Significance
Unlike all differentiated cells, pluripotent stem cells do not elicit a productive antiviral response when infected by a pathogen. This observation seems at odds with the importance of pluripotent stem cells given their absolute requirement for the development of life. Here we investigate why this antiviral response is not utilized in these unique cells. We find that the factors required to maintain pluripotency are incompatible with those involved in eliciting the canonical interferon-based response to virus infection.
Keywords: pluripotency, interferon, virus, IRF7, KLF4
Abstract
Upon virus infection, pluripotent stem cells neither induce nor respond to canonical type I interferons (IFN-I). To better understand this biology, we characterized induced pluripotent stem cells (iPSCs) as well as their differentiated parental or rederived counterparts. We confirmed that only iPSCs failed to respond to viral RNA, IFN-I, or viral infection. This lack of response could be phenocopied in fibroblasts with the expression of a reprogramming factor which repressed the capacity to induce canonical antiviral pathways. To ascertain the consequences of restoring the antiviral response in the context of pluripotency, we engineered a system to engage these defenses in iPSCs. Inducible expression of a recombinant virus-activated transcription factor resulted in the successful reconstitution of antiviral defenses through the direct up-regulation of IFN-I–stimulated genes. Induction of the antiviral signature in iPSCs, even for a short duration, resulted in the dysregulation of genes associated with all three germ layers despite maintaining pluripotency markers. Trilineage differentiation of these same cells showed that engagement of the antiviral defenses compromised ectoderm and endoderm formation and dysregulated the development of mesodermal sublineages. In all, these data suggest that the temporal induction of the antiviral response primes iPSCs away from pluripotency and induces numerous aberrant gene products upon differentiation. Together these results suggest that the IFN-I system and pluripotency may be incompatible with each other and thus explain why stem cells do not utilize the canonical antiviral system.
The cellular response to virus infection is of fundamental importance for organismal survival. From bacteria to mammals, cells that comprise every form of life have evolved defense systems to minimize infection. This evolutionary arms race has generated immense diversity in all three domains of life (1). In mammals, this process has resulted in a stratified defense platform composed of type I IFN (IFN-I)-mediated effector proteins (intrinsic), a population of specialized cells that react nonspecifically to pathogens (innate), and a multifaceted and highly specific response (adaptive). While the innate and adaptive immune systems are enabled by distinct cellular populations, the intrinsic response is thought to be relatively uniform in vertebrate cells. In response to virus infection, unique hallmarks of replication such as: double-stranded RNA (dsRNA), RNA containing 5′ di- or triphosphates, or cytoplasmic DNA can all be recognized by specific pattern recognition receptors (PRRs) (2). Engagement of these so-called pathogen-associated molecular patterns (PAMPs) to their cognate PRRs induces a cascade of events that ultimately culminate in the transcriptional activation of the IFN response factors (IRFs)—most notably IRF3 and IRF7. Activation of these IRF members induces the transcriptional activation of interferon beta (IFNB), a key member of the IFN-I system, as well as many antiviral effector proteins directly (3). Production and secretion of IFNB functions to prime uninfected cells through a second transcriptional pathway involving both IRF- and STAT-based transcription factors (2). Together, this complex, denoted as IFN-Stimulated Gene Factor 3 (ISGF3) is responsible for the up-regulation of hundreds of genes that interfere with transcription, translation, and transport in an effort to slow and/or inhibit virus replication (2). Collectively, these genes are referred to as IFN-stimulated genes (ISGs). This vertebrate-specific ISG response is effective enough to completely protect cells from the cytopathic effects of many viruses and is broadly conserved among vertebrates (4).
In stark contrast to differentiated cells, pluripotent stem cells lack the capacity to respond to extracellular viral or bacterial PAMPs (5–7). While this phenotype has been proposed to be, in part, due to low levels of PRRs, a comprehensive understanding as to why the IFN-I system is not utilized in these cells still remains incomplete (6). Moreover, iPSCs not only fail to induce IFN-I in response to infection, IFN-I treatment of these cells also yields little transcriptional response (8, 9). Despite this phenotype, iPSCs do not demonstrate hypersusceptability to virus infection, an activity that has been credited to a wide variety of mechanisms including: iPSC-specific factors, RNA interference (RNAi), and the high baseline levels of a subset of ISGs (10–12).
As the precise mechanisms underlying why pluripotent stem cells fail to induce a robust transcriptional response to virus remain somewhat unclear, here we focused on how reprogramming induces this phenotype and why the canonical IFN-I system is not utilized. To this end, we established a primary cell model in which differentiated fibroblasts could be made pluripotent and then redifferentiated back to a fibroblast lineage. We demonstrate that there is an inverse correlation between a functioning IFN-I system and the maintenance of pluripotency. Moreover, we find that of the four factors required for the generation of iPSCs, Krüppel-like factor 4 (KLF4), could repress antiviral induction through the direct engagement of IFN-stimulated response elements (ISREs). To further understand the basis for this phenotype, we established an iPSC model in which we could induce the expression of a constitutively active version of IRF7, a strong transactivator of ISREs (13). In the absence of induction, these cells were indistinguishable from untreated or GFP-expressing cells but, upon activation, a significant subset of ISGs could be observed. In addition to ISGs, temporal expression of this IRF7Δ transcriptional program resulted in dysregulation of many pluripotency and lineage-specific genes in both resting and differentiated iPSCs. Taken together, these data suggest that the IFN-I system is incompatible with maintaining a pluripotent state and may explain why these cells fail to utilize such a successful intrinsic defense system.
Results
Induced Pluripotent Stem Cells Demonstrate a Diminished Response to Virus-Related Signals.
To investigate the ability of stem cells to respond to virus infection, we utilized a model system from which human primary foreskin fibroblasts (FFs) were reprogrammed to induced pluripotent stem cells (iPSCFF) utilizing lentiviruses expressing OCT4 (POU class 5 homeobox 1), SOX2 (SRY-box 2), KLF4, and c-MYC (MYC protooncogene) (referred to as OSKM) (SI Appendix, Fig. S1 A–D) (14). In addition, iPSCFF were redifferentiated back into fibroblasts (FFiPSC), allowing us to directly compare pluripotent and differentiated cells from identical genetic backgrounds. The transcriptional landscape of these three cellular populations was subsequently defined by multidimensional scaling (MDS) based on next generation sequencing (Fig. 1A and SI Appendix, Table S1). These data confirmed a high degree of transcriptional similarity between FF and FFiPSC and illustrated a significant variation between these cell populations and iPSCFF. Moreover, iPSCFF expressed stem cell-specific markers including: endogenous NANOG (Nanog homeobox), SOX2, TRA-1-60, and OCT4 and maintained the capacity to form ectoderm, mesoderm, and endoderm in vitro through directed differentiation (SI Appendix, Fig. S1 B–D).
