Abstract
Histone methylation plays an important regulatory role in chromatin restructuring and RNA transcription. Arginine methylation that is enzymatically catalyzed by the family of protein arginine methyltransferases (PRMTs) can either activate or repress gene expression depending on cellular contexts. Given the strong correlation of PRMTs with pathophysiology, great interest is seen in understanding molecular mechanisms of PRMTs in diseases and in developing potent PRMT inhibitors. Herein, we reviewed key research advances in the study of biochemical mechanisms of PRMT catalysis and their relevance to cell biology. We highlighted how a random binary, ordered ternary kinetic model for PRMT1 catalysis reconciles the literature reports and endorses a distributive mechanism that the enzyme active site utilizes for multiple turnovers of arginine methylation. We discussed the impacts of histone arginine methylation and its biochemical interplays with other key epigenetic marks. Challenges in developing small-molecule PRMT inhibitors were also discussed.
Keywords: arginine methylation, histone code, inhibitors, enzyme kinetics, PRMT
Graphical Abstract
Histone arginine methylation is a high-profile epigenetic mark, and the enzymes that catalyze arginine methylation, protein arginine methyltransferases (PRMTs), have become attractive drug targets for different diseases including cancer. We discuss the different kinetic mechanisms, histone crosstalk, and progress with inhibitor development.
1. Protein Arginine Methylation
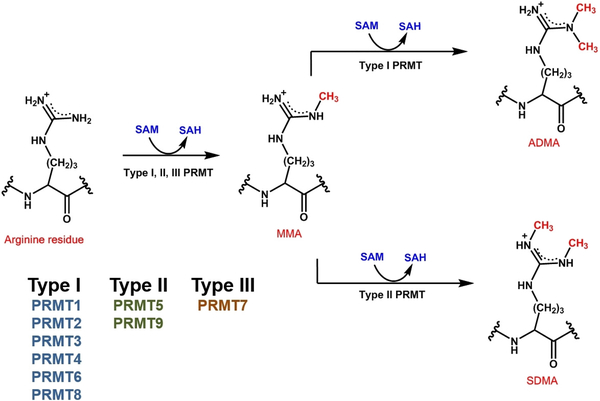
Arginine methylation is a universal post-translational modification (PTM) conserved in all eukaryotic organisms, from yeasts to humans.[1] Biochemically, arginine methylation in mammalian systems is mediated by the family of N-arginine methyltransferases (PRMTs) that catalyze the transfer of a methyl group from S-adenosyl-L-methionine (SAM or AdoMet) to one or both omega nitrogens located at the terminus of the side-chain guanidino group on an arginine residue.[2] PRMTs belong to the class I of SAM-dependent methyltransferases, which bind SAM with a Rossmann fold-like region.[3] Thus far, 9 PRMT members are found in mammalian cells,[4] among which PRMT1, –2, –3, –4, –6, and –8 are grouped into type I enzymes that catalyze methylation of the arginine residue to NG-monomethylarginine (MMA, Rme1) and further methylate MMA to produce asymmetric NG,NG-dimethylarginine (ADMA, Rme2a). PRMT4 is also well known as coactivator associated arginine methyltransferase 1 (CARM1). PRMT5 and PRMT9 are grouped into type II enzymes that produce MMA and symmetric NG, N’G-dimethylarginine (SDMA, Rme2s).[5] Also, while not a PRMT family member, it is noteworthy to recognize there is another mammalian class I methyltransferase, NDUFAF7, which has been observed to serve a critical role in the symmetric dimethylation of Arg-85 in complex I subunit NDUFS2 of the mitochondrial electron transport chain.[6] PRMT7 is grouped as a type III enzyme that only catalyzes the conversion of an arginine residue to MMA (Figure 1).[7] Among the PRMT members, PRMT1 and PRMT5 play the most prominent contributions to the arginine methylation levels in mammalian cells. Biochemical tests showed that PRMT1 is the major enzyme for arginine methylation in the RAT1 fibroblast cells.[8] Knockdown of PRMT1 gene in mouse embryonic fibroblast (MEF) cells leads to ~58% loss of the normal steady-state levels of ADMA.[9] On the other hand, Knockdown of PRMT5 in MEF cells largely abolished the bulk of SDMA signals (~ 95%), supporting that PRMT5 is the predominant enzyme responsible for SDMA formation.[10]
Figure 1.
Chemical reaction of arginine methylation.
The activity of PRMT9 seems to be unique. In MEF cells, PRMT9 is only responsible for a small percentage of SDMA production (~5%), including the methylation of the splicing factor SF3B2 (SAP145).[10–11] Chromatographic analysis showed that the main product of PRMT9 is MMA, with SDMA being a very small fraction.[10] Therefore, one may regard PRMT9 as a predominantly type III enzyme, with tangible type II activity. This argument is rational in that the phylogenetic sequence analysis showed that PRMT9 is most close to the type III member PRMT7, and both PRMT9 and PRMT7 contain two SAM binding domains.[12] As such, it can be envisaged that PRMT9 would exhibit similar enzymatic activity as PRMT7. Furthermore, human PRMT9 (hPRMT9) was suggested to be an ortholog of C. elegans PRMT3 (UniProt O02325) which was shown to methylate recombinant human histone H2A protein to form MMA only.[10,13]
Proteomic screens show that arginine methylation occurs in numerous cellular proteins, which implicates that PRMTs regulate multifaceted biological processes in cell function.[14] Thus far, biological research in arginine methylation is particularly concentrated on three types of protein targets:[15] DNA-binding proteins, RNA-binding proteins, and signaling proteins. Methylation of many DNA-interacting proteins, histones in particular, showcases the importance of PRMTs in epigenetic regulation of DNA-templated pathways including RNA transcription, DNA replication, and DNA damage repair. Secondly and interestingly, many RNA-binding proteins are heavily arginine methylated, such as PABPN1, Sam68, hnRNPs, and Lsm4, which highlights that arginine methylation is likely extensively involved in various RNA post-transcription processes such as RNA processing and mRNA-templated protein translation.[16] Last, but not least, many signaling proteins are also arginine methylated, such as ERa,[17] Smad6,[18] and NF-kB.[19] Therefore, arginine methylation could resemble protein phosphorylation, acting as a biochemical switch to turn on or off signal transduction.
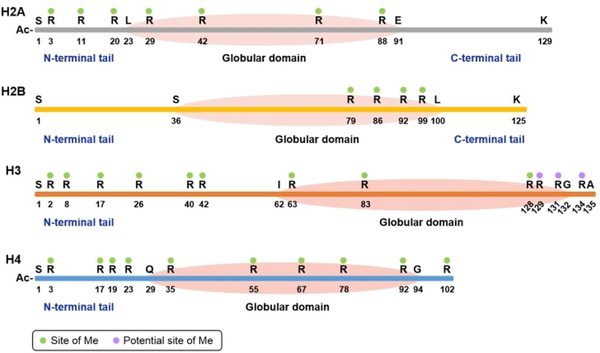
Histone arginine methylation has become a high-profile epigenetic mark since the outcome is directly associated with chromatin structure remodeling and gene transcription regulation.[20] Multiple arginine methylation sites have been identified based on proteomics and biochemical analysis (Figure 2).[14,21] The best-studied methylations are on the N-terminal regions of histones H3 and H4, including H3R2, H3R17, and H4R3. The authenticity and functions of the other methylated arginine residues, particularly in the globular and C-terminal regions, have yet to be elucidated. Many studies on histone modifications have revealed intricate crosstalks between arginine methylation and certain lysine acetylation/methylation.[22] Yet, since quite a few other histone post-translational modifications (PTMs) such as phosphorylation, ubiquitination, glycosylation, and glutamate methylation, are in proximal distance to arginine methylation sites, we project that arginine methylation-involved crosstalk pathways will continue to be intensely investigated in order to crack the molecular language of histone codes.[21b,23]
Figure 2.
Summary of arginine methylation sites in nucleosomal core histones.
2. The Kinetic Model of PRMT Catalysis
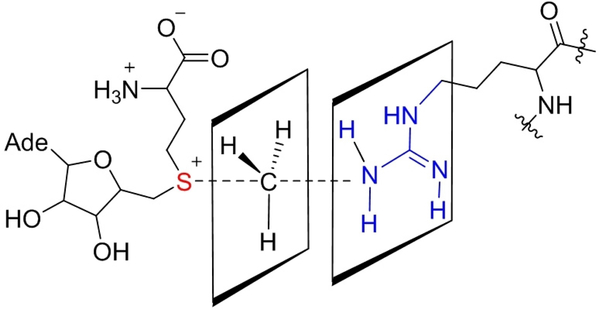
In respect to the organic reaction, PRMTs utilize a sequential ternary complex mechanism to catalyze arginine methylation, in which both SAM and the arginine substrate are required to bind to the active pocket of PRMTs to form a tri-molecular complex. Upon binding, the guanidino group on the arginine residue is within spatial proximity to the reacting methyl group of SAM for nucleophilic attack. The methyl transfer is a SN2 reaction with a guanidino nitrogen attacking the methyl carbon and SAH being the leaving group.[24] In principle, the transition state of arginine methylation adopts a sp2 hybridized planar geometry at the carbon center of the reacting methyl group (Figure 3). X-ray crystal structures showed that the planar guanidino group is approximately perpendicular to the NCS axis.[25] This indicates that the parallel guanidino plane likely stabilizes the sp2-hybrized planar methyl group in the transition state through a π-π stacking interaction (Figure 3). A neighboring allyl chemical group that stabilizes such a planar configuration in the transition state can potentially facilitate the carbon transfer rate.[26]
Figure 3.
A proposed transition-state structure of arginine methylation reaction. Although not yet experimentally verified, in principle, CH3 should be in planar geometry due to the sp2 hybridization. Based on the crystal structure, we hypothesize that the guanidino plane is in parallel to the CH3 plane in the transition state.
Determining the binding order of SAM and protein/peptide substrate in the pathway of forming the PRMTSAM-substrate ternary complex is an important issue in PRMT biochemistry. From an enzymology perspective, it would be interesting to see if PRMT methyltransferase stands alone as a unique enzyme class or has similar propensities to other types of methyltransferases. From a cell biology perspective, enzyme kinetics will help to elucidate the pathway of monomethylarginine and dimethylarginine formation at the molecular level. Further, from the drug discovery perspective, the order of cofactor and substrate binding to the enzyme is of great value in providing mechanistic insight into designing enzyme inhibitors. It can be envisaged that one may design inhibitors that bind the apo, cofactor-bound, or substrate-bound form of the enzyme, depending on which forms exist in cells. Inhibitors with different modes of action likely will trigger different biological responses in disease models.
