TCP-1 Ring Complex chaperonins were present in regions with low Rac1 activity (SI Appendix, Fig. S7B).
To define how persistent the two distinct morphologies are and how they relate to the dynamics of Rac1 activity, we first screened all of the regions that displayed continuous high levels of Rac1 activation throughout the entire time (300 s) of our fluorescence movies (SI Appendix, Fig. S8). This screen resulted only in the characteristic Rac1-induced short, anisotropic single actin filament morphology, with no evidence of long filaments, ribosomes, or other discernable macromolecular structures within these volumes. Next, we screened regions where high Rac1 activity only appeared within a short time window, less than 150 s before halting via fixation. Here again, in regions that only recently started to show high Rac1 activity, the same characteristic morphology was observed as in the longer exposure times. The fact that the parameters we measured showed no change suggests that there is no cumulative effect of sustained high Rac1 activity. In contrast, in cell regions where high Rac1 activity dropped to low levels immediately before fixation (e.g., red to blue in Fig. 3B), there were no traces of the morphology induced by high Rac1 activity. We only observed the longer, aligned filaments characteristic of low Rac1 activity, even at the shortest onset of low Rac1 activity levels (150 s before fixation). The very high correlation between Rac1 activation levels and the presence of characteristic morphology, in combination with our knowledge of the history of Rac1 activity with nanometer resolution, strongly suggests that high Rac1 activation levels cause the short filament morphology we observed.
Discussion
Up to this point, it has not been possible to directly explore the localized structural changes that Rho GTPase activation produces in intact cells. Rac1 activation, which is critically important in regulation of cell polarization and migration, is promoted by integrin engagement and acts through downstream effectors (e.g., p21-activated kinases) that regulate leading edge cytoskeletal rearrangements and adhesion dynamics (1, 5). FRET-based biosensors have revealed the spatiotemporal dynamics of Rac1 activation state. Patterns of activity extend up to a few micrometers and last a few minutes during protrusion. However, correlating the dynamics of Rac1 signaling with underlying functional outputs requires determining the nanoarchitecture of these regions.
Here, we employed a cryogenic Correlative Light and Electron Microscopy (cryo-CLEM) workflow that combines biosensor imaging of Rac1 dynamics with cryo-ET−based reconstruction of the nanoorganization of intact cells. We centered our analysis within 2 μm to 4 μm from the periphery of protruding or retracting cells growing on cryo-EM amenable substrates, consistent with spatial localization of time-lapse image sequences of peak activity fluctuations in Rac1 GTPase activity reported by us and others.
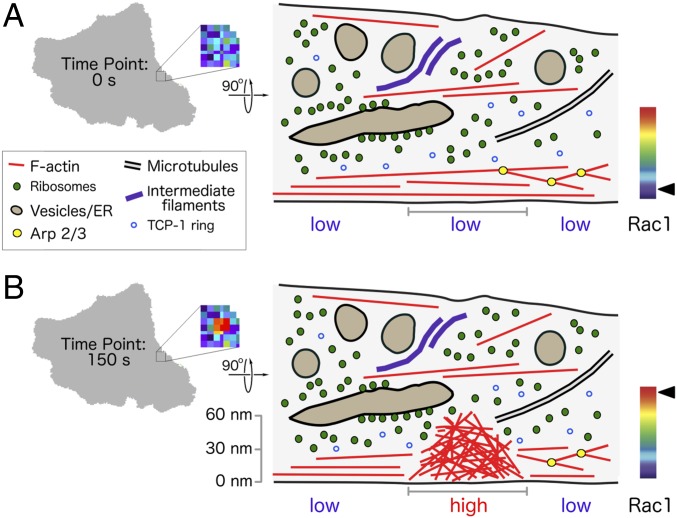
In regions with low Rac1 activity, we identified actin filaments, ribosomes, microtubules, intermediate filaments, vesicles, endoplasmic reticulum (ER), and tentative TCP-1 Ring Complexes (Fig. 5A). The quality and nanometer resolution of the 3D cryotomographic reconstructions allowed us also to identify a unique structural regime with densely packed, directionally anisotropic, short actin filaments in regions of high Rac1 activity, starting at the ventral membrane up to a height of ∼60 nm (Fig. 5B).
