Abstract
Proteasome-mediated degradation of proteins is a vital cellular process and is performed by the ubiquitin-dependent proteasome system (UPS) and the ubiquitin-independent proteasome system (UIPS). While both systems are necessary to maintain healthy cell function, many disease states are characterized by reduced activity of the UPS, and the UIPS cannot by itself maintain proper protein levels. It has been suggested that the 20S core particle (20S CP), the isoform of the proteasome in the UIPS that can degrade proteins without a ubiquitin tag, can be stimulated with a small molecule to assist the 20S CP to accept and hydrolyze substrates more rapidly. Several small molecule stimulators of the 20S CP have since been discovered, including AM-404, an arachidonic acid derivative. AM-404 has previously been shown to inhibit fatty acid amide hydrolase activity. We wished to evaluate what structural components of AM-404 are required to stimulate the 20S CP with the long-term goal of using this information to design a stimulator with better drug-like qualities. We synthesized numerous derivatives of AM-404, varying the chain length, substitutions, and degree of unsaturation. Through this endeavor, we obtained several molecules capable of stimulating the 20S CP to various degrees. We discovered that though chain length is important, the presence of a cis-alkene in a specific location in the aliphatic chain has the greatest impact on the ability to stimulate the 20S CP. Two of the derivatives maintain modest stimulatory activity, and have improved toxicity over AM-404.
Keywords: Proteasome, Synthesis, Stimulator, AM-404
Graphical Abstract
The proteasome is a large protein complex capable of degrading proteins that a cell no longer requires or that have become damaged.1 There are two established mechanisms by which the proteasome can recognize and degrade proteins.2 The first is through the ubiquitin-dependent pathway, commonly abbreviated as UPS. In this pathway, a protein is tagged with a chain of ubiquitin moieties, which the 19S regulatory particle (19S RP) can recognize. The 19S RP caps one or both of the ends of the core particle of the proteasome, called the 20S CP, Figure 1A.3 After the 19S RP interacts with the 20S CP, the gate of the 20S CP is opened and can accept unfolded or disordered proteins. The 19S RP is also responsible for removing the ubiquitin chain from the protein substrate and denaturing the protein through an ATP-dependent mechanism so it can fit inside the catalytic chamber of the 20S CP to be hydrolyzed.
