Abstract
Epigenetic modifications confer stable transcriptional patterns in the brain and both normal and abnormal brain function involve specialized brain regions. We examined DNA methylation by whole genome bisulfite sequencing in neuronal and non-neuronal populations from four brain regions (anterior cingulate gyrus, hippocampus, prefrontal cortex, and nucleus accumbens) as well as chromatin accessibility in the latter two. We find pronounced differences in CpG and non-CpG differentially methylated regions (CG- and CH-DMRs) only in neuronal cells across regions. While neuronal CH-DMRs were highly associated with differential gene expression, CG-DMRs were consistent with chromatin accessibility and enriched for regulatory regions. These CG-DMRs comprise ~12 Mb of the genome that is highly enriched for genomic regions associated with heritability of neuropsychiatric traits including addictive behavior, schizophrenia, and neuroticism, suggesting a mechanistic link between pathology and differential neuron-specific epigenetic regulation in distinct brain regions.
INTRODUCTION
Distinct cognitive functions and behaviors are often correlated with particular regions and/or cell types in the brain; much of disease-based brain research is focused on identifying the anatomical structures that mediate normal function, e.g. the hippocampus in memory, the prefrontal cortex in cognition, and the nucleus accumbens in addictive behavior. Further, many neuropsychiatric diseases preferentially affect individual neuronal subpopulations present in particular brain regions. The epigenome is particularly important in maintaining cellular identity and responding to environmental perturbations1; therefore, mapping the cell type- and brain region-specific transcriptional and epigenetic landscapes is necessary for identifying functional genomic differences that contribute to disease phenotypes. Recent transcriptome analyses have revealed extensive differences between non-cerebellar human brain regions2 and between distinct neuronal3 and glial4 subpopulations in mice. Substantial transcriptional heterogeneity also exists among single cells profiled in mammalian cortical regions5–7. Single cell DNA methylation8 within a single brain region has only recently been measured and the specific locations and functional consequences of methylation changes among cell types remain underexplored.
In contrast to gene expression, few if any DNA methylation differences among phenotypically normal non-cerebellar human brain regions have been reported 9–11. DNA methylation is known to be altered in patients with neuropsychiatric disease, including schizophrenia12, Alzheimer’s13, and major depressive disorder14. The only whole genome bisulfite (WGBS) analyses across multiple human brain regions9,15 found very few differences among cortical tissues. The apparent lack of epigenetic diversity among brain regions is surprising given the known transcriptional diversity. Importantly, large-scale –omics12,16 as well as case-control17 studies are predominately conducted using bulk tissues comprised of neuronal and non-neuronal cell populations at variable proportions. While the authors of these studies acknowledge the confounding potential due to cellular heterogeneity, a robust reference of sorted populations from multiple brain regions would greatly improve current computational deconvolution strategies.
In contrast to mice3,18,19, few studies have examined DNA methylation between cellular subpopulations isolated from human brain tissue20. Neurons are easily distinguished from non-neurons (astrocytes, oligodendrocytes, microglia, and epithelial cells) using the nuclear neuronal marker, NeuN. DNA methylation differences between these two broadly defined populations have been widely reported21,22 and have been assessed genome-wide within a single brain region across development22 and in the context of several neurodegenerative diseases23. Still, a comprehensive analysis of brain region-specific DNA methylation has not been performed using sorted nuclei isolated from human tissues.
Here, we addressed this knowledge gap by analyzing the DNA methylation landscape (both CpG and non-CpG) using WGBS in fractionated neuronal and non-neuronal nuclei (n=45) and bulk tissues (n=27) isolated from four post-mortem brain regions: dorsolateral prefrontal cortex (BA9), anterior cingulate gyrus (BA24), hippocampus (HC), and nucleus accumbens (NAcc). We also examined both gene expression (n=20) and chromatin accessibility (n=22) in NAcc and BA9 nuclei. Importantly, we find that regions of differential methylation, specifically within the neuronal population, are highly enriched for heritability of schizophrenia, addictive behavior, and neuroticism.
RESULTS
Cell type heterogeneity obscures epigenetic differences between brain regions
We mapped the DNA methylation landscape using whole-genome bisulfite sequencing (WGBS) of four human post-mortem brain regions: dorsolateral prefrontal cortex (BA9), anterior cingulate cortex (BA24), hippocampus (HC), and nucleus accumbens (NAcc) (Supplementary Table 1). We isolated neuronal and non-neuronal nuclei based on the neuronal marker NeuN using fluorescence-activated nuclear sorting (FANS) followed by WGBS in a total of 45 samples from 6 donors (Supplementary Table 2; Supplementary Figure 1a,b) and also generated data from 27 bulk tissue samples. We observed substantial variation in the proportion of NeuN+ nuclei among brain regions, within the same brain region between individuals, and even among samplings from the same tissue specimen (Supplementary Figure 1c). Several factors contribute to this variability including nuclei loss or damage during sample processing, unequal subsampling of tissue specimens, and differences in post-mortem intervals though only within HC and NAcc (Supplementary Figure 1d; p = 0.025 from linear mixed model). This finding emphasizes the inherent challenge in accounting for cellular heterogeneity in complex solid tissues that, unlike blood and other peripheral tissues, cannot be repeatedly sampled.
Neuronal DNA methylation (CpG [mCG] and non-CpG [mCH]) in the NAcc was distinct from BA9 and HC (Supplementary Figure 1e). We also confirmed previous observations21,22 of higher global mCG and mCH (p < 2.2 × 10−16, for both) in neuronal compared to non-neuronal nuclei (Supplementary Figure 1e,f). Interestingly, NAcc neuronal nuclei had higher global levels of mCG, but lower levels of mCH than the other brain regions. Principal component analysis (PCA) of autosomal DNA methylation reveals clear segregation of cell types (neuronal from non-neuronal nuclei) and brain regions within the neuronal population (Figure 1a) that is not observed in analysis of bulk tissue (Supplementary Figure 2a, Supplementary Table 3). Segregation by brain region becomes even more apparent upon separate analysis of neuronal nuclei (Supplementary Figure 2b) and joint analysis of bulk tissues and sorted nuclei allows better resolution of brain region-specific differences among the bulk samples (compare Supplementary Figures 2a,c). In contrast, separate analysis of non-neuronal nuclei reveals segregation of samples by donor rather than tissue (Supplementary Figure 2d) similar to previous findings10.
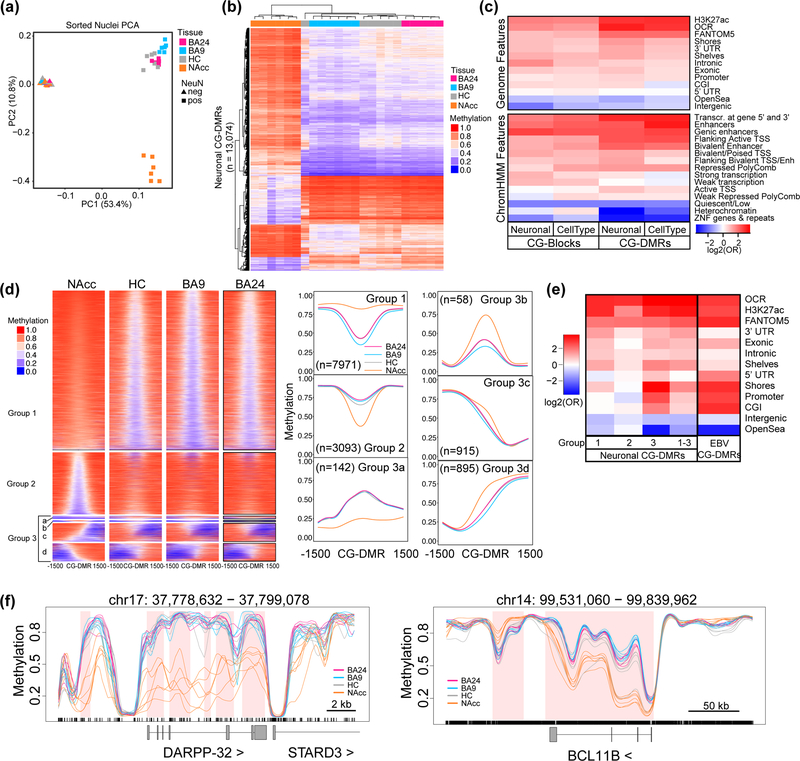
Figure 1. Neuronal nuclei isolated from different brain regions display widespread differences in CpG methylation.
DNA methylation was assessed in samples from four tissues: anterior cingulate cortex (BA24; pink), prefrontal cortex (BA9; blue), nucleus accumbens (NAcc; orange), and hippocampus (HC; grey). (a) Principal component analysis (PCA) of distances derived from average autosomal CpG methylation (in 1 kb intervals) in NeuN+ and NeuN- nuclei from each brain region; each point is a sample (n=6 for BA9,HC,NAcc in both NeuN+ and NeuN-; for BA24, n=5 for NeuN+ and n=4 for NeuN-). (b) Hierarchical clustering of samples based on the average methylation per sample of the neuronal CG-DMRs (n=13,074). (c) Log odds ratios for the enrichment of CpGs within cell type (NeuN+ vs NeuN-) and neuronal (NeuN+) CG-DMRs and blocks compared to the rest of the genome for genomic features. Gene models from GENCODE (promoters, intronic, exonic, 5’UTR, 3’UTR, intergenic), CpG islands (CGIs) and related features from UCSC (shores, shelves, OpenSea), putative enhancer regions (H3K27ac30, FANTOM531), open chromatin regions (OCRs) from this study, and brain-specific ChromHMM15 annotations (core 15-state model). (d) Average methylation for NeuN+ nuclei over a 3 kb window centered on the neuronal CG-DMRs. CG-DMRs were grouped by k-means clustering based on their methylation patterns. Metagene plots for each group are shown to the right with the number of CG-DMRs in each group indicated. (e) Heatmap as in (c) showing enrichment within neuronal CG-DMRs from groups 1–3 and CG-DMRs from EBV transformed B cells33. (f) Example CG-DMR (top) and CG-block (bottom) showing average methylation values for NeuN+ nuclei from each tissue as indicated. Regions of differential methylation are shaded in pink.
Consistent with the PCA results, we find a high correlation among bulk tissues and non-neuronal nuclei (Supplementary Figure 2e). Further, correlation within an individual is much lower than within sample type (NeuN+, NeuN-, or bulk tissue) (Supplementary Figure 3a). While the non-neuronal population is also composed of several distinct cell types, we found that these samples had the least between-sample variability and smallest effect size (1%, proportion of CpGs with absolute mean difference >10%) with bulk tissue samples having the greatest variability and the neuronal population having the greatest effect size (3–12%) (Supplementary Figure 3b,c). While our results cannot exclude the possibility of brain region-specific methylation in rare non-neuronal populations, they indicate that sample-to-sample variability in cell type composition is not confounding our assertion that non-neuronal samples show little brain region-specific methylation. This finding contrasts with the regional transcriptome differences in astrocytes that have been observed in mice24, but is consistent with the relatively consistent composition of glial subpopulations across multiple brain regions25. Our data indicate that the neuronal populations contain the relevant functional variability in DNA methylation between brain regions and are consistent with the recent characterizations of cell type-specific gene expression, and methylation signatures of sorted neuronal nuclei and among distinct neuronal populations in both mouse and humans5–8.
