Abstract
Protein arginine deiminases (PADs) catalyze the post-translational deimination of peptidyl arginine to form peptidyl citrulline. This modification is increased in multiple inflammatory diseases and in certain cancers. PADs regulate a variety of signaling pathways including apoptosis, terminal differentiation, and transcriptional regulation. Activity-based protein profiling (ABPP) probes have been developed to understand the role of the PADs in vivo and to investigate the effect of protein citrullination in various pathological conditions. Furthermore, these ABPPs have been utilized as a platform for high-throughput inhibitor discovery. This review will showcase the development of ABPPs targeting the PADs. In addition, it provides a brief overview of PAD structure and function along with recent advances in PAD inhibitor development.
1. Introduction
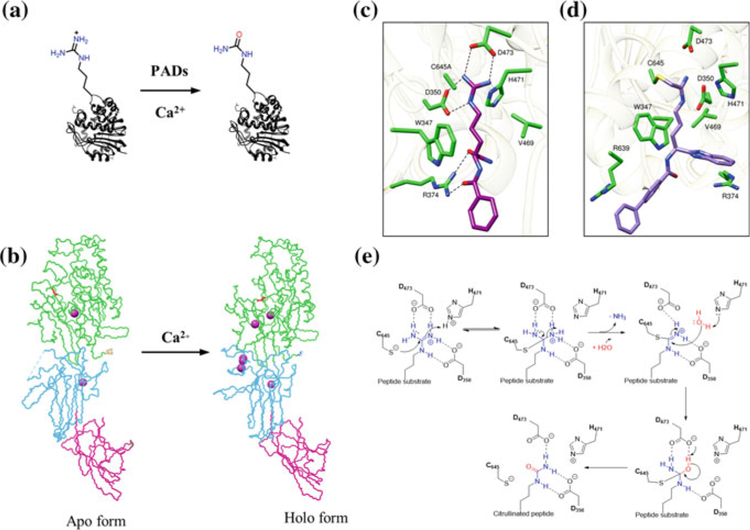
Protein citrullination was first described by Rogers and Simmonds in 1958 (Fig. 1a) (Rogers and Simmonds 1958). This post-translational modification (PTM) converts an arginine residue into a citrulline via hydrolysis of the guanidinium group in a so-called deimination reaction.
Fig. 1.
a PAD-catalyzed hydrolysis of peptidyl arginine to peptidyl citrulline. b Backbone conformation of PAD2 showing both the apo and holo forms that is generated upon calcium binding. The structural change due to calcium binding is clearly evident in the catalytic domain (green), which harbors the catalytic cysteine C647 (shown in red in the catalytic domain). c Crystal structure of PAD4 C654A protomer bound to the substrate BAA (PDB code 1WDA). d Co-crystal structure of BB-F-amidine (5a) bounds to PAD4 (PDB code 5N0 M). e Proposed catalytic mechanism for PAD4
This PTM is generated by a family of calcium-dependent enzymes called protein arginine deiminases (PADs) (Bicker and Thompson 2013; Fuhrmann et al. 2015; Fuhrmann and Thompson 2016). There are five human PADs, i.e., PAD1, PAD2, PAD3, PAD4, and PAD6, of which only PADs 1–4 are catalytically active (Raijmakers et al. 2007; Taki et al. 2011; Witalison et al. 2015). These isozymes, which share 50–55% sequence identity within the same species (Arita et al. 2004), are found in a myriad of cells and tissue types. PAD1 is highly expressed in epidermis and uterus and is thought to citrullinate keratins and filaggrins during skin keratinization (Ishida-Yamamoto et al. 2002; Senshu et al. 1996). PAD2 is distributed in various tissues and is abundant in muscles and brain (Moscarello et al. 2002). PAD3 is found in hair follicles and epidermis (Rogers et al. 1997), whereas PAD4 is expressed in neutrophils, granulocytes and macrophages (Nakashima et al. 1999). PAD6 is only found in oocytes and embryos (Esposito et al. 2007). In addition to their localization in the cytoplasm, PAD1, PAD2, and PAD4 also localize to the nucleus where they citrullinate histones and other chromatin-associated proteins (Cherrington et al. 2010; Fuhrmann et al. 2015; Jang et al. 2011; Kan et al. 2012; Lewallen et al. 2015).
Citrullination can have profound effects on the primary, secondary, and tertiary structures of proteins due to the loss of a positive charge. Additionally, this PTM can result in the loss of protein–protein or protein–DNA interactions with consequent effects on cell signaling (Clancy et al. 2017; Fuhrmann et al. 2015; Lewis and Nacht 2016). Notably, the PADs regulate various biological processes, including myelination, cell differentiation, gene regulation, and the innate immune response (Bicker and Thompson, 2013; Christophorou et al. 2014; Li et al. 2010; Nauseef and Borregaard, 2014; Senshu et al. 1996; Slade et al. 2014). Additionally, aberrant PAD activity can lead to protein hypercitrullination, which is a hallmark of various inflammatory and neurodegenerative disorders (Jang et al. 2008; Jones et al. 2009; Khandpur et al. 2013; Knight et al. 2013, 2014; Leffler et al. 2012; Musse et al. 2008).
Specifically, citrullination occurs during NETosis, a proinflammatory form of cell death that is aberrantly upregulated in numerous autoimmune diseases including RA, atherosclerosis, and lupus (Khandpur et al. 2013; Knight et al. 2013, 2014). Protein citrullination is also elevated in luminal breast cancer, multiple sclerosis and certain inflammatory diseases (Jones et al. 2009; McElwee et al. 2012; Moscarello et al. 2002; Zhang et al. 2012). Importantly, inhibition of PAD2 in breast cancer cell lines decreases disease progression by increasing apoptosis (McElwee et al. 2012). Recently, PAD1 was shown to be overexpressed in human triple-negative breast cancer lines (e.g., MDA-MB-231 cells) as well as in xenograft mouse models and its inhibition resulted in reduced cell proliferation and metastasis (Qin et al. 2017). PAD2 and PAD4 are also activated in the central nervous system (CNS) during neurodegenerative processes and are observed to be co-localized in regions of degraded neurons in Alzheimer’s patients (Acharya et al. 2012; Ishigami et al. 2005). The distinct roles of the PADs in various pathophysiologic states are quite alarming, and there is a pressing need to develop isozyme-specific inhibitors to be used as therapeutics and probes to decipher the physiological roles of these enzymes in various disease states. This review will focus on the discovery and development of PAD-targeted ABPPs. It will also provide a brief overview of the structure and function of the PADs and recent progress in PAD inhibitor development.