Several research groups previously have conducted steady-state kinetic studies of PRMT catalysis.[5b,27] Based on the inhibition patterns of products and dead-end analogs on the Michaelis-Menten data curves, Thompson et al. proposed a rapid equilibrium random Bi Bi mechanism for PRMT1,[27a] cPRMT5,[27b] and PRMT6,[28] all of which catalyze H4 methylation. Under such a kinetic schematic, SAM and substrate would bind PRMT randomly, and after the methyl transfer, the products SAH and methyl-substrate are released from the enzyme pocket in a random fashion. Jacques et al. also proposed the similar random kinetic mechanism for the activity of human CARM1 in methylating histone H3.[27c]
On the other hand, steady-state kinetic studies performed by others support that an ordered sequential kinetic mechanism for PRMT1,[29] PRMT2,[27f] and PRMT6.[27g] In this model, SAM binds the enzyme active pocket first, followed by substrate binding and, after the methyl transfer, the methylated arginine product is the first to dissociate from the enzyme followed by SAH release. The mandatory ordered sequential mechanism typically applies to those cases that binding of the first substrate triggers a conformational change in the enzyme which is a prerequisite for the second substrate to bind, or possibly the first substrate provides essential contact points for the second substrate to bind.
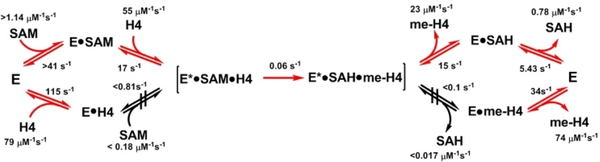
Recently, we used stopped-flow fluorescence together with global kinetic simulation to dissect the transient kinetics of PRMT1 catalysis.[30] Histone H4 peptides were used as the methyl acceptor as histones are important arginine methylation substrates. From our studies, we found that the fluorophores on the substrate (i.e., fluorescein labeled peptide) or on the enzyme (i.e., intrinsic tryptophan residues) are highly sensitive to the events of substrate-enzyme interaction and enzyme conformational changes.[30–31] By careful examination and modeling of the transient kinetic data, we were able to calculate the microscopic rate constants for virtually all individual elementary steps of the binary complex formation/decomposition and ternary complex formation/decomposition. Availability of these individual kinetic rate constants allows us to generate a complete kinetic model of PRMT1 catalysis. In this model, PRMT1 catalysis follows a catalytic schematic that we termed random binary, ordered ternary kinetic mechanism in which the cofactor or the peptide can bind PRMT1 randomly to form a binary complex; however, subsequent formation of the catalytically competent E-SAM-H4 ternary complex follows a kinetically ordered pathway, with SAM binding first and substrate binding second. The product release follows a sequential order: peptide product is released first and then SAH (Figure 4). It is worth stressing that steady-state kinetic studies are valuable to define the overall catalytic properties of an enzyme, especially the Km and kcat values, as well as the effect of product and dead-end inhibitors. However, steady-state kinetic experiments are of very limited utility in identification of enzyme conformational changes and rate-limiting steps in the catalytic pathway, and understanding the formation and reactivity of enzyme intermediates.
Figure 4.
A proposed kinetic model of PRMT1-catalyzed arginine methylation. The mechanism was collectively determined based on the stopped-flow measurements and global fitting of the time courses of the intrinsic tryptophan fluorescence of PRMT1 upon binding to SAM, SAH, or H4 peptide at different orders, as well as the fluorescence of fluorophore labeled H4 peptides during methylation reaction. No-Go symbols denote that those steps are kinetically hindered. See more details in ref.[30]
Several important mechanistic insights were revealed from our transient kinetics studies. The most important discovery is on the order of SAM and peptide substrate binding to PRMT1. Both our biochemical binding data and others showed that the cofactor SAM/SAH and the peptide substrate can bind to PRMT1 independently.[27d,32] Although this phenomenon at first glance would be in agreement with a random kinetic model of PRMT1 catalysis, our transient kinetic studies dig further into the details of the ternary complex formation, methyl transfer, and decomposition of the ternary complex, which ultimately leads to a conclusion that contradicts random sequential mechanism. An interesting finding from the transient kinetic study is that binding of the H4 substrate to PRMT1 does not appear to be catalytically competent or productive because the PRMT1-H4 binary complex is strongly hindered to interact with SAM to form the ternary complex (Figure 4). Considering this kinetic impediment, we predict that the enzyme conformation in the PRMT1-H4 binary complex would be different from that in the ternary complex. Importantly, the dissociation rate of PRMT1-H4 complex is much faster than the ternary complex formation rate. Therefore, the PRMT1-H4 complex, even formed, is doomed to dissociate back to free peptide and apo enzyme, which will bind SAM and then H4 peptide to form active ternary complex undergoing methylation. Reciprocally, the proposed mechanism is also supported by our finding that product release follows an ordered fashion, with peptide dissociation followed by release of the byproduct SAH (Figure 4).
We want to stress that our random binary and ordered ternary kinetic mechanism matches well with the steady-state kinetic data reported in the literature.[27a–c,28] On the one hand, the patterns of product and dead-end inhibition in steady-state kinetics experiments for our random binary, ordered ternary kinetic model should be very similar to that of classic rapid equilibrium random mechanism because SAM/SAH and substrate/product bind to the enzyme randomly in both models. Purely by measuring product and dead-end inhibition pattern in steady-state kinetics experiments, it would be difficult, if not impossible, to distinguish the rapid equilibrium random kinetic model from the random binary, ordered ternary mechanism that we proposed. On the other hand, when low-affinity peptide substrates are used, the contribution of enzyme-substrate binding (i.e., E-H4 formation) would be negligible and can be ignored in the kinetic scheme, in which case the steady-state kinetics would behave like a standard ordered sequential mechanism. This may explain why certain steady-state kinetic studies showed that PRMT catalysis follows an ordered sequential mechanism.[27f,g,29] Together, we believe that our transient kinetic model provides a mechanistic explanation that unifies the seemingly contradictory steady-state kinetic studies reported in the literature.
Our kinetic model in which efficient formation of the ternary complex requires an ordered binding of SAM (first) and peptide substrate (second) agrees well with the reported PRMT structural studies. The catalytic cores of all PRMT members share a ~320-residue-long, highly similar structural architecture that comprises an N-terminal Rossmann fold-like region and a C-terminal seven-beta strand barrel region.[33] The Rossmann fold and the b-barrel are connected by a conserved Z-shaped PX pucker (Pro175 in hPRMT1). The active site of the PRMTs, identified by the location of bound SAH and the methyl-accepting arginine, is located in a cleft between the Rossmann fold and the β-barrel domain. Importantly, all the X-ray structures show that the cofactor is buried underneath the N-terminal αX and αY-helices of PRMTs.[34] The reported X-ray crystal PRMT structures showed that cofactor binding induces salient changes of the enzymes’ conformation. In CARM1 crystal structures, motif I is shifted significantly upon SAH binding to interact with the cofactor.[34–35] Two residues of motif II (Gly195 and Ser196) and of the loop between helix F and G’ of the dimerization arm also change. Side chains of Arg169, Glu258, Gln160 and Met269 rearrange to recognize the methionine moiety of SAH. Prominently, binding of SAH renders the αX-helix sequence from an unstructured state to an ordered α-helix state.[34–35] As a matter of fact, this shift generates a T-shaped π-π interaction of Phe150 in the αX-helix region with the purine base of the cofactor.[34–36] Cofactor binding also saliently stabilizes αY-helix sequence, as seen in the PRMT8 structure in which αY-helix sequence is ordered in the presence of SAH but unstructured without SAH.[37] Overall, the structural reorganizations in PRMTs induced by cofactor binding seem to be an essential step for the subsequent recruitment of peptide substrate to the enzyme’s active pocket, which concludes with our kinetic model of ordered ternary complex formation. This is further reflected in the fact that none of PRMT-peptide binary complex structures have been reported. All the peptide-containing PRMT structures contain SAH or cofactor analogs. In regards to ordered product release, the structural evidence is also clear: SAH is deeply buried and its entrance is blocked by both the substrate and α-helix. Thus, the substrate release seems to be a prerequisite for SAH to dissociate from the active pocket.[36] Given that all type I PRMT structural folds are highly conserved, the mechanism of kinetics we proposed should be applicable to all the family members.
3. Processivity of Arginine Methylation
Arginine residues in proteins can be modified into monomethylated state and then to dimethylated state. In theory, PRMTs can maneuver the formation of dimethylarginines through either a processive or distributive manner. In a processive arginine dimethylation mechanism, the arginine substrate remains bound within the active site of PRMT1 to consecutively fulfill two rounds of methylation. In this process, the monomethylated intermediate is not released into the bulk solution before the last turnover is completed. In contrast, in a distributive arginine dimethylation mechanism, the monomethylated substrate is released into the bulk solution after the first turnover reaction and rebinds the enzyme active site for the second round of methylation. Our kinetic model strongly supports that the active site of PRMT1 utilizes a distributive mechanism for the production of dimethylarginines. The first evidence is that the dissociation rate (9.1 s–1) of the monomethylated intermediate H4me1 from the ternary complex is much faster than the methyl transfer (0.034 s–1).[30] Thus, the monomethylated intermediate easily dissociates from the enzyme active site before SAH-SAM exchange and methyl transfer can occur. The second and more important support for distributive dimethylation comes from the finding that both the formation and decomposition of the ternary complex are an ordered process (Figure 4): after the chemistry step of methyl transfer, the monomethylarginine has to be dissociated from the enzyme active site, followed by SAH release. SAH cannot be released first from the ternary complex because it is kinetically hindered (<0.1 s–1). This mechanistic restriction unequivocally governs that PRMT1 utilizes a distributive mechanism for the production of dimethylarginines. Understanding this principle from the molecular level, removal of monomethylarginine from the enzyme active site is a prerequisite step for SAH-SAM exchange before the next round of turnover. Some researchers proposed that PRMT1 catalyzes dimethylarginine formation in a partially processive fashion and that the degree of processivity is affected by the affinity of substrate.[27d,38] Nevertheless, in our view, the processivity is an intrinsic propensity that is dictated by the kinetic and structural mechanism of PRMT catalysis, but not by environmental contexts. An ordered formation of the ternary complex and ordered release of products demands that monomethylated substrate must be evacuated from the active pocket so that SAH can be substituted with SAM prior to the next round of methylation. Therefore, different substrates and protein binding partners may alter the relative distributions of mono- and dimethylarginine formation along the progression course, but they cannot change the nature of the processivity of the catalysis in the active site.[39]
The nonprocessive feature of PRMT activity in forming dimethylarginines has been experimentally substantiated for several members: PRMT1,[27e] PRMT2,[27f] PRMT3,[27e] hPRMT4,[40] cPRMT5,[41] hPRMT5,[39] and PRMT6.[27g] Of note, MEP50 simply accelerates the reaction rate of arginine methylation by hPRMT5 and does not alter the nonprocessive propensity.[39] The mechanism of distributive arginine methylation provides valuable insights for understanding the dynamics and function of arginine methylation in biological systems. The distributive methylation mechanism of PRMTs would dictate that the genesis of monomethylarginines and dimethylarginines in cells are two independent, discrete biochemical reactions. Under a processive mechanism, MMA would be a transiently existing intermediate in the course of dimethylarginine formation, and thus MMA levels would be limited. Nevertheless, high MMA levels have been observed in different cells.[9b,14] Bedford et al. found that when PRMT1 was knocked out in MEF cells, ADMA levels greatly decreased while MMA levels significantly increased, which would argue that PRMT1 is the major enzyme for ADMA formation, but not for MMA formation, which is a concept that coincides with our proposal that formation of MMA and ADMA represents two uncoupled processes.[9b] The biological function of monomethylated histone H3R2 (H3R2me) opposes that of H3R2me2a,[42] which is evidence that the type I member PRMT6 mediates the monomethylation and dimethylation in separate processes at the same arginine residue site, coinciding with a distributive process.
