Abstract
Understanding the topology adopted by individual G-quadruplex (GQ)-forming sequences in vivo and targeting a specific GQ motif among others in the genome will have a profound impact on GQ-directed therapeutic strategies. However, this remains a major challenge as most of the tools poorly distinguish different GQ conformations and are not suitable for both cell-free and in-cell analysis. Here, we describe an innovative probe design to investigate GQ conformations and recognition in both cell-free and native cellular environments by using a conformation-sensitive dual-app nucleoside analogue probe. The nucleoside probe, derived by conjugating fluorobenzofuran at the 5-position of 2′-deoxyuridine, is composed of a microenvironment-sensitive fluorophore and an in-cell NMR compatible 19F label. This noninvasive nucleoside, incorporated into the human telomeric DNA oligonucleotide repeat, serves as a common probe to distinguish different GQ topologies and quantify topology-specific binding of ligands by fluorescence and NMR techniques. Importantly, unique signatures displayed by the 19F-labeled nucleoside for different GQs enabled a systematic study in Xenopus laevis oocytes to provide new structural insights into the GQ topologies adopted by human telomeric overhang in cells, which so far has remained unclear. Studies using synthetic cell models, immunostaining on fixed cells, and crystallization conditions suggest that parallel GQ is the preferred conformation of telomeric DNA repeat. However, our findings using the dual-app probe clearly indicate that multiple structures including hybrid-type parallel-antiparallel and parallel GQs are formed in the cellular environment. Taken together, our findings open new experimental strategies to investigate topology, recognition, and therapeutic potential of individual GQ-forming motifs in a biologically relevant context.
Introduction
Noncanonical four-stranded GQ-forming sequences are prevalent in the telomeres and in the promoter DNA and untranslated regions of mRNA of several genes that are known to cause cancer.1,2 The location and conservation of putative GQ-forming sequences across various eukaryotes,3,4 and extensive biophysical investigations point out that GQ is an important structural element in maintaining the stability of genome and in the regulation of core cellular processes like replication, transcription, and translation.1,5,6 Further, small molecule ligands that stabilize GQ structure have provided compelling evidence linking expression levels of proto-oncogenes and GQ structure.7–11 Hence, controlling the ensuing function of GQ structure by using ligands is viewed as an alternative therapeutic strategy for cancer and aging related diseases.12,13 Despite a flurry of interest in this direction,12–16 the discovery of clinically viable GQ-binders remains a major challenge.17,18 This is because GQ exhibits a high degree of structural polymorphism (e.g., parallel, antiparallel, hybrid-type mixed parallel–antiparallel stranded GQs), which depends on the sequence, ionic conditions, confinement, and molecular crowding,19–23 and most GQ binders/probes poorly differentiate different GQs topologies.24 Hence, recent studies are directed toward developing biophysical platforms to understand the topology adopted by a G-rich sequence inside the cell and identify binders that specifically target a GQ motif among other such motifs in the genome.25
Circular dichroism (CD), fluorescence, and NMR techniques are commonly used to study the structure, dynamics, and binding affinities of GQs in synthetic models mimicking the crowded and confined environment of the cell.26–29 One well-studied system is the human telomeric (H-Telo) DNA repeat (TTAGGG)n. In an aqueous buffer containing K+ ions, H-Telo DNA oligonucleotide (ON) repeat forms multiple GQ structures,30 whereas in the presence of crowding agents like PEG it adopts a parallel conformation, which exhibits slower folding dynamics and reduced ligand-binding affinities.28,31 In contrast, by using optical tweezers, it was identified that H-Telo DNA in a confined environment of DNA origami nanocages formed a GQ with higher stability and faster folding rates.32 While these studies provide valuable structural information, non-native conditions bias the ONs to adopt a certain GQ structure as in the case of PEG due to its dehydrating effect.26,28 Alternatively, electron microscopy,33 and GQ-specific antibodies34–37 and fluorescent ligands38–41 have facilitated the visualization of GQ structures in different cell types. Although these methods are quantitative, they do not provide the much-needed information on the topology of GQs. Very recently, by using a combination of antibodies D1 and BG4, which specifically bind to parallel GQs and “all types” of GQ structures, respectively, it was identified that ∼77% of H-Telo repeats exist in parallel conformation, and the remaining are presumed to adopt other GQ structures.42 EPR analysis of frozen Xenopus laevis oocytes microinjected with nitroxide radical-labeled H-Telo DNA ON repeat also showed the presence of a mixture of parallel- and antiparallel-stranded GQs.43 However, an important limitation of EPR and immunofluorescence GQ detection methods is that they are restricted to frozen or fixed and permeabilized cells wherein morphological integrity is compromised.38a On the contrary, NMR analysis in oocyte egg extract suggested that parallel conformation is not the preferred conformation of the telomeric repeat.44 However, severe broadening of NMR signal hampered the structural elucidation in live oocytes.45
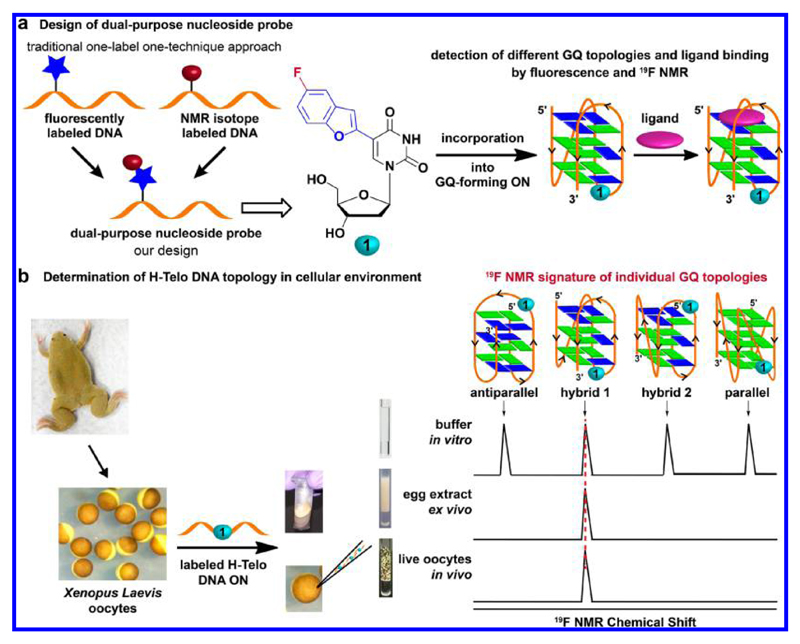
Amidst these conflicting results and limitations in currently available tools, it is hypothesized that understanding the structure of individual GQ-forming sequences in native cellular environment and devising discovery platforms to identify topology-specific GQ binders are of high priority. This, we envisioned, can be achieved by developing a GQ-sensing biophysical platform, which (i) is compatible to both cell-free and in-cell analysis, (ii) reports the formation as well as distinguishes different GQ topologies in cell-free and cellular systems, and (iii) supports discovery assays to identify topology-specific GQ binders. Such a platform would not only allow the profiling of the topology of various GQ-forming sequences in cells but also would have a profound impact on harnessing the potential of GQ-directed therapeutic strategies. Here, we describe an innovative approach to investigate GQ structures and their ligand binding in vitro and in live cells by using a conformation-sensitive dual-purpose nucleoside analogue probe (Figure 1). The nucleoside probe is composed of a microenvironment-sensitive fluorophore and 19F NMR label, which is derived by attaching fluorobenzofuran at the 5-position of 2′-deoxyuridine. This minimally perturbing nucleoside, when incorporated into the loop region of H-Telo DNA ON repeat, serves as a common probe to detect different GQ topologies and quantify topology-specific binding of ligands to GQ structures by fluorescence and NMR techniques. Importantly, through systematic 19F NMR studies in intraoocyte buffer, Xenopus laevis oocyte lysate, egg extract, and live oocytes, we successfully determined the GQ topologies adopted by the H-Telo overhang in live cells.
Figure 1.
Probe design and determination of GQ topologies in live cells. (a) The dual-purpose nucleoside probe 1 is designed by combining the environment-sensitivity of a fluorescent 5-benzofuran-modified nucleoside analogue and high responsiveness of 19F NMR signal to its local environment. Nucleoside 1, incorporated into a G-rich ON sequence, reports and distinguishes different GQ conformations and facilitates the estimation of topology-specific binding of ligands to different GQs by fluorescence and NMR spectroscopy techniques. (b) A schematic diagram showing the experimental design for the determination of GQ structure in cells by using Xenopus laevis oocyte as the model. The sensitivity of 19F-labeled nucleoside 1 to subtle differences in the conformation produces a distinct NMR signature for different GQ conformations. A comparison of signatures in the in vitro (buffer), ex vivo (egg extract), and in vivo (live oocytes) conditions provides insights into the GQ topologies adopted by H-Telo DNA overhang in cells.
