Abstract
The chromosomes within the eukaryotic cell nucleus are highly dynamic and adopt complex hierarchical structures. Understanding how this three-dimensional (3D) nuclear architectureaffects gene regulation, cell cycle progression and disease pathogenesis are important biological questions in development and disease. Recently, many genome-wide technologies including chromosome conformation capture (3C) and 3C-based methodologies (4C, 5C, and Hi-C) have been developed to investigate 3D chromatin structure. In this review, we introduce 3D genome methodologies, with a focus on their application for understanding the nuclear architecture of the human malaria parasite, Plasmodium falciparum. An increasing amount of evidence now suggests that gene regulation in the parasite is largely regulated by epigenetic mechanisms and nuclear reorganization. Here, we explore the 3D genome architecture of P. falciparum, including local and global chromatin structure. In addition, molecular components important for maintaining 3D chromatin organization including architectural proteins and long non-coding RNAs are discussed. Collectively, these studies contribute to our understanding of how the plasticity of 3D genome architecture regulates gene expression and cell cycle progression in this deadly parasite.
Keywords: Plasmodium, Malaria, Genome architecture, 3D genome, Chromatin
1. The organization of the eukaryotic nucleus
The coordinated expression of genes is a fundamental feature in all living cells including eukaryotic pathogens such as the human malaria parasite, Plasmodium falciparum. Specific gene expression signatures can be found depending on different cell conditions such as cell growth, division and response to stimuli. For this reason, it is not surprising that atypical gene expression patterns often correlate with development and pathology. Mechanistically, different components such as promoters, enhancers and their target genes need to be organized and brought into close spatial proximity to warrant a transcriptional outcome. However, for a transcriptional unit to form, two problems need to be overcome. First, as DNA inside the nucleus is tightly wrapped around histone proteins and assembled into nucleosomes, which are then coiled and packaged together to form a fiber also known as chromatin, this packaging system introduces issues for the transcriptional machinery to access DNA when needed. Second, promoter regions and their distal regulatory elements need to physically interact to allow for gene expression, yet promoters and corresponding regulatory elements are often separated linearly along the chromosome.
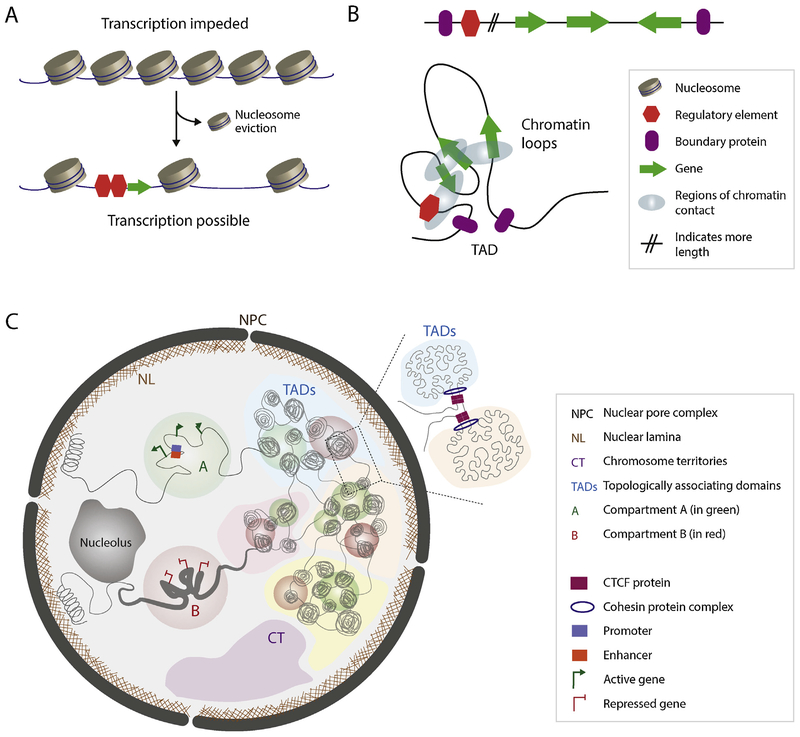
Recent genome-wide nucleosome landscape studies in organisms such as yeast and human revealed patters of nucleosome organization, including lower nucleosome density in intergenic regions, nucleosome-depleted regions next to promoters, and strongly positioned nucleosomes (i.e −1 and +1 nucleosomes) at the transcription start site (TSS) containing the variant histone H2A.Z [1–4]. Additionally, histone post-translational modifications (PTMs) such as acetylation of histone H3 at lysine 9 or 14 (H3K9ac and H3K14ac) can change the stability of nucleosomes, causing an open chromatin structure and promoting transcription [5], while trimethylation of histone H3 at lysine 9 and 27 (H3K9me3 and H3K27me3) can promote heterochromatin states and repress gene expression [6]. These findings suggest that transcriptional regulation can be largely affected by specific positioning of nucleosomes (Fig. 1A).
Fig. 1.
Hierarchical chromatin organization within the eukaryotic nucleus. (A) DNA is tightly wrapped around histone proteins and assembled into nucleosomes. This packaging system impedes transcription as transcriptional machinery cannot access the DNA. Local displacement of nucleosomes allows transcription machinery to access the genes. (B) Linear representation of a hypothetical genomic region. Possible chromatin loops that can form to specially connect promoters and their regulatory elements are depicted. (C) Model of 3D genome organization at different resolutions as described for animal models. During interphase chromosomes are organized into discrete chromosome territories (CTs). Within CTs, a particular locus can be surrounded by an active (A compartment) or repressive environment (B compartment). Different topologically-associating domains (TADs) with similar epigenetic signatures are characterized by strong inter-domain interactions and are organized into compartments. On the right, two different TADs are schematically represented. Insulator proteins (CTCF) are shown in purple rectangles and cohesin protein complexes are shown in blue circles.
To overcome the hierarchical organization of chromatin and the spatial separation of promoters and their regulatory elements for transcriptional activation or repression, chromatin loops can be formed (Fig. 1B). One well-known example in mammalian genomes is the locus control region (LCR) of beta-globin ([H9252]-globin) genes, which strongly interacts with its target genes via long-range chromatin contacts [7–11]. Chromatin loops can also be used for transcription repression as seen with the maternal copy of the insulin-like growth factor 2 (Igf2), where placement of this gene within an inactive chromatin loop prevents enhancer-promoter contact [12].
Another interesting discovery in the field of chromatin and nuclear organization is that chromosomes are spatially arranged into megabase-sized sub-domains termed topologically-associating domains (TADs) (Fig. 1C), where genomic regions within the same TAD interact more frequently compared to the regions located in adjacent domains [13–15]. TAD boundaries are enriched for insulator proteins such as CTCF, and transcription activating marks such as H3K4me3 [13]. Using recently developed chromosome conformation (3C) technologies (see methodologies below), data revealed that TADs are organized into two types of compartments corresponding to open (compartment A) and closed chromatin (compartment B) (Fig. 1C) [16]. At an even larger scale, chromatin is organized into specific chromosome territories that rarely intermix. This observation was made by both fluorescence in situ hybridization (FISH) studies [17,18] and genome-wide chromosome conformation capture-based (Hi-C) data [16], where interactions between loci on the same chromosome occurred at a much higher frequency than contacts between different chromosomes. Collectively, these observations conclude that nuclear architecture is formed in a hierarchical fashion. In recent years, many new technologies including chromosome conformation capture (3C) have been instrumental in investigating the role of the 3D genome in gene regulation and cellular functions in many cell types including the human malaria parasite, P. falciparum.