2. PAD Structure and Function
2.1. PAD Structures
One of the major advances that aided the development of PAD inhibitors and activity-based probes was the determination of high-resolution X-ray crystal structures for PAD1, PAD2, and PAD4 (Arita et al. 2004; Saijo et al. 2016; Slade et al. 2015). PADs contain an α/β propeller domain located in the C-terminal lobe that harbors the active site. The N-terminal domain is comprised of two immunoglobulin-like subdomains that are proposed to be important for protein–protein interactions and for substrate selection (Fig. 1b) (Arita et al. 2004; Fuhrmann et al. 2015). Crystal structures of both the inactive (apo, calcium-free) and active (holo, calcium-bound) states of PAD2 and PAD4 have been reported confirming that calcium binding is required to form a catalytically competent active site (Fig. 1b). A significant difference between the apo and holo states is the position of the catalytic cysteine (C647 in PAD2 and C645 in PAD4), which moves ≥ 10 Å into the active site upon calcium binding (Fig. 1b–d). From a careful analysis of the crystal structures, we know that PAD1, PAD2, and PAD4 bind four, six, and five calcium ions, respectively—PAD1 lacks Ca5 and PAD2 has a unique sixth site, i.e., Ca6. Based on calcium titration experiments with PAD2, we know that Ca1 and Ca6 are tightly and constitutively bound to PAD2. Ca3, Ca4, and Ca5 bind next and trigger a conformational change that generates the Ca2-binding site. Ca2 binding then triggers a conformational change that moves C647, as well as W347, into catalytically competent conformations (Slade et al. 2015).
2.2. PAD Substrate Recognition and Catalytic Mechanism
PADs are highly selective in citrullinating peptidyl arginine over free arginine (Knuckley et al. 2010a). However, not all arginine residues are equally citrullinated by the PADs. Evidence suggests that PADs preferentially catalyze the citrullination of arginine residues present in a β-turn or in specific loops as compared to other secondary structure elements in a protein (Knuckley et al. 2010a; Tarcsa et al. 1996). PADs are also known to catalyze the citrullination of arginine containing small peptides. A structure of the PAD4 (C645A) bound to benzoyl-L-arginine amide (BAA) depicts the key residues that interact with the substrate (Arita et al. 2004). As shown in Fig. 1c, D473 and D350 make salt bridges with the guanidinium group of the substrate, C645A is positioned for nucleophilic attack, and H471 is involved in general acid/base catalysis. The methylene groups of the substrate side chain make hydrophobic interactions with W347 and V469. R374 makes hydrogen bonding interactions with the backbone carbonyl group of the substrate. This is a notable interaction as it confers specificity toward peptidyl arginine as opposed to free arginine, which lacks an amide bond. This observation was further confirmed by crystal structures of PAD4 bound to various histone substrate peptides, which illustrated that most of the interactions occur between the backbone carbonyl groups of the substrate and the side chain of the active-site residues (Arita et al. 2006). The lack of relevant interactions between the side chain of protein substrates and PAD4 is consistent with the broad substrate scope of PAD4. The substrate scope of the other PADs is similarly broad suggesting that substrate recognition is driven primarily by sequence context rather than key side chain interactions.
Based on the available crystal structures and biochemical experiments (Arita et al. 2004; Knuckley et al. 2007, 2010a, b), we proposed that PAD4 catalyzes citrulli-nation via attack on the guanidinium carbon by a nucleophilic cysteine thiolate, which results in the formation of a tetrahedral intermediate (Fig. 1e). Proton transfer from H471 stabilizes the positively charged intermediate via interactions with D350 and D473 (Knuckley et al. 2010b). Collapse of the intermediate generates an S-alkylthiouronium intermediate that is ultimately hydrolyzed by water via a second tetrahedral intermediate. The thiol group in the catalytic cysteine needs to be deprotonated for optimal enzyme activity. Based on pH-dependent kinetic inactivation studies, the pKa of this cysteine was calculated to be ~8.3 when iodoacetamide was used as the thiol-reactive reagent (Dreyton et al. 2014b; Knuckley et al. 2007, 2010a, b). This data coupled with the observation of an inverse solvent isotope effect on kcat/Km supports the notion that PAD4 (as well as PADs 1 and 3) uses a reverse protonation mechanism. Further supporting this mechanism is the fact that 2-chloroacetamidine, a positively charged thiol-reactive compound, does not depress the pKa of C645 in PAD4 (Knuckley et al. 2010b). By contrast, 2-chloroacetamidine does depress the pKa of C647 in PAD2 (pKa ~ 7.2) (Dreyton et al. 2014b). This data along with a normal solvent isotope effect indicates that PAD2 uses a substrate assisted mechanism of thiol deprotonation (Dreyton et al. 2014b).
3. PAD Inhibitors
Over the past several years, numerous PAD inhibitors have been developed. These compounds include both reversible and irreversible inhibitors with decent potencies and selectivities. The mechanisms of action have been extensively studied using biochemical and X-ray crystallographic techniques. This section will provide an overview of both reversible (non-covalent) and irreversible (covalent) PAD inhibitors.
3.1. Reversible PAD Inhibitors
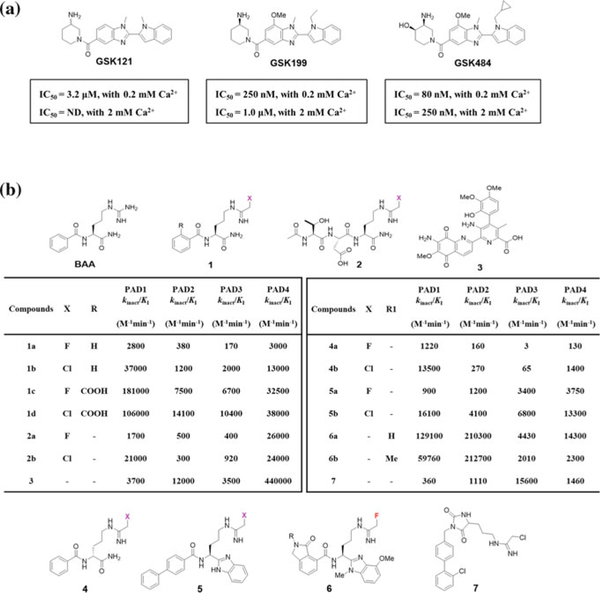
Taxol (pacitaxel) was discovered as a weak non-competitive inhibitor (Ki = 4–10 mM) of PAD4 by Pritzker and Moscarello two decades ago. This finding sparked the beginning of further PAD inhibitor development (Pritzker and Moscarello 1998). Knuckley et al. later discovered streptomycin (Ki ~ 0.56 mM), minocycline (Ki ~ 0.63 mM), and chlorotetracycline (Ki ~ 0.11 mM) as relatively potent inhibitors of PAD4 (Knuckley et al. 2008). Recently, Lewis et al. reported the development of highly potent reversible inhibitors that are selective for the apo form of PAD4 (Lewis et al. 2015). A DNA-encoded small molecule library screen for PAD inhibitors in the presence and absence of calcium yielded GSK121 as a modest inhibitor of apo PAD4. A detailed SAR for the primary hit resulted in optimized leads GSK199 and GSK484, which inhibited PAD4 with IC50 values of 250 nM and 80 nM, respectively, in the presence of 0.2 mM Ca2+ (Fig. 2a). The potencies of these compounds were, however, reduced by >fivefold at higher calcium concentrations. Detailed kinetic analyses and a co-crystal structure indicate that these inhibitors are competitive with respect to calcium binding and preferentially bind the apo form of PAD4, thereby preventing the calcium-induced movement of Cys645 into the active site. Not only did these inhibitors demonstrate more than 15-fold selectivity for PAD4, but they also revealed a unique approach for targeting the apo state of a specific PAD isoform. Importantly, GSK199 shows efficacy in the murine collagen induced arthritis model of RA (Willis et al. 2017).