It is worthwhile to emphasize that the mechanism that PRMTs utilize a distributive mechanism for arginine methylation does not contradict in any way some experimental observations that ADMA is more abundant than MMA in certain substrates and in certain cellular contexts.[43] Multiple factors can influence the abundance of MMA and ADMA in substrates. First of all, any chemical reaction, including arginine methylation, is a time-dependent process. If given a sufficiently lengthy period of time, substrate arginine residues will ultimately be fully dimethylated regardless of processive or distributive mechanism. Second, we and others have found that monomethyl arginine is a moderately better substrate than arginine.[27d,32] Therefore, at comparable concentrations, monomethylarginines can effectively compete with unmethylated arginines for the active site of PRMTs. Third, PRMTs and their substrates could be confined within a subcellular location or organelle. Such confinement would lead to a rapid increase in the local concentrations of PRMTs and monomethylated substrates, which speed up the conversion of MMA to ADMA. Also, liquid-liquid phase separation in cells may create enzyme-substrate droplets that greatly accelerate arginine methylation.[44] Fourth, the protein-protein interaction may introduce additional and enhanced binding for speedy transition of MMA to ADMA. In this regard, PRMTs have a general tendency to self- or cross-assemble into oligomeric architectures,[31a,37,45] a feature that can favorably increase local abundance of PRMTs to maximize their capacity to fully methylate their substrates. Furthermore, it is often seen that multiple methylating arginine residues are clustered at a Gly-Arg rich region in a target of PRMTs.[1b,46] Such spatial proximity of arginine residues would physically tether methylation sites around the PRMT active site, thus expediting multiple turnovers of arginine methylation. Clustering of multiple positively charged guanidino groups can also generate repulsive forces among them, the consequence of which would lead to decreased pKa of arginines’ side chain guanidino groups, thereby increasing their nucleophilicity in methyl transfer reaction.
4. Interplay of Arginine Methylation with Other PTM Marks on Histones
The major PRMT members (i.e. PRMT1 and PRMT5) are ubiquitously expressed in the cells, and so far there is no general mechanism reported for PRMT activity regulation. Expressed PRMTs likely are constitutively active. As such, regulation at the substrate level via PTM crosstalks can be an important mechanism for modulating arginine methylation levels. Nucleosomal histones are important substrates of PRMTs.[20,22,47] Like many other histone PTMs, arginine methylation is involved in multiple histone crosstalk events that comprise the histone code.[1a,22,48] The histone code hypothesis is stated as “multiple histone modifications, acting in a combinatorial or sequential fashion on one or multiple histone tails, specify unique downstream functions.”[49] In many cases, these downstream events involve the repression or activation of gene transcription as a result of one or a combination of histone PTMs promoting the recruitment or inhibition of other chromatin modifying enzymes that can subsequently remove or add PTMs to the histone N-terminal tails.[50] Since the bulk of the PTM crosstalk events that involve arginine methylation have been observed on histones H3 and H4, these are highlighted in the following sections with the focus on mammalian cell systems.
4.1. H3R2me2a
While an earlier report describes CARM1 methylating H3R2, PRMT6 is the main enzyme that deposits the H3R2me2a mark in vivo and in vitro.[21c,51] H3R2me2a is a mark of transcriptional repression and there is antagonistic crosstalk between H3R2me2a and a transcription activating mark, H3K4me3.[51a,b] The presence of H3K4me3/me2 is inhibitory to PRMT6 arginine methylation of H3R2, while the presence of H3R2me2a inhibits the methylation of H3K4 in vivo and in vitro by the MLL1 complex.[51] When both marks are present on the H3 N-terminal tail (H3R2me2aK4me3), the H3R2me2a mark prevents effector proteins PHF2, DATF1, BPTF, and WDR5 from recognizing H3K4me3.[51c] Thus, it is reasonable to conclude the crosstalk between H3R2me2a and H3K4me3 is mutually exclusive. There are likely other PTMs on H3 lysine residues that are affecting the methylation of H3R2 by PRMT6. Hyllus and colleagues observed an increase in arginine methylation by PRMT6 when the peptide substrate had H3K27me2/me3 while the presence of H3K4me2, H3K4me3, H3K9me2, and H3K9me3 inhibited arginine methylation by PRMT6 (Hyllus 2007 supplemental material).[51a] It is not clear if this crosstalk exists in vivo and how the combination of these marks contributes to regulating gene transcription.
4.2. H3R2me2s
H3R2me2s is a highly conserved mark from Drosophila melanogaster to humans that localizes with areas of euchromatin.[52] Immunoprecipitates of PRMT5 and PRMT7 can catalyze H3R2me2s on recombinant histone H3;[52a] however, PRMT5 may be more likely the enzyme candidate to catalyze this mark since PRMT7 catalyzes only MMA.[7a,53] In contrast to H3R2me2a, the H3R2me2s mark has a supportive crosstalk relationship with H3K4me3 that promotes transcriptional activation. H3R2me2s coexists with H3K4me3 predominantly at highly transcribed genes, and the presence of both marks (H3R2me2sK4me3) enhances the binding affinity of an H3 peptide by at least 23-fold to the V(D)J recombinase subunit, RAG2, in comparison H3K4me3 modified or H3 unmodified peptides (Yuan 2012 supplemental table 1).[52b] H3R2me2s also has high binding affinity (KD =0.1 μM) with WDR5, which is a subunit of the coactivator complexes ATAC, SET1A, SET1B, and MLL.[52a] The presence of H3R2me2s stimulates the lysine methyltransferase activity of MLL complex to deposit H3K4me/me2.[52a] In contrast, H3R2me2s prevents the recognition of H3K4me3 by RBBP7, which is a major component of co-repressor complexes PRC2, NURD, and SIN3 A.[52a]
4.3. H3R8me2s & H3R8me2a
PRMT5 catalyzes arginine methylation of H3R8, and the H3R8me2s mark is important for activating myogenic gene expression[54] yet it is also associated with promoting gene repression of tumor suppressor genes.[55] PTMs that appear to crosstalk with H3R8me2s are H3K9ac and H3K14ac. Using H3 peptides and recombinant PRMT5, Pal and colleagues demonstrated that H3K9ac and H3K14ac inhibit methylation of H3R8 by PRMT5 in vitro.[55a] Overexpression of PRMT5 in NIH 3T3 cells results in increased H3R8me2s and reduced H3K9ac, while the knockdown of PRMT5 results in abolishment of H3R8me2s and increased H3K9ac.[55a] While these results support the crosstalk between H3R8 arginine methylation and H3 lysine acetylation, there is also crosstalk between H3R8 methylation and H3K9 methylation. The lysine methyltransferase G9a can catalyze the methylation of H3K9 to produce H3K9me1/2/3, though H3K9me3 requires overnight incubation.[56] Interestingly, the presence of H3R8me2s significantly inhibits G9a methylation of H3K9 while substitution of H3R8 with another amino acid or the presence of H3R8me2a abolishes G9a activity.[57] The H3R8me2a mark appears to be deposited by PRMT2, since immunoprecipitation of PRMT2 from Xenopus embryos was able to methylate histone H3.3 while substitution of H3R8 for Ala resulted in reduced methylation of H3.3.[58] Though, it remains to be examined how H3K9me3 or the other methylated states could modulate PRMT5 or PRMT2 activity towards H3R8.
4.4. H3R17me2a & H3R26me2a
The major sites of CARM1 arginine methylation are H3R17 and H3R26, while H3R2 is a minor site for CARM1.[21c,59] Similar to H3R8, there is crosstalk between arginine methylation at H3R17 and H3 lysine acetylation. Acetylation of H3K18 and H3K23 by CBP recruits CARM1 to H3, and the H3K18ac and H3K23ac marks are recognized by CARM1 and promote arginine methylation at H3R17.[60] This crosstalk is associated with estrogen induced transcriptional activation.[59–60] Methylation at arginine 17 of H3 on promoters was later found to be linked with estrogen-receptor-regulated pS2 gene activation and with steroid-hormone dependent activation.[61] Arginine methylation by CARM1 was linked to lysine acetylation.[59–60] H3K18 and H3K23 acetylations by CBP precede and favor the high-affinity binding of CARM1/PRMT4 to chromatin. H3R17 is subsequently methylated, and this is followed by enhanced gene transcription. Once again, the ordered deposition of these modifications was suggested to cooperate in transcriptional activation.