Results and Discussion
Design and Synthesis of Dual-Purpose Nucleoside Probe
We recently developed a conformation-sensitive fluorescent nucleoside probe by conjugating benzofuran moiety at the 5-position of uracil.46 The nucleoside analogue is structurally minimally invasive and, importantly, serves as an excellent GQ sensor, wherein it photophysically discriminates duplex and various GQ forms of H-Telo DNA ON repeat. While being highly sensitive to subtle differences in conformations, its fluorescence properties, particularly excitation maximum in the UV region (λmax = 330 nm), precluded its use in cell-based analysis. So, to retain the high conformation sensitivity of the fluorophore and still determine the structure of GQ in cells, we came up with the idea of introducing a 19F atom as an NMR label to expand the proficiency of benzofuran-modified nucleoside analogue in probing the structure and recognition of GQ structures in both cell-free and cellular environments (Figure 1). The choice of 19F isotope label was based on the following reasons.47 It is 100% naturally abundant and is highly sensitive to changes in its local environment. Its signal is highly dispersed as compared to proton signal and, importantly, the absence of endogenous 19F label eliminates the background signal from the cells. Further, 19F NMR spectroscopy, which has emerged as a powerful tool in discovering protein binders, has recently been used to study noncanonical nucleic acid structures.48,49 Akin to antibodies, these studies predicted the formation of GQs, but did not provide information on the topology as these systems were not designed to distinguish different GQ topologies.
On the basis of the responsiveness of benzofuran-modified nucleoside analogue and useful properties of 19F isotope, a dual-purpose nucleoside probe (1) was assembled by attaching fluorobenzofuran moiety at the 5-position of uracil (Figure 1). 5-Fluorobenzofuran was first stanylated, which upon cross-coupling with 5-iodo-2′-deoxyuridine in the presence of a palladium catalyst gave 5-fluorobenzofuran-2′-deoxyuridine 1 (Scheme S1). An important aspect of this design is that the fluorophore and 19F atom are intentionally integrated into the same electronic system so that the effect of microenvironment will be similar on both of the labels. Hence, the signature of individual GQ structures obtained using fluorescence and NMR techniques could be efficiently correlated with one another.
Nucleoside 1 Is Highly Sensitive to Its Microenvironment
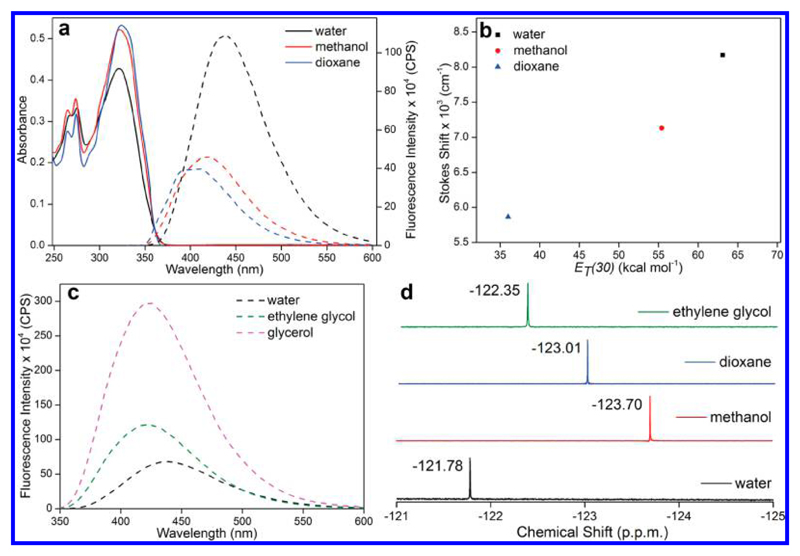
The ground-state electronic spectrum of nucleoside 1 in solvents of different polarity and viscosity revealed only minor changes in absorption maxima (Figure 2a, Table 1). However, excited-state properties like emission maximum, quantum yield, and lifetime were significantly affected. The nucleoside in water exhibited a strong emission band centered at 437 nm with a quantum yield and lifetime of 0.11 and 0.84 ns, respectively (Figure 2a, Table 1, Figure S1). In less polar solvents like methanol and dioxane, the emission band was considerably blue-shifted, and the quantum yield and lifetime were found to be significantly lower as compared to those in water. A plot of Stokes shift in solvents of different polarity versus ET(30) (Reichardt’s microscopic solvent polarity parameter) gave nearly a linear correlation, which further confirmed the responsiveness of the nucleoside to changes in microenvironment (Figure 2b). By design, the nucleoside analogue contains a rotatable aryl–aryl bond between fluorobenzofuran and uracil rings. This could invoke conformation sensitivity as the relative conformation of two rings will influence π-conjugation, and hence the fluorescence properties. As the viscosity of the medium was increased from water to ethylene glycol to glycerol, the nucleoside exhibited a progressive increase in quantum yield and lifetime due to rigidification of the fluorophore in a more viscous medium (Figure 2c, Table 1, and Figure S1). Further, significantly higher fluorescence anisotropy displayed by the nucleoside in a viscous medium confirmed the conformation sensitivity of the system (Table 1).
Figure 2.
Effect of microenvironment on the fluorescence and 19F NMR signal of nucleoside 1. (a) UV absorption (25 μM, solid lines) and fluorescence (5 μM, dashed line) spectra of nucleoside 1 in solvents of different polarity. In fluorescence study, samples were excited at respective lowest energy absorption maximum (Table 1) with excitation and emission slit widths of 2 and 4 nm, respectively. (b) Stokes shift versus ET(30) plot of nucleoside 1 in solvents of different polarity. (c) Fluorescence spectra (5 μM, dashed line) of nucleoside 1 in solvents of different viscosity. Excitation and emission slit widths were kept at 2 and 3 nm, respectively. (d) 19F NMR spectra of nucleosides 1 in solvents of different polarity and viscosity. All samples contained 15% d6-DMSO, and each spectrum was referenced relative to an external standard (trifluorotoluene (TFT) –63.72 ppm).
Table 1. Photophysical Properties of Nucleoside 1 in Different Microenvironments.
| solvent | λmaxa (nm) | λem (nm) | Stokes shift (cm−1) | Irelb | Φc | τavc (ns) | rc |
|---|---|---|---|---|---|---|---|
| water | 322 | 437 | 8.17 × 103 | 1.00 | 0.11 | 0.84 | 0.03 |
| methanol | 322 | 418 | 7.13 × 103 | 0.43 | 0.04 | 0.33 | n.d. |
| dioxane | 324 | 400 | 5.86 × 103 | 0.36 | 0.03 | 0.31 | n.d. |
| ethylene glycol | 325 | 420 | 6.96 × 103 | 1.79 | 0.20 | 0.94 | 0.20 |
| glycerol | 326 | 424 | 7.09 × 103 | 4.39 | 0.52 | 2.36 | 0.34 |
Lowest energy absorption maximum is given.
Relative emission intensity at λem is given with respect to intensity in water.
Standard deviation for quantum yield (Φ), average lifetime (τav), and anisotropy (r) in different solvents is ≤0.005, ≤0.02 ns, and ≤0.003, respectively. n.d. = not determined.
Akin to fluorescence, the nucleoside exhibited a well-resolved 19F chemical shift in solvents of different polarity and viscosity (Figure 2d). Depending on the solvent polarity, the dipole of a fluorinated molecule can polarize the surrounding solvent molecules or align itself with the dipole of the solvent molecules. This could either shield or deshield the fluorinated molecule and alter the 19F chemical shift.50a Similarly, changes in solvent viscosity can alter the relative conformation between the fluorobenzofuran and uracil rings, which can also lead to shielding or deshielding of the 19F label.50b Hence, a distinct NMR signal displayed by nucleoside 1 in different solvents is due to a combination of polarity and viscosity effects. Collectively, these studies indicate that environment-sensitive nucleoside 1 could serve as a two-channel probe to study nucleic acids by fluorescence and 19F NMR techniques.