Fig. 2.
a Structure of reversible PAD4 inhibitors. The IC50 values for each compound with PAD4 are shown underneath. These compounds are >15-fold selective for PAD4. b Structure of irreversible PAD inhibitors. The generic warhead on the irreversible inhibitors is colored in pink. The second-order rate constants for PAD inactivation by covalent inhibitors are shown in the table
3.2. Irreversible PAD Inhibitors
The search for irreversible covalent inhibitors of PAD isoforms began with the discovery of 2-chloroacetamidine, a time-dependent inactivator of PAD4 with an IC50 > 0.5 mM. In parallel, Luo et al. replaced the guanidinium group in the PAD substrate benzoyl-L-arginine amide (BAA) with either a fluoro- or chloroacetamidine-based warhead, thereby generating F-amidine (1a) or Cl-amidine (1b), respectively (Fig. 2b) (Luo et al. 2006a, c). Kinetic studies revealed that both F-amidine and Cl-amidine function as mechanism-based inhibitors that irreversibly inactivate PAD4 by modifying the catalytic cysteine in a time-dependent manner. Interestingly, Cl-amidine was more potent than F-amidine when evaluated against all PAD isoforms (except PAD6, which is not active), consistent with the increased reactivity of the chloro-warhead over the fluoro-warhead. More importantly, Cl-amidine showed efficacy in treating a variety of animal models, including mouse models of RA, lupus, atherosclerosis, ulcerative colitis, neuron injury, and breast cancer (Willis et al. 2011; Knight et al. 2013, 2014, 2015; Lange et al. 2011) (Chumanevich et al. 2011; McElwee et al. 2012).
Optimization of F-amidine and Cl-amidine resulted in the identification of the more potent ortho-carboxylate derivatives o-F-amidine (1c) and o-Cl-amidine (1d) (Causey et al. 2011). The increased potency of 1c and 1d against PAD4 was found to be due to enhanced interactions between the o-carboxylate groups and the indole NH of W347 as well as a water-mediated hydrogen bond with the side chain of Q346 (Causey et al. 2011). From an extensive tripeptide library screen, the haloacetamidine-based peptides, TDFA (2a) and TDCA (2b) (Fig. 2b), were discovered to be potent and selective PAD4 inactivators (Jones et al. 2012). The improved inhibition of these peptides was attributed to their interaction with Q346, R374, and R639 in PAD4. Interaction with R639 was looked upon as key for selectivity against PAD4 as this residue is unique to this isozyme. From a fluorescence polarization activity-based high-throughput screen (HTS), assay streptonigrin (3) was also found to be a selective inhibitor of PAD4 (Dreyton et al. 2014a; Knuckley et al. 2010c).
Despite these early advances, the peptidic nature of these first-generation haloacetamidine-based inhibitors possessed various limitations, including reduced metabolic stability due to proteolytic cleavage and poor membrane permeability (Knight et al. 2015). This led to the development of second-generation inhibitors that were predicated on improving the lipophilicity of the core structure by substituting the C-terminal carboxamide in 1a and 1b with a benzimidazole moiety and the N-terminal phenyl ring with a biphenyl substituent resulting in BB-F-amidine (5a) and BB-Cl-amidine (5b) (Figs. 1d and 2b) (Knight et al. 2015; Muth et al. 2017). This led to a dramatic increase in the lipophilicity of BB-Cl-amidine (CLogP = 4.17) as compared to Cl-amidine (CLogP = −0.23), which was predicted to aid cell entry (Knight et al. 2015). Consistent with this prediction, BB-Cl-amidine was far superior when compared to Cl-amidine in a variety of cell-based assays and animal models but showed similar in vitro efficacy (Ghari et al. 2016; Horibata et al. 2015; Kawalkowska et al. 2016; Knight et al. 2015). Recently, Muth et al. reported a detailed SAR for 5a and 5b with a methyl benz-imidazole scaffold that included a lactam ring in place of N-terminal phenyl group (Muth et al. 2017). The most potent inhibitors 6a and 6b exhibited excellent PAD2 selectivity (up to 106-fold) and proved to be potent in several cell-based assays (Fig. 2b) (Muth et al. 2017).
The search for other PAD isoform-selective inhibitors led to the discovery of D-F-amidine (4a) and D-Cl-amidine (4b) (Fig. 2b), the D-ornithine derivatives of 1a and 1b, which exhibited remarkable selectivity against PAD1 (400-fold by 4a and 200-fold by 4b) (Bicker et al. 2012a). D-Cl-amidine also demonstrated improved metabolic stability as compared to Cl-amidine (Bicker et al. 2012a). Furthermore, Jamali et al. identified a novel scaffold (7) through a substrate-based fragment screen, inhibitor, which was potent and selective for PAD3 (Fig. 2b) (Jamali et al. 2015, 2016). In addition to the in vitro selectivity, the hydantoin-based inhibitor (7) also demonstrated efficacy in a cellular model of thapsigargin induced cell death in PAD3-expressing HEK293T cells (Jamali et al. 2016).
4. Activity-Based Proteomic Probes Targeting the PADs
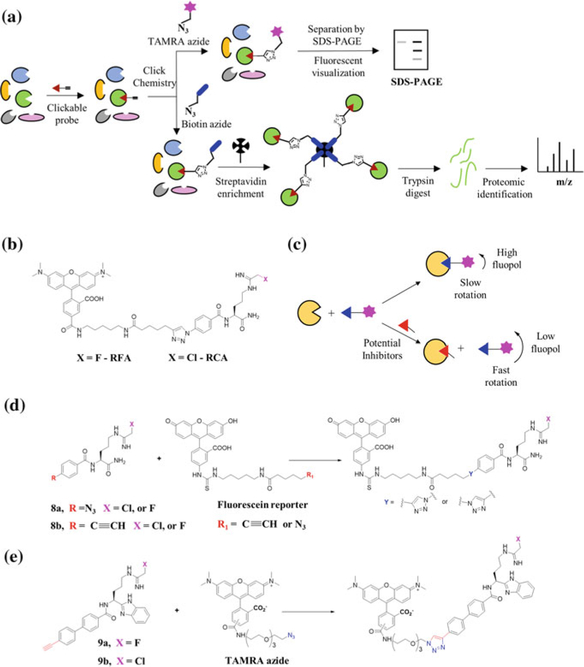
The ability of haloacetamidine-based inhibitors to covalently modify PADs in an irreversible manner was considered to be a prerequisite for the development of activity-based probes. In addition to a chemical warhead that reacts with functional sites in proteins, activity-based probes also bear chemical handles, such as fluorophores, biotin, or alkynes for subsequent fluorescent visualization, streptavidin enrichment and mass spectrometry-based quantification or bio-orthogonal conjugation, respectively, for subsequent analysis of protein activities (Fig. 3a). An overview of the development of PAD-targeted ABPPs is described below.