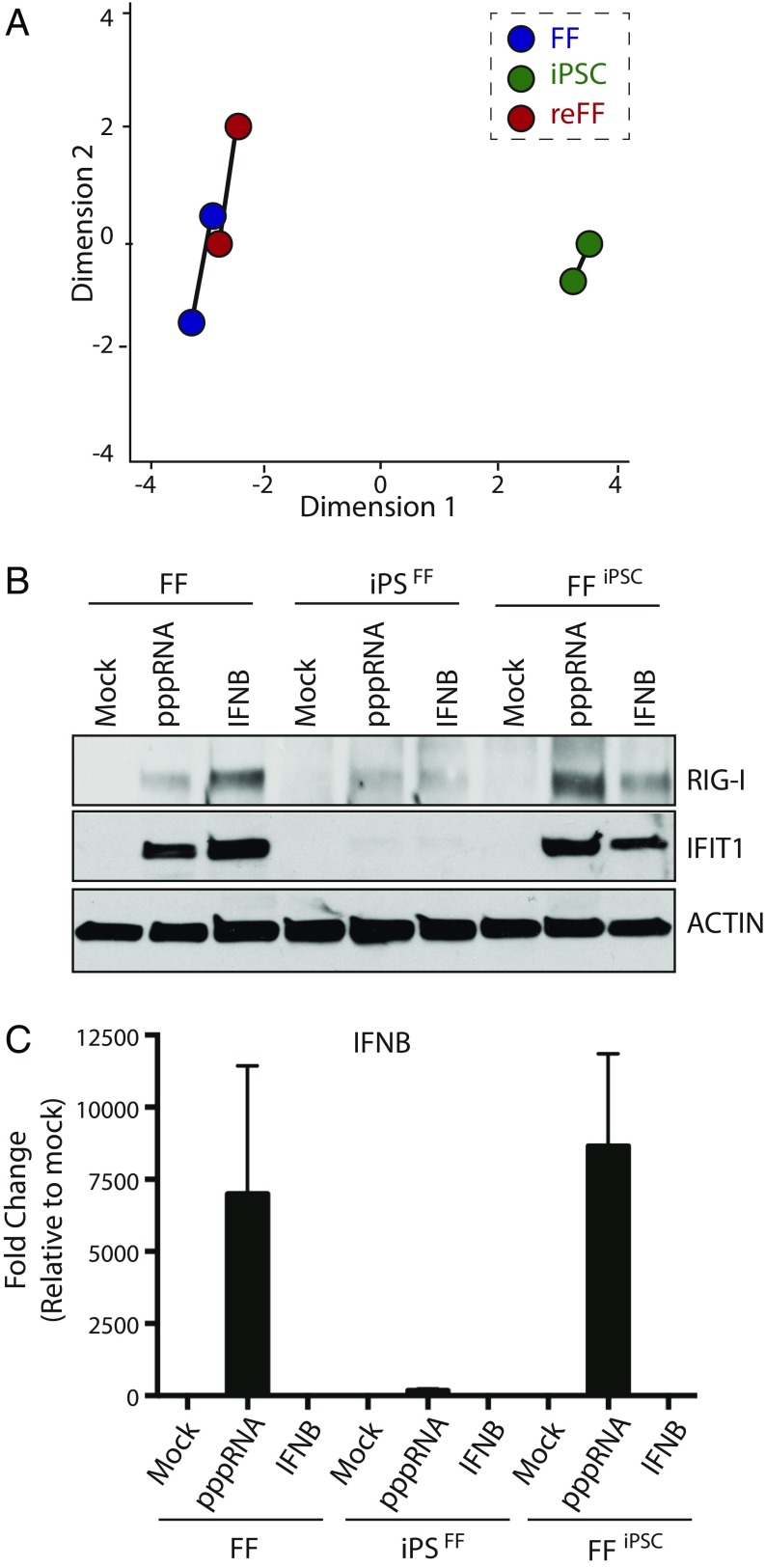
Fig. 1.
Pluripotent cells lack the IFN-I response observed in differentiated cells. (A) Analysis of the transcriptional variation between FFs, iPSCs, and rederived foreskin fibroblasts (reFFs) by MDS. (B) Western blot of cells described in A either mock treated or transfected with viral RNA (pppRNA) or treated with IFN-beta (IFNB). Blots depict levels of RIG-I, IFIT1, and ACTIN. (C) Same conditions as B in which RNA was analyzed by quantitative real-time PCR for IFNB mRNA expression, represented as fold change and normalized to mock-treated samples.
Having established a model system to compare pluripotent and differentiated cells directly, we next treated them with either viral-derived RNA containing 5′ triphosphates (herein denoted as pppRNA) or recombinant IFNB (Fig. 1B). Total whole cell extract was collected 12 h posttreatment and protein levels of the IFN-stimulated gene IFIT1 (IFN-Induced Protein with Tetratricopeptide Repeats 1) and RIG-I (Retinoic Acid-Inducible Gene 1 Protein) were determined. These data demonstrated that in contrast to fibroblasts, iPSCs failed to induce a response to either pppRNA or IFNB. These dynamics were also reflected in the small RNA profiles of resting or pppRNA-treated cellular populations, suggesting neither miRNAs, nor virus-induced small RNAs, were involved in this biology (SI Appendix, Table S2). Moreover, we could confirm that this phenotype was not the result of clonal selection or loss of genetic material as redifferentiation of our iPSCs back to fibroblasts successfully restored responsiveness to pppRNA and IFNB. Evaluation of the IFNB transcript itself demonstrated a robust induction in response to pppRNA in both fibroblast models but showed little to no induction in iPSCs consistent with previous findings (Fig. 1C) (8, 9). Taken together, these data suggest that pluripotency renders cells relatively nonresponsive with regards to the canonical antiviral defenses.
Characterizing the iPSC Transcriptome in Response to IFN-I and Virus Infection.
To further evaluate whether the inability to induce IFIT1 following pppRNA treatment reflected a general lack of type I IFN responsiveness in iPSCs, we next treated FFs and additional clones of iPSCs, these generated by Sendai delivery of OSKM (Methods), with recombinant IFNB, and determined the transcriptional landscape by RNA-Seq. Differential expression at 8 h post-IFNB treatment demonstrated FFs to have significant enrichment in genes involved in the cellular response to virus and IFN-I signaling, as would be expected (Fig. 2A and SI Appendix, Table S3). Included in the response to IFNB, we observed a significant induction of IRF7 (∼30-fold induction in FF cells compared with no induction in iPSCs), MX1 (MX Dynamin-Like GTPase 1) (∼300-fold induction in FF cells compared with ∼4-fold induction in iPSCs), and IFIT1 (∼200-fold induction in FF cells compared with ∼5-fold induction in iPSCs). These results could all be independently corroborated by qRT-PCR (SI Appendix, Fig. S2A).
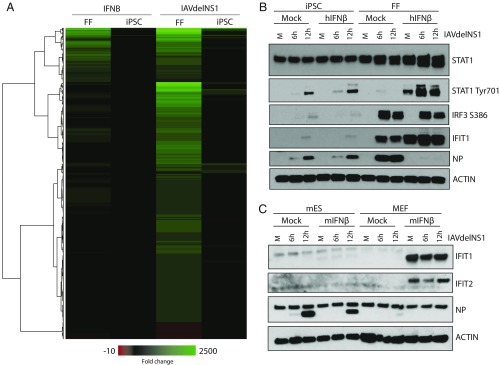
Fig. 2.
