Abstract
Acute myelogenous leukemia (AML) is a heterogeneous disease and often relapses after standard chemotherapy. Recently, the neddylation (NEDD8) and the mammalian target of rapamycin (mTOR) signaling pathways have emerged as promising pharmaceutical targets for AML therapy. However, the interaction of these two pathways remains unclear. Here we evaluated the effects of pevonedistat, an inhibitor of the NEDD8 activating enzyme (NAE), and sapanisertib (TAK-228), an inhibitor of mTORC1 and mTORC2 as single agents or in combination on AML cell lines. We found that inhibition of neddylation with pevonedistat partially inhibited mTOR signaling transduction and vice versa, inhibition of mTOR signaling with sapanisertib partially inhibited neddylation in AML cell lines. Pevonedistat alone was able to induce cytotoxicity in most AML cell lines as well as in primary AML, whereas sapanisertib alone decreased cell metabolic activity, reduced cell size and arrested cells in G0 phase with only minimal induction of cell death. In addition, pevonedistat was able to induce cell differentiation, arrest cells in G2/M cell cycle phases, and induce DNA re-replication and damage. However, co-treatment with sapanisertib suppressed pevonedistat induced apoptosis, differentiation, S/G2/M arrest, and DNA damage. Taken together, our data demonstrate that pevonedistat and sapanisertib exhibit distinct anti-tumor effects on AML cells, i.e. cytotoxic and cytostatic effects, respectively; however, sapanisertib can attenuate pevonedistat-induced cellular responses in AML cells. Understanding mTOR and neddylation pathway interaction could provide therapeutic strategies for treatment of AML and other malignancies.
Introduction
Acute myelogenous leukemia (AML) is a heterogeneous disease which often relapses after standard chemotherapy or proves refractory to available treatments. Therefore, novel therapies for AML are urgently needed. In AML, many signaling pathways are abnormally activated and lead to uncontrolled proliferation/survival of immature myeloid progenitors [[1], [2], [3], [4], [5]]. Recently, the NEDD8 (neural precursor cell-expressed, developmentally down-regulated 8) conjugation pathway has emerged as an important regulatory pathway for cancer therapy [6]. NEDD8 is a small ubiquitin (Ub)-like molecule which is linked to cullin ring E3 ligases (CRLs), a type of E3 Ub ligase. Conjugation of Nedd8 to cullin assists CRLs to recruit Ub-conjugating E2 enzyme via the RING (Really Interesting New Gene) domain and facilitates the transfer of Ub from E2 to a bound substrate. Therefore CRLs aid in the ubiquitination of certain proteins which are then degraded by the proteasome [7]. CRL1 or SCF (Skp1-Cul1-F-box protein, the best characterized CRL complex) neddylation increases the degradation of the inhibitors of cell cycle progression such as p130, the cyclin-dependent kinase (CDK) inhibitors p27 Kip1 and p21Cip1, the pro-apoptotic BH3-only tumor suppressor protein (BimEL), and the NF-κB inhibitor IκBα [8], [9]. Other CRLs also promote the degradation of a variety of cancer relevant targets such as those involved in DNA replication and nucleotide excision repair including chromatin licensing and DNA replication factor 1 (CDT1, CRL1/4) [10], in the response to hypoxia transcription factor hypoxia-inducible factor 1-alpha (HIF1a, CRL2) [11], in oxidative responses such as nuclear factor E2-related factor 2 (NRF2, CRL3) [12], in mTOR signaling such as the mTOR inhibitor tuberous sclerosis complex 2 (TSC2, CRL4) [13] and in tumor suppression such as P53 (CRL5/7) [14]. Moreover, aberrant activation of the neddylation pathway has been reported in human cancers where overactive CRLs confer a survival advantage [15].
Pevonedistat (TAK-924, MLN4924) is a small molecule which specifically inhibits NEDD8-activating enzyme E1 (NAE) activity, blocks the neddylation pathway, and subsequently increases the stability of CRL substrates [16]. Pevonedistat has been shown to prevent tumor cell growth through inducing tumor cell apoptosis and has entered into several early phase as well as phase III trials for various solid tumors and hematological malignancies [17], [18], [19].
