Abstract
Introduction
Notch receptor family dysregulation can be tumor promoting or suppressing depending on cellular context. Our studies shed light on the mechanistic differences that are responsible for NOTCH1’s opposing roles in lung adenocarcinoma and lung squamous cell carcinoma.
Methods
We integrated transcriptional patient-derived datasets with gene co-expression analyses to elucidate mechanisms behind NOTCH1 function in subsets of non-small cell lung cancer. Differential co-expression was examined using hierarchical clustering and principal component analysis. Enrichment analyses was used to examine pathways associated with the underlying transcriptional networks. These pathways were validated in vitro and in vivo. Endogenously epitope-tagged NOTCH1 was used to identify novel interacting proteins.
Results
NOTCH1 co-expressed genes in lung adenocarcinoma and squamous carcinoma were distinct, and associated with either angiogenesis and immune system pathways or cell cycle control and mitosis pathways, respectively. Tissue culture and xenograft studies of lung adenocarcinoma and lung squamous models with NOTCH1 knockdown demonstrated growth differences and opposing effects on these pathways. Differential NOTCH1 interacting proteins were identified as potential mediators of these differences.
Conclusions
Recognition of the opposing role of NOTCH1 in lung cancer, downstream pathways, and interacting proteins in each context may help direct the development of rational NOTCH1 pathway-dependent targeted therapies for specific tumor subsets of non-small cell lung cancer.
Keywords: Lung cancer, Notch, co-expression, immune function, genomics, AP-MS/MS
Introduction:
Notch signaling plays a fundamental role during embryonic development and adult tissue homeostasis1. In cancer, abnormal regulation of Notch signaling can occur through multiple types of genomic alterations2, and changes to various regulatory pathways3–11. Indeed, the Notch pathway is involved in most of the hallmarks of cancer11 and its role can vary widely Depending on cancer subtype. NOTCH1, in particular, has cell autonomous and non-autonomous roles resulting in oncogenesis or tumor suppression in different contexts, cancer subtypes, and specific gene backgrounds3–11. Unfortunately the mechanistic details of these various roles are not well understood11.
The evidence that NOTCH1 can be an oncogene or a tumor suppressor comes from mutational analysis of tumors and limited in vitro studies. Over 50% of human T-cell acute lymphoblastic leukemia have activating mutations in NOTCH112,13. In vitro and in vivo studies have shown that NOTCH1 acts as an oncogene in lung adenocarcinoma (AD) where it plays a critical role in invasion, metastasis, and malignant transformation3–5,14. In contrast, a tumor suppressive role of Notch has been claimed across different squamous cell carcinoma (SCC) tumors based on the loss of function mutations commonly found in cutaneous SCC15.
Mutational analysis has not given us the full picture of how Notch signaling functions in different cancer types and another approach is needed. Notch mutations have been identified in less than 10% of lung tumors, but aberrant Notch signaling has been reported in 33% of non-small cell lung cancers (NSCLCs)19. In the absence of mutations, the role of Notch in cancer progression can be probed by determining the phenotypic response to perturbation of Notch signaling11. The importance of Notch expression is reflected in the fact that NOTCH1 expression levels in non-mutated tumors have opposite prognostic effects in AD and SCC20–23.
While a number of Notch-targeted therapies have been tried (Supplemental Table 1), none has resulted in significant clinical benefit in unselected patient populations. Defining the functional roles of Notch in different tumor subtypes is essential to understand its biology and to provide better therapeutic options for cancer patients. Recent papers suggest that Notch plays a key role maintaining the balance of immune cells within the tumor microenvironment24–27. The gap in our understanding of NOTCH1’s role in regulating the tumor microenvironment adds to the complexity of predicting the outcome of therapeutic modulation of NOTCH110.
One method of inferring gene function is co-expression analysis. Differential co-expression networks have been used to identify disease associated genes and gene modules in solid and hematologic tumors28. It can be used to define tumor intrinsic and extrinsic biological processes associated with a gene of interest in a particular disease or disease subtype24,26,29. To date, no studies have examined or compared the vector of correlation coefficients between the Notch family of transcription factors and the transcriptome in an unbiased manner in human solid tumors.
Here we have identified differential co-expression networks of Notch gene expression. In our analysis, we revealed a pattern of gene co-expression with NOTCH1 in lung AD that is very different from lung SCC and identified pathways that could underlie the observed differences in Notch function. We confirmed these observed differences in vitro and in vivo. We further identified potential links associated with these differences by proteomic examination of the NOTCH1 interactome using CRISPR-tagged endogenous models. We feel that this combined bioinformatics and proteomics approach provides insights revealing possible novel mechanisms underlying the opposing roles of NOTCH1 in lung cancer.
Materials and Methods:
Cell culture
Two lung AD cell lines (A549, H358) and two lung SCC cell lines (HCC15, HCC95) were used. Cells were grown in standard culture medium under standard conditions at 37°C in a humidified atmosphere, 5% CO2. Additional details about cell culture including authentication and mycoplasma testing are provided in Supplemental Methods.
Preparation of The Cancer Genome Atlas Clinical and Transcriptional Datasets
The top 13 solid epithelial cancers ranked by estimated deaths in 2017 were identified using data from the American Cancer Society (https://cancerstatisticscenter.cancer.org/#/)30. TCGA has 38 cancer datasets with publicly available RNAseqV2 data (http://firebrowse.org/). RSEM normalized data from 14 datasets that matched categories with the top 13 solid epithelial cancers (Table 1) were used. Selected tumor subtypes were confirmed by a clinical pathologist (K. Shilo). Additional details regarding data sets and analysis are provided in Supplemental Methods.
Table 1.