Fig. 5.
Schematic model of actin structure triggered by high Rac1 activity. (Left) A gray overview of a cell with a region of interest marked by a box that is enlarged to show the Rac1 activity. (Right) A schematic side view of the region of interest. (A) When Rac1 activity is low, the region contains a thin layer (about 30 nm) of actin structures near the ventral membrane that consist of either long actin filaments or branched dendritic networks. Vesicles, ER, intermediate filaments, ribosomes, some arranged as ribosomes clusters, microtubules, vesicles, tentative TCP-1 Ring Complexes, and occasional long actin filaments can be identified on top of that layer. (B) In regions with high Rac1 activity, an actin structure composed of densely packed, short actin filaments with little or no mutual alignment emerges. The height of the structure is up to 60 nm from the ventral membrane and does not contain any of the features that can be typically seen in regions with low Rac1 activity.
This actin architecture we observed in regions of high Rac1 activity has not been observed before. Its supramolecular organization was strikingly different from that described for lamellipodia. Examination of platinum replicas, negatively stained preparations, and cryopreparations have all suggested that, in the lamellipodia, actin filaments form dendritic networks by extending short filaments from the sides of preexisting actin filaments, due to activation of the Arp2/3 complex’s nucleation-promoting factors (NPFs) (29, 50–52). In contrast to this dendritic network architecture, high Rac1 activity generated a local high concentration of poorly aligned short individual filaments, with average filament length below 150 nm, and a packing density of 20 to 25%, significantly denser than the actin filament structures in regions with low Rac1 activity (packing density 10 to 20%). Similar to dense fiber networks in the polyglutamine (polyQ) neurodegenerative disorder diseases inclusions of mammalian cells that were recently investigated by cryo-ET (53), the actin filament structures in regions of high Rac1 activity were devoid of large macromolecules such as ribosomes or other filament types, which were abundant within the remainder of the imaged volumes.
Numerous polymerization nucleation factors play a role in lamellipodia formation, but the main mode of filament assembly is via the Arp2/3 complex activated by a specific NPF, the Wiskott–Aldrich syndrome protein family verprolin-homologous protein (WAVE) complex (54). The WAVE complex is itself intrinsically inactive and needs Rac1 and lipids to become functional for activating the Arp2/3 complex (55). The lack of branches and long actin filaments in the regions of high Rac1 activation suggested the presence of discrete compartmentalized membrane-bound organelles that allowed for the very tight control of Arp2/3 complex-based polymerization.
The time and place of actin polymerization in a cell is determined by local de novo nucleation of filaments with free barbed ends, as well as by local severing or uncapping of existing longer filaments. The fact that the actin filament polymerization mediated by Rac1 appeared rapidly and did not contain branches suggested that the filaments were nucleated de novo, without the involvement of canonical Arp2/3-dependent nucleation from existing fibers. This most likely occurred through processive barbed-end elongation by formins, consistent with the fact that Rac1 binds to formins and is believed to activate them (6), while other actin nucleators such as Spire or Cobl have not been linked to Rac1 activity (54). Another possibility is noncanonical Arp2/3-dependent nucleation utilizing the Rac1 effector SPIN90 (56), which has been shown to be capable of activating Arp2/3 in the absence of existing actin structures (57).
Another feature of these scaffold systems is that they contain very short actin filaments (<150 nm) with the same lengths distribution maintained over a time frame of minutes. Because there is no evidence for filament severing (which would initially result in short segments but with high mutual alignment), the length of an actin filament in these structures can be considered as the net result of an interplay between elongation and capping activities at the barbed end. Capping protein (CP) is considered one of the key factors limiting filament elongation (58). CP primarily incorporates at the cell edges within about 2 µm (59), coinciding with the regions of high Rac1 activity investigated here. Cells depleted of CP form large star-like actin filament patterns at the ventral membrane emanating from a dense nodule of about 1 µm in diameter (59), consistent with the Rac1-mediated scaffolds we observe. Furthermore, constitutively active Rac1 activates uncapping of actin filaments (60). These observations support the notion that CP is involved in the formation of the Rac1-mediated scaffold systems.
