1. INTRODUCTION
Nuclear factor kappa B (NF-κB) plays a key role in coordinating inflammatory responses through the regulation of genes, such as CXCL10, CCL5, and IL6, encoding pro-inflammatory cytokines, chemokines and growth factors (1–3). Chronic inflammation promotes tumorigenesis by increasing cellular stress and DNA damage (4), and the appropriate regulation and control of NF-κB activity has long been considered a promising avenue for treating inflammatory diseases and cancer. Efforts in developing NF-κB pathway specific therapies have been largely unsuccessful due to its simultaneous influence in a vast number of physiological functions (5–7).
NF-κB is activated by a variety of extracellular stimuli including interleukin 1 beta (IL-1β) and tumor necrosis factor alpha (TNF-α), both playing roles in tumor progression and inflammation (8–10). In resting cells, NF-κB is maintained in the cytoplasm by inhibitory kappa B (IkB) proteins (11, 12). Upon stimulation, IκBs are phosphorylated by the IκB kinase complex (IKK), which causes its rapid degradation and releases NF-κB for nuclear translocation and subsequent control of gene transcription. IkB subunit beta (IKKβ) is one of the three kinases in IKK which is a convergence point for many NF-κB signaling pathways (11, 13). Alterations of NF-kB signaling dynamics has been demonstrated as a function of inhibiting IKK action (14–17). Additionally, dynamical changes of NF-κB activation have been shown to result in altered gene expression profiles (2, 14, 18). However, the extent to which pathway perturbations effect signal dynamics and the relationship between NF-κB signaling dynamics and transcriptional output have not been fully elucidated. 1
In this contribution, we test the hypotheses that 1) inhibitors acting on pleiotropic pathway components can have stimulus-specific effects on signal dynamics, and 2) alterations in signal dynamics caused by small molecule inhibitors affect downstream gene transcription. To this end, we treated 3T3 fibroblasts with SC-514, an IKKβ selective inhibitor, and characterized changes in the temporal profile of NF-κB activity and corresponding gene expression in response to TNF-α or IL-1β stimulation. Taken together, the results in this study demonstrate the value of protein-specific small molecule inhibitors as tools for probing signaling pathways, and also highlight their potential for altering signal dynamics affecting gene expression as a result.
2. MATERIALS and METHODS
2.1. Cell Culture
3T3 mouse fibroblast cells were a gift from Dr. Laura Suggs (Department of Biomedical Engineering, University of Texas, Austin, TX) and were maintained in glucose-containing Dulbecco’s modified Eagle’s medium (DMEM, VWR International, LLC, Radnor, PA) supplemented with 10% bovine calf serum and penicillin-streptomycin (100 units/ml). One day before the stimulation experiments, cells were seeded in 8-well chambered cover glass (Cellvis, Mountain View, CA) in 200 μl of culture medium at an estimated 35,000 cells/well. Cell count was determined by the Countess II FL automated cell counter (Thermo Fisher Scientific Inc., Waltham, MA).
2.2. Stimulation experiments
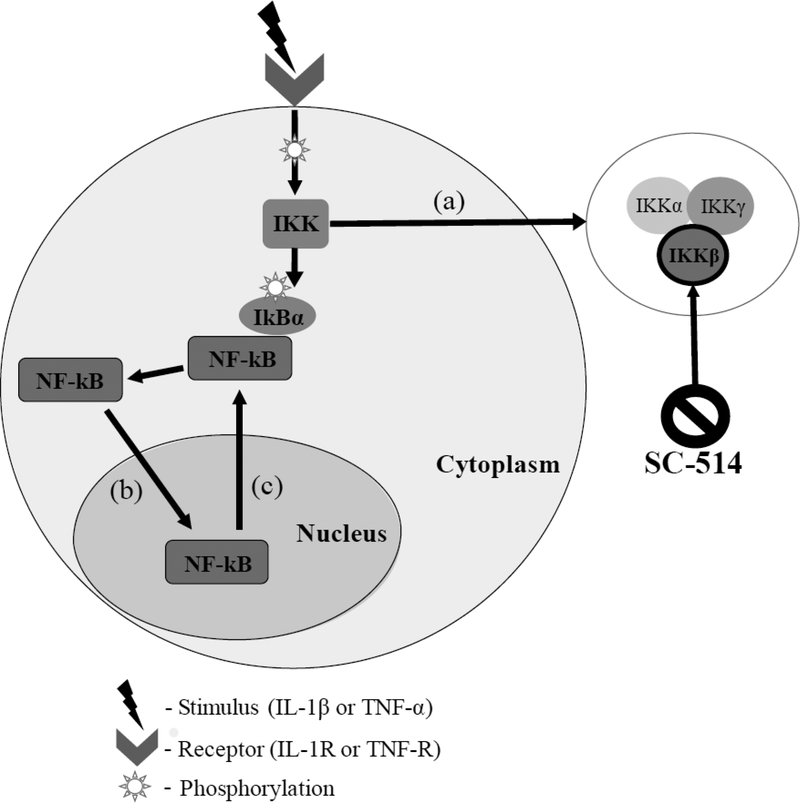
3T3 fibroblasts in each 8-well chamber were pretreated with culture medium containing SC-514 (0, 10, 25, 50 or 100 μM) (Selleck Chemicals, Houston, TX) 1 hour prior to stimulation. SC-514 is a selective inhibitor of IKKβ action, inhibiting IkBα phosphorylation (when treated with SC-514 above 100 μM, cell dissociation occurred). Cells were stimulated with either 1 ng/ml of IL-1β, 10 ng/ml of TNF-α, or control saline for 5, 10, 15, 20, 25, 30, 45, 60, 75, or 90 min. An initial dose response experiment for each stimuli was conducted. The lowest concentration of stimuli that achieved the fastest rate of peak NF-κB activation was chosen for each stimuli. Cells were fixed and permeabilized with 100% methanol at −20 °C for 5 min and blocked in phosphate buffered saline (PBS, Caisson Laboratories, Inc., Smithfield, UT) + 2% bovine serum albumin (BSA, Fisher Scientific