Highlights
-
•
Screening of bacteria producing carotenoid in sub Antarctic region.
-
•
Identification of 10 carotenoids by chromatographic and spectroscopic analysis.
-
•
Salinibacterium sp. and Cryobacterium sp. were identified as new carotenoid sources.
-
•
C.p.450 monoglucoside was identified in Planoccous isolates.
-
•
All isolates could be potential sources for carotenoid production.
Keywords: Bioprospection, Bacteria, Antarctica, Carotenoids, HPLC-PDA-APCI-MS
Abstract
Carotenoids are isoprenoid pigments used by pharmaceutical, cosmetic, food and feed industry as antioxidants and colorants. Although traditional sources of carotenoids are fruits, vegetables and chemical synthesis, prospecting for alternative sinks of common and/or unusual carotenoids is important for the development of natural carotenoid industry.
In this work, 30 pigmented bacterial strains from Fildes Peninsula in King George Island, Antarctica, were isolated and identified by 16S rRNA gene sequencing and classified in three phyla, Bacteroidetes, Firmicutes and Actinobacteria. After cells extraction, ten different carotenoids were identified based on the chromatographic and spectroscopic characteristic obtained by HPLC-PDA and HPLC-PDA-APCI-MS analyses. Strains assigned to Bacteroidetes affiliated to Flavobacterium, Chryseobacterium and Zobellia genera, presented a pigment profile composed of zeaxanthin, β-cryptoxanthin and β-carotene. Firmicutes strains of Planococcus genus produced a C50 carotenoid, identified as C.p. 450 glucoside. Actinobacteria isolates were mainly assigned to Arthrobacter genus, and few to Salinibacterium and Cryobacterium genera. Arthrobacter strains produced C50 carotenoids such as decaprenoxanthin and its glucosylated derivatives, as well as some C40 carotenoids such as lycopene which is used as synthesis precursors of the C50 carotenoids. Salinibacterium and Cryobacterium genera produced C.p. 450 free form and its glucosylated derivatives.
Although most isolates produce carotenoids similar in diversity and quantity than those already reported in the literature, novel sources for C50 carotenoids results from this work. According to their carotenoid content, all isolates could be promising candidates for carotenoids production.
1. Introduction
Carotenoids are the most diverse and widespread pigments found in nature [1]. All share a backbone structure of isoprene units, and they are classified as carotenes if they are hydrocarbons, and as xanthophylls if they are oxygenated derivatives. The most frequent carotenoids are based on symmetrical C40 carbon chains, e.g. carotenes such as β-carotene and lycopene, and xanthophyll as astaxanthin, zeaxanthin and β-cryptoxanthin. Less frequent carotenoids are those of C30 and C50 chain length, which only can be found in non-photosynthetic bacteria and archaea [2].
Animals are not able to synthesize carotenoids de novo, so they must incorporate them from diet. Traditional sources of carotenoids are fruits and vegetables, but they are also distributed among bacteria, algae and fungi [3]. Carotenoids are used by food and feed industry, as well as in pharmaceutics and cosmetics products, and their demand is growing not only due to their utilization as food colorants, but also because of their biologic and physiologic roles. Most of the chemical compounds used to color food are produced by chemical synthesis, because of their lower cost. However, the negative perception of the synthetic colorants by consumers has increased the demand of natural pigments, such as carotenoids. Besides, carotenoids are involved in numerous metabolic functions [3], and epidemiological studies support their protective role in prevention of certain diseases. There is evidence of the effect of β-carotene and lycopene as chemo-protective agents against some kinds of cancer, and the role of lutein and zeaxanthin against macular degeneration, as they are highly concentrated in the retina and protect from blue light [4]. C30 and C50 carotenoids have been proposed to have strong singlet oxygen quenching activity and high antioxidant activities [5,6]. The pharmaceutical potential and application of these carotenoids have recently been examined, being available several patents such as the application of C50 carotenoids in sunscreens [7].
Bacteria have immense potential to produce carotenoids. In heterotrophic bacteria, carotenoids are secondary metabolites that play fundamental roles in cell adaptability. Carotenoids protect cells from UV radiation and oxidative damage [8,9] and are involved in the mechanisms of membrane fluidity [10,11]. The correct maintenance of the fluidity and structure of the cell membrane is essential for growth at low temperatures and for the regulation of nutrient transport.
The aim of bioprospection in extreme environments is to find organisms poorly studied as potential new sources of chemicals for biotechnological applications. Microbiological and biotechnological research in Antarctica has been increasing in the last years, focusing on new microorganisms with biotechnological interest [[12], [13], [14]]. Bacteria from cold environments, like Antarctica, must survive in extreme conditions of temperature, freezing-thawing cycles, drastic light conditions, high UV-B doses and low humidity. Carotenoids provide protection in these harsh conditions, so it is expected to find efficient carotenoid-producing bacteria. A great diversity of microorganisms exists in Antarctica, most of them belonging to the phyla Proteobacteria, Bacteroidetes, Actinobacteria, Fimicutes and Deinococcus-Thermus [15,16]. Furthermore, studies on bacterial species from Antarctica showed preponderance of pigmented bacteria [17]. Screening of Antarctic microorganisms could provide new potential strains for carotenoids production.
In the present study, pigmented strains from Fildes Peninsula, Antarctica, were isolated and identified by 16S rRNA gene sequencing. The isolates were evaluated for their potential to produce carotenoids. The identification and quantification of the carotenoid profile was carried out to evaluate the isolates as an alternative source of natural carotenoids.