Summary of solid epithelial cancer datasets
| Mortality Data | Estimated Deaths1 |
TCGA DATASET | TCGA Subtype | ||||
|---|---|---|---|---|---|---|---|
| Rank | Cancer | Dataset [Abbreviation] | # samples | Subtype [Abbreviation] | # samples | N>50 | |
| 1 | Lung and bronchus |
155,870 | Lung Adenocarcinoma [LUAD] | 521 | Lung Adenocarcinoma [LUAD] | 517 | Yes |
| Lung Squamous Cell Carcinoma [LUSC] |
504 | Lung Squamous Cell Carcinoma [LUSC] |
501 | Yes | |||
| 2 | Colorectum | 50,260 | Colon Adenocarcinoma [COAD] | 461 | Colon [Colon] Rectal [Rectal] |
247 94 |
Yes Yes |
| 3 | Pancreas | 43,090 | Pancreatic Adenocarcinoma [PAAD] |
185 | Pancreatic Adenocarcinoma [PAAD] |
170 | Yes |
| 4 | Breast | 41,070 | Breast Invasive Carcinoma [BRCA] |
1097 | Breast Invasive Lobular Carcinoma [BRCA_lobular] |
206 | Yes |
| Breast Invasive Ductal Carcinoma [BRCA_ductal] |
812 | Yes | |||||
| 5 | Liver and intrahepatic bile duct |
28,920 | Cholangiocarcinoma [CHOL] | 36 | Cholangiocarcinoma [CHOL] | 35 | No |
| 6 | Prostate | 26,730 | Prostate Adenocarcinoma [PRAD] |
498 | Prostate [PRAD] | 498 | Yes |
| 7 | Urinary bladder |
16,870 | Bladder Urothelial Carcinoma [BLCA] |
412 | Bladder [BLCA] | 408 | Yes |
| 8 | Esophagus | 15,690 | Esophageal Carcinoma [ESCA] | 185 | Esophageal Adenocarcinoma [ESCA_AD] |
89 | Yes |
| Esophageal Squamous Cell Carcinoma [ESCA_SCC] |
96 | Yes | |||||
| 9 | Ovary | 14,080 | Ovarian Serous Cystadenocarcinoma [OV] |
586 | Papillary Serous Carcinoma or Serous Ovarian Cancer [OV] |
303 | Yes |
| 10 | Stomach | 10,960 | Stomach Adenocarcinoma [STAD] |
443 | Stomach Adenocarcinoma [STAD] |
230 | Yes |
| 11 | Uterine corpus |
10,920 | Uterine Corpus Endometrial Carcinoma [UCEC] |
548 | Uterine Endometrial Carcinoma [UEC] |
119 | Yes |
| Uterine Serous Carcinoma/Uterine Papillary Serous Carcinoma [USC] |
58 | Yes | |||||
| 12 | Oral cavity and pharynx |
9,700 | Head and Neck Squamous Cell Carcinoma [HNSC] |
528 | Head and Neck Squamous Cell Carcinoma [HNSCC] |
522 | Yes |
| 13 | Cervix | 4,210 | Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma [CESC] |
307 | Cervical Squamous Cell Carcinoma [CESC_SCC] |
253 | Yes |
| Endocervical Adenocarcinoma or Mucinous Carcinoma [CESC_AD] |
44 | No | |||||
Top 13 solid epithelial tumors ranked by estimated deaths in 2017. TCGA datasets corresponding to the top cancers were matched and the total number of samples with RNAseqV2 data available is indicated. Using clinical annotations, the TCGA datasets were refined to reflect predominant subtypes. The number of samples utilized is indicated for each subtype. The 17 cancer subtypes with n ≥ 50 samples were used in downstream solid tumor analysis.
Source: Estimated 2017 deaths based on 2000–2014 United States mortality data. ACS. https://cancerstatisticscenter.cancer.org/#/
Data Visualization
The ClustVis tool (http://biit.cs.ut.ee/clustvis/)31 was used for principal component analysis (PCA) with gene scaling using the SVDimpute algorithm. Global transcriptional patterns were visualized using the heatmaply32 package in R3.4.0 (2017–04-21) using clustering determined by Euclidean distance with complete linkage. Descriptive statistics, unsupervised hierarchical clustering with distances defined by Euclidian distance and Ward’s linkage, and Venn diagrams were produced in JMP-Pro 12.2.0.
Gene list enrichment analysis
Enrichment analysis was performed using Entrez gene identifiers. The biological relevance of the network modules used the ToppGene Suite (Division of Biomedical Informatics, Cincinnati Children’s Hospital Medical Center, http://toppgene.cchmc.org)33 and was performed using probability density function method, false discovery rate (FDR) p-value cutoff 0.05 and verified with DAVID34,35 and Ingenuity Pathway Analysis36. Additional details, raw gene input and results from ToppGene provided in Supplemental Methods and Supplemental Dataset 1. Enrichment analysis of RNA-sequencing data from human xenografts samples was performed in Ingenuity Pathway Analysis (IPA) and further described in Supplemental Methods.
shRNA vectors
shRNA knockdown (KD) experiments were performed using the pLKO.1-puro vector. To knockdown NOTCH1 expression we used NOTCH1 Mission shRNA’s (Sigma,SHCLNG-NM_017617:TRCN0000003359,TRCN0000003362,TRCN0000350254) or non-targeting control (NTC) (Sigma,SHCO16). We refer to each NOTCH1 knockdown by the last four TRCN digits.
NOTCH1 shRNA virus production and cell line transduction
Lentiviral particles were produced by transfecting 293FT packaging cells with packaging-psPAX2, envelope-pMD2.G and shRNA plasmid. Experiments were begun 24h after infection. Knockdown efficiency was assessed by immunoblot analysis of NOTCH1 protein expression after 72h. Additional details are provided in Supplemental Methods.
Metabolic Proliferation Assay
Cells were seeded (2500 cells/well) in a 96-well plate in culture and incubated for 72h under standard conditions. Assays were run using a standard alamarBlue (Thermo#DAL1100) assay protocol, using 3h incubation, 3 technical replicates, 3 biologic replicates, and repeated in triplicate. Error bars indicate the mean±SEM. Immunoblots were harvested and run in parallel to verify NOTCH1 expression levels on the day metabolic readouts were assessed (72h).
Xenografts
Female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained from The Ohio State University (OSU) Target Validation Shared Resource. Six-week-old mice were implanted with 10×106 cells. Four tumor models (A549, H358, HCC15, and HCC95) were used. For each tumor model, 4 mice were implanted using paired NTC and NOTCH1 KD#0254 constructs on bilateral flanks. All tumors harvested were assessed by ex vivo analysis. Studies were blinded to group assignment. Additional details are provided in Supplemental Methods.
Immunohistochemistry (IHC) staining and analysis
Formalin-fixed paraffin-embedded (FFPE) xenograft tumor sections from paired NTC and NOTCH1 knockdown mice were stained by OSU Solid Tumor Shared Resource using the Leica Bond RX system. IHC staining was performed for Anti-CD31 (Abcam#ab28364,1:50) and pHH3 (CST#9701,1:200). Biological replicates from in vivo studies were used; for each mouse 1NTC and knockdown xenograft tumor sections were stained (4 cell lines, 4 mice/cell line). Additional details regarding sample analysis are provided in Supplemental Methods.
RNA-Sequencing
RNA from flash frozen tissue from paired NTC and NOTCH1 knockdown xenograft tumor sections lung AD model (A549) and in the lung SCC model (HCC15) were sequenced. 8 tumors were sequenced (2 cell lines, 2 mice/cell line). Additional details regarding data generation and analysis are provided in Supplemental Methods.
Immunoblots
Cell lysates were harvested while cells were in exponential growth phase or from flash frozen tissue in lysis buffer, homogenized and run on precast gels (BioRad#4561083). Standard LiCor techniques were used for antibody staining using primary antibodies. Additional details are provided in Supplemental Methods.