Figure 1.
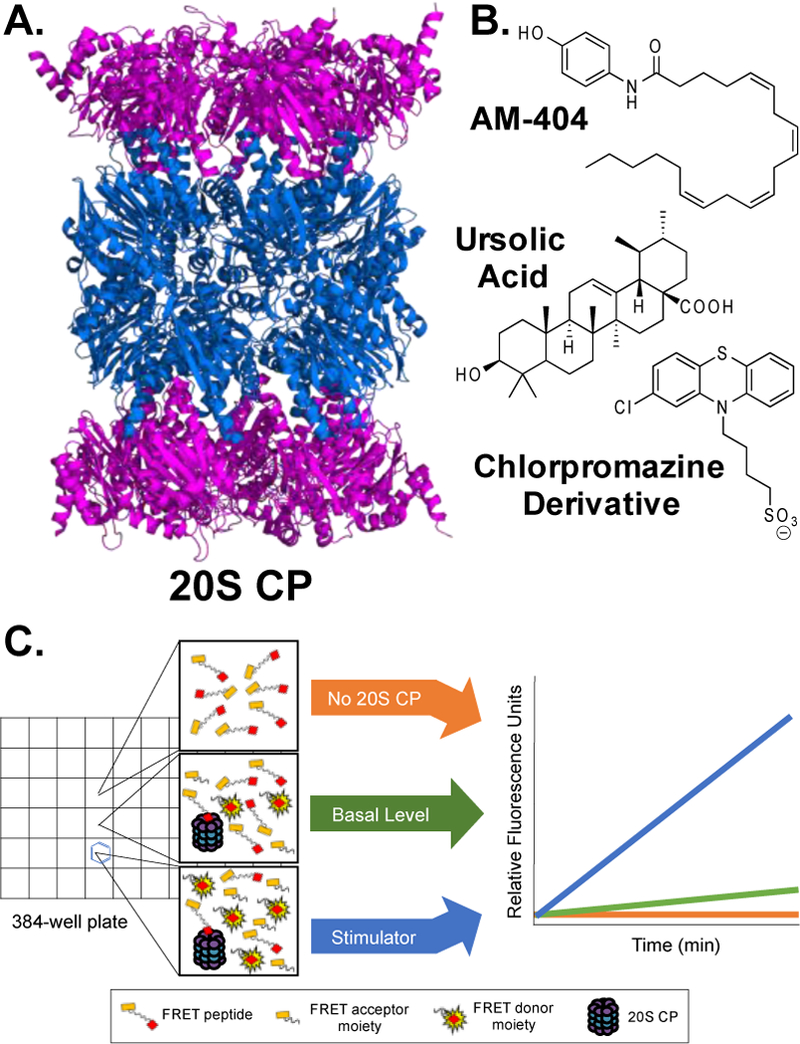
(A) The 20S CP is composed of two alpha rings (purple) and two beta rings (blue). The N-termini of the alpha ring protein’s form a “gate,” limiting the amount of protein that can enter the core particle to be hydrolyzed. (B) Previously discovered small molecules that are believed to interact with the alpha ring proteins to open the gate, allowing more substrates to enter to be hydrolyzed. (C) 20S CP activity is monitored using a peptide that upon being hydrolyzed by the 20S CP generates a fluorescent signal. The increase in stimulation is determined by comparing the rate of hydrolysis of the basal level of activity to that when a stimulator is added
The 20S CP alone can degrade proteins without the 19S RP and does so through a ubiquitin-independent proteasome system, or the UIPS.2 Only proteins that have a large region of disorder can fit past the closed-gate conformation to reach the catalytic core for hydrolysis. A substantial amount of 20S CP is present in healthy cells, uncapped to rapidly degrade misfolded proteins as they are synthesized by the ribosome. The balance of the UPS and the UIPS maintains healthy protein levels in cells.4
During the normal aging process, the expression levels of the 19S RP subunits diminishes, which leads to a decrease in the ability of the UPS to process proteins.5–8 This leaves the UIPS as the major proteasome pathway to degrade proteins. The UIPS alone cannot regulate the amount of protein in a cell effectively. This then leads to an accumulation of unwanted proteins in a cell, increased cellular stress, and eventually apoptosis. A number of diseases, including Parkinson’s and Alzheimer’s, are associated with the accumulation of toxic proteins and have an impaired UPS.
It has previously been shown that small molecules can help to stimulate the UIPS by interacting with the 20S CP. This can be done through two mechanisms of action. The first is through an interaction with one of the active sites to help the 20S CP turn over a protein substrate faster. The second is through a gate-opening mechanism, where a small molecule disrupts the protein-protein interactions of the N-termini of the alpha-subunits to allow larger or more substrates to enter at a faster rate.