It is conceivable that activated Rac1 is, in turn, activating formins, possibly in a ternary complex with CP (61, 62), leading to fast de novo polymerization of actin filaments. Elongation would then be quickly arrested by CP, and the scaffold system would keep a sustained morphology of short actin filaments, unless Rac1 activity subsides. Once Rac1 signaling ceases, RhoA suppression is likely released.
The prominent presence of cytoplasmic ribosomes and possibly TCP-1 Ring Complex chaperonins, which fold ∼10% of newly synthesized cytosolic proteins, including actin, raises the possibility that low Rac1 activity may promote the recruitment of the cytoplasmic translation machinery associated with protein synthesis. Consistent with this idea, translation initiation factors eIF4E and eIF4GI and active sites of protein synthesis were previously colocalized at the leading edge of migrating fibroblasts, where actively translating ribosomes have also been visualized (63). This would allow actin synthesis to take place in these regions, consistent with the physiological importance of β-actin mRNA presence at the leading edge of fibroblast cells (64). Combining local biogenesis, with the supply of proteins provided through active transport or diffusion (65, 66), would quickly support the formation of more complex, on-demand nanoarchitecture of actin filaments.
It is tempting to speculate that this highly dynamical scaffold system is sensitive to local spatial perturbations that can either propagate to large changes to form more complex architecture or completely dissipate. Indeed, the observations described here, where cessation of Rac1 activity leads rapidly (<150 s) to a complete structural transition, provide direct structural evidence for rapid network turnover leading to the absence of detectable remnants of such structures. It was proposed that RhoA activity is rapidly suppressed as Rac1 reaches its maximum activation (18). This suggestion is consistent with our observation that Rac1 activity is strongly correlated with the rapid formation of unique scaffolds that are incompatible with actin behavior mediated by RhoA, such as formation of stress fibers and contractility (1).
Rapid actin morphology turnover in response to Rho-GTPase signaling has been associated with integral parts of biological processes. One such example in which a fast transition of actin morphology is required and where Rho GTPases are involved is epithelial−mesenchymal transition (EMT). Mechanical regulation of EMT mandates rapid cytoskeletal rearrangements, protein relocalization, and tissue-level reorganization. The flexible, signal-dependent actin structure we observe in this study would allow the fast and spatially localized cytoskeletal transitions necessary for such processes. Understanding how mechanical cues integrate with other signals present in the complex in vivo microenvironment will be key to fully understanding the mechanisms involved in guiding embryonic development or inducing disease states. It is believed that cells maintain a dynamic actin cytoskeleton by carefully balancing the activities of a diverse collection of actin regulators, thereby forming the right structure at the right place and time within the cell in response to cellular signals.
Here, we provided a quantitative nanoscale description of an architecture that directly correlates with a defined signaling event. The ability to assign spatial details and associated numerical values for the interior of a cell at the nano scale and to correlate them with signaling events is of particular importance for biophysical modeling. Many mathematical models focusing on different aspects of understanding the propulsion mechanism process have been helpful in rationalizing and interpreting experimental results. However, they are frequently challenged by the lack of direct observations such as those provided here. The technology described here offers a powerful descriptive tool and can be used to develop hypotheses about cellular organization and dynamics.
Materials and Methods
Time series of Rac1 activity were measured in living MEF cells, using genetically encoded fluorescent biosensors. After the final frame, cells were fixed to arrest movement and then plunge-frozen for correlative cryo-ET. Selected regions with defined Rac1 activity levels were imaged by cryo-ET and analyzed using automated software to extract the underlying architecture of the actin cytoskeleton as well as the distributions of other macromolecular complexes. Details are given in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
National Institutes of Health (NIH) Program Project Grants P01-GM098412 (to D.H.), R01 GM115972 (to D.H.), P01 GM121203 (to N.V.), P41-EB002025 (to K.M.H.), and R35GM122596 (to K.M.H.) supported this work. NIH Grants S10-OD012372 (to D.H.) and P01-GM098412-S1 (to D.H.) funded the purchase of the Titan Krios TEM (Thermo Fisher Scientific; FEI Company) and Falcon II direct detector (Thermo Fisher Scientific; FEI Company). D.J.M. was supported by funding from The Leukemia and Lymphoma Society (LLS).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.K.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808830116/-/DCSupplemental.