International Inc., Pittsburgh, PA) for 1 hour. Cells were incubated with NF-kB-p65 antibody Alexa Fluor 488 (AF488) conjugate (Cell Signaling Technology, Beverly, MA) in PBS+2% BSA for 1 hour followed by a 5-min incubation with 4’,6-diamidino-2-phenylindole (DAPI) in PBS. This experiment was replicated for each drug treatment four times at each time point. A simplified diagram of the NF-kB signaling pathway can be seen in Figure 1. NF-kB is activated by both IL-1β and TNF-α. When IL-1β and TNF-α bind to their receptors, (IL-1R and TNF-R, respectively), a series of phosphorylation events take place, eventually converging at the IKK complex which consists of three isoforms; IKKα, IKKγ, and IKKβ (Figure 1a). IKKβ phosphorylates an inhibitory protein IkBα, subsequently allowing NF-kB to translocate to the nucleus where gene transcription takes place (Figure 1b). NF-kB translocates back into the cytoplasm (Figure 1c) due to negative feedback in signaling.
Figure 1.
High level representation of the NF-κB signaling pathway and SC-514 mechanism of action. Stimuli IL-1β and TNF-α bind IL-1R and TNF-R, respectively, triggering downstream phosphorylation. NF-κB is sequestered in the cytoplasm by IkBα until activation of the IKK complex, which consists of three isoforms, IKKα, IKKγ, and IKKβ (a). IKKβ phosphorylates IkBα, allowing NF-κB to translocate to the nucleus for gene transcription (b) until eventually translocating back to the cytoplasm (c). SC-514 is a selective inhibitor of IKKβ action.
2.3. Microscopy and Image Analysis.
Cells were imaged in the green fluorescent protein (GFP) (Ex. 450–49 nm Em. 500–550 nm) and DAPI (Ex. 325–375 nm Em. 435485 nm) channels with the Leica DMI6000 microscope (Leica Microsystems, Wetzlar, Germany) at 20X resolution. Image analysis was performed using CellProfiler (www.cellprofiler.org). An image analysis pipeline was constructed to measure DAPI-labeled nuclei and AF488-labeled NF-kB. Specifically, raw images were first corrected for uneven illumination (vignetting) (19–21). An illumination function was calculated using the intensity of each pixel (regular) and convex hull smoothing method of an empty field image was applied. Each image was divided with this illumination function and then background intensity was subtracted. All images were converted to grayscale. Nuclei were identified with the DAPI stain using Otsu two-class global thresholds. ‘Shape’ which identifies indentations on edges of touching cells was used to distinguish the number of objects in a clump. Cell boundaries were drawn using AF488-labeled NF-kB intensity, where boundaries are defined to be the dimmest points between two objects (19, 21). The nucleus was masked from the whole cell to define the cytoplasm region. Intensity of AF488-labeled NF-kB was measured in the nucleus, cytoplasm, and whole cell. Each image was analyzed independently. Two 4.19 megapixel images were taken per well in each experiment with ~100–200 cells in view in each image. Mean fluorescent intensity was calculated for each cell and averaged together for each well. Values were normalized to the averaged time zero intensity. Images where cell boundaries could not be accurately distinguished were eliminated from analysis.
2.4. RNA sample preparation.
24 hours before RNA purification, 3T3 fibroblasts were plated in a 24 well plate at 140,000 cells/well. Cells were pretreated with culture medium containing SC-514 (0, 10, 25, 50 or 100 μM) 1 hour prior to stimulation. Cells were stimulated with either 1 ng/ml of IL-1β, or 10 ng/ml of TNF-α for 15, 30, 60, 120, and 360 min. All reagents for RNA purification were part of the RNeasy Mini kit (Quiagen, Hilden, Germany). Briefly, cells were lysed directly in the 24 well plate and homogenized by vortexing. Precipitate was formed using 70% ethanol. All samples were treated with DNase and eluted into 30 μl of RNase free water. RNA concentration and purity was determined using the Qubit 4 Fluorometer (Thermo Fisher Scientific Inc., Waltham, MA). This process was replicated for each drug treatment at each time point four times.