Response to IFNB and viral infection in pluripotent cells is attenuated. (A) Heatmap derived from RNA-Seq gene expression profiles from FFs and iPSCs in response to IFNB or influenza A virus (IAV) lacking the IFN-I antagonist NS1 (IAVdelNS1), represented as fold change. Heatmap represents genes that are significant (adjusted P value <0.05) and have an abs(log2fold change) >1 in any of the four conditions. (B) Human iPSCs and FFs were mock or pretreated with 300 units of recombinant human IFNB for 12 h before infection with IAVdelNS1. Whole cell extract was obtained at 6 and 12 h postinfection and immunoblotted against STAT1, IFIT1, NP, phosphorylated STAT1, phosphorylated IRF3, and ACTIN as indicated. (C) mESCs and MEFs were treated with 200 units of recombinant murine IFNB and analyzed as in B. Immunoblots depict the protein levels of IFIT1, IFIT2, NP, and ACTIN.
Next, to define the transcriptional response to virus infection we treated FFs and iPSCs with an H1N1 influenza A virus lacking its IFN-I antagonist NS1 (herein referred to as IAVdelNS1) (15). Sequence and cluster analyses of IAVdelNS1-infected cells at 8 h postinfection demonstrated a robust antiviral response in FFs as well as a dramatically diminished induction of a small subset of ISGs in iPSCs (SI Appendix, Table S3). In contrast to the canonical ISG profile following IFNB stimulation, infection with IAVdelNS1 in FF cells also induced many other transcriptional changes that were equally absent in iPSCs—further demonstrating their lack of response to virus infection (Fig. 2A). Interestingly, when examining virus levels, we find that iPSCs maintained a relatively effective response, given the lack of antiviral employment, a phenotype likely due to the high baseline levels of ISGs in these cells as reported elsewhere (SI Appendix, Fig. S2 B and C) (12).
Given the previous results, we predicted that upon pretreatment with IFNB and subsequent viral challenge, we would see limited protection in iPSCs compared with our fibroblasts. To test this, we treated cells with IFNB for 12 h before infection with IAVdelNS1 at a multiplicity of infection (MOI) of two for 6 and 12 h. As anticipated, IFNB treatment resulted in the up-regulation of STAT1 (Signal Transducer and Activator of Transcription 1) and IFIT1 in fibroblasts, and complete protection from IAVdelNS1 challenge (Fig. 2B). In contrast, iPSCs treated with IFNB failed to control IAVdelNS1-mediated production of viral nucleoprotein (NP), correlating with a lack of STAT1 and IFIT1 up-regulation. Upon examination of the phosphorylation status of IRF3 and STAT1, corresponding to their activated states, we find diminished levels of both S386 and Y701, respectively, suggesting iPSCs may harbor a dominant negative factor that represses this response (Fig. 2B).
To validate this phenotype across species and to ensure this phenotype was not the product of the nonphysiological process of “dedifferentiation,” we also looked at sensitivity of murine embryonic stem cells (mESCs) to virus infection and IFNB (Fig. 2C). These analyses revealed that, comparable to our iPSC model, mESCs failed to control virus levels of NP or induce IFIT1 or IFIT2 in response to either IFNB or virus infection. This is in contrast to primary murine embryonic fibroblasts (MEFs), which demonstrated a robust IFNB response. Moreover, while IAVdelNS1 does not replicate well in MEFs, we do observe a complete loss of NP signal following the treatment of IFNB. These data corroborate our RNA-Seq analyses and further suggest that pluripotent stem cells do not respond to virus infection using the canonical IFN-I response. Moreover, the shared phenotypes between iPSCs and mESCs suggest a generalizability to this phenomenon and point to a greater underlying reason as to why it is implemented.
Defining the Interplay Between Cellular Reprogramming and Induction of the Antiviral State.
IFNB induction relies on the activation of IRF3 and IRF7 which bind to two adjacent ISREs just upstream of the transcriptional start site (16). IRF7, like IRF3, requires phosphorylation by the IKK-related kinases to relieve its autoinhibition—a process which can be bypassed through the deletion of residues 247–467, a construct herein referred to as IRF7Δ (17). In contrast to IRF3, which is expressed ubiquitously and only engages perfect ISREs, IRF7 levels are induced in response to virus and can activate ISREs that deviate from the consensus by two to three bases, increasing its transcriptional footprint. This promiscuous activity of IRF7 ultimately results in generating a greater array of transcriptional targets and is responsible for the stochastic expression of IFNB as its ISREs deviate from the canonical sequence (18–21). For this reason, IRF7 is often referred to as the “master regulator” of the IFN-I system as it is sufficient to both induce IFN-I and protect cells from virus infection by directly up-regulating a large subset of ISGs in an IFN-I–independent manner (SI Appendix, Fig. S3 A–C and Table S4) (3, 13, 22).