Fig. 3.
a Schematic representation depicting the use of clickable ABPPs. b Chemical structures of RFA and RCA. c Schematic overview of the fluorescence polarization assay using RFA as an ABPP for high-throughput inhibitor discovery. d Structures of first-generation ‘clickable’ PAD-targeted ABPPs. Labeled probes are subjected to click-chemistry using fluorescein reporters through a post-inactivation strategy. e Structures of second-generation ‘clickable’ PAD-targeted ABPPs. Probe-labeled proteins are subjected to click-chemistry using TAMRA azide reporter through a post-inactivation strategy
4.1. First-Generation PAD-Targeted ABPPs
First-generation PAD-targeted ABPPS were based on the structure of Cl-amidine and F-amidine. Replacement of the benzoyl group with a p-benzylic azide facilitated their conjugation to rhodamine–alkyne via the formation of a triazole linker to generate rhodamine-conjugated F-amidine (RFA) and rhodamine-conjugated Cl-amidine (RCA) (Fig. 3b) (Luo et al. 2006b). RFA and RCA preferentially labeled the calcium-bound holo form of PAD4 and do not modify the C645S active-site mutant (Luo et al. 2006b). These probes are as potent as the parent compounds suggesting that the fluorophore does not alter enzyme binding (Luo et al. 2006b). The versatility of RFA and RCA was further demonstrated through the development of a competitive gel-based ABPP-assay, which was used to identify novel PAD inhibitors from diverse chemical libraries (Knuckley et al. 2008). Using this strategy, Knuckley et al. discovered streptomycin as a competitive inhibitor and chlorotetracycline as a mixed inhibitor of PAD4 (Knuckley et al. 2008).
The success of this RFA-based screening strategy led to the development of an RFA-based high-throughput screen (HTS), termed a fluorescence polarization activity-based protein profiling (fluopol-ABPP)-based HTS assay (Fig. 3c) (Knuckley et al. 2010c). This assay monitors changes in fluorescence polarization (fluopol) as a result of RFA binding to PAD4. Since the RFA-PAD4 complex rotates slowly, there is a larger change in the fluorescent polarization signal. In contrast, unbound RFA rotates faster and leads to a smaller change in the fluorescence polarization signal. In the case of an inhibitor bound to PAD4, RFA-PAD4 binding will be hampered, resulting in a lower fluopol signal (Knuckley et al. 2010c). Using this strategy, Knuckley et al. screened more than 2000 compounds and identified 10 inhibitors, from which streptonigrin (3) was discovered as a potent and selective inhibitor of PAD4. Despite the utility of this RFA-based HTS assay, the methodology suffered limitations including a strong bias toward irreversible inhibitors that preferentially targeted the active holo form of PAD4.
To overcome this limitation, Lewallen et al. developed a modified approach to identify inhibitors that targeted the apo, calcium-free, form of PAD2 (Lewallen et al. 2014). Specifically, they performed the screen at lower Ca2+ concentrations (350 μM CaCl2) and screened the 1280 compound LOPAC library (Sigma-Aldrich Library of Pharmacologically Active Compounds). Using this approach, Lewallen et al. identified ruthenium red as a calcium competitive inhibitor that binds to the apo form of PAD2 (Lewallen et al. 2014). In the future, this RFA-based screening strategy could be used to identify isoform-specific PAD inhibitors from various compound libraries. Furthermore, this approach will be useful for the discovery of allosteric inhibitors that target alternate sites in PAD isoforms, similarly to the PAD4-selective inhibitors GSK199 and GSK484.
4.2. Second-Generation PAD-Targeted ABPPs
A major drawback of the rhodamine-based probes RFA and RCA is that they are not ideal cellular probes due to the presence of a bulky fluorophore, which may alter their cell permeability. To circumvent this issue, Slack et al. employed a bio-orthogonal strategy that involved the conjugation of fluorescein and biotin-based reporters to F-amidine and Cl-amidine using the copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC) reaction (Slack et al. 2011). Development of fluorescein-based ABPPs, in addition to the rhodamine-based ABPPs, in principle, generates tools that allow for the optimal visualization of labeled proteins across the entire pH spectrum. To achieve this, Slack et al. synthesized azide and alkyne containing F-amidine and Cl-amidine (8a and 8b) and used them to label recombinant PAD4 and PAD4 in Escherichia coli cell extracts (Fig. 3d). The labeled samples were then subjected to click-chemistry using the corresponding azide or alkyne fluorescein reporter tags to obtain post-inactivated ABPPs bound to PAD4 that were visualized by fluorescence. F-amidine and Cl-amidine containing an alkyne handle demonstrated efficient post-inactivation click-chemistry as compared to their azide counterparts and proved to be selective for PAD4 in E. coli cell lysates (Fig. 3a, d). Despite their utility in the efficient detection of PAD4 by fluorescent visualization, these fluorescein-based ABPPs were not useful for isolating PAD4 or to identify PAD4-interacting proteins in biological samples (Slack et al. 2011).
To demonstrate efficient isolation of PAD4 from bacterial (E. coli) and mammalian (e.g., MCF-7 and HL60) cells, Slack et al. treated the cell lysates with F-amidine-Y-ne and Cl-amidine-Y-ne and employed a TEV-tagged biotin–azide reporter to perform post-inactivation click-chemistry (Slack et al. 2011). The subsequent pulldown of the probe-modified proteins using streptavidin–agarose and Western blot established the utility of these biotin–azide reporter tags in isolating the active form of PAD4 and identifying several known PAD4-binding proteins, including HDAC1, p53, and histone H3. The approach was also used to isolate PAD4 from HL60 granulocytes, suggesting a potential future role for these ABPPs in identifying the post-translational modifications that occur to the PADs under various physiological conditions.
Building on the success of the second-generation inhibitors BB-F-amidine and BB-Cl-amidine as cell-permeable and metabolically stable pan-PAD inhibitors, we next developed clickable probes based on this scaffold (Nemmara et al. 2018). Specifically, Nemmara et al. developed alkyne containing derivatives of BB-F-amidine and BB-Cl-amidine, which are termed BB-F-Yne (9a) and BB-Cl-Yne (9b). These probes are as potent as the parent compounds (5a and 5b) and can label PAD2 in HEK cells (Fig. 3a, e). The versatility of these probes was verified by their ability to pull down PAD2 from HEK cell lysates after post-inactivation click-chemistry with biotin–azide. Furthermore, using a chemoproteomic approach, we demonstrated that BB-F-Yne, which possesses a fluoroacetamidine warhead, is remarkably selective for PAD2 in HEK cells—the only protein isolated was PAD2. By contrast, BB-Cl-Yne has a few ‘off-targets’ in addition to PAD2. Most of the off-targets are highly abundant proteins with reactive cysteines, i.e., ‘the usual suspects’ that are found in other proteomic screens that use cysteine-reactive electrophiles (Weerapana et al. 2007, 2010). Projecting forward, it will be worthwhile to assess the potential of future ABPPs harboring a fluoroacetamidine warhead, especially, for the selective detection of PAD isoforms in complex biological systems as well as in identifying novel proteins and signaling pathways associated with PAD biology.