2.5. TagSeq profiling.
RNA TagSeq is a 3’ Tag-based approach to full RNA sequencing that has been proven highly reliable for differential gene expression analysis in well annotated genomes (22, 23). RNA samples were sent to the Genome Sequence and Analysis Facility at The University of Texas at Austin where libraries were constructed using a modified protocol from Lohman et al. (22). Modifications from Lohman et al. included: i) Fragmentation of RNA was done using 5X First Strand Buffer (Takara Bio USA, Inc., Mountain View, CA) for 2.5 min ii) Titanium tag polymerase (Takara Bio USA, Inc., Mountain View, CA) was used in cDNA amplification master mix. Samples were sequenced with the Illumina HiSeq 2500 1 × 100 with an average of 3.5 × 106 raw reads per sample.
2.6. TagSeq processing.
TagSeq clipping was carried out using FASTX-Toolkit’s fastx-clipper with parameters to remove non-template bases introduced by reverse transcriptase on the 5’ end (23, 24). Quality assurance of processed reads was done using dispersion calculations implemented in DESeq2. Poor quality reads due to low amounts of sample (yield < 10 Mbp) for sequencing were removed from analyses (n = 2). Reads were aligned against an assembly of cDNA sequences for the GRCm38 (mm10) assembly genome using Bowtie2 (25). The output of processing was a count matrix of raw reads per gene for each experimental condition.
2.7. Differential gene expression analysis.
Differential expression analyses were carried out via DESeq2 from the Bioconductor suite of R tools (26) using count matrices generated from the processing pipeline. Normalization of reads was done by modeling read counts for each sample as following a negative binomial distribution with a mean that is proportional to the concentration of cDNA fragments from each gene scaled by a factor based on sequencing depth and library size. Expression differences were calculated by comparing coefficients in a generalized linear model (GLM) between experimental conditions. The GLM returns a coefficient matrix indicating overall expression strength of all genes in the sample and the shrunken (minimized dispersion) log2 fold change between two experimental conditions. Expression at each inhibitor concentration (25, 50, and 100 μM) for both IL-1β and TNF-α at each longitudinal time point was individually compared against the no inhibitor condition at that same time point. All code for this portion of the analysis, including visualization of results, was written in R statistical software (27).
2.8. Statistical analysis.
Analyses for fluorescent images were performed with Matlab (MathWorks Inc., Natick, MA). Error bars in Fig. 2–6 represent 95% confidence intervals. Comparison of the mean fluorescent intensity between control and inhibitor treated data sets was determined with a two-way analysis of variance test (ANOVA). If the interaction term (time and concentration of inhibitor) exhibited P < 0.05, then the difference was considered statistically significant. A pairwise t-test was used to determine significant differences in the fold change of peak intensity values. For cell measurements from the image analysis, outlier samples were determined statistically through the Grubbs outlier test and were eliminated from the data set. Analyses for gene expression were completed in R. Standard error for the GLM coefficient based log2 fold change estimates are calculated using the curvature of each coefficient’s posterior probability at the maximum of a density plot of coefficient estimates for log2 fold change. In DESeq2, p-values are generated using a Wald test and adjusted using Benjamini and Hochberg corrections (26).
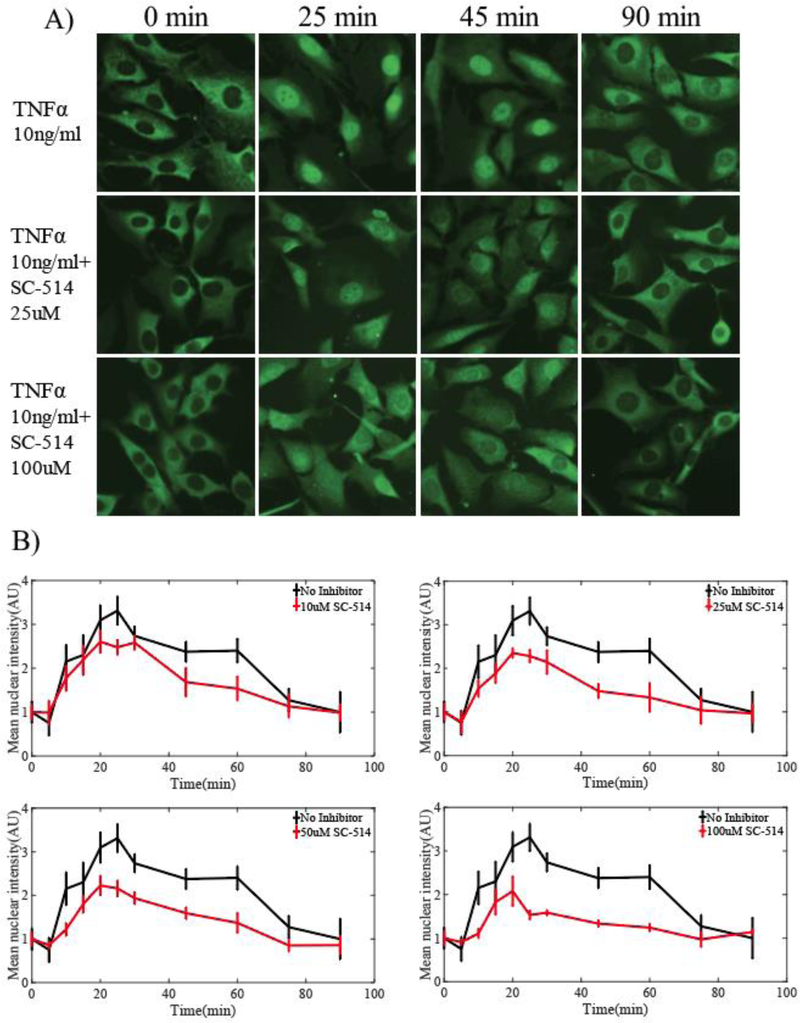
Figure 2.