Previous reports have shown that the mTOR signaling pathway is activated in 50% to 80% of AML cases [20]. mTOR is an evolutionarily conserved serine/threonine protein kinase that senses signals of growth factors, nutrients, energy status and metabolic stresses [21]. mTOR exists in two distinct multi-factor complexes: mTOR complex 1 (mTORC1) and 2 (mTORC2). mTORC1 controls protein synthesis, ribosome biogenesis, cell growth, and cell cycle progression through phosphorylation of its substrates such as ribosome protein S6 kinase 1 (S6K1) and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1). mTORC2 regulates cell proliferation, cell survival, and the cytoskeleton through its downstream effectors such as AKT and protein kinase C (PKC) [22]. The first generation of mTORC1 inhibitors, such as rapamycin, have had minimal impact on AML [23]. Negative feedback loops between mTORC1 and mTORC2 as well as failure to inhibit the phosphorylation of the translation repressor 4E-BP1 limited the efficacy of rapamycin in AML treatment [24]. Dual mTORC1/2 inhibitors may overcome these limitations.
Sapanisertib (TAK-228, MLN0128) is a selective, highly potent, and orally bioavailable ATP competitor of both mTORC1 and mTORC2, which is currently in phase I and II clinical trials as a single agent and in combination with other therapeutic agents in patients with advanced malignancies [25], [26]. Since DEPTOR, a naturally occurring inhibitor of mTORC1/2 is ubiquitinated by CRL/SCF E3 ubiquitin ligase [27] and several other negative regulators of the mTOR pathway are also substrates of CRLs (TSC2, REDD1, IRS1, and HIF1α) [28], targeting the neddylation pathway is therefore expected to cause the accumulation of mTOR negative regulators with resulting blockade of the mTOR pathway. Thus we postulated that simultaneous inhibition of the neddylation and mTOR pathways by pevonedistat and sapanisertib, respectively might have effects on leukemia cell growth beyond those agents used singly.
Materials and Methods
Cell Lines
The human AML cell lines HL-60, MV4–11, THP-1 and U937 were purchased from ATCC (American Type Culture Collection) and Molm-13 was purchased from AddexBio and cultured in RPMI 1640 (Invitrogen) containing 10% heat-inactivated fetal bovine serum (HI-FBS) and 100 U/mL penicillin/streptomycin (P/S, Sigma-Aldrich). The KG1a cell line was also purchased from ATCC and cultured in IMDM (Invitrogen) containing 10% HI-FBS and P/S. The HEK293TN Cell Line (transformed with the SV40 large T antigen to promote robust growth) was purchased from System Bioscience and cultured in DMEM containing 10% HI-FBS, 1xGlutamax, and 50 ng/ml of G418 for selection. All cells were incubated in a humidified atmosphere at 37 °C in 5% CO2. All tissue culture adapted cell lines were mycoplasma free by MycoAlert™ PLUS kit testing (Lonza).
Primary AML Cells
Primary bone marrow aspirates or peripheral blood samples were collected from AML patients after informed consent or from excess samples obtained from de-identified AML subjects at diagnosis on a protocol approved by the Research Subjects Review Board of the University of Rochester. Mononuclear cells were isolated using Ficoll-Paque Plus (GE Healthcare Bio-Science AB) following the manufacturer's procedure and stored frozen in liquid N2. Primary AML cells were cultured in IMDM supplied with 10% HI-FBS and recombinant human (rh) stem-cell factor (SCF, 10 ng/mL), rh interleukin-3 (IL-3, 10 ng/mL), and rh FLT3-Ligand (10 ng/mL) (all from PeproTech). All cells were incubated in a humidified atmosphere at 37 °C in 5% CO2.
Reagents
Pevonedistat and sapanisertib were obtained from Millennium Pharmaceuticals, Inc., a subsidiary of Takeda Pharmaceutical Company Limited, under a materials transfer agreement. IKKB inhibitor IV was purchased from Santa Cruz. All of the compounds were dissolved in dimethyl sulfoxide (DMSO, AMRESCO) at 10 mM stock at −20 °C. The final DMSO (with or without drugs) concentration added in cell culture medium was 0.1% (v/v).