A number of gate-opening 20S CP stimulators have been reported, Figure 1B.9–12 Many of these stimulators are molecules with long hydrophobic tails. For example, sodium dodecyl sulfate (SDS) is known to be a 20S CP gate opener.13 While SDS can serve as a positive control in a biochemical assay with purified 20S CP, dosing cells would not work. Another molecule recently described as a 20S CP stimulator, called AM-404, can aid SH-SY5Y cells to degrade α-synuclein at 32 µM.12 AM-404 is another molecule with a polar head group and a long aliphatic tail, but its tail is unsaturated, providing some secondary structure to the tail portion. AM-404 has been described as an inhibitor of the endocannabinoid transport pathway affecting neuropathic pain, and its metabolite has been shown to disrupt prostaglandin biosynthesis.14,15
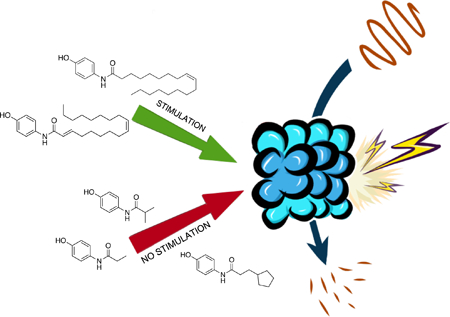
We wished to investigate if molecules with shorter aliphatic tails or with different substitution patterns from that of AM-404 could act as 20S CP stimulators. Here, we report a series of molecules that contain the aminophenol head group linked to aliphatic chains of varying length and unsaturation and their effect on the activity of the 20S CP. Our results indicate that it is not the chain length that is the most important structural component but a cis-double bond in the carbon chain that leads to effective 20S CP stimulation.
AM-404 was initially discovered as a 20S CP stimulator through a mass spectrometry-based screening technique and then verified using a GFP-α-synuclein fusion assay in HEK-293T cells.12 Since the scaffold of AM-404 is already known to penetrate the blood-brain barrier, we decided to try and retain the aminophenol head group and modify the length of the aliphatic chain. We recently have developed an effective assay specifically designed to monitor the stimulation abilities of a small molecule on the 20S CP. Briefly, this assay is performed in a 96- or 384-well plate with purified human 20S CP and a reporter peptide that is 11 amino acids in length with a fluorescence resonance energy transfer (FRET) pair, Figure 1C.9 The 20S CP can slowly degrade the FRET peptide without any stimulation, but upon adding a small molecule stimulator, the rate of hydrolysis increases substantially. The rate of hydrolysis of the FRET peptide is then calculated, with and without a stimulator, to generate a percent increase in hydrolysis over basal level. This assay was used to test all newly synthesized derivatives.
Our initial set of studies tested AM-404-like molecules that retained the aminophenol head group of AM-404 but contained aliphatic tails ranging from two to nine carbons in length. Based on the results from molecules 1–6, the aminophenol head group with a saturated chain cannot stimulate the 20S CP, Table 1. Ortho-substituted and biphenyl derivatives with a range of aliphatic tail lengths also showed no stimulation, Supporting Information Table S1 and S2. Neither of these results is surprising since AM-404 has four cis-double bonds, which can induce significant structure beyond what can be obtained with a saturated aliphatic tail. An additional set of molecules was synthesized and tested where the long aliphatic tail was exchanged with moieties that have substitution, Table 2. We hypothesized that these molecules could potentially fill the same binding pocket as AM-404. Again, none of these molecules, with the exception of molecule 9 showed any significant stimulation (>20%) over basal level. This was a surprising result as we had hypothesized that adding more steric bulk on the aliphatic chain would increase the binding contacts with the 20S CP, eliciting a stimulation result. We hypothesized that even though this series of molecules had more bulk on the aliphatic chain, as compared to the series in Table 1, they probably do not reach as far into the same binding pocket that AM-404 occupies.
Table 1.
Molecules with the same head group as AM-404 but with saturated carbon tails ranging from three to ten carbons are not effective 20S CP stimulators.
| Molecule | Structure | Stimulation |
|---|---|---|
| 1 | 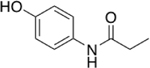 |
0% |
| 2 | 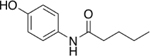 |
14% |
| 3 | 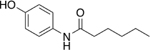 |
9% |
| 4 | 8% | |
| 5 | 7% | |
| 6 | 0% |
Table 2.