A) Representative images of NF-κB translocation over time. Row 1 shows untreated 3T3 fibroblast cells, while rows 2 and 3 display cells treated with 25 and 100 μM of SC-514, respectively. All cells were stimulated with IL-1β (1ng/ml) and fixed at 0, 25, 45, and 90 min. Visual differences in nuclear NF-κB translocation (stained green) can be seen between untreated cells and cells treated with 25 and 100 μM of SC514 at 25 and 45 min. B) Quantification of the mean nuclear NF-κB fluorescent intensity per each experimental condition. Nuclear and cytoplasmic regions were segmented from original immunofluorescence images and intensity values were averaged at each time point. Error bars represent 95% confidence intervals. A two-way analysis of variance test determined significant differences (P < 0.05) between time series of untreated cells and cells treated with 25, 50 and 100 μM of SC-514 (red).
2.9. Data availability.
The CellProfiler image analysis pipeline is available upon request. All processing code used in TagSeq processing including RMarkdown files containing session information used in differential expression analysis is available at https://github.com/sachitsaksena/nfkb-tag-seq.
2.10. Accession numbers.
Mouse genome (GRCm38 (mm10)): GCA_000001635.8
RNA TagSeq dataset deposited on NIH Sequence Read Archive: SRP163157
3. RESULTS
3.1. IKKβ inhibition causes significant differences in temporal profiles of nuclear NF-κB translocation when stimulated with IL-1β.
Representative images of NF-κB translocation after IL-1β stimulation at 0, 25, 45, and 90 min in three experimental conditions shown in Figure 2A. A difference in nuclear accumulation of NF-κB can be seen between untreated cells and cells treated with 25 μM of SC-514 at 25 and 45 min. An increased difference is apparent in cells treated with 100 μM of SC-514 and untreated cells at 25 and 45 min. At 90 min, NF-κB has cycled out of the nucleus and back into the cytoplasm in all experimental conditions. Statistical analyses revealed significant differences in time series between untreated cells and cells treated with 25, 50 and 100 μM of SC-514 (P < 0.001) (Figure 2B, red). Time series data of cells treated with 10 μM of SC-514 were also significantly different from cells treated with 25, 50, and 100 μM of SC-514 (P < 0.001) but not so when compared to untreated cells (P = 0.224). Significant differences were also observed between cells treated with 25 μM and those treated with 50 or 100 μM of inhibitor (P = 0.027 and P < 0.001, respectively). Significant differences were also significant between cells treated with 50 μM and 100 μM of inhibitor (P = 0.003). No concentration of inhibitor completely blocked nuclear translocation.
3.2. IKKβ inhibition causes significant differences in temporal profiles of nuclear NF-κB translocation when stimulated with TNF-α.
Representative images of NF-κB translocation after TNF-α stimulation at 0, 25, 45, and 90 min in three experimental conditions are shown in Figure 3A. A difference in the nuclear accumulation of NF-κB can be seen between untreated cells and cells treated with 25 μM of SC-514 at 25 and 45 min. An increased difference is apparent in cells treated with 100 μM of SC-514 at 25 and 45 min. At 90 min, NF-κB has cycled out of the nucleus and back into the cytoplasm in all experimental conditions. Statistical analyses revealed significant differences in time series between untreated cells and cells treated with 10, 25, 50 and 100 μM SC-514 (P < 0.001) (Figure 3B, red). Time series data of cells treated with 10 μM of SC-514 were also significantly different from cells treated with 50 and 100 μM of SC-514 (P = 0.012 and P < 0.001, respectively). Cells treated with 25 μM and 50 μM of SC-514 had significant differences when compared to cells treated with 100 μM of inhibitor (P < 0.001). No significant differences were seen between 10 μM and 25 μM conditions or 25 μM and 50 μM conditions (P = 0.585 and P = 0.091 respectively). No concentration of inhibitor completely blocked nuclear translocation.
Figure 3.
A) Representative images of NF-κB translocation over time. Row 1 shows untreated 3T3 fibroblast cells, while Rows 2 and 3 display cells treated with 25 μM and 100 μM of SC-514, respectively. All cells were stimulated with TNF-α (10 ng/ml) and fixed at 0, 25, 45, and 90 min. Visual differences in nuclear NF-κB translocation (stained green) can be seen between untreated cells and cells treated with 25 and 100 μM of SC514 at 25 and 45 min. B) Quantification of the mean nuclear NF-κB fluorescent intensity per each experimental condition. Nuclear and cytoplasmic regions were segmented from original immunofluorescence images and intensity values were averaged at each time point. Error bars represent 95% confidence intervals. A two-way analysis of variance test determined significant differences (P < 0.05) between time series of untreated cells and cells treated with 10, 25, 50 μM and 100 μM of SC-514 (red).