Neuronal nuclei display brain region-specific DNA methylation profiles
To identify differentially methylated regions (DMRs), we extended our previously published statistical method26 to accommodate multi-group comparisons while accounting for the variation between biological replicates. This new method allows us to simultaneously compare all 45 samples and identify regions of differential methylation between any two groups.
mCG
We found substantial (>10%) mean methylation differences among all four brain regions in the NeuN+ population (Figure 1b) in addition to the previously reported21,22 “cell type” differences between neuronal and non-neuronal nuclei (Supplementary Figure 4a). We identified 97,924 autosomal cell type CG-DMRs of which 21,802 are novel21,22,27, and 19,072 large blocks of differential mCG [family-wise error rates (FWER) ≤ 5%] with neurons being primarily hypermethylated, consistent with their global hypermethylated status21,22 (Supplementary Figure 1e, Supplementary Tables 4–7). These cell type CG-DMRs clearly distinguish NeuN+ from NeuN- nuclei and are present at the promoters of many cell type-specific genes (Supplementary Figure 4a,b).
Given the large methylation differences among NeuN+ nuclei, we repeated our differential methylation analysis on the NeuN+ and NeuN- samples separately and compared the results to those generated from bulk tissues. We identified 13,074 autosomal neuronal (NeuN+) CG-DMRs between brain regions containing >1% of all CpGs analyzed (FWER <5%) (Figure 1b). These neuronal CG-DMRs, as for the cell type CG-DMRs, were enriched in regulatory regions that have been 1) defined by H3K27ac in human brain regions28, 2) identified as permissive enhancers across many cell types and tissues29, and 3) identified using a map of chromatin states in 4 brain regions15 and in regions of open chromatin (Figure 1c). In contrast, we found few autosomal CG-DMRs among NeuN- nuclei (114 CG-DMRs) or bulk tissues (71 CG-DMRs) from these brain regions (Supplementary Tables 4, 8–9). These findings demonstrate that methylation differences between neurons from distinct brain regions are masked by the substantial variation in cellular heterogeneity across samples.
Several patterns emerged upon hierarchical clustering of methylation levels over neuronal CG-DMRs, similar to those seen in cancer/normal comparisons30 (Figure 1d). The largest clusters (group 1 and 2) consisted of symmetric methylation differences enriched for regulatory regions (Figure 1e). The other group consisted primarily of CG-DMRs representing shifts in methylation boundaries and was enriched in promoters, CpG islands (CGI), and shores. These CG-DMRs (group 3) represent the smallest fraction of our neuronal CG-DMRs, but their enrichment is similar to CG-DMRs we previously identified as a consequence of oncogenic transformation of B cells using the Epstein-Barr virus31. These data suggest that methylation is altered in specific ways depending on the genomic feature, with focal increases/decreases in methylation at regulatory regions and more subtle spreading/shrinking of methylation boundaries at promoter-associated features.
Recent single cell analyses have clearly shown that epigenetic and transcriptional profiles reflect distinct neuronal subpopulations with distinct functions5,8. Therefore, it is unsurprising that the neuronal compositions of the brain regions we have analyzed are reflected in their DNA methylation profiles (Supplementary Figure 2b). Unlike NAcc [composed primarily of inhibitory GABAergic medium spiny neurons (MSNs)], HC, BA9, and BA24 are very heterogeneous containing both excitatory and inhibitory subpopulations5,8. The majority of neuronal CG-DMRs (11,895) distinguish the NAcc from the other brain regions. Using the Genomic Regions Enrichment of Annotations Tool (GREAT)32 we found that regions of hypomethylation in NAcc were enriched in categories highly relevant to MSN function: dopamine receptor signaling pathway (q-value = 2.59 × 10−24), adenylate cyclase-activating dopamine receptor signaling pathway (q-value = 3.55 × 10−18), and synaptic transmission – dopaminergic (q-value = 5.21 × 10−5) (Supplementary Table 10). Examples of hypomethylated genes include those encoding markers of GABAergic neurons (BCL11B and DARPP-32) (Figure 1F) and dopamine receptors (DRD1 and DRD3) (Supplementary Figure 4c). In contrast, regions of hypermethylation were enriched in multiple brain development categories (q-values < 1 × 10−11) (Supplementary Table 10).
Given the overwhelming differences between the NAcc and the other brain regions, we hypothesized that neuronal CG-DMRs among BA9, BA24, and HC could be obscured. Therefore, we repeated our analysis using only the neuronal samples from these regions resulting in identification of 208 autosomal neuronal CG-DMRs including 25 CG-DMRs unique to this analysis (Supplementary Figure 4d), Supplementary Tables 4, 11). Again, we find clusters of CG-DMRs representing boundary shifts and focal changes such as that shown in the promoter of SATB2 (Supplementary Figure 4c).
Previous studies have identified large blocks of differential methylation during brain development and cancer/normal comparisons16,30. We also identify 1,808 blocks of differential mCG among neurons from our 4 brain regions; the majority (964) of which have ≥ 10% mean methylation difference (Supplementary Table 4; Supplementary Table 12). Interestingly, 23% of these blocks cover the entirety of a protein coding gene including BCL11B (Figure 1f) and GABRB2 (Supplementary Figure 4f; Supplementary Table 12).
mCH
In addition to mCG, neurons and embryonic stem cells also have extensive non-CpG methylation (mCH). While mCH and mCG are spatially correlated33,34, there is evidence to suggest they can have independent functions in the brain19,22. In agreement with previous reports, we detected the highest methylation levels in the CA context (~10%) of our NeuN+ samples (Supplementary Figure 1f). mCH clearly distinguished NAcc from the other tissues by PCA, similar to mCG (Figure 2a). In addition, we observed segregation of CA and CT contexts but high concordance of signal between strands and methylation levels in the two contexts are linearly related (Supplementary Figure 5). This suggests a single underlying signal in mCH which is reflected in both contexts and strands.
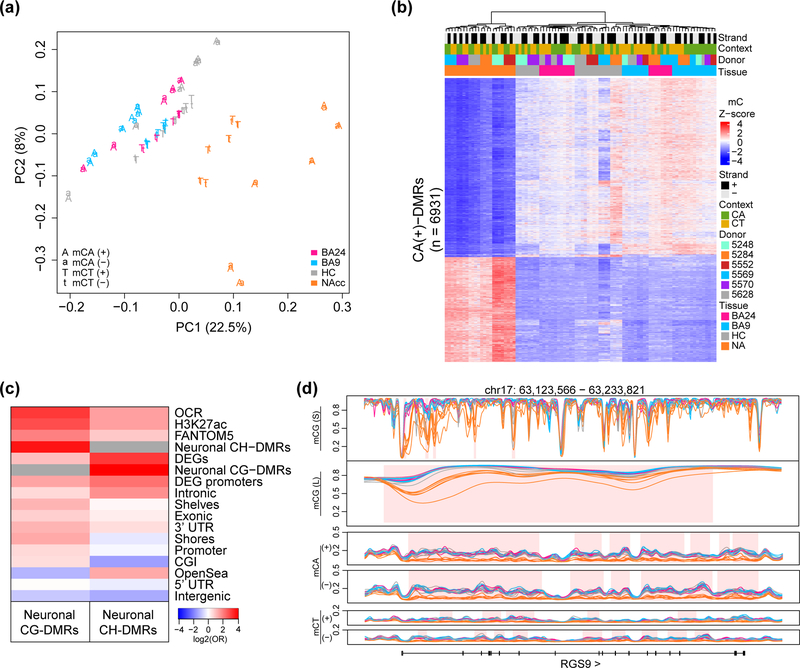
Figure 2. Differential non-CpG methylation among neuronal nuclei from different brain regions.
(a) Principal component analysis of autosomal non-CpG methylation in NeuN+ nuclei; each point is a sample, colored by brain region and character denoting strand and context (n=6 individuals for BA9, HC, NAcc and n=5 for BA24). (b) Hierarchical clustering of samples based on the z-score of methylation over CA(+) DMRs. (c) Log odds ratios for the enrichment of cytosines within CG-DMRs and CH-DMRs compared to the rest of the genome in different genomic features. (d) Example CH DMR over RGS9 with all strands and contexts depicted in addition to CG-DMRs (mCG(S); obtained from small smoothing window) and blocks (mCG(L); obtained from large smoothing window). Average methylation values calculated from NeuN+ nuclei isolated from BA24, BA9, HC and NAcc. Regions of differential methylation are shaded in pink.
We further extended our previously published statistical method used to identify CG-DMRs26 to map differential mCH. Using this method, which for the first-time accounts for biological variability between samples, we identified a large number of CH-DMRs (covering a total of 40 Mb) (Supplementary Table 13). We visualized the similarities between mCH in different contexts and strands by generating a mC z-score for each CH-DMR (Figure 2b shows CA-DMRs on the forward strand, Supplementary Figure 6 for other strands and contexts). This analysis indicates that strand and context are paired within an individual for NAcc samples, with some separation in the other brain regions, presumably due to the larger overall similarity of methylation. Finally, we observe 93% of CT-DMRs overlap CA-DMRs. Given these findings, we merged contexts and strands and termed these CH-DMRs for the analyses below.
CH-DMRs are highly enriched in neuronal CG-DMRs (with 15% overlapping a neuronal CG-DMR or block) and the two marks are highly concordant in these regions (Supplementary Figure 6d). However, CH-DMRs are more enriched over differentially expressed genes (DEGs) (identified between NeuN+ samples from NAcc and BA9, and discussed in a later section) and their promoters compared to neuronal CG-DMRs (Figure 2c). RGS9, which encodes a member of the regulator of G-protein signaling family, is shown as an example gene that is highly upregulated in NAcc neurons (60-fold) and has CH-DMRs covering the majority of the gene body along with multiple CG-DMRs (Figure 2d).
Differential chromatin accessibility and methylation intersect at brain region-specific neuronal regulatory regions
We measured gene expression (via RNA-seq) and chromatin accessibility (via ATAC-seq35) in neuronal and non-neuronal populations between BA9 and NAcc from 6 additional donors (Supplementary Table 1). We identified 2,952 differentially expressed genes (DEGs) between these neuronal populations and only one between non-neuronal nuclei (FDR < 5%) (Figure 3a, b; Supplementary Tables 14–16). Genes expressed at a higher level in NAcc (n = 1,473) were enriched in gene ontology categories of particular relevance to MSN function: regulation of catecholamine secretion (q-value = 8.6 × 10−4), striatum development (q-value = 1.8 × 10−3), and opioid signaling pathway (q-value = 5.4 × 10−5) (Supplementary Table 17). In contrast, genes expressed at a higher level in BA9 (n = 1,479) were enriched in categories related to more general neuronal functions: neuron development (q-value = 9.53 × 10−30), small GTPase mediated signal transduction (q-value = 1.57 × 10−11), and axon guidance pathway (q-value = 4.12 × 10−13).