AP-MS/MS
Descriptions of molecular cloning work, CRISPR-Cas9 gene editing, DNA constructs, stable and transient transductions, co-immunoprecipitation validations and the proteomic procedure and analyses for the discovery dataset are described in the Supplemental Methods and Supplemental Dataset 2. Briefly, the protocol for mass spectrometry (MS) based investigation of protein-protein interactions were based on in-solution (single) affinity purification (AP) followed by ultra-performance liquid chromatography-tandem mass spectrometry. For each of the 4 cell line samples (A549, H358, HCC15 and HCC95) 2 controls and 3 biological replicates were analyzed. Samples were prepared at OSU and analyzed at the University of Michigan (Proteomics Resource Facility, Department of Pathology).
Statistics
Detailed methods for statistical evaluation regarding all components of the described studies are contained in Supplemental Methods.
Study Approval
All animal studies were performed in accordance with the protocols approved by OSU Institutional Animal Care and Use Committee (IACUC, Protocol#2014A00000116) and in accordance with the accepted standard of humane animal care, American Association for Laboratory Animal Care Institutional Guidelines.
Results and Discussion:
Unique role of NOTCH1 signaling in solid epithelial cancers
The Notch signaling pathway, with multiple receptors and ligands, is very complex and plays important roles in almost every aspect of cancer11. As such, Notch is a potential target for cancer therapy37. However, Notch can be oncogenic or tumor suppressive depending on the cancer type2,11. Unfortunately, the individual role of each Notch receptor or ligand is cancer-specific and the behavior of each Notch family member in different cancer types is unclear.
To gain a better understanding of how Notch works in various cancers, we analyzed TCGA transcriptional datasets associated with solid epithelial cancers with the highest mortality rates in the United States (Table 1). A correlation matrix was built on pairwise correlations between the expression of each of the 4 Notch receptors or 5 ligands and all other genes for each dataset. Two-way hierarchical clustering with Euclidian distance and Wards linkage rule was performed to help detect the existence of multivariate structures (Supplemental Figure 1). Patterns of gene co-expression with NOTCH2, 3, and 4 receptors were consistent across tumor types suggesting a conserved function. However, the NOTCH1 receptor showed a very different clustering pattern between tumors (Figure 1A and Supplemental Figure 1). For example, as shown in Figure 1A, both SCC datasets (LUSC and HNSCC) clustered together while NOTCH1 in the LUAD dataset clustered at the opposite end of the dendrogram (farthest distance away) indicating that different sets of genes are co-expressed with NOTCH1 in this subtype.
Figure 1. Patterns of NOTCH1 co-expression in squamous type cancers are distinct from most adenocarcinomas.
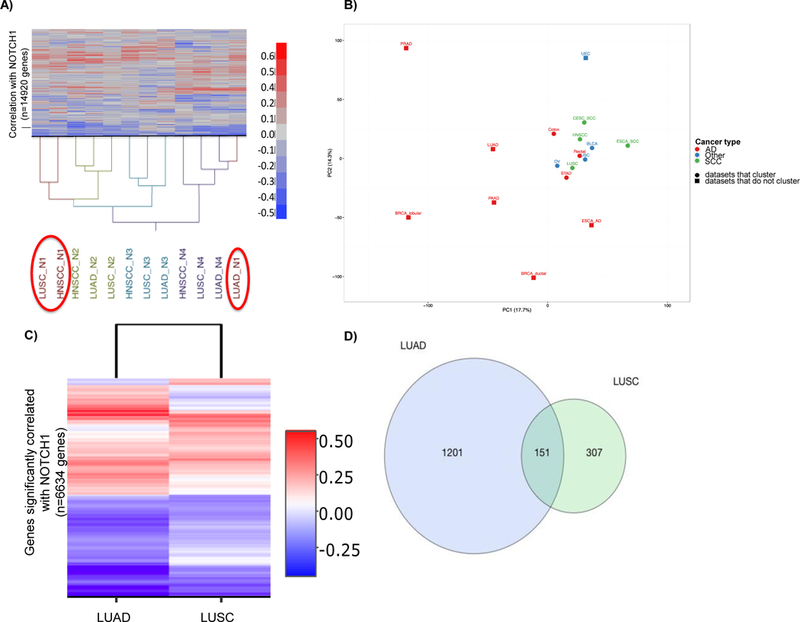
(A) Unsupervised hierarchical clustering of gene (n=14920) correlation with NOTCH1–4 receptors (n=4) in 3 TCGA datasets. (B) Principal component analysis (PCA) of NOTCH1 co-expression with 15518 genes in TCGA solid epithelial cancer datasets (n=17). Using datasets from Table 1 (n>50), PC1 and PC2 account for 17.7% and 14.3% of the total variance, respectively. Cancers were classified as adenocarcinoma (AD) squamous cell carcinoma (SCC) or Other. Circles indicate datasets with similar patterns of NOTCH1 correlated genes that clustered together. (C) Unsupervised hierarchical clustering of genes (n=6634) correlated with NOTCH1 that are statistically significant (n=2 datasets). (D) Venn diagram of genes that were significantly different between LUAD and LUSC datasets (n=1659). The size of the Venn diagrams are proportional to the number of genes significantly correlated with NOTCH1 in LUAD (blue) or LUSC (green) datasets. Statistical significance of genes in (C) determined by Beta-uniform mixture model FDR <0.001 and in (D) by FDR <0.002. Clustering in (A) determined by Euclidian distance and Wards linkage and in (C) determined by Euclidean distance with the complete linkage. PCA analysis in (B) was generated with gene scaling using the SVDimpute algorithm in ClustVis (http://biit.cs.ut.ee/clustvis_large/). LUAD=TCGA lung adenocarcinoma dataset; LUSC=TCGA lung squamous cell carcinoma dataset; HNSCC = TCGA head and neck squamous cell carcinoma dataset; N1=NOTCH1; N2=NOTCH2; N3=NOTCH3; N4=NOTCH4; PC=principal component. See Table 1 for complete dataset names from (B).
Global differences or similarities of NOTCH1 co-expression in solid epithelial cancers was assessed by principal component analysis (PCA) and revealed a group of cancers that clustered (circles, Figure 1B). A pooled statistic was calculated to identify genes associated with these clusters (circles). Enrichment analysis in ToppGene on genes significantly correlated (false discovery rate (FDR) ≤1.0%) within these clusters indicated gene ontology (GO) biological processes involved in RNA processing, cell cycle, and cellular macromolecule localization (Supplemental Table 2).
Our PCA results also show that histology alone may not be sufficient. For example, colorectal cancer is classified histologically as an AD, but in our analysis the colon and rectal datasets clustered near the SCCs. The recent failure of a NOTCH1 inhibitor in a colorectal Phase1b clinical trial are consistent with our in silico predictions and suggest that NOTCH1 may not act as an oncogene in unselected colorectal carcinomas38. As datasets get larger, functionally important NOTCH1 subgroups within each histologic type might be better defined.