Fluorescence Detection of H-Telo GQ Topologies
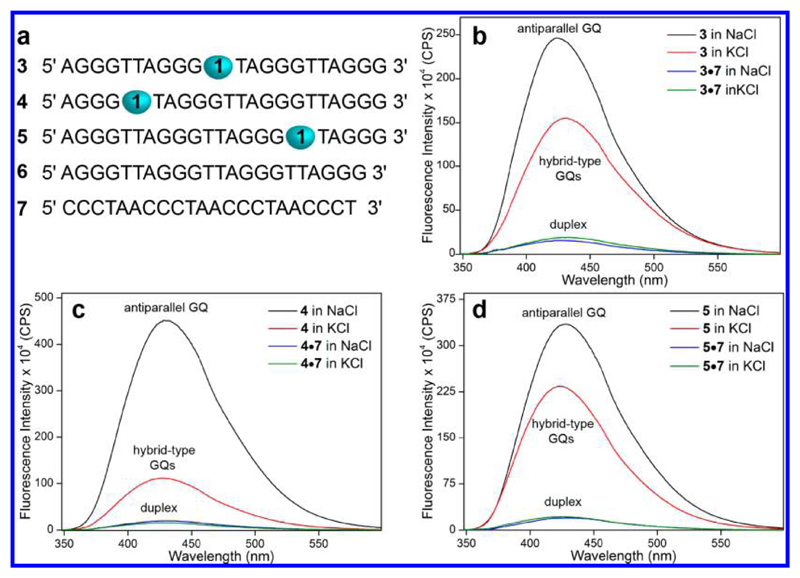
The nucleoside probe was incorporated into the loop regions formed by TTA residues for two reasons: (i) The conformation of loop residues is distinctly different in different H-Telo GQ structures,19 and (ii) modifications on the G-tetrads could potentially affect the formation as well as the stability of GQ.51 One of the T residues in each of the three loops was replaced with the dual-purpose nucleoside analogue 1 by solid-phase ON synthesis protocol using phosphoramidite substrate 2 to produce H-Telo DNA ONs 3–5 (Figure 3a, Scheme S1). HPLC and MALDI-TOF mass analyses of PAGE purified ONs confirmed the purity and integrity of the modified ONs (Figures S2 and S3 and Table S1).
Figure 3.
Fluorophore component of nucleoside 1 reports the formation of different GQ structures. (a) Sequence of fluorobenzofuran-labeled H-Telo DNA ONs 3–5. One of the T residues of loop 2 (3), loop 1 (4), and loop 3 (5) was replaced with nucleoside 1. Sequence of control unmodified H-Telo DNA ON 6 and complementary ON 7 is shown. (b–d) Spectrum of GQs of ON 3–5 and the corresponding duplexes 3·7, 4·7, and 5·7 in different ionic conditions. Samples (0.5 μM) were excited at 330 nm with excitation and emission slit widths of 4 and 6 nm, respectively.
The effect of modification on the formation and stability of GQ structure was examined by CD and thermal-melting experiments. Modified (3–5) and control unmodified (6) H-Telo DNA ONs were annealed in different ionic conditions using either potassium phosphate buffer containing 100 mM KCl or sodium phosphate buffer containing 100 mM NaCl. Both control and modified ONs in K+ ionic conditions showed similar CD profiles characteristic of hybrid type mixed parallel–antiparallel stranded GQ structures (a positive peak at ∼290 nm and a shoulder at ∼270 nm, Figure S4a).30 In the presence of NaCl, the ONs exhibited a positive peak at ∼293 nm and a strong negative peak at ∼263 nm characteristic of an antiparallel GQ structure (Figure S4b).30 UV-thermal melting analysis of the modified and native ONs in different ionic conditions gave a characteristic melting profile for the GQ structure at 295 nm with similar Tm values (Figure S5 and Table S2).52 Consistent with the literature reports, these results clearly prove that the modified H-Telo DNA ONs form respective GQ structures in different ionic conditions, and the fluorobenzofuran modification has only a minor impact on the GQ stability.
Samples of labeled ONs, annealed into GQs and duplexes (3·7, 4·7, and 5·7), in a buffer containing K+ or Na+ ions were excited at 330 nm. Steady-state fluorescence of duplexes in both of the ionic conditions showed similar emission bands, which were very weak (Figure 3b–d and Figure S6). The telomeric DNA ONs, which form multiple structures with hybrid-type GQs as the predominant conformation in K+ conditions, displayed significantly higher fluorescence intensity (6–11-fold) as compared to respective perfect duplexes. Remarkably, in the presence Na+ ions, which favor the antiparallel conformation, the ONs exhibited further enhancement in fluorescence intensity as compared to the hybrid-type GQs (Figure 3b–d). Excited-state decay kinetic analysis showed that the antiparallel conformation has discernibly higher lifetime as compared to the hybrid-type GQs formed in K+ ionic conditions (Table 2 and Figure S7). It is important to mention here that changes in ionic conditions did not affect the fluorescence profile of the free nucleoside probe (Figure S8). Hence, the changes in fluorescence properties (intensity, quantum yield, and lifetime) are due to the differences in the microenvironment of the nucleoside probe in different G-quadruplex conformations.
Table 2. Fluorescence Properties of H-Telo DNA ONs 3–5 and Respective Duplexes in Aqueous Buffer.
| ON sample |
λem (nm) |
Φa | Φrb |
τavc (ns) |
ON sample |
λem (nm) |
Φa | Φrb |
τavc (ns) |
ON sample |
λem (nm) |
Φa | Φrb |
τav c (ns) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 in NaCl | 424 | 0.053 | 27 | 1.09 | 4 in NaCl | 428 | 0.093 | 93 | 1.40 | 5 in NaCl | 428 | 0.062 | 31 | 0.89 |
| 3 in KCl | 429 | 0.030 | 15 | 0.60 | 4 in KCl | 428 | 0.022 | 11 | 0.79 | 5 in KCl | 423 | 0.043 | 22 | 0.73 |
| 3·7 in NaCl | 426 | 0.002 | 1 | n.d. | 4·7 in NaCl | 426 | 0.001 | 1 | n.d. | 5·7 in NaCl | 428 | 0.002 | 1 | n.d. |
| 3·7 in KCl | 432 | 0.002 | 1 | n.d. | 4·7 in KCl | 428 | 0.002 | 1 | n.d. | 5·7 in KCl | 426 | 0.002 | 1 | n.d. |
Standard deviation for quantum yield (Φ) of GQs is ≤0.006, and that of duplexes is ≤0.0002.
Φr = relative quantum yield with respect to the quantum yield of the duplex in the respective ionic conditions.
Standard deviation for lifetime (τav) is ≤0.06 ns. n.d. = not determined. Reliable lifetime values could not be obtained for duplexes as they were very weakly fluorescent.
The ability of nucleoside probe 1 to photophysically distinguish different GQ topologies from the duplex form is due to the distinct conformation and microenvironment of the probe in these structures. In the duplex state, the base-paired nucleoside analogue is likely to experience a strong stacking interaction with adjacent bases, which is known to promote nonradiative decay pathway.53 The presence of guanine residue next to the emissive analogue can further quench the fluorescence as guanine is very well-known to quench the fluorescence of several dyes by electron transfer process.54 However, in the GQ structures, the nucleoside analogue placed in the loop region is not H-bonded and is also away from the G-tetrad. Hence, enhancement in fluorescence exhibited by different GQs is likely due to reduced (i) stacking interaction between the fluorophore and adjacent bases and (ii) electron transfer process between the fluorophore and guanine residues. Further, solvation–desolvation and rigidification–derigidification of the fluorophore could have also influenced the fluorescence outcome of the probe in duplex and GQ structures.