References
- 1.Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40:1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 3.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt A, Hall MN. Signaling to the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- 6.Kühn S, Geyer M. Formins as effector proteins of Rho GTPases. Small GTPases. 2014;5:e29513. doi: 10.4161/sgtp.29513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard O. Lim kinases, regulators of actin dynamics. Int J Biochem Cell Biol. 2007;39:1071–1076. doi: 10.1016/j.biocel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Faix J, Weber I. A dual role model for active Rac1 in cell migration. Small GTPases. 2013;4:110–115. doi: 10.4161/sgtp.23476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–112. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen WE, Jones GE, Pollard JW, Ridley AJ. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J Cell Sci. 1997;110:707–720. doi: 10.1242/jcs.110.6.707. [DOI] [PubMed] [Google Scholar]
- 11.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 12.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 13.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 14.Zawistowski JS, Sabouri-Ghomi M, Danuser G, Hahn KM, Hodgson L. A RhoC biosensor reveals differences in the activation kinetics of RhoA and RhoC in migrating cells. PLoS One. 2013;8:e79877. doi: 10.1371/journal.pone.0079877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh RE, et al. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang HW, Collins SR, Meyer T. Locally excitable Cdc42 signals steer cells during chemotaxis. Nat Cell Biol. 2016;18:191–201. doi: 10.1038/ncb3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komatsu N, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampton CM, et al. Correlated fluorescence microscopy and cryo-electron tomography of virus-infected or transfected mammalian cells. Nat Protoc. 2017;12:150–167. doi: 10.1038/nprot.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sartori A, et al. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Schorb M, et al. New hardware and workflows for semi-automated correlative cryo-fluorescence and cryo-electron microscopy/tomography. J Struct Biol. 2017;197:83–93. doi: 10.1016/j.jsb.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asano S, Engel BD, Baumeister W. In situ cryo-electron tomography: A post-reductionist approach to structural biology. J Mol Biol. 2016;428:332–343. doi: 10.1016/j.jmb.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 23.Beck M, Baumeister W. Cryo-electron tomography: Can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 2016;26:825–837. doi: 10.1016/j.tcb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2012;45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoshouei M, Pfeffer S, Baumeister W, Förster F, Danev R. Subtomogram analysis using the Volta phase plate. J Struct Biol. 2017;197:94–101. doi: 10.1016/j.jsb.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Schur FK, Hagen WJ, de Marco A, Briggs JA. Determination of protein structure at 8.5Å resolution using cryo-electron tomography and sub-tomogram averaging. J Struct Biol. 2013;184:394–400. doi: 10.1016/j.jsb.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Schur FK, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 28.Medalia O, et al. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 29.Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- 30.Anderson KL, et al. Nano-scale actin-network characterization of fibroblast cells lacking functional Arp2/3 complex. J Struct Biol. 2017;197:312–321. doi: 10.1016/j.jsb.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jasnin M, et al. Three-dimensional architecture of actin filaments in Listeria monocytogenes comet tails. Proc Natl Acad Sci USA. 2013;110:20521–20526. doi: 10.1073/pnas.1320155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller J, et al. Electron tomography and simulation of baculovirus actin comet tails support a tethered filament model of pathogen propulsion. PLoS Biol. 2014;12:e1001765. doi: 10.1371/journal.pbio.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigort A, et al. Automated segmentation of electron tomograms for a quantitative description of actin filament networks. J Struct Biol. 2012;177:135–144. doi: 10.1016/j.jsb.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Xu XP, Page C, Volkmann N. Efficient extraction of macromolecular complexes from electron tomograms based on reduced representation templates. Comput Anal Images Patterns. 2015;9256:423–431. doi: 10.1007/978-3-319-23192-1_35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson KL, Page C, Swift MF, Hanein D, Volkmann N. Marker-free method for accurate alignment between correlated light, cryo-light, and electron cryo-microscopy data using sample support features. J Struct Biol. 2018;201:46–51. doi: 10.1016/j.jsb.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 37.Page C, Hanein D, Volkmann N. Accurate membrane tracing in three-dimensional reconstructions from electron cryotomography data. Ultramicroscopy. 2015;155:20–26. doi: 10.1016/j.ultramic.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volkmann N. A novel three-dimensional variant of the watershed transform for segmentation of electron density maps. J Struct Biol. 2002;138:123–129. doi: 10.1016/s1047-8477(02)00009-6. [DOI] [PubMed] [Google Scholar]