2. Materials and methods
2.1. Sample processing, conservation and strains characterization
A total of 32 liquid and solid samples (in 50 mL sterile tubes) were collected from Fildes Peninsula, King George Island, during the expedition organized by IAU (Uruguayan Antarctic Institute) on December 2014. Approximately 100 mg of each sample was suspended in 900 μl of sterile NaCl 0.9% (w/v) solution, serially diluted and plated on adequate media. The isolation medium for samples of organic matter, sediments and ice water was Tryptic Soy Agar (TSA, Sigma Aldrich), and for sea water was TSA complemented with 20 g/L of sea salts (Sigma). Plates were incubated at 10 °C for 7–10 days, and colored colonies were selected for strain isolation by streak-plating technique. Once purity was verified, strains were conserved at −80 °C on glass beads with 20% glycerol in Tryptic Soy Broth (TSB, Oxoid) and sea salts when needed. Strains were characterized by colony and cell morphology, Gram staining and pigment composition.
2.2. Amplification of the 16S rRNA gene and sequencing
Genomic DNA extraction was performed with a commercial kit according manufacturer’s instructions (Genomic DNA Purification Kit, Thermo Fisher). Amplification of the 16S rRNA gene fragments were done in a Palm-1870 Cycler TM (Corbett Research UK Ltd) as follows: initial denaturation 3 min at 95 °C, then 35 cycles of 45 s at 94 °C, 45 s at 58 °C, 60 s 72 °C, and a final extension step 9 min at 72 °C. Reaction mixtures contained: 2.5 U polymerase (Mango Taq, Bioline), 10 μL of buffer solution, 2.5 μL of 50 mM MgCl2, and 2.5 μL each of forward primer 27 F (5´-AGAGTTTGATC MTGGCTCAG-3´) and reverse primer 1492R (5´-TACGGYTACC TTGTTACGACTT-3´), genomic DNA, and water to 50 μL final volume. The PCR products were analyzed by electrophoresis with 1% agarose gels. DNA sequencing was carried out by Macrogen Inc. (Korea) using universal primers. DNA sequences obtained from each isolate were aligned by CLUSTALW [18] using MEGA 7 [19]. Assembled DNA sequence data were analyzed by BLASTn [20] and compared with the 16S RNA gene sequences (bacteria and archaea) of the National Centre for Biotechnology Information database (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.3. Nucleotide sequence accession numbers
The nucleotide sequences were deposited in the NCBI GeneBank database under accession numbers: MF288792 - MF288795, MF288807 - MF288826, MF288828 - MF288831 and MF288833 - MF288834.
2.4. Strain culture for pigment production
Strains were cultured in 1 L Erlenmeyer flasks with 250 mL medium in an orbital shaker at 15 °C and 200 rpm. The culture media was Tryptic Soy Broth and when sample origin required, it was supplemented with 20 g/L sea salts. After 48 h of growth, cells were harvested by centrifugation. Pellets were washed with distilled water, frozen at −80 °C, and lyophilized (VirTis BenchTop 2 K Freeze Dryer, SP Industries Inc.).
2.5. Pigment extraction and characterization
Approximately 0.1 g of lyophilized biomass was extracted with 3 mL of methanol until bleaching. The solvent was evaporated to dryness under stream of nitrogen and the dry extract was dissolved in acetone for chromatographic analysis. If saponification was required, the methodology applied was described previously [21]. The presence of flexirubin-type pigments was determined using Fautz test methodology with KOH [22]. The bacteria colonies were covered with an aqueous solution of 20% KOH. A reversible colour shift from yellow/orange to red or brown, indicates the presence of flexirrubin-type pigments.
Carotenoid identification was based on the chromatographic behavior and spectroscopic characteristics (UV–vis and mass spectra) obtained by HPLC-DAD and HPLC-PDA-APCI-MS. Additionaly, UV–vis spectra of total carotenoid extracts were recorded from 300 to 600 nm on a Spectrophotometer Genesys 10S UV–vis (Thermo Scientific). Data was compared with those of standards and literature values [23]. HPLC-DAD analysis was carried out using a Waters e2695 Alliance chromatograph fitted with a Waters 2998 photodiode array detector and controlled with Empower2 software (Waters Cromatografía, SA, Barcelona, Spain). The separation was done in a reversed-phase C18 (200 mm × 4.6 mm i.d., 3 μm, Mediterranea SEA18; Teknokroma, Barcelona, Spain) fitted with a guard column of the same material (10 mm × 4.6 mm). The chromatographic method used was previously described [21]. Briefly, carotenoid separation was carried out by a binary-gradient elution using an initial composition of 75% acetone and 25% deionised water, which was increased linearly to 95% acetone in 10 min, then hold for 7 min and raised to 100% in 3 min, and maintained constant for 10 min. Initial conditions were reached in 5 min. The temperature of column was kept at 25 °C and the sample compartment was refrigerated at 15 °C. An injection volume of 10 μL and a flow rate of 1 mL/min were used. Detection was performed at 450 nm, and the online spectra were acquired in the 330–700 nm wavelength range with a resolution of 1.2 nm.
HPLC-DAD-APCI-MS was performed on a Dionex Ultimate 3000RS U-HPLC (Thermo Fisher Scientific) fitted with a DAD detector and linked to a micrOTOF-QII high-resolution TOF mass spectrometer (UHR-TOF) with quadrupole (qQ)-TOF geometry (Bruker Daltonics) equipped with atmospheric pressure chemical ionization (APCI) source. Chromatographic conditions were same as described above for HPLC-DAD. A split postcolumn of 0.4 mL/min was introduced directly onto the mass spectrometer ion source. The MS instrument was operated in positive ion mode, with a scan range of m/z 50-1200. Mass spectra were acquired through the broadband collision-induced dissociation mode, providing MS and MS/MS spectra simultaneously. The instrument control was performed using Bruker Daltonics Hystar 3.2. Data evaluation was performed with Bruker Daltonics DataAnalysis 4.0.