2. Chromosome conformation capture technologies
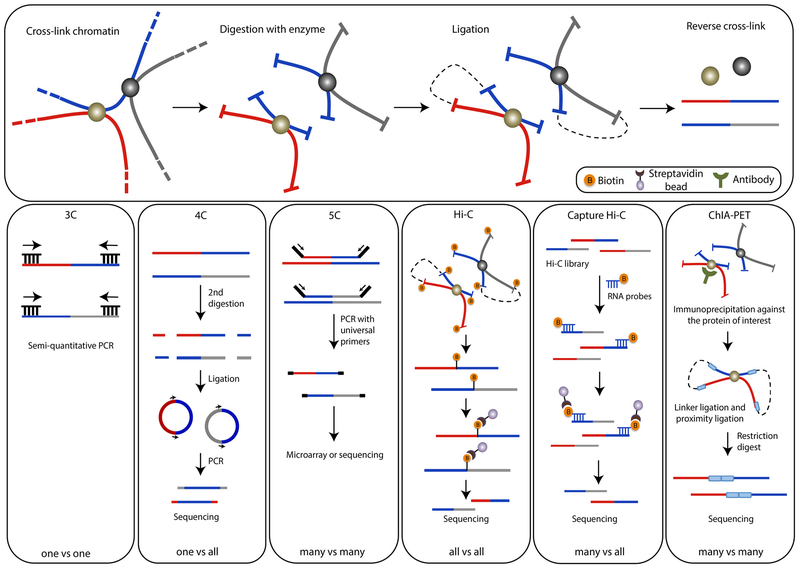
The development of chromosome conformation (3C) technologies has been critical for investigating chromatin spatial organization and its role in biological function with more detail. Chromosome conformation capture (3C), developed by Job Dekker and colleagues in 2002, allows for the capture of long-range chromatin interactions between two genomic sites [19]. During the 3C analysis, the samples undergo a series of experimental steps (Fig. 2), including in situ fixation with formaldehyde, enzymatic digestion, ligation of DNA ends in spatial proximity and finally detection by quantitative polymerase chain reaction (qPCR) using primers corresponding to the two chromatin loci in question. A major limitation of this assay is that it can only be used to investigate chromatin interactions between predefined loci. In 2006, chromosome conformation capture on chip (4C) was developed [20,21] to detect chromatin interactions between one locus and multiple loci. Additionally, a high-throughput 3C approach, named 3C-Carbon Copy (5C), was developed to address the capture of multiple-to-multiple loci interactions [22] and map cis- and trans-chromatin interactions.
Fig. 2.
Chromosome conformation capture (3C)-based technologies to study chromatin architecture. Experimental procedures of 3C and 3C-based methodologies. Cross-linking with formaldehyde is followed by enzymatic digestion and proximity-based ligation. Ligation products are enriched and contact frequencies are determined subsequently for all methodologies discussed.
In 2009, with the advancement of next-generation sequencing (NGS) technology, Dekker and colleagues developed a method, termed Hi-C, to identify ‘all-vs-all’ chromatin interactions in a given genome [16]. Hi-C combines DNA digestion and ligation with massively parallel sequencing to map chromatin interactions within spatial proximity at the level of the entire genome. Since its introduction in 2009, the Hi-C methodology has been used in numerous organisms and cell types to explore chromosome interactions within the nucleus. In 2014, in situ Hi-C was applied to construct kilobase-resolution chromatin interaction maps for nine mammalian cell types to identify the presence of chromatin loops between enhancers and promoters [23]. More recently, single-cell Hi-C methodologies have enabled the detection of chromatin interactions within single cells and their heterogeneity within a population of cells [24,25].
Hi-C, although a powerful tool for capturing chromosome organization within the nucleus, comes at the cost of two problems. First, low resolution due to sequencing depth does not allow for enough spatial resolution to interrogate specific chromosomal contracts. Second, the sequencing of invalid read pairs does not reflect authentic chromatin interactions. To address these limitations, Capture Hi-C methodology was developed, which enables the capture of targeted genomic regions by hybridizing ligation products to biotin labeled probes [26]. Using this approach, it is possible to deep sequence specific loci and achieve high-resolution mapping of local chromatin interactions at a much-improved level compared to Hi-C.
All the above-mentioned technologies, 3C, 4C, 5C, Hi-C and Capture Hi-C, aim to capture long-range chromatin interactions involved in transcriptional regulation irrespective of the proteins facilitating the interactions. Chromatin interaction analysis by paired-end tag (ChIA-PET), a methodology that combines 3C with chromatin-immunoprecipitation (ChIP), was established to capture chromatin interactions mediated by a protein of interest such as transcription factors or regulators [27]. Since the introduction of ChIA-PET, more sensitive and cost-effective methods have been developed to analyze chromatin interactions on a genome level in an unbiased manner. Proximity ligation-assisted ChIP-seq (PLAC-seq) [28] greatly improves the efficiency and accuracy of detecting long-range chromatin interactions over ChIA-PET, while protein-centric chromatin conformation assay (HiChIP) [29] greatly improves the efficiency of DNA contact capture and minimizes the possible false-positive chromatin interactions.
Collectively, since the development of the first chromosome conformation capture assay in the early 2000’s, these 3C-based technologies have generated vast amounts of genome-wide interaction data and have greatly advanced our understanding of 3D chromatin organization. However, it is important to keep in mind that initial 3C-based methods only provided steady-state chromo-some conformations as measured across a population of cells and could not be used to investigate chromatin dynamics on a cell-by-cell basis. Microscopy-based techniques such as DNA-FISH (DNA fluorescence in situ hybridization), while limited to a few regions of interest, have provided important information about cell cycle timing and spatial DNA organization at the single-cell level. Therefore, until single-cell Hi-C methodologies become more accessible and more affordable, chromatin architecture is most effective and informative when studied using a combination of complementary approaches. Some of these approaches have been successfully used to understand mechanisms regulating gene expression in human malaria parasites throughout their infectious life cycles.
3. The malaria parasite
Malaria, caused by a protozoan parasite of the genus Plasmodium is responsible for an estimated 200 million cases of disease and over 400,000 deaths each year [30]. Of the Plasmodium species that infect humans, Plasmodium falciparum is the most prevalent and most deadly. P. vivax, while less virulent than P. falciparum, can remain in the body in a dormant state and can cause relapses weeks to months after the initial infection.