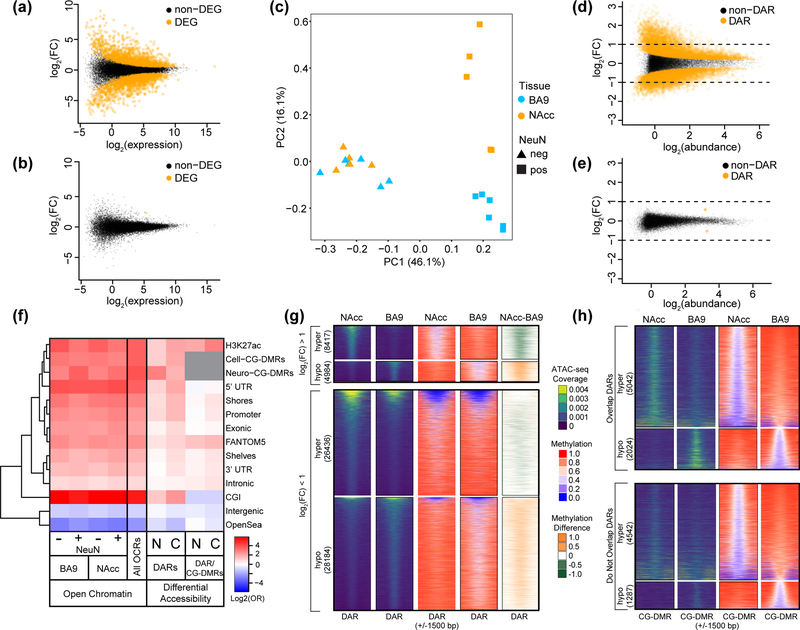
Figure 3. Chromatin accessibility landscape differs between neuronal nuclei isolated from the nucleus accumbens and prefrontal cortex.
Mean-difference plots of gene expression data comparing NAcc to BA9 in (a) NeuN+ nuclei and (b) NeuN-. Differentially expressed genes (DEGs) are shown in orange. (c) Principal component analysis of open chromatin regions [log2(counts per million)] in NeuN+ and NeuN- nuclei from NAcc or BA9. For (a-c), NeuN+ nuclei were from: NAcc, n = 5 individuals and BA9, n = 6 individuals; for NeuN-: NAcc, n = 4 individuals and BA9, n = 5 individuals. (d,e) Mean-difference plots of peak accessibility data comparing: NAcc to BA9 in either (d) NeuN+ nuclei, or (e) NeuN- nuclei. Differentially accessible regions (DARs) are shown in orange. Data shown in (d,e) were randomly sampled (20% of DARs, 10% of non-DARs). (f) Log odds ratios for the enrichment of open chromatin regions (OCRs), DARs, or the intersection of DARs and differentially methylated regions (CG-DMRs) (cell type or neuronal as indicated) in different genomic features. OCRs and DAR/CG-DMR intersection enrichments were calculated in comparison to the rest of the genome while DARs were compared to non-DARs. Gene models from GENCODE (promoter, intronic, exonic, 3’UTR, 5’UTR, intergenic), CpG islands and related features from UCSC (CGI, shores, shelves, OpenSea), and putative enhancer regions (brain-specific H3K27ac30, FANTOM531). (g) ATAC-peak coverage for NeuN+ nuclei from NAcc or BA9 over a 3 kb window centered on neuronal DARs (n=68,201). Neuronal DARs were annotated based on log fold change (greater or less than one) and direction (“hyper” - more accessible in NAcc; “hypo” – more accessible in BA9). The number of neuronal DARs in each category is indicated. Average CpG methylation values and the methylation difference (NAcc minus BA9) for these regions are also shown. (h) As in (g), but centered on neuronal CG-DMRs (n=12,895) identified between NeuN+ nuclei from NAcc and BA9. Neuronal CG-DMRs are separated by those that overlap neuronal DARs and those that do not as indicated. The number of neuronal CG-DMRs in each category is indicated.
Chromatin accessibility distinguished cell types and brain regions via PCA (Figure 3c); although, in contrast to DNA methylation, clustering of samples based on correlation distance across all open chromatin regions (OCRs) did not distinguish brain regions within the neuronal cell type (compare Supplementary Figure 2e to Supplementary Figure 7a). Similar to previous reports36, 163,026 of the 283,812 open chromatin regions tested were differentially accessible (FDR < 5%) between NeuN+ and NeuN- nuclei, termed cell type differentially accessible regions (DARs) (Supplementary Figure 7b; Supplementary Table 18). We further identified 68,021 “neuronal” DARs (only 13 identified among non-neuronal nuclei) between NAcc and BA9 (FDR < 5%) (Figure 3d, e; Supplementary Tables 19–21). OCRs were enriched over multiple features including gene centric features such as promoters and exons and depleted in intergenic and “open sea” regions (Figure 3f). In contrast, neuronal DARs lacked strong enrichment for any specific genomic feature, with the exception of neuronal CG-DMRs.
Given this enrichment of neuronal CG-DMRs in DARs, we further investigated the overlap between these features using the 12,895 CG-DMRs identified between NAcc and BA9 neurons. Only 10% of DARs overlapped CG-DMRs, and conversely 55% of CG-DMRs overlapped DARs. This was true even when restricted to the most divergent DARs (absolute fold change > 2, 12% overlap CG-DMRs). The direction of change in DARs and CG-DMRs is highly concordant with 99.9% of neuronal CG-DMRs having higher methylation when the region is less accessible, consistent with earlier reports in mice3. Further, we observed consistent methylation differences near all DARs (Figure 3g) and some degree of change in accessibility surrounding CG-DMRs (Figure 3h). Together, these data suggest that these distinct epigenetic modalities are weakly dependent, and that which of these modalities exhibit the strongest change can vary, likely due to genome location.
Differential mCH is more strongly correlated with differential gene expression than either mCG or chromatin accessibility
We next investigated the relationship between differential expression of protein coding genes, differential methylation, and differential accessibility. An inverse relationship between global genic cytosine methylation and gene expression in neurons is well-established (Supplementary Figure 8a) and this relationship is maintained when correlating differential methylation and differential expression between NAcc and BA9 (Supplementary Figure 8b). Likewise, differential accessibility and expression are positively correlated (Supplementary Figure 8c). CG-DMRs that overlap DARs revealed a stronger correlation with gene expression than CG-DMRs alone, which was not the case with CA-DMRs (Supplementary Figure 8d,e).
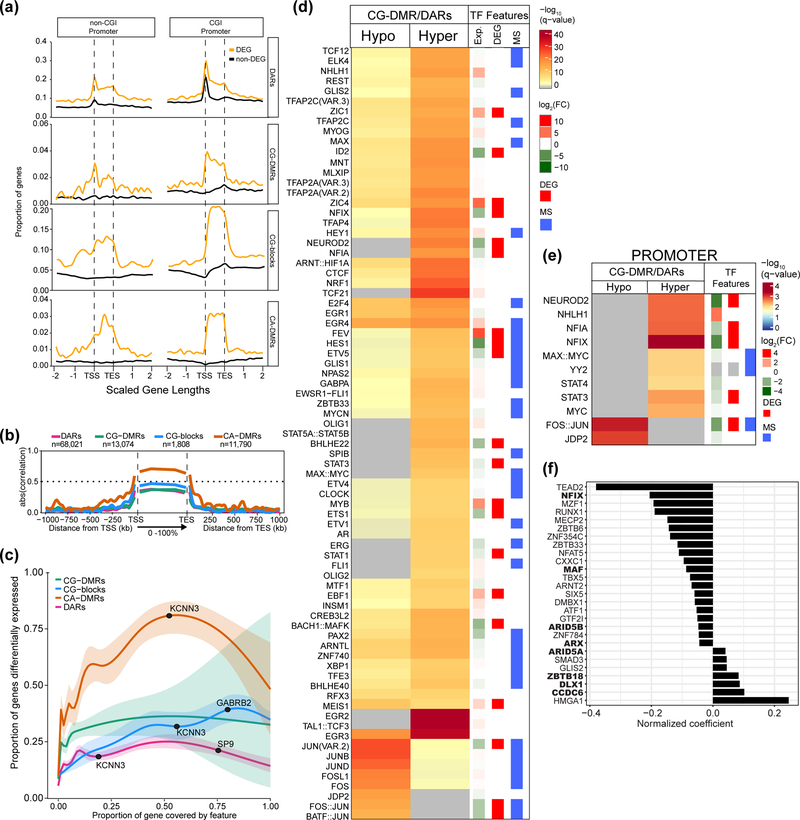
Both DARs and DMRs preferentially occur within DEGs, with CG-DMRs and DARs near the TSS and CA-DMRs and CG-blocks are distributed across the gene body (Figure 4a). Additionally, CA-DMRs have the strongest correlation with expression over gene bodies followed by CG-blocks (Figure 4b). Interestingly, as a larger portion of the gene body is covered by a CA-DMR, that gene is more likely to be differentially expressed (Figure 4c). This is not the case for DARs, CG-DMRs, or CG-blocks. We also find that protein coding genes with CA-DMRs tend to have a lower average expression across all neuronal samples (0.58 vs 1.83 RPKM, respectively; p-value < 2.2 × 10−16; Mann-Whitney) even when differentially expressed (0.78 vs 2.03 RPKM, respectively; p-value < 2.2 × 10−16; Mann-Whitney). This finding is consistent with a previous report19, and our own data, showing that lowly expressed genes have higher levels of mCA than highly expressed genes (Supplementary Figure 8f). Together, these data support and extend previous assertions that among accessibility, mCG and mCH, mCH is the best predictor of gene expression3 and further, that differential mCH specifically occurs over the gene bodies of DEGs consistent with the recent finding that mCA serves to fine-tune gene expression of lowly expressed genes19.
Figure 4. Differential gene expression is strongly associated with neuronal CA-DMRs.
Regions where neuronal CG-DMRs and DARs intersect are enriched in binding sites for transcription factors (TFs) associated with synaptic activity. (a) Proportion of protein coding genes (n=19,823) with a differential epigenetic feature in each bin around gene bodies. Each gene length is split into 100 bins and data extend two gene lengths up and downstream. Differentially expressed genes (DEGs) are indicated. (b) Absolute Pearson correlation values of differential epigenetic features with protein coding gene expression across scaled gene bodies with each gene length split into 100 bins (0–100%); data extend 1 Mb up and downstream in 1 kb bins. The sample for the Pearson correlation in each bin is the subset of features that overlap that bin and, therefore, the sample size for which Pearson’s correlations were determined varies for each bin and feature. (c) Estimated probability (with 95% CI) that a gene is differentially expressed in terms of the proportion of the gene covered by differential epigenetic features (Methods). Example DEGs are annotated. (d) Enrichment of TFs whose motifs were enriched in DARs that overlap hypo- (n=11,734) or hypermethylated CG-DMRs (n=14,463) in NAcc compared to BA9. Differentially expressed TFs in NAcc vs. BA9 are shown by red bars (see Methods for description of RNA-seq analysis). TFs whose binding is influenced by DNA methylation (as determined in47) are indicated with a blue bar. (e) As in (d), showing TFs with motifs enriched where DAR/CG-DMRs overlap promoters (hyper, n=2,618; hypo, n=1,435). Significance determined using one-sided Fisher’s test in (d,e). (f) Bar plot showing the predicted impact of TFs on gene expression (normalized coefficient) for each TF with a binding site within a DAR located within 5 kb of a gene. Large values denote a higher impact of the TF on differential gene expression. Bold type indicates that the TF is differentially expressed between NAcc and BA9.