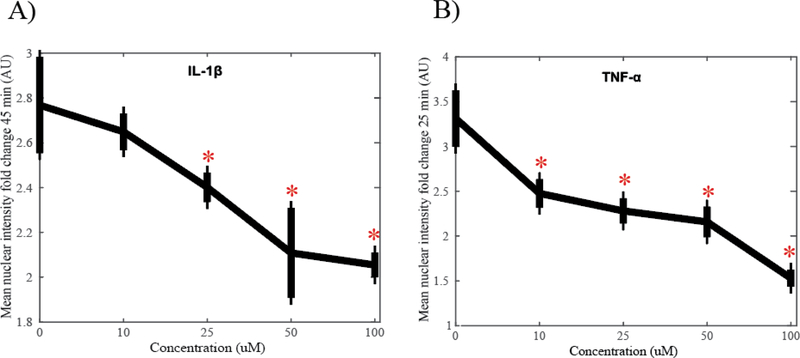
3.3. Degree of fold change in NF-κB nuclear translocation at peak activation is stimulus specific.
The dose response for fold change in the mean NF-κB nuclear intensity at peak time points is shown in Figure 4. Cells stimulated with IL-1β reach peak activation at 45 min with a maximum fold change of 2.77 with no inhibitor (Figure 4A). There is a significant difference in intensity fold change between untreated cells and cells treated with 25 μM (P = 0.013) as well as 50 and 100 μM (P < 0.001) of SC-514 indicated with asterisks. Maximum difference in fold change is achieved with 50 μM of SC-514, reaching a fold change of 2.11, a 0.66 decrease from peak activation. There is no significant difference in peak activation between cells treated with 50 μM of inhibitor and 100 μM of inhibitor (P = 0.554) suggesting saturation of the inhibitor effect. Cells stimulated with TNF-α reach peak activation at 25 min with a maximum fold change of 3.31 with no inhibitor (Figure 4B). There is a significant difference in intensity fold change between untreated cells and cells treated with 10, 25, 50 and 100 μM (P < 0.001) of SC-514 indicated with asterisks. Maximum difference in fold change is achieved with 100 μM of SC-514, reaching a fold change of 1.53, a 1.78 decrease from peak activation.
Figure 4.
A) Mean nuclear intensity fold change of NF-κB translocation stimulated with IL-1β (1 ng/ml) at 45 min. Significant differences (P < 0.05) between untreated cells and cells treated with 25, 50, and 100 μM of SC-514 indicated with asterisk. The maximum fold change difference in peak activation from untreated cells was achieved with 50 μM SC-514. B) Mean nuclear intensity fold change of NF-κB translocation stimulated with TNFα (10 ng/ml) at 25 min. Significant differences (P < 0.05) between untreated cells and cells treated with 10, 25, 50, and 100 μM of SC-514 indicated with asterisk. Maximum fold change difference in peak activation from untreated cells achieved with 100 μM of SC-514.
3.4. RNA TagSeq data reveals temporal alterations in gene expression in response to stimuli.
Total number of significantly (P < 0.01) differentially expressed genes in cells treated with 25, 50 and 100 μM of SC-514 compared to untreated cells after stimulation with IL-1β are shown in Figure 5A. For each individual time point, the number of differentially expressed genes increases with inhibitor concentration. Log2 fold changes in expression compared to untreated cells after stimulation with IL-1β of six inflammatory genes treated with 25, 50 and 100 μM of SC-514 are shown in Figure 5B. Significant differences in expression of cells treated with inhibitor from untreated cells are shown in (Figure 5B, red).
Figure 5.
A) Total number of significantly differentially expressed genes (P < 0.01) in cells treated with 25, 50 and 100 μM of SC-514 compared to untreated cells at 15, 30, 60, 120, and 360 min after IL-1β (1ng/ml) stimulation. An increasing number of differentially expressed genes is seen with increasing concentration of SC-514 for each individual time point. B) Log2 fold change in gene expression in cells treated with 25, 50 and 100 μM of SC-514 compared to untreated cells for six inflammatory genes: Il1r1, Il6, Cox2, Nfkbia, Tnfaip3 and Egr1 at 15, 30, 60, 120, and 360 min after IL-1β (1ng/ml) stimulation. Significance (P < 0.05) indicated in red. Note vertical axis are different for each gene.
Total number of significantly (P < 0.01) differentially expressed genes in cells treated with 25, 50 and 100 μM of SC-514 compared to untreated cells after stimulation with TNF-α are shown in Figure 6A. For each individual time point, the number of differentially expressed genes increases with inhibitor concentration. Log2 fold changes in expression compared to untreated cells after stimulation with TNF-α of six inflammatory genes treated with 25, 50 and 100 μM of SC-514 are seen in Figure 6B. Significant differences in expression of cells treated with inhibitor from untreated cells are shown in (Figure 6B, red).
Figure 6.