Co-expression analysis as a valuable tool for identifying novel NOTCH1 pathway susceptibilities in lung cancer
NSCLC is an example of a tumor type that demonstrates both AD and SCC differentiation. The clinical characteristics of patients in the LUAD and LUSC datasets were similar (Supplemental Table 3). The TCGA esophageal and cervical datasets were used for validation of our methods since they represent both AD and SCC histology. The HNSCC dataset was also evaluated as another upper aerodigestive tumor type with similar etiology to lung SCC. Statistical analyses were performed to identify genes that were significantly correlated with NOTCH1. To correct for multiple hypotheses, we used the Beta-Uniform Mixture (BUM) model39 to estimate the p-value distribution and Benjamin-Hochberg method controlling FDR at alpha=0.00140,41. A high percentage, 44.5% (n=6634) of genes were significantly correlated with NOTCH1 in either the LUAD or LUSC dataset. A heatmap of genes with Pearson correlation coefficients that reached significance is shown in Figure 1C. Analysis of NOTCH1 co-expressed genes (n=6634) with the validation datasets recapitulated distinct clustering of AD or SCC cancer subtypes (Supplemental Figure 2A,B). We performed a Fisher z-transformation to rescale the correlation coefficients to a standard normal assuming the null hypothesis that no correlation is true. We used Fisher z-scores to identify genes differentially correlated with NOTCH1 in the LUAD or LUSC dataset (FDR, alpha=0.002). The number of genes differentially correlated within each dataset is depicted in the Venn diagram (Figure 1D), shown in l̥C with the validation datasets and listed in Supplemental Dataset 1. The top 10 genes that were uniquely associated with either dataset (Table 2A) illustrate differences in NOTCH1 co-expression between the datasets (Supplemental Figure 3,4).
Table 2A.
Top ten lung adenocarcinoma and squamous cell carcinoma genes that are differentially correlated with NOTCH1 in the LUAD and LUSC datasets
| LUAD dataset (Top 10) | ||
|---|---|---|
| Gene Symbol | Full Name | Function |
| MMRN2 | Multimerin 2 | Negative regulator of blood vasculature and angiogenesis, cell motility |
| VWF | Von Willebrand factor | Hemostasis, response to wound healing, innate immune system, cell adhesion, macromolecular complex binding, platelet activation |
| CDH5 | Cadherin 5, type 2 (vascular endothelium) | Vasculature and blood vessel development, negative regulation of cell proliferation, cell adhesion |
| ARHGEF15 | Rho guanine nucleotide exchange factor (GEF) 15 |
GEF for RhoA activation, regulator of vascular development GTPase regulator, and guanyl nucleotide exchange factor |
| CD93 | CD93 molecule | Intercellular adhesion, regulation of immune and defense response, leukocyte activation, and immune effector process |
| GIMAP8 | GTPase, IMAP family member 8 | Anti-apoptotic effect in immune system, mitochondrion |
| SH2D3C | SH2 domain containing 3C | Cell migration, GTPase, guanyl-nucleotide exchange factor, kinase binding, immune function |
| NOTCH4 | NOTCH4 | Vasculature and blood vessel development, angiogenesis, innate immune system |
| CLEC14A | C-type lectin domain family 14, member A | Cell-cell adhesion, angiogenesis, lymphangiogenesis |
| ROBO4 | Roundabout, axon guidance receptor, homolog 4 (Drosophila) |
Angiogenesis, vascular patterning, blood vessel development, [inhibition of] cell migration, modulate vascular response to inflammatory cytokines |
| LUSC dataset (Top 10) | ||
| Gene Symbol | Full Name | Function |
| B4GALNT4 | beta-1,4-N-acetyl-galactosaminyl transferase 4 |
Metabolism, transferase activity, mitotic spindle |
| LHX2 | LIM homeobox 2 | Macromolecular complex binding, chromatin binding, cell differentiation |
| ASPM | Asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
Mitotic cell cycle regulation and coordination, cell cycle, nuclear division, microtubule cytoskeleton |
| NUSAP1 | Nucleolar and spindle associated protein 1 | Spindle microtubule organization, cell cycle, mitotic cell cycle and process, cell division, microtubule cytoskeleton, macromolecular complex binding, RNA binding, chromosome organization and segregation |
| DTL | Denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
Cell cycle control, microtubule cytoskeleton and organizing center, chromosome, centrosome, DNA damage response |
| TFAP2A | Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha |
Microtubule cytoskeleton, centrosome, macromolecular complex binding, chromatin binding |
| TOP2A | Topoisomerase (DNA) II alpha | Makes double strand breaks, segregation of daughter chromosomes, macromolecular complex binding, cell cycle, chromosome, mitotic cell cycle, RNA binding, chromatin |
| KNTC1 | Kinetochore associated 1 | Component of mitotic checkpoint, cell cycle, chromosome, nuclear division, cell division, chromosomal segregation, microtubule cytoskeleton |
| KIF14 | Kinesin family member 14 | Cell cycle, mitosis, nuclear and cell division, cytokinesis, chromosomal segregation, cell proliferation, apoptosis, microtubule, macromolecular complex binding, sister chromatid segregation |
| HNRNPK | Heterogeneous nuclear ribonucleoprotein K |
Pre-mRNA binding protein, response to DNA damage |
Functional enrichment analyses provided insights on the biological pathways associated with differential biomolecular networks (co-expressed genes) (n=1659). In lung AD, NOTCH1 co-expression was distinctly enriched for pathways associated with angiogenesis and vascular development, immune system, and Rho GTPase activity (Table 2B; Supplemental Figure 5 and Supplemental Table 4). For example, higher expression of NOTCH1 was positively correlated with the expression of the vascular pathway genes (MMRN2, VWF, NOTCH4) in the LUAD dataset (pink, Supplemental Figure 6A). In contrast there was no clear correlation between these vascular genes and NOTCH1 in the LUSC dataset (teal). Likewise, higher expression of NOTCH1 was positively correlated with the expression of immune markers including innate immune cells, macrophages, dendritic cells and immune stem cells in the LUAD dataset (pink) but not LUSC dataset (teal, Supplemental Figure 6B). In the LUAD dataset we observed a positive correlation between the expression of myeloid/macrophage biology genes as exemplified by CD9342,43. This is consistent with the literature that suggests Notch signaling plays a role in myeloid/macrophage biology where CD93 plays a key role in differentiation of monocytes to macrophages42. In addition, chemokines such as CCL14 and chemokine receptors involved in the innate immune system were related to NOTCH1 expression in LUAD (data not shown).