Recently, crosstalk between H3R26me2a, H3K27me3, and H3K27ac was reported to be important for regulating HIV-1 LTR transcription in resting CD4+T cells.[62] The presence of the transcriptionally active mark, H3K27ac, stimulates asymmetric arginine methylation of H3R26 by CARM1, yet H3K27ac does not appear to have a significant effect on changing the levels of H3R17me2a.[62] In fact, H3R17 is still the preferred site of CARM1 since the lack of any acetylation on H3 results in predominantly H3R17me2a.[62] On the other hand, the presence of the transcriptionally repressive mark, H3K27me3, results in reduced H3R26me2a levels.[62] Hence, it appears that CARM1 prefers to methylate arginine residues that are adjacent (+1) to neutral charged, mainly hydrophobic residues. Since other acylation modifications (e.g., butyrylation, crotonylation, 2-hydroxyisobutyrylation) exist on H3K27 as well as H3K18,[21b] it would be interesting to see how CARM1 activity responds to different acylations at +1 position in the substrate.
4.5. H4R3me2a
Methylation of H4R3 is a well-conserved PTM in eukaryotic organisms, from yeast to humans, with Tetrahymena as an exception to this since there is a Gly3 instead of Arg3 in its histone H4 protein sequence.[63] PRMT1 is the main enzyme that methylates H4R3 in vivo.[63–64] PRMT1 deposits H4R3me and H4R3me2a, and this reaction is reproducible in vitro with recombinant PRMT1, SAM, and core histones or histone H4 peptides H4(1–20) and H4(1–21).[65] PRMT1-catalyzed methylation of H4R3 recruits other chromatin modifying enzymes that deposit PTMs on histones to promote transcriptional activation. A classic example is the relationship between PRMT1 and p300. Wang and colleagues first demonstrated that H4R3 methylation by PRMT1 is activating for p300 lysine acetylation on H4, specifically H4K8 and H4K12.[66] Additionally, there is evidence for a trans mechanism of crosstalk between H4R3me2a and H3 lysine acetylation. ChIP studies performed by Huang and colleagues on the chicken β-globin locus in the chicken bone marrow cell line, 6C2, demonstrated in vivo that PRMT1 is important for the overall acetylation of H4 and H3 at the β-globin locus.[67] Knockdown of PRMT1 in 6C2 cells decreases dimethylation at H4R3 and decreases acetylation at H3K9, H3K14, H4K5, H4K12, and to a lesser degree H4K8.[68] Similarly in MEL cells, knockdown of PRMT1 results in reduced enrichment of H4 and H3K9/K14 acetylation at the βmaj-promoter.[69] This is complemented by the in vitro acetyltransferase activity assays demonstrating that PCAF, and partially p300, prefer to acetylate H3K9 and H3K14 after PRMT1 has catalyzed arginine methylation of nucleosomes.[69] Hence, simply the presence of the methylated arginine residues (H4R3me/me2a) on the nucleosomes is enough to stimulate PCAF and p300 acetyltransferase activity, though the molecular mechanism is still unclear.
While asymmetric methylation of H4R3me stimulates histone acetylation, specific sites of lysine acetylation on H4 can be inhibitory to arginine methylation by PRMTs. H4K5ac, H4K5acK8acK12ac, and H4K5acK8acK12acK16ac inhibit arginine methylation of H4 by PRMT1.[65b,70] Furthermore, all H4K5 acylations (H4K5ac/pr/bu/cr/hib) are inhibitory to arginine methylation by PRMT1.[70b] Interestingly, knockdown of PRMT1 increases the population of transcriptionally repressive PTMs, H3K27me3 and H3K9me2, and decreases expression of the erythroid-specific folate receptor.[68]
4.6. H4R3me2s
PRMT5 is responsible for catalyzing symmetric dimethylarginine methylation of H4R3.[55a] Early studies with PRMT5 hint to a potential crosstalk between symmetric arginine methylation and DNA methylation in that PRMT5 activity was connected with transcriptional repression along with the recruitment of methyl CpG binding protein 2 (MBD2) to CpG islands.[55a,71] Notably, the studies of the human β-globin locus by Zhao and colleagues demonstrated that this crosstalk exists between PRMT5 and DNA methyltransferase 3A (DNMT3A).[72] PRMT5 deposits the H4R3me2s mark, and this mark serves as a docking site for DNMT3A to methylate CpG islands and silence gene transcription.[72] The PRMT5-activity dependent recruitment of DNMT3A also includes casein kinase 2α (CK2α) and the histone lysine N-methyltransferase SUV4–20h1, which together form a repressive protein complex that promotes DNA methylation of CpG islands and the repressive mark H4K20me3.[73] In addition, unlike H4R3me2a and unmethylated H4R3, H4R3me2s inhibits the binding of the tandem plant homeodomain (PHD4–6) of MLL4, and thereby prevents the docking of MLL4 to H3K4 for subsequent deposit of the gene activating mark H3K4me3.[74]
There is also crosstalk with lysine acetylation and methylation. H4K5ac promotes H4 arginine methylation by PRMT5 in vivo and in vitro, while H4K8ac and H4K12ac do not have as strong of an activating effect on PRMT5 activity.[65b,70b,75] Interestingly, including H4K20me3 with H4K5ac or H4K12me3 with H4K16ac are strongly activating PTM combinations for PRMT5 activity.[75] It is not clear though how H4R3me2s may affect HAT activity on the H4 lysine residues.
4.7. H4R17me
H4R17 is a site for monomethylation by PRMT7.[5a,7a,53] Recently, Jain and colleagues have observed in vitro with H4(1–21) peptides that the H4R17me mark strongly stimulates PRMT5 activity in comparison to the unmodified peptide substrate (~4.9-fold increase in catalytic efficiency, kcat/K0.5).[76] H4R17me is also activating for PRMT1, albeit to a lesser extent (~2.4-fold increase in catalytic efficiency, kcat/K0.5). This raises the question of how and when cells use PRMT7 to promote the type I mark H4R3me2a vs. type II mark H4R3me2s on histones. These results may explain why others have observed PRMT7 to be important for regulating the levels of H4R3me2s in cells.[74] Since this is a recent key discovery, there is still need to examine this crosstalk in cells, especially in regards to its role in transcriptional regulation.
4.8. Histone PTM Crosstalk Beyond Arginine Methylation
While we have largely focused on the major crosstalk events between two chromatin modifying enzymes (e.g., PRMT-PRMT or PRMT-HAT) that impact arginine methylation on histone tails (Figure 5), these highlighted cases are just snippets of a larger picture of histone PTM interplay. For example, we have mentioned in the above that H3K4me3 and H3R2me2a are mutually exclusive marks. Yet, what regulates the methylation state of H3K4? H3K4 is trimethylated by the MLL1 complex, and H3K4me3/2 is demethylated by lysine demethylase 5 family members (KDM5A/B/C/D) while H3K4me2/1 is demethylated by lysine specific demethylase 1 (LSD1).[51a,77] However, when PKCb1 phosphorylates Thr6 on H3 (H3T6ph) then the presence of H3T6ph changes the substrate preference of LSD1 to target methylated H3K9 instead of H3K4me1/2.[78] Furthermore, the presence of H3T6ph inhibits KDM5B from demethylating H3K4me3 to H3K4me in vitro, which results in H3K4me2 as the predominant product.[78] On the other hand, dimethylated H3R2 is reported to be a site for demethylation by Jumonji domain containing 6 protein (JMJD6), and H3R2 is a potential site for deimination by peptidylarginine deiminase 1 (PADI1) to produce citrulline.[79] Hence, while we have highlighted major crosstalk events between arginine methylation and other histone PTMs, there is a larger picture to appreciate.
Figure 5.
Crosstalk of arginine methylation with other PTMs on the histone N-terminal tails.
There are also some differences in outcomes of the crosstalk events between higher and lower eukaryotes. The mutual exclusivity of H3K4me3 and H3R2me2a is conserved from yeast to humans.[51b,80] In contrast, while acetylated H4 is inhibitory to PRMT1 catalyzed methylation of H4R3 in mammalian cells,[65b,70] acetylated H4 (particularly H4K8ac) is a preferred substrate for the PRMT1 functional homologue in yeast, hnRNP arginine N-methyl methyltransferase 1 (HMT1), and the resulting H4R3me2a mark is transcriptionally repressive in yeast.[81] These and other crosstalk events have been well reviewed by others.[1a,20,22,47,82]
The diversity of histone PTMs that have been discovered is remarkable, and yet there is still much to learn about the functional significance of the crosstalk between histone PTMs. For example, human biotinidase catalyzes the biotinylation of histones at H4K12, H4K8, H3K4, H3K9, and H3K18.[83] Thus far, H3R2me2R8me2, H3R8me2, H3Ornithine8, and H3R17me2 promote biotinylation of H3 and H3S10ph is inhibitory to H3 biotinylation.[83a] While it is not clear whether the dimethylated arginine residues that activate biotinidase activity are ADMA or SDMA, there is still more to learn in regards to how biotinylated histones affect chromatin structure and gene transcription. Also of note is the crosstalk at the tip of the H4 N-terminal between phosphorylation, N-terminal acetylation, and arginine methylation. Based on a histone peptide assay data, H4S1ph dominates at inhibiting PRMT5 activity in vitro despite the presence of other PTMs such as H4R3me and various H4 lysine methylation and acetylation marks.[75] Yet, in chromatin fractions collected during the later embryonic stages of Xenopus laevis, high levels of H4S1ph have been observed in the midst of high levels of H4R3me, H4Rme2s, and even H4R3me2a in vivo.[84] In these cases when both PTMs are present on the same H4 N-terminal tail, arginine methylation may have preceded phosphorylation. However, the presence of H4S1ph may be influenced by the acetylation state of the alpha amino group of H4S1, since recent evidence supports that CK2α prefers to bind to an H4 peptide without N-terminal acetylation versus an H4 peptide that is N-terminally acetylated.[85] While we have not observed a preference for the predominant PRMTs (PRMT1 and PRMT5) to methylate H4 with or without N-terminal acetylation, the presence of N-terminal acetylation may serve to prevent phosphorylation of H4S1 and thereby provide a better substrate for PRMTs to methylate H4. In contrast, the lack of N-terminal H4 acetylation by NatD can promote phosphorylation of H4S1 and may be overall inhibitory to PRMT catalyzed methylation of H4R3. This would be interesting to study in context to R17me that appears to allosterically activate PRMT5 and PRMT1 activity towards H4. Finally, while we have covered the main histone PTMs that involve arginine methylation on H4 and H3, the histone PTMs of H2A and H2B are less well studied, yet important for elucidating the complex language of the histone code.