Total carotenoid content was estimated by UV–vis spectrophotometry at 450 nm, with a specific absorbance coefficient of 2500 [23].
3. Results and discussion
3.1. Isolation and identification of sub-Antarctic pigmented bacteria
The screening methodology applied resulted in the isolation of 30 pigmented strains. Table 1 presents their origin, colony color and strain identification by 16S rRNA gene analysis and GenBank accession numbers. All isolates presented round colonies with smooth edge, and color ranged from yellow to orange. By analysis of almost complete sequences of their 16S rRNA genes, the isolates were assigned to different genera corresponding to three phyla: Actinobacteria, Firmicutes and Bacteroidetes. Among isolated strains, 12 phylotypes of pigmented bacteria were identified using a unique isolation medium. To increase the resulting taxonomic richness, application of other isolation media with different nutrients sources, selective culture media or other enrichment techniques would be recommended.
Table 1.
Samples origin, color and identification by 16S rRNA gene analysis of 30 strains isolated from Fildes Peninsula, King George Island (62°11´S, 58°54´W).
| Clone name | Accession number | Sample source | Color | Database microorganism with highest similarity (Accession number) | Identity (%) |
|---|---|---|---|---|---|
| P7 | MF288829 | Sea water | orange | Zobellia amurskyensis KMM 3526 (NR_024826) | 99 |
| P8 | MF288830 | Sea water | orange | Flavobacterium frigidarium A2i (NR_025020) | 99 |
| P14 | MF288831 | Sediment | orange | Flavobacterium weaverense AT1042 (NR_042999) | 99 |
| P15 | MF288808 | Algal mat | yellow | Salinibacterium amurskyense KMM 3673(NR_041932) | 99 |
| P16 | MF288807 | Penguin feathers | yellow | Salinibacterium amurskyense KMM 3673(NR_041932) | 99 |
| P19 | MF288809 | Penguin dung | yellow | Cryobacterium arcticum SK1 (NR_108605) | 99 |
| P20 | MF288820 | Sand | yellow | Arthrobacter psychrochitiniphilus GP3 (NR_104702) | 99 |
| P21 | MF288795 | Penguin feathers | orange | Planococcus halocryophilus Or1 (JF742665) | 99 |
| P22 | MF288819 | Stagnant water | yellow | Arthrobacter alpinus S6-3 (NR_117254) | 99 |
| P23 | MF288821 | Snowbreak | yellow | Arthrobacter cryoconiti Cr6-08 (NR_108846) | 99 |
| P24 | MF288815 | Snowbreak | yellow | Arthrobacter antarcticus SPC 26 (AM931709) | 99 |
| P25 | MF288818 | Sand | yellow | Arthrobacter psychrochitiniphilus GP3(NR_104702) | 99 |
| P26 | MF288817 | Penguin dung | yellow | Arthrobacter antarcticus SPC 26 (AM931709) | 100 |
| P27 | MF288816 | Penguin dung | yellow | Arthrobacter antarcticus SPC 26 (AM931709) | 99 |
| P28 | MF288810 | Sediment | yellow | Arthrobacter antarcticus SPC 26 (AM931709) | 99 |
| P30 | MF288811 | Sediment | yellow | Arthrobacter antarcticus SPC 26 (AM931709) | 99 |
| P31 | MF288813 | Deposited sediment | yellow | Arthrobacter antarcticus SPC 26 (AM931709) | 99 |
| P32 | MF288812 | Penguin feathers | yellow | Arthrobacter antarcticus SPC 26 (AM931709) | 99 |
| P33 | MF288834 | Snowbreak | orange | Flavobacterium antarcticum DSM 19726 (AY581113) | 96 |
| P34 | MF288793 | Sediment | orange | Planococcus halocryophilus Or1 (JF742665) | 99 |
| P36 | MF288828 | Sediment | orange | Chryseobacterium marinum NBRC 103143 (NR_114212) | 99 |
| P39 | MF288824 | Sediment | yellow | Arthrobacter psychrochitiniphilus GP3 (NR_104702) | 99 |
| P40 | MF288814 | Sediment | yellow | Arthrobacter antarcticus SPC 26 (AM931709) | 99 |
| P43 | MF288822 | Snowbreak | yellow | Arthrobacter psychrochitiniphilus GP3 (NR_104702) | 99 |
| P44 | MF288826 | Sediment | yellow | Arthrobacter alpinus S6-3 (NR_117254) | 99 |
| P45 | MF288825 | Sediment | yellow | Arthrobacter psychrochitiniphilus GP3 (NR_104702) | 99 |
| P46 | MF288792 | Sediment | orange | Planococcus halocryophilus Or1 (JF742665) | 99 |
| P47 | MF288823 | Dry seaweed | yellow | Arthrobacter psychrochitiniphilus GP3 (NR_104702) | 99 |
| P48 | MF288794 | Algal mat | orange | Planococcus halocryophilus Or1 (JF742665) | 99 |
| P50 | MF288833 | Sediment | orange | Flavobacterium antarcticum DSM 19726 (AY581113) | 99 |
Actinobacteria was the most numerous group with 20 isolates, and 17 strains belonged to Arthrobacter genus. Arthrobacter are generally mesophilic, but several strains have been isolated form Arctica, Antarctica and glaciers, being psychrotolerant or psychrophilic [24]. A. antarcticus and A. psychrochitiniphilus were found in Antarctic sediments and penguin guano, and A. alpinus from alpinus soil, and they were also described as yellow colonies [25]. The other Actinobacteria strains were affiliated to Salinibacterium and Cryobacterium genera. Both genera were previously reported in Antarctica and in other cold regions [26,27]. They were described previously as yellow pigmented strains [26,28] although to the author’s knowledge up to the date, there are no studies on pigment identification.