Table 2B. Summary of the top three distinct pathways correlated with NOTCH1 in the LUAD and LUSC datasets.
Gene list enrichment analysis based on functional annotations was performed using probability density function method, FDR P-value cutoff 0.05; Toppgene (https://toppgene.cchmc.org/). P-values are shown in Supplemental Table 4 and 5.
| Distinct pathways in LUAD dataset (Top 3) | ||
|---|---|---|
| Pathway | Terms (Top 3) | GO/BioSystems ID (Top 3) |
| Angiogenesis and vasculature development |
vasculature development, blood vessel development, response to wounding |
GO: 0001944, 0001568, 0009611 |
| Immune System | regulation of defense response, regulation of immune system process, leukocyte activation |
GO: 0031347, 0002682, 0045321 |
| Rho/GTPase | signaling by Rho GTPases, Rho GTPase cycle, RHO GTPases Activate Formins |
BioSystems: REACTOME: 1269507, 1269508, 1269519 |
| Distinct pathways in LUSC dataset (Top 3) | ||
| Pathway | Terms (Top 3) | GO/BioSystems ID (Top 3) |
| Cell cycle | cell cycle, cell cycle process, mitotic cell cycle |
GO: 0007049, 0022402, 0000278 |
| Macromolecular complex binding |
microtubule cytoskeleton, macromolecular complex binding, spliceosomal complex |
GO: 0015630, 0044877, 0005681 |
| Chromosome | chromosome organization, sister chromatid segregation, chromatin binding |
GO: 0051276, 0000819, 0003682 |
In lung SCC, NOTCH1 co-expression was uniquely enriched for genes related to cell cycle, macromolecular complex binding, and chromosome pathways (Table 2B; Supplemental Figure 7 and Supplemental Table 5). For example, the expression levels of mitotic genes (ex:BUB1B, ASPM, and TOP2A) were positively correlated in the LUSC dataset, but negatively correlated in the LUAD dataset (Supplemental Figure 6C). Interestingly, enrichment analysis on genes correlated with the clustered datasets from multiple tumor types (Figure 1B), which included all of the SCC datasets, also indicated that cell cycle pathways were associated with NOTCH1 co-expression (Supplemental Table 2).
Finally, lineage-specific biomarkers for lung AD and lung SCC were not differentially correlated with NOTCH1. The lung AD biomarker, cytokeratin 7 (KRT7) and lung SCC biomarker, tumor protein p63 (TP63), showed the expected differences in expression, but not in correlations with NOTCH1 in the LUAD and LUSC datasets. Thus, the differences in correlations observed were not just markers of specific lineages (Supplemental Figure 8).
NOTCH1 shows opposite functional effects in lung adenocarcinoma and lung squamous cell carcinoma in vitro and in vivo
The role of Notch perturbation in lung cancer is unclear. Wael, et al. demonstrated that NOTCH1 has a tumor suppressive role in the lung AD (A549) model and no effect in the lung SCC (H2170) model17. In contrast, many other studies have demonstrated an oncogenic role for NOTCH1 in lung AD3–5,14 but have not experimentally demonstrated the role of NOTCH1 in lung SCC. There have been no direct comparisons between lung AD and lung SCC in well controlled experiments in vitro and in vivo.
We knocked down NOTCH1 in lung AD (A549, H358) and lung SCC (HCC15, HCC95) cell lines with three different lentiviral shRNA constructs. All four cell lines endogenously express NOTCH1 (Supplemental Figure 9) and do not have Notch mutations. In the lung AD cell lines, all three lentiviral shRNA constructs resulted in decreased NOTCH1 expression and reduced cell proliferation relative to non-targeting control (NTC) (Figure 2A,B). In contrast, NOTCH1 knockdown resulted in decreased NOTCH1 expression but increased cell proliferation in lung SCC cell lines. These results demonstrate an opposing phenotypic response to NOTCH1 knockdown in NSCLC. Paired immunoblots demonstrated that that the strength of the phenotype correlated with the degree of knockdown in most cell lines (Figure 2B).
Figure 2. NOTCH1 knockdown (KD) reduces cell viability in lung adenocarcinoma and increases cell viability in lung squamous cell carcinoma in vitro.

(A) Two lung adenocarcinoma (A549 and H358) and two lung squamous cell carcinoma (HCC15 and HCC95) cell lines were infected with lentivirus-based shRNAs targeting NOTCH1 (#3359, #3362, #0254) or non-targeting control (NTC) and 72h later, proliferation was evaluated by alamarBlue, a substrate that measures metabolic activity (n=3). (B) Gene knockdown efficiency was assessed in parallel to (A) with immunoblot analysis of NOTCH1. Statistical significance determined by Kruskal-Wallis test, with Dunn’s post-test for multiple comparisons. Data are presented as mean ± SEM. *P<0.05; ***P<0.001. Experiments are representative of three independent studies.
Knockdown of NOTCH1 in vivo supported a pro-growth role in lung AD and tumor suppressive role in lung SCC. Cell lines infected with either NOTCH1 knockdown construct (shRNA#0254) or NTC control were implanted in bilateral flanks as subcutaneous xenografts in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Knockdown of NOTCH1 in the lung AD cell lines (A549, H358) resulted in tumors with significantly reduced tumor volume relative to NTC, supporting a pro-growth role of NOTCH1 in AD (Figure 3A,B). In contrast, knockdown of NOTCH1 in lung SCC models resulted in significant increase in tumor growth, supporting a tumor suppressive role of NOTCH1 in SCC (Figure 3C,D). The relative levels of NOTCH1 knockdown at the beginning and end of our study are shown in Supplemental Figures 10 and 11.
Figure 3. NOTCH1 knockdown (KD) has opposing effects on tumor growth, vasculature, and mitotic markers in lung adenocarcinoma and lung squamous cell carcinoma in vivo.

NSG mice were bilaterally implanted subcutaneously with cell lines infected with either the NOTCH1 shRNA (#0254) or a non-targeting control (NTC) shRNA. (A,B) In lung adenocarcinoma cell lines (A549 and H358) NOTCH1 KD reduced mean tumor volume relative to NTC (oncogenic phenotype). (C,D) In lung squamous cell carcinoma cell lines (HCC15 and HCC95) NOTCH1 KD increased mean tumor volume as compared to NTC (tumor suppressor phenotype). (E) Representative (magnified 20x) regions show pHH3 and (F) PCAM-1/CD31 staining on A549 (lung adenocarcinoma) and HCC95 (lung squamous cell carcinoma) xenografts under NTC or NOTCH1 KD conditions. (G,H) Quantification of vasculature and mitotic markers represented in (E,F) (per cell line n=4 NTC, n=4 KD#0254). Tumor volume in (A-D) was monitored by caliper measurement on the indicated days following tumor implantation (n=4 mice per cell line). NOTCH1 KD efficiency was assessed at the beginning and completion of the study by immunoblot analysis of NOTCH1 (Supplemental Figure 10, 11). Statistical significance (A-D) determined by RM Anova analysis. Data are represented as mean ± SEM of tumor volumes (mm3). A549 P=0.0053; H358 P=0.0346; HCC15 P=0.0089, HCC95 P=0.0490. Statistical significance (G,H) was determined by Wilcox signed-rank test for comparison between paired NTC and KD#0254 samples. P*<0.05; P**<0.01; P***<0.001.