5. Isoform-Selective PRMT1 Inhibitors
With an ability to modify gene expression as well as other protein activities, PRMTs have been found in numerous studies to be associated with pathology of various human diseases, such as cancer and inflammation.[86] Aberrant expression of PRMT1 is found in breast, prostate, lung, colon, and bladder cancers, neuroblastoma, and leukemia.[86e,87] Expression of PRMT1 variant 2 in colon cancer patients or high expression of PRMT1 in breast cancer patients is associated with poor prognosis.[88] PRMT1 is an essential component of the mixed lineage leukemia (MLL) oncogenic transcriptional complex and confers an aberrant transcriptional activation property critical for the induction of leukemia.[89] Specific knockdown of PRMT1 expression suppresses MLL-mediated transformation. PRMT1 is also upregulated in pulmonary diseases such as pulmonary fibrosis, pulmonary hypertension, chronic obstructive pulmonary disease (COPD), and asthma.[90] Further, PRMT1 plays regulatory roles in cardiovascular disease,[91] diabetes,[92] and renal disease.[93] Therefore, the development of PRMT inhibitors has emerged as an imperative task to provide novel therapeutic agents to treat diseases and to find chemical probes to investigate the biological functions of PRMTs.[12b,94]
Biochemical assays for PRMT inhibitor discovery and characterization generally involves radiometric assays in which radiolabeled methyl groups from SAM are transferred to the substrate followed by the separation of labeled substrate via gel electrophoresis or filtering on glass fiber or phosphocellulose paper discs. Products can be quantified by either fluorography or liquid scintillation counting.[94a] Washing steps are unnecessary in the format of scintillation proximity assay (SPA) in which the substrate is labeled with biotin and the scintillants are encapsulated in streptavidin-coated microsphere beads.[94a,95] The scintillation signal requires the close proximity of the radiolabel and the scintillant, and the binding of biotin to the streptavidin brings the substrate within micrometer ranges to produce optical signals while the free SAM in the bulk solution is not tethered to the bead and remains distant from the scintillant, thereby resulting in low interference with sample signal. In addition to radioactive assays, we have been actively investigating fluorescent methods to study PRMT activity and inhibitors. In particular, we found that fluorophore-labeled histone H4 peptides are highly sensitive to PRMT1 association and catalysis.[31,96] Either fluorescence intensity or anisotropy signals can be measured to determine the KD of enzyme binding with substrate or substrate-competitive inhibitors.[31b,96] Moreover, we used fluorophore-labeled histone H4 peptides in transient kinetic experiments to dissect the rate constants of substrate/product interactions with PRMT1 and methyl transfer reaction.[31a] Our data showed that methyl transfer is the rate-limiting step in PRMT1 catalysis. Of interest, we recently found that the stopped-flow fluorescence can be effectively applied to quantify inhibitory potency (IC50) of PRMT1 inhibitors as well as examine their mode of inhibition.[97] Our efforts were also applied to create fluorophore-labeled SAM analogs to set up fluorescent-based binding assays to study cofactor competitive inhibitors.[98] For unknown reasons, we did not detect any salient binding of the fluorescent SAM analogs with PRMTs (unpublished data). It may be that the PRMTs cannot accommodate the fluorescein group, which is supported by previous observations that certain SAM analogs of lesser bulk have also been poor cofactors in the absence of active site engineering.[99]
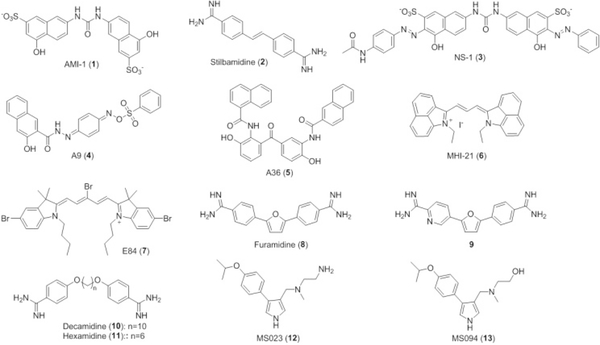
The first set of small molecule inhibitors were developed by the Bedford group in 2004, the most potent of which was AMI-1 (1).[96,100] More inhibitors soon followed such as stilbamidine (2) and allantodapsone.[96] Armed with this knowledge, our group thus set out to inhibit PRMT enzymes in a variety of fashions. The first compounds were discovered from a virtual screening against rat PRMT1 the most potent of which (IC50 =12.7 μM) were noted to resemble AMI-1 (NS-1, 3).[96] These compounds all contained ridged, planar, conjugated systems with naphthalene groups along with charged sulfonate and hydroxyl moieties. Further kinetic assays showed that 3 was competitive with respect to H4 and noncompetitive with respect to SAM; however, with its negative charges, 3 was not predicted to fit into the substrate arginine binding pocket, so its mode of inhibition had to be different.[96] It was then revealed that 3 inhibits PRMT function not by binding to the enzyme itself, but by forming a complex with its substrate thus preventing substrate binding.[96] This finding led our group to subsequently perform a screening of compounds based on the pharmacophore established by the previous work. The resulting six hits could be divided into two groups based on similar structures with group I containing naphthalene groups and phenolic hydroxyls, and group II containing a heterocyclic thiazole.[101] Group I compounds were found to be more effective than group II compounds, and the two compounds A9 (4) and A36 (5) had IC50 values of 42 μM and 12 μM, respectively which are stronger than 1 (77 μM) and comparable to 3.[101] Kinetic and fluorescent studies also showed that 4 and 5 appeared to inhibit in the same manner as 3; however with its charged sulfonate groups, 3 is predicted to have a lower bioavailability than either 4 or 5.[101] In vivo studies using LNCaP C81 cells showed that 4 had a marked reduction in cell proliferation at 10 μM in both normal and steroid reduced media; 5 showed a much weaker biological activity which was proposed to be due in part by its size.[101]
While this approach differs from a more classical enzyme binding approach, it does have both pros and cons in regards to pharmacological impacts. The advantage is that these compounds are potentially histone substrate-specific chemical probes. The shortcoming is that the bound peptides may contain multiple modification sites that are otherwise blocked by inhibitor binding; in other words, this method may not be selective for just PRMT1. For example, the histone acetyltransferase p300 was inhibited by both 3 and 5 with potency similar to their respective PRMT1 inhibition.[96,101]
Following this, our group sought to inhibit the enzyme itself in a more selective fashion. A series of cyanine compounds were developed for PRMT1 inhibition, with two representative leads (compounds 6 and 7) shown in Figure 6.[102] Compound 7 was found to be the most selective inhibitor for PRMT1 with specificity for it over CARM1, PRMT5, and PRMT8 ranging from 6- to-25 fold.[102b] Cell proliferation assays with 7 resulted in significantly reduced the growth of three leukemia cell lines: Meg01, MOLM13, and HEL cells with HEL cells requiring a higher concentration (200 nM vs 100 nM) due to their mutation in JAK2 that makes them less dependent on PRMT1 signaling.[103] The cyanine molecules typically possess long-wavelength absorption and fluorescence properties which allow them to be used for visualization in cells and tissues. By using the optical and fluorescent microscopy techniques, we found that MHI-21 (6) is capable to cross the plasma membrane and localize in the nucleus.[102a] Therefore, the cyanine-type of compounds provide unique photoactive chemical probes for both PRMT inhibition and microscopic imaging.
Figure 6.
Representative small molecule PRMT1 inhibitors.
Another class of PRMT inhibitors is the diamidine compounds like 2. With its positive charge and planarity, the amidine moiety is a mimic of the guanidino moiety on the substrate arginine residue and is theorized to be structurally competitive for its binding site.[102a] We screened a focused set of diamidine compounds against PRMT1 and identified furamidine (DB75, 8) as a lead inhibitor.[103] It was found that the addition of alkyl or phenyl substituents onto the amidine reduced its activity.[103] Substitution of the oxygen on the furan core with an isostere such as sulfur or selenium had no effect, while substitution to nitrogen reduced activity.[103] Finally, ortho-phenyl substituents resulted in a drop in activity, whereas the meta position was less sensitive.[103] 8 was shown to have IC50 of 9.4 μM for PRMT1, with 17-, 30-, and >42-fold selectivity against PRMT5, PRMT6, and CARM1 respectively.[103] Molecular modeling studies showed that the binding of compound 8 is promoted by one amidine group extending into an acidic channel in the substrate arginine binding site and interacts with Glu144 and Glu153, and the other amidine extends into the SAM binding site to interact with Glu129 while Try35, Phe36, and Tyr39 form π-π interactions with the aromatic regions.[103] The selectivity against PRMT5 is also explained by a partially, solvent-exposed binding, pocket that disfavors binding of 8.[103] Another inhibitor, compound 9, was also found in this study with comparable potencies (7.2 mM) and selectivity to 8.[103] Furamidine was also shown to be cell permeable via its inhibition of GFP-ALY methylation in 293T cells, and studies in leukemia cells lines showed that growth was inhibited in most of the cell lines at 20 μM with those derived from Down’s Syndrome patients (such as MOLM13 and CMY) being the most sensitive.[103] Furamidine was applied as a chemical probe to illuminate the functions of PRMT1 in bone morphogenetic proteins (BMP)-induced Smad signaling[18] and RNA-binding motif protein 15 (RBM15)-controlled, RNA splicing pathway.[104]
In a recent study, our group made additional diamidine compounds and showed that the potency could be increased by increasing the length of the middle linker.[105] Compound 10, decamidine, was identified as having the more potent PRMT1 inhibition than 8; however, this increased potency came at a cost of selectivity over PRMT5.[105] Another compound in the study 11 had a decreased potency (52 μM), but increased selectivity.[105] Molecular docking studies revealed different binding modes for 10 and 11. While 11 has a binding mode similar to 8, one of the amidines on 10 extends into the cofactor methionine binding site. When docked with PRMT5, compound 11 spans both the cofactor and arginine sites for PRMT1, but only the cofactor site for PRMT5. For compound 10, it binds to PRMT1 and PRMT5 similarly. This may partially explain the low selectivity of 10.[105]
Another type of inhibitor, MS023 (12), was developed by Eram et al. using a scaffold from two previously developed inhibitors: CMPD-1 and EPZ020411.[106] Through SAR analysis, the authors found that using a pyrrole ring over a 1,2,3-triazole ring had a 70-fold increase in inhibition for all type I PRMTs except PRMT6 where it showed a 10-fold increase. Substitution of a meta-trifluoromethyl group to a para-isopropoxy substituent increased inhibition potency against all type I PRMTs (30 nM for PRMT1, 119 nM for PRMT3, 83 nM for PRMT4, 4 nM for PRMT6, and 5 nM for PRMT8).[106] The exchange of the terminal amino group for a hydroxyl group afforded MS094 (13) which completely abolished activity and served as a great control compound in assays.[106] Increasing concentrations of peptide substrate and SAM showed no change in the IC50 values, indicating that 12 is noncompetitive for both substrate and cofactor across all human PRMTs, with the exception of PRMT3 in which the authors found that it was noncompetitive with substrate and uncompetitive with SAM.[106] Another unique quality of 12 was found during selectivity assays in which it was shown that 12 had unprecedented selectivity for type I PRMTs against type II/III PRMTs, protein lysine methyltransferases, DNA methyltransferases, and three histone lysine demethylases.[106] In cellular assays, 12 was shown to decrease H4R3me2a marks in MCF7 cells in a concentration dependent manner with 13 having no effect.[106] After treating both the MCF7 and HEK293 cells for two days with 12, the authors noted a global decrease in ADMA and concurrent increase in SDMA and MMA marks. Plus, after a ten-day treatment with 12 at 10 μM, both MCF7 and HEK293 cells showed significant reductions in cell growth.[106]
Overall, because of its association with a multitude of diseases, PRMT1 is a viable drug target. Developing selective PRMT1 inhibitors has been a flourishing endeavor in the field.[94a,107] Our group has screened and designed compounds with different structural scaffolds to target and selectively inhibit PRMT1, and in one case unexpectedly achieving PRMT inhibition via binding of the inhibitor to the substrate pool. Many compounds made thus far have reached the low micromolar or submicromolar range of inhibition with a good understanding of different pharmacophores associated with their binding activities. Future work directed at modifying and diversifying these pharmacophores could lead to the development of the next-generation of potent and specific inhibitors for this major arginine methyltransferase.