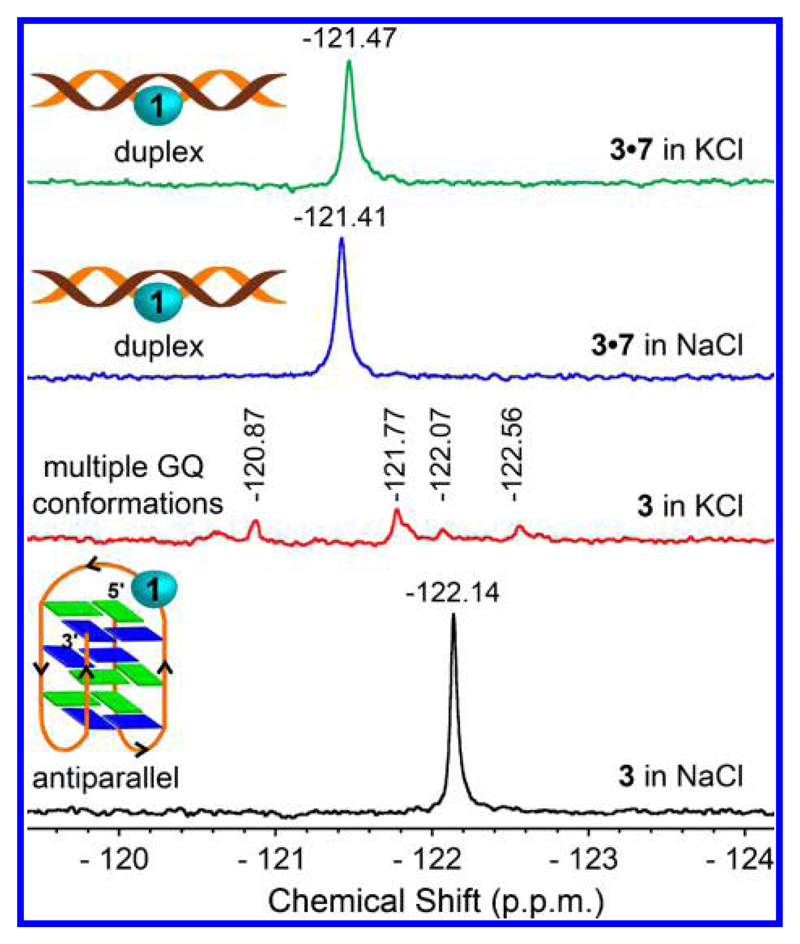
19F Label Exhibits a Distinct and Resolved Signature for Different GQ Topologies
Unlike the 1H NMR spectrum of a GQ structure, which shows multiple signals for the imino protons between 10 and 12 ppm,55 the proton-decoupled 19F NMR spectrum of modified H-Telo DNA ONs should give a single and distinct peak for each and every GQ structure if the 19F-labeled nucleoside 1 is sensitive to subtle differences in the conformation (Figure 1b). H-Telo DNA ON 3, containing modified nucleoside in the middle loop, was chosen as the study model. In consensus with fluorescence data, the duplex form (3·7) in the presence of NaCl or KCl gave very similar chemical shifts (Figure 4). Interestingly, ON 3 in K+ ionic conditions produced at least four well-resolved peaks corresponding to potentially four different GQ topologies as has been predicted in the literature for this sequence.30 Although ON 3 in the presence of K+ ions showed higher fluorescence and lifetime as compared to the duplex form, we could not ascertain the formation of multiple structures as in the case of 19F NMR measurements. ON 3, which adopts only an antiparallel conformation in the presence of Na+ ions,56 gave only one peak with a distinct chemical shift. Collectively, these results clearly underscore the bifunctionality of the nucleoside probe in faithfully reporting the formation as well as discriminating different GQ topologies by fluorescence and NMR techniques.
Figure 4.
19F label of nucleoside 1 gives a distinct NMR signal for different H-Telo DNA GQ topologies. 19F NMR spectrum (100 μM) of H-Telo DNA ON 3 and corresponding duplex (3·7) in sodium phosphate buffer containing 100 mM NaCl or potassium phosphate buffer containing 100 mM KCl at 25 °C. All samples (100 μM) contained 20% D2O and each spectrum was referenced relative to an external standard (TFT).
Estimation of Ligand Binding to Different GQ Topologies
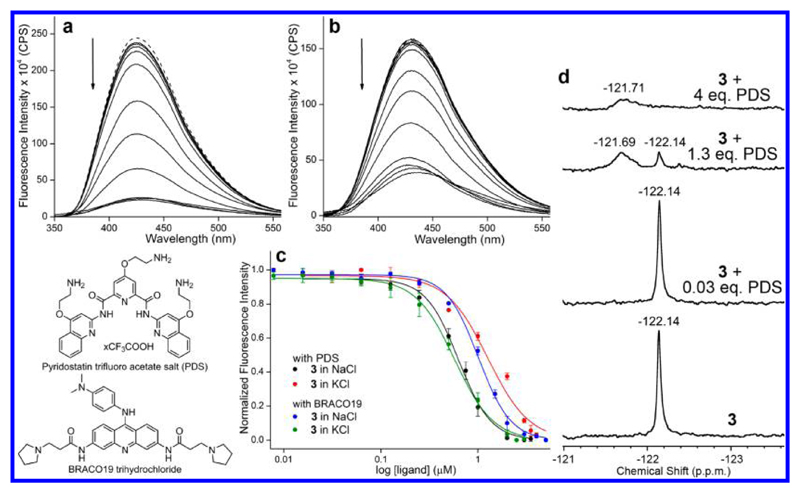
The conformation-sensitivity of the dual-purpose probe was put to use in estimating the binding affinity of ligands to different H-Telo GQ structures of ON 3 by titrating with pyridostatin (PDS) and BRACO19, which are known functional GQ binders (Figure 5).57,58 As the concentration of the ligand was increased, a dose-dependent quenching in fluorescence intensity, with no apparent change in emission maximum, was observed (Figure 5a,b and Figure S9). The fluorescence quenching upon ligand binding could be due to the distinct conformation attained by the nucleoside probe, which is less rigid and favors a nonradiative decay pathway as a result of its proximity to the polyaromatic ligands.59 The apparent Kd values determined from fluorescence experiment revealed that PDS binds to the antiparallel conformation with higher binding affinity as compared to hybrid-type structures formed in KCl conditions (Figure 5c and Table 3). On the contrary, BRACO19 shows higher binding affinity for hybrid-type structures as compared to the antiparallel GQ topology. Further, the suitability of 19F label in detecting the ligand binding event was tested by using H-Telo DNA ON 3 in NaCl, wherein it forms only an antiparallel conformation with an intense peak at –122.14 ppm. Upon increasing PDS concentration, the peak intensity of free GQ structure reduced, and a new broad peak at –121.69 ppm corresponding to the ligand-bound GQ appeared (Figure 5d).
Figure 5.
Nucleoside probe 1 enables the monitoring of ligand binding to different GQ topologies. (a, b) Representative emission spectra for the titration of labeled H-Telo DNA ON 3 (0.5 μM) in sodium phosphate buffer (10 mM, pH 7.0) containing 100 mM NaCl and potassium phosphate buffer (10 mM, pH 7.0) containing 100 mM KCl, respectively, with increasing concentrations of PDS. Samples were excited at 330 nm with excitation and emission slit widths of 4 and 6 nm, respectively. The dashed line represents the spectrum of ON 3 without PDS. (c) Curve fits for the binding of PDS and BRACO19 to GQ structures of H-Telo DNA ON 3 in different ionic conditions. (d) Changes in 19F NMR signal of the antiparallel GQ structure of ON 3 (100 μM) in sodium phosphate buffer containing 100 mM NaCl as a function of increasing PDS concentration.
Table 3. Dissociation Constant (Kd) of PDS and BRACO19 Binding to H-Telo DNA ON 3.
|
Kd (μM) |
||
|---|---|---|
| ligand | in NaCl | in KCl |
| PDS | 0.63 ± 0.03 | 1.26 ± 0.06 |
| BRACO19 | 1.01 ± 0.03 | 0.57 ± 0.05 |
Small molecule ligands tested against ONs in screening assays are usually soluble in DMSO. To test the amount of DMSO that would not affect the prevalent GQ topology in solution, CD, fluorescence, and 19F NMR spectrum of ON 3 were recorded in a buffer containing an increasing percentage of DMSO. The results indicate that up to 2.5% of DMSO can be used in binding assays without affecting the GQ topology (Figure S10).
Structural Insights into the H-Telo DNA Overhang in Live Cells
We took advantage of the sensitivity of the 19F label, which provides distinct NMR signatures for different GQs, in determining the topology of H-Telo overhang in live cells. Xenopus laevis oocyte was chosen as the model cell as it is large in size, easily injectable, and is also commonly used in the in-cell NMR analysis of proteins60 and, more recently, nucleic acids.45,61 These NMR studies rely on transfection or microinjection of isotope-enriched proteins and oligonucleotides as endogenous proteins and nucleic acids do not contain intrinsic isotope labels. Similarly, a notable number of GQ-specific light-up and fluorogenic small molecule probes require transfection of the ON sequence of interest into the cells for efficient visualization.38b,40,62 Unlike proteins, isotope labeling of nucleic acids with 15N/13C is laborious and very expensive. In this context, the fluorobenzofuran-modified nucleoside analogue has an added advantage as it can be easily incorporated into various DNA ON sequences by the solid-phase method.