Lentiviral Production
The lentiviral cloning, high titer lentiviral production and lentiviral titer determination were described previously [29]. Briefly, short hairpin RNA (shRNA) was designed using Invitrogen Block-it RNAi Designer. The shRNA oligonucleotides against mTOR (sh-mTOR#1: GCAAAGATCTCATGGGCTTCG; sh-mTOR#2: GCTATGTAGTAGAGCCCTACA) were constructed into pLKO.1-GFP vector to generate lentiviral particles. The lentivector pLKO.1-GFP construct DNA, pPax2 (packaging plasmid) and pMD2.G (vesicular stomatitis virus-g) envelope plasmid were transfected into HEK293TN cells using TransIT-LT1 Transfection Reagent (Mirus Bio) according to manufacturer's instructions. Lentiviral particles were harvested on the following 2 days and filtered through 0.45 μm PES syringe filter (Nalgene). Pooled lentiviral supernatants were PEG concentrated, aliquoted, and stored at −80 °C. Lentiviral titer quantitation was performed using HEK293TN cells. Percentages of GFP positive cells were determined by flow cytometry.
Lentiviral Infection
To infect human leukemic cell lines, 1 × 107 lentiviral particles plus 8 μg/ml polybrene (AmericanBio) were added to 1 × 106 cells at the ratio of 10 MOI (Multiplicity of infection). To infect primary AML cells, 5 × 106 primary cells were cultured in IMDM with 10% HI-FBS with cytokines and mixed with 10 MOI of lentivirus and 8 μg/ml polybrene. The infection was carried out at 37 °C in 5% CO2 overnight. The medium was changed after overnight infection. The infection efficiency was evaluated by GFP expression using flow cytometry 3 or 4 days after infection. The knockdown efficiency was confirmed by mTOR western blot analysis.
NFκB Reporter Cell Line Generation and Luciferase Assay
KG1a and U937 cell lines were transduced with lentivirus containing pGreenfire-NF-κB-responsive transcriptional elements-GFP/luciferase gene (Systems Bioscience) and selected with 2 μg/ml puromycin. Then the NFκB reporter cells were treated with pevonedistat or sapanisertib or both for timed exposures, lysed in passive lysis buffer (Promega) and analyzed for luciferase activity using Steady-GLO luciferase reagent (Promega) with a plate reader (Synergy 2, BioTek).
Flow Cytometry
For cell cycle analysis, cells were stained with 10 μg/ml HOECHST 33342 (Thermo Scientific) for 45 min at 37 °C, then pyronin Y (Alfa Aesar) was added to cell culture at final concentration of 0.5 μg/ml and the cells were continually cultured at 37 °C for another 15 min before flow analysis. For apoptosis analysis, the cells were washed with 0.5% FBS/PBS and stained with Annexin V/7-AAD (BD Biosciences) according to the manufacturer's procedure. Combination index (CI) for drug cytotoxicity was calculated using Compusyn software. For CD11b staining, the cells were washed with 0.5% FBS/PBS and stained with FITC conjugated CD11b (BD Biosciences) on ice for 30 min, and followed by AnnexinV/7-AAD staining. The samples were analyzed using a flow cytometer (FACS LSR II, BD Biosciences), and data were analyzed using FlowJo software version 10.2 (Tree Star). Antibodies to CD11c and CD36 were also used for cell staining as described for CD11b.
Cell Count and Viability Assays
After the AML cells were treated with DMSO control alone or with pevonedistat and/or sapanisertib, cells were stained with trypan blue and the live cell number and viability were determined by using TC20 Automated Cell Counter (Bio-Rad).
Western Blotting
Cells were lysed in 62.5 mM Tris pH 6.8, 2% SDS, 10% Glycerol, with protease-inhibitor cocktail (Cell Signaling). Protein concentration was measured using DC Protein Assay (Bio-Rad), and Western blot analysis was performed. The primary antibodies used are listed in Table 1 (in supplement). Secondary horse radish peroxidase conjugated anti-mouse and anti-rabbit antibodies were from Jackson ImmunoResearch or BioRad. GAPDH (Am4300) (Ambion) was used as an internal control. Antibodies were diluted 1:1000 in TBS/1%BSA/0.05% Tween20 buffer except for GAPDH using a 1:5000 dilution.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software). For comparison between groups of three or more, an analysis of variance (ANOVA) with a Newman–Keuls multiple comparison test was used to determine differences between treatments and results given as means +/− standard deviation. For all statistical analysis, P < .05 was considered significant.