A) Total number of significantly differentially expressed genes (P < 0.01) in cells treated with 25, 50 and 100 μM of SC-514 compared to untreated cells at 15, 30, 60, 120, and 360 min after TNF-α (10 ng/ml) stimulation. An increasing number of differentially expressed genes is seen with increasing concentration of SC-514 for each individual time point. B) Log2 fold change in gene expression in cells treated with 25, 50 and 100 μM of SC-514 compared to untreated cells for six inflammatory genes Il1r1, Il6, Cox2, Nfkbia, Tnfaip3 and Egr1 at 15, 30, 60, 120, and 360 min after TNF-α (10ng/ml) stimulation. Significance (P < 0.05) indicated in red Note vertical axis are different for each gene.
Expression of Interleukin 1 receptor, type I (Il1r1) is downregulated in a dose dependent manner in cells stimulated with both IL-1β and TNF-α and has a consistent trend through time and between stimulation conditions. Early growth response 1 (Egr1) has a similar change in expression in both stimulation conditions, with the highest upregulation occurring at 15 and 120 min in a dose dependent manner. However, tumor necrosis factor alpha-induced protein 3 (Tnfaip3) is significantly upregulated in cells stimulated with TNF-α at 15 min which is not the case in cells stimulated with IL-1β. Cytochrome c oxidase II, mitochondrial (Cox2) and Interleukin 6 (Il6) exhibit varying degrees of up- and down- regulation through time. Particular differences in Cox2 expression between stimuli are seen at 15 min where IL-1β stimulated cells treated with 50 μM of inhibitor exhibit significant downregulation of expression while TNF-α stimulated cells do not and in expression of Il6 at 360 min where IL-1β stimulated cells treated with 25 μM exhibit an upregulation in expression and TNF-α stimulated cells do not.
4. DISCUSSION
Regulation of NF-κB is a critical step in inflammatory diseases and cancer. Pathway analyses are necessary to understand NF-κB transcriptional activity to aid in the development of targeted treatments. This study quantitatively evaluated the effect of SC514, a selective inhibitor of IKKβ, on NF-κB transcriptional activity using two stimuli, IL-1β and TNF-α. Results demonstrated significant temporal differences in NF-κB nuclear translocation between untreated cells and cells treated with various concentrations of SC-514 when stimulated with both IL-1β and TNF-α. Furthermore, this study identifies changes in inflammatory gene expression due to differing temporal profiles of NF-κB nuclear translocation of cells treated with IL-1β and TNF-α.
Protein- specific small molecules have been used to investigate signaling pathways and have proved valuable in developing targeted treatments (28). These tools have allowed researchers to understand how specific signaling pathway perturbations alter genetic profiles and cellular function in disease states (14, 15, 28, 29). Previous studies have shown altering upstream signaling events can effect NF-κB transcriptional activity (14–17),(30–32). Our results corroborate this and further quantify these differences on a dose dependent level. A decrease in peak activation of NF-κB was seen even at low concentrations (i.e., 10 and 25 μM); however, there was never a complete blockade. Our results are in agreement with studies that show decreased activation of NF-κB due to perturbations of the IKK complex (30–32); however, the timing and degree of NF-κB inhibition varies in studies depending on the cell type and stimulus (14, 33). This demonstrates how various cells can interpret signals independently and yield different responses (34). Cellular function must be taken into consideration when conducting pathway analyses in targeted drug development, as one pathway of interest may be present in multiple cell types relevant to the disease state.
The results of the present study show a stimulus specific response of NF-κB activation between cells stimulated with IL-1β and TNF-α supporting preliminary studies which investigated how targeting signaling dynamics through pharmacological intervention can produce stimulus-selective responses (5). When stimulated with IL-1β, cells exhibit peak NF-κB activation at 45 min, a significant change in temporal profile starting with concentrations of 25 μM SC-514, and a maximum decrease in peak intensity with 50 μM of SC-514. These results show SC-514 blunts, in a concentration-dependent manner the amplitude of the transient peak of NF-κB activity induced by IL-1β stimulation. Interestingly, even at the highest dose, SC-514 does not change the timing of the signal decay. When stimulated with TNF-α, cells exhibit peak NF-κB activation at 25 min, a significant change in temporal activation profile starting with 10 μM of SC-514, and maximum decrease in peak activation with 100 μM of SC-514. These results show SC-514 substantially re-modulates, in a concentration-dependent manner, the temporal dynamics of the NF-κB signal induced by TNF-α stimulation. In contrast to the IL-1β case, IKK inhibition by SC-514 seem to affect both peak amplitude as well as the decay of the signal induced by TNF-α. These findings warrant further analyses quantifying the effect of SC-514 on additional pathway molecules, including IKK itself in this specific cell type, to determine other possible factors contributing to dynamical changes.
Results from genetic sequencing yield results in agreement from others showing that manipulation of IKK leads to distinct NF-κB activation profiles and changes in gene expression profiles (2, 35), however our findings offer a further in depth analysis using longitudinal gene expression data with varying degrees of IKK inhibition in response to IL-1β and TNF-α. Overall, there is an SC-514 dose dependent trend of increasing differentially expressed genes through time. When looking at individual gene expression profiles through time, vast expression differences are observed in certain genes between experimental conditions.
Pathway perturbations can have differing effects on transcriptional activity depending on the stimuli present and is important to consider when designing therapeutic targets; in this application, especially in diseases where the inflammatory stimuli levels can vary such as cancer and rheumatoid arthritis (36, 37). Our results not only elucidate differences in inflammatory gene expression between stimuli but also show how expression levels can change in a matter of minutes, demonstrating the need for further, highly time-resolved gene expression studies.