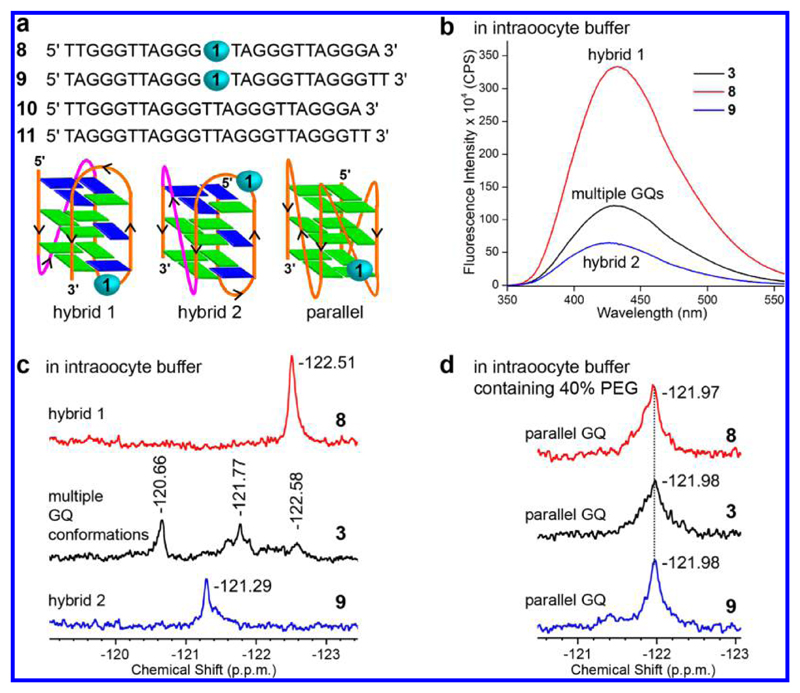
To obtain a progressive understanding of the GQ structure in cell-free and cellular environments, the 19F NMR spectrum was recorded in intraoocyte buffer conditions, oocyte clear lysate, egg extract (ex vivo model), and live oocyte (in vivo model). Particularly for these experiments, fluorobenzofuran-labeled H-Telo DNA ONs 8 and 9, which predominantly form hybrid-type 1 and 2 GQ structures, respectively, in K+ ionic conditions were additionally synthesized (K+ concentration is significantly higher than Na+ concentration in the intraoocyte environment).63,64 While both of these sequences form a 3 + 1 GQ structure (three strands parallel and one strand antiparallel), they show a difference in loop arrangement as the double chain reversal loop in hybrid 1 and hybrid 2 is located at the 5′-end and 3′-end, respectively (Figure 6a).
Figure 6.
19F NMR signature of individual H-Telo DNA GQ topologies in intraoocyte buffer conditions. (a) Sequence of labeled H-Telo DNA ONs 8 and 9 and respective control unmodified ONs 10 and 11, which predominantly adopt hybrid 1 and hybrid 2 GQ conformation, respectively. The double chain reversal loop (marked in magenta) in hybrid 1 and hybrid 2 is located at the 5′-end and 3′-end, respectively. (b) Nucleoside analogue 1 photophysically distinguishes hybrid 1 and hybrid 2 GQ structures of ONs 8 and 9. ON samples (0.5 μM) were excited at 330 nm with excitation and emission slit widths of 4 and 6 nm, respectively. (c) 19F NMR signature of H-Telo DNA ONs 3, 8, and 9 (100 μM) in intraoocyte buffer at 18 °C. (d) 19F NMR signature of the parallel GQ conformation of ONs 3, 8, and 9 (100 μM). ONs 3, 8, and 9 convert into parallel GQ conformation in the presence of PEG (see Figure S11 for CD profiles).26
Fluorescence and NMR analyses in intraoocyte buffer (25 mM HEPES, pH = 7.5, 10.5 mM NaCl, 110 mM KCl, 130 nM CaCl2, 1 mM MgCl2, 0.1 mM EDTA)45a clearly revealed the ability of the nucleoside probe to distinguish different GQ structures of H-Telo ONs 3, 8, and 9 (Figure 6). Hybrid 1 conformation of 8 exhibited significantly higher fluorescence intensity (∼5-fold) as compared to hybrid 2 conformation of 9 (Figure 6b). Similarly, these GQs showed sharp and distinct 19F NMR signals (Figure 6c). Consistent with the structural polymorphism of ON 3, we observed intermediate fluorescence and multiple 19F signals. In intraoocyte buffer, containing 40% of a synthetic crowding agent (PEG), all three ONs gave a similar chemical shift corresponding to the parallel conformation (Figure 6d and Figure S11). PEG, due to its dehydrating effect, is known to bias the H-Telo DNA ONs to adopt a parallel GQ structure in solution as well as in the solid state.28 It is important to mention here that imino proton signals of labeled ONs were marginally affected by the 5-fluorobenzofuran label (Figure S12). Further, CD and Tm measurements revealed that the control unmodified and modified ONs formed respective GQ structures with little difference in CD and melting profiles (Figure S13 and Table S3). From these studies, we successfully determined the 19F NMR signature of individual H-Telo GQ structures for further conformational analysis in oocytes (Figure S14).
Mechanically crushed oocyte suspension was heated at 95°C to denature the proteins, and the clear lysate thus obtained after centrifugation was used in NMR experiments. In case of measurements in egg extract, the oocytes were allowed to mature, which were then crushed, and without further manipulations the mixture was centrifuged. The crude interphase egg extract, apart from providing molecular crowding, is also known to maintain most of the biological activities of an intact cell.44 The 19F NMR spectrum of ON 3 in clear lysate almost resembled the spectra obtained in intraoocyte buffer conditions (Figure S15). Although in egg extract there was an indication for the formation of multiple GQ structures, the 19F signals were broad and not well resolved. This observation is not surprising as molecular crowding and inhomogeneity of the extract can reduce the tumbling rate and increase the relaxation process.65 It is important to mention here that the 1H NMR spectrum of 3 in lysate and egg extract was poorly resolved to provide structural information, which is in consensus with the reported NMR spectrum of the native H-Telo DNA ON of the same sequence.45b However, the 19F NMR experiment indicates that telomeric ON repeat 3 is not completely converted into the parallel topology in cellular environment as has been predicted by using synthetic molecular crowding agents and cosolvents (Figure S15).26,28
A spectrum of ON 8 in clear lysate displayed a sharp peak at –122.47 ppm corresponding to the signature of hybrid 1 GQ structure obtained in intraoocyte buffer conditions (Figure 7a). Interestingly, a small but visible peak at –121.97 ppm potentially corresponding to the parallel GQ conformation was also observed. Further analysis in egg extract produced a broader peak most probably encompassing both hybrid 1 and parallel conformations. In the absence of experimental proof, it is believed that the merging of signals could be due to dynamic interconversion between hybrid 1 and parallel conformations.66,67 Unfortunately, 19F NMR signatures could not be matched in live oocytes due to line broadening. 1H NMR spectrum of this ON sequence indicated the formation of hybrid 1 structure in clear lysate, but failed to provide structural information in egg extract due to severe line broadening (Figure 7a). These results support the notion that parallel GQ is not the only conformation adopted by a H-Telo DNA repeat in the cellular environment.
Figure 7.
19F label of nucleoside 1 serves as a useful tool to determine the GQ structure of H-Telo overhang in cellular environment. (a) 19F and 1H NMR spectra of ON 8 in intraoocyte buffer, oocyte clear lysate, egg extract, and intraoocyte buffer containing 40% PEG. (b) 19F and 1H NMR spectra of ON 9 in intraoocyte buffer, oocyte clear lysate, egg extract, live oocytes, and intraoocyte buffer containing 40% PEG. NMR measurements were performed at 18 °C.