Adding steric bulk to the aliphatic side chain off the AM-404 head group does not induce any simulation of the 20S CP.
| Molecule | Structure | Stimulation |
|---|---|---|
| 7 | 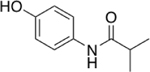 |
0% |
| 8 | 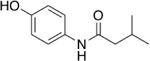 |
7% |
| 9 | 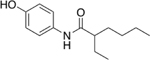 |
20% |
| 10 | 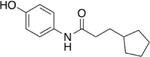 |
0% |
| 11 | 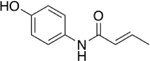 |
9% |
| 12 | 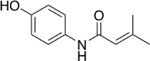 |
3% |
Based on these results, we decided that molecules with aliphatic chains substantially longer than the ones we had previously tested would be needed and that alkenes should be included in the aliphatic chain to mimic the structure of AM-404. AM-404 is derived by combining arachidonic acid with aminophenol, but arachidonic acid is not very stable as it has been shown to oxidize readily.16 We tested other naturally occurring acids such as docosahexaenoic acid (DHA) and oleic acid, which are known to be more stable in aqueous solutions, for their ability to stimulate the 20S CP. Both DHA and oleic acid substantially increased the hydrolysis activity of the 20S CP, Supporting Information Table S3. With these results in hand and with the development of recent allyl-palladium catalyzed carboxylic acid α,β-dehydrogenation reactions, we generated a series of unnatural fatty acids.17 The first series varied the aliphatic tail length, and the second series examined how the positioning of cis- or trans-alkenes affected 20S CP stimulation. After determining which fatty acids were the most active, they would be converted into the phenolic amide to limit micelle formation and would be more applicable for future cell dosing studies.
The first series contained molecules with aliphatic tails ranging in length from eight to sixteen carbons. Molecules with tails of thirteen carbons or less weakly stimulated the 20S CP. A significant jump in stimulation of 20S CP activity was observed with molecules with aliphatic tails of fourteen and sixteen carbons, Supporting Information Table S4. None of these were as effective as oleic acid. Three molecules, 13–15, were next synthesized and tested. These three molecules contained the cis-alkene between carbons nine and ten, similar to oleic acid. Oleic acid has 18 carbons in its aliphatic tail; these new molecules contained 16 or 18 carbons with an additional trans-alkene as compared to oleic acid, while molecule 15 had a total chain length of 18 carbons with another cis-alkene between carbons 12 and 13. Molecule 14 showed the best ability to stimulate the 20S CP, which is not surprising as its structure is the most similar to oleic acid, Table 3.
Table 3.
Fatty acids with a cis-alkene between carbons nine and ten can increase the activity of the 20S CP.
| Molecule | Structure | Stimulation |
|---|---|---|
| 13 | 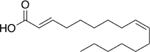 |
191% |
| 14 | 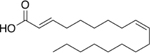 |
264% |
| 15 | 160% |
Oleic acid and four other acids that showed the ability to stimulate the 20S CP were converted to the phenolic amide derivatives, molecules 16–20, and were tested for their ability to stimulate purified 20S CP, Table 4. The oleic acid derivative, 16, stimulated the 20S CP 201% over basal level. Molecules 17 and 18, which do not have the cis-alkene at position nine to ten, showed substantially less ability to stimulate the 20S CP. Molecule 19, which has an extra trans-alkene alpha to carbonyl, was the best derivative, stimulating the hydrolysis activity of the 20S CP by 260%. Molecule 20 has two trans alkenes, and again, is not a very effective 20S CP stimulator. These results highlight that a cis-alkene is required to be an effective stimulator. Molecules 16 and 19 are possibly not as effective as AM-404 because their alkene is between other cis-alkenes or because the additional cis-alkenes of AM-404 may induce additional contacts with the 20S CP. Three derivatives of molecule 16 were synthesized by changing the phenolic amide to other substituted aryl groups and were found to have a greatly diminished capacity to stimulate the 20S CP, Supporting Information Table S5. The combination of all these results highlight that the phenolic amide and at least one cis-alkene in the aliphatic tail are required for the molecule to be able to stimulate the 20S CP.