6. Summary and Perspective
This account is focused on some important perspectives of biochemical mechanisms of arginine methylation which is enzymatically catalyzed by different PRMT members. We discussed the findings of enzymatic kinetics of arginine methylation from steady-state and pre-steady-state experiments, processivity in multi-arginine methylation, key structural features of PRMTs, and crosstalks of methyl arginine marks with adjacent PTMs on the core histones. We also recapitulated some typical examples of PRMT1 inhibitors. These findings and advances pave the foundation for further understanding of PRMT functions in cell development, differentiation, and proliferation, as well as in pathological processes. Although great progress has been made in PRMT research, many challenging issues remain to be determined. Although thousands of arginine methylation substrates and sites have been identified, tremendous efforts are needed to delineate detailed functions of individual arginine methylations in various cellular pathways. It is conceivable that not all arginine methylation marks have essential functions; many marks could be just biochemical noise without indispensable effects. It is challenging to discriminate essential from nonessential arginine methylation sites. PRMT isoforms have different sub-methylomes. It remains to be determined how the arginine methylome was constructed by different PRMT members. Current efforts are focused on the enyzmatic activities of PRMTs; on the other end, it remains to be determined whether PRMTs have non-enzymatic or other functions (i.e., moonlight functions) in cell biology beyond their methyltransferase activity. In this regard, recent work unexpectedly showed that PRMT8 has phospholipase function, which is attributed to its HKD motif (HxKxxxxDxxxxxxGG/S) that is distinct from the other PRMTs yet characteristic of phospholipase D enzymes.[108] PRMT1 has been found to exist as several isoforms, at least two of which are enzymatically inactive.[45a,109] Exact functions of these inactive PRMT1 isoforms remain to be determined. Future research will provide a better understanding of PRMT function in biological and disease processes from a broader and deeper scope. In regards of drug discovery, most PRMT1 inhibitors have micromolar or submicromolar potency. Both affinity and selectivity need to be improved significantly. No PRMT inhibitors have made it to the clinic yet. It would be thrilling to see in the near future some PRMT inhibitors can be translated into potent leads with clinical efficacy.
Acknowledgements
We are thankful to the National Institutes of Health grants R01GM126154 and R01GM086717 for financial support of our work.
Biographies
 Melody D. Fulton earned her B.S. degree in biochemistry and biology from Washington State University in 2013. She is a Ph.D. candidate in pharmaceutical and biomedical sciences under the supervision of Prof. Y. George Zheng at the University of Georgia. Her research focuses on understanding the regulatory mechanisms and functional consequences of PRMT activity.
Melody D. Fulton earned her B.S. degree in biochemistry and biology from Washington State University in 2013. She is a Ph.D. candidate in pharmaceutical and biomedical sciences under the supervision of Prof. Y. George Zheng at the University of Georgia. Her research focuses on understanding the regulatory mechanisms and functional consequences of PRMT activity.
 Tyler Brown obtained his B.S. in pharmaceutical and biomedical sciences from the University of Georgia in 2017. He is currently a Ph.D. student whose research interests include the use of medicinal chemistry to design and synthesize inhibitors of protein arginine methyltransferases.
Tyler Brown obtained his B.S. in pharmaceutical and biomedical sciences from the University of Georgia in 2017. He is currently a Ph.D. student whose research interests include the use of medicinal chemistry to design and synthesize inhibitors of protein arginine methyltransferases.
 Y. George Zheng received his B.S. in chemistry at Peking University, Ph.D. at University of Miami, and postdoctoral training at Johns Hopkins University School of Medicine. From 2006 to 2013, he was a faculty member in the Department of Chemistry at Georgia State University. Since 2013, he has been a faculty member in the Department of Pharmaceutical and Biomedical Science at University of Georgia. His research interest is on developing chemical probes and drug leads to interrogate functions of epigenetic enzymes in biology and disease.
Y. George Zheng received his B.S. in chemistry at Peking University, Ph.D. at University of Miami, and postdoctoral training at Johns Hopkins University School of Medicine. From 2006 to 2013, he was a faculty member in the Department of Chemistry at Georgia State University. Since 2013, he has been a faculty member in the Department of Pharmaceutical and Biomedical Science at University of Georgia. His research interest is on developing chemical probes and drug leads to interrogate functions of epigenetic enzymes in biology and disease.
References
- [1].a) Migliori V, Phalke S, Bezzi M, Guccione E, Epigenomics 2010, 2, 119–137 [DOI] [PubMed] [Google Scholar]; b) Larsen SC, Sylvestersen KB, Mund A, Lyon D, Mullari M, Madsen MV, Daniel JA, Jensen LJ, Nielsen ML, Sci. Signaling 2016, 9, rs9. [DOI] [PubMed] [Google Scholar]
- [2].a) Bedford MT, Clarke SG, Mol. Cell 2009, 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Morales Y, Cáceres T, May K, Hevel JM, Arch. Biochem. Biophys 2016, 590, 138–152 [DOI] [PubMed] [Google Scholar]; c) Fuhrmann J, Clancy KW, Thompson PR, Chem. Rev 2015, 115, 5413–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Schubert HL, Blumenthal RM, Cheng X, Trends Biochem. Sci 2003, 28, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang Y, Bedford MT, Nat. Rev. Cancer 2013, 13, 37–50. [DOI] [PubMed] [Google Scholar]
- [5].a) Hadjikyriacou A, Clarke SG, Biochemistry 2017, 56, 2612–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang M, Fuhrmann J, Thompson PR, Biochemistry 2014, 53, 7884–7892 [DOI] [PubMed] [Google Scholar]; c) Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, Kelly R, Guo A, Li W, Clarke SG, Bedford MT, Nat. Commun 2015, 6, 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Zurita Rendon O, Silva Neiva L, Sasarman F, Shoubridge EA, Hum. Mol. Genet 2014, 23, 5159–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rhein VF, Carroll J, Ding S, Fearnley IM, Walker JE, J. Biol. Chem 2013, 288, 33016–33026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG, J. Biol. Chem 2012, 287, 7859–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Feng Y, Hadjikyriacou A, Clarke SG, J. Biol. Chem 2014, 289, 32604–32616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR, J. Biol. Chem 2000, 275, 7723–7730. [DOI] [PubMed] [Google Scholar]
- [9].a) Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE, Mol. Cell. Biol 2000, 20, 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dhar S, Vemulapalli V, Patananan AN, Huang GL, Di Lorenzo A, Richard S, Comb MJ, Guo A, Clarke SG, Bedford MT, Sci. Rep 2013, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Hadjikyriacou, Yang Y, Espejo A, Bedford MT, Clarke SG, J. Biol. Chem 2015, 290, 16723–16743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, Kelly R, Guo A, Li W, Clarke SG, Bedford MT, Nat. Commun 2015, 6, 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Wang YC, Wang JD, Chen CH, Chen YW, Li C, Mol. Phylogenet. Evol 2015, 84, 101–111 [DOI] [PubMed] [Google Scholar]; b) Qian K, Zheng YG, Epi-Informatics: Discovery and Development of Small Molecule Epigenetic Drugs and Probes 2016, 231–256. [Google Scholar]
- [13].Takahashi Y, Daitoku H, Yokoyama A, Nakayama K, Kim JD, Fukamizu A, Recept J Signal Transduction 2011, 31, 168–172. [DOI] [PubMed] [Google Scholar]
- [14].Larsen SC, Sylvestersen KB, Mund A, Lyon D, Mullari M, Madsen MV, Daniel JA, Jensen LJ, Nielsen ML, Sci. Signaling 2016, 9, rs9. [DOI] [PubMed] [Google Scholar]
- [15].Blanc RS, Richard S, Mol. Cell 2017, 65, 8–24. [DOI] [PubMed] [Google Scholar]
- [16].Arribas-Layton M, Dennis J, Bennett EJ, Damgaard CK, Lykke-Andersen J, Mol. Cell. Biol 2016, 36, 2226–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Le Romancer M, Treilleux I, Leconte N, RobinLespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L, Mol. Cell 2008, 31, 212–221. [DOI] [PubMed] [Google Scholar]