4.3. Phenylglyoxal-Based Probes to Detect Protein Citrullination
Given that aberrant citrullination is a hallmark of various autoimmune diseases and certain tumor types, it is important to develop protein detection methods to identify citrullinated proteins in complex biological systems. Due to the low abundance of this PTM in biological samples, the enrichment of citrullinated proteins by chemical derivatization improves detection. It should be noted that the detection of citrullinated proteins by mass spectrometry is also challenging because the 1 Da mass change is easily confused with a deamidation event or an isotope effect. To improve the MS-based detection of citrullinated proteins, Holm et al. showed that selective derivatization of the urea group in citrulline is possible with 2,3-butanedione alone or in combination with antipyrine (De Ceuleneer et al. 2011; Holm et al. 2006). These modifications resulted in a mass increase of 50 or 238 Da, which is readily detected by MS, thereby differentiating citrullinated peptides from other arginine containing ones. However, this methodology does not facilitate the enrichment of citrullinated proteins or peptides.
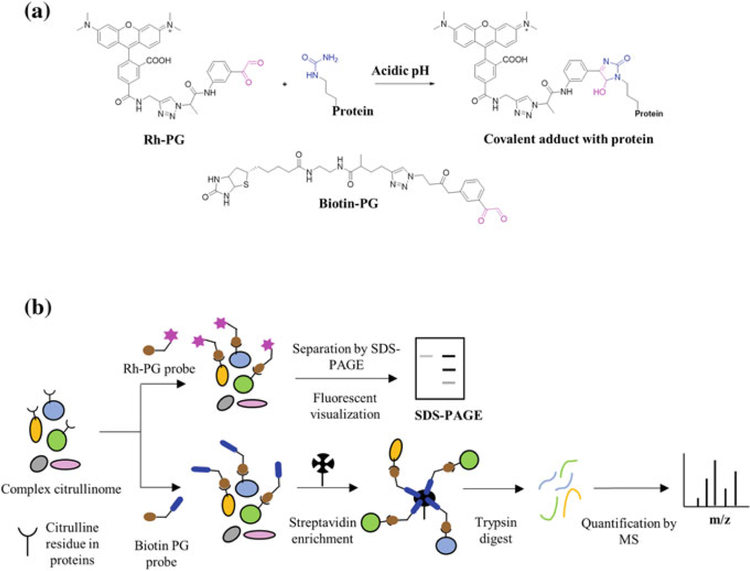
Building upon this concept, Bicker et al. developed a citrulline-specific probe by linking rhodamine and phenylglyoxal via a triazole linker. This compound rhodamine–phenylglyoxal (Rh-PG) specifically labels citrullinated proteins under acidic conditions (Bicker et al. 2012b) (Fig. 4a, b). Rh-PG was able to monitor the kinetics of protein citrullination of PAD4 substrates (e.g., histone H3) due to its low limit of detection (LOD ~ 600 fmol for citrullinated histone H3). In addition, Bicker et al. used Rh-PG to detect citrullinated proteins in a mouse model of ulcerative colitis treated with and without a PAD inhibitor. Notably, several proteins showed reduced citrullination in the presence of inhibitor, suggesting they could be useful biomarkers of ulcerative colitis (Fig. 4a). Using Rh-PG, Bawadekar et al. also quantified citrullinated proteins in lysates in a mouse model of TNF-α induced lung inflammation and showed that PADs other than PAD4 are responsible for inflammation-mediated protein citrullination in lungs (Bawadekar et al. 2016). Moreover, Carmona-Rivera et al. used Rh-PG to show that citrullinated proteins are abundantly generated during NET formation induced by either IgM or rheumatoid factor (Carmona-Rivera et al. 2017). In total, Rh-PG is a powerful probe for the robust quantification of citrullinated proteins and can be used to characterize unique disease biomarkers in various autoimmune diseases.
Fig. 4.
a Chemical structures of Rh-PG and BPG and the chemoselective reaction of Rh-PG with citrulline under acidic conditions. b Schematic representation of the strategy for the chemical derivatization of peptidyl citrulline using Rh-PG and biotin PG
To extend the versatility of these phenylglyoxal-based probes, Lewallen et al. developed biotin-conjugated phenylglyoxal (BPG). This probe serves as an antibody surrogate for Western blotting and as a chemical handle to isolate citrullinated proteins from biological mixtures (Fig. 4a, b) (Lewallen et al. 2015). Using this probe, numerous citrullinated proteins were identified for the first time from PAD2-expressing HEK293T cells treated with ionomycin. These novel PAD substrates included multiple proteins involved in RNA splicing, suggesting a potential role for the PADs in this process (Lewallen et al. 2015). More recently, Tilvawala et al. used BPG to map the RA-associated citrullinome in serum, synovial fluid, and synovial tissue samples (Tilvawala et al. 2018). In addition to identifying more than 150 novel citrullinated proteins, they developed a BPG-based ELISA assay to validate the proteomic data. Among the various proteins identified, there were numerous SERPINs (SERine Protease INhibitors) and metabolic enzymes. Fascinatingly, Tilvawala et al. demonstrated that citrullination inactivates a subset of SERPINs and modulates the activity of a range of metabolic enzymes. Specifically, citrullination of antiplasmin, C1-inhibitor and tissue plasminogen inhibitor (t-PAI) abolished their inhibitory activity against their cognate proteases, i.e., plasmin, kallikrein, and tissue plasminogen (t-PA).
An important limitation of this probe is that in its current form, it is not possible to identify the exact site of citrullination in target proteins. Moreover, it does not distinguish between citrullination and lysine carbamylation. In the future, development of phenylglyoxal-based probes that can identify endogenous sites of citrullination should be a priority.
5. Conclusions
Over the past several years, chemical labeling has produced a wealth of novel technologies to identify the biological roles of a variety of proteins. Activity-based protein profiling (ABPP) is one such technique that has become a centerpiece of chemical biology. The discovery of RFA, a haloacetamidine-based ABPP, has led to the development of high-throughput assay platforms for identifying isoform-selective PAD inhibitors. Moreover, the first-generation PAD-targeted ABPPs, F-amidine-Yne (8a) and Cl-amidine-Yne (8b), were able to label PAD4 in cells and isolate PAD4-binding proteins. Remarkably, the second-generation PAD-targeted ABPPs, BB-F-Yne (9a) and BB-Cl-Yne (9b), showed enhanced cellular uptake and efficient labeling of PADs in cells (Fig. 3e). Perhaps the most exciting discovery is the superior selectivity of fluroacetamidine-based probes toward PADs in cell systems, a finding that is counterintuitive to the historical perception that covalent inhibitors are non-selective in nature. We predict that these fluoroacetamidine-based probes will be used to detect the PADs in complex biological milieus and in identifying pathways that regulate PAD activity. Recent advances in chemical labeling techniques have also led to the development of citrulline-specific probes to detect protein citrullination in a variety of disease states. The citrulline-specific probes Rh-PG and BPG have proved to be powerful tools to detect and quantify protein citrullination in complex systems. We predict that these probes have the potential to transform our understanding of PAD biology by uncovering novel citrullinated biomarkers associated with a range of pathophysiological states. Finally, we expect the next-generation citrulline-specific probes will enable the identification of specific sites of citrullination.