- 39.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 40.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber CH, Stewart M, Duke T. Simulation of cell motility that reproduces the force-velocity relationship. Proc Natl Acad Sci USA. 2010;107:9141–9146. doi: 10.1073/pnas.1002538107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodney D, Fivel M, Dendievel R. Discrete modeling of the mechanics of entangled materials. Phys Rev Lett. 2005;95:108004. doi: 10.1103/PhysRevLett.95.108004. [DOI] [PubMed] [Google Scholar]
- 43.ben-Avraham D, Tirion MM. Dynamic and elastic properties of F-actin: A normal-modes analysis. Biophys J. 1995;68:1231–1245. doi: 10.1016/S0006-3495(95)80299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Isambert H, et al. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 46.Claessens MM, Bathe M, Frey E, Bausch AR. Actin-binding proteins sensitively mediate F-actin bundle stiffness. Nat Mater. 2006;5:748–753. doi: 10.1038/nmat1718. [DOI] [PubMed] [Google Scholar]
- 47.Brandt F, Carlson LA, Hartl FU, Baumeister W, Grünewald K. The three-dimensional organization of polyribosomes in intact human cells. Mol Cell. 2010;39:560–569. doi: 10.1016/j.molcel.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Mahamid J, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 49.Miller OL, Jr, Hamkalo BA, Thomas CA., Jr Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- 50.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: Mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Small JV, Herzog M, Anderson K. Actin filament organization in the fish keratocyte lamellipodium. J Cell Biol. 1995;129:1275–1286. doi: 10.1083/jcb.129.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauerlein FJB, et al. In situ architecture and cellular interactions of PolyQ inclusions. Cell. 2017;171:179–187.e10. doi: 10.1016/j.cell.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 54.Rottner K, Faix J, Bogdan S, Linder S, Kerkhoff E. Actin assembly mechanisms at a glance. J Cell Sci. 2017;130:3427–3435. doi: 10.1242/jcs.206433. [DOI] [PubMed] [Google Scholar]
- 55.Chen Z, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teodorof C, et al. SPIN90-IRSp53 complex participates in Rac-induced membrane ruffling. Exp Cell Res. 2009;315:2410–2419. doi: 10.1016/j.yexcr.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Wagner AR, Luan Q, Liu SL, Nolen BJ. Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr Biol. 2013;23:1990–1998. doi: 10.1016/j.cub.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DiNubile MJ, Cassimeris L, Joyce M, Zigmond SH. Actin filament barbed-end capping activity in neutrophil lysates: The role of capping protein-beta 2. Mol Biol Cell. 1995;6:1659–1671. doi: 10.1091/mbc.6.12.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mejillano MR, et al. Lamellipodial versus filopodial mode of the actin nanomachinery: Pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 60.Hartwig JH, et al. Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis in permeabilized human platelets. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 61.Bombardier JP, et al. Single-molecule visualization of a formin-capping protein ‘decision complex’ at the actin filament barbed end. Nat Commun. 2015;6:8707. doi: 10.1038/ncomms9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shekhar S, et al. Formin and capping protein together embrace the actin filament in a ménage à trois. Nat Commun. 2015;6:8730. doi: 10.1038/ncomms9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willett M, Brocard M, Davide A, Morley SJ. Translation initiation factors and active sites of protein synthesis co-localize at the leading edge of migrating fibroblasts. Biochem J. 2011;438:217–227. doi: 10.1042/BJ20110435. [DOI] [PubMed] [Google Scholar]
- 64.Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- 65.Singer RH, Lawrence DS, Ovryn B, Condeelis J. Imaging of gene expression in living cells and tissues. J Biomed Opt. 2005;10:051406. doi: 10.1117/1.2103032. [DOI] [PubMed] [Google Scholar]
- 66.Liao G, Mingle L, Van De Water L, Liu G. Control of cell migration through mRNA localization and local translation. Wiley Interdiscip Rev RNA. 2015;6:1–15. doi: 10.1002/wrna.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.