Table 4.
After appending the phenolic amide, the best stimulators still required the cis-alkene at carbon nine.
| Molecule | Structure | Stimulation |
|---|---|---|
| 16 | 201% | |
| 17 | 85% | |
| 18 | 60% | |
| 19 | 260% | |
| 20 | 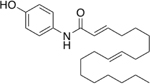 |
63% |
AM-404, molecule 16, and molecule 19 were then tested to determine their EC50 and how much stimulation can be obtained at their EC50 value, Figure 2A. AM-404, not surprisingly, stimulates the 20S CP the most at its EC50 (28 µM) to nearly 1000% over basal level. Molecules 16 and 19 have similar EC50 values to AM-404 at 35 µM and 24 µM, but they increase 20S CP stimulation over basal level 400% and 500%, respectively. These molecules were also tested for their toxicity in HEK-293T cells. Not surprisingly, AM-404 is the most toxic, which is why we wanted to synthesize derivatives that were still able to stimulate the 20S CP but would lead to potentially less off-target effects. Molecules 16 and 19 were less toxic than AM-404 at high concentrations, but this could be due to their lower ability to stimulate the 20S CP, Figure 2B. More studies are required to determine if there is a correlation between level of 20S CP stimulation and toxicity. We tested only the toxicity of the aminophenol capped derivatives (molecules 16 and 19) and not the unsaturated fatty acid derivatives (molecules 13 and 14) because of their lack of solubility in the amount of DMSO that the cell assay could tolerate before unwanted cytotoxic effects were observed.
Figure 2.

(A) EC50 curves for AM-404, 16, and 19. Their EC50 values are similar, but AM-404 still is the most effective stimulator. (B) HEK-293T cells were treated for 16 hrs with varying concentrations of the stimulators. Toxicity was not observed until 50 µM.
Molecules 16, 19, and AM-404 were tested to determine if their toxicity was elicited from disrupting the integrity of the cell’s membrane. We used a colorimetric assay to detect the amount of lactate dehydrogenase (LDH) that had been released into the media, indicating a damaged membrane. After dosing for 24 hrs with our molecules of interest, the results indicate that cytotoxicity is not related to disruption of the cell’s membrane, Supporting Information Figure S1. AM-404 has previously been reported that it can affect NF-kB activation, COX-2 expression, and NFAT signaling.18,19 We are currently evaluating molecules 16 and 19 to determine if they also affect these cellular targets.
This study has demonstrated that aliphatic stimulators of the 20S CP work the best when there is a cis-alkene present and that molecules with long saturated chains are generally not able to stimulate the activity of the 20S CP. While AM-404 still remains the most active stimulator, it may be too potent and lead to significant off-target effects. A molecule such as 16 or 19, which has half of the activity of AM-404 in the biochemical assay, may still provide enough stimulation to help the 20S CP degrade accumulated toxic proteins but lead to less cytotoxicity. Studies to understand how much stimulation is required to degrade a protein such as α-synuclein are currently ongoing. We are also working towards generating structures of the interaction of AM-404 and the 20S CP using a variety of biophysical techniques in order to rationally design better molecules.
Supplementary Material
Acknowledgments.
The work by the Trader laboratory was supported through a start-up package from Purdue University School of Pharmacy, the Purdue University Center for Cancer Research NIH grant P30 CA023168 and a grant from the Ralph W. and Grace M. Showalter Research Trust. The work by the Newhouse laboratory was supported by a NSF-CAREER Grant (CHE-1653793).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References.