A limitation of this study is that analyses were performed at fixed times on populations of cells. Real time, single cell imaging data would better capture cell to cell heterogeneity and assess the signaling consequences from these differences. This study focuses specifically on NF-κB dynamical changes from IKK inhibition; monitoring pathway proteins upstream of NF-κB would offer additional insight on the mechanisms of these changes. Further studies should also investigate dynamical changes from pathway perturbations in the NF-κB network at later time scales and the differences that arise in inflammatory gene expression. Additionally, our results would be further generalized if evaluated in other cell types. There may be differences between cell responses in the 3T3 cells used in these studies and primary cell lines. Studying the changes in signal dynamics due to pathway perturbations in vivo will increase biological insight of cellular behavior in disease states and prove valuable in pharmacology.
5. CONCLUSIONS
In summary, we demonstrate novel differences in early NF-κB transcriptional activity from IKKβ selective inhibition in response to IL-1β and TNF-α. Significant differences of NF-κB nuclear translocation were quantified at a high temporal resolution to both stimuli and revealed stimulus-specific responses to perturbation. Furthermore, gene expression profiles revealed an increase in significantly differentially expressed genes with increasing concentration of IKKβ inhibition. Individual inflammatory genes were up and down regulated with varying degrees calling for further analysis of specific genes of interest. This work analyzes changes in signaling and transcriptional dynamics necessary to develop effective NF-κB pathway targeted drugs and delineates the importance of pathway analyses in therapeutic design.
Highlights.
NF-κB exhibits stimulus specific signaling dynamics from IKKβ inhibition
Alterations in NF-κB signaling dynamics affects inflammatory gene expression
Higher inhibitor concentration results in more differentially expressed genes
Changes in individual inflammatory genes are inhibitor and concentration dependent
ACKNOWLEDGEMENTS
We thank the National Cancer Institute for support through U01CA174706 and R01CA186193. We thank the Cancer Prevention and Research Institute of Texas (CPRIT) for funding through RR160005. T.E.Y. is a CPRIT Scholar of Cancer Research. Anna Sorace was supported in part by a Research Scholar Grant, RSG-18–006-01-CCE from the American Cancer Society.
Abbreviations:
- NF-kB
nuclear factor kappa B
- IL-1β
interleukin 1 beta
- TNF-α
tumor necrosis factor alpha
- IkB
inhibitory kappa B
- IKK
IκB kinase complex
- IKKβ
IkB subunit beta
- Il1r1
interleukin 1 receptor type I
- Il6
interleukin 6
- Cox2
cytochrome c oxidase II mitochondrial
- Nfkbia
NF-κB inhibitor alpha
- Tnfaip3
tumor necrosis factor, alpha-induced protein 3
- Egr1
early growth response 1
Footnotes
DECLARATIONS OF INTEREST: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Baltimore D 2011. NF-κB is 25. Nat Immunol 12:683–685. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann A, Levchenko A, Scott ML, Baltimore D. 2002. The IκB-NF-κB Signaling Module: Temporal Control and Selective Gene Activation. Science 298:1241–1245. [DOI] [PubMed] [Google Scholar]
- 3.1999. Activators and target genes of Rel/NF-κB transcription factors., Published online: 22 November 1999; | doi:101038/sj.onc120323918.
- 4.Hanahan D, Weinberg RA. 2011. Hallmarks of Cancer: The Next Generation. Cell 144:646–674. [DOI] [PubMed] [Google Scholar]
- 5.Behar M, Barken D, Werner SL, Hoffmann A. 2013. The Dynamics of Signaling as a Pharmacological Target. Cell 155:448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiDonato JA, Mercurio F, Karin M. 2012. NF-κB and the link between inflammation and cancer. Immunol Rev 246:379–400. [DOI] [PubMed] [Google Scholar]
- 7.Berger SI, Iyengar R. 2011. Role of systems pharmacology in understanding drug adverse events. Wiley Interdiscip Rev Syst Biol Med 3:129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo B, Fu S, Zhang J, Liu B, Li Z. 2016. Targeting inflammasome/IL-1 pathways for cancer immunotherapy. Scientific Reports 6:srep36107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis AM, Varghese S, Xu H, Alexander HR. 2006. Interleukin-1 and cancer progression: the emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment. J Transl Med 4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ham B, Fernandez MC, D’Costa Z, Brodt P. 2016. The diverse roles of the TNF axis in cancer progression and metastasis. Trends Cancer Res 11:1–27. [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden MS, Ghosh S. 2004. Signaling to NF-κB. Genes Dev 18:2195–2224. [DOI] [PubMed] [Google Scholar]
- 12.Hoesel B, Schmid JA. 2013. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer 12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheidereit C 2006. IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene 25:6685–6705. [DOI] [PubMed] [Google Scholar]