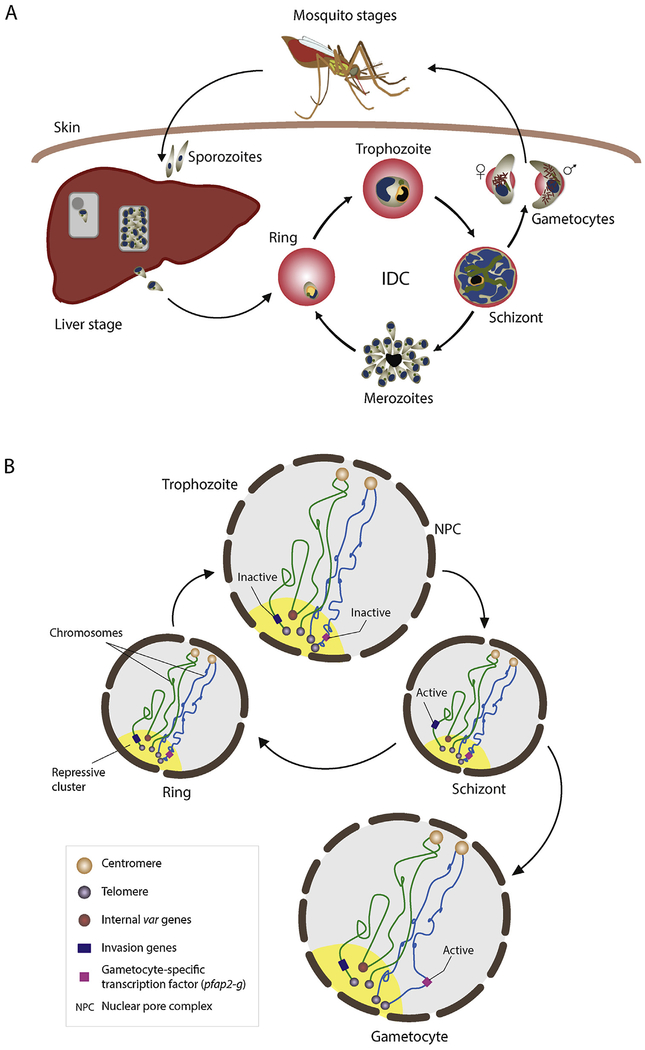
All Plasmodium species have similar, complex life cycles that involve two hosts: the Anopheles mosquito and the human host (Fig. 3A). The human infection starts as a female mosquito takes a blood meal and injects sporozoites into the host bloodstream. The sporozoites travel into the liver, invade hepatocytes and replicate extensively to release thousands of merozoites into the blood stream. The merozoites invade red blood cells (RBCs) where they begin a 48-hour replication cycle and develop asexually [31,32]. During the intra-erythrocytic developmental cycle (IDC) the parasite progresses through three distinct stages termed ring, trophozoite and schizont and multiply into 16–32 daughter cells by a processed known as schizogony. These daughter parasites burst out of the host cell and invade new healthy erythrocytes.
Fig. 3.
Parasite life cycle (A) and dynamic remodeling of the parasite nuclear architecture (B) during the asexual and sexual life stages. A) Sporozoites enter humans through an infected mosquito bite and subsequently establish an infection in the liver. Upon egress from the liver, the parasite replicates asexually inside red blood cells (intraerythrocytic developmental cycle; IDC). During this replication cycle, a small proportion of parasites will commit to sexual differentiation into male and female gametocytes that can be taken up by a mosquito. Sexual reproduction takes place inside the mosquito midgut and ultimately results in the formation of sporozoites that can be transmitted to a new human host. B) Centromeric (orange circles) and telomeric (grey circles) clusters remain localized to the nuclear periphery at all stages. Telomere regions as well as the internally located var genes (red circle) are located within the repressive center(s) (yellow). The remaining genome (light grey) is largely present in a transcriptionally permissive euchromatin state. Extensive remodeling of the nucleus takes place as the parasite progresses through the ring, trophozoite, schizont and gametocyte stages. In the transition from the relatively inactive ring stage to the transcriptionally active trophozoite stage, the nuclear size and the number of nuclear pores increase. Invasion genes that localize to the parasite repressive center become active at the schizont stage and are moved away from the heterochromatin environment. Loss of interaction between the repressive cluster and pfap2-g transcription factor is observed at the gametocyte stages.
During the IDC, environmental stress, such as limited nutrient levels, can trigger sexual development of parasites into male and female gametocytes. The mature gametocytes can be ingested by a feeding mosquito, undergo sexual replication in the mosquito midgut and develop further into salivary gland sporozoites that can be transmitted to a new human host.
Understanding this tightly regulated multi-stage life cycle of the parasite still remains an important goal in malaria research, especially since the development of therapeutics that could halt parasite differentiation or transmission can lead to disruption of the cycle of infection. Developmental stage transitions are regulated by coordinated changes in gene expression; however, the nature and contribution of mechanisms regulating gene expression [33–38], including the role of chromatin structure and three-dimensional (3D) genome organization in the parasite are only now starting to emerge.
4. The Plasmodium falciparum genome
The genome of P. falciparum is relatively compact, contains twenty-three million base pairs per haploid genome and is organized into fourteen chromosomes [39]. The parasite genome is extremely rich in adenine and thymine with an average of ~80% AT that can reach ~90 to 95% AT within introns and intergenic regions of the genome. This unusual nucleic acid bias has caused significant hindrance for identifying regulatory DNA elements in the P. falciparum genome. While components of the general transcription machinery (RNA polymerase II and general transcription factors) have been identified in the parasite [40,41], a surprisingly low number of specific transcription factors have been identified and validated [41–48]. Therefore, the coordinated cascade of gene expression observed throughout the parasite life cycle is unlikely to be regulated by the unusually low number of specific transcription factors that have been identified. This points towards additional mechanisms such as post-transcriptional [49–53], translational, and post-translational [52,54–56] mechanisms as well as epigenetic features and 3D genome organization regulating the expression of the 6372 parasite genes. In the following sections, we provide an overview of our current understanding of genome organization and its role in gene expression and disease pathogenesis of malaria.
5. P. falciparum local chromatin structure
5.1. Histone and nucleosome landscape
The nucleosome landscape of P. falciparum is, in some ways, similar to other eukaryotes. First, nucleosome depleted regions are observed in the promoters of genes [57–59], likely to allow for binding of the transcriptional machinery. Second, intergenic regions show lower levels of nucleosome occupancy as compared to coding regions [57,59,60]. While some studies suggest that sequencing biases introduced by the high AT-content in the intergenic regions contribute to the differences in nucleosome occupancy [61], alternative methodologies that enrich for nucleosome depleted regions such as FAIRE-Seq [60] or ATAC-Seq [62,63] validate this initial coverage. Additionally, lower nucleosome occupancy in intergenic regions is also observed in other eukaryotes [64–67], including Tetrahymena thermophila, an organism with an AT-rich genome [68]. Finally, several studies have also demonstrated that promoter regions of highly transcribed genes display more open chromatin structure compared to repressed gene regions [57,58,61].
While the parasite shares some nucleosome landscape features with other eukaryotes, P. falciparum displays several unique characteristics. First, the parasite genome lacks a strongly positioned +1 nucleosome that marks the TSS and instead, harbors strongly positioned nucleosomes at the start and end of coding regions [57,58]. Next, histone variants H2A.Z and H2B.Z exclusively occupy intergenic regions of the parasite genome [69,70], instead of being restricted to active promoters similar to other eukaryotes. These histone variants are also known to bind weakly but more efficiently to AT-rich DNA sequences, and are likely an adaptation by the parasite to allow for nucleosome assembly in AT-rich regions.