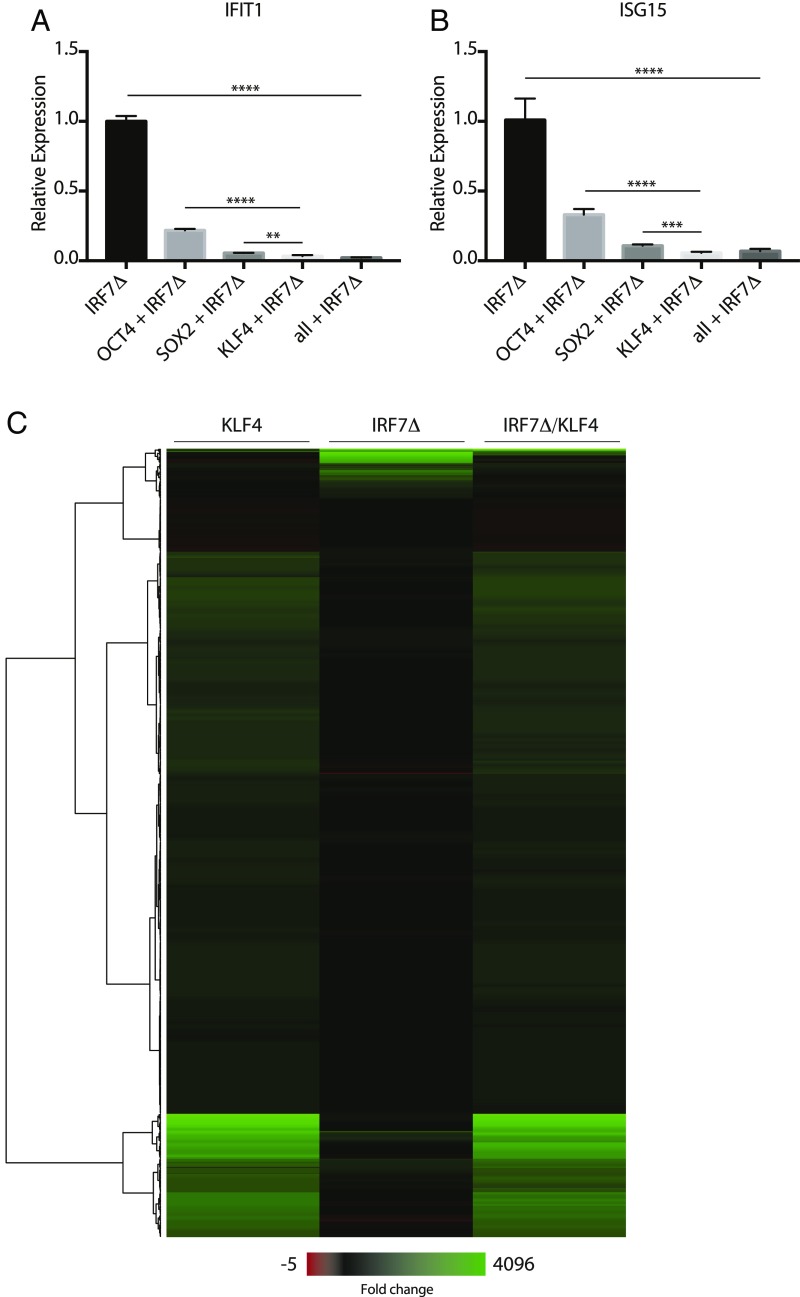
To better understand the molecular basis for the lack of response to virus in iPSCs, we next utilized the reprogramming factors that enable their biology. To this end, we coexpressed OCT4, SOX2, and KFL4 together with IRF7Δ and monitored endogenous expression of IFIT1 and ISG15 (IFN-Stimulated Gene 15) (Fig. 3 A and B). Interestingly, these data revealed that each of the three factors could repress IRF7Δ with KLF4 being the most potent. These data are consistent with recent reports that these three transcription factors can cause significant chromatin remodeling and can compact the DNA thus restricting transcriptional induction (23). Given the potent transcriptional repression observed upon KLF4 expression, we performed RNA-Seq on fibroblasts expressing KLF4 with or without IRF7Δ (Fig. 3C and SI Appendix, Table S4). KLF4 alone showed a distinct transcriptional footprint that did not include the canonical ISGs induced by IRF7Δ alone. When expressed together, KLF4 maintained its transcriptional signature while neutralizing the output of IRF7Δ supporting the idea that KLF4 prevents DNA binding of ISREs and, in so doing, also condenses the chromatin around these areas consistent with the findings of others (23, 24).
Fig. 3.
Reprogramming factors inhibit IRF7Δ response in fibroblasts. (A and B) Overexpression of OCT4, SOX2, and KLF4 with IRF7Δ in 293T cells. RNA was analyzed by qRT-PCR for IFIT1 and ISG15 mRNA expression, represented as fold change and normalized to IRF7Δ alone. Error bars depict the SD of the mean. Significance was determined by unpaired Student’s t test, where ** to **** denote P values of 0.005 to <0.0001, respectively. Horizontal bars denote comparison groups. (C) Heatmap depicting gene expression profiles from 293T cells transfected with KLF4, IRF7Δ, or KLF4 and IRF7Δ. Heatmap represents genes that are significant (adjusted P value <0.05) and have an abs(log2fold change) >1 in any of the three conditions.
Eliciting an Antiviral Response in iPSCs Through the Constitutive Activation of IRF7.
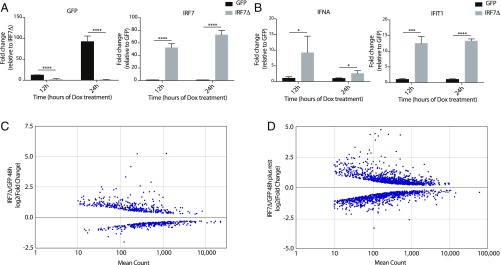
Given the effectiveness of IRF7Δ in inducing the IFN-I system, we set to determine whether extended expression of this construct would be sufficient to overcome the KLF4 block and induce an antiviral defense in iPSCs in the absence of virus infection or PAMP stimulation. To this end, we developed a lentivirus-based system that allowed for the expression of either GFP or IRF7Δ following culture in media containing doxycycline (Dox) (SI Appendix, Fig. S4A). Transduction and puromycin selection of iPSCs yielded multiple heterogenous lines expressing this inducible cassette. In response to Dox treatment, we observed the inducible expression of either GFP or IRF7Δ in each of the respective lines (Fig. 4A). Moreover, initial characterization of IRF7Δ-expressing cells at 12 and 24 h post-Dox treatment demonstrated a significant induction of IFNA (IFN alpha 1) and IFIT1 as well as distinct morphological changes (Fig. 4B and SI Appendix, Fig. S4B). To ascertain the consequences of eliciting an antiviral response in iPSCs, we performed RNA-Seq on these cells which revealed dramatic transcriptional changes to the IRF7Δ-expressing iPSCs, including many ISGs [e.g., IFIT1, IFITM3 (IFN Induced Transmembrane Protein 3), and TRIM22 (Tripartite Motif Containing 22)] (Fig. 4C and SI Appendix, Fig. S4C, and Table S5). Given that we could successfully engage the antiviral response in iPSCs, we next wanted to determine whether there were long-term consequences for this action.
Fig. 4.
Expression of IRF7Δ in iPSCs leads to short- and long-term transcriptional changes. (A) qRT-PCR analyses of GFP- or IRF7Δ-inducible iPSCs following 12 or 24 h of Dox. Data display levels of GFP and IRF7 as relative fold change. Error bars depict the SD of the mean. Significance was determined by unpaired Student’s t test, where **** denotes P values of <0.0001. (B) qRT-PCR analyses as in A for IFNA and IFIT1, where * to **** denote P values of 0.05 to <0.0001, respectively. (C) Graph depicting differential expression of genes as determined by log2(fold change) of mRNA-Seq data derived from GFP- vs. IRF7Δ-induced conditions at 48 h post-Dox treatment. The x axis denotes the mean read counts from biological replicates. Genes with low read counts or those that did not reach statistical significance are not represented. (D) Graph as in C of mRNA-Seq data derived from GFP- vs. IRF7Δ-induced conditions at 7 d posttreatment.