Unlike prior studies44 that have extrapolated a tumor suppressor function for NOTCH1 in lung SCC from the existence of loss of function mutations, these experiments show the pro-growth and tumor suppressive role of NOTCH1 in vitro and in vivo in controlled lung models.
NOTCH1 has different effects on mitotic, vascular, and immune pathways in lung adenocarcinoma and lung squamous cell carcinoma
To explore how NOTCH1 knockdown experimentally correlates with changes in mitotic, vasculature, and immune process predicted by our in silico studies, we performed immunohistochemistry (IHC) staining and RNA-sequencing on the human lung AD and SCC xenograft tumors collected at the end of the study. For IHC analysis, Formalin-Fixed, Paraffin-Embedded (FFPE) sections from all tumors were stained for phosphohistone H3 (pHH3; mitotic marker) and cluster of differentiation 31 (CD31; vascular marker) (Figure 3E,F). Flash frozen tumor xenografts were evaluated for changes in human and mouse mRNA following NOTCH1 knockdown in two of the lung AD model (A549) and lung SCC model (HCC15) xenografts.
Gene enrichment analysis revealed that genes distinctly co-expressed with NOTCH1 in the LUSC dataset were enriched in pathways involved in cell cycle, specifically mitosis, macromolecular complex binding, and chromosomal organization and binding (Table 2B; Supplemental Table 5). Experimentally we observed that NOTCH1 inhibition had different effects on cell cycle pathways in lung AD and lung SCC. NOTCH1 knockdown corresponded with a significant increase in pHH3 expression in tumors generated from the lung SCC but no significant change in pHH3 expression in the lung AD cell lines (Figure 3G) as predicted by our in silico analysis. Furthermore, pathway analysis of the RNA sequencing data supported our IHC results and predicted activation of the G2 phase of cell cycle progression in the SCC model (HCC15) but decreased function in the lung AD model (A549) with NOTCH1 knockdown (Supplemental Table 6). Together, our in silico, in vitro, in vivo, IHC and RNA sequencing studies in lung SCC suggest that anti-mitotic therapies as single agents or in combination with NOTCH1 agonists may be useful in the treatment lung SCC patients. Our hypothesis is supported by the findings of Ye and collaborators who reported that a specific inhibitor of CDC7 (LY3177833), which plays a role in cell cycle, centromere cohesion, and chromosome segregation, resulted in tumor stasis or regression in the majority of lung SCC cancer patient derived tumor models45. The comparable co-expression patterns we observed in other types of SCC (Figure 1A) suggest that this hypothesis may be generalizable to additional SCC cancers. Future studies may bring additional insights to these intriguing observations.
Vascular pathways were distinctly associated with the LUAD dataset but not in the LUSC dataset (Table 2B; Supplemental Figure 6A). In lung AD tumors NOTCH1 knockdown resulted in a significant increase in CD31 expression, but was unchanged or decreased in lung SCC tumors (Figure 3H). While it is counterintuitive that we should see increased CD31 staining, since that might imply a pro-growth phenotype, other studies have observed an increase of endothelial proliferation following NOTCH1 inhibition46 and the production of non-functional vessels resulting in vascular leakage and dysfunction46. Studies have also shown that the expression of NOTCH1 and VEGFA in lung AD have a significant negative prognostic impact in the lung AD cohort20.
NOTCH1 has been implicated in the regulation of the innate immune system and involvement of myeloid/macrophage biology11,49. In our study, NOTCH1 inhibition had opposing effects on the transcription levels of mouse immune and inflammatory response genes. Our RNA sequencing pathway analysis of in vivo NOTCH1 knockdown tumors in lung AD (A549) predicted an increase in the recruitment of the innate immune and inflammatory system components and a decrease in lung SCC (HCC15) (Supplemental Table 7).
Other studies have proposed that DLL1 ligand treatment results in increased Notch activity and may be beneficial by enhancing T-cell infiltration in tumors48. Our studies were carried out using human cell lines with NOTCH1 knockdown that were implanted in NSG mice. These animals lack T and B cells, but the mouse innate immune system is at least partially intact. Recent studies have shown that NSG mouse monocytes and granulocytes exert a host versus graft response that can lead to rejection of human tissue50,51.
While preliminary, our RNA sequencing results support our in silico findings. Additional studies are required to validate the association of NOTCH1 function with cell cycle, vascular, and immune pathways in subsets of human solid tumors. New ex vivo autologous models are in development that may also enable the investigation of primary human tumors in the presence of human microenvironment components52.
NOTCH1 has distinct protein interactions in lung adenocarcinoma and lung squamous cell carcinoma.
Comparison of Notch interacting proteins in lung AD and SCC could yield clues about the environment in which NOTCH1 exerts its opposing effects. Prior studies have relied on artificial overexpression systems potentially resulting in unintended and/or non-specific interactions due to higher, non-physiological levels of NOTCH1 protein. We inserted the FLAG epitope sequence into the last coding exon of the NOTCH1 gene using CRISPR Cas-9 gene editing to prevent these potential issues (Figure 4A). Using AP-MS/MS at endogenous expression levels of NOTCH1, we demonstrated both novel as well as known (ex: RBPJ, MAML) NOTCH1 protein interactions.
Figure 4. Identifying and validating novel NOTCH1 interacting proteins.
(A) Immunoblot demonstrating that FLAG-epitope tagged endogenous NOTCH1 is present in each clone and can be immunoprecipitated (IP) with anti-FLAG magnetic beads. Immunoprecipitations were performed using anti-FLAG beads on a negative control cell line (Vector) and three individual clones with a FLAG-epitope on the C-terminus of the NOTCH1 protein. Input lane is whole cell lysate. NOTCH1 was detected by anti-NOTCH1 antibody. (B) Venn diagram comparing the number of NOTCH1 interacting proteins identified in each cell line (n=669). Black circles highlight unique protein interactions with lung adenocarcinoma and lung squamous cell carcinoma. Protein identifications are listed in Supplemental Table 8. (C-I) Co-immunoprecipitation experiments of transiently co-transfected NOTCH1-ICD with V5 tag (Bait) and the indicated FLAG or Myc tagged proteins (Prey) in the COS7 cell line. Cell lines transiently expressing NOTCH1-ICD (V5-tagged) or empty vector control were subjected to immunoprecipitation using anti-V5 beads. (C,D) Myc and FLAG-epitope tagged RBPJ was used as a positive control showing strong interaction. (E) FLAG-epitope tagged GFP was used as a negative control. (F-I) Interaction of SDHA, SDHB, IDH2, and AATF with NOTCH1-ICD was detected by anti-FLAG or anti-Myc. Lung adenocarcinoma cell lines (A549, H358), lung squamous cell carcinoma cell lines (HCC15, HCC95). L=Left; R=Right.