Differential epigenomics identify transcription factors driving tissue-specific gene expression
We hypothesized that neuronal DAR/CG-DMRs could identify transcription factor (TF) binding motifs actively involved in regulating brain region-specific neuronal function. Using Haystack37 we find that these regions were enriched in immediate-early genes (IEGs), a class of TFs influenced by synaptic activity with important roles in regulating neuronal function38 (Figure 4d; Supplementary Table 22). Several of these IEGs have been implicated in schizophrenia39 and bipolar disorder40 or play known roles in addiction41. DNA methylation influences binding of a diverse set of TFs42, many of which were enriched in regions of differential methylation and accessibility (Figure 4d, blue bar). Additionally, many TFs whose motifs were enriched were also differentially expressed between NAcc and BA9 neurons (Figure 4d, red bar). When we restrict our analysis to those neuronal DAR/CG-DMRs that overlap promoters, we again detect enrichment for IEGs, specifically those encoding AP-1 family members (JUN, FOS, JDP2) (Figure 4e).
Using our neuronal DARs, we employed a method that uses the accessibility of known TF binding sites in gene promoters and enhancers to calculate the importance of each TF for the observed gene expression profile43,44. A positive regression coefficient indicates that the presence of a binding site for a particular TF predicts the target gene to be upregulated in NAcc vs BA9, and vice versa (Figure 4f; Supplementary Table 23). MECP2, in particular, has a negative correlation coefficient (Figure 4f) and is recruited to gene bodies by mCA to restrain transcription of lowly expressed genes19. Several TFs identified are known regulators of neuronal function (NFIX, MECP2, ARX, DLX1, ZBTB33/KAISO) while others have not been previously implicated in adult neuronal regulation (TEAD2, ARID5A/B, ZNF354C/hKID3) (Figure 4f). Eight of these TFs were also differentially expressed between NAcc and BA9 (Figure 4f, bold). Taken together, our data show that differential methylation and accessibility mark regions of the genome that are regulated by synaptic activity-responsive TFs and suggest that these TFs regulate different targets in neurons from distinct brain regions.
Differential epigenomics identify regions of genetic importance for psychiatric disorders and behavioral-cognitive traits
The regions we identified using differential epigenomics, particularly the CG-DMRs and DARs, have characteristics typical of regulatory elements, as expected21,22,36. Multiple studies have provided evidence that disease-associated genetic variation is enriched in regulatory elements active in physiologically relevant cell types45,46. We therefore asked whether our differential epigenetic features were linked with neurological, psychiatric, and behavioral-cognitive phenotypes (brain-linked traits).
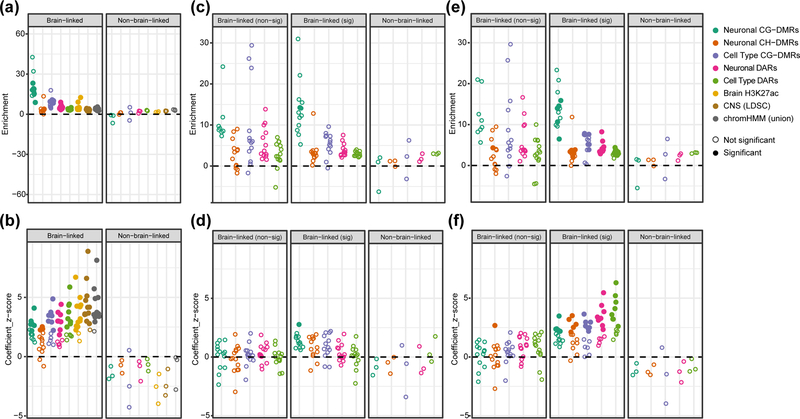
We used stratified linkage disequilibrium score regression (SLDSR45) to identify genomic features strongly associated with brain-linked traits. SLDSR partitions the heritability of a trait across a set of overlapping genomic features using summary statistics from a genome-wide association study (GWAS). We considered eight brain-derived genomic features: five ‘differential’ features identified in our above analyses and three previously published ‘non-differential’ features (Methods). For each feature-trait combination, SLDSR reports a ‘z-score’, indicating whether the feature explains heritability beyond that explained by other features in the model, and an ‘enrichment score’ relating the heritability explained by the feature relative to that expected for a feature containing an equal number of SNPs. At least one of the eight brain-derived features explained a significant proportion of heritability for 13 of 27 brain-linked traits while none explained a significant proportion of heritability for any negative control trait (Figure 5b; Supplementary Figure 9; Supplementary Tables 24, 25). The neuronal CG-DMRs have much higher enrichment scores than any of the non-differential features (Figure 5a), explaining additional heritability in 6/13 traits including schizophrenia and neuroticism, although their size (20–40× smaller) results in more uncertain estimates (Supplementary Figure 9b).
Figure 5. Neuronal CG-DMRs and DARs are highly enriched for explained heritability of multiple psychiatric, neurological, and behavioral-cognitive traits.
Results from running stratified linkage disequilibrium score regression (SLDSR) using 30 GWAS traits with 5 brain-specific differential features, 3 brain-specific non-differential features (Brain H3K27ac44, CNS51, chromHMM26), and 53 baseline features. Traits are stratified by whether they are viewed as brain-related and by whether any of the brain-specific features explained additional heritability of the trait above the 53 baseline features (see Methods and Supplementary Table 24). Feature-trait combinations with a z-score significantly larger than 0 (one-sided z-test with alpha = 0.05, P-values corrected within each trait using Holm’s method) are shown by filled-in circles. (a) Enrichment score and (b) coefficient z-score from running SLDSR for each of the 8 brain specific features separately combined with the 53 baseline features. (c) Enrichment score and (d) Coefficient z-score from running SLDSR for each of the 5 brain-specific differential features separately combined with baseline features. Baseline features for this analysis included the 3 brain-specific non-differential features. (e) Enrichment score and (f) coefficient z-score from running SLDSR for each of the 5 brain specific differential features separately combined with the baseline features. For this analysis the baseline features included only the unique portions of the 3 brain-specific non-differential features (non-differential feature regions that are also present in the differential features were excluded from the baseline).
We then performed a more stringent analysis of the five differential features by testing them against a baseline model that included the three non-differential brain-specific features. We found that when using this stringent approach, only CG-DMRs still contributed significantly (with a 14-fold enrichment) to the explained heritability of a brain-linked trait (schizophrenia; z-score adjusted p-value = 0.013; Figure 5c,d; Supplementary Figure 10a,b).
Finally, we removed regions found by our differential approach from the non-differential features and repeated the analysis. This ensured that the heritability associated with a region common to two sets of brain-specific features is exclusively assigned to the differential feature. This approach highlights the trade-offs of using the highly specific differential features compared to the more general maps of brain regulatory regions, as evidenced by the much larger size of the non-differential features. Based on this analysis, the neuronal CG-DMRs are significantly associated with the heritability of schizophrenia, ADHD, BMI, IQ, and neuroticism (Figure 5f; Supplementary Figure 11a), with a 10–16-fold enrichment (Figure 5e; Supplementary Figure 11b).
Together, these data support the hypothesis that the genetic signal associated with neuropsychiatric traits is mediated through epigenetically distinct regions among neurons from diverse brain regions, particularly CG-DMRs. These analyses further demonstrate the power of using differential methods to precisely identify key regulatory regions, while also highlighting that, from a genetic perspective, CH-DMRs may be less interesting than CG-DMRs.
DISCUSSION
This work represents the most comprehensive dataset to date of whole genome bisulfite sequencing across neuronal and non-neuronal populations and bulk tissues from multiple human brain regions. We have identified 12 Mb of differential mCG and 40 Mb of differential mCH among neuronal nuclei isolated from four brain regions implicated in neuropsychiatric disorders. We have further correlated these methylation changes to changes in both chromatin accessibility (118 Mb of differential accessibility) and gene expression (2,952 DEGs) in two of the tissues examined (NAcc and BA9). These data are made available as the ‘BrainEpigenomeHub’ UCSC track hub, available from http://genome.ucsc.edu/cgi-bin/hgHubConnect (direct link: http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=chmalee&hgS_otherUserSessionName=hg19_brainEpigenome) that can be used as a resource of normal epigenetic states and variation in the human brain to advance neuroepigenetics.
Our findings have four main implications important for the field of neuroepigenetics. First, differential methylation among brain regions appears to be driven by the neuronal cell population. While the non-neuronal fraction can be divided into distinct subpopulations, we show through our analysis of sample-to-sample variability that this heterogeneity cannot account for the remarkable consistency of non-neuronal methylation across the human brain. Importantly, we have demonstrated that the ratio of neuronal to non-neuronal nuclei is highly variable both among brain regions and between samples taken from a single brain region, even within a single individual. While the need to account for cellular heterogeneity is widely appreciated, these results should be of immense value in improving and benchmarking current computational deconvolution strategies applied to studies of human brain tissues.
Second, this study illustrates the power and specificity of differential epigenetics across brain regions, particularly the ability of differential mCG to identify regulatory elements and the ability of differential mCH to identify differential transcription. While differential chromatin accessibility alone showed little enrichment in any particular genome feature, the specificity for regulatory elements (particularly for regions marked by brain-specific H3K27ac) increased nearly five-fold when combined with differential mCG.
Third, we find that regions of differential mCG and chromatin accessibility in neurons are enriched in binding sites for transcription factors regulated by synaptic activity (e.g. FOS, JUN, MEF2C) (reviewed in47), several possessing methylation sensitivity42. These data are consistent with a link between synaptic activity and epigenetic modification of transcription factor binding sites. In addition, some transcription factors we identified play known roles in neurodevelopment, but have not previously been implicated in adult neuronal function (e.g. HMGA1). These findings further illustrate the importance of differential epigenetic analysis in understanding normal neuronal function and provide novel targets for further investigation.
Finally, we show that tissue-specific differential mCG in the neuronal population has a five-fold greater enrichment for the heritability of neuropsychiatric diseases than the more generally defined regulatory genomic fraction identified in the human brain by ChromHMM, and is contained within only 12 Mb of the human genome. While the majority of neuronal CG-DMRs (9,033/13,074; 69%) are found within genes, they primarily occur in intronic regions (7,669; 58.6%) that frequently harbor regulatory elements. Importantly, many of these differentially methylated genes have been repeatedly implicated in schizophrenia and other neuropsychiatric disorders. For example, we identified differential methylation in 19 of 36 schizophrenia GWAS-derived genes analyzed in a previous expression study48 (CACNA1C, CACNB2, CACNA1I, GPM6A, GRAMD1B, SATB2, MEF2C, GRIN2A, MAD1L1, BCL11B, TCF4, TLE1, TLE3, PODXL, ZNF536, KCNV1, MMP16, MAN2A1, and GALNT10). Further, CG-DMRs are present in 56 (Supplementary Table 26) of the 237 genes recently shown to have differentially expressed features in prefrontal cortex schizophrenia vs control samples49. Clearly, these 12 Mb of regional differential methylation constitute critical sequences for future epigenetic analyses of neuropsychiatric diseases. It will be interesting to determine whether a subset of these regions are particularly associated with individual disorders and whether these associations are brain region-specific. These observations are consistent with the idea50 that tissue-specific epigenetic patterning is frequently disrupted in human disease.