References
- Acharya NK, Nagele EP, Han M, Coretti NJ, DeMarshall C, Kosciuk MC, Boulos PA, Nagele RG (2012) Neuronal PAD4 expression and protein citrullination: possible role in production of autoantibodies associated with neurodegenerative disease. J Autoimmun 38(4):369–380. 10.1016/j.jaut.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M (2004) Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol 11(8):777–783. 10.1038/nsmb799 [DOI] [PubMed] [Google Scholar]
- Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M (2006) Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci U S A 103(14):5291–5296. 10.1073/pnas.0509639103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawadekar M, Gendron-Fitzpatrick A, Rebernick R, Shim D, Warner TF, Nicholas AP, Lundblad LK, Thompson PR, Shelef MA (2016) Tumor necrosis factor alpha, citrullination, and peptidylarginine deiminase 4 in lung and joint inflammation. Arthritis Res Ther 18(1):173 10.1186/s13075-016-1068-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker KL, Anguish L, Chumanevich AA, Cameron MD, Cui X, Witalison E, Subramanian V, Zhang X, Chumanevich AP, Hofseth LJ et al. (2012a) D-amino acid based protein arginine deiminase inhibitors: Synthesis, pharmacokinetics, and in cellulo efficacy. ACS Med Chem Lett 3(12):1081–1085. 10.1021/ml300288d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker KL, Subramanian V, Chumanevich AA, Hofseth LJ, Thompson PR (2012b) Seeing citrulline: development of a phenylglyoxal-based probe to visualize protein citrullination. J Am Chem Soc 134(41):17015–17018. 10.1021/ja308871v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker KL, Thompson PR (2013) The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers 99(2):155–163. 10.1002/bip.22127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Rivera C, Carlucci PM, Moore E, Lingampalli N, Uchtenhagen H, James E, Liu Y, Bicker KL, Wahamaa H, Hoffmann V et al. (2017) Synovial fibroblast-neutrophil interactions promote pathogenic adaptive immunity in rheumatoid arthritis. Sci Immunol 2(10). 10.1126/sciimmunol.aag3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causey CP, Jones JE, Slack JL, Kamei D, Jones LE, Subramanian V, Knuckley B, Ebrahimi P, Chumanevich AA, Luo Y et al. (2011) The development of N-alpha-(2-carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-L-ornithine amide (o-F-amidine) and N-alpha-(2-carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)- L -ornithine amide (o-Cl-amidine) as second generation protein arginine deiminase (PAD) inhibitors. J Med Chem 54(19):6919–6935. 10.1021/jm2008985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington BD, Morency E, Struble AM, Coonrod SA, Wakshlag JJ (2010) Potential role for peptidylarginine deiminase 2 (PAD2) in citrullination of canine mammary epithelial cell histones. PLoS ONE 5(7):e11768 10.1371/journal.pone.0011768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JC, Zernicka-Goetz M et al. (2014) Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 507(7490):104–108. 10.1038/nature12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, Davis T, Matesic LE, Thompson PR, Hofseth LJ (2011) Suppression of Colitis in Mice by Cl-Amidine: a novel Peptidylarginine Deiminase (Pad) Inhibitor. Am J Physiol Gastrointest Liver Physiol 300(6):G929–G938. 10.1152/ajpgi.00435.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy KW, Russell AM, Subramanian V, Nguyen H, Qian Y, Campbell RM, Thompson PR (2017) Citrullination/methylation crosstalk on Histone H3 regulates ER-target gene transcription. ACS Chem Biol 12(6):1691–1702. 10.1021/acschembio.7b00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ceuleneer M, De Wit V, Van Steendam K, Van Nieuwerburgh F, Tilleman K, Deforce D (2011) Modification of citrulline residues with 2,3-butanedione facilitates their detection by liquid chromatography/mass spectrometry. Rapid Commun Mass Spectrom 25(11):1536–1542. 10.1002/rcm.5015 [DOI] [PubMed] [Google Scholar]
- Dreyton CJ, Anderson ED, Subramanian V, Boger DL, Thompson PR (2014a) Insights into the mechanism of streptonigrin-induced protein arginine deiminase inactivation. Bioorg Med Chem 22(4):1362–1369. 10.1016/j.bmc.2013.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyton CJ, Knuckley B, Jones JE, Lewallen DM, Thompson PR (2014b) Mechanistic studies of protein arginine deiminase 2: evidence for a substrate-assisted mechanism. Biochemistry 53 (27):4426–4433. 10.1021/bi500554b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Vitale AM, Leijten FP, Strik AM, Koonen-Reemst AM, Yurttas P, Robben TJ, Coonrod S, Gossen JA (2007) Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol 273(1–2):25–31. 10.1016/j.mce.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Fuhrmann J, Clancy KW, Thompson PR (2015) Chemical biology of protein arginine modifications in epigenetic regulation. Chem Rev 115(11):5413–5461. 10.1021/acs.chemrev.5b00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann J, Thompson PR (2016) Protein arginine methylation and citrullination in epigenetic regulation. ACS Chem Biol 11(3):654–668. 10.1021/acschembio.5b00942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghari F, Quirke AM, Munro S, Kawalkowska J, Picaud S, McGouran J, Subramanian V, Muth A, Williams R, Kessler B et al. (2016) Citrullination-acetylation interplay guides E2F-1 activity during the inflammatory response. Sci Adv 2(2):e1501257 10.1126/sciadv.1501257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm A, Rise F, Sessler N, Sollid LM, Undheim K, Fleckenstein B (2006) Specific modification of peptide-bound citrulline residues. Anal Biochem 352(1):68–76. 10.1016/j.ab.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Horibata S, Vo TV, Subramanian V, Thompson PR, Coonrod SA (2015) Utilization of the soft agar colony formation assay to identify inhibitors of tumorigenicity in breast cancer cells. J Vis Exp (99):e52727 10.3791/52727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida-Yamamoto A, Senshu T, Eady RA, Takahashi H, Shimizu H, Akiyama M, Iizuka H (2002) Sequential reorganization of cornified cell keratin filaments involving filaggrin-mediated compaction and keratin 1 deimination. J Invest Dermatol 118(2):282–287. 10.1046/j.0022-202x.2001.01671.x [DOI] [PubMed] [Google Scholar]
- Ishigami A, Ohsawa T, Hiratsuka M, Taguchi H, Kobayashi S, Saito Y, Murayama S, Asaga H, Toda T, Kimura N et al. (2005) Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer’s disease. J Neurosci Res 80(1):120–128. 10.1002/jnr.20431 [DOI] [PubMed] [Google Scholar]