Five isolates were affiliated to the phylum Bacteroidetes, and belong to the genera Zobellia, Chryseobacterium and Flavobacterium, and the other four isolates were affiliated to the phylum Firmicutes and assigned to Planococcus genus. All genera mention above have been previously reported in Antarctica, and they were characterized as pigmented [[29], [30], [31]]. In particular, strain P33 was assigned to Flavobacterium genus with a 96% similarity, indicating that it could be a new specie. This reinforces Antarctic environment as source of novel species of pigment bacteria.
3.2. Characterization of the carotenoid profile
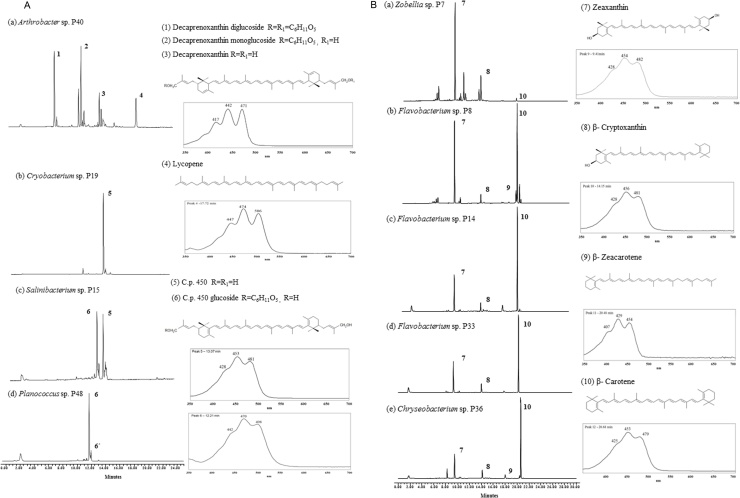
All UV–vis spectra obtained from crude methanol extracts from the isolated strains, presented three maxima around 450 nm, which is characteristic of carotenoid compounds [32]. To characterize and identified the carotenoid composition of the extracts, chromatographic behavior, UV–vis spectra and mass spectroscopy data were analyzed as presented in Table 2. Fig. 1 shows the chromatograms obtained for each bacteria strain, the UV–vis spectra of the major carotenoids and the chemical structures.
Table 2.
Identification of the carotenoids in the bacterial extracts and chromatographic and mass spectroscopy properties.
| Strain | Peak | Rt (min) | Carotenoid | UV-visible spectrum (λmax nm) | Characteristic APCI(+) MS pattern |
|---|---|---|---|---|---|
| Arthrobacter sp. P40 | 1 | 6.36 | Decaprenoxanthin diglucoside | 417, 442, 471 | 1029.6 [M+H]+, 1011.6 [M+H-18]+, 849.6 [M+H-180]+ |
| 2 | 10.07 | Decaprenoxanthin monoglucoside | 417, 442, 471 | 867.6 [M+H]+, 849.6 [M+H-18]+, 687.5 [M+H-180]+ | |
| 3 | 12.61 | Decaprenoxanthin | 417, 442, 471 | 705.6 [M+H]+, 687.5 [M+H-18]+, 595.5[M+H-18-92]+ | |
| 4 | 17.72 | Lycopene | 447, 474, 506 | 537.4 [M+H]+ | |
| Cryobacterium sp. P19, Salinibacterium sp. P15, Planococcus sp. P48 | 5 | 13.07 | C.p. 450 | 428, 453, 481 | 705.6 [M+H]+, 687.5 [M+H-18]+, 669.5 [M+H-18-18]+, 613.5 [M+H-92]+ |
| 6 | 12.21 | C.p. 450 glucoside | 442, 470, 498 | 849.6 [M+H-18]+, 831.6 [M+H-18-18]+ | |
| Flavobacterium sp.P33, Chryseobacterium sp. P36 and Zobellia sp. P7 | 7 | 9.41 | Zeaxanthin | 428, 454, 482 | 569.4 [M+H]+, 551.4 [M+H-18]+, 477.4 [M+H-92]+ |
| 8 | 14.15 | β-Cryptoxanthin | 428, 456, 481 | 553.4 [M+H]+, 535.4 [M+H-18]+, 461.4 [M+H-92]+ | |
| 9 | 20.68 | β-Carotene | 425, 453, 479 | 537.4 [M+H]+ | |
| 10 | 20.48 | β-Zeacarotene | 407, 429, 454 | – |
Fig. 1.
Reversed-phase HPLC-DAD chromatograms at 450 nm and UV–vis spectra of the carotenoid extracts of the bacteria strains. (A) C50 carotenoids, (B) C40 carotenoids.
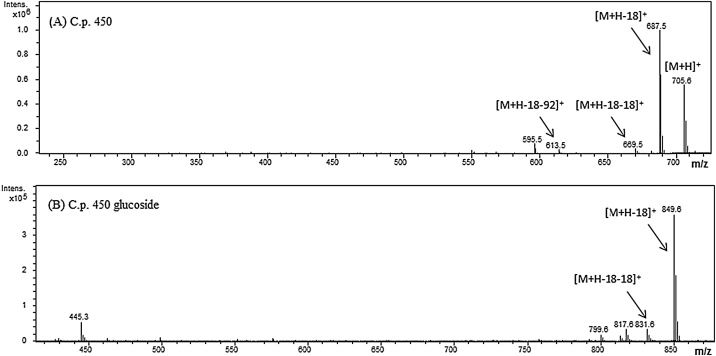
Arthrobacter strain P40 presented four peaks. The UV–vis spectra of the three major carotenoids presented three maxima at 417, 442 and 471 nm, which agrees with a chromophore composed of nine conjugated double bonds. Peak 1 and 2 were assigned to the glycosylated derivatives of decaprenoxanthin. For decaprenoxanthin diglucoside the MS-APCI(+) characteristic fragment pattern presented the protonated molecule [M+H]+ at m/z 1029.6 corresponding to the formula C62H92O12 and the MS spectrum of decaprenoxanthin monoglucoside presented [M+H]+ at m/z 867.6 corresponding to C56H82O7. Both compounds also showed the fragments [M+H-18]+ and [M+H-180]+ revealing the presence of a hydroxyl group and a glucose molecule. Peak 3 was assigned to decaprenoxanthin, as it presented the protonated molecule [M+H]+ at m/z 705.6 corresponding to the formula C50H72O2. It also produced the fragments ions [M+H-18]+ at m/z 687.5 and [M+H-18-92]+ at m/z 595.5 confirmed the loss of water (presence of a hydroxyl group) and toluene. The MS-APCI(+) profile obtained for decaprenoxanthin in this work is in accordance with those reported by Giufrida et al. [6]. Peak 4 was identified as lycopene, presenting a molecular ion [M+H]+ at m/z 537.4 which corresponds to C40H56, UV–vis spectra (447, 474, 506 nm) and coeluted with standard. Biosynthesis of C50 carotenoids proceeds via lycopene elongation, which explains its presence in the profile obtained [33]. Besides, the other compounds identified in this work, are aligned with those reported previously in the Arthrobacter genus containing C50 carotenoids such as decaprenoxanthin, bacterioruberin, A.g 470, sarcinoxanthin, and corresponding derivatives [6,34,35]. The carotenoid profile Cryobacterium sp. P19 showed the presence of a main compound with λmax at 428, 453 and 481 nm in the chromatogram presented in Fig. 1. The mass spectrum showed in Fig. 2A presented a protonated molecule [M+H]+ at m/z 705.6, according to the formula C50H72O2. It also presented fragments at m/z of 687.5 [M+H-18]+, 669.5 [M+H-18-18]+ and 613.5 [M+H-92]+. These fragments indicate the presence of two hydroxyl groups and the loss of toluene, which conforms to the presence of extensive conjugation in the molecule [36]. The UV–vis characteristics and the MS-APCI(+) corresponded to those reported for the carotenoid C.p. 450 [32]. This carotenoid was previously reported in Cornybacterium poinsettiae [37] and in metabolically engineered Corynebacterium glutamicum [33]. However, the nature of the pigments produced by Cryobacterium species has not been previously identified.
Fig. 2.
Mass spectra of (A) C.p. 450 and (B) C.p. 450 glucoside in atmospheric pressure chemical ionization (APCI) in positive mode.
The chromatographic analysis of Salinibacterium sp. P15 showed the presence of two peaks. The first compound (peak 6) was identified as C.p. 450 glucoside based on the UV–vis spectra and the mass spectrum shown in Fig. 2B. This peak presented three maxima at 442, 470 and 498 nm. The MS fragmentation pattern presented the fragments at m/z 849.6 and 831.6, that could be assigned to [M+H-18]+ and [M+H-18-18]+. These fragments indicate the presence of two hydroxyl groups in the molecule. The molecular ion [M+H]+ at m/z 867.5 was not detected, as reported by Britton et al. [32]. The second peak (peak 5) was identified as C.p. 450, as it showed the same UV–vis characteristics and MS-APCI(+) pattern of the C.p. 450 mentioned above for Cryobacterium sp. P19. To the authors knowledge, there are not reports about pigment identification in strains of Salinibacterium genus, although it has been reported as yellow pigmented strain [28].
Planococcus sp. P48 presented two main carotenoids, as shown in Fig. 1a. The first compound resembles the pattern obtained for peak 6, which corresponds to C.p. 450 glucoside, since it exhibit the same UV–vis spectra, retention time and MS-APCI(+) pattern. Peak 6´ was assigned to the cis isomer of peak 6, since UV–vis spectra presented shorter λmax (6 nm), compared with those obtained for the trans compound and the presence of a maximum (cis-peak) at 357 nm. Even though strains belonging to Planococcus genus have been widely reported as pigmented strains, the identification of carotenoids produced by Planococcus is scarce in the literature. Shindo et al. [5] reported that P. maritimus produced methyl glucosyl-3,4-dehydro-apo-8′-lycopenoate, and Kim et al. [29] that P. faecalis produced glycosyl-4,4′-diaponeurosporen-4′-ol-4 oic acid as major pigment, and both are C30 carotenoids. However, C.p. 450 and its glycosilated derivate identified in Planococcus sp. P48 are C50 carotenoids, evidencing the diversity of carotenoids that the genus may present.
Flavobacterium, Chryseobacterium and Zobellia isolates presented similar HPLC carotenoid profile. Only Zobellia sp. P7 and Flavobacterium sp. P8 produced flexirrubin-type pigments. Therefore, in these cases the extracts were saponificated with 20% KOH in MeOH in order to remove flexirubins before chromatographic analysis. Flexirrubins are common pigments in Flavobacterium, Chryseobacterium, Zobiella [38]. All the strains produced zeaxanthin, β-cryptoxanthin and β-carotene as identified by their chromatographic and spectroscopic behavior. The mass spectrum for the three carotenoids, presented as the most abundant fragment the molecular ion [M+H]+ at m/z 569.4 (C40H56O2), 553.4 (C40H56O) and 537.4 (C40H56), respectively. In addition, the zeaxanthin and β-cryptoxanthin peaks showed the presence of two characteristic fragments [M+H-18]+ and [M+H-92]+ which are in agreement with the loss of a hydroxylated group and toluene, respectively. The identification was completed with the comparison of the chromatographic and UV–vis properties with authentic samples and data in literature. The strains Flavobacterium sp. P8 and P14 and Chryseobacterium sp. P36 presented peak 10, tentatively identified as β-zeacarotene by comparison with authentic samples and data in literature. The carotenoids identified are part of those involved in the biochemical pathway of zeaxanthin production, in which the hydroxylation of β-carotene and β-cryptoxanthin leads to accumulation of zeaxanthin. β-zeacarotene is a precursor of β-carotene, and results from the cyclation of neurosporene, producing γ-carotene, prior to the synthesis of β-carotene [39].
The identification of the carotenoids of the isolated strains, not only contributed to further knowledge of the metabolic capacities of the species, but also to evaluate new potential sources of carotenoids of biotechnological production.
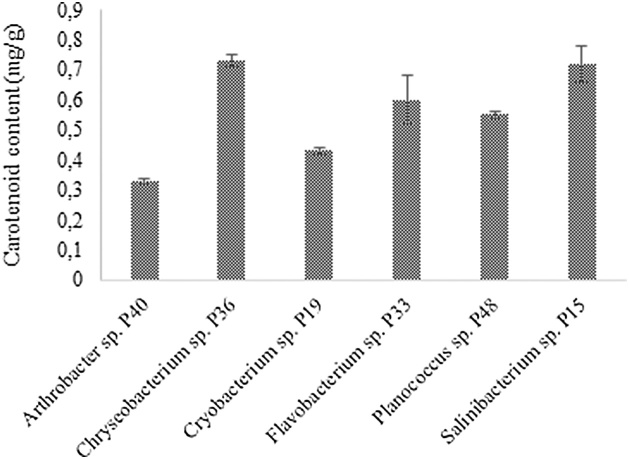
3.3. Total carotenoid content
Total carotenoid contents of the isolates are shown in Fig. 3. Among bacterial sources of carotenoids, Flavobacterium spp. have been widely reported as zeaxanthin producer. A mutant culture of Flavobacterium sp. ATCC 21588 was reported to reach in a specific optimized medium presented a carotenoid content of 16 mg/g [40]. On the other hand, lower values were reported for Flavobacterium multivorum, with a zeaxanthin content of 0.7–1.2 μg/g dry biomass in an optimized medium [41]. In this study, Flavobacterium sp. P33 reached a total carotenoid content of 0.60 mg/g. This carotenoid content could be improved by optimization of the culture medium and operational conditions such as the oxygen supplied. Arthrobacter sp. P40 reached a carotenoid content of 0.33 mg/g, similar to values obtained for Arthrobacter arilaitensis of 0.14-0.25 mg/g [42].
Fig. 3.
Total carotenoid content for different genus isolates. The total carotenoid content is expressed per gram of dry biomass.
Regarding to the other genera study in this work, there are no reports available in literature about quantification of carotenoids produced. As shown in Fig. 3, the total carotenoid contents of the isolates were similar, ranging between 0.33-0.73 mg/g dry biomass.
4. Conclusions
Thirty heterotrophic bacterial strains from sub-Antarctic region were isolated, identified and characterized as carotenoids producers. Arthrobacter, Flavobacterium, Chryseobacterium, and Zobellia isolates produce carotenoids similar in diversity and quantity than those already reported. On the other hand, Cryobacterium sp. P19, Salinibacterium sp. P15 and Planococcus sp. 48 are presented as novel C50 carotenoid sources. Then, this work increased the information on heterotrophic bacteria as promising source of carotenoids for biotechnological production, a field with actual scarce development.
Conflict of interest
No conflict of interest are declared.
Funding
This work was supported by “Comisión Sectorial de Investigación Científica” [project CSIC I+D 2014 219], and “Agencia Nacional de Investigación e Innovación” [grant POS_NAC_2014_1102321] and [grant MOV_CA__2017_1_138162].
Acknowledgements
The authors thank the “Instituto Antártico Uruguayo” for logistic support during the stay in Base Artigas. Authors thank to José Julian Rios, at the Mass Spectrometry Service of Instituto de la Grasa (IG-CSIC, Spain), for technical assistance on mass spectrometry.
References
- 1.Varela J.C., Pereira H., Vila M., León R. Production of carotenoids by microalgae: achievements and challenges. Photosyn. Res. 2015;125(3):423–436. doi: 10.1007/s11120-015-0149-2. [DOI] [PubMed] [Google Scholar]
- 2.Rodríguez-Concepción M., Avalos J., Bonet M.L., Boronat A., Gomez-Gomez L., Hornero-Méndez D., Limon M.C., Melendez-Martinez A.J., Olmedilla - Alonso B., Palou A., Rodrigo M.J., Zacarias L., Zhu C. A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018;70:62–93. doi: 10.1016/j.plipres.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Carle R., Schweiggert R., editors. Handbook on Natural Pigments in Food and Beverages: Industrial Applications for Improving Food Color. Woodhead Publishing; 2016. [Google Scholar]
- 4.Johnson E.J. The role of carotenoids in human health. Nutr. Clin. Care. 2002;5(2):56–65. doi: 10.1046/j.1523-5408.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- 5.Shindo K., Endo M., Miyake Y., Wakasugi K., Morritt D., Bramley P.M., Fraser P.D., Kasai H., Misawa N. Methyl Glucosyl-3,4-dehydro-apo-8′-lycopenoate, a novel antioxidative glyco-C30-carotenoic acid produced by a marine bacterium Planococcus maritimus. J. Antibiot. (Tokyo) 2008;61(12):729–735. doi: 10.1038/ja.2008.86. [DOI] [PubMed] [Google Scholar]
- 6.Giuffrida D., Sutthiwong N., Dugo P., Donato P., Cacciola F., Girard-Valenciennes E., Le Mao Y., Monnet C., Fouillaud M., Caro Y., Dufossé L. Characterisation of the C50 carotenoids produced by strains of the cheese-ripening bacterium Arthrobacter arilaitensis. Int. Dairy J. 2016;55:10–16. [Google Scholar]
- 7.Goksøyr, A. (2013). U.S. Patent Application No. 13/701,249.
- 8.Krinsky N.I. Non-photosynthetic functions of carotenoids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1978;284(1002):581–590. [Google Scholar]
- 9.Miller N.J., Sampson J., Candeias L.P., Bramley P.M., Rice-Evans C.A. Antioxidant activities of carotenes and xanthophylls. FEBS Lett. 1996;384(3):240–242. doi: 10.1016/0014-5793(96)00323-7. [DOI] [PubMed] [Google Scholar]
- 10.Jagannadham M.V., Chattopadhyay M.K., Subbalakshmi C., Vairamani M., Narayanan K., Mohan Rao C., Shivaji S. Carotenoids of an Antarctic psychrotolerant bacterium, Sphingobacterium antarcticus, and a mesophilic bacterium, Sphingobacterium multivorum. Arch. Microbiol. 2000;173(5–6):418–424. doi: 10.1007/s002030000163. [DOI] [PubMed] [Google Scholar]
- 11.Subczynski W.K., Markowska E., Gruszecki W.I., Sielewiesiuk Effects of polar carotenoids on dimyristoylphosphatidylcholine membranes: a spin-label study. Acta Biochim. Biophys. 1992;1105(1):97–108. doi: 10.1016/0005-2736(92)90167-k. [DOI] [PubMed] [Google Scholar]
- 12.Bianchi A.C., Olazábal L., Torre A., Loperena L. Antarctic microorganisms as source of the omega-3 polyunsaturated fatty acids. World J. Microbiol. Biotechnol. 2014;30(6):1869–1878. doi: 10.1007/s11274-014-1607-2. [DOI] [PubMed] [Google Scholar]
- 13.Godinho V.M., Furbino L.E., Santiago I.F., Pellizzari F.M., Yokoya N.S., Pupo D., Alves T.M.A., Junior P.A.S., Romanha A.J., Zani C.L., Cantrell C.L., Rosa C.A., Rosa L.H. Diversity and bioprospecting of fungal communities associated with endemic and cold-adapted macroalgae in Antarctica. Microb. Ecol. 2013;7(7):1434–1451. doi: 10.1038/ismej.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnan A., Convey P., Gonzalez-Rocha G., Alias S.A. Production of extracellular hydrolase enzymes by fungi from King George Island. Polar Biol. 2016;39(1):65–76. [Google Scholar]
- 15.Peeters K., Verleyen E., Hodgson D.A., Convey P., Ertz D., Vyverman W., Willems A. Heterotrophic bacterial diversity in aquatic microbial mat communities from Antarctica. Polar Biol. 2012;35(4):543–554. [Google Scholar]
- 16.Wilmotte A., Vyverman W., Willems A., Verleyen E., Peeters K., Obbels D., Souffreau C., DeCarvalho-Maalouf P. Belgian Science Pilicy; Brussels: 2012. Antacrtoc Microbial Diversity: the Importance of Geographical and Ecological Factors. Research Programme Science for a Sustainable Development; p. 98. [Google Scholar]
- 17.Chauhan S., Shivaji S. Growth and pigmentation in Sphingobacterium antarcticus, a psychrotrophic bacterium from Antarctica. Polar Biol. 1994;14(1):31–36. [Google Scholar]
- 18.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Delgado-Pelayo R., Hornero-Méndez D. Identification and quantitative analysis of carotenoids and their esters from sarsaparilla (Smilax aspera L.) berries. J. Agric. Food Chem. 2012;60(33):8225–8232. doi: 10.1021/jf302719g. [DOI] [PubMed] [Google Scholar]
- 22.Fautz E., Reichenbach H. A simple test for flexirubin-type pigments. FEMS Microbiol. Lett. 1980;8(2):87–91. [Google Scholar]
- 23.Britton G. General carotenoid methods. Meth. Enzymol. 1985;111:113–149. doi: 10.1016/s0076-6879(85)11007-4. [DOI] [PubMed] [Google Scholar]
- 24.Busse H., Wieser M. Springer; Berlin, Heidelberg: 2014. The Genus Arthrobacter." The Prokaryotes; pp. 105–132. [Google Scholar]
- 25.Zhang D.C., Schumann P., Liu H.C., Xin Y.H., Zhou Y.G., Schinner F., Margesin R. Arthrobacter alpinus sp. nov., a psychrophilic bacterium isolated from alpine soil. Int. J. Syst. Evol. Microbiol. 2010;60(9):2149–2153. doi: 10.1099/ijs.0.017178-0. [DOI] [PubMed] [Google Scholar]
- 26.Lee J., Cho A., Yang J.Y., Woo J., Lee H.K., Hong S.G., Kim O.S. Complete genome sequence of Cryobacterium arcticum strain PAMC 27867, isolated from a sedimentary rock sample in northern Victoria land, Antarctica. Genome Announc. 2016;4(5):e00885–16. doi: 10.1128/genomeA.00885-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin S.C., Kim S.J., Ahn D.H., Lee J.K., Lee H., Lee J., Hong S.G., Lee T.M., Park H. Genome sequence of a Salinibacterium sp. isolated from antarctic soil. J. Bacteriol. 2012;108(2010):1889–1902. doi: 10.1128/JB.00235-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leiva S., Alvarado P., Huang Y., Wang J., Garrido I. Diversity of pigmented gram-positive bacteria associated with marine macroalgae from Antarctica. FEMS Microbiol. Lett. 2015;362(24) doi: 10.1093/femsle/fnv206. [DOI] [PubMed] [Google Scholar]
- 29.Kim J.H., Kang H.J., Yu B.J., Kim S.C., Lee P.C. Planococcus faecalis sp. nov., a carotenoid-producing species isolated from stools of Antarctic penguins. Int. J. Syst. Evol. Microbiol. 2015;65(10):3373–3378. doi: 10.1099/ijsem.0.000423. [DOI] [PubMed] [Google Scholar]
- 30.Van Trappen S., Vandecandelaere I., Mergaert J., Swings J. Algoriphagus antarcticus sp. nov., a novel psychrophile from microbial mats in Antarctic lakes. Int. J. Syst. Evol. Microbiol. 2004;54(6):1969–1973. doi: 10.1099/ijs.0.02973-0. [DOI] [PubMed] [Google Scholar]
- 31.Yi H., Chun J. Flavobacterium weaverense sp. nov. and Flavobacterium segetis sp. nov., novel psychrophiles isolated from the Antarctic. Int. J. Syst. Evol. Microbiol. 2006;56(6):1239–1244. doi: 10.1099/ijs.0.64164-0. [DOI] [PubMed] [Google Scholar]
- 32.Britton G., Liaaen-Jensen S., Pfander H. vol. 1B. Springer; 1995. (Carotenoids: Spectroscopy). [Google Scholar]
- 33.Heider S.A., Peters-Wendisch P., Netzer R., Stafnes M., Brautaset T., Wendisch V.F. Production and glucosylation of C 50 and C 40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014;98(3):1223–1235. doi: 10.1007/s00253-013-5359-y. [DOI] [PubMed] [Google Scholar]
- 34.Arpin N., Fiasson J.L., Norgard S., Borch G., Liaaen-Jensen S. Bacterial carotenoids, XLVI. C50-Carotenoids, 14. C50-Carotenoids from Arthrobacter glacialis. Acta Chem. Scand. 1975;29(9):921–926. doi: 10.3891/acta.chem.scand.29b-0921. [DOI] [PubMed] [Google Scholar]
- 35.Fong N.J.C., Burguess M.L., Barrow K.D., Glenn D.R. Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 2001;56(5–6):750–756. doi: 10.1007/s002530100739. [DOI] [PubMed] [Google Scholar]
- 36.Rivera S.M., Christou P., Canela‐Garayoa R. Identification of carotenoids using mass spectrometry. Mass Spectrom. Rev. 2014;33(5):353–372. doi: 10.1002/mas.21390. [DOI] [PubMed] [Google Scholar]
- 37.Britton G., Mundy A.P., Englert G. Revised structures of the two C 50 carotenoids Cp 450 and Cp 473 from Corynebacterium poinsettiae. J. Chem. Soc. Perkin Trans. 1985;1:601–603. [Google Scholar]
- 38.Bernardet J.F., Bowman J.P. The Prokaryotes. Springer New York; New York, NY: 2006. The genus Flavobacterium; pp. 481–531. [Google Scholar]
- 39.Britton G., Brown D.J., Goodwin T.W., Leuenberger F.J., Schocher A.J. The carotenoids of Flavobacterium strain R1560. Arch. Microbiol. 1977;113(1–2):33–37. doi: 10.1007/BF00428576. [DOI] [PubMed] [Google Scholar]
- 40.Shepherd, D., Dasek, J., and Carels, M. S. C. (1976). U.S. Patent No. 3,951,743. Washington, DC: U.S. Patent and Trademark Office.
- 41.Bhosale P., Larson A.J., Bernstein P.S. Factorial analysis of tricarboxylic acid cycle intermediates for optimization of zeaxanthin production from Flavobacterium multivorum. J. Appl. Microbiol. 2004;96(3):623–629. doi: 10.1111/j.1365-2672.2004.02197.x. [DOI] [PubMed] [Google Scholar]
- 42.Sutthiwong N., Fouillaud M., Valla A., Caro Y., Dufossé L. Bacteria belonging to the extremely versatile genus Arthrobacter as novel source of natural pigments with extended hue range. Food Res. Int. 2014;65:156–162. [Google Scholar]