ON 9 presented an identical 19F NMR signal (–121.29 ppm) in clear lysate and egg extract corresponding to the hybrid 2 conformation obtained in intraoocyte buffer conditions, albeit little line broadening in egg extract (Figure 7b). Following this observation, a concentrated solution of ON 9 (3.6 mM) in water was microinjected (50 nL) into live oocytes. Nearly 200 oocytes each containing nearly 180 μM of the DNA ON were used in the in-cell NMR experiments. Rewardingly, a broad peak at –121.29 ppm characteristic of a hybrid 2 GQ conformation was obtained in live oocytes (Figure 7b). Unlike ON 8, the hybrid 2 conformation of ON 9 did not show detectable conversion into other structures. Notably, under these conditions, there was no apparent cell death. Because of severe line broadening, 1H NMR analysis in egg extract and live oocytes could not be used in deducing the structure, which again substantiates the limited use of 1H NMR in the in-cell structural analysis (Figure 7b). To confirm if the observed 19F signal associated with the folded GQ structure of ON 9 is indeed emanating from inside the live cells, the following control experiments were performed. After the NMR spectrum was recorded, the extracellular buffer was subjected to 19F NMR and fluorescence analyses. While no signal was seen in 19F NMR spectrum, an emission band at 430 nm indicated only a small fraction (6–10%) of the microinjected sample leaked into the extracellular buffer during the NMR acquisition time (∼10 h, Figure S16). The integrity of labeled ON 9 in egg extract and live oocytes after NMR measurement was also examined. The egg extract and live oocytes were heat denatured, and the clear lysate obtained was subjected to HPLC and mass analyses. The retention time of extracted ON 9 and its mass as compared to the authentic sample confirmed the intactness of the ON in the cellular environment (Figure S17 and Table S4). It is important to mention here that in-cell concentration of the dual-app probe-labeled ON is considerably higher than the concentrations of nucleic acids in cells. However, performing NMR analysis using ultrasensitive cryogenic 19F probe will significantly bring down the working concentration of labeled DNA ONs in cells.68
Among the various GQ-forming sequences predicted to be present in the genome, the ones present in proto-oncogenes have attracted significant attention.1,2 Many of these sequences have been individually studied in vitro to assess their propensity to adopt different GQ structures.12 However, it is very important to understand what GQ topology or topologies an individual GQ-forming sequence will form in the cell and how they interact with ligands so that screening platforms can be designed to identify GQ motif-specific binders. In this regard, fluorobenzofuran-modified nucleoside probe is highly beneficial as its fluorescence and NMR labels could be used to set up screening assays and detect GQ topology in cells, respectively. Further, high throughput parallel and conventional solid-phase DNA synthesizers have similar reaction cycles. Hence, access to different GQ-forming sequences labeled with the dual-app probe should not be difficult.
Conclusions
We have used an innovative yet a simple probe design approach to develop a new nucleoside probe functioning both as conformation-sensitive fluorescent and as NMR labels. The fluorophore component of the dual-purpose probe provides an in vitro platform to photophysically distinguish different GQ topologies and estimate topology-specific binding of ligands. The 19F-label of the nucleoside exhibits unique signatures for individual GQ conformations, thereby facilitating the direct correlation of GQ structures formed in vitro and in native cellular environment. Our findings using the dual-purpose nucleoside probe indicate that H-Telo DNA ON repeat adopts multiple conformations, namely the hybrid-type and parallel GQ structures in cellular environment. The telomeric repeat did not outrightly get converted into a parallel conformation in cellular environment. This is contrary to the GQ studies using immunofluorescence staining in fixed cells, synthetic crowding agents, and under crystallization conditions, which strongly support the predominant formation of a parallel conformation.28,42 The utility of our probe and new findings demonstrated using telomeric DNA repeat are highly important on two counts. The conformations adopted by a nucleic acid motif in non-native conditions need not be favored under complex cellular environment, and the structural information obtained using synthetic cell models cannot be straightforwardly extended to native cells. Hence, understanding the topology adopted by individual GQ-forming sequences in cells and then identifying small molecule ligands that can specifically target a GQ motif among others in the genome will have a profound impact on GQ-directed therapeutic strategies. In this context, our bifunctional nucleoside probe represents a unique tool, which will not only enables the determination of GQ structures adopted by individual GQ-forming sequences in cells by NMR, but also harness GQ-directed therapeutic strategies by supporting fluorescence-based discovery platforms to identify topology-specific binders.
Methods
Detailed synthetic procedure and characterization of 5-fluorobenzofuran-2′-deoxyuridine 1 and its phosphoramidite substrate 2 are described in the Supporting Information. Photophysical and NMR analyses of the nucleoside probe can be found in the Supporting Information. Incorporation of nucleoside analogue into GQ-forming H-Telo DNA ONs by solid-phase method and their characterization by MALDI-TOF mass, CD, and UV-thermal melting measurements are provided in the Supporting Information.
Steady-State and Time-Resolved Fluorescence of Labeled H-Telo DNA ONs
GQs of ONs 3–5 (0.5 μM) were formed by heating the ONs at 90 °C for 3 min either in sodium phosphate buffer (10 mM, pH 7.0) containing 100 mM NaCl or in potassium phosphate buffer (10 mM, pH 7.0) containing 100 mM KCl. The samples were allowed to come to room temperature over a period of 2 h and were kept at 4 °C for 30 min. ON duplexes were made by heating a 1:1.1 mixture of ONs 3–5 and complementary DNA ON 7 at 90 °C for 3 min in buffers as mentioned above. Similarly, ONs 3, 8, and 9 were annealed to form respective GQ structures in intraoocyte buffer (25 mM HEPES (pH = 7.5), 10.5 mM NaCl, 110 mM KCl, 130 nM CaCl2, 1 mM MgCl2, 0.1 mM EDTA). Steady-state fluorescence analysis of GQs (0.5 μM) and duplexes (0.5 μM) was performed by exciting the samples at 330 nm with excitation and emission slit widths of 4 and 6 nm, respectively, at 20 °C. Time-resolved fluorescence analysis was performed by exciting the samples (0.5 μM) using a 339 nm LED source (IBH, UK, NanoLED-339L) on a TCSPC instrument (Horiba Jobin Yvon, Fluorolog-3) at 20 °C, and collecting the fluorescence signal at respective emission maximum. All measurements were performed in triplicate, and lifetimes were calculated by fitting the decay profile using IBH DAS6 software. The χ2 value for all of the curve fits was found to be nearly 1. Quantum yield determination is provided in the Supporting Information.
Fluorescence Binding Assay
H-Telo DNA ON 3 was annealed to form respective GQs in different buffers as mentioned above. A series of ON 3 samples (0.5 μM) containing increasing concentration of the ligand (PDS or BRACO19; from 8 nM to 5 μM) was prepared and incubated for 30 min before fluorescence analysis. Samples were excited at 330 nm with excitation and emission slit widths of 4 and 6 nm, respectively. The measurements were made in triplicate at 20 °C. The spectrum corresponding to a blank without any ON but containing the particular ligand concentration was subtracted from each spectrum. From the dose-dependent quenching curves, the apparent dissociation constants (Kd) for the binding of ligands to the H-Telo DNA ON were determined by fitting the normalized fluorescence intensity (FN) versus log of ligand concentration plot to the Hill equation (Origin 8.5).46
Fi is the fluorescence intensity at each ligand concentration. F0 and Fs are the fluorescence intensities in the absence of ligand (L) and at saturation point, respectively. n is the Hill coefficient or degree of cooperativity associated with the binding.
Sample Preparation for 19F NMR Analysis of H-Telo DNA ONs
Phosphate and Intraoocyte Buffers
H-Telo DNA ONs (100 μM) were annealed into GQs in phosphate buffers (containing NaCl or KCl) and intraoocyte buffer by heating the samples at 90 °C for 3 min. All samples contained 20% D2O. The samples were cooled to room temperature and were kept at 4 °C for 30 min. Samples were transferred into a Shigemi tube (5 mm advance NMR microtube) for NMR analysis.
Clear Lysate45
Around 250 healthy stage VI oocytes were selected and kept in a Petri dish containing Ori-Ca2+ buffer (5 mM HEPES (pH = 7.6), 110 mM NaCl, 5 mM KCl, 2 mM CaCl2, and 1 mM MgCl2). The Petri dish was kept on an ice bath for 15 min, and the oocytes were transferred to another Petri dish containing ice cold intraoocyte buffer. Finally, oocytes were transferred into an Eppendorf tube, buffer above the oocytes was removed carefully, and 200 μL of ice cold intraoocyte buffer supplemented with 20% D2O was added. Oocytes were mechanically crushed in ice, and insoluble mixture was removed by centrifuging at 20 000g for 20 min at 4 °C. The interphase extract was taken in another Eppendorf tube and heated at 95 °C for 10 min. The precipitated protein fraction was removed by centrifuging at 20 000g for 10 min at 4 °C. The supernatant (∼270 μL) was taken in an Eppendorf tube, and 30 μL of 1 mM preannealed DNA ON stock in water (see above) was added to it so that the final concentration of the DNA ON was 100 μM.
Egg Extract44
850–900 oocytes were transferred into a Petri dish containing 1 μM progesterone in Ori Ca2+ buffer and incubated for 12 h. Matured eggs were then washed thoroughly with Ori buffer (5 × 15 mL). The eggs in Ori Ca2+ buffer were kept on an ice bath for 20 min. The eggs were then transferred to another Petri dish containing ice cold intraoocyte buffer, and finally to an Eppendorf tube containing intraoocyte buffer (200 μL) composed of 20% D2O. The buffer just above the eggs was removed, and again intraoocyte buffer (200 μL) containing 20% D2O was added. This step was repeated two more times. The eggs were then packed by centrifuging at 400g for 1 min (4 °C), and the buffer just above the eggs was removed very carefully. Afterward, the eggs were centrifuged for 5 min at 12 000g (4 °C) and were mechanically crushed on an ice bath. The extract was centrifuged for 30 min at 12 000g (4 °C). Around 280 μL of crude interphase extract was obtained, and 15 μL of 5 mM preannealed DNA ON stock in water was added. The prepared mixture was incubated at 37 °C for 30 min and subjected to NMR analysis.