The intrinsic antiviral response in cells generally does not exceed 48 h postinfection. By this time, either the cell will have died in response to viral replication or the antiviral program will have successfully inhibited the infection and subsequently restored the cellular transcriptome to a baseline level (25). Given the morphological and transcriptional changes to iPSCs following just 48 h of expression, we next focused on whether the nonutilization of the antiviral program was the result of long-term changes that may be incompatible with pluripotency. To ascertain whether this was indeed the case, we stimulated iPSCs for 48 h and then allowed them to reset to baseline over a 5-d time period in culture conditions that should maintain stemness. Interestingly, RNA-Seq data from this experiment demonstrated ∼2,000 differentially expressed genes for which a randomly selected subset of transcripts could be further corroborated by qRT-PCR (Fig. 4D and SI Appendix, Fig. S4D and Table S6). In contrast to our 48-h Dox treatment, in these cells we neither observe differential expression of IRF7Δ nor any of the canonical ISGs associated with its initial expression (SI Appendix, Fig. S4E). It is noteworthy that this phenotype could be recapitulated in mESCs using transient transfection of a different IRF-inducing transcript whose expression resulted in temporal ISG induction that was followed by the loss of pluripotency markers (KLF4) and other chromatin remodeling enzymes (SI Appendix, Table S7). Taken together, these data suggest that the antiviral response can be engaged in iPSCs with use of IRF7Δ. Moreover, when IRF7Δ is removed, while the direct transcriptional targets return to baseline, our data support the idea that the cells may have undergone long-lasting changes as a consequence.
IRF7Δ Impairs the Differentiation Potential of Pluripotent Stem Cells.
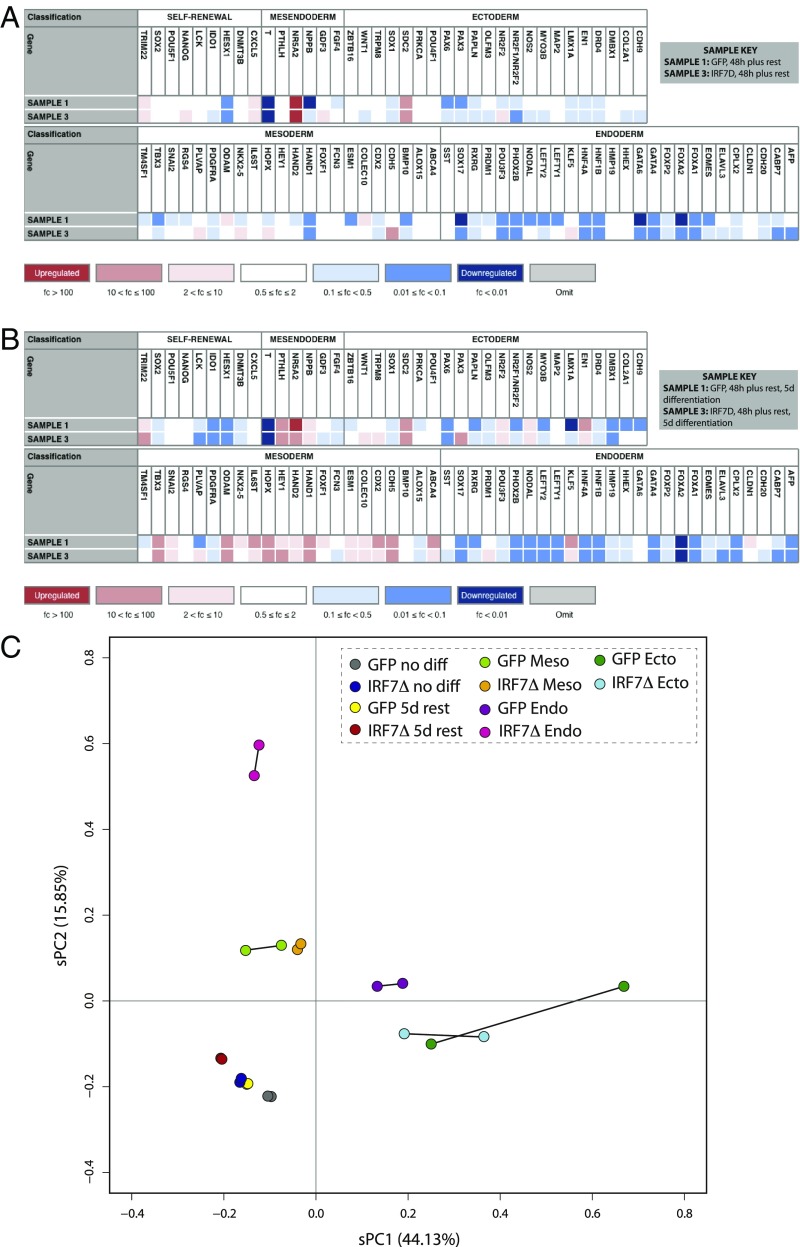
By definition, a cell is functionally pluripotent if it retains the capacity to differentiate into cellular subtypes derived from all three embryonic germ layers: ectoderm, mesoderm, and endoderm. The propensity of a cell to do this can be measured molecularly by both the expression of self-renewing factors at resting state as well as the expression of germ layer-specific genes upon directed differentiation. To determine how engagement of the antiviral response influenced iPSC differentiation potential, we induced cells to express GFP or IRF7Δ for 48 h and then allowed them to rest for 5 d before measuring the relative expression levels of a panel of self-renewal and germ layer-specific markers using the TaqMan hPSC Scorecard assay (26). These data demonstrated that while the temporal engagement of the antiviral response did not formally result in loss of pluripotency, at least two of the markers [i.e., NANOG and IDO1 (indoleamine 2,3-dioxygenase 1)] were dysregulated (Fig. 5A). Moreover, we can observe additional changes to the iPSC transcriptional landscape with markers for mesendoderm (a precursor to mesoderm and endoderm), ectoderm, mesoderm, and endoderm following IRF7Δ induction (Fig. 5A).
Fig. 5.
Transient IRF7Δ expression results in lasting transcriptional changes to iPSCs and affects differentiation potential. (A and B) hPSC ScoreCard results for Dox-treated (A) and differentiated (B) iPSCs. Each sample represents biological duplicates. Colors correlate to the fold change in expression of the indicated gene relative to the undifferentiated reference set, where blue is down-regulated, white is unchanged, and red is up-regulated. (C) Analysis of the transcriptional variation between duplicate samples of untreated iPSCs, Dox-treated iPSCs, and trilineage differentiations by sPCA for GFP and IRF7Δ lines.
Given the transcriptional changes that result following the temporal induction of IRF7Δ, we next investigated how these cells would behave following undirected differentiation (Fig. 5B). For this we treated 48-h pulsed GFP and IRF7Δ iPSCs with FBS and allowed them to spontaneously form cell types of the ectoderm, mesoderm, and endoderm. After 48 h of stimulation, 5 d of rest, followed by an additional 5 d of FBS treatment, we observed a complete departure from pluripotency and a general tendency to form mesoderm in both cellular populations (Fig. 5B). However, while GFP-expressing cells also showed low levels of ectoderm and endoderm following undirected differentiation, these lineages appeared significantly dysregulated in IRF7Δ iPSCs. These data suggest that even short-term induction of ISGs is impairing ectoderm and endoderm while seemingly not affecting mesoderm formation (Fig. 5B).
In an effort to better define the impact the antiviral response has on developmental potential, we performed a directed differentiation into each of the three germ layers following pulsed induction of GFP or IRF7Δ. Following the removal of Dox and 5 d of rest, we performed differentiations into the endoderm, mesoderm, and ectoderm using a monolayer protocol (Fig. 5C and SI Appendix, Table S8). After treatments, cells were sequenced, and the resulting transcriptional output was analyzed by sparse principal component analysis (sPCA). These data found that, in agreement with our ScoreCard results, GFP- or IRF7Δ-induced cells had minimal impact at resting state showing that biological duplicates were positioned in an overlapping region on the sPCA with their untreated pluripotent counterparts (Fig. 5C). In contrast, differentiation of IRF7Δ-induced cells showed variable distances on the sPCA compared with the GFP-induced population. While mesoderm differentiation showed a tight cluster on the sPCA denoting comparable gene signatures, ectoderm, and mesoderm differed more dramatically. While ectoderm signatures still maintained some overlap between GFP and IRF7Δ, endoderm differentiation following temporal engagement of the antiviral defenses resulted in a dramatic departure from the control with the biological replicates appearing a great distance from the GFP samples on the sPCA (Fig. 5C). These results suggest that the induction of the antiviral state causes significant transcriptional changes that are evident well beyond the direct induction of IRF7Δ target genes.
IRF7Δ Induces Aberrant Gene Expression Despite Enhanced Cardiomyocyte Formation.
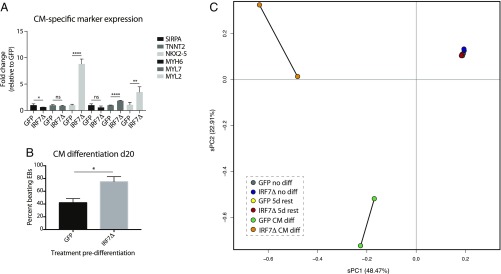
Upon finding that IRF7Δ expression predominantly impacted endoderm and ectoderm formation, we next determined whether induction of the antiviral response in iPSCs perhaps influenced the differentiation of mesoderm sublineages. As cardiomyocyte development is a well-characterized mesoderm derivative, we chose to focus on this differentiation event to ascertain whether there were consequences of IRF7Δ expression. Interestingly, we first noted that IRF7Δ-induced iPSCs which did not undergo directed differentiation showed significantly elevated expression levels for a number of cardiomyocyte markers at baseline (Fig. 6A) (27). To further test cardiomyocyte formation, we treated embryoid bodies (EBs) derived from GFP- and IRF7Δ-pulsed cells with activin A and BMP4 (SI Appendix, Fig. S6A). At 10 and 20 d postinduction, cardiac-specific markers were detected by qRT-PCR analysis. In support of our earlier findings, IRF7Δ-pulsed cells exhibited a significant increase in TNNT2, SIRPA, MYH6, and MYL7 transcript expression levels compared with GFP-pulsed cells (SI Appendix, Fig. S6B). Interestingly, whereas baseline induction of MYL2 was high following IRF7Δ, levels relative to GFP were significantly reduced following cardiomyocyte differentiation. At the completion of this differentiation process, we also could observe significantly increased beating in IRF7Δ-expressing cells (Fig. 6B and Movie S1 and S2).
Fig. 6.
IRF7Δ dysregulates gene expression during cardiomyocyte differentiation. (A) qRT-PCR analysis of cardiomyocyte (CM)-specific markers from GFP- or IRF7Δ- expressing iPSC clones treated with Dox for 48 h before a 5-d rest. Levels of SIRPA, TNNT2, NKX2-5, MYH6, MYL7, and MYL2 mRNA are depicted as fold change over GFP. Error bars depict the SD of the mean. Significance was determined by unpaired Student’s t test where * to **** denote P values of 0.05 to <0.0001, respectively. (B) Graph quantifying the beating behavior of GFP- vs. IRF7Δ-pulsed iPSCs on day 20. (C) Analysis of the transcriptional variation between duplicate samples of untreated iPSCs, Dox-treated iPSCs, and cardiomyocyte differentiations for GFP and IRF7Δ lines.
Given the aberrant pattern among the cardiomyocyte gene markers in IRF7Δ-pulsed cells, we next analyzed whether, despite the increased beating phenotype, these cardiomyocytes demonstrated other anomalies. To this end, we performed RNA-Seq and analyzed the data by sPCA (Fig. 6C and SI Appendix, Table S9). These data revealed dramatic transcriptional changes in IRF7Δ-pulsed cardiomyocytes compared with the control GFP cells, suggesting that, despite successful differentiation, temporal induction of the antiviral state leads to lasting effects (denoted in this experiment by ∼5,000 differentially expressed genes).
Discussion
Every multicellular organism derives from progenitor stem cells (28). These specialized cells maintain the capacity to differentiate into all of the cellular lineages that comprise the organism. Stem cell potency itself exists as a gradient, with totipotent cells having the greatest potential, followed by pluripotent, multipotent, unipotent, and finally differentiated cells (29). Pluripotent cells maintain the capacity to differentiate into ectoderm, mesoderm, and endoderm, whereas multipotent stem cells are limited to self-renewal and the capacity to develop into multiple cell types present in a specific tissue or organ (30).
The cellular genes involved in both differentiation and defense are vast in number and show a high propensity of duplication events exemplified by the family of hox or IFN genes, respectively (31, 32). Perhaps even more noteworthy is the fact that these two systems, development and defense, often borrow from each other over an evolutionary time course. For example, Toll like receptors, while required for development in many invertebrates, were repurposed to be PRRs in vertebrates (33). Conversely, antiviral RNAi systems of early eukaryotes were independently duplicated to generate developmental miRNA systems in plants and animals (34). As biology tends to repurpose the old rather than generate de novo material (35), one can observe many gene duplications for cellular components involved in one or even both of these two systems [i.e., bone marrow stromal antigen 2 (BST2/tetherin) which also serve to aggregate viruses at the cells lipid bilayer] (36). Given this relationship, it is tempting to speculate that these two systems maintain a complex relationship which renders pluripotency and defense genes at odds with each other. This concept is supported by the fact that in contrast to pluripotent stem cells, multipotent adult stem cells which have enabled development expression profiles become responsive to IFN-I, suggesting that lineage commitment rapidly restores their innate antiviral program (37, 38).
Here we show that KLF4, and to a lesser extent SOX2 and OCT4, are incompatible with the transcriptional potential of IRF7, a master regulator of the antiviral defenses. Given that the binding motifs for these reprogramming factors do not overlap with that of an ISRE, we can assume that this phenotype is an indirect consequence of downstream target genes. Given that RNA-Seq data show no overt loss of read numbers to transcripts associated with initiating the antiviral response, we postulate that the accessibility to ISREs must be restricted as a result of chromatin remodeling, a concept supported by Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-Seq) data (23). However, while this idea explains the lack of transcriptional induction, it fails to address the low levels of phosphorylation of IRF3, NF-κB, or STAT1 observed by us, and others (6, 39). In this regard it is noteworthy that despite comparable levels of DDX58 read numbers, the baseline levels of RIG-I (the product of the DDX58 gene) are significantly different (Fig. 1B) suggesting that pluripotent cells may express unique proteases and/or microRNAs responsible for the observed low levels of select antiviral proteins, work that has also been independently suggested by others (40, 41).
The induction of pluripotency is directly associated with a complete loss of responsiveness to IFN-I, viral RNA, or direct infection. While pluripotent cells are physically protected within a blastocyst, vertical transmission of pathogens to these cells can occur by several routes including infection of the surrounding trophoblasts as recently exemplified by Zika virus (42). Given this, the threat of vertical transmission suggests there would be value in having the capacity to elicit an antiviral response in pluripotent cells and, indeed two independent groups have found pluripotent stem cells harbor high baseline levels of specific ISGs to help minimize infection (10, 12). However, as effective as a handful of constitutive ISGs may be at combating a subset of viruses, this minimal defense strategy cannot substitute for the full breadth of the IFN-I response.
Here we sought to determine what the consequences would be should this defense be artificially induced. Our logic was that the consequences of induction may illustrate why the intrinsic defenses are not utilized in what are arguably the most important cells. To this end, we engineered iPSCs to express IFN-I–related genes through a constitutively active transcript of IRF7. We found this strategy to successfully induce an antiviral-like response in iPSCs—a phenotype that we could not accomplish through any other means. Induction of IRF7 successfully induced many ISGs in agreement with its capacity to engage a wide variety of ISREs and was sufficient to protect cells from the cytotoxicity of virus infection. However, even with only a partial antiviral response, we observed immediate effects on the morphology of iPSCs in response to IRF7 expression. Moreover, changes in morphology also coincided with extensive transcriptional activity that was indirect from IRF7 that seemingly pushed cells away from pluripotency and, in the case of our iPSCs, toward the mesoderm lineage. Based on our data, it is clear that IRF7 primes cells to exit pluripotency, but it remains to be determined whether the bias toward mesoderm is specific or is additionally influenced by the baseline tendency of a given iPSC line.
Taken together, the data presented here support the hypothesis that engagement of any aspect of the antiviral response may be incompatible with maintaining pluripotency. These observations not only aid in our understanding as to why these specialized cells remain inert with regards to intrinsic defenses, but it also may explain why developmentally they are compartmentalized within trophoblasts which harbor a number of unique antiviral defense mechanisms (43). In all, it would appear that the complexities of maintaining pluripotency are incompatible with the induction of the many antiviral effectors that comprise the IFN-based system. While the basis for this incompatibly is likely multifactorial, it is tempting to speculate that the crosstalk that has occurred between developmental and defense systems over the past 2.5 million years of vertebrate evolution may preclude the simultaneous engagement of these two programs under any circumstance.
Methods
iPSC Cell Lines.
Human-induced pluripotent stem cells (hiPSCs) used in Fig. 1 were generated from primary FFs using a lentivirus expressing NANOG, SOX2, OCT4, and cMYC and selected for TRA-1–60 expression. Human iPSCs used in Figs. 2–6 were generated from healthy patient fibroblasts using a Sendai virus expressing NANOG, SOX2, OCT4, and cMYC and selected for TRA-1–60 expression. To validate pluripotency in these clonal iPSC populations, cells were differentiated into the three embryonic germ layers in vitro: mesoderm, endoderm, and ectoderm. Additional details regarding cells and culturing can be found in SI Appendix, SI Materials and Methods.
RNA-Seq and Analysis.
Deep sequencing was performed on indicated samples, with each condition containing two biological replicates. Libraries were prepared using the TruSeq RNA Library Preparation Kit version 2 (Illumina) according to the manufacturer’s instructions. In brief, 1 μg of total mRNA was purified using oligo-dT RNA purification beads. Eluted RNA was fragmented and reverse transcribed, followed by a second strand synthesis, end repair, A-tailing, adaptor ligation, and PCR amplification. Libraries were quantified using a universal complete KAPA Library Quantification Kit (KAPA Biosystems). Pooled libraries were sequenced on an Illumina MiSeq (Figs. 1, 2, and 4) or an Illumina NextSeq (Figs. 5 and 6) platform. Sequencing reads were aligned to the human genome (hg19) combined with differential expression analyses using the RNA Express application (Illumina, BaseSpace). Global transcriptional profiles were visualized by MDS on normalized gene counts using plotMDS from the limma package in R (44). Heatmap plots with hierarchical clustering were performed on log2(fold change) gene values using heatmap.2 of the gplots package in R. sPCA was performed on reads per kilobase per million (RPKM) counts for selected samples using the SPC function {[k = 4, sumabsv = FLOOR(SQRT(#samples))]} from the PMA package in R (45). All sequencing data can be found at the GEO accession no. GSE122794 (46).
Cloning and Expression Analyses.
Expression of GFP and IRF7Δ is under the control of the TRE3G promoter (repeat tet operator sequences). This vector also expresses BFP and PuroR under the control of the hPGK promoter. FUW-M2rtTA plasmid is a lentiviral vector that expresses a reverse tetracycline-controlled transactivator (rtTA) under the control of the ubiquitin C (UBC) promoter. FUW-M2rtTA was a gift from Rudolf Jaenisch, Whitehead Institute, Cambridge, MA (Addgene, plasmid 20342) (47). Transduction conditions and methods to determine expression can be found in SI Appendix, SI Materials and Methods.
Trilineage and Undirected Differentiations.
iPSCs were differentiated to the three germ layers using a directed Trilineage Differentiation Kit (05230, STEMdiff) according to the manufacturer’s protocol. In short, iPSCs were plated as single cells on Matrigel in either ectoderm single-cell plating medium or mesoderm/endoderm single-cell plating medium. Every 24 h, media were replaced with STEMdiff trilineage ectoderm medium, STEMdiff trilineage mesoderm medium, or STEMdiff trilineage endoderm medium for the duration of 5 d (mesoderm and endoderm lineages) or 7 d (ectoderm lineages). Undirected differentiation of iPSCs was performed in reduced hiPSC media (DMEM/F12, 1× NEAA, 1× GlutaMAX, 1× Anti/Anti, 1% B-mercaptoethanol) supplemented with 10% FBS as a monolayer culture for the duration of 5 d. Lineage development was assessed by hPSC Scorecard analysis (SI Appendix, SI Materials and Methods).
Cardiomyocyte Differentiations.
iPSCs were differentiated to the cardiomyocyte lineage using an EB differentiation protocol (27, 48, 49). EBs were generated at day 0 by culture in base media (RPMI, 0.5× B27 + insulin supplement, 10 μL/mL ascorbic acid, 10 μL/mL glutamine, and 3 μL/mL monothioglycerol) supplemented with 2 ng/μL BMP4 and 0.3 μL/mL thiazovivin. On day 1, EBs were harvested and resuspended in base media supplemented with 20 ng/mL BMP4, 20 ng/mL activin A, 5 ng/mL bFGF, and 0.1 μL/mL thiazovivin) for the induction of primitive streak/mesoderm. On day 3, EBs were harvested and resuspended in base media supplemented with 5 μM XAV and 5 ng/mL VEGF for cardiac mesoderm induction. On days 5 and 7, EBs were again harvested and resuspended in base media supplemented with 5 ng/mL VEGF for expansion of cardiovascular lineages. On day 10, and as needed thereafter until day 20, EBs were harvested and resuspended in base media. Cultures were maintained in a 5% CO2, 37 °C incubator for the duration of the experiment.
Cardiomyocyte Dissociations.
EBs generated from iPSC differentiation experiments at days 10 and 20 were incubated in collagenase type II (1 mg/mL; Worthington) in Hanks’ solution (HBSS) overnight at 37 °C. The following day, the equivalent amount of wash buffer (DMEM supplemented with BSA and DNase) was added to the cell suspension and the EBs were pipetted to dissociate the cells. After dissociation, cells were centrifuged for 3 min at 300 × g and resuspended in TRIzol.
Statistical Analysis.
Statistical analysis was performed on indicated samples using a two-tailed, unpaired Student’s t test. Data are considered significant if P value is <0.05.
Supplementary Material
Acknowledgments
We thank Dr. Nicole Dubois and Evan Bardot (Icahn School of Medicine at Mount Sinai, ISMMS) for the cardiomyocyte differentiation protocol and helpful discussions; Seok-Man Ho (a former graduate student of Dr. K. Brennand, ISMMS) for gifting us the pTRE3G backbone; Dr. Ivan Marazzi and Yesai Fstkchyan for gifting us murine ES cells; Dr. Garcia-Sastre for the RIG-I antibody; and the Stem Cell Core at ISMMS for supplying us with reagents. J.E. is supported in part by the New York University–ISMMS Mechanisms of Virus–Host Interactions National Institutes of Health T32 Training Grant (AI007647-09). D.B.-M. is supported by a Life Science Research Fellowship. This work was also supported by the National Institutes of Health (Grants R21 AI132913-01A1 and R01 MH101454) and the New York Stem Cell Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE122794).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812449116/-/DCSupplemental.
References
- 1.tenOever BR. The evolution of antiviral defense systems. Cell Host Microbe. 2016;19:142–149. doi: 10.1016/j.chom.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: A complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid S, Mordstein M, Kochs G, García-Sastre A, Tenoever BR. Transcription factor redundancy ensures induction of the antiviral state. J Biol Chem. 2010;285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AE, et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017;15:e2004086. doi: 10.1371/journal.pbio.2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zampetaki A, Xiao Q, Zeng L, Hu Y, Xu Q. TLR4 expression in mouse embryonic stem cells and in stem cell-derived vascular cells is regulated by epigenetic modifications. Biochem Biophys Res Commun. 2006;347:89–99. doi: 10.1016/j.bbrc.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 6.Wang R, et al. Mouse embryonic stem cells are deficient in type I interferon expression in response to viral infections and double-stranded RNA. J Biol Chem. 2013;288:15926–15936. doi: 10.1074/jbc.M112.421438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Rossi R, Hale C, Goulding D, Dougan G. Interaction of enteric bacterial pathogens with murine embryonic stem cells. Infect Immun. 2009;77:585–597. doi: 10.1128/IAI.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke DC, Graham CF, Lehman JM. Appearance of interferon inducibility and sensitivity during differentiation of murine teratocarcinoma cells in vitro. Cell. 1978;13:243–248. doi: 10.1016/0092-8674(78)90193-9. [DOI] [PubMed] [Google Scholar]
- 9.Hong XX, Carmichael GG. Innate immunity in pluripotent human cells: Attenuated response to interferon-β. J Biol Chem. 2013;288:16196–16205. doi: 10.1074/jbc.M112.435461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maillard PV, et al. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X, et al. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172:423–438.e25. doi: 10.1016/j.cell.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 14.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 16.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 18.Lin R, Génin P, Mamane Y, Hiscott J. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol Cell Biol. 2000;20:6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin R, Mamane Y, Hiscott J. Multiple regulatory domains control IRF-7 activity in response to virus infection. J Biol Chem. 2000;275:34320–34327. doi: 10.1074/jbc.M002814200. [DOI] [PubMed] [Google Scholar]
- 20.Génin P, Lin R, Hiscott J, Civas A. Differential regulation of human interferon A gene expression by interferon regulatory factors 3 and 7. Mol Cell Biol. 2009;29:3435–3450. doi: 10.1128/MCB.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao M, Zhang J, Phatnani H, Scheu S, Maniatis T. Stochastic expression of the interferon-β gene. PLoS Biol. 2012;10:e1001249. doi: 10.1371/journal.pbio.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid S, Sachs D, tenOever BR. Mitogen-activated protein kinase-mediated licensing of interferon regulatory factor 3/7 reinforces the cell response to virus. J Biol Chem. 2014;289:299–311. doi: 10.1074/jbc.M113.519934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D, et al. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell. 2017;21:819–833 e816. doi: 10.1016/j.stem.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Luo WW, Lian H, Zhong B, Shu HB, Li S. Krüppel-like factor 4 negatively regulates cellular antiviral immune response. Cell Mol Immunol. 2016;13:65–72. doi: 10.1038/cmi.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heaton NS, et al. Long-term survival of influenza virus infected club cells drives immunopathology. J Exp Med. 2014;211:1707–1714. doi: 10.1084/jem.20140488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois NC, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sánchez Alvarado A, Yamanaka S. Rethinking differentiation: Stem cells, regeneration, and plasticity. Cell. 2014;157:110–119. doi: 10.1016/j.cell.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weissman IL. Stem cells: Units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 30.Seydoux G, Braun RE. Pathway to totipotency: Lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 31.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 32.Puigbò P, Makarova KS, Kristensen DM, Wolf YI, Koonin EV. Reconstruction of the evolution of microbial defense systems. BMC Evol Biol. 2017;17:94. doi: 10.1186/s12862-017-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leulier F, Lemaitre B. Toll-like receptors–Taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 34.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: From protists to man. Curr Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguado LC, et al. RNase III nucleases from diverse kingdoms serve as antiviral effectors. Nature. 2017;547:114–117. doi: 10.1038/nature22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanco-Melo D, Venkatesh S, Bieniasz PD. Origins and evolution of tetherin, an Orphan antiviral gene. Cell Host Microbe. 2016;20:189–201. doi: 10.1016/j.chom.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 38.Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Angelo W, et al. The molecular basis for the lack of inflammatory responses in mouse embryonic stem cells and their differentiated cells. J Immunol. 2017;198:2147–2155. doi: 10.4049/jimmunol.1601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buckley SM, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Celià-Terrassa T, et al. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nat Cell Biol. 2017;19:711–723. doi: 10.1038/ncb3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coyne CB, Lazear HM. Zika virus–Reigniting the TORCH. Nat Rev Microbiol. 2016;14:707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- 43.Delorme-Axford E, et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci USA. 2013;110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witten DM, Tibshirani R, Hastie T. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009;10:515–534. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eggenberger JA, et al. 2018 The Type I interferon response impairs differentiation potential of pluripotent stem cells. Gene Expression Omnibus (GEO) database. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122793 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122791. Deposited on December 20, 2018.
- 47.Hockemeyer D, et al. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 49.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.