- 14.Kishore N, Sommers C, Mathialagan S, Guzova J, Yao M, Hauser S, Huynh K, Bonar S, Mielke C, Albee L, Weier R, Graneto M, Hanau C, Perry T, Tripp CS. 2003. A Selective IKK-2 Inhibitor Blocks NF-κB-dependent Gene Expression in Interleukin-1β-stimulated Synovial Fibroblasts. J Biol Chem 278:32861–32871. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Wu H, Chim SM, Zhou L, Zhao J, Feng H, Wei Q, Wang Q, Zheng MH, Tan RX, Gu Q, Xu J, Pavlos N, Tickner J, Xu J. 2013. SC-514, a selective inhibitor of IKKβ attenuates RANKL-induced osteoclastogenesis and NF-κB activation. Biochemical Pharmacology 86:1775–1783. [DOI] [PubMed] [Google Scholar]
- 16.Cheong R, Bergmann A, Werner SL, Regal J, Hoffmann A, Levchenko A. 2006. Transient IκB Kinase Activity Mediates Temporal NF-κB Dynamics in Response to a Wide Range of Tumor Necrosis Factor-α Doses. J Biol Chem 281:2945–2950. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan RK, Nolte H, Sun T, Kaur H, Sreenivasan K, Looso M, Offermanns S, Krüger M, Swiercz JM. 2015. Quantitative analysis of the TNF-α-induced phosphoproteome reveals AEG-1/MTDH/LYRIC as an IKKβ substrate. Nat Commun 6:6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee REC, Walker SR, Savery K, Frank DA, Gaudet S. 2014. Fold-change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol Cell 53:867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamprecht MR, Sabatini DM, Carpenter AE. 2007. CellProfiler: free, versatile software for automated biological image analysis. BioTechniques 42:71–75. [DOI] [PubMed] [Google Scholar]
- 21.Bray M-A, Vokes MS, Carpenter AE. 2015. Using CellProfiler for Automatic Identification and Measurement of Biological Objects in Images. Curr Protoc Mol Biol 109:14.17.1–14.17.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohman BK, Weber JN, Bolnick DI. Evaluation of TagSeq, a reliable low-cost alternative for RNAseq. Molecular Ecology Resources 16:1315–1321. [DOI] [PubMed] [Google Scholar]
- 23.Meyer E, Aglyamova GV, Matz MV. 2011. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Molecular Ecology 20:3599–3616. [DOI] [PubMed] [Google Scholar]
- 24.Hannon lab. FASTX Toolkit.
- 25.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team. 2011. R: A Language and Environment for Statistical Computing. Vienna Austria, : the R Foundation for Statistical Computing. [Google Scholar]
- 28.Castoreno AB, Eggert US. 2011. Small Molecule Probes of Cellular Pathways and Networks. ACS Chem Biol 6:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewan K, Pajak B, Stubbs M, Todd H, Barbeau O, Quevedo C, Botfield H, Young R, Ruddle R, Samuel L, Battersby A, Raynaud F, Allen N, Wilson S, Latinkic B, Workman P, McDonald E, Blagg J, Aherne W, Dale T. 2010. A useful approach to identify novel small-molecule inhibitors of Wnt-dependent transcription. Cancer Res 70:5963–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanters E, Pasparakis M, Gijbels MJJ, Vergouwe MN, Partouns-Hendriks I, Fijneman RJA, Clausen BE, Förster I, Kockx MM, Rajewsky K, Kraal G, Hofker MH, de Winther MPJ. 2003. Inhibition of NF-κB activation in macrophages increases atherosclerosis in LDL receptor–deficient mice. J Clin Invest 112:1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z-W, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M. 1999. The IKKβ Subunit of IκB Kinase (IKK) is Essential for Nuclear Factor κB Activation and Prevention of Apoptosis. Journal of Experimental Medicine 189:1839–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke JR, Pattoli MA, Gregor KR, Brassil PJ, MacMaster JF, McIntyre KW, Yang X, Iotzova VS, Clarke W, Strnad J, Qiu Y, Zusi FC. 2003. BMS-345541 Is a Highly Selective Inhibitor of IκB Kinase That Binds at an Allosteric Site of the Enzyme and Blocks NF-κB-dependent Transcription in Mice. J Biol Chem 278:1450–1456. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto Y, Yin M-J, Lin K-M, Gaynor RB. 1999. Sulindac Inhibits Activation of the NF-κB Pathway. J Biol Chem 274:27307–27314. [DOI] [PubMed] [Google Scholar]
- 34.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. General Principles of Cell Communication.
- 35.Werner SL, Barken D, Hoffmann A. 2005. Stimulus Specificity of Gene Expression Programs Determined by Temporal Control of IKK Activity. Science 309:1857–1861. [DOI] [PubMed] [Google Scholar]
- 36.Culig Z 2011. Cytokine disbalance in common human cancers. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1813:308–314. [DOI] [PubMed] [Google Scholar]
- 37.Mateen S, Moin S, Shahzad S, Khan AQ. 2017. Level of inflammatory cytokines in rheumatoid arthritis patients: Correlation with 25-hydroxy vitamin D and reactive oxygen species. PLOS ONE 12:e0178879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The CellProfiler image analysis pipeline is available upon request. All processing code used in TagSeq processing including RMarkdown files containing session information used in differential expression analysis is available at https://github.com/sachitsaksena/nfkb-tag-seq.