METHODS
Experimental Methods
Human Postmortem Brain Samples
Fluorescence-activated nuclear sorting (FANS) was performed on flash-frozen postmortem dorsolateral prefrontal cortex (BA9), hippocampus (HC), nucleus accumbens (NAcc), and anterior cingulate gyrus (BA24) from six individuals not affected with neurological or psychiatric disease. These samples underwent nuclei extraction and sorting as described below for subsequent DNA methylation analysis. Additionally, neuronal nuclei were isolated from the nucleus accumbens (NAcc) and dorsolateral prefrontal cortex (BA9) of 6 different individuals for RNA-seq and ATAC-seq analysis. To underscore the importance of cell sorting, we also prepared DNA from unsorted material from the four brain regions above (BA9, n=9; HC, n=7; NAcc, n=7; BA24, n=5). The majority of individuals were matched between sorted and unsorted, but not all. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications20,24. All samples were obtained from the University of Maryland Brain and Tissue Bank which is a Brain and Tissue Repository of the NIH NeuroBioBank (Supplementary Table 1). We have complied with all relevant ethical regulations. The research described here was classified as not human subjects research (NHSR) by the Johns Hopkins Institutional Review Board (IRB00061004). Data collection and analyses were not performed blind to tissue of origin and randomization was performed at the library preparation stage.
Nuclei Extraction, FANS, and DNA Isolation
Total nuclei were extracted via sucrose gradient centrifugation as previously described29 with the following changes. For WGBS analysis, a total of 2 × 250 mg of frozen tissue per sample was homogenized in 5 mL of lysis buffer (0.32 M sucrose, 10 mM Tris pH 8.0, 5 mM CaCl2, 3 mM Mg acetate, 1 mM DTT, 0.1 mM EDTA, 0.1% Triton X-100) by douncing 50 times in a 40 mL dounce homogenizer. Lysates were combined and transferred to a 38 mL ultracentrifugation tube and 18 mL of sucrose solution (1.8 M sucrose, 10 mM Tris pH 8.0, 3 mM Mg acetate, 1 mM DTT) was dispensed to the bottom of the tube. The samples were then centrifuged at 28,600 rpm for 2 h at 4°C (Beckman Optima XE-90; SW32 Ti rotor). After centrifugation, the supernatant was removed by aspiration and the nuclear pellet was resuspended in 500 uL staining mix (2% normal goat serum, 0.1% BSA, 1:500 anti-NeuN conjugated to AlexaFluor488 (Millipore, cat#: MAB377X) in PBS) and incubated on ice. Unstained nuclei and nuclei stained with only secondary antibody served as negative controls. The fluorescent nuclei were run through a Beckman Coulter MoFlo Cell Sorter with proper gate settings using Summit v4.3.02 software (Supplementary Figure 1). A small portion of the NeuN+ and NeuN− nuclei were re-run on the sorter to validate the purity which was greater than 95%. Immuno-negative (NeuN−) and -positive (NeuN+) nuclei were collected in parallel. For DNA extraction, sorted nuclei were pelleted by adding 2 mL of sucrose solution, 50 uL of 1 M CaCl2, and 30 uL of Mg acetate to 10 mL of nuclei in PBS. This solution was incubated on ice for 15 min, then centrifuged at 3,000 rpm for 20 min. The nuclear pellets were flash frozen in liquid nitrogen and stored at −80°C. DNA was extracted from the frozen nuclear pellets using the MasterPure DNA Extraction kit (Epicentre, Madison, Wisconsin, USA) following the manufacturer’s instructions. For ATAC-seq and RNA-seq, nuclei were processed as described below.
Whole genome bisulfite sequencing (WGBS)
WGBS single indexed libraries were generated using NEBNext Ultra DNA library Prep kit for Illumina (New England BioLabs, Ipswich, MA, USA) according to the manufacturer’s instructions with modifications. 400 ng gDNA was quantified by Qubit dsDNA BR assay (Invitrogen, Carlsbad, CA, USA) and 1% unmethylated lambda DNA (cat#: D1521, Promega, Madison, WI, USA) was spiked in to measure bisulfite conversion efficiency. Samples were fragmented to an average insert size of 350 bp using a Covaris S2 sonicator. Size selection was performed using AMPure XP beads and insert sizes of 300–400 bp were isolated (0.4x and 0.2x ratios). Samples were bisulfite converted after size selection using EZ DNA Methylation-Gold Kit (cat#: D5005, Zymo, Irvine, CA, USA) following the manufacturer’s instructions. Amplification was performed after the bisulfite conversion using Kapa Hifi Uracil+ (cat#: KK282, Kapa Biosystems, Boston, USA) polymerase using the following cycling conditions: 98°C 45s / 8cycles: 98°C 15s, 65°C 30s, 72°C 30s / 72°C 1 min. Final libraries were run on 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) High-Sensitivity DNA assay; samples were also run on Bioanalyzer after shearing and size selection for quality control purposes. Libraries were quantified by qPCR using the Library Quantification Kit for Illumina sequencing platforms (cat#: KK4824, KAPA Biosystems, Boston, USA), using 7900HT Real Time PCR System (Applied Biosystems). Libraries were sequenced with the Illumina HiSeq2500 using 125 bp paired-end single indexed run and 10% PhiX spike-in.
Assay for transposase-accessible chromatin using sequencing (ATAC-seq)
NeuN+ and NeuN− nuclei were isolated as previously described and 100,000 nuclei were used for ATAC-seq library preparation as per standard protocols38. Briefly, 2X lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) was added to sorted nuclei to reach a final concentration of 1X and incubated 20 min on ice followed by centrifugation for 10 min as previously described. The transposition reaction was incubated for 1 h at 37°C (Nextera DNA library prep kit; cat #:FC-121–1031, Illumina). After PCR amplification of libraries and column clean up via the Qiagen MinElute PCR purification kit (cat#:28004, Qiagen, Valencia, CA, USA), an additional clean up with AMPure XP beads (0.8x ratio) was performed twice with 80% ethanol washes before quantification using a DNA High Sensitivity chip on a 2100 BioAnalyzer (Agilent, Santa Clara, CA, USA). Libraries were sequenced with the Illumina HiSeq4000 using 70 bp paired-end single indexed run with a 5% PhiX spike-in.
RNA sequencing (RNA-seq)
RNA isolated from bulk tissue was assessed and only tissues with a RIN ≥ 4 were used for nuclei isolation. NeuN+ and NeuN− nuclei were isolated as previously described with the addition of 20 U/mL RNAse Inhibitors (cat#: N8080119, Applied Biosystems) to the lysis buffer, sucrose solution, and antibody solution while protease inhibitor cocktail (cat#: 50-751-7359, Amresco) was added to the lysis buffer only. Approximately 200,000 nuclei were sorted directly into RLT buffer + 150 mM 2-mercaptoethanol and RNA was isolated using the Qiagen RNeasy Kit (cat #:74106, Qiagen, Valencia, CA, USA). Nuclear RNA quality was assessed by running samples on a Total RNA Pico Chip on a 2100 BioAnalyzer (Agilent, Santa Clara, CA, USA). RNA-seq libraries were created using 2.5 ng input RNA with the SMARTer® Stranded Total RNA-Seq Kit - Pico Input Mammalian (cat#: 635005, Takara Bio, Mountain View, CA, USA) following the manufacturer’s instructions for degraded RNA samples. Libraries were sequenced with the Illumina HiSeq4000 using 70 bp paired-end single indexed run with 5% PhiX spike-in.
Computational Methods
Annotation
The hg19 build of the human reference genome was used for all analyses. Only analyses of autosomal data are reported. Genes, exons, introns, and UTRs were taken from GENCODE v19 (http://www.gencodegenes.org/releases/19.html)51. Gene bodies were defined by taking the union over all transcripts (transcription start site to transcription end site) for each gene. Promoters were defined as 4 kb centered on the transcription start site.
CpG islands were downloaded from UCSC (http://genome.ucsc.edu/)52,53. A GRanges object was created defining CpG shores as 2 kb flanking CpG islands and CpG shelves as 2 kb flanking CpG island shores. Open sea regions are parts of the genome which are neither CpG islands, shores or shelves.
The 15-state ChromHMM model for 7 adult brain tissues (E071, E074, E068, E069, E072, E067, E073) from the Roadmap Epigenomics Project15 was downloaded using the R/Bioconductor AnnotationHub package (v2.6.4).
Whole genome bisulfite sequencing (WGBS)
Mapping and quality control of WGBS reads
We trimmed reads of their adapter sequences using Trim Galore! (v0.4.0) (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and quality-trimmed using the following parameters: trim_galore -q 25 --paired ${READ1} ${READ2}. We then aligned these trimmed reads to the hg19 build of the human genome (including autosomes, sex chromosomes, mitochondrial sequence, and lambda phage (accession NC_001416.1) but excluding non-chromosomal sequences) using Bismark54 (v0.14.3) with the following alignment parameters: bismark --bowtie2 -X 1000 −1 ${READ1} −2 ${READ2}. Supplementary Tables 2 and 3 summarize the alignment results. Using the reads aligned to the lambda phage genome, we estimated that all libraries had a bisulfite conversion rate > 99%.
We then used bismark_methylation_extractor to summarize the number of reads supporting a methylated cytosine and the number of reads supported a unmethylated cytosine for every cytosine in the reference genome. Specifically, we first computed and visually inspected the M-bias28 of our libraries. Based on these results, we decided to ignore the first 5 bp of read1 and the first 10 bp of read2 in the subsequent call to bismark_methylation_extractor with parameters: --ignore 5 --ignore_r2 10. The final cytosine report file summarizes the methylation evidence at each cytosine in the reference genome.
Smoothing WGBS
We used BSmooth to estimate CpG methylation levels as previously described28. Specifically, we ran a ‘small’ smooth to identify small DMRs (smoothing over windows of at least 1 kb containing at least 20 CpGs) and a ‘large’ smooth to identify large-scale blocks (smoothing over windows of at least 20 kb containing at least 500 CpGs). Following smoothing, we analyzed all CpGs that had a sequencing coverage of at least 1 in all samples (n = 45 for sorted data, n = 27 for unsorted data).
We also adapted BSmooth to estimate CpA and CpT methylation levels in NeuN+ samples. Unlike CpGs, CpAs and CpTs are not palindromic, so were analyzed separately for each strand, for a total of 4 strand/dinucleotide combinations:
mCA (forward strand)
mCA (reverse strand)
mCT (forward strand)
mCT (reverse strand)
For each dinucleotide/strand combination we ran a single ‘small-ish’ smooth to identify DMRs (smoothing over windows of at least 3 kb containing at least 200 CpAs or CpTs). Following smoothing, we analyzed all CpAs and CpTs regardless of sequencing coverage.
Identification of small DMRs and large-scale blocks
We ran separate analyses to identify 6 types of differentially methylated regions:
CG-DMRs: Using data from the ‘small’ smooth of CpG methylation levels
CG-blocks: Using data from the ‘large’ smooth of CpG methylation levels
CA-DMRs (forward strand): Using data from the ‘small-ish’ smooth of CpA methylation levels on the forward strand
CA-DMRs (reverse strand): Using data from the ‘small-ish’ smooth of CpA methylation levels on the reverse strand
CT-DMRs (forward strand): Using data from the ‘small-ish’ smooth of CpT methylation levels on the forward strand
CT-DMRs (reverse strand): Using data from the ‘small-ish’ smooth of CpT methylation levels on the reverse strand
Previously, we have used BSmooth to perform pairwise (two-group) comparisons33. In the present study, we had up to 8 groups to compare: 4 brain regions (BA9, BA24, HC, NAcc) and, for the sorted data, 2 cell types (NeuN+, NeuN−). Rather than running all 28 pairwise comparisons, we extended the BSmooth method to handle multi-group comparisons, which we refer to as the F-statistic method.