Another unusual feature of the nucleosome landscape in P. falciparum, which remains an area of debate, is the variability of nucleosome levels during the parasite life cycle [57,60]. At the transcriptionally active trophozoite stage, nucleosome levels drop in a genome-wide fashion, and as the asexual cycle progresses towards the schizont stage, nucleosomes are reassembled and DNA is condensed in preparation for egress and invasion of new RBCs. Other studies suggest a transcription-independent nucleo-some positioning that is driven by sequence specificity [61]. The reason for these conflicting observations is likely linked to data normalization and cell cycle progression. When nucleosome occupancy datasets are normalized by number of parasite nuclei at the trophozoite stage, these datasets display nucleosome depletion in a genome-wide fashion. Additionally, change in nucleosome profile throughout the parasite life cycle has been confirmed by alternative methodologies including western blots [71], mass spec-trometry [57,72,73], MNase-Seq, FAIRE-Seq [60], and ChIP-seq [57]. It is therefore likely that genome-wide nucleosome eviction drives the massive transcriptional event at the trophozoite stage. At the schizont stage, nucleosomes are reassembled and restored to levels detected before transcriptional activation in preparation for re-invasion. At the early and late stages of the parasite erythrocytic cycle, transcriptional regulation is more likely driven via classical mechanisms such as stage-specific transcription factors and his-tone PTMs. Recent work provided a mechanism for coordinated regulation of invasion genes by a transcription factor bromodomain protein (PfBDP1) [74]. PfBDP1 enrichment was observed at the transcription start sites of invasion genes at the late schizont stage and was shown to regulate invasion gene expression by binding to acetylated histone H3. Furthermore, conditional PfBDP1 knockdown caused dramatic defects in parasite invasion and growth, confirming the essentiality of this chromatin-associated factor for the coordinated expression of invasion genes in the parasite and indicating the importance of histone modifications in parasite development.
A recent mass spectrometry experiment has indeed identified a total of 232 different histone PTMs during the P. falciparum IDC stages, including methylations, acetylations, phosphorylations, ubiquitylations and sumoylations [73]. Many of these PTMs are novel in both Plasmodium and other organisms and their exact functions remain to be determined. Interestingly, the majority of the parasite genome remains in a constitutively active state marked by activating histone marks (H3K9ac and H3K4me3), while a small subset including subtelomeric regions and a few internal loci, remain in a condensed heterochromatin environment marked by silencing histone marks (H3K9me3 and H3K36me3) and heterochromatin protein 1 (PfHP1) [75,76]. These heterochromatin environments harbor gene families encoding clonally variant antigens (var, rifin, stevor and pfmc2tm), invasion genes (eba and clag) and several other loci including pfap2-g, a gametocyte-specific transcription factor [77–81]. In mammalian genomes, H3K9ac and H3K4me3 localization is restricted to active promoters [82–86], while in P. falciparum these modifications extend to promoters, 5’ coding regions of highly transcribed genes [87,88], intergenic regions and ‘silenced’ promoters [81,87,89].
Most recently, ChIP-seq experiments demonstrated that heterochromatin features marked by H3K9me3 and HP1 are stage-specific, restricted to a limited amount of parasite-specific genes and conserved in several Plasmodium species, and that epigenome patterns are linked to distinct stages of parasite development [90,91]. Both of these studies nicely demonstrated that these features are essential and are likely to be connected to transcriptional activity of several parasite-specific gene families i.e those involved in pathogenesis, liver cell invasion, erythrocyte remodeling, and regulation of sexual differentiation.
6. Global nuclear architecture in P. falciparum
6.1. Organization of the parasite 3D genome
An important question in P. falciparum genome biology is how the structural features of 3D chromatin organization are established, maintained and altered during cell cycle progression and development of the parasite. Similar to complex metazoans, the 3D genome structure of P. falciparum plays important roles in gene expression regulation. For decades, numerous microscopic imaging technologies have been the preferred methods for visualizing nuclear architecture in many different organisms [92–94]. Initially, immunofluorescence and FISH experiments were performed to observe global chromatin organization within the parasite nucleus [95,96]. Earlier FISH experiments revealed that repressed gene regions, subtelomeric and a few internal loci harboring antigenic variation genes, localized to a few clusters around the parasite nucleus [76,80] [90,97]. These observations were further confirmed using Hi-C experiments that capture intra- and interchromosomal interactions in a genome-wide manner [75,90].
Hi-C experiments demonstrated that within the P. falciparum nucleus, chromosomes are arranged into folded structures, which are anchored at the centromere with both chromosome arms folding over parallel to each other (Fig. 3B) [75]. Much like the 3D chromosome organization in the similarly sized budding and fission yeast, centromeres and telomeres cluster in opposite regions of the nucleus [98,99]. Compared to the yeast genome, the parasite nucleus displays an additional level of complexity, mostly as a result of genes involved in antigenic variation (var genes) located internally on five out of the fourteen chromosomes [75]. These internal var genes colocalize with the subtelomeric regions at the periphery of the nucleus by forming additional loops in the chromosomes. The clonally variant var gene expression and clustering of var genes in a condensed heterochromatin region is much like the epigenetic signature of the olfactory receptor genes in mice, where all but one are located in a heterochromatin environment enriched in H3K9me3 and H4K20me3 histone marks [100].
6.2. Regulation of virulence genes
Disease pathogenesis in malaria is caused by the ability of the parasite to escape the host immune response by expressing variants of antigens on the surface of the infected RBC. The var gene family, encoding erythrocyte membrane protein 1 (PfEMP1), remains the best characterized multigene family in the parasite to date [101]. There are approximately 60 var genes present in the parasite haploid genome but only one var gene is expressed at any given time [102]. This process of allelic exclusion contributes to constant antigenic variation and enables the parasite to evade attacks by the host immune system. Extensive research has been conducted to explore the mechanism of var gene regulation in vitro and increasing evidence indicates that these genes are regulated at the epigenetic and chromatin structure level. Silent var genes are marked by H3K9me3 and PfHP1 and are localized to repressed regions of the genome at the periphery of the nucleus [75,76,78–81,103,104]. Several studies highlight proteins that are critical for var gene activation and silencing, and disruption of these proteins results in the loss of monoallelic var gene expression [80,105–110]. Together, these results emphasize the relationship between precise nuclear organization and regulation of antigenic variation in the parasite.
While the parasite genome architecture is relatively simple and does not contain well-defined TADs, it adopts drastically different forms during the numerous life cycle stages. For example, the heterochromatin region expands significantly during sexual differentiation [90]. Genes encoding for proteins exported to the surface of RBCs and proteins that are critical for parasite invasion to new erythrocytes such as Eba, rhoph1/clag, acbp and PfRH, which are no longer needed, move towards the parasite repressive center (Fig. 3B). Repression of these genes is also mediated by the H3K9me3 heterochromatin mark. While an association of the invasion genes to the heterochromatin cluster during sexual differentiation is observed, the master regulator of sexual differentiation, the pfap2-g transcription factor, disassociates from the repressive center to euchromatin regions [90]. Hi-C data generated during asexual and sexual stages of the erythrocytic cycle also identified the formation of a superdomain on chromosome 14 in sexual stage parasites. One arm of chromosome 14 moves away from the rest of the chromosomes in a more compact structure close to the nuclear membrane [90]. This structure is reminiscent of the barr body structure within nuclei of female mammals, which consists of a compacted inactive X chromosome [111,112]. The domain boundary of these super domain observed in the malaria parasite during sexual differentiation is relatively close to a transcription factor, pfap2, known to be essential during sexual reproduction in the mosquito. Using a transgenic P. berghei strain that expressed a GFP-tagged pfap2 protein, this ApiAP2 TF was shown to localize to the nucleus of female gametocytes. Significant changes of the chromatin structure surrounding genes involved in hepatocyte invasion were also observed at the sporozoite stage, the stage responsible for transmission of the disease from mosquitoes to human. These results demonstrate that large-scale chromatin structure rearrangement may control specific regulation of gene expression in the parasite and highlight a strong connection between heterochromatin, genome organization, and stage-specific gene expression. Collectively, these results emphasize the importance of understanding the molecular components regulating dynamic nuclear organization and its role in regulating gene expression in Plasmodium.