Results
Pevonedistat Induced Cytotoxicity in Human AML Cell Lines and Primary AML Cells
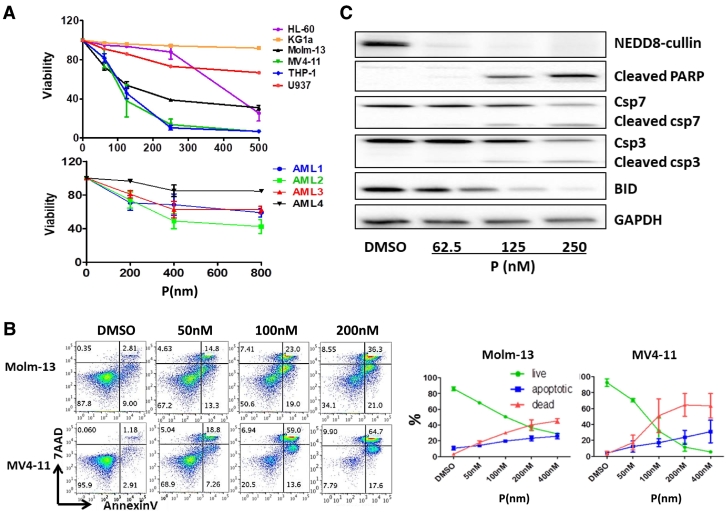
We first investigated the effects of pevonedistat on various AML cell lines. Six human AML cell lines (HL-60, KG1a, Molm-13, MV4–11, THP-1 and U937) were treated with varying concentrations of pevonedistat from 62.5 nM to 500 nM for 2 days and cell viability was determined using flow cytometry. Cells considered viable were annexinV and 7AAD double negative. There was a dose-dependent decrease in cell viability (Figure 1A and Fig. S1) in most of the cell lines. MV4–11 and THP-1 were the most sensitive cell lines with the EC50 as low as 100 nM; whereas KG1a was the most resistant cell line. A variable level of cytotoxicity induced by pevonedistat was also observed in primary AML blast cells as shown in the lower portion of Figure 1A. Characteristics of the AML cases are shown in Table 2 in the supplementary materials.
Figure 1.
Pevonedistat (P) induced AML cell death, decreased cell viability and promoted cell apoptosis. A. P induced cytotoxicity in AML cells. Six AML cell lines HL-60, KG1a, Molm-13, MV4–11, THP-1 and U937 cells and four primary AML blasts from different patients were treated with various concentrations of P for 48 hours and the cell viability was determined by percentage AnnexinV/7AAD negative staining using flow cytometry. The dose response pattern of cell viability is expressed as percentage of vehicle-treated cells (control). B. AnnexinV/7AAD apoptosis flow cytometry assay shows the percentage of live, apoptotic and dead cells in AML cell lines Molm-13 and MV4–11 treated with vehicle DMSO only or 62.5 nM, 125 nM or 250 nM of P for 48 hours. Flow cytometry figures shown are representative of three independent experiments of Molm-13 and MV4–11 treated with P, and the percentage of live, dead or apoptotic cell are shown in the right panel. C. Western blot analysis shows P suppressed neddylation and induced apoptosis in AML cells. MV4–11 cells were treated with different concentrations of P for 24 hours, and the apoptotic protein markers cleaved PARP, cleaved caspase 3 and 7, anti-apoptotic protein Bid and NEDD8-cullin expression levels were tested using immunoblotting.
Flow cytometry analysis showed that pevonedistat significantly induced AML cell death via apoptosis in a dose-dependent manner (Figure 1B). Compared to vehicle control which had less than 5% cell death and apoptosis; at a concentration of 250 nM, about 75% of MV4–11 cells were dead; more than 16% cells underwent apoptosis and less than 10% of cells were alive. Molm-13 had the same response pattern, but was not as sensitive as MV4–11.
Immunoblotting assays confirmed the apoptotic effects of pevonedistat on AML cells (Figure 1C). Since pevonedistat is a NEDDylation inhibitor, we first detected NEDD8-cullin expression in MV4–11 cells. In the presence of pevonedistat, the neddylation of cullins was markedly inhibited. Apoptosis markers such as cleaved Caspase7/3 and cleaved poly ADP ribose polymerase (PARP) showed a dose-dependent increase, and BID (BCL-2 homology 3 (BH3)-interacting domain death agonist), a caspase 8-substrate, showed dose-dependent decreases, suggesting that AML cells actively underwent apoptosis upon pevonedistat treatment.