For the F-statistic method, we constructed a design matrix with a term for each group (e.g., BA9_neg for NeuN− cells from BA9, BA9_pos for NeuN+ cells from BA9, etc.). For each CpX (CpG, CpA, or CpT), we then fitted a linear model of the smoothed methylation levels against the design matrix. To improve standard error estimates, we thresholded the residual standard deviations at the 75% percentile and smoothed these using a running mean over windows containing 101 CpXs. We then combined the estimated coefficients from the linear model, their estimated correlations, and the smoothed residual standard deviations to form F-statistics to summarize the evidence that methylation differs between the groups at each of the CpXs.
Next, we identified runs of CpXs where the F-statistic exceeded a cutoff and where each CpX was within a maximum distance of the next. Specifically, we used cutoffs of F = 4.62 for CG-DMRs, F = 22 for CG-blocks (following32), and F = 42 for CA-DMRs and CT-DMRs, and required that the CpXs were within 300 bp of one another for DMRs and 1000 bp of one another for blocks. For blocks, we also required that the average methylation in the block varied by at least 0.1 across the groups. These runs of CpXs formed our candidate DMRs and blocks. Each candidate DMR and block was summarized by the area under the curve formed when treating the F-statistic as a function along the genome (areaStat).
We used permutation testing to assign a measure of statistical significance to each candidate DMR/block. We randomly permuted the design matrix, effectively permuting the sample labels, and repeated the F-statistic analysis with the same cutoffs using the permuted design matrix, resulting in a set of null DMRs/blocks for each permutation. We performed 1000 such permutations. We then asked, for each candidate DMR/block, in how many permutations did we see a null DMR/block anywhere in the genome with the same or better areaStat as the candidate DMR/block; dividing this number by the total number of permutations gives a permutation P-value for each DMR/block. Since we are comparing each candidate block/DMR against anything found anywhere in the genome in the permutation set, we are also correcting for multiple testing by controlling the family-wise error rate. Those candidates DMRs/blocks with a permutation P-value ≤ 0.05 form our set of DMRs/blocks.
Annotation of small CG-DMRs and CG-blocks
The F-statistic approach allows us to jointly use all samples for the identification of differentially methylated regions. However, it does not tell us which group(s) are hypomethylated or hypermethylated for the region. To assign such labels to our F-statistic CG-DMRs and CG-blocks, we used a post-hoc analysis for specific pairwise comparisons of interest: NeuN+ vs. NeuN−; NeuN+ cells in NAcc vs. NeuN+ cells in BA9, BA24, and HC; NeuN+ cells in NAcc vs. NeuN+ cells in BA9. We identified CG-DMRs and CG-blocks using the original t-statistic method of BSmooth; an F-statistic CG-DMR was assigned a label (e.g., hypermethylated in NeuN+ and hypomethylated in NeuN−) if the corresponding t-statistic CG-DMR or CG-block overlapped at least 50% of the F-statistic CG-DMR or CG-block. This procedure does not change the coordinates of the CG-DMR/CG-block and means an F-statistic CG-DMR/CG-block may be assigned multiple labels. No such analysis was performed for CA-DMRs or CT-DMRs due to computational costs.
Subset analyses of CG-DMRs and CG-blocks
We found, as expected, that the differences between NeuN+ and NeuN− samples dominated our results (98,420 / 100,875 F-statistic CG-DMRs and 19,072 / 20,373 F-statistic CG-blocks were assigned the label, NeuN+ vs NeuN−; Supplementary Tables 4, 5 and 6). To better focus on the differences between brain regions within a given cell type (NeuN+ or NeuN−), we repeated the F-statistic analysis using just the NeuN+ or NeuN− samples (Supplementary Tables 8, 9, 12). We again found that one group dominated: 11,895 / 13,074 F-statistic NeuN+ CG-DMRs were specific to NAcc. To better focus on the differences between the remaining brain regions, we repeated the analysis using just the BA9, BA24, and HC NeuN+ samples (Supplementary Table 11).
Novel NeuN+ vs NeuN− CG-DMRs
Three published datasets of NeuN+ vs NeuN− methylation differences23,24,29 were used for comparison with our CG-DMRs. Data from Montano et al. was generated from our own lab and is accessible through GEO series accession number GSE48610. Differentially methylated sites from Kozlenkov et al. were obtained by request directly from the authors. CG-DMRs from Lister et al. were obtained from http://brainome.ucsd.edu/BrainMethylomeData/CG_DMR_lists.tar.gz and converted to hg19 using the UCSC liftOver tool55. As three different platforms were used to measure methylation (CHARM, 450K, and WGBS, respectively), we combined the differentially methylated CpGs from the autosomes of each study and compared to the differentially methylated CpGs within our NeuN+ vs NeuN− CG-DMRs. Sites that were unique to our NeuN+ vs NeuN− CG-DMRs were reported as novel.
Assay for transposase-accessible chromatin using sequencing (ATAC-seq)
Mapping and quality control of ATAC-seq reads
We trimmed reads of their adapter sequences using trimadap (v0.1, https://github.com/lh3/trimadap/archive/0.1.zip) with the following parameters: trimadap-mt −3 CTGTCTCTTATACACATCTCCGAGCCCACGAGA ${READ1}; trimadap-mt −3 CTGTCTCTTATACACATCTGACGCTGCCGACGA ${READ2}. We then aligned these trimmed reads to the hg19 build of the human genome (including autosomes, sex chromosomes, mitochondrial sequence, unplaced sequence, and unlocalized sequence) using Bowtie256 (v2.2.5) with alignment parameters: bowtie2 -X 2000 --local --dovetail. Potential PCR duplicate reads were marked using MarkDuplicates from the Picard library (http://broadinstitute.github.io/picard/; v2.2.1). Supplementary Table 21 summarizes the alignment results for the 22 libraries.
Identifying differentially accessibly ATAC-seq regions (DARs)
Peaks were called in each condition (NAcc_pos, NAcc_neg, BA9_pos, and BA9_neg) using MACS57 (v2.1.0). Specifically, data from each condition were combined into a metasample formed by using all non-duplicate-marked reads with a mapping quality > 30 and then processed using: macs2 callpeaks --nomodel --nolambda --call-summits -t ${BAMS[@]}. We took the ‘narrowPeaks’ produced by MACS and filtered out those regions overlapping the ENCODE mappability consensus blacklist regions (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/) and the blacklist for ATAC-seq created by Buenrostro et al.38 (https://sites.google.com/site/atacseqpublic/atac-seq-analysis-methods/mitochondrialblacklists-1). We took this filtered list as our condition-specific sets of open chromatin regions (OCRs).
To perform the differential analysis, we first took the union of condition-specific OCRs on the autosomes to construct an ‘overall’ set of OCRs. This ‘overall’ set of OCRs contained 853,053 regions (630 Mb). For each sample, we then counted the number of fragments (fragment = start of read1 to end of read2) overlapping each of the ‘overall’ OCRs using the summarizeOverlaps() function in the GenomicAlignments R/Bioconductor package58 (v1.10.0). Specifically, we only counted those fragments where both reads had a mapping-quality score > 30, reads not marked as potential PCR duplicates, and those where any part of the fragment overlapped exactly one peak.
We then analyzed these data using the voom method, originally designed for differential expression analysis of RNA-seq data59. Briefly, the read counts were transformed to counts per million (cpm) and only those 283,812 / 853,053 peaks with at least 1 cpm for at least 5 samples (the size of the smallest group of samples) were retained. These 283,812 peaks were used in all downstream analyses of differential accessibility described in the main text. We normalized these counts using TMM60, then used edgeR61 (v3.16.5) and limma62 (v3.30.7) to transform these counts to log2-cpm, estimate the mean-variance relationship, and compute appropriate observation-level weights ready for linear modelling.
In our design matrix, we blocked on donor (donor1, …, donor6) and included a term for each group (e.g., BA9_neg for NeuN− cells from BA9, BA9_pos for NeuN+ cells from BA9, etc.). We ran surrogate variable analysis63 using the sva (v3.22.0) R/Bioconductor package and identified 4 surrogate variables. We ultimately decided to include all 4 surrogate variables in the linear model. Using the empirical Bayes shrinkage method implemented in limma, we tested for differential accessibility of peaks in three comparisons: (1) NAcc vs. BA9 in NeuN+ cells; (2) NAcc vs. BA9 in NeuN− cells; (3) NeuN+ cells vs NeuN− cells. For an ATAC-seq peak to be called a differentially accessible region (DAR), it had to have a Benjamini-Hochberg adjusted P-value < 0.05.
RNA sequencing (RNA-seq)
Mapping and quality control of RNA-seq reads
We trimmed the first 3 bp of read1, which were derived from template switching oligos and not the cDNA of interest, using seqtk (https://github.com/lh3/seqtk; v1.2-r94) with the following parameters: seqtk trimfq -b 3 ${READ1}. We then quasi-mapped these trimmed reads to a FASTA file of protein-coding and lncRNA genes from GENCODE v19 (http://www.gencodegenes.org/releases/19.html)51 and performed transcript-level quantification using Salmon64 (v0.7.2). Supplementary Table 16 summarizes these results for the 20 libraries.
Identifying differentially expressed genes (DEGs)
We used tximport65 (v1.2.0) to compute normalized gene-level counts from the transcript-level abundance estimates (scaling these using the average transcript length over samples and the library size) for autosomal genes (33,351 genes). Only autosomal genes with at least 1 cpm in at least 4 libraries (the size of the smallest group of samples) were retained for downstream analysis (24,161 / 33,351 genes). We normalized these counts using TMM60 then used edgeR61 (v3.16.5) and limma62 (v3.30.7) to transform these counts to log2-cpm, estimate the mean-variance relationship, and compute appropriate observation-level weights ready for linear modelling.
In our design matrix, we blocked on donor (donor1, …, donor6) and included a term for each group (e.g., BA9_neg for NeuN− cells from BA9, BA9_pos for NeuN+ cells from BA9, etc.). We ran surrogate variable analysis63 using the sva (v3.22.0) R/Bioconductor package and identified 5 surrogate variables, some of which correlated with the date on which these samples were flow-sorted. We ultimately decided to include all 5 surrogate variables in the linear model. Using the empirical Bayes shrinkage method implemented in limma, we tested for differential expression of genes in three comparisons: (1) NAcc vs. BA9 in NeuN+ cells; (2) NAcc vs. BA9 in NeuN− cells; (3) NeuN+ cells vs NeuN− cells. For a gene to be called a differentially expressed gene (DEG), it had to have a Benjamini-Hochberg adjusted P-value < 0.05 with no minimum log2 fold change cutoff.
Enrichment of DMRs, ATAC peaks, and DARs in genomic features
Enrichment odds ratios and P-values
We formed a 2×2 contingency table of (n11, n12, n21, n22); specific values of (n11, n12, n21, n22) are described below. The enrichment log odds ratio was estimated by log2(OR) = log2(n11) + log2(n22) - log2(n12) - log2(n21), its standard error was estimated by se(log2(OR)) = sqrt(1 / n11 + 1 / n12 + 1 / n21 + 1 / n22), and an approximate 95% confidence interval formed by [log2(OR) – 2 × se(log2(OR)), log2(OR) + 2 × se(log2(OR))]. We also report the P-value obtained from performing Fisher’s exact test for testing the null of independence of rows and columns in the 2×2 table (i.e. the null of no enrichment or depletion) using the fisher.test()function from the ‘stats’ package in R66.