As a whole, it appears that the nuclear architecture in P. falciparum shares features with both unicellular and multicellular organisms. While unicellular throughout most of its life cycle, the parasite needs to survive in a variety of cell types in both human and mosquito hosts. Most importantly, P. falciparum has to evade the host immune system. This strategy helps control expression of variable but distinct proteins at the different life-cycle stages of the parasite. For this reason, the parasite has likely developed a more complex nuclear architecture to regulate cell cycle progression, differentiation and survival compared to other unicellular organisms with a small genome size.
A combination of Hi-C experiments and advanced microscopy methodologies have revealed that parasite nuclear architecture undergoes distinct changes during its life cycle progression. Throughout the asexual, sexual and transmission stages of the parasite, the nucleus and chromatin are drastically remodeled, most likely to allow for the changes in transcriptional activity that takes place during these stages (Fig. 3B). First, after invading erythrocytes, Plasmodium ring-stage parasites settle and grow slowly inside the RBCs. After 18 to 24 h, the nucleus expands in size [113], reaching its maximum size and volume at the trophozoite stage, which can easily be observed using microscopy images of Giemsa stained par asites [75]. Second, the number of nuclear pores increases from 3 to 7 clustered pores at the ring stage to 12–58 pores evenly distributed around the nucleus at the trophozoite stage [113]. The increased number of nuclear pores suggests the need to facilitate the transcriptionally active trophozoite stage. Finally, along with the increased nuclear volume, the chromatin structure opens up [75] together with nucleosome eviction [57,60], chromosome intermingling [75] and increased transcriptional activity. As the parasite reaches schizogony, the contents of the nucleus along with the nuclear pores are distributed between the daughter nuclei, nucleosomes are reassembled, chromosome territories are re-established and the chromatin structure recompacts.
6.3. Proteins involved in P. falciparum nuclear organization
The structure of the eukaryotic nucleus is organized by a network of proteins, known as nucleoskeleton that anchor the contents of the nucleus to the nuclear envelope and help mediate the movement of chromosomes. It is now apparent that the nucleoskeleton confers shape and functionality to the eukaryotic nucleus [114]. Some of these proteins directly interact with chromatin by binding to DNA, histones and chromatin-remodeling complexes. Not all the proteins forming the metazoan nucleoskeleton are present in all unicellular eukaryotes including P. falciparum. Specifically, P. falciparum seems to lack lamin proteins that form the nuclear lamina on the inside of the nuclear envelope [115]. In addition, the parasite lacks CTCF proteins that insulate the boundaries of TADs and tether chromatin to the nuclear lamina [116]. However, proteins such as actin, myosin and kinesin, motor components of the nucleoskeleon, are present in the parasite and are most likely critical for maintaining chromatin structure. They have also recently gained attention as possible drug targets as a small molecule inhibitor against Kinesin-5 in P. falciparum and P. vivax showed no cross-reactivity in human cell lines [117].
Similar to other eukaryotes, the parasite centromeres are marked by PfCENH3, a special isoform of histone H3 [118,119]. PfCENH3 has also been shown to interact with PfCENP-C to form the functional centromeric complex that facilitates kinetochore assembly [118–120]. Furthermore mitotic spindle integrity was lost upon disruption of the dimerization domain of PfCENP-C, suggesting that this protein is essential for chromosome segregation and cell cycle progression much like in other eukaryotes [120]. Several SMCs (structural maintenance of chromosome) critical in the assembly of cohesin and condensin complexes, which are essential for chromosome assembly and segregation in all eukaryotic organisms [121,122], have also been identified in the P. falciparum genome. However, the characterization of these proteins in the parasite has yet to be completed.
Proteins involved in the maintenance of the heterochromatin cluster(s) in the parasite nucleus, have been subjected to extensive research. The most critical structural element of the parasite repressive center identified thus far has been heterochromatin protein-1 (PfHP1), which is structurally and functionally homologous to HP1 in other eukaryotes. PfHP1 specifically binds to the repressive histone mark H3K9me3. Knockdown of this protein disrupts the monoallelic expression of genes involved in antigenic variation, the var genes, known to cluster together in heterochromatin environments [123]. A recent study, using Hi-C methodology, also demonstrated that the depletion of PfHP1 results in the loss of chromatin structure and parasite death [90], highlighting the essentiality of PfHP1 in controlling chromatin structure and parasite development. SPE2- interacting protein (PfSIP2), a member of the ApiAP2 transcription factor family in P. falciparum, has also been shown to be involved in organization of subtelomeric heterochromatin [124]. PfSIP2 interacts with SPE2 DNA motifs present on telomeric and subtelomeric regions. DNA/RNA binding proteins PfAlba1–4 (Acetylation lowers binding affinity) family of proteins have also been implicated in heterochromatin maintenance. PfAlba 1,2 and 4 bind to TARE6 (telomere-associated repetitive element 6) [125] and PfAlba3 has been shown to bind to telomeric and subtelomeric regions as well as var gene promoters [126]. PfSIR2A, a histone deacetylase (HDAC) also present in the repressive center, is thought to increase DNA-binding affinity by deacetylating lysine residues in its N-terminus via interaction with PfAlba3 [126].
It is now increasingly evident that P. falciparum nuclear organization and chromosome dynamics are quite complex, and it is likely that many other proteins are regulating these processes. Identification and characterization of these molecular components will likely contribute to a better understanding of the molecular machinery regulating chromatin structure, gene regulation and parasite development.
6.4. Long non-coding RNAs
Emerging evidence confirms that non-protein coding transcripts, long non-coding RNAs (lncRNAs), also play a role in transcriptional regulation and 3D genome activity by affecting chromatin-remodeling events such as chromatin looping and nucleosome positioning [127]. A well-studied lncRNA, Xist, mediates X-chromosome inactivation during zygotic development in placental mammals [128]. The expression of the Xist lncRNA on the X chromosome recruits histone-modifying enzymes that place repressive histone marks, such as H3K9 and H3K27 methylation at the Xist locus, leading to gene silencing and heterochromatin formation.
In Plasmodium, a variety of long non-coding RNAs (lncRNAs) transcribed from telomere-associated repetitive elements (TAREs) have been identified [129–131]. These TARE-lncRNAs reach their highest expression levels at the schizont stage. While the exact role of these lncRNAs has yet to be determined, it is likely that these transcripts are part of a network regulating and maintaining the heterochromatin environment within the parasite nucleus. Interestingly, the lncRNA TARE regions harbor ApiAP2 transcription factor PfSIP2 binding sites. Since PfSIP2 has been implicated in heterochromatin formation around subtelomeric regions, these TARE-lncRNAs may play a role in regulating var gene expression. The TARE-lncRNAs in P. falciparum are functionally similar to the eukaryotic family of non-coding RNA called telomeric repeat-containing RNA (TERRA) important for telomere maintenance and heterochromatin assembly [132], which further validates the importance of lncRNA-TAREs for regulating the repressive centers within the parasite nucleus.