- [18].Xu J, Wang AH, Oses-Prieto J, Makhijani K, Katsuno Y, Pei M, Yan L, Zheng YG, Burlingame A, Bruckner K, Derynck R, Mol. Cell 2013, 51, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lu T, Stark GR, Cancer Res. 2015, 75, 3692–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].D Lorenzo A, Bedford MT, FEBS Lett. 2011, 585, 2024–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].a) Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, Ren J, Beausoleil SA, Silva JC, Vemulapalli V, Bedford MT, Comb MJ, Mol. Cell. Proteomics 2014, 13, 372–387 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y, Cell 2014, 159, 458–458 e451 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, HenschenEdman A, Mackay DR, Stallcup MR, Aswad DW, Biochemistry 2001, 40, 5747–5756 [DOI] [PubMed] [Google Scholar]; d) Sylvestersen KB, Horn H, Jungmichel S, Jensen LJ, Nielsen ML, Mol. Cell. Proteomics 2014, 13, 2072–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Zeeshan M, Kaur I, Joy J, Saini E, Paul G, Kaushik A, Dabral S, Mohmmed A, Gupta D, Malhotra P, J. Proteome Res 2017, 16, 368–383 [DOI] [PubMed] [Google Scholar]; f) Bremang M, Cuomo A, Agresta AM, Stugiewicz M, Spadotto V, Bonaldi T, Mol. BioSyst 2013, 9, 2231–2247. [DOI] [PubMed] [Google Scholar]
- [22].Molina-Serrano D, Schiza V, Kirmizis A, Biochem. Soc. Trans 2013, 41, 751–759. [DOI] [PubMed] [Google Scholar]
- [23].Zhang M, Xu JY, Hu H, Ye BC, Tan M, Proteomics 2018, 18. [DOI] [PubMed] [Google Scholar]
- [24].Zhang RH, Li X, Liang ZJ, Zhu KK, Lu JY, Kong XQ, Ouyang SS, Li L, Zheng YG, Luo C, PLoS One 2013, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang X, Cheng XD, Structure 2003, 11, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guo H, Wang R, Zheng W, Chen Y, Blum G, Deng H, Luo M, ACS Chem. Biol 2014, 9, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].a) Obianyo O, Osborne TC, Thompson PR, Biochemistry 2008, 47, 10420–10427 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang M, Xu R-M, Thompson PR, Biochemistry 2013, 52, 5430–5440 [DOI] [PubMed] [Google Scholar]; c) Jacques SL, Aquino KP, Gureasko J, Boriack-Sjodin PA, Porter Scott M, Copeland RA, Riera TV, Biochemistry 2016, 55, 1635–1644 [DOI] [PubMed] [Google Scholar]; d) Gui S, Wooderchak-Donahue WL, Zang T, Chen D, Daly MP, Zhou ZS, Hevel JM, Biochemistry 2013, 52, 199–209 [DOI] [PubMed] [Google Scholar]; e) Kolbel K, Ihling C, Bellmann-Sickert K, Neundorf I, Beck-Sickinger AG, Sinz A, Kuhn U, Wahle E, J. Biol. Chem 2009, 284, 8274–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Lakowski T, Frankel A, Biochem. J 2009, 421, 253–261 [DOI] [PubMed] [Google Scholar]; g) Lakowski TM, Frankel A, J. Biol. Chem 2008, 283, 10015–10025. [DOI] [PubMed] [Google Scholar]
- [28].Obianyo O, Thompson PR, J. Biol. Chem 2012, 287, 6062–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brown JI, Koopmans T, van Strien J, Martin NI, Frankel A, ChemBioChem 2017. [DOI] [PubMed] [Google Scholar]
- [30].Hu H, Luo C, Zheng YG, J. Biol. Chem 2016, 291, 26722–26738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].a) Feng Y, Xie N, Jin M, Stahley MR, Stivers JT, Zheng YG, Biochemistry 2011, 50, 7033–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Feng Y, Xie N, Wu J, Yang C, Zheng YG, Biochem. Biophys. Res. Commun 2009, 379, 567–572. [DOI] [PubMed] [Google Scholar]
- [32].Feng Y, Xie N, Jin M, Stahley MR, Stivers JT, Zheng YG, Biochemistry 2011, 50, 7033–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schapira M, Ferreira de Freitas R, MedChemComm 2014, 5, 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH, EMBO J. 2007, 26, 4402–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J, EMBO J. 2007, 26, 4391–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Boriack-Sjodin PA, Jin L, Jacques SL, Drew A, Sneeringer C, Scott MP, Moyer MP, Ribich S, Moradei O, Copeland RA, ACS Chem. Biol 2016, 11, 763–771. [DOI] [PubMed] [Google Scholar]
- [37].Lee WC, Lin WL, Matsui T, Chen ES, Wei TY, Lin WH, Hu H, Zheng YG, Tsai MD, Ho MC, Biochemistry 2015, 54, 7514–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR, Biochemistry 2007, 46, 13370–13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, Han B, Jungheim LN, Qian Y, Rauch C, Russell M, Sauder JM, Wasserman SR, Weichert K, Willard FS, Zhang A, Emtage S, Proc. Natl. Acad. Sci. USA 2012, 109, 17960–17965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jacques SL, Aquino KP, Gureasko J, Boriack-Sjodin PA, Porter Scott M, Copeland RA, Riera TV, Biochemistry 2016, 55, 1635–1644. [DOI] [PubMed] [Google Scholar]
- [41].Wang M, Xu RM, Thompson PR, Biochemistry 2013, 52, 5430–5440. [DOI] [PubMed] [Google Scholar]
- [42]. Kirmizis, Santos-Rosa H, Penkett CJ, Singer MA, Green RD, Kouzarides T, Nat. Struct. Mol. Biol 2009, 16, 449–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Herrmann F, Bossert M, Schwander A, Akgun E, Fackelmayer FO, J. Biol. Chem 2004, 279, 48774–48779. [DOI] [PubMed] [Google Scholar]
- [44].Dolgin E, Nature 2018, 555, 300–302. [DOI] [PubMed] [Google Scholar]
- [45].a) Patounas O, Papacharalampous I, Eckerich C, Markopoulos GS, Kolettas E, Fackelmayer FO, J. Cell. Biochem 2018, 119, 2110–2123 [DOI] [PubMed] [Google Scholar]; b) Toma-Fukai S, Kim JD, Park KE, Kuwabara N, Shimizu N, Krayukhina E, Uchiyama S, Fukamizu A, Shimizu T, J. Mol. Biol 2016, 428, 1197–1208. [DOI] [PubMed] [Google Scholar]
- [46].Thandapani P, O’Connor TR, Bailey TL, Richard S, Mol. Cell 2013, 50, 613–623. [DOI] [PubMed] [Google Scholar]
- [47].Wysocka J, Allis CD, Coonrod S, Front. Biosci 2006, 11, 344–355. [DOI] [PubMed] [Google Scholar]
- [48].a) Schwammle V, Sidoli S, Ruminowicz C, Wu X, Lee CF, Helin K, Jensen ON, Mol. Cell. Proteomics 2016, 15, 2715–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pal S, Sif S, J. Cell. Physiol 2007, 213, 306–315. [DOI] [PubMed] [Google Scholar]
- [49].Strahl BD, Allis CD, Nature 2000, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- [50]. Lothrop P, Torres MP, Fuchs SM, FEBS Lett. 2013, 587, 1247–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].a) Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM, Genes Dev. 2007, 21, 3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B, Nature 2007, 449, 933–937 [DOI] [PubMed] [Google Scholar]; c) Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT, J. Biol. Chem 2008, 283, 3006–3010. [DOI] [PubMed] [Google Scholar]
- [52].a) Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, Cecatiello V, De Marco A, Blackstock W, Kuznetsov V, Amati B, Mapelli M, Guccione E, Nat. Struct. Mol. Biol 2012, 19, 136–144 [DOI] [PubMed] [Google Scholar]; b) Yuan CC, Matthews AG, Jin Y, Chen CF, Chapman BA, Ohsumi TK, Glass KC, Kutateladze TG, Borowsky ML, Struhl K, Oettinger MA, Cell Rep. 2012, 1, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Li Z, Zurita-Lopez C, Al-Hadid Q, Clark AT, Bedford MT, Masson JY, Clarke SG, J. Biol. Chem 2013, 288, 37010–37025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN, Mol. Cell. Biol 2007, 27, 384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].a) Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S, Mol. Cell. Biol 2004, 24, 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang L, Pal S, Sif S, Mol. Cell. Biol 2008, 28, 6262–6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Patnaik D, Chin HG, Esteve PO, Benner J, Jacobsen SE, Pradhan S, J. Biol. Chem 2004, 279, 53248–53258. [DOI] [PubMed] [Google Scholar]
- [57].Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A, Nat. Chem. Biol 2008, 4, 344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS, Dev. Cell 2010, 19, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T, EMBO Rep. 2002, 3, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T, Curr. Biol 2002, 12, 2090–2097. [DOI] [PubMed] [Google Scholar]
- [61].Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR, Curr. Biol 2001, 11, 1981–1985. [DOI] [PubMed] [Google Scholar]
- [62].Zhang Z, Nikolai BC, Gates LA, Jung SY, Siwak EB, He B, Rice AP, O’Malley BW, Feng Q, Nucleic Acids Res. 2017, 45, 9348–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, Allis CD, Curr. Biol 2001, 11, 996–1000. [DOI] [PubMed] [Google Scholar]
- [64].a) Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE, Mol. Cell. Biol 2000, 20, 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR, J. Biol. Chem 2000, 275, 7723–7730. [DOI] [PubMed] [Google Scholar]
- [65].a) Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR, Biochemistry 2007, 46, 13370–13381 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Feng Y, Wang J, Asher S, Hoang L, Guardiani C, Ivanov I, Zheng YG, J. Biol. Chem 2011, 286, 20323–20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E, Genes Dev. 2003, 17, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Huang SM, Litt M, Felsenfeld G, Genes Dev. 2005, 19, 1885–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Huang SM, Litt M, Felsenfeld G, Genes Dev. 2005, 19, 1885–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Li XG, Hu X, Patel B, Zhou Z, Liang S, Ybarra R, Qiu Y, Felsenfeld G, Bungert J, Huang SM, Blood 2010, 115, 2028–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].a) Wang HB, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong JM, Tempst P, Zhang Y, Science 2001, 293, 853–857 [DOI] [PubMed] [Google Scholar]; b) Fulton MD, Zhang J, He M, Ho MC, Zheng YG, Biochemistry 2017, 56, 3539–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].a) Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, Sardet C, EMBO Rep. 2002, 3, 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG, Mol. Cell. Biol 2006, 26, 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhao Q, Rank G, Tan YT, Li HT, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM, Nat. Struct. Mol. Biol 2009, 16, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q, Blood 2010, 116, 1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, Reinberg D, Lee MG, Genes Dev. 2012, 26, 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ho MC, Wilczek C, Bonanno JB, Xing L, Seznec J, Matsui T, Carter LG, Onikubo T, Kumar PR, Chan MK, Brenowitz M, Cheng RH, Reimer U, Almo SC, Shechter D, PLoS One 2013, 8, e57008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Jain K, Jin CY, Clarke SG, Proc. Natl. Acad. Sci. USA 2017, 114, 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Verrier L, Vandromme M, Trouche D, Biol. Cell 2011, 103, 381–401. [DOI] [PubMed] [Google Scholar]