DMRs and blocks
For DMRs and blocks, we computed the enrichment of CpXs (CpGs, CpAs, or CpTs, as appropriate) within DMRs inside each genomic feature (e.g., exons, FANTOM5 enhancers, etc.). Specifically, for each genomic feature, we constructed the 2×2 table (n11, n12, n21, n22), where:
n11 = Number of CpXs in DMRs/blocks that were inside the feature
n12 = Number of CpXs in DMRs/blocks that were outside the feature
n21 = Number of CpXs not in DMRs/blocks that were inside the feature
n22 = Number of CpXs not in DMRs/blocks that were outside the feature
The total number of CpXs, n = n11 + n12 + n21 + n22, was the number of autosomal CpXs in the reference genome. We counted CpXs rather than the number of DMRs or bases because this accounts for the non-uniform distribution of CpXs along the genome and avoids double-counting DMRs that are both inside and outside the feature.
Open chromatin regions (OCRs)
For OCRs, we computed the enrichment of bases within OCRs inside each genomic feature. Specifically, for each genomic feature, we constructed the 2×2 table (n11, n12, n21, n22), where:
n11 = Number of bases in OCRs that were inside the feature
n12 = Number of bases in OCRs that were outside the feature
n21 = Number of bases in the rest of the genome that were inside the feature
n22 = Number of bases in the rest of the genome that were outside the feature
The total number of bases, n = n11 + n12 + n21 + n22, was the number of autosomal bases in the reference genome. We counted bases rather than number of OCRs to account for the variation in OCR width, the large variation in width for the ‘rest of the genome’ features, and to avoid double-counting OCRs that were both inside and outside the feature.
Differentially accessible regions (DARs)
For DARs, we computed the enrichment of bases within DARs inside each genomic feature in two ways. Firstly, for each genomic feature, we constructed the 2 × 2 table (n11, n12, n21, n22), where:
n11 = Number of bases in DARs that were inside the feature
n12 = Number of bases in DARs that were outside the feature
n21 = Number of bases in the rest of the genome that were inside the feature
n22 = Number of bases in the rest of the genome that were outside the feature
Secondly, for each genomic feature, we constructed the 2 × 2 table (n11, n12, n21, n22), where:
n11 = Number of bases in DARs that were inside the feature
n12 = Number of bases in DARs that were outside the feature
n21 = Number of bases in null-regions that were inside the feature
n22 = Number of bases in null-regions that were outside the feature
‘Null-regions’ were those OCRs that were not differentially accessible between the relevant condition (NAcc and BA9 in NeuN+ cells) based on the peak having a Benjamini-Hochberg adjusted P-value > 0.05 in the analysis of differential accessibility. By comparing to null-regions rather than the rest of the genome, we account for the non-uniform distribution of OCRs along the genome.
We counted the number of bases rather than the number of DARs to account for the variation in DAR width and to avoid double-counting DARs that were both inside and outside the feature.
Association of gene body methylation and chromatin accessibility with gene expression
We analyzed the relationship between gene body methylation and chromatin accessibility with gene expression using protein coding genes (n = 19,823). We focused on data from NAcc (NeuN+) and BA9 (NeuN+) samples because for these brain regions we had WGBS, ATAC-seq, and RNA-seq, as well as a substantial number of ‘DiffEpi’ marks (‘DiffEpi’ being a collective abbreviation for DMRs, blocks, and DARs).
To examine the spatial distribution of DiffEpi marks around gene bodies, we took 100 bins across each gene and recorded whether that bin overlapped a DiffEpi mark. We extended this for 2 gene body equivalents upstream of the transcription start site (TSS) and downstream of the transcription end site (TES) (i.e. if a gene was 1 kb long then we extended it 2 kb upstream of TSS and 2 kb downstream of TES) and similarly recorded whether each bin overlapped a DiffEpi mark. Figure 4a plots the proportion of genes with a DiffEpi mark in each bin as we move from upstream of the TSS, across the gene body, and downstream of the TES, stratified by whether the gene was differentially expressed between NAcc (NeuN+) and BA9 (NeuN+) samples.
We performed various analyses of the relationship between mCA (taking estimates from the same strand as the gene), mCG (aggregated over strand and so ‘unstranded’), and average chromatin accessibility (ATAC-seq reads-per-kilobase mapped (RPKM)) with gene expression (RNA-seq RPKM) for all protein coding genes for NAcc (NeuN+) and BA9 (NeuN+) samples. We examined these features over gene bodies and gene promoters, as well as in bins along and surrounding each gene. Different types of bins were used for different analyses, as noted in figure captions. For some analyzes we used a fixed number of bins for each gene so that bin width varied according to gene width (e.g., 100 bins covering the gene body in Figure 4a and Supplementary Figure 8a). For other analyses, we used a fixed bin size (e.g., 1 kb bins upstream of TSS and downstream of TES in Figure 4b). Furthermore, some analyses used scaled distances upstream of the TSS and downstream of the TES (e.g., Figure 4a and Supplementary Figures 8a) while others used fixed distances (e.g., Figure 4b).
We then focused on correlating DiffEpi marks with changes in gene expression. We again took all protein coding genes and created 100 bins across each gene body. We identified all DiffEpi marks that overlapped each bin and correlated the change in methylation (mCG from small smooth for CG-DMRs; mCG from large smooth for CG-blocks; mCA on the same strand as the gene for CA-DMRs) or change in chromatin accessibility (log2FC using RPKM for DARs) with the change in expression of the gene (log2FC). To emphasize, only genes with a DiffEpi mark in that bin contribute to the correlation estimate for that bin. We performed a similar procedure upstream of the TSS and downstream of the TES, but here using a fixed bin size (1 kb).
Finally, we used a binomial generalized additive model to estimate the probability that a protein coding gene is differentially expressed given that x% of the gene body is covered by a DiffEpi mark. The ‘gam’ function in the ‘mgcv’ package67 (v1.8–23) was used to fit the generalized additive model with the formula y ~ bs(x, bs = “cs”) and additional argument family = “binomial”. This estimated probability and its standard error are shown in Figure 4c, annotated with example genes.
Stratified linkage disequilibrium score regression (SLDSR)
We used stratified linkage disequilibrium score regression (SLDSR), implemented in the LDSC68 software, to evaluate the enrichment of common genetic variants from genome-wide association study (GWAS) signals to partition trait heritability within functional categories represented by our DMRs, OCRs, and DARs51. SLDSR estimates the proportion of genome-wide single nucleotide polymorphism (SNP)-based heritability that can be attributed to SNPs within a given genomic feature by a regression model that combines GWAS summary statistics with estimates of linkage disequilibrium from an ancestry-matched reference panel. Links to GWAS summary statistics are available in Supplementary Table 24. Additional files needed for the SLDSR analysis were downloaded from https://data.broadinstitute.org/alkesgroup/LDSCORE/ following instructions at https://github.com/bulik/ldsc/wiki.
We ran LDSC (v1.0.0; https://github.com/bulik/ldsc) to estimate the proportion of genome-wide SNP-based heritability of 30 traits (Supplementary Table 24) across 53 ‘baseline’ genomic features (24 main annotations, 500 bp windows around of each of the 24 main annotations, and 100 bp windows around 5 sets of ChIP-seq peaks; described in51) and eight brain-specific genomic features:
CG-DMRs (NeuN+): CG-DMRs between brain regions in NeuN+ samples (11.8 Mb) (Supplementary Table 8)
CH-DMRs (NeuN+): Union of CA-DMRs and CT-DMRs between brain regions in NeuN+ samples (39.6 Mb) (Supplementary Table 13)
CG-DMRs (NeuN+ vs. NeuN−): CG-DMRs between NeuN+ and NeuN− samples (70.0 Mb) (Supplementary Table 5)
DARs (NeuN+): DARs between brain regions in NeuN+ samples (118.1 Mb) (Supplementary Table 19)
DARs (NeuN+ vs. NeuN−): DARs NeuN+ and NeuN− samples (275.8 Mb) (Supplementary Table 18)
Brain H3K27ac: A set of regions marked by H3K27ac in human brain30 (215.4 Mb)
CNS (LDSC): A union of regulatory regions active in brain, previously considered by the authors of LDSC51 (338.8 Mb)
ChromHMM (union): A union of regulatory regions found by ChromHMM using Roadmap Epigenomics15 data (498.1 Mb). The selected brain regions and their Roadmap Epigenomics codes were: Brain Angular Gyrus (E067); Brain Anterior Caudate (E068); Brain Cingulate Gyrus (E069); Brain Germinal Matrix (E070); Brain Hippocampus Middle (E071); Brain Inferior Temporal Lobe (E072); Brain Dorsolateral Prefrontal Cortex (E073); Brain Substantia Nigra (E074); Fetal Brain Male (E081); Fetal Brain Female (E082). The ChromHMM states selected as ‘regulatory regions’ were: Bivalent Enhancer; Bivalent/Poised TSS; Genic enhancers; Flanking Active TSS; Active TSS; Strong transcription; Enhancers. Processed ChromHMM tracks were downloaded using the AnnotationHub R/Bioconductor package (v1.36.2).
Collectively, we refer to features 1–5 as ‘differential features’, being the product of differential analyses, and features 6–8 as ‘non-differential features’, each being the union of regions marked by various active histone modifications present in different brain regions.
We performed three rounds of analysis:
‘Baseline’: A standard SLDSR analysis, as suggested by the LDSC authors, whereby each of the eight brain-specific features was added one at a time to a ‘full baseline model’ that included the 53 ‘baseline’ categories that capture a broad set of genomic annotations.
‘Stringent’: Add each of the 5 differential features one at a time to a model that included the 53 baseline features and 3 brain-specific non-differential features.
‘Adjusting for non-differential features (ndf) excluding differential features (df)’: Add each of the 5 differential features on at a time to a model that included the 3 brain-specific non-differential features (after having excluded any regions shared with the brain-specific differential feature) and the 53 baseline features.
For each round of analysis, we used SLDSR to estimate a ‘coefficient z-score’ and an ‘enrichment score’ for each feature-trait combination. A brief description of their interpretation is given below; we refer the interested reader to the Online Methods of Finucane, H.K. et al. 201527 for the complete mathematical derivation.
A coefficient z-score statistically larger than zero indicates that adding the feature to the model increased the explained heritability of the trait, beyond the heritability explained by other features in the model.
The enrichment score is defined as (proportion of heritability explained by the feature) / (proportion of SNPs in the feature). The enrichment score is unadjusted for the other features in the model, but is more readily interpretable as an effect size. It should be noted, however, that the enrichment score depends on the terms included in the model. Although the denominator of the enrichment score is constant, the numerator depends on the set of features in the model and how much the feature of interest overlaps the other features in the set. In particular, the “proportion of heritability explained by the feature” will decrease when another feature that overlaps it is added to the model (e.g., the enrichment scores of the differential features in the ‘stringent’ analysis are lower than in the ‘baseline’ analysis due to the differential features overlapping the non-differential features that are added to the baseline in the ‘stringent’ analysis).
Particularly interesting are those feature-trait combinations with statistically significant z-score coefficients and large enrichment scores. P-values within each trait were post-hoc adjusted for multiple testing using Holm’s method69 (Supplementary Table 25).