Live Oocytes
The chemically synthesized ONs, which were PAGE purified, were again precipitated using butanol (400 μL of butanol for 40 μL of DNA sample). ONs were dissolved in water and filtered two times using a 0.45 μm spin filter tube. A stock solution of DNA ONs (3.6 mM) in autoclaved water was heated at 90 °C for 3 min and slowly cooled to room temperature. 50 nL of DNA ON stock was microinjected into each oocyte using a NARISHIGE micromanipulator equipped with a manual oocyte microinjection pipet (Drummond Scientific Co.). Around 200 oocytes were injected with the labeled DNA ON over a period of 3.5 h. Each injected oocyte will contain ∼180 μM of the labeled DNA ON based on the injected volume and size of an oocyte (∼1 μL). Microinjected oocytes were transferred to a Petri dish containing Ori Ca2+ buffer and incubated for 1 h. The oocytes were thoroughly washed with Ori Ca2+ buffer (5 × 15 mL) and transferred to a Shigemi NMR tube prefilled with 1 mL of Ori Ca2+ buffer containing 20% D2O and 10% Ficoll 400. Ficoll was used in the extracellular buffer so that the oocytes are not crushed during the NMR acquisition time.61
19F NMR Analysis of H-Telo DNA ONs
19F NMR spectra were recorded at a frequency of 564.9 MHz on a Bruker AVANCE III HD ASCEND 600 MHz spectrometer equipped with BB(F) Double Channel Probe. All spectra were referenced relative to an external standard trifluorotoluene (TFT, –63.72 ppm). In phosphate buffers, NMR studies were performed at 25 °C, and in intraoocyte buffer, lysate, egg extract, and live oocytes, spectra were recorded at 18 °C. The following spectral parameters were used:48,69 19F excitation pulse, 11 μs; spectral width, 21.28 ppm; transmitter frequency offset, –121.14 ppm; acquisition time, 1.4 s in phosphate buffer, 80 ms in intraoocyte buffer and lysate, 50 ms in egg extract and live oocytes; relaxation delay, 1.5 s in phosphate buffer, 1 s in intraoocyte buffer, lysate, egg extract, and live oocytes; number of scans 28 000 or 39 000. Using these parameters, 19F NMR spectra were obtained in 8–10 h. Each spectrum was processed with an exponential window function using lb = 10 Hz in case of studies in buffer, lysate, and egg extract and lb = 50 Hz in case of in-cell study.69
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b08436.
Experimental procedure, characterization data, and supporting figures and tables (PDF)
Acknowledgments
We thank Dr. Jeet Kalia for generously providing the oocytes for NMR experiments from his frog oocytes facility at IISER Pune. We thank Dr. Jeetender Chugh and Mr. Harshad Paithankar for their kind help in performing NMR experiments. We thank DST-FIST program for funding the IISER Pune high-field NMR facility. S.M. thanks UGC, India, for a graduate research fellowship. S.G.S. thanks Wellcome Trust-DBT India Alliance (IA/S/16/1/502360) and DST, India (EMR/2014/000419), for research grants.
Footnotes
ORCID
Seergazhi G. Srivatsan: 0000-0001-5765-3967
Notes
The authors declare no competing financial interest.
References
- (1).Hänsel-Hertsch R, Di Antonio M, Balasubramanian S. Nat Rev Mol Cell Biol. 2017;18:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- (2).Bedrat A, Lacroix L, Mergny J-L. Nucleic Acids Res. 2016;44:1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Meyne J, Ratliff RL, Moyzis R. Proc Natl Acad Sci U S A. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rhodes D, Lipps H. J Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Halder K, Wieland M, Hartig JS. Nucleic Acids Res. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc Natl Acad Sci U S A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Gomez D, Guédin A, Mergny J-L, Salles B, Riou J-F, Teulade-Fichou M-P, Calsou PA. Nucleic Acids Res. 2010;38:7187–7198. doi: 10.1093/nar/gkq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Alzeer J, Vummidi BR, Roth PJC, Luedtke NW. Angew Chem, Int Ed. 2009;48:9362–9365. doi: 10.1002/anie.200903685. [DOI] [PubMed] [Google Scholar]
- (10).Wang X-D, Ou T-M, Lu Y-J, Li Z, Xu Z, Xi C, Tan J-H, Huang S-L, An L-K, Li D, Gu L-Q, et al. J Med Chem. 2010;53:4390–4398. doi: 10.1021/jm100445e. [DOI] [PubMed] [Google Scholar]
- (11).McLuckie KIE, Waller ZAE, Sanders DA, Alves D, Rodriguez R, Dash J, McKenzie GJ, Venkitaraman AR, Balasubramanian S. J Am Chem Soc. 2011;133:2658–2663. doi: 10.1021/ja109474c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Collie GW, Parkinson GN. Chem Soc Rev. 2011;40:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- (13).Neidle S. Nat Rev Chem. 2017;1 doi: 10.1038/s41570-023-00551-5. 0041. [DOI] [PubMed] [Google Scholar]
- (14).Micco M, Collie GW, Dale AG, Ohnmacht SA, Pazitna I, Gunaratnam M, Reszka AP, Neidle S. J Med Chem. 2013;56:2959–2974. doi: 10.1021/jm301899y. [DOI] [PubMed] [Google Scholar]
- (15).Dai J, Carver M, Hurley LH, Yang D. J Am Chem Soc. 2011;133:17673–17680. doi: 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).(a) Dhamodharan V, Harikrishna S, Bhasikuttan AC, Pradeepkumar PI. ACS Chem Biol. 2015;10:821–833. doi: 10.1021/cb5008597. [DOI] [PubMed] [Google Scholar]; (b) Mohanty J, Barooah N, Dhamodharan V, Harikrishna S, Pradeepkumar PI, Bhasikuttan AC. J Am Chem Soc. 2013;135:367–376. doi: 10.1021/ja309588h. [DOI] [PubMed] [Google Scholar]
- (17).Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JKC, et al. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- (18).Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, Santos ND, Silvester J, Wei V, Garcia J, Kabeer F, et al. Nat Commun. 2017;8 doi: 10.1038/ncomms14432. 14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chen Y, Yang D. Curr Protoc Nucleic Acid Chem. 2012;50:17.5.1–17.5.17. doi: 10.1002/0471142700.nc1705s50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Marušič M, Plavec J. Angew Chem, Int Ed. 2015;54:11716–11719. doi: 10.1002/anie.201505348. [DOI] [PubMed] [Google Scholar]
- (21).Vialet B, Gissot A, Delzor R, Barthélémy P. Chem Commun. 2017;53:11560–11563. doi: 10.1039/c7cc05693a. [DOI] [PubMed] [Google Scholar]
- (22).Miyoshi D, Fujimoto T, Sugimoto N. Top Curr Chem. 2012;330:87–110. doi: 10.1007/128_2012_335. [DOI] [PubMed] [Google Scholar]
- (23).Zhou J, Wei C, Jia G, Wang X, Feng Z, Li C. Chem Commun. 2010;46:1700–1702. doi: 10.1039/b925000j. [DOI] [PubMed] [Google Scholar]
- (24).Vummidi BR, Alzeer J, Luedtke NW. ChemBioChem. 2013;14:540–558. doi: 10.1002/cbic.201200612. [DOI] [PubMed] [Google Scholar]
- (25).Panda D, Saha P, Das T, Dash J. Nat Commun. 2017;8 doi: 10.1038/ncomms16103. 16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Miller MC, Buscaglia R, Chaires JB, Lane AN, Trent JO. J Am Chem Soc. 2010;132:17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- (27).Lannan FM, Mamajanov I, Hud NV. J Am Chem Soc. 2012;134:15324–15330. doi: 10.1021/ja303499m. [DOI] [PubMed] [Google Scholar]