- Jamali H, Khan HA, Stringer JR, Chowdhury S, Ellman JA (2015) Identification of multiple structurally distinct, nonpeptidic small molecule inhibitors of protein arginine deiminase 3 using a substrate-based fragment method. J Am Chem Soc 137(10):3616–3621. 10.1021/jacs.5b00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamali H, Khan HA, Tjin CC, Ellman JA (2016) Cellular activity of new small molecule protein arginine deiminase 3 (PAD3) inhibitors. ACS Med Chem Lett 7(9):847–851. 10.1021/acsmedchemlett.6b00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang B, Kim E, Choi JK, Jin JK, Kim JI, Ishigami A, Maruyama N, Carp RI, Kim YS, Choi EK (2008) Accumulation of citrullinated proteins by up-regulated peptidylarginine deiminase 2 in brains of scrapie-infected mice: a possible role in pathogenesis. Am J Pathol 173(4):1129–1142. 10.2353/ajpathajpath.2008.080388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang B, Shin HY, Choi JK, du Nguyen PT, Jeong BH, Ishigami A, Maruyama N, Carp RI, Kim YS, Choi EK (2011) Subcellular localization of peptidylarginine deiminase 2 and citrullinated proteins in brains of scrapie-infected mice: nuclear localization of PAD2 and membrane fraction- enriched citrullinated proteins. J Neuropathol Exp Neurol 70(2):116–124. 10.1097/NEN.0b013e318207559e [DOI] [PubMed] [Google Scholar]
- Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR (2009) Protein arginine deiminase 4 (PAD4): current understanding and future therapeutic potential. Curr Opin Drug Discov Devel 12(5):616–627 [PMC free article] [PubMed] [Google Scholar]
- Jones JE, Slack JL, Fang P, Zhang X, Subramanian V, Causey CP, Coonrod SA, Guo M, Thompson PR (2012) Synthesis and screening of a haloacetamidine containing library to identify PAD4 selective inhibitors. ACS Chem Biol 7(1):160–165. 10.1021/cb200258q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan R, Jin M, Subramanian V, Causey CP, Thompson PR, Coonrod SA (2012) Potential role for PADI-mediated histone citrullination in preimplantation development. BMC Dev Biol 1219 10.1186/1471-213x-12-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawalkowska J, Quirke AM, Ghari F, Davis S, Subramanian V, Thompson PR, Williams RO, Fischer R, La Thangue NB, Venables PJ (2016) Abrogation of collagen–induced arthritis by a peptidyl arginine deiminase inhibitor is associated with modulation of T cell-mediated immune responses. Sci Rep 626430 10.1038/srep26430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS, Friday S, Li S, Patel RM, Subramanian V et al. (2013) NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med 5(178):178ra140 10.1126/scitranslmed.3005580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Luo W, O’Dell AA, Yalavarthi S, Zhao W, Subramanian V, Guo C, Grenn RC, Thompson PR, Eitzman DT et al. (2014) Peptidylarginine deiminase inhibition reduces vascular damage and modulates innate immune responses in murine models of atherosclerosis. Circ Res 114(6):947–956. 10.1161/CIRCRESAHA.114.303312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Subramanian V, O’Dell AA, Yalavarthi S, Zhao W, Smith CK, Hodgin JB, Thompson PR, Kaplan MJ (2015) Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann Rheum Dis 74(12):2199–2206. 10.1136/annrheumdis-2014-205365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Zhao W, Luo W, Subramanian V, O’Dell AA, Yalavarthi S, Hodgin JB, Eitzman DT, Thompson PR, Kaplan MJ (2013) Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J Clin Invest 123(7):2981–2993. 10.1172/JCI67390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B, Bhatia M, Thompson PR (2007) Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry 46(22):6578–6587. 10.1021/bi700095s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR (2010a) Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry 49(23):4852–4863. 10.1021/bi100363t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR (2010b) Haloacetamidine- based inactivators of protein arginine deiminase 4 (PAD4): evidence that general acid catalysis promotes efficient inactivation. ChemBioChem 11(2):161–165. 10.1002/cbic.200900698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B, Jones JE, Bachovchin DA, Slack J, Causey CP, Brown SJ, Rosen H, Cravatt BF, Thompson PR (2010c) A fluopol-ABPP HTS assay to identify PAD inhibitors. Chem Commun (Camb) 46(38):7175–7177. 10.1039/c0cc02634d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B, Luo Y, Thompson PR (2008) Profiling Protein Arginine Deiminase 4 (PAD4): a novel screen to identify PAD4 inhibitors. Bioorg Med Chem 16(2):739–745. 10.1016/j.bmc.2007.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Gogel S, Leung KY, Vernay B, Nicholas AP, Causey CP, Thompson PR, Greene ND, Ferretti P (2011) Protein deiminases: new players in the developmentally regulated loss of neural regenerative ability. Dev Biol 355(2):205–214. 10.1016/j.ydbio.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler J, Martin M, Gullstrand B, Tyden H, Lood C, Truedsson L, Bengtsson AA, Blom AM (2012) Neutrophil extracellular traps that are not degraded in systemic lupus erythematosus activate complement exacerbating the disease. J Immunol 188(7):3522–3531. 10.4049/jimmunol.1102404 [DOI] [PubMed] [Google Scholar]
- Lewallen DM, Bicker KL, Madoux F, Chase P, Anguish L, Coonrod S, Hodder P, Thompson PR (2014) A FluoPol-ABPP PAD2 high-throughput screen identifies the first calcium site inhibitor targeting the PADs. ACS Chem Biol 9(4):913–921. 10.1021/cb400841k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewallen DM, Bicker KL, Subramanian V, Clancy KW, Slade DJ, Martell J, Dreyton CJ, Sokolove J, Weerapana E, Thompson PR (2015) Chemical proteomic platform to identify citrullinated proteins. ACS Chem Biol 10(11):2520–2528. 10.1021/acschembio.5b00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HD, Liddle J, Coote JE, Atkinson SJ, Barker MD, Bax BD, Bicker KL, Bingham RP, Campbell M, Chen YH et al. (2015) Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat Chem Biol 11(3):189–191. 10.1038/nchembio.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HD, Nacht M (2016) iPAD or PADi-’tablets’ with therapeutic disease potential? Curr Opin Chem Biol 33169–178. 10.1016/j.cbpa.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Li P, Wang D, Yao H, Doret P, Hao G, Shen Q, Qiu H, Zhang X, Wang Y, Chen G et al. (2010) Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene 29(21):3153–3162. 10.1038/onc.2010.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Sato M, Thompson PR (2006a) Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry 45(39):11727–11736. 10.1021/bi061180d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Knuckley B, Bhatia M, Thompson PR (2006b) Activity based protein profiling reagents for Protein Arginine Deiminase 4 (PAD4): synthesis and in vitro evaluation of a fluorescently-labeled probe. J Am Chem Soc 128(45):14468–14469. 10.1021/ja0656907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR (2006c) A fluoro-acetamidine based inactivator of protein arginine deiminase 4 (PAD4): design, synthesis, and in vitro and in vivo evaluation. J Am Chem Soc 128(45):1092–1093. 10.1021/ja0576233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JL, Mohanan S, Griffith OL, Breuer HC, Anguish LJ, Cherrington BD, Palmer AM, Howe LR, Subramanian V, Causey CP et al. (2012) Identification of PADI2 as a potential breast cancer biomarker and therapeutic target. BMC Cancer 12(1):500 10.1186/1471-2407-12-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscarello MA, Pritzker L, Mastronardi FG, Wood DD (2002) Peptidylarginine deiminase: a candidate factor in demyelinating disease. J Neurochem 81(2): 335–343. 10.1191/1352458502ms776oa [DOI] [PubMed] [Google Scholar]