Transcription Factor Motif Enrichment
We used the Haystack (v0.4)40 haystack_motifs module to scan for vertebrate JASPAR (2016)70 transcription factor binding motifs enriched in our datasets. A list of neuronal DARs that overlapped hypo- or hypermethylated neuronal CG-DMRs identified between NAcc and BA9 in NeuN+ nuclei were input into Haystack as a BED file (hyper CG-DMR/DARs, n = 14,463; hypo CG-DMR/DARs, n = 11,734). A subset of these neuronal DAR/CG-DMRs found in promoters was analyzed separately (hyper CG-DMR/DARs, n = 2,618; hypo CG-DMR/DARs, n = 1,435). All autosomal promoters were input as background for the neuronal DAR/CG-DMRs that overlapped promoters. For the complete list of neuronal DAR/CG-DMRs, Haystack selected a random, CG content-matched subset of the input background to use for enrichment calculations. Significance was determined using a one-sided Fisher’s test.
To identify novel transcriptional regulators for the differentially expressed genes found between NeuN+ populations from NAcc and BA9, we first generated transcription factor-gene scores using TEPIC49. This software utilizes epigenetic information along with transcription factor binding sites to generate these scores which can then be used by DYNAMITE48 to infer potentially important transcriptional regulators by predicting up/down-regulation for differentially expressed genes. Using the combined TEPIC/DYNAMITE pipeline, transcription factor affinities were computed within the DARs identified between NeuN+ nuclei from NAcc and BA9 (n = 68,021) using the provided human_jaspar_hoc_kellis.PSEM position weight matrix. The affinities per gene were calculated over a 5 kb window around a gene’s TSS incorporating the signal abundance (cpm) within a peak into the transcription factor annotation. We also provided the log2 gene expression ratios for differentially expressed genes (DEGs) between NeuN+ nuclei from NAcc and BA9 (n = 2,952). Other input parameters used: outerCV=6, innerCV=10, alpha_Step_Size=0.01. We reported only those transcription factors that were expressed in our NeuN+ nuclei and had a correlation coefficient > |0.04| in Figure 4f. The full output (including transcription factors not expressed in our samples) is reported in Supplementary Table 23.
Gene Ontology Annotation
We utilized the Genomic Regions Enrichment of Annotations Tool (GREAT; v3.0.0)35 to assess nearest gene enrichment for hypo- and hypermethylated CG-DMRs. We used the hg19 assembly, reduced the default input parameter for max extension to 100 kb, and kept all other default parameters the same (settings available at the GREAT website: http://great.stanford.edu/). Gene Ontology (GO) terms returned must be significant by both the binomial and hypergeometric tests using the multiple hypothesis correction false discovery rate (FDR) ≤ 0.05 whose binomial fold enrichment is at least 2.0.
Metascape71 (http://metascape.org) was used to perform gene ontology (GO), Reactome, and KEGG pathway analyses using lists of gene symbols for differentially expressed genes (either up- or downregulated in NAcc as compared to BA9 NeuN+ nuclei) as input. The gene lists were generated by matching GENCODE gene IDs to gene symbols (“external_gene_id”) using biomaRt72,73 (v2.30.0).
Supplementary Software
All statistical analyses were performed using R66 (v3.3.x) and made use of packages contributed to the Bioconductor project74,75. In addition to those R/Bioconductor packages specifically referenced in the above, we made use of several other packages in preparing results for the manuscript:
bsseq (v1.14)
AnnotationHub (v2.6.4)
GenomicAlignments58 (v1.10.0)
GenomicFeatures58 (v1.26.2)
GenomicRanges58 (v1.26.2)
ggplot276 (v2.2.1)
Hmisc (v4.0–2)
Matrix (v1.2–8)
rtracklayer77 (v1.34.1)
SummarizedExperiment (v1.4.0)
EnrichedHeatmap (v1.4.0)
mgcv (v1.8–23)67
sva (v3.22.0)
EnrichedHeatmap (v1.4.0)
Picard (v2.2.2)
seqtk (v1.2-r94)
Salmon (v0.7.2)
tximport (v1.2.0)
Metascape (http://metascape.org)
GREAT (v3.0.0; http://great.stanford.edu)
Supplementary Material
Acknowledgements
This work was supported by funding awarded to A.P.F. (U01MH104393n) through the enhanced Genotype-Tissue Expression (eGTEx) project supported by the Common Fund of the Office of the Director of NIH. This work was supported by National Cancer Institute under U24CA180996. We would like to thank H. Zhang from the Flow Cytometry Cell Sorting Core Facility at Johns Hopkins School of Public Health for flow sorting. The core facility is supported by CFAR: 5P30AI094189-04, 1S10OD016315-01, and 1S10RR13777001. Brain tissues were received from the NIH NeuroBioBank at the University of Maryland and University of Pittsburgh.
Footnotes
Accession Codes
All data generated in this study are deposited in NCBI GEO under accession code GSE96615.
Competing Interests Statement
The authors declare no competing financial interests.
Code Availability
The code used in this manuscript is available through Github (hansenlab/BrainEpigenomicsNN) and is archived at Zenodo (DOI:10.5281/zenodo.1469577).
Data Availability Statement
Raw and processed data generated are available through NCBI GEO under accession number GSE96615. [Reviewer link for GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96615.]
Processed data is available through a UCSC hub, at http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&hubUrl=https://s3.us-east-2.amazonaws.com/brainepigenome/hub.txt.
Reporting Summary
Further information is available in the Life Sciences Reporting Summary linked to in this article.
REFERENCES
- 1.Hoffmann A, Sportelli V, Ziller M & Spengler D Epigenomics of Major Depressive Disorders and Schizophrenia: Early Life Decides. Int J Mol Sci 18(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Negi SK & Guda C Global gene expression profiling of healthy human brain and its application in studying neurological disorders. Sci Rep 7, 897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mo A et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marques S et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lake BB et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tasic B et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeisel A et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–42 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Luo C et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies MN et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol 13, R43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Illingworth RS et al. Inter-individual variability contrasts with regional homogeneity in the human brain DNA methylome. Nucleic Acids Res 43, 732–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladd-Acosta C et al. DNA methylation signatures within the human brain. Am J Hum Genet 81, 1304–15 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viana J et al. Schizophrenia-associated methylomic variation: molecular signatures of disease and polygenic risk burden across multiple brain regions. Hum Mol Genet (2016). [DOI] [PMC free article] [PubMed]
- 13.Watson CT et al. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer’s disease. Genome Med 8, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaut O et al. Aberrant NMDA receptor DNA methylation detected by epigenome-wide analysis of hippocampus and prefrontal cortex in major depression. Eur Arch Psychiatry Clin Neurosci 265, 331–41 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Roadmap Epigenomics C. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffe AE et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 19, 40–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis SE, Gupta S, Moes A, West AB & Arking DE Exaggerated CpH methylation in the autism-affected brain. Mol Autism 8, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mo A et al. Epigenomic landscapes of retinal rods and cones. Elife 5, e11613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stroud H et al. Early-Life Gene Expression in Neurons Modulates Lasting Epigenetic States. Cell 171, 1151–1164 e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozlenkov A et al. Substantial DNA methylation differences between two major neuronal subtypes in human brain. Nucleic Acids Res 44, 2593–612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozlenkov A et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res 42, 109–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lister R et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Mut JV et al. Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. Transl Psychiatry 6, e718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morel L et al. Molecular and Functional Properties of Regional Astrocytes in the Adult Brain. J Neurosci 37, 8706–8717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Bartheld CS, Bahney J & Herculano-Houzel S The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J Comp Neurol 524, 3865–3895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen KD, Langmead B & Irizarry RA BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol 13, R83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montano CM et al. Measuring cell-type specific differential methylation in human brain tissue. Genome Biol 14, R94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vermunt MW et al. Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep 9, 767–79 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Andersson R et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen KD et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43, 768–75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen KD et al. Large-scale hypomethylated blocks associated with Epstein-Barr virus-induced B-cell immortalization. Genome Res 24, 177–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean CY et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JU et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 17, 215–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziller MJ et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet 7, e1002389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fullard JF et al. Open chromatin profiling of human postmortem brain infers functional roles for non-coding schizophrenia loci. Hum Mol Genet 26, 1942–1951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinello L, Farouni R & Yuan GC Haystack: systematic analysis of the variation of epigenetic states and cell-type specific regulatory elements. Bioinformatics (2018). [DOI] [PMC free article] [PubMed]
- 38.Fukuchi M & Tsuda M Convergence of neurotransmissions at synapse on IEG regulation in nucleus. Front Biosci (Landmark Ed) 22, 1052–1072 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Hu TM, Chen CH, Chuang YA, Hsu SH & Cheng MC Resequencing of early growth response 2 (EGR2) gene revealed a recurrent patient-specific mutation in schizophrenia. Psychiatry Res 228, 958–60 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Pfaffenseller B et al. Differential expression of transcriptional regulatory units in the prefrontal cortex of patients with bipolar disorder: potential role of early growth response gene 3. Transl Psychiatry 6, e805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larson EB et al. Striatal regulation of DeltaFosB, FosB, and cFos during cocaine self-administration and withdrawal. J Neurochem 115, 112–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin Y et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durek P et al. Epigenomic Profiling of Human CD4+ T Cells Supports a Linear Differentiation Model and Highlights Molecular Regulators of Memory Development. Immunity 45, 1148–1161 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Schmidt F et al. Combining transcription factor binding affinities with open-chromatin data for accurate gene expression prediction. Nucleic Acids Res 45, 54–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okuno H Regulation and function of immediate-early genes in the brain: beyond neuronal activity markers. Neurosci Res 69, 175–86 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Hertzberg L, Katsel P, Roussos P, Haroutunian V & Domany E Integration of gene expression and GWAS results supports involvement of calcium signaling in Schizophrenia. Schizophr Res 164, 92–9 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Jaffe AE et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci 21, 1117–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feinberg AP Phenotypic plasticity and the epigenetics of human disease. Nature 447, 433–40 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Harrow J et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardiner-Garden M & Frommer M CpG islands in vertebrate genomes. J Mol Biol 196, 261–82 (1987). [DOI] [PubMed] [Google Scholar]
- 53.Rosenbloom KR et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 43, D670–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krueger F & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuhn RM, Haussler D & Kent WJ The UCSC genome browser and associated tools. Brief Bioinform 14, 144–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawrence M et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Law CW, Chen Y, Shi W & Smyth GK voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson MD & Oshlack A A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leek JT & Storey JD Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3, 1724–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon provides accurate, fast, and bias-aware transcript expression estimates using dual-phase inference. bioRxiv (2016).
- 65.Soneson C, Love MI & Robinson MD Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Team, R.C. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: 2016 (2016). [Google Scholar]
- 67.Wood SN Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. Journal of the Royal Statistical Society (B) 73, 3–36 (2011). [Google Scholar]
- 68.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holm S A Simple Sequentially Rejective Multiple Test Procedure. Scandnavian Journal of Statistics 6, 65–70 (1979). [Google Scholar]
- 70.Mathelier A et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44, D110–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tripathi S et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 18, 723–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durinck S et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–40 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Durinck S, Spellman PT, Birney E & Huber W Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4, 1184–91 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gentleman RC et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huber W et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12, 115–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wickham H ggplot2: elegant graphics for data analysis, (Springer, 2016). [Google Scholar]
- 77.Lawrence M, Gentleman R & Carey V rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25, 1841–2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.