- [78].Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Muller JM, Greschik H, Kirfel J, Ji S, Kunowska N, Beisenherz-Huss C, Gunther T, Buettner R, Schule R, Nature 2010, 464, 792–U175. [DOI] [PubMed] [Google Scholar]
- [79].a) Chang BS, Chen Y, Zhao YM, Bruick RK, Science 2007, 318, 444–447 [DOI] [PubMed] [Google Scholar]; b) Zhang XQ, Liu XQ, Zhang M, Li TT, Muth A, Thompson PR, Coonrod SA, Zhang XS, Sci. Rep 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Kirmizis, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T, Nature 2007, 449, 928–U917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kuo MH, Xu XJ, Bolck HA, Guo DW, Biochim. Biophys. Acta Gene Regul. Mech 2009, 1789, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].a) Litt M, Qiu Y, Huang S, Biosci. Rep 2009, 29, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Low JKK, Wilkins MR, The FEBS J. 2012, 279, 4423–4443. [DOI] [PubMed] [Google Scholar]
- [83].a) Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J, The FEBS J. 2005, 272, 4249–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J, Eur. J. Biochem 2004, 271, 2257–2263. [DOI] [PubMed] [Google Scholar]
- [84].Wang WL, Anderson LC, Nicklay JJ, Chen H, Gamble MJ, Shabanowitz J, Hunt DF, Shechter D, Epigenet. Chromatin 2014, 7, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ju J, Chen A, Deng Y, Liu M, Wang Y, Wang Y, Nie M, Wang C, Ding H, Yao B, Gui T, Li X, Xu Z, Ma C, Song Y, Kvansakul M, Zen K, Zhang CY, Luo C, Fang M, Huang DCS, Allis CD, Tan R, Zeng CK, Wei J, Zhao Q, Nat. Commun 2017, 8, 928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].a) Morettin A, Baldwin RM, Cote J, Mutagenesis 2015, 30, 177–189 [DOI] [PubMed] [Google Scholar]; b) Copeland RA, Moyer MP, Richon VM, Oncogene 2013, 32, 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Michalak EM, Visvader JE, Mol. Oncol 2016, 10, 1497–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kim JH, Yoo BC, Yang WS, Kim E, Hong S, Cho JY, Mediators Inflammation 2016, 2016, 4028353. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Yang Y, Bedford MT, Nat. Rev. Cancer 2013, 13, 37–50. [DOI] [PubMed] [Google Scholar]
- [87].a) Copeland RA, Clin. Cancer Res 2013, 19, 6344–6350 [DOI] [PubMed] [Google Scholar]; b) Hansen JN, Li X, Zheng YG, Lotta LT Jr., Dedhe A, Schor NF, ACS Chem. Neurosci 2017, 8, 2118–2123. [DOI] [PubMed] [Google Scholar]
- [88].a) Mathioudaki K, Scorilas A, Ardavanis A, Lymberi P, Tsiambas E, Devetzi M, Apostolaki A, Talieri M, Tumor Biol. 2011, 32, 575–582 [DOI] [PubMed] [Google Scholar]; b) Papadokostopoulou A, Mathioudaki K, Scorilas A, Xynopoulos D, Ardavanis A, Kouroumalis E, Talieri M, Anticancer Res. 2009, 29, 1361–1366. [PubMed] [Google Scholar]
- [89].Cheung N, Chan LC, Thompson A, Cleary ML, So CWE, Nat. Cell Biol 2007, 9, 1208–1215. [DOI] [PubMed] [Google Scholar]
- [90].a) Zakrzewicz D, Zakrzewicz A, Preissner KT, Markart P, Wygrecka M, Int. J. Mol. Sci 2012, 13, 12383–12400 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sun Q, Yang X, Zhong B, Jiao F, Li C, Li D, Lan X, Sun J, Lu S, J. Immunol 2012, 188, 3506–3512 [DOI] [PubMed] [Google Scholar]; c) Sun Q, Liu L, Roth M, Tian J, He Q, Zhong B, Bao R, Lan X, Jiang C, Sun J, Yang X, Lu S, J. Immunol 2015, 195, 298–306. [DOI] [PubMed] [Google Scholar]
- [91].a) Vallance P, Leiper J, Arterioscler. Thromb. Vasc. Biol 2004, 24, 1023–1030 [DOI] [PubMed] [Google Scholar]; b) Bouras G, Deftereos S, Tousoulis D, Giannopoulos G, Chatzis G, Tsounis D, Cleman MW, Stefanadis C, Curr. Top. Med. Chem 2013, 13, 180–200. [DOI] [PubMed] [Google Scholar]
- [92].a) Lajer M, Tarnow L, Jorsal A, Teerlink T, Parving H-H, Rossing P, Diabetes Care 2008, 31, 747–752 [DOI] [PubMed] [Google Scholar]; b) Iwasaki H, Life Sci. 2009, 85, 161–166. [DOI] [PubMed] [Google Scholar]
- [93].a) Baylis C, Nat. Clin. Pract. Nephrol 2006, 2, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schwedhelm E, Böger RH, Nat. Rev. Nephrol 2011, 7, 275–285 [DOI] [PubMed] [Google Scholar]; c) Cvetković T, Pavlović R, Ɖorđević V, Stojanović I, Veličković-Radovanović R, Ignjatović A, Stefanović N, Živanović S, Ɖorđević V, J. Med. Biochem 2012, 31, 301–308 [Google Scholar]; d) Raptis V, Kapoulas S, Grekas D, Nephrology 2013, 18, 11–21. [DOI] [PubMed] [Google Scholar]
- [94].a) Hu H, Qian K, Ho MC, Zheng YG, Expert Opin. Invest. Drugs 2016, 25, 335–358 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li KK, Luo C, Wang D, Jiang H, Zheng YG, Med. Res. Rev 2012, 32, 815–867 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Smith E, Zhou W, Shindiapina P, Sif S, Li C, Baiocchi RA, Expert Opin. Ther. Targets 2018, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zheng YG, Wu J, Chen Z, Goodman M, Med. Res. Rev 2008, 28, 645–687 [DOI] [PubMed] [Google Scholar]; e) Richters A, Future Med. Chem 2017, 9, 2081–2098. [DOI] [PubMed] [Google Scholar]
- [95].Wu J, Xie N, Feng Y, Zheng YG, Biomol J Screening 2012, 17, 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Feng Y, Li M, Wang B, Zheng YG, J. Med. Chem 2010, 53, 6028–6039. [DOI] [PubMed] [Google Scholar]
- [97].Qian K, Hu H, Xu H, Zheng YG, Signal Transduction Targeted Ther. 2018, 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Luan Y, Blazer LL, Hu H, Hajian T, Zhang J, Wu H, Houliston S, Arrowsmith CH, Vedadi M, Zheng YG, Org. Biomol. Chem 2015, 14, 631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]; Blazer LL, Li F, Kennedy S, Zheng YG, Arrowsmith CH, Vedadi M, SLAS Discovery 2017, 22, 32–39. [DOI] [PubMed] [Google Scholar]
- [99].a) Guo H, Wang R, Zheng W, Chen Y, Blum G, Deng H, Luo M, ACS Chem. Biol 2014, 9, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang R, Ibanez G, Islam K, Zheng W, Blum G, Sengelaub C, Luo M, Mol. BioSyst 2011, 7, 2970–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang R, Zheng W, Yu H, Deng H, Luo M, J. Am. Chem. Soc 2011, 133, 7648–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Cheng DH, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT, J. Biol. Chem 2004, 279, 23892–23899. [DOI] [PubMed] [Google Scholar]
- [101].Wang JX, Chen LM, Sinha SH, Liang ZJ, Chai HF, Muniyan S, Chou YW, Yang C, Yan LL, Feng Y, Li KK, Lin MF, Jiang HL, Zheng YG, Luo C, J. Med. Chem 2012, 55, 7978–7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].a) Sinha SH, Owens EA, Feng Y, Yang Y, Xie Y, Tu Y, Henary M, Zheng YG, Eur. J. Med. Chem 2012, 54, 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hu H, Owens EA, Su H, Yan L, Levitz A, Zhao X, Henary M, Zheng YG, J. Med. Chem 2015, 58, 1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yan L, Yan C, Qian K, Su H, Kofsky-Wofford SA, Lee WC, Zhao X, Ho MC, Ivanov I, Zheng YG, J. Med. Chem 2014, 57, 2611–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhang L, Tran NT, Su H, Wang R, Lu Y, Tang H, Aoyagi S, Guo A, Khodadadi-Jamayran A, Zhou D, Qian K, Hricik T, Cote J, Han X, Zhou W, Laha S, AbdelWahab O, Levine RL, Raffel G, Liu Y, Chen D, Li H, Townes T, Wang H, Deng H, Zheng YG, Leslie C, Luo M, Zhao X, Elife 2015, 4, e07938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhang J, Qian K, Yan CL, He MM, Jassim BA, Ivanov I, Zheng YJG, MedChemComm 2017, 8, 440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Eram MS, Shen Y, Szewczyk M, Wu H, Senisterra G, Li F, Butler KV, Kaniskan HU, Speed BA, Dela Sena C, Dong A, Zeng H, Schapira M, Brown PJ, Arrowsmith CH, Barsyte-Lovejoy D, Liu J, Vedadi M, Jin J, ACS Chem. Biol 2016, 11, 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].a) Luo M, Epigenomics 2015, 7, 1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kaniskan HU, Konze KD, Jin J, J. Med. Chem 2015, 58, 1596–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kim JD, Park KE, Ishida J, Kako K, Hamada J, Kani S, Takeuchi M, Namiki K, Fukui H, Fukuhara S, Hibi M, Kobayashi M, Kanaho Y, Kasuya Y, Mochizuki N, Fukamizu A, Sci. Adv 2015, 1, e1500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].a) Goulet I, Gauvin G, Boisvenue S, Cote J, J. Biol. Chem 2007, 282, 33009–33021 [DOI] [PubMed] [Google Scholar]; b) Baldwin RM, Morettin A, Paris G, Goulet I, Côté J, Cell Cycle 2012, 11, 4597–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]