- Musse AA, Li Z, Ackerley CA, Bienzle D, Lei H, Poma R, Harauz G, Moscarello MA, Mastronardi FG (2008) Peptidylarginine deiminase 2 (PAD2) overexpression in transgenic mice leads to myelin loss in the central nervous system. Dis Model Mech 1(4–5):229–240. 10.1242/dmm.000729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth A, Subramanian V, Beaumont E, Nagar M, Kerry P, McEwan P, Srinath H, Clancy K, Parelkar S, Thompson PR (2017) Development of a selective inhibitor of protein arginine deiminase 2. J Med Chem 60(7):3198–3211. 10.1021/acs.jmedchem.7b00274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Hagiwara T, Ishigami A, Nagata S, Asaga H, Kuramoto M, Senshu T, Yamada M (1999) Molecular characterization of peptidylarginine deiminase in HL-60 cells induced by retinoic acid and 1-alpha,25-dihydroxyvitamin D(3). J Biol Chem 274(39):27786–27792. 10.1074/jbc.274.39.27786 [DOI] [PubMed] [Google Scholar]
- Nauseef WM, Borregaard N (2014) Neutrophils at work. Nat Immunol 15(7):602–611. 10.1038/ni.2921 [DOI] [PubMed] [Google Scholar]
- Nemmara VV, Subramanian V, Muth A, Mondal S, Salinger AJ, Maurais AJ, Tilvawala R, Weerapana E, Thompson PR (2018) The Development of benzimidazole-based clickable probes for the efficient labeling of cellular Protein Arginine Deiminases (PADs). ACS Chem Bioldoi. 10.1021/acschembio.7b00957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritzker LB, Moscarello MA (1998) A novel microtubule independent effect of paclitaxel: the inhibition of peptidylarginine deiminase from bovine brain. Biochim Biophys Acta 1388 (1):154–160. 10.1016/S0167-4838(98)00175-7 [DOI] [PubMed] [Google Scholar]
- Qin H, Liu X, Li F, Miao L, Li T, Xu B, An X, Muth A, Thompson PR, Coonrod SA et al. (2017) PAD1 promotes epithelial-mesenchymal transition and metastasis in triple-negative breast cancer cells by regulating MEK1-ERK1/2-MMP2 signaling. Cancer Lett 40930–41. 10.1016/j.canlet.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen PA, Drijfhout JW, Pruijn GJ (2007) Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol 367(4):1118–1129. 10.1016/j.jmb.2007.01.054 [DOI] [PubMed] [Google Scholar]
- Rogers G, Winter B, McLaughlan C, Powell B, Nesci T (1997) Peptidylarginine deiminase of the hair follicle: characterization, localization, and function in keratinizing tissues. J Invest Dermatol 108(5):700–707. 10.1111/1523-1747.ep12292083 [DOI] [PubMed] [Google Scholar]
- Rogers GE, Simmonds DH (1958) Content of citrulline and other amino-acids in a protein of hair follicles. Nature 182(4629):186–187. 10.1038/182186a0 [DOI] [PubMed] [Google Scholar]
- Saijo S, Nagai A, Kinjo S, Mashimo R, Akimoto M, Kizawa K, Yabe-Wada T, Shimizu N, Takahara H, Unno M (2016) Monomeric Form of peptidylarginine deiminase Type I revealed by X-ray crystallography and small-angle X-ray scattering. J Mol Biol 428(15):3058–3073. 10.1016/j.jmb.2016.06.018 [DOI] [PubMed] [Google Scholar]
- Senshu T, Kan S, Ogawa H, Manabe M, Asaga H (1996) Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochem Biophys Res Commun 225(3):712–719. 10.1006/bbrc.1996.1240 [DOI] [PubMed] [Google Scholar]
- Slack JL, Causey CP, Luo Y, Thompson PR (2011) Development and use of clickable activity based protein profiling agents for protein arginine deiminase 4. ACS Chem Biol 6(5):466–476. 10.1021/cb1003515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade DJ, Fang P, Dreyton CJ, Zhang Y, Fuhrmann J, Rempel D, Bax BD, Coonrod SA, Lewis HD, Guo M et al. (2015) Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design. ACS Chem Biol 10(4):1043–1053. 10.1021/cb500933j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade DJ, Subramanian V, Thompson PR (2014) Pluripotency: citrullination unravels stem cells. Nat Chem Biol 10(5):327–328. 10.1038/nchembio.1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki H, Gomi T, Knuckley B, Thompson PR, Vugrek O, Hirata K, Miyahara T, Shinoda K, Hounoki H, Sugiyama E et al. (2011) Purification of enzymatically inactive peptidylarginine deiminase type 6 from mouse ovary that reveals hexameric structure different from other dimeric isoforms. Adv Biosci Biotechnol 02(04):304–310. 10.4236/abb.2011.24044 [DOI] [Google Scholar]
- Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM (1996) Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem 271(48):30709–30716. 10.1074/jbc.271.48.30709 [DOI] [PubMed] [Google Scholar]
- Tilvawala R, Nguyen SH, Maurais AJ, Nemmara VV, Nagar M, Salinger AJ, Nagpal S, Weerapana E, Thompson PR (2018) The rheumatoid arthritis-associated citrullinome. Cell Chem Biol 25(6):691–704. 10.1016/j.chembiol.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Speers AE, Cravatt BF (2007) Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)—A general method for mapping sites of probe modification in proteomes. Nat Protoc 2(6):1414–1425. 10.1038/nprot.2007.194 [DOI] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468(7325):790–795. 10.1038/nature09472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis VC, Banda NK, Cordova KN, Chandra PE, Robinson WH, Cooper DC, Lugo D, Mehta G, Taylor S, Tak PP et al. (2017) PAD4 inhibition is sufficient for the amelioration of collagen-induced arthritis. Clin Exp Immunoldoi. 10.1111/cei.12932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, Luo Y, Levitt B, Glogowska M, Chandra P et al. (2011) N-alpha;-Benzoyl-N5-(2-Chloro-1-Iminoethyl)- L-Ornithine Amide, a protein arginine deiminase inhibitor, reduces the severity of murine collagen-induced arthritis. J Immunol 186(7):4396–4404. 10.4049/jimmunol.1001620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witalison EE, Thompson PR, Hofseth LJ (2015) Protein arginine deiminases and associated citrullination: physiological functions and diseases associated with dysregulation. Curr Drug Targets 16(7):700–710. 10.2174/1389450116666150202160954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, Slade DJ, Dreyton CJ, Subramanian V, Bicker KL et al. (2012) Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc Natl Acad Sci U S A 10913331–13336. 10.1073/pnas.1203280109 [DOI] [PMC free article] [PubMed] [Google Scholar]