Our comparison of interacting proteins identified 15 proteins that uniquely interacted with NOTCH1 in lung AD cell lines (A549, H358) and 10 proteins that only interacted with NOTCH1 in lung SCC cell lines (HCC15, HCC95) (Supplemental Table 8; Figure 4B). A number of mitochondrial proteins were identified to interact with NOTCH1 (Supplemental Table 9). We validated the potential for NOTCH1 to interact with four of the novel interacting proteins: SDHA, SDHB, IDH2 and AATF. NOTCH1 interactions were specific; we detected NOTCH1 interaction with the positive control RBPJ-MYC or RBPJ-FLAG and NOTCH1-ICD V5 (Figure 4C,D) but not in our negative control experiment with GFP-FLAG (Figure 4E). Anti-V5 co-immunoprecipitation experiments demonstrated that SDHA-MYC, SDHB-FLAG, IDH2-MYC, and AATF-MYC interacted with NOTCH1-ICD V5 but not vector control (Figure 4F-I), signifying that the interactions were dependent on the presence of the bait (NOTCH1-ICD). Taken together these results support our proteomics findings.
These results suggest that NOTCH1 may engage in context dependent protein interactions. Three mitochondrial proteins SDHB, IDH2, and MRPS22 that we identified interacting in lung AD but not lung SCC models are known to have key roles in cancer53 including effects on metabolism54, HIF1A stabilization, and impairment of DNA methylation55. IDH2 is a very interesting link given that the genetic variant rs11540478 is a risk factor for lung cancer56. Additionally, high serum levels of IDH2 correlate with poor survival in NSCLC57. Additional experiments are needed to further validate how and if these interactions contribute to the opposing phenotypes of NOTCH1 in NSCLC.
Conclusion
The current study provides insights into the molecular differences that may underlie the diverse effects of NOTCH1 in different lung cancer subtypes. Our findings extend the results of prior loss of function and mutation studies and importantly demonstrate that NOTCH1 has a tumor suppressor role in lung SCC that is independent of mutations. Specific downstream pathway targeting with angiogenesis inhibitors, cell cycle inhibitors, and immune modulators may be particularly effective in the NOTCH1 subsets defined here. Together, our findings shed new light on the complex Notch signaling pathway, provide leads for potentially important functional subgroups of tumors and novel differential interacting proteins for future molecular studies.
Supplementary Material
Acknowledgements
Konstantin Shilo (KS) for consultation; Yung-Mae M. Yao, Bernice A. Agana, and Tiffany D. Talabere for support. We thank Dr. Daniel C. Liebler for discussions of protein-protein interaction studies. We thank Venkatesha Basrur’, Alexey Nesvizhski and the University of Michigan Proteomics Resource Facility for running our samples. We recognize the contribution of the appropriate specimen donors and research groups who contributed to the TCGA database.
This work was supported by the Schnipke Family Endowment Fund; the National Institutes of Health/National Library of Medicine [3 T15 LM 11270–4 S1]; the University of Texas Lung SPORE [P50 CA070907]; and the OSU Cancer Center Support Grant [P30 CA016058].
Abbreviations:
- AD
Adenocarcinoma
- SCC
squamous cell carcinoma
- LUAD
the TCGA lung adenocarcinoma dataset
- LUSC
the TCGA lung squamous cell carcinoma dataset
- PCA
principal component analysis
- GO
gene ontology
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nature reviews Cancer 2003;3(10):756. [DOI] [PubMed] [Google Scholar]
- 2.Previs RA, Coleman RL, Harris AL, Sood AK. Molecular pathways: translational and therapeutic implications of the Notch signaling pathway in cancer. Clinical Cancer Research 2015;21(5):955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgart A, Mazur P, Anton M, et al. Opposing role of Notch1 and Notch2 in a KrasG12D-driven murine non-small cell lung cancer model. Oncogene 2014. [DOI] [PubMed]
- 4.Licciulli S, Avila JL, Hanlon L, et al. Notch1 is required for Kras-induced lung adenocarcinoma and controls tumor cell survival via p53. Cancer research 2013;73(19):5974–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X, Huang L, Futtner C, et al. The cell of origin and subtype of K-Ras-induced lung tumors are modified by Notch and Sox2. Genes & development 2014;28(17):1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radtke F, Fasnacht N, MacDonald HR. Notch signaling in the immune system. Immunity 2010;32(1):14–27. [DOI] [PubMed] [Google Scholar]
- 7.Nicolas M, Wolfer A, Raj K, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet 2003;33(3):416–421. [DOI] [PubMed] [Google Scholar]
- 8.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood 2006;107(6):2223–2233. [DOI] [PubMed] [Google Scholar]
- 9.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. Journal of Experimental Medicine 2011;208(10):1931–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan L, Yao J, Wu X, Fan M. Growth suppression induced by Notch1 activation involves Wnt—β‐catenin down‐regulation in human tongue carcinoma cells. Biology of the Cell 2006;98(8):479–490. [DOI] [PubMed] [Google Scholar]
- 11.Aster JC, Pear WS, Blacklow SC. The varied roles of notch in cancer. Annual Review of Pathology: Mechanisms of Disease 2017;12:245–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aster JC, Blacklow SC, Pear WS. Notch signalling in T‐ cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. The Journal of pathology 2011;223(2):263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paganin M, Ferrando A. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood reviews 2011;25(2):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen TD, Rodriguez EM, Jones KD, Bishop JM. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer research 2011;71(18):6010–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Biswas S, Qin X, Gong W, Deng W, Yu H. Does Notch play a tumor suppressor role across diverse squamous cell carcinomas? Cancer medicine 2016;5(8):2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo L, Zhang T, Xiong Y, Yang Y. Roles of NOTCH1 as a Therapeutic Target and a Biomarker for Lung Cancer: Controversies and Perspectives. Disease markers 2015;2015. [DOI] [PMC free article] [PubMed]
- 17.Wael H, Yoshida R, Kudoh S, Hasegawa K, Niimori-Kita K, Ito T. Notch1 signaling controls cell proliferation, apoptosis and differentiation in lung carcinoma. Lung Cancer 2014;85(2):131–140. [DOI] [PubMed] [Google Scholar]
- 18.Brooks YS, Ostano P, Jo S-H, et al. Multifactorial ERβ and NOTCH1 control of squamous differentiation and cancer. The Journal of clinical investigation 2014;124(5):2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westhoff B, Colaluca IN, D’Ario G, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A 2009;106(52):22293–22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnem T, Andersen S, Al‐Shibli K, Al‐Saad S, Busund LT, Bremnes RM. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer. Cancer 2010;116(24):5676–5685. [DOI] [PubMed] [Google Scholar]