The Cytotoxic Effects of Pevonedistat Were Attenuated by the mTOR Inhibitor Sapanisertib
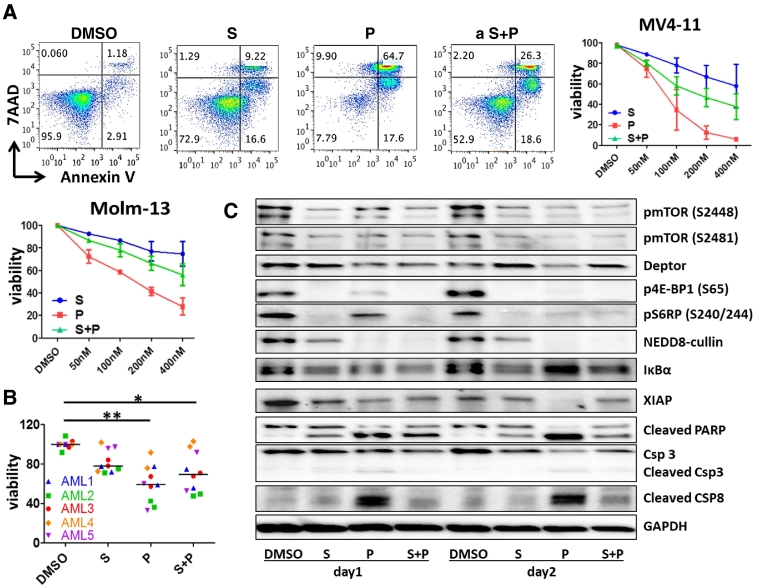
Previous reports showed that mTOR signaling is significantly up-regulated in AML [24], and pevonedistat inhibits AKT and mTOR activation in human myeloma cells [30]. Thus we hypothesized that mTOR inhibitors might have additive or synergistic effects with pevonedistat. To study the effects of the combination, we first treated the AML cell lines, MV4–11 and Molm-13 with increasing concentrations of pevonedistat with the simultaneous addition of the mTOR inhibitor, sapanisertib for 48 hours. By flow cytometry apoptosis analysis using Annexin V staining, we found that there was no additive or synergistic interaction between these two compounds; and in fact, sapanisertib attenuated pevonedistat induced cytotoxicity with the CI value >42, indicating that the two compounds have antagonistic effects [31](Figure 2A). Pevonedistat alone induced cytotoxicity in AML cells in a dose dependent manner, while sapanisertib alone had minimal effect on AML cell death. In MV4–11 cells at the dose of 200 nM, pevonedistat treated cells were <10% viable, while sapanisertib treated cells were about 80% viable. When both compounds were added simultaneously, the cytotoxicity effect was intermediate. Other AML cell lines and primary AML blasts were analyzed similarly, and the results showed the same pattern (Figure 2B), suggesting that the two compounds are not synergistic, but antagonistic regarding effects on apoptosis. To examine whether this observation would also be seen in primary AML blasts not supplemented with growth factors, 6 additional cases were examined (Fig. S2) at 24 and 48 hours of exposure with variability in inhibition between cases noted but with similar overall trends noted, suggesting lack of an additive effect of the drug combination.
Figure 2.
Pevonedistat (P) induced AML cell apoptosis was attenuated by the mTOR inhibitor sapanisertib (S). A. AML cell lines MV4–11 or Molm-13 cells were incubated with 50 nM, 100 nM 200 nM or 400 nM of P, S, or combination (P + S) for 48 hours. Flow cytometry figures shown are representative of three independent experiments of MV4–11 treated with 200 nM of P, S or P + S. The dose response pattern of cell viability is expressed as percentage of vehicle-treated cells (control). B. AML blasts from different patients (n = 5) were incubated with 400 nM P, S or combination for 48 hours, or with DMSO as a vehicle control. Apoptosis was detected using Annexin V and 7-AAD staining by flow cytometry. Results shown are representative of two independent experiments. * P < .05; **P < .01 using one-way ANOVA followed by Newman–Keuls Multiple Comparison Test. C. Western blot shows that the S attenuated P induced AML apoptosis. After 24 or 48 hours of 400 nM of S, P or S + P exposure, expression of apoptosis mediators, NEDD8-cullin, and various mTOR pathway components in MV4–11 were examined by Western blotting.
Immunoblotting analysis confirmed effects of these two compounds on apoptosis (Figure 2C). After 24 or 48 hour exposures in the MV4–11 cell line, pevonedistat treatment alone strongly induced PARP and caspases 8/3 cleavage and decreased the levels of the anti-apoptosis protein XIAP (X-linked mammalian inhibitor of apoptosis protein). Sapanisertib alone induced the cleavage of PARP and caspase 8 to some extent, but the intensity was much less than pevonedistat alone. The apoptosis mediator protein levels with combination treatment were intermediate between the agