Significance
Due to its severity and early onset, X-linked retinitis pigmentosa (XLRP) is a particularly devastating form of retinal degeneration. Most males with XLRP come to medical attention before the age of 20. Approximately 15% of all RP is X-linked, and more than 70% of these cases are caused by mutations in the RPGR gene. Currently, there is no known assay to evaluate the nature of RPGR missense variants functionally. In this study, we developed an in vitro assay to examine how different missense variations in RPGR cause XLRP. Our assay is based on the RPGR protein interaction network. Our method provides a cost-effective test for RPGR functional mutation analysis.
Keywords: RPGR, cilia, retinal degeneration, PDE6D, INPP5E
Abstract
Retinitis pigmentosa (RP) is an inherited retinal degenerative disease with severe vision impairment leading to blindness. About 10–15% of RP cases are caused by mutations in the RPGR gene, with RPGR mutations accounting for 70% of X-linked RP cases. The mechanism by which RPGR mutations cause photoreceptor cell dysfunction is not well understood. In this study, we show that the two isoforms of RPGR (RPGR1−19 and RPGRORF15) interact with endogenous PDE6D, INPP5E, and RPGRIP1L. The RPGR1−19 isoform contains two PDE6D binding sites with the C-terminal prenylation site being the predominant PDE6D binding site. The C terminus of RPGR1−19 that contains the prenylation site regulates its interaction with PDE6D, INPP5E, and RPGRIP1L. Only the RPGR1−19 isoform localizes to cilia in cultured RPE1 cells. Missense variations found in RPGR patients disrupt the interaction between RPGR isoforms and their endogenous interactors INPP5E, PDE6D, and RPGRIP1L. We evaluated a RPGR missense variation (M58K) found in a family with X-linked retinitis pigmentosa (XLRP) and show that this missense variation disrupts the interaction of RPGR isoforms with their endogenous interactors. The M58K variation also disrupts the ciliary localization of the RPGR1−19 isoform. Using this assay, we also show that some of the RPGR missense variants reported in the literature might not actually be disease causing. Our data establishes an in vitro assay that can be used to validate the potential pathogenicity of RPGR missense variants.
Retinitis pigmentosa (RP) is an inherited retinal degenerative disease that results in the primary loss of rod photoreceptor cells followed by the secondary loss of cone photoreceptor cells (1). The disorder may be nonsyndromic occurring with retina dysfunction alone (1), or syndromic with other neurosensory disorders, developmental abnormalities, or complex clinical findings such as in Bardet-Biedl syndrome and Usher syndrome (2–4).
Mutations in any of dozens of different genes can cause the clinical phenotypes of retinitis pigmentosa, and these mutations can be inherited in an autosomal dominant, autosomal recessive, X-linked dominant, X-linked recessive, mitochondrial, or digenic fashion (5–9). Approximately 15% of all RP is X-linked, and more than 80% of these patients (i.e., more than 12% of the total) have mutations in the gene RPGR (10). About 60% of all RP-causing mutations in RPGR occur in exon 15 of the gene, and two-thirds of these (more than 5% of all RP) occur in a highly repetitive region between codons 801 and 1070 (10).
There are multiple isoforms of RPGR. Two major isoforms of RPGR were detected in the retina (11, 12): RPGR1−19, which has 19 exons encoding an 815-aa protein, and RPGRORF15, which has 15 exons plus part of intron 15 encoding a 1,152-aa protein. Both isoforms share exons 1–14. The N-terminal half of RPGR protein contains a tandem repeat structure highly similar to the regulator of chromosome condensation (RCC1) named RCC1-like domain (RLD), which regulates the RAN GTPase. The RPGRORF15 protein has a Glu-Gly–rich low complexity region in the C-terminal domain. The C terminus of the RPGR1−19 protein contains a cluster of basic residues and a consensus prenylation site. Rescue experiments in Rpgr knockout mice demonstrate that the RPGRORF15 isoform is functionally sufficient to rescue photoreceptor degeneration (13, 14).
RPGR localizes to connecting cilia of rod and cone photoreceptors, the transitional zone of motile cilia, and primary cilia both in vivo and in vitro (15–18). RPGR has been shown to be involved in microtubule organization or regulation of transport in primary cilia and actin stability (19–23). Likewise, RPGR is known to interact with numerous proteins including Whirlin (24), Gelsolin (20), SMC, IFT88, KIF3A, KAP3, NPM1, RAB8A, CEP290, RPGRIP1, RPGRIP1L, NPHP4, PDE6D, and INPP5E (17, 21, 22, 25–34). However, the importance of these interactions relative to the pathophysiology of RPGR is not known. Numerous mutations in RPGR associated with X-linked retinitis pigmentosa (XLRP) have been identified (35–41). These variants are found across the entire gene, with the majority of mutations occurring in the low complexity region of RPGRORF15. How these variants cause XLRP is not well understood. Here, we show that RPGR missense variations that cause XLRP disrupt the interaction of RPGR with its interactors and prevent ciliary localization of RPGR1−19. Our study establishes a practical in vitro assay to evaluate RPGR missense variations in the RLD region in a cell culture system.
Results
Both Isoforms of RPGR Interact with Endogenous PDE6D, INPP5E, and RPGRIP1L, but only RPGR1−19 Localizes to Cilia.
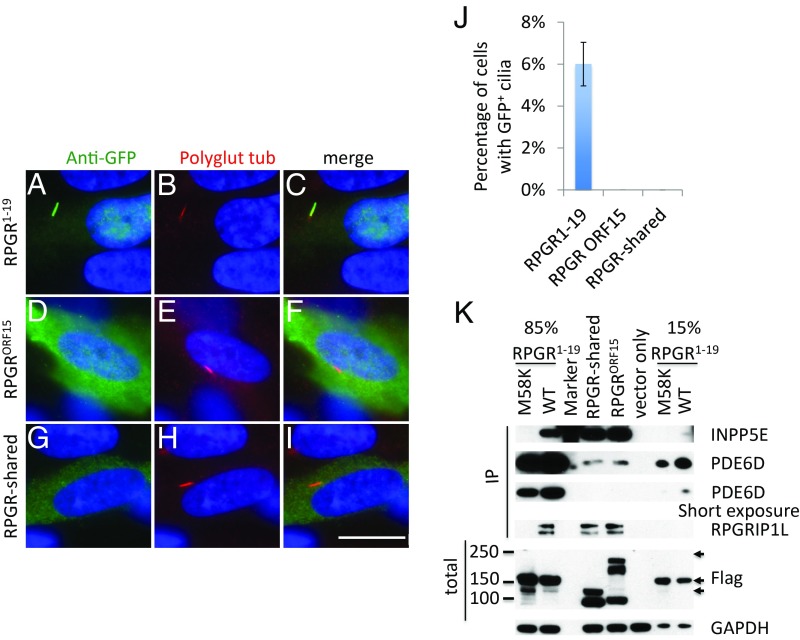
Two major RPGR isoforms are present in tissues and cells that arise through differential splicing (SI Appendix, Fig. S1): RPGR1−19 and the retina-specific isoform RPGRORF15 (11, 12). It has been shown that the RPGR1−19 isoform interacts with PDE6D, INPP5E, and RPGRIP1L and localizes to primary cilia including connecting cilia of the retina (17, 27, 41). However, it is not known whether the RPGRORF15 isoform also binds to these interactors. We first examined ciliary localization of RPGRORF15 by transfecting GFP-tagged RPGRORF15 into human retinal pigmented epithelial (RPE1) cells. In contrast to the RPGR1−19 isoform (Fig. 1 A–C), the RPGRORF15 isoform (Fig. 1 D–F) did not localize to cilia, instead, it was detected in the cytoplasm. Similar results were obtained in mouse IMCD3 cells (SI Appendix, Fig. S2 A–C). To determine if the RPGRORF15 can interact with endogenous PDE6D, INPP5E, and RPGRIP1L, we generated a Flag-S-tag–tagged construct and expressed it in HEK293T cells. Using pulldown with anti-Flag beads, we show that RPGRORF15 also interacts with endogenous PDE6D, INPP5E, and RPGRIP1L (Fig. 1K). Since both isoforms of RPGR share the RLD region (SI Appendix, Fig. S3), these data suggest that the binding domain for PDE6D, INPP5E, and RPGRIP1L resides in the RLD region. To test this, we generated a RPGR construct that contains the shared region of the two isoforms (RPGR-shared) and tested its interaction with PDE6D, INPP5E, and RPGRIP1L. The results demonstrate that the shared region of the two RPGR isoforms contain the binding domains for endogenous PDE6D, INPP5E, and RPGRIP1L (Fig. 1K). However, the shared region cannot be targeted to primary cilia (Fig. 1 G–I, also see SI Appendix, Fig. S2C).
Fig. 1.
Both isoforms of RPGR interact with endogenous PDE6D, INPP5E, and RPGRIP1L, but only RPGR1−19 localizes to cilia. (A–I) GFP-tagged RPGR isoforms were transfected into RPE1 cells, the cells were stained with anti-GFP antibody to enhance and visualize GFP signal, and anti-polyglutamylated tubulin antibody GT335 was used as the cilia marker. Only GFP-tagged RPGR1−19 isoform localizes to the primary cilia (A–C), while RPGRORF15 isoform (D–F) and RPGR-shared region (G–I) do not localize to cilia. (J) Quantification of percentage of cells with GFP+ cilia in transfected RPE1 cells. (K) Flag-S-tag–conjugated RPGR isoforms were transfected into HEK293T cells and RPGR was pulled down by Flag beads. Western blots were used to detect endogenous RPGR interaction proteins using antibodies against PDE6D, RPGRIP1L, and INPP5E. RPGR1−19 M58K and vector only transfected cells were used as negative controls. Detailed analysis of RPGR M58K variant can be found in Fig. 5. To avoid oversaturation of PDE6D by RPGR1−19 IP, we loaded 15% of RPGR1−19 IP at one end of the gel and 85% of RPGR1−19 IP at another end of the gel. The anti-INPP5E antibody nonspecifically binds to the Precision Plus Protein Dual Color Stardards from Bio-Rad at 75 kDa. Arrows indicate Flag-tagged RPGR isoforms. Each transfection was done three times. (Scale bar: 10 μm.)
The C-Terminal of the RPGR1−19 Isoform That Contains the Prenylation Site Regulates Its Interaction with Endogenous PDE6D, INPP5E, and RPGRIP1L.
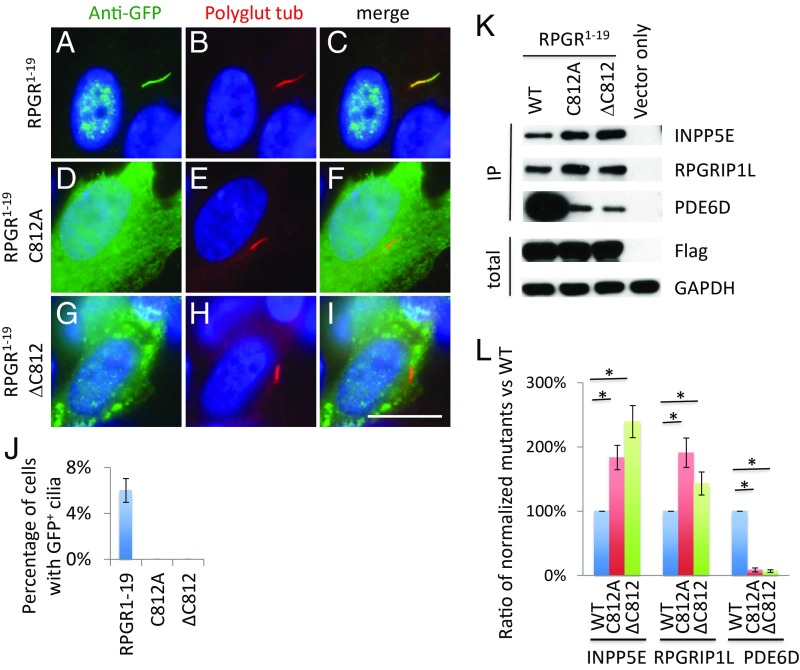
The RPGR1−19 isoform interacts with PDE6D much more strongly than the RPGRORF15 isoform, indicating that the RPGR1−19 isoform has additional binding sites for PDE6D. Altering the prenylation site of cysteine 812 to alanine (Fig. 2 D–F) or deleting cysteine 812 (Fig. 2 G–I) abolish RPGR1−19 isoform ciliary localization, indicating that prenylation of cysteine 812 is required for RPGR1−19 ciliary localization. Similar results were obtained in mouse IMCD3 cells (SI Appendix, Fig. S2). Changing the prenylation site of cysteine 812 to alanine or deleting cysteine 812 also greatly decreases but does not fully abolish RPGR1−19 binding to PDE6D (Fig. 2K), indicating that the C-terminal of the RPGR1−19 is the stronger binding site for PDE6D and that there is another binding site for PDE6D, likely in the RLD region. Interestingly, mutating the prenylation site of cysteine 812 to alanine or deleting cysteine 812 increases RPGR1−19 isoform binding to endogenous INPP5E and RPGRIP1L (Fig. 2K). This finding suggests that prenylation of RPGR1−19 modulates its protein structure, which in turn affects the RLD region of the protein, the binding region for INPP5E and RPGRIP1L, which leads to stronger binding of the RPGR1−19 to INPP5E and RPGRIP1L.
Fig. 2.
Prenylation of the C terminus of RPGR1−19 regulates its interaction with endogenous PDE6D, INPP5E, and RPGRIP1L. The C terminus of RPGR1−19 contains a cysteine residue for prenylation. Mutation (D–F) or deletion (G–I) of the cysteine residue abolish the ability of the RPGR1−19 isoform to localize to cilia compared with WT control (A–C). (J) Quantification of percentage of cells with GFP+ cilia in transfected RPE1 cells. (K) Disruption of RPGR prenylation affects its interaction with endogenous PDE6D, INPP5E, and RPGRIP1L. Flag-S-tag–conjugated WT and mutant RPGR1−19 were transfected into HEK293T cells, and RPGR was pulled down by Flag beads. Western blots were used to detect RPGR1−19 interaction proteins using antibodies against PDE6D, INPP5E, and RPGRIP1L. (L) The ratio of mutants to WT was calculated after quantifying each band using ImageJ and normalized to Flag and GAPDH. Each transfection was done three times. *P < 0.05. (Scale bar: 10 μm.)
The Ciliary Localization of RPGR1−19 Is Regulated by PDE6D and RPGRIP1L, but Not by INPP5E.
Although both isoforms of RPGR bind to endogenous PDE6D, INPP5E, and RPGRIP1L, only the RPGR1−19 isoform localizes to primary cilia in cultured cells (Fig. 1, also see SI Appendix, Fig. S2), although the RPGRORF15 isoform was also shown to localize to connecting cilia in the mouse retina. To determine the importance of RPGR binding with PDE6D, INPP5E, and RPGRIP1L, mutations in all of which cause retinal degeneration, we knocked out PDE6D, INPP5E, and RPGRIP1L in RPE1 cells using the CRISPR-Cas9 genome editing system. We confirmed the knockout of these genes by direct sequencing, Western blotting, and/or immunofluorescent staining (Fig. 3 D and J′ and SI Appendix, Fig. S4). While knockout of RPGRIP1L and PDE6D affect cilia number, knockout of RPGR and INPP5E do not have apparent effects on ciliogenesis (Fig. 3Q). It is known that PDE6D is required for the ciliary localization of prenylated proteins (42). In the absence of PDE6D, RPGR (Fig. 3 N–P) and INPP5E (Fig. 3 N′–P′), two proteins containing prenylation sites, cannot localize to cilia. In contrast to a previous report, our data indicate that RPGR is not required for INPP5E ciliary localization (Fig. 3 D′–F′). To confirm this result, we also used siRNA to knock down RPGR expression. Treating RPE1 cells with RPGR siRNA greatly decreases RPGR protein levels (SI Appendix, Fig. S5N) and effectively abolishes ciliary localization of RPGR (SI Appendix, Fig. S5 D–F). To rule out the possibility that there might be unknown RPGR isoforms that may not be targeted by the CRISPR guides, we treated RPGR knockout RPE1 cells or WT RPE1 cells with RPGR siRNA. siRNA against RPGR does not affect INPP5E ciliary localization (SI Appendix, Fig. S5 J–M). Similarly, loss of INPP5E does not affect ciliary localization of RPGR (Fig. 3 J–M). To further confirm these findings, we knocked down INPP5E expression using siRNA. Treating RPE1 cells with INPP5E siRNA effectively abolishes ciliary localization of INPP5E (SI Appendix, Fig. S6 D–F). However, knocking down INPP5E with siRNA does not affect RPGR ciliary localization (SI Appendix, Fig. S6 J–M). Of note, ciliary localization of RPGR and INPP5E do not absolutely require, but are regulated by, RPGRIP1L (Fig. 3 R and S), suggesting that the integrity of the cilia transition zone regulates RPGR and INPP5E cilia localization. However, the transition zone localization of RPGRIP1L is not affected by loss of RPGR (SI Appendix, Fig. S7).
Fig. 3.
Ciliary localization of the RPGR1−19 isoform is regulated by PDE6D and RPGRIP1L, but not by INPP5E. RPGR, INPP5E, PDE6D, and RPGRIP1L knockout RPE1 cells were generated by CRISPR-Cas9 and stained with RPGR (A–P) and INPP5E (A′–P′) antibodies. All RPE1 knockout cell lines have normal cilia (stained by anti-polyglutamylated tubulin GT335) except for PDE6D and RPGRIP1L, which have fewer ciliated cells. PDE6D (N–P and N′–P′) is required for ciliary localization of RPGR and INPP5E, while RPGRIP1L (G–I and G′–I′) is not absolutely required for ciliary localization of RPGR and INPP5E, but regulate ciliary localization of RPGR and INPP5E compared with WT (A–C and A′–C′). Cilia localization of RPGR is not affected by loss of INPP5E (J–M) and ciliary localization of INNPP5E is not affected by loss of RPGR (D′–F′). (Q) Quantification of cilia frequency in CRISPR knockout cells. (R) Quantification ciliary RPGR frequency in CRISPR knockout cells. (S) Quantification ciliary INPP5E frequency in CRISPR knockout cells. Three different clone lines for each gene knockout were analyzed. *P < 0.05. (Scale bar: 10 μm.)
Most RP-Causing Missense Variations in the RLD Region Disrupt the Interaction Between RPGR Isoforms and Endogenous PDE6D, INPP5E, and RPGRIP1L.
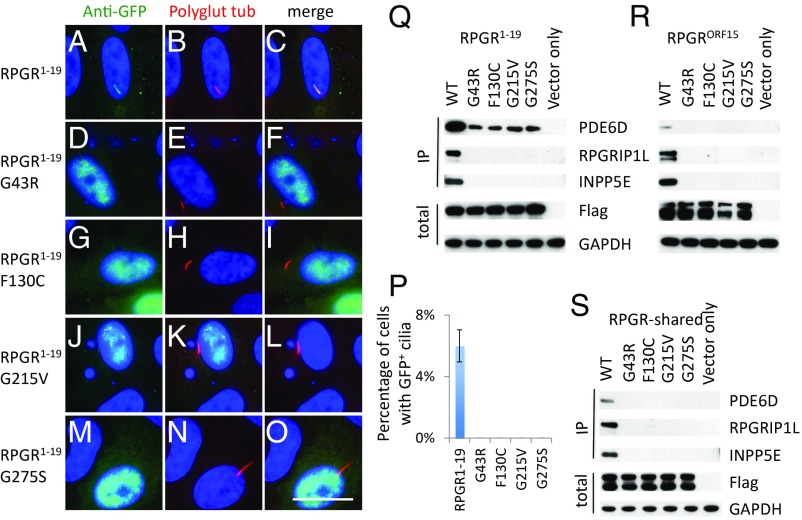
About 200 mutations in RPGR have been reported to cause XLRP. Most of them are deletions or insertions leading to premature stops in the low complexity region of the RPGRORF15 isoform. Several RPGR missense variations have also been identified, the majority of which are located within the shared RLD region. These missense variants are rarely found in the gnomAD database (SI Appendix, Table S1). How these missense variations cause XLRP is not well understood. Since these variations locate to the RLD domain, a region that interacts with endogenous PDE6D, INPP5E, and RPGRIP1L, we hypothesized that these missense mutations disrupt the interaction of RPGR with these interactors. To test this hypothesis, we chose to evaluate G43R, F130C, G215V, and G275S variants, which are predicted to be damaging to RPGR protein function by Polyphen-2, SIFT, and Provean (SI Appendix, Tables S2–S4). To test if these variations affect RPGR1−19 isoform ciliary localization, we transfected GFP-tagged WT and mutant RPGR1−19 into RPE1 cells. In contrast to WT RPGR1−19 (Fig. 4 A–C), mutant RPGR1−19 (Fig. 4 D–O) cannot localize to the primary cilia. Similar results were obtained in mouse IMCD3 cells (SI Appendix, Fig. S2 F–I). To test if these variations affect the interaction between RPGR isoforms and their interactors, we transfected Flag-S-tag–tagged WT and mutant RPGR and RPGR-shared region into HEK293T cells and pulled down RPGR isoforms by Flag beads. Indeed, known mutations G43R, F130C, G215V, and G275S disrupt the interaction between RPGR isoforms (RPGR1−19 and RPGRORF15) and their endogenous interactors PDE6D, INPP5E, and RPGRIP1L (Fig. 4 Q–S). Our data indicate that disruption of the RPGR protein network is probably a common feature for most RPGR missense variations in the RLD region that cause XLRP.
Fig. 4.
Most RP causing missense mutations in the RLD region disrupt the interaction between RPGR and PDE6D, INPP5E, and RPGRIP1L. GFP-tagged WT (A–C), G43R (D–F), F130C (G–I), G215V (J–L), and G275S (M–O) mutated RPGR1−19 transfected RPE1 cells show that these mutations disrupt the RPGR1−19 ciliary localization. (P) Quantification of percentage of cells with GFP+ cilia in transfected RPE1 cells. Flag-S-tag conjugated WT and mutant RPGR isoforms, and RPGR-shared region were transfected into HEK293T cells, and RPGR was pulled down by Flag beads. Western blots were used to detect RPGR interacting proteins. These mutations disrupt the interaction between the RPGR1−19 (Q), RPGRORF15 (R), and RPGR-shared region (S) with endogenous PDE6D, INPP5E, and RPGRIP1L. Each transfection was done three times. (Scale bar: 10 μm.)
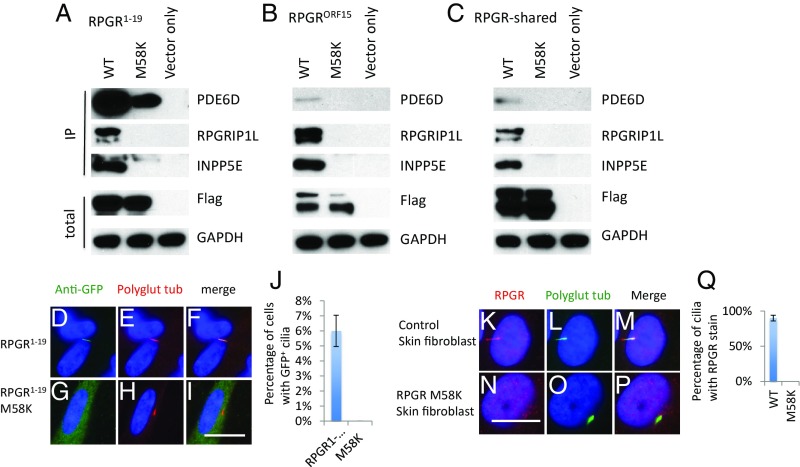
An RPGR M58K Variation Identified in a Family with XLRP Disrupts the Interaction of RPGR Isoforms with Their Interactors and Ciliary Localization of RPGR1−19 Isoform.
We recently identified an M58K RPGR variation in a family with XLRP (10). The M58K variation is located in the first RCC1 domain of the RLD region (SI Appendix, Fig. S3) and is predicted to be damaging to RPGR protein function by Polyphen-2, SIFT, and Provean (SI Appendix, Tables S2–S4). To determine how this missense variation causes XLRP, we performed a pulldown assay as described above. The M58K variation disrupts the interaction between RPGR isoforms (RPGR1−19 and RPGRORF15) and their endogenous interactors PDE6D, INPP5E, and RPGRIP1L (Fig. 5 A–C). The M58K variation also disrupts ciliary localization of the RPGR1−19 isoform (Fig. 5 G–I). Analysis of skin fibroblast cells from one of the patients with M58K variation confirms the lack of ciliary localization of mutant RPGR (Fig. 5 N–P). The M58K mutant RPGR protein level is also decreased (SI Appendix, Fig. S8), indicating the mutant RPGR protein is not stable. Skin fibroblast cells from the patient still have normal ciliary localization of INPP5E (SI Appendix, Fig. S9), consistent with data from CRISPR-Cas9 knockout RPE1 cells. Collectively, these data indicate that the M58K variation is the causative mutation in this XLRP family.
Fig. 5.
An M58K variation found in RP patients disrupts the interaction of RPGR with its interactors and ciliary localization of RPGR1−19. Flag-S-tag–conjugated WT and mutant RPGR isoforms and RPGR-shared region were transfected into HEK293T cells, and RPGR was pulled down by Flag beads. Western blots were used to detect RPGR interacting proteins. M58K variation disrupts the interaction between RPGR1−19 (A), RPGRORF15 (B), and RPGR-shared region (C) with endogenous PDE6D, INPP5E, and RPGRIP1L. GFP-tagged WT (D–F) and M58K (G–I) mutated RPGR1−19 transfected RPE1 cells show that M58K variation disrupts the RPGR1−19 ciliary localization. (J) Quantification of percentage of cells with GFP+ cilia in transfected RPE1 cells. Skin fibroblast cells from a patient with the M58K mutation lack RPGR cilia localization (N–P) compared with control skin fibroblast cells (K–M). (Q) Quantification of RPGR frequency of skin fibroblast cells. Each transfection was done three times. (Scale bars: 10 μm.)
The RPGR Variation V36F That Causes Stationary Night Blindness Decreases the RPGR1−19 Isoform, but Not RPGRORF15 Isoform, Interaction with Endogenous INPP5E.
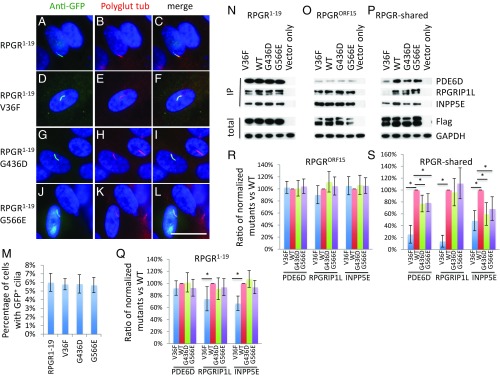
Three additional RPGR missense variations were investigated: G436D that causes XLRP, G566E that is likely a nondisease-causing polymorphism (36, 37, 40, 43, 44), and V36F that causes X-linked congenital stationary night blindness (CSNB) (44, 45). These variations locate outside of the RLD region (SI Appendix, Fig. S3). Polyphen-2, SIFT, and Provean analysis (SI Appendix, Tables S2–S4) indicates that the V36F mutation is predicted to be damaging, but to a lesser degree compared with other missense variations in the RLD region. The G566E variation is predicted to be damaging by Polyphen-2 but are predicted to be likely benign by SIFT and Provean, while the G436D variation is listed as benign (SI Appendix, Tables S2–S4). To determine the nature of these three missense variations, we first examined whether these missense variations affect RPGR1−19 isoform ciliary localization. None of the three variations affects RPGR1−19 ciliary localization (Fig. 6 D–L). Similar results were obtained in mouse IMCD3 cells (SI Appendix, Fig. S2 J–M). To determine if these missense variations affect the interaction between RPGR isoforms and their endogenous interactors PDE6D, INPP5E, and RPGRIP1L, we performed pulldown assays. Of note, these missense variations do not fully disrupt the interaction between RPGR isoforms and their endogenous interactors PDE6D, INPP5E, and RPGRIP1L, unlike missense variations located in the RLD region (Fig. 6 N–P). Further analysis revealed that the V36F missense variation decreases the interaction of the RPGR1−19 isoform with INPP5E and RPGRIP1L, while the interaction between the RPGRORF15 isoform with PDE6D, INPP5E, and RPGRIP1L is not affected (Fig. 6O). The decreased interaction of the RPGR V36F variant with PDE6D, INPP5E, and RPGRIP1L is more evident when using the RPGR-shared region (Fig. 6O). The mild effects on the interaction with endogenous interactors PDE6D, INPP5E, and RPGRIP1L by the V36F missense variation may explain the less severe CSNB phenotypes caused by this variation. The interaction between the RPGR1−19 isoform and RPGRORF15 isoform with their endogenous interactors PDE6D, INPP5E, and RPGRIP1L is not affected by the G436D and G566E missense variations. The G436D and G566E missense variations mildly affect the interaction between the RPGR-shared and endogenous PDE6D and INPP5E. Our data suggest that the G566E missense variation is probably a polymorphism and the G436D missense variation is not a RP causative mutation. It is possible that there is another mutation residing within the RPGR genome region, considering the difficulty of sequencing exon 15 of RPGR due to the high GC content of this region.
Fig. 6.
RPGR V36F variation that causes stationary night blindness decreases the RPGR1−19 isoform and RPGR-shared region, but not RPGRORF15 isoform interaction with INPP5E, PDE6D, and RPGRIP1L. Similar to WT GFP-tagged RPGR1−19 (A–C), the V36F (D–F), G436D (G–I), and G566E (J–L) variants localize to cilia. (M) Quantification of percentage of cells with GFP+ cilia in transfected RPE1 cells. (N–P) Flag-S-tag conjugated WT and mutant RPGR isoforms and RPGR-shared region were transfected into HEK293T cells and RPGR was pulled down by Flag beads. Western blots were used to detect RPGR interaction proteins. Compared with WT controls, V36F mutation greatly decreases RPGR1−19 (N) and RPGR-shared region (P) interaction with endogenous INPP5E and RPGRIP1L. It also greatly decreases RPGR-shared interaction with endogenous PDE6D (P). V36F variation does not affect RPGRORF15 isoform interaction with endogenous PDE6D, INPP5E, or RPGRIP1L (O). Reported RP-causing RPGR variation G436D and a polymorphism variation G566E have mild effect on the interaction between RPGR-shared region and PDE6D, INPP5E, or RPGRIP1L, but have no effects on RPGR1−19 and RPGRORF15 isoforms interaction with endogenous INPP5E, PDE6D, and RPGRIP1L. (Q–S) The ratio of mutants to WT was calculated after quantifying each band using ImageJ and normalized to Flag and GAPDH. Each transfection was done three times. *P < 0.05. (Scale bar: 10 μm.)
Discussion
With the development of next generation sequencing, it is more cost-effective and easier to identify gene variants in patients than ever before. However, identified variants are not necessarily disease causing and functional predictive algorithms are not fully accurate. Verification of the causative nature of identified gene variants is important for appropriate genetic counseling and recurrence risk assessment for patients and families. In addition, development of gene-specific treatments such as gene and cell therapy requires accurate diagnoses and understanding of disease mechanisms. Therefore, there is a need to develop rapid, cost-effective functional assays to verify or refute the causative nature of identified gene variations.
Due to its severity and early onset, XLRP is a particularly devastating form of retinal degeneration. Most males with XLRP come to medical attention before the age of 20. Approximately 15% of all RP is X-linked, and more than 70% of these cases are caused by mutations in the RPGR gene (10). There are estimated to be about 9,000 RPGR-associated RP patients in the United States, with just over 100 new cases identified each year (10). As the majority of RPGR mutations fall within the difficult-to-sequence low complexity region of the gene, to accurately diagnose and treat the disease, it is important to develop a practical assay to accurately evaluate the pathogenicity of RPGR variants.
In this study, we developed an in vitro assay to examine how different missense variations in RPGR cause XLRP. Our assay is based on the RPGR protein interaction network. We demonstrated that XLRP causative missense mutations disrupt the interaction between RPGR isoforms and their endogenous interactors PDE6D, INPP5E, and RPGRIP1L. Using this assay, we can distinguish variants that cause XLRP, CSNB, and nondisease-causing polymorphisms. Furthermore, we also developed an assay based on the cilia localization of the RPGR1−19 isoform to distinguish among variants that cause XLRP, which disrupt cilia localization, and CSNB or nondisease-causing polymorphisms, which have normal cilia localization of the RPGR1−19 isoform. These methods provide a cost-effective test for RPGR functional mutation analysis.
The existence of multiple isoforms in cells and tissues makes the study of RPGR function challenging. While the RPGRORF15 isoform is mainly expressed in the retina, the RPGR1−19 isoform has a broader expression pattern (12, 16, 46). The shared N-terminal RLD region suggests that these isoforms have some functions in common, whereas the unique C termini of these isoforms suggest each isoform may also have different functional properties. Several lines of evidence support this hypothesis. First, the RPGR1−19 isoform, which has a C-terminal prenylation site, localizes to primary cilia both in human RPE1 cells and mouse IMCD3 cells (17, 27, 41). In contrast, the RPGRORF15 isoform does not localize to cilia in cultured cells. Second, rescue experiments in Rpgr knockout mice demonstrated that the RPGRORF15 isoform, but not the RPGR1−19 isoform, is the functionally significant isoform in photoreceptor cells (13, 14, 47). Third, some RPGR patients have other phenotypes including respiratory tract infections, hearing loss, and primary ciliary dyskinesia in addition to XLRP (15, 46, 48). Given the broader expression of the RPGR1−19 isoform, this might reflect the function of RPGR1−19 in nonretinal tissues.
The functional importance of RPGR isoforms in the retina is not fully understood. It has been shown that the level of the RPGR1−19 isoform is gradually decreased while the level of the RPGRORF15 isoform is gradually increased during eye development, implying the RPGR1−19 isoform plays a role during the early stage of eye development, while the RPGRORF15 isoform has an important function in the mature retina (14). The fact that both RPGR isoforms interact with the same endogenous interactors suggests that they may compete with each other for the availability of endogenous binding partners, therefore fine tuning each isoforms’ function. Consistent with this notion, truncated RPGR has been shown to have dominant effects on endogenous RPGR proteins (39, 49). This may explain why overexpression of the RPGR1−19 isoform in mice causes retinal degeneration.
The discrepancy between our data and published data regarding the effect of RPGR on INPP5E ciliary localization may be due to species and cell-type differences. We used human RPE1 cells in our study, while data in the literature was generated using mouse fibroblast cells (27). Another possibility is the existence of modifying genes, which, in combination with loss of RPGR, regulates INPP5E ciliary localization. When we generate stable RPGR knockout cell lines in RPE1 cells using CRISPR-Cas9, we selected two gRNAs targeting RPGR. Ten stable cell lines from one guide all show ciliary localization of INPP5E. Interestingly, four of eight stable cell lines generated using a second guide show the lack of ciliary localization of INPP5E, suggesting there may be genes whose functions are disturbed by the guide. Interestingly, it has been shown that variations in RPGRIP1L and NPHP5 serve as modifiers for RPGR (32).
Although our in vitro assay raises questions about the nature of some known RPGR missense variations, we cannot completely rule out the possibility that these missense variations are causative mutations. These missense variations could affect the protein stability of RPGR. We cannot distinguish this possibility with our assay.
Materials and Methods
Study Approval.
All experiments involving human skin biopsy specimens were conducted with the approval of and under the supervision of the University of Iowa's Internal Review Board (IRB, application no. 200202022).
Reagents and Antibodies.
The following antibodies were used in this study: RPGR rabbit polyclonal antibody (HPA001593, 1:200 for IF; Sigma), mouse monoclonal anti-polyglutamylated tubulin [clone GT335, AG-20B-0020, 1:1,000 for immunofluorescence (IF), 1:1,000 for Western blot (WB); AdipoGen], PDE6D rabbit polyclonal antibody (MBS7005086, 1:1,000 for WB; MyBiosources), INPP5E rabbit polyclonal antibody (17797-1-AP, 1:500 for IF, 1:500 for WB; Proteintech), RPGRIP1L rabbit polyclonal antibody (55160–1-AP, 1:300 for IF, 1:500 for WB; Proteintech), RPGRIP1L rabbit polyclonal antibody (HPA039405, 1:300 for WB; Sigma), ARL13B mouse monoclonal antibody (N295B/66, 1:500 for IF; NeuroMab), FLAG mouse monoclonal antibody (F1804, 1:200 for IF; Sigma), HRP-conjugated FLAG mouse monoclonal antibody (A8592, 1:20,000 for WB; Sigma), GAPDH mouse monoclonal antibody (MA5-15738, 1:10,000 for WB; Invitrogen), and GFP rabbit monoclonal antibody (G10362, 1:500 for IF; Invitrogen). Blasticidin was from InvivoGen. SMARTpool siRNA against human RPGR and INPP5E were from Dharmacon.
Cell Cultures.
hTERT-RPE1 cells and mouse IMCD3 cells were maintained in DMEM/F12 (Invitrogen) with 10% FBS (Gibco) and penicillin/streptomycin. Human HEK293T cells were maintained in DMEM (Invitrogen) with 10% FBS and penicillin/streptomycin. Skin fibroblasts were maintained in MEM alpha (Invitrogen) with 10% FBS and primocin (InvivoGen).
Generation of RPE1 Knockout Cell Lines Using CRISPR-Cas9.
We generated RPGR, PDE6D, INPP5E, and RPGRIP1L knockout cell lines in RPE1 cells using CRISPR-Cas9. The guide sequences for RPGR is as follows: guide 1 GAAGTGAAATTAGCTGCCTG. Guide 2 GTCCCTGTACATCTTTCATG. The guide sequence for PDE6D is as follows: GGACCTGTCTGTCCCTGGTG. The guide sequence for INPP5E is as follows: GAGGGTTACCCCCCGAGGAC. The guide sequence for RPGRIP1L is as follows: GGTGTCACGTGTCAGTCGTG. Stable clones were selected with blasticidine and indels were identified by Sanger sequencing. Stable clones were also verified by immunofluorescent staining or Western blotting whenever antibodies were available.
Generation of Human RPGR Missense Variations and RPGR Shared Region.
RPGR missense variations were generated by site-directed mutagenesis (Stratagene) using N-terminal Flag-S–tagged or GFP-S–tagged wild-type RPGR as template and mutations were verified by Sanger sequencing. RPGR shared region was generated by PCR and verified by Sanger sequencing.
Immunoprecipitation.
RPGR WT and missense variations were transfected into HEK293T cells in a six-well plate. Forty-eight hours after transfection, the cells were lysed in lysis buffer (1× PBS, 1% Triton X-100, and protease inhibitor; Roche) and spun at 20,000 × g for 15 min at 4 °C. Cleared lysates were incubated with anti-Flag beads (Biotool) for 4 h. The beads were washed four times with lysis buffer, and the interactions were detected by Western blotting.
Immunofluorescence Microscopy.
Cells were fixed in 4% paraformaldyhyde in PBS for 10 min and permeabilized with 0.2% Triton X-100 at room temperature for 7 min or fixed with cold methanol for 5 min (for γ-tubulin stain). Cells were washed three times with PBS and blocked with blocking buffer (1% BSA in PBS). Primary antibodies were diluted in blocking buffer and incubated at room temperature for 1 h. Cells were washed 3× PBS, blocked with blocking buffer, then incubated with Alexa 488- or Alexa 568-labeled secondary antibodies (1:1,500 for IF; Invitrogen). Slides were mounted with VectaShield mounting medium with DAPI (Vector Laboratories).
Image Quantification and Statistical Analysis.
To quantify GFP+ cilia in GFP-tagged RPGR WT and mutant-transfected RPE1 cells and IMCD3 cells, 10–15 random fields (10 fields for IMCD3 cells and 12–15 fields for RPE1 cells) under 60× objective were chosen to count GFP+ cilia and DAPI to count for total cells. To quantify cilia number frequency, 5–10 random fields (10 fields for RPGRIP1L knockout cells, 5 fields for the rest gene knockout cells) under 60× objective were chosen to count cilia number and DAPI to count for total cells. To quantify RPGRIP1L stain, 10 random pictures were taken under 100× objective, anti-polyglutamyalted tubulin was used to count cilia, and γ-tubulin was used to count basal body. To quantify RPGR and INPP5E frequency, about 100 cells under 100× objective were counted and anti-polyglutamyalted tubulin was used to count cilia. For Western blots quantification, images were quantified using NIH ImageJ. We performed statistical analysis using unpaired t tests. Mean values ± SEM are reported.
Supplementary Material
Acknowledgments
We thank the patients and family members for their willingness to participate in this study. This work is supported by NIH Grants EY-011298, EY-017168 (to V.C.S.), R01-EY026008 (to B.A.T.), and R01-EY026008 (to E.M.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.C.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817639116/-/DCSupplemental.
References
- 1.Fahim AT, Daiger SP, Weleber RG. Nonsyndromic retinitis pigmentosa overview. In: Adam MP, et al., editors. GeneReviews. University of Washington; Seattle: 1993–2018. [Google Scholar]
- 2.Mockel A, et al. Retinal dystrophy in Bardet-Biedl syndrome and related syndromic ciliopathies. Prog Retin Eye Res. 2011;30:258–274. doi: 10.1016/j.preteyeres.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg T, Haim M, Hauch AM, Parving A. The prevalence of Usher syndrome and other retinal dystrophy-hearing impairment associations. Clin Genet. 1997;51:314–321. doi: 10.1111/j.1399-0004.1997.tb02480.x. [DOI] [PubMed] [Google Scholar]
- 4.Williams DS. Usher syndrome: Animal models, retinal function of Usher proteins, and prospects for gene therapy. Vision Res. 2008;48:433–441. doi: 10.1016/j.visres.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andréasson S, et al. Phenotypes in three Swedish families with X-linked retinitis pigmentosa caused by different mutations in the RPGR gene. Am J Ophthalmol. 1997;124:95–102. doi: 10.1016/s0002-9394(14)71649-6. [DOI] [PubMed] [Google Scholar]
- 6.Churchill JD, et al. Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2013;54:1411–1416. doi: 10.1167/iovs.12-11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daiger SP, Bowne SJ, Sullivan LS. Genes and mutations causing autosomal dominant retinitis pigmentosa. Cold Spring Harb Perspect Med. 2014;5:a017129. doi: 10.1101/cshperspect.a017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge Z, et al. NGS-based molecular diagnosis of 105 eyeGENE(®) probands with retinitis pigmentosa. Sci Rep. 2015;5:18287. doi: 10.1038/srep18287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- 10.Stone EM, et al. Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology. 2017;124:1314–1331. doi: 10.1016/j.ophtha.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He S, et al. Retinitis pigmentosa GTPase regulator (RPGR) protein isoforms in mammalian retina: Insights into X-linked retinitis pigmentosa and associated ciliopathies. Vision Res. 2008;48:366–376. doi: 10.1016/j.visres.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirschner R, et al. RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Hum Mol Genet. 1999;8:1571–1578. doi: 10.1093/hmg/8.8.1571. [DOI] [PubMed] [Google Scholar]
- 13.Hong DH, Pawlyk BS, Adamian M, Sandberg MA, Li T. A single, abbreviated RPGR-ORF15 variant reconstitutes RPGR function in vivo. Invest Ophthalmol Vis Sci. 2005;46:435–441. doi: 10.1167/iovs.04-1065. [DOI] [PubMed] [Google Scholar]
- 14.Wright RN, Hong DH, Perkins B. Misexpression of the constitutive Rpgr(ex1-19) variant leads to severe photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2011;52:5189–5201. doi: 10.1167/iovs.11-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukowy-Bieryłło Z, et al. RPGR mutations might cause reduced orientation of respiratory cilia. Pediatr Pulmonol. 2013;48:352–363. doi: 10.1002/ppul.22632. [DOI] [PubMed] [Google Scholar]
- 16.Hong DH, et al. RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Invest Ophthalmol Vis Sci. 2003;44:2413–2421. doi: 10.1167/iovs.02-1206. [DOI] [PubMed] [Google Scholar]
- 17.Lee JJ, Seo S. PDE6D binds to the C-terminus of RPGR in a prenylation-dependent manner. EMBO Rep. 2015;16:1581–1582. doi: 10.15252/embr.201541220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patil SB, Verma R, Venkatareddy M, Khanna H. Expression and localization of the ciliary disease protein retinitis pigmentosa GTPase regulator in mammalian kidney. Kidney Int. 2010;78:622–623. doi: 10.1038/ki.2010.252. [DOI] [PubMed] [Google Scholar]
- 19.Gakovic M, et al. The role of RPGR in cilia formation and actin stability. Hum Mol Genet. 2011;20:4840–4850. doi: 10.1093/hmg/ddr423. [DOI] [PubMed] [Google Scholar]
- 20.Megaw R, et al. Gelsolin dysfunction causes photoreceptor loss in induced pluripotent cell and animal retinitis pigmentosa models. Nat Commun. 2017;8:271. doi: 10.1038/s41467-017-00111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murga-Zamalloa CA, Swaroop A, Khanna H. RPGR-containing protein complexes in syndromic and non-syndromic retinal degeneration due to ciliary dysfunction. J Genet. 2009;88:399–407. doi: 10.1007/s12041-009-0061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rachel RA, Li T, Swaroop A. Photoreceptor sensory cilia and ciliopathies: Focus on CEP290, RPGR and their interacting proteins. Cilia. 2012;1:22. doi: 10.1186/2046-2530-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao KN, et al. Loss of human disease protein retinitis pigmentosa GTPase regulator (RPGR) differentially affects rod or cone-enriched retina. Hum Mol Genet. 2016;25:1345–1356. doi: 10.1093/hmg/ddw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright RN, Hong DH, Perkins B. RpgrORF15 connects to the usher protein network through direct interactions with multiple whirlin isoforms. Invest Ophthalmol Vis Sci. 2012;53:1519–1529. doi: 10.1167/iovs.11-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murga-Zamalloa CA, Atkins SJ, Peranen J, Swaroop A, Khanna H. Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: Implications for cilia dysfunction and photoreceptor degeneration. Hum Mol Genet. 2010;19:3591–3598. doi: 10.1093/hmg/ddq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto EA, et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 27.Rao KN, Zhang W, Li L, Anand M, Khanna H. Prenylated retinal ciliopathy protein RPGR interacts with PDE6δ and regulates ciliary localization of Joubert syndrome-associated protein INPP5E. Hum Mol Genet. 2016;25:4533–4545. doi: 10.1093/hmg/ddw281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roepman R, et al. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet. 2000;9:2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 29.Roepman R, Wolfrum U. Protein networks and complexes in photoreceptor cilia. Subcell Biochem. 2007;43:209–235. doi: 10.1007/978-1-4020-5943-8_10. [DOI] [PubMed] [Google Scholar]
- 30.Shu X, et al. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum Mol Genet. 2005;14:1183–1197. doi: 10.1093/hmg/ddi129. [DOI] [PubMed] [Google Scholar]
- 31.Wätzlich D, et al. The interplay between RPGR, PDEδ and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep. 2013;14:465–472. doi: 10.1038/embor.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahim AT, et al. Polymorphic variation of RPGRIP1L and IQCB1 as modifiers of X-linked retinitis pigmentosa caused by mutations in RPGR. Adv Exp Med Biol. 2012;723:313–320. doi: 10.1007/978-1-4614-0631-0_41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khanna H, et al. RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J Biol Chem. 2005;280:33580–33587. doi: 10.1074/jbc.M505827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang B, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breuer DK, et al. A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet. 2002;70:1545–1554. doi: 10.1086/340848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buraczynska M, et al. Spectrum of mutations in the RPGR gene that are identified in 20% of families with X-linked retinitis pigmentosa. Am J Hum Genet. 1997;61:1287–1292. doi: 10.1086/301646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita R, et al. Analysis of the RPGR gene in 11 pedigrees with the retinitis pigmentosa type 3 genotype: Paucity of mutations in the coding region but splice defects in two families. Am J Hum Genet. 1997;61:571–580. doi: 10.1086/515523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guevara-Fujita M, et al. Five novel RPGR mutations in families with X-linked retinitis pigmentosa. Hum Mutat. 2001;17:151. doi: 10.1002/1098-1004(200102)17:2<151::AID-HUMU7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 39.Hong DH, Pawlyk BS, Adamian M, Li T. Dominant, gain-of-function mutant produced by truncation of RPGR. Invest Ophthalmol Vis Sci. 2004;45:36–41. doi: 10.1167/iovs.03-0787. [DOI] [PubMed] [Google Scholar]
- 40.Shu X, et al. RPGR mutation analysis and disease: An update. Hum Mutat. 2007;28:322–328. doi: 10.1002/humu.20461. [DOI] [PubMed] [Google Scholar]
- 41.Fansa EK, O’Reilly NJ, Ismail S, Wittinghofer A. The N- and C-terminal ends of RPGR can bind to PDE6δ. EMBO Rep. 2015;16:1583–1585. doi: 10.15252/embr.201541404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baehr W. Membrane protein transport in photoreceptors: The function of PDEδ: The Proctor lecture. Invest Ophthalmol Vis Sci. 2014;55:8653–8666. doi: 10.1167/iovs.14-16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharon D, et al. X-linked retinitis pigmentosa: Mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Invest Ophthalmol Vis Sci. 2000;41:2712–2721. [PubMed] [Google Scholar]
- 44.Shu X, McDowall E, Brown AF, Wright AF. The human retinitis pigmentosa GTPase regulator gene variant database. Hum Mutat. 2008;29:605–608. doi: 10.1002/humu.20733. [DOI] [PubMed] [Google Scholar]
- 45.Linari M, et al. The retinitis pigmentosa GTPase regulator, RPGR, interacts with the delta subunit of rod cyclic GMP phosphodiesterase. Proc Natl Acad Sci USA. 1999;96:1315–1320. doi: 10.1073/pnas.96.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iannaccone A, et al. Increasing evidence for syndromic phenotypes associated with RPGR mutations. Am J Ophthalmol. 2004;137:785–786, author reply 786. doi: 10.1016/j.ajo.2003.11.050. [DOI] [PubMed] [Google Scholar]
- 47.Pawlyk BS, et al. Photoreceptor rescue by an abbreviated human RPGR gene in a murine model of X-linked retinitis pigmentosa. Gene Ther. 2016;23:196–204. doi: 10.1038/gt.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iannaccone A, et al. Clinical and immunohistochemical evidence for an X linked retinitis pigmentosa syndrome with recurrent infections and hearing loss in association with an RPGR mutation. J Med Genet. 2003;40:e118. doi: 10.1136/jmg.40.11.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rozet JM, et al. Dominant X linked retinitis pigmentosa is frequently accounted for by truncating mutations in exon ORF15 of the RPGR gene. J Med Genet. 2002;39:284–285. doi: 10.1136/jmg.39.4.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.