The monoallelic var gene expression can also be regulated via transcription of lncRNAs. Two lncRNAs are transcribed from a bidirectional promoter within the intron of var genes [133]. These transcripts are incorporated into chromatin after being capped but not polyadenylated. It is likely that the sense lncRNA functions to silence var gene expression, while the antisense lncRNA is associated with the single active var gene [133]. Thus, the presence of these lncRNAs adds an additional layer of complexity to epigenetic mechanisms regulating var gene expression.
7. Conclusions
An increasing amount of evidence emphasizes the importance of the epigenetic landscape and nuclear architecture in regulating gene expression in P. falciparum and higher eukaryotes such as human and mouse. Here we discuss the local and global genome architecture of P. falciparum including similarities with other eukaryotic organisms and differences that contribute to the unique biology of the parasite. As outlined above, the Plasmodium 3D nuclear architecture points toward a binary structure where a majority of the genome is maintained in a transcriptionally permissive euchromatin state and a small subset of genes are harbored within a transcriptionally repressed heterochromatin state. The asexual cycle of the parasite reflects large changes in genome organization that are characterized by nucleosome landscape and global histone levels. The overall genome organization in gametocytes is similar to IDC stages with a few exceptions including the localization of gametocyte-specific transcription factor, pfap2-g, erythrocytic remodeling and invasion genes. These results suggest that transcriptional regulation in the parasite is controlled at different layers and these layers shape the overall organization of the nucleus. However, our understanding of the parasite nucleome is far from complete. It is important to understand the extent to which nuclear reorganization controls gene expression in P. falciparum. In particular, molecular components that are likely involved in regulating genome architecture in the parasite could serve as potential drug targets that can disrupt parasite development with high specificity and low toxicity to the host.
Acknowledgements
This work was financially supported by the National Institutes of Health (grants R01 AI06775–01 and R01 AI136511 to KLR) and the University of California, Riverside (NIFA-Hatch-225935 to KGLR).
References
- [1].Guillemette B, et al. , Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning, PLoS Biol. 3 (12) (2005) e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jiang C, Pugh BF, A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome, Genome Biol. 10 (10) (2009) R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Raisner RM, et al. , Histone variant H2A.Z marks the 5’ ends of both active and inactive genes in euchromatin, Cell 123 (2) (2005) 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tolstorukov MY, et al. , Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes, Genome Res. 19 (6) (2009) 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Karmodiya K, et al. , H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells, BMC Genomics 13 (2012) 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kouzarides T, Chromatin modifications and their function, Cell 128 (4) (2007) 693–705. [DOI] [PubMed] [Google Scholar]
- [7].Carter D, et al. , Long-range chromatin regulatory interactions in vivo, Nat. Genet 32 (4) (2002) 623–626. [DOI] [PubMed] [Google Scholar]
- [8].Deng W, et al. , Reactivation of developmentally silenced globin genes by forced chromatin looping, Cell 158 (4) (2014) 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Palstra RJ, et al. , The beta-globin nuclear compartment in development and erythroid differentiation, Nat. Genet 35 (2) (2003) 190–194. [DOI] [PubMed] [Google Scholar]
- [10].Ragoczy T, et al. , The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation, Genes Dev. 20 (11) (2006) 1447–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tolhuis B, et al. , Looping and interaction between hypersensitive sites in the active beta-globin locus, Mol. Cell 10 (6) (2002) 1453–1465. [DOI] [PubMed] [Google Scholar]
- [12].Dean A, In the loop: long range chromatin interactions and gene regulation, Brief. Funct. Genomics 10 (1) (2011) 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dixon JR, et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions, Nature 485 (7398) (2012) 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nora EP, et al. , Spatial partitioning of the regulatory landscape of the X-inactivation centre, Nature 485 (7398) (2012) 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sexton T, et al. , Three-dimensional folding and functional organization principles of the Drosophila genome, Cell 148 (3) (2012) 458–472. [DOI] [PubMed] [Google Scholar]
- [16].Lieberman-Aiden E, et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome, Science (80-) 326 (5950) (2009) 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lichter P, et al. , Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries, Hum. Genet 80 (3) (1988) 224–234. [DOI] [PubMed] [Google Scholar]
- [18].Pinkel D, et al. , Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4, Proc. Natl. Acad. Sci. U. S. A 85 (23) (1988) 9138–9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dekker J, et al. , Capturing chromosome conformation, Science (80-) 295 (5558) (2002) 1306–1311. [DOI] [PubMed] [Google Scholar]
- [20].Simonis M, et al. , Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C), Nat. Genet 38 (11) (2006) 1348–1354. [DOI] [PubMed] [Google Scholar]
- [21].van de Werken HJ, et al. , Robust 4C-seq data analysis to screen for regulatory DNA interactions, Nat. Methods 9 (10) (2012) 969–972. [DOI] [PubMed] [Google Scholar]
- [22].Dostie J, et al. , Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements, Genome Res. 16 (10) (2006) 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rao SS, et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping, Cell 159 (7) (2014) 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ramani V, et al. , Massively multiplex single-cell Hi-C, Nat. Methods 14 (3) (2017) 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stevens TJ, et al. , 3D structures of individual mammalian genomes studied by single-cell Hi-C, Nature 544 (7648) (2017) 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mifsud B, et al. , Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C, Nat. Genet 47 (6) (2015) 598–606. [DOI] [PubMed] [Google Scholar]
- [27].Fullwood MJ, et al. , An oestrogen-receptor-alpha-bound human chromatin interactome, Nature 462 (7269) (2009) 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fang R, et al. , Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq, Cell Res. 26 (12) (2016) 1345–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mumbach MR, et al. , HiChIP: efficient and sensitive analysis of protein-directed genome architecture, Nat. Methods 13 (11) (2016) 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].WHO, World Malaria Report, 2016, 2016. http://www.who.int/malaria/publications/world-malaria-report-2016/report/en/.
- [31].Rosenberg R, et al. , An estimation of the number of malaria sporozoites ejected by a feeding mosquito, Trans. R. Soc. Trop. Med. Hyg 84 (2) (1990) 209–212. [DOI] [PubMed] [Google Scholar]
- [32].Yuda M, Ishino T, Liver invasion by malarial parasites–how do malarial parasites break through the host barrier? Cell Microbiol. 6 (12) (2004) 1119–1125. [DOI] [PubMed] [Google Scholar]
- [33].Bozdech Z, et al. , The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum, PLoS Biol. 1 (1) (2003) E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bunnik EM, et al. , Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum, Genome Biol. 14 (11) (2013) R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Le Roch KG, et al. , Discovery of gene function by expression profiling of the malaria parasite life cycle, Science (80-) 301 (5639) (2003) 1503–1508. [DOI] [PubMed] [Google Scholar]
- [36].Lopez-Barragan MJ, et al. , Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum, BMC Genomics 12 (2011) 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Otto TD, et al. , New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq, Mol. Microbiol 76 (1) (2010) 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rovira-Graells N, et al. , Transcriptional variation in the malaria parasite Plasmodium falciparum, Genome Res. 22 (5) (2012) 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gardner MJ, et al. , Genome sequence of the human malaria parasite Plasmodium falciparum, Nature 419 (6906) (2002) 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Callebaut I, et al. , Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes, BMC Genomics 6 (2005) 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Coulson RM, Hall N, Ouzounis CA, Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum, Genome Res. 14 (8) (2004) 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Balaji S, et al. , Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains, Nucleic Acids Res. 33 (13) (2005) 3994–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Campbell TL, et al. , Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite, PLoS Pathog. 6 (10) (2010) e1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kafsack BF, et al. , A transcriptional switch underlies commitment to sexual development in malaria parasites, Nature 507 (7491) (2014) 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sinha A, et al. , A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium, Nature 507 (7491) (2014) 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Young JA, et al. , In silico discovery of transcription regulatory elements in Plasmodium falciparum, BMC Genomics 9 (2008) 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yuda M, et al. , Transcription factor AP2-Sp and its target genes in malarial sporozoites, Mol. Microbiol 75 (4) (2010) 854–863. [DOI] [PubMed] [Google Scholar]
- [48].Yuda M, et al. , Identification of a transcription factor in the mosquito-invasive stage of malaria parasites, Mol. Microbiol 71 (6) (2009) 1402–1414. [DOI] [PubMed] [Google Scholar]
- [49].Balu B, et al. , CCR4-associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins, Eukaryot. Cell 10 (9) (2011) 1257–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bunnik EM, et al. , The mRNA-bound proteome of the human malaria parasite Plasmodium falciparum, Genome Biol. 17 (1) (2016) 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Eshar S, et al. , PfSR1 controls alternative splicing and steady-state RNA levels in Plasmodium falciparum through preferential recognition of specific RNA motifs, Mol. Microbiol 96 (6) (2015) 1283–1297. [DOI] [PubMed] [Google Scholar]
- [52].Kirchner S, Power BJ, Waters AP, Recent advances in malaria genomics and epigenomics, Genome Med. 8 (1) (2016) 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vembar SS, et al. , The PfAlba1 RNA-binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages, Genome Biol. 16 (2015) 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Caro F, et al. , Genome-wide regulatory dynamics of translation in the Plasmodium falciparum asexual blood stages, Elife (2014) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Foth BJ, et al. , Quantitative protein expression profiling reveals extensive post-transcriptional regulation and post-translational modifications in schizont-stage malaria parasites, Genome Biol. 9 (12) (2008) R177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Vaurijoux A, et al. , Detection of partial-body exposure to ionizing radiation by the automatic detection of dicentrics, Radiat. Res 178 (4) (2012) 357–364. [DOI] [PubMed] [Google Scholar]
- [57].Bunnik EM, et al. , DNA-encoded nucleosome occupancy is associated with transcription levels in the human malaria parasite Plasmodium falciparum, BMC Genomics 15 (2014) 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ponts N, et al. , Nucleosome occupancy at transcription start sites in the human malaria parasite: a hard-wired evolution of virulence? Infect. Genet. Evol 11 (4) (2011) 716–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Westenberger SJ, et al. , Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes, BMC Genomics 10 (2009) 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ponts N, et al. , Nucleosome landscape and control of transcription in the human malaria parasite, Genome Res. 20 (2) (2010) 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kensche PR, et al. , The nucleosome landscape of Plasmodium falciparum reveals chromatin architecture and dynamics of regulatory sequences, Nucleic Acids Res. 44 (5) (2016) 2110–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Schep AN, et al. , Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions, Genome Res. 25 (11) (2015) 1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Toenhake CG, et al. , Chromatin accessibility-based characterization of the Gene regulatory network underlying Plasmodium falciparum blood-stage development, Cell Host Microbe 23 (4) (2018) 557–569, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee CK, et al. , Evidence for nucleosome depletion at active regulatory regions genome-wide, Nat. Genet 36 (8) (2004) 900–905. [DOI] [PubMed] [Google Scholar]
- [65].Mavrich TN, et al. , Nucleosome organization in the Drosophila genome, Nature 453 (7193) (2008) 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Pokholok DK, et al. , Genome-wide map of nucleosome acetylation and methylation in yeast, Cell 122 (4) (2005) 517–527. [DOI] [PubMed] [Google Scholar]
- [67].Valouev A, et al. , A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning, Genome Res. 18 (7) (2008) 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Beh LY, et al. , DNA-guided establishment of nucleosome patterns within coding regions of a eukaryotic genome, Genome Res. 25 (11) (2015) 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hoeijmakers WA, et al. , H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome, Mol. Microbiol 87 (5) (2013) 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Petter M, et al. , H2A.Z and H2B.Z double-variant nucleosomes define intergenic regions and dynamically occupy var gene promoters in the malaria parasite Plasmodium falciparum, Mol. Microbiol 87 (6) (2013) 1167–1182. [DOI] [PubMed] [Google Scholar]
- [71].Le Roch KG, et al. , Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle, Genome Res. 14 (11) (2004) 2308–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Oehring SC, et al. , Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum, Genome Biol. 13 (11) (2012) R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Saraf A, et al. , Dynamic and combinatorial landscape of histone modifications during the intraerythrocytic developmental cycle of the malaria parasite, J. Proteome Res. 15 (8) (2016) 2787–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Josling GA, et al. , A Plasmodium Falciparum bromodomain protein regulates invasion gene expression, Cell Host Microbe 17 (6) (2015) 741–751. [DOI] [PubMed] [Google Scholar]
- [75].Ay F, et al. , Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression, Genome Res. 24 (6) (2014) 974–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Freitas-Junior LH, et al. , Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum, Nature 407 (6807) (2000) 1018–1022. [DOI] [PubMed] [Google Scholar]
- [77].Chookajorn T, et al. , Epigenetic memory at malaria virulence genes, Proc. Natl. Acad. Sci. U. S. A 104 (3) (2007) 899–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Crowley VM, et al. , Heterochromatin formation in bistable chromatin domains controls the epigenetic repression of clonally variant Plasmodium falciparum genes linked to erythrocyte invasion, Mol. Microbiol 80 (2) (2011) 391–406. [DOI] [PubMed] [Google Scholar]
- [79].Freitas-Junior LH, et al. , Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites, Cell 121 (1) (2005) 25–36. [DOI] [PubMed] [Google Scholar]
- [80].Lopez-Rubio JJ, Mancio-Silva L, Scherf A, Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites, Cell Host Microbe 5 (2) (2009) 179–190. [DOI] [PubMed] [Google Scholar]
- [81].Salcedo-Amaya AM, et al. , Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum, Proc. Natl. Acad. Sci. U. S. A 106 (24) (2009) 9655–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Barski A, et al. , High-resolution profiling of histone methylations in the human genome, Cell 129 (4) (2007) 823–837. [DOI] [PubMed] [Google Scholar]
- [83].Bernstein BE, et al. , Genomic maps and comparative analysis of histone modifications in human and mouse, Cell 120 (2) (2005) 169–181. [DOI] [PubMed] [Google Scholar]
- [84].Kim TH, et al. , A high-resolution map of active promoters in the human genome, Nature 436 (7052) (2005) 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nishida H, et al. , Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell, Chromosome Res. 14 (2) (2006) 203–211. [DOI] [PubMed] [Google Scholar]
- [86].Wang Z, et al. , Combinatorial patterns of histone acetylations and methylations in the human genome, Nat. Genet 40 (7) (2008) 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Bartfai R, et al. , H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3, PLoS Pathog. 6 (12) (2010) e1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Cui L, et al. , PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum, Eukaryot Cell 6 (7) (2007) 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Trelle MB, et al. , Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum, J. Proteome Res 8 (7) (2009) 3439–3450. [DOI] [PubMed] [Google Scholar]
- [90].Bunnik EM, et al. , Changes in genome organization of parasite-specific gene families during the Plasmodium transmission stages, Nat. Commun 9 (1) (2018) 1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Fraschka SA, et al. , Comparative heterochromatin profiling reveals conserved and unique epigenome signatures linked to adaptation and development of malaria parasites, Cell Host Microbe 23 (3) (2018) 407–420, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cremer T, et al. , Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture, Crit. Rev. Eukaryot. Gene Expr 10 (2) (2000) 179–212. [PubMed] [Google Scholar]
- [93].Misteli T, Beyond the sequence: cellular organization of genome function, Cell 128 (4) (2007) 787–800 [DOI] [PubMed] [Google Scholar]
- [94].Takizawa T, Meaburn KJ, Misteli T, The meaning of gene positioning, Cell 135 (1) (2008) 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Horrocks P, et al. , Control of gene expression in Plasmodium falciparum -ten years on, Mol. Biochem. Parasitol 164 (1) (2009) 9–25. [DOI] [PubMed] [Google Scholar]
- [96].Segal E, Widom J, Poly(dA:dT) tracts: major determinants of nucleosome organization, Curr. Opin. Struct. Biol 19 (1) (2009) 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Dahan-Pasternak N, et al. , PfSec13 is an unusual chromatin-associated nucleoporin of Plasmodium falciparum that is essential for parasite proliferation in human erythrocytes, J. Cell. Sci 126 (Pt 14) (2013) 3055–3069. [DOI] [PubMed] [Google Scholar]
- [98].Duan Z, et al. , A three-dimensional model of the yeast genome, Nature 465 (7296) (2010) 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Tanizawa H, et al. , Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation, Nucleic Acids Res 38 (22) (2010) 8164–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Magklara A, et al. , An epigenetic signature for monoallelic olfactory receptor expression, Cell 145 (4) (2011) 555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Miller LH, Good MF, Milon G, Malaria pathogenesis, Science (80-) 264 (5167) (1994) 1878–1883. [DOI] [PubMed] [Google Scholar]
- [102].Scherf A, Lopez-Rubio JJ, Riviere L, Antigenic variation in Plasmodium falciparum, Annu. Rev. Microbiol 62 (2008) 445–470. [DOI] [PubMed] [Google Scholar]
- [103].Flueck C, et al. , Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors, PLoS Pathog. 5 (9) (2009) e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Perez-Toledo K, et al. , Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes, Nucleic Acids Res. 37 (8) (2009) 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Coleman BI, et al. , A Plasmodium falciparum histone deacetylase regulates antigenic variation and gametocyte conversion, Cell Host Microbe 16 (2) (2014) 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Duraisingh MT, et al. , Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum, Cell 121 (1) (2005) 13–24. [DOI] [PubMed] [Google Scholar]
- [107].Tonkin CJ, et al. , Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum, PLoS Biol. 7 (4) (2009) e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Jiang L, et al. , PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum, Nature 499 (7457) (2013) 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Ukaegbu UE, et al. , Recruitment of PfSET2 by RNA polymerase II to variant antigen encoding loci contributes to antigenic variation in P. falciparum, PLoS Pathog. 10 (1) (2014) e1003854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Volz JC, et al. , PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division, Cell Host Microbe 11 (1) (2012) 7–18. [DOI] [PubMed] [Google Scholar]
- [111].Deng X, et al. , Bipartite structure of the inactive mouse X chromosome, Genome Biol 16 (2015) 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Giorgetti L, et al. , Structural organization of the inactive X chromosome in the mouse, Nature 535 (7613) (2016) 575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Weiner A, et al. , 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum, Cell Microbiol. 13 (7) (2011) 967–977. [DOI] [PubMed] [Google Scholar]
- [114].Simon DN, Wilson KL, The nucleoskeleton as a genome-associated dynamic’ network of networks’, Nat. Rev. Mol. Cell Biol 12 (11) (2011) 695–708. [DOI] [PubMed] [Google Scholar]
- [115].Batsios P, et al. , A lamin in lower eukaryotes? Nucleus 3 (3) (2012) 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Heger P, et al. , The chromatin insulator CTCF and the emergence of metazoan diversity, Proc. Natl. Acad. Sci. U.S. A 109 (43) (2012) 17507–17512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Liu L, et al. , Small molecule screen for candidate antimalarials targeting Plasmodium Kinesin-5, J. Biol. Chem 289 (23) (2014) 16601–16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hoeijmakers WA, et al. , Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony, Cell Microbiol. 14 (9) (2012) 1391–1401. [DOI] [PubMed] [Google Scholar]
- [119].Verma G, Surolia N, Plasmodium falciparum CENH3 is able to functionally complement Cse4p and its, C-terminus is essential for centromere function, Mol. Biochem. Parasitol 192 (1–2) (2013) 21–29. [DOI] [PubMed] [Google Scholar]
- [120].Verma G, Surolia N, The dimerization domain of PfCENP-C is required for its functions as a centromere protein in human malaria parasite Plasmodium falciparum, Malar. J 13 (2014) 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Freeman L, Aragon-Alcaide L, Strunnikov A, The condensin complex governs chromosome condensation and mitotic transmission of rDNA, J. Cell Biol 149 (4) (2000) 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Strunnikov AV, Jessberger R, Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions, Eur. J. Biochem 263 (1) (1999) 6–13. [DOI] [PubMed] [Google Scholar]
- [123].Brancucci NM, et al. , Heterochromatin protein 1 secures survival and transmission of malaria parasites, Cell Host Microbe 16 (2) (2014) 165–176. [DOI] [PubMed] [Google Scholar]
- [124].Flueck C, et al. , A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology, PLoS Pathog 6 (2) (2010) e1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Chene A, et al. , PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites, Nucleic Acids Res. 40 (7) (2012) 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Goyal M, et al. , Identification and molecular characterization of an Alba-family protein from human malaria parasite Plasmodium falciparum, Nucleic Acids Res. 40 (3) (2012) 1174–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Bohmdorfer G, Wierzbicki AT, Control of chromatin structure by Long noncoding RNA, Trends Cell Biol. 25 (10) (2015) 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Maclary E, et al. , Long nonoding RNAs in the X-inactivation center, Chromosome Res. 21 (6–7) (2013) 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Broadbent KM, et al. , A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs, Genome Biol. 12 (6) (2011) R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Raabe CA, et al. , A global view of the nonprotein-coding transcriptome in Plasmodium falciparum, Nucleic Acids Res. 38 (2) (2010) 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Sierra-Miranda M, et al. , Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum, Mol. Biochem. Parasitol 185 (1) (2012) 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Luke B, Lingner J, TERRA: telomeric repeat-containing RNA, EMBO J. 28 (17) (2009) 2503–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Amit-Avraham I, et al. , Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum, Proc. Natl. Acad. Sci. U. S. A 112 (9) (2015) E982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]