- 1.Lecker SH. Protein Degradation by the Ubiquitin-Proteasome Pathway in Normal and Disease States. J Am Soc Nephrol 2006;17(7):1807–1819. [DOI] [PubMed] [Google Scholar]
- 2.Opoku-Nsiah KA, Gestwicki JE. Aim for the core: suitability of the ubiquitin-independent 20S proteasome as a drug target in neurodegeneration. Transl Res 2018; 198; 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, Luan B, Wu J, Shi Y. An atomic structure of the human 26S proteasome. Nat Struct Mol Biol 2016;23(9):778–785. [DOI] [PubMed] [Google Scholar]
- 4.Coleman R, Trader D. All About the Core: A Therapeutic Strategy to Prevent Protein Accumulation. ACS Pharmacol Transl Sci 2018;1(2):140–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeg S, Grune T. Protein Oxidation in Aging: Does It Play a Role in Aging Progression? Antioxid Redox Signal 2015;23(3):239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Husom AD, Peters EA, Kolling EA, Fugere NA, Thompson LV, Ferrington DA. Altered proteasome function and subunit composition in aged muscle. Arch Biochem Biophys 2004;421(1):67–76. [DOI] [PubMed] [Google Scholar]
- 7.McNaught KS, Jenner P. Proteasomal function is impaired in substantia nigra in Parkinson’s disease. Neurosci Lett 2001;297(3):191–194. [DOI] [PubMed] [Google Scholar]
- 8.Huang Q, Figueiredo-Pereira ME. Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications. Apoptosis 2010;15(11):1292–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman R, Trader D. Development and Application of a Sensitive Peptide Reporter to Discover 20S Proteasome Stimulators. ACS Comb Sci 2018;20:269–276. [DOI] [PubMed] [Google Scholar]
- 10.Jones CL, Njomen E, Sjögren B, Dexheimer TS, Tepe JJ. Small Molecule Enhancement of 20S Proteasome Activity Targets Intrinsically Disordered Proteins. ACS Chem Biol 2017;12(9):2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witkowska J, Giżyńska M, Grudnik P, et al. Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome – insights into the enzyme activation mechanism. Sci Rep 2017;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trader D, Simanski S, Kodadek T. Establishment of A Suite of Assays That Support the Discovery of Proteasome Agonists. Biochim Biophys Acta 2017;1861:892–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibatani T, Ward WF. Sodium dodecyl sulfate (SDS) activation of the 20S proteasome in rat liver. Arch Biochem Biophys 1995;321(1):160–166. [DOI] [PubMed] [Google Scholar]
- 14.Costa B, Siniscalco D, Trovato AE, et al. AM404, an inhibitor of anandamide uptake, prevents pain behaviour and modulates cytokine and apoptotic pathways in a rat model of neuropathic pain: AM404 relieves neuropathic pain. Br J Pharmacol 2009;148(7):1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saliba SW, Marcotegui AR, Fortwängler E, et al. AM404, paracetamol metabolite, prevents prostaglandin synthesis in activated microglia by inhibiting COX activity. J Neuroinflammation 2017;14(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyashita K, Nara E, Ota T. Oxidative Stability of Polyunsaturated Fatty Acids in an Aqueous Solution. Biosci Biotechnol Biochem 1993;57(10):1638–1640. [Google Scholar]
- 17.Zhao Y, Chen Y, Newhouse TR. Allyl-Palladium-Catalyzed α,β-Dehydrogenation of Carboxylic Acids via Enediolates. Angew Chem Int Ed 2017;56(42):13122–13125. [DOI] [PubMed] [Google Scholar]
- 18.Caballero FJ, Soler-Torronteras R, Lara-Chica M, et al. AM404 inhibits NFAT and NF-κB signaling pathways and impairs migration and invasiveness of neuroblastoma cells. Eur J Pharmacol 2015;746:221–232. [DOI] [PubMed] [Google Scholar]
- 19.Högestätt ED, Jönsson BAG, Ermund A, et al. Conversion of Acetaminophen to the Bioactive N - Acylphenolamine AM404 via Fatty Acid Amide Hydrolase-dependent Arachidonic Acid Conjugation in the Nervous System. J Biol Chem 2005;280(36):31405–31412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