- 21.Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis 2013:bgt127. [DOI] [PMC free article] [PubMed]
- 22.Chen C-Y, Chen Y-Y, Hsieh M-S, et al. Expression of notch gene and its impact on survival of patients with resectable non-small cell lung cancer. Journal of Cancer 2017;8(7):1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong J, Zhang X, Chen X, et al. Prognostic roles of mRNA expression of notch receptors in non-small cell lung cancer. Oncotarget 2017;8(8):13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fazio C, Ricciardiello L. Inflammation and Notch signaling: a crosstalk with opposite effects on tumorigenesis. Cell death & disease 2016;7(12):e2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faruki H, Mayhew GM, Serody JS, Hayes DN, Perou CM, Lai-Goldman M. Lung adenocarcinoma and squamous cell carcinoma gene expression subtypes demonstrate significant differences in tumor immune landscape. Journal of Thoracic Oncology 2017;12(6):943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang Y, Smith S, Hu X. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein & cell 2016;7(3):159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Q, Cohen B, Zheng W, et al. Notch shapes the innate immunophenotype in breast cancer. Cancer Discovery 2017:CD-17–0037. [DOI] [PubMed]
- 28.Gov E, Arga KY. Differential co-expression analysis reveals a novel prognostic gene module in ovarian cancer. Scientific reports 2017;7(1):4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serin EA, Nijveen H, Hilhorst HW, Ligterink W. Learning from co-expression networks: possibilities and challenges. Frontiers in plant science 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Statistics Center, Estimated deaths, 2017. 2017 https://cancerstatisticscenter.cancer.org/module/yg6E0ZLc. Accessed 09/10/2017.
- 31.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic acids research 2015;43(W1):W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galili T heatmaply: Interactive Heat Maps Using’plotly’ 2016. R package version 0.6. 0. In.
- 33.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic acids research 2009;37(suppl 2):W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 2009;4(1):44. [DOI] [PubMed] [Google Scholar]
- 35.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research 2008;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2013;30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platonova N, Lesma E, Basile A, et al. Targeting Notch as a therapeutic approach for human malignancies. Current pharmaceutical design 2017;23(1):108–134. [DOI] [PubMed] [Google Scholar]
- 38.Corral M OncoMed’s Phase 2 Trial of Tarextumab in Small Cell Lung Cancer Does Not Meet Endpoints (None:OMED) 2017; http://www.oncomed.com/invest/releasedetail.cfm?ReleaseID=1021452. Accessed 22 September, 2017.
- 39.Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics 2003;19(10):1236–1242. [DOI] [PubMed] [Google Scholar]
- 40.Morris JS, Wu C, Coombes KR, Baggerly KA, Wang J, Zhang L. Alternative probeset definitions for combining microarray data across studies using different versions of affymetrix oligonucleotide arrays. Meta-Analysis in Genetics 2006:1–214.
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological) 1995:289–300.
- 42.Jeon J-W, Jung J-G, Shin E-C, et al. Soluble CD93 induces differentiation of monocytes and enhances TLR responses. The Journal of Immunology 2010;185(8):4921–4927. [DOI] [PubMed] [Google Scholar]
- 43.Bohlson SS, Silva R, Fonseca MI, Tenner AJ. CD93 is rapidly shed from the surface of human myeloid cells and the soluble form is detected in human plasma. The Journal of Immunology 2005;175(2):1239–1247. [DOI] [PubMed] [Google Scholar]
- 44.Wang NJ, Sanborn Z, Arnett KL, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proceedings of the National Academy of Sciences 2011;108(43):17761–17766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye XS. A selective CDC7 inhibitor (LY3177833) impacts chromosome dynamics and has robust and durable activity in PDX tumor models. Paper presented at: AACR Annual Meeting 2016; April 17, 2016, 2016; New Orleans Theater C, Morial Convention Center, New Orleans, Louisiana. [Google Scholar]
- 46.Trindade A, Djokovic D, Gigante J, Mendonça L, Duarte A. Endothelial Dll4 overexpression reduces vascular response and inhibits tumor growth and metastasization in vivo. BMC cancer 2017;17(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kangsamaksin T, Murtomaki A, Kofler NM, et al. NOTCH decoys that selectively block DLL/NOTCH or JAG/NOTCH disrupt angiogenesis by unique mechanisms to inhibit tumor growth. Cancer discovery 2015;5(2):182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biktasova AK, Dudimah DF, Uzhachenko RV, et al. Multivalent forms of the Notch ligand DLL-1 enhance antitumor T-cell immunity in lung cancer and improve efficacy of EGFR-targeted therapy. Cancer research 2015;75(22):4728–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathern D, Laitman L, Hovhannisyan Z, et al. Mouse and human Notch-1 regulate mucosal immune responses. Mucosal immunology 2014;7(4):995. [DOI] [PubMed] [Google Scholar]
- 50.Racki WJ, Covassin L, Brehm M, et al. NOD-scid IL2rγnull (NSG) Mouse Model of Human Skin Transplantation and Allograft Rejection. Transplantation 2010;89(5):527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirkiles-Smith NC, Harding MJ, Shepherd BR, et al. Development of a humanized mouse model to study the role of macrophages in allograft injury. Transplantation 2009;87(2):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radhakrishnan P, Sekar V, Brijwani N, et al. A patient derived ex vivo platform CANScriptTM predicts distinct therapeutic outcomes to multiple PD-1 checkpoint inhibitors in single tumor biopsies. In: AACR; 2017.
- 53.Yen K, Bittinger M, Su S, Fantin V. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene 2010;29(49):6409. [DOI] [PubMed] [Google Scholar]
- 54.Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Molecular and cellular biology 2008;28(2):718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pecqueur C, Oliver L, Oizel K, Lalier L, Vallette FM. Targeting metabolism to induce cell death in cancer cells and cancer stem cells. International journal of cell biology 2013;2013. [DOI] [PMC free article] [PubMed]
- 56.Li J, Lu J, He Y, et al. A new functional IDH2 genetic variant is associated with the risk of lung cancer. Molecular carcinogenesis 2017;56(3):1082–1087. [DOI] [PubMed] [Google Scholar]
- 57.Li Jj, Li R, Wang W, et al. IDH 2 is a novel diagnostic and prognostic serum biomarker for non‐small‐cell lung cancer. Molecular oncology 2018;12(5):602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