- (28).Heddi B, Phan AT. J Am Chem Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- (29).Manna S, Panse CH, Sontakke VA, Sangamesh S, Srivatsan SG. ChemBioChem. 2017;18:1604–1615. doi: 10.1002/cbic.201700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Chen Z, Zheng K-W, Hao Y-H, Tan Z. J Am Chem Soc. 2009;131:10430–10438. doi: 10.1021/ja9010749. [DOI] [PubMed] [Google Scholar]
- (32).Shrestha P, Jonchhe S, Emura T, Hidaka K, Endo M, Sugiyama H, Mao H. Nat Nanotechnol. 2017;12:582–588. doi: 10.1038/nnano.2017.29. [DOI] [PubMed] [Google Scholar]
- (33).Artusi S, Perrone R, Lago S, Raffa P, Di Iorio E, Palu G, Richter SN. Nucleic Acids Res. 2016;44:10343–10353. doi: 10.1093/nar/gkw968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Plückthun A. Proc Natl Acad Sci U S A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM, Jr, Sen D, Lansdrop PM. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. Nat Chem. 2014;6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).(a) Laguerre A, Hukezalie K, Winckler P, Katranji F, Chanteloup G, Pirrotta M, Perrier-Cornet J-M, Wong JMY, Monchaud D. J Am Chem Soc. 2015;137:8521–8525. doi: 10.1021/jacs.5b03413. [DOI] [PubMed] [Google Scholar]; (b) Tseng T-Y, Wang Z-F, Chien C-H, Chang T-C. Nucleic Acids Res. 2013;41:10605–10618. doi: 10.1093/nar/gkt814. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Laguerre A, Wong JMY, Monchaud D. Sci Rep. 2016;6:32141. doi: 10.1038/srep32141. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Doria F, Nadai M, Zuffo M, Perrone R, Freccero M, Richter SN. Chem Commun. 2017;53:2268–2271. doi: 10.1039/c6cc08492c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Huang W-C, Tseng T-Y, Chen Y-T, Chang C-C, Wang Z-F, Wang C-L, Hsu T-N, Li P-T, Chen C-T, Lin J-J, Lou P-J, et al. Nucleic Acids Res. 2015;43:10102–10113. doi: 10.1093/nar/gkv1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Chen S-B, Hu M-H, Liu G-C, Wang J, Ou T-M, Gu L-Q, Huang Z-S, Tan J-H. J Am Chem Soc. 2016;138:10382–10385. doi: 10.1021/jacs.6b04799. [DOI] [PubMed] [Google Scholar]
- (41).Shivalingam A, Izquierdo MA, Marois AL, Vyšniauskas A, Suhling K, Kuimova MK, Vilar R. Nat Commun. 2015;6:8178. doi: 10.1038/ncomms9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Liu H-Y, Zhao Q, Zhang T-P, Wu Y, Xiong Y-X, Wang S-K, Ge Y-L, He J-H, Lv P, Ou T-M, Tan J-H, et al. Cell Chem Biol. 2016;23:1261–1270. doi: 10.1016/j.chembiol.2016.08.013. [DOI] [PubMed] [Google Scholar]
- (43).Azarkh M, Singh V, Okle O, Dietrich DR, Hartig JS, Drescher M. ChemPhysChem. 2012;13:1444–1447. doi: 10.1002/cphc.201100980. [DOI] [PubMed] [Google Scholar]
- (44).Hänsel R, Löhr F, Foldynoá-Trantírková S, Bamberg E, Trantírek L, Dötsch V. Nucleic Acids Res. 2011;39:5768–5775. doi: 10.1093/nar/gkr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).(a) Hänsel R, Foldynoá-Trantírková S, Löhr F, Buck J, Bongartz E, Bamberg E, Schwalbe H, Dötsch V, Trantárek L. J Am Chem Soc. 2009;131:15761–15768. doi: 10.1021/ja9052027. [DOI] [PubMed] [Google Scholar]; (b) Hänsel R, Löhr F, Trantárek L, Dötsch V. J Am Chem Soc. 2013;135:2816–2824. doi: 10.1021/ja312403b. [DOI] [PubMed] [Google Scholar]
- (46).Tanpure AA, Srivatsan SG. Nucleic Acids Res. 2015;43:e149. doi: 10.1093/nar/gkv743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Chen H, Viel S, Ziarelli F, Peng L. Chem Soc Rev. 2013;42:7971–7982. doi: 10.1039/c3cs60129c. [DOI] [PubMed] [Google Scholar]
- (48).Bao H-L, Ishizuka T, Sakamoto T, Fujimoto K, Uechi T, Kenmochi N, Xu Y. Nucleic Acids Res. 2017;45:5501–5511. doi: 10.1093/nar/gkx109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ishizuka T, Zhao P-Y, Bao H-L, Xu Y. Analyst. 2017;142:4083–4088. doi: 10.1039/c7an00941k. [DOI] [PubMed] [Google Scholar]
- (50).(a) Giam CS, Lyle JL. J Am Chem Soc. 1973;95:3235–3239. [Google Scholar]; (b) Sinkeldam RW, Wheat AJ, Boyaci H, Tor Y. ChemPhysChem. 2011;12:567–570. doi: 10.1002/cphc.201001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).(a) Gros J, Rosu F, Amrane S, De Cian A, Gabelica V, Lacroix L, Mergny J-L. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sproviero M, Fadock KL, Witham AA, Manderville RA. ACS Chem Biol. 2015;10:1311–1318. doi: 10.1021/cb5009926. [DOI] [PubMed] [Google Scholar]
- (52).Rachwal PA, Fox KR. Methods. 2007;43:291–301. doi: 10.1016/j.ymeth.2007.05.004. [DOI] [PubMed] [Google Scholar]
- (53).Jean JM, Hall KB. Proc Natl Acad Sci U S A. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Doose S, Neuweiler H, Sauer M. ChemPhysChem. 2009;10:1389–1398. doi: 10.1002/cphc.200900238. [DOI] [PubMed] [Google Scholar]
- (55).Feigon J, Koshlap KM, Smith FW. Methods Enzymol. 1995;261:225–255. doi: 10.1016/s0076-6879(95)61012-x. [DOI] [PubMed] [Google Scholar]
- (56).Wang Y, Patel D. J Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- (57).Rodriguez R, Müller S, Yeoman JA, Trentesaux C, Riou J-F, Balasubramanian S. J Am Chem Soc. 2008;130:15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Moore MJB, Schultes CM, Cuesta J, Cuenca F, Gunaratnam M, Tanious FA, Wilson WD, Neidle S. J Med Chem. 2006;49:582–599. doi: 10.1021/jm050555a. [DOI] [PubMed] [Google Scholar]
- (59).Le DD, Di Antonio M, Chan LKM, Balasubramanian S. Chem Commun. 2015;51:8048–8050. doi: 10.1039/c5cc02252e. [DOI] [PubMed] [Google Scholar]
- (60).(a) Selenko P, Serber Z, Gadea B, Ruderman J, Wagner G. Proc Natl Acad Sci U S A. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Selenko P, Frueh DP, Elsaesser SJ, Haas W, Gygi SP, Wagner G. Nat Struct Mol Biol. 2008;15:321–329. doi: 10.1038/nsmb.1395. [DOI] [PubMed] [Google Scholar]; (c) Serber Z, Selenko P, Hänsel R, Reckel S, Löhr F, Ferrell JE, Jr, Wagner G, Dötsch V. Nat Protoc. 2006;1:2701–2709. doi: 10.1038/nprot.2006.181. [DOI] [PubMed] [Google Scholar]
- (61).Salgado GF, Cazenave C, Kerkour A, Mergny J-L. Chem Sci. 2015;6:3314–3320. doi: 10.1039/c4sc03853c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Xu Y, Suzuki Y, Ito K, Komiyama M. Proc Natl Acad Sci U S A. 2010;107:14579–14584. doi: 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. J Am Chem Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Phan AT, Luu KN, Patel D. J Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).(a) Selenko P, Wagner G. J Struct Biol. 2007;158:244–253. doi: 10.1016/j.jsb.2007.04.001. [DOI] [PubMed] [Google Scholar]; (b) Ye Y, Liu X, Zhang Z, Wu Q, Jiang B, Jiang L, Zhang Xu, Liu M, Pielak GJ, Li C. Chem - Eur J. 2013;19:12705–12710. doi: 10.1002/chem.201301657. [DOI] [PubMed] [Google Scholar]
- (66).Buer BC, Chugh J, Al-Hashimi HM, Marsh ENG. Biochemistry. 2010;49:5760–5765. doi: 10.1021/bi100605e. [DOI] [PubMed] [Google Scholar]
- (67).Kleckner IR, Foster MP. Biochim Biophys Acta, Proteins Proteomics. 2011;1814:942–968. doi: 10.1016/j.bbapap.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Giassa I-C, Rynes J, Fessl T, Foldynova-Trantirkova S, Trantirek L. FEBS Lett. 2018;592:1997–2011. doi: 10.1002/1873-3468.13054. [DOI] [PubMed] [Google Scholar]
- (69).Riedl J, Pohl R, Rulíšek L, Hocek M. J Org Chem. 2012;77:1026–1044. doi: 10.1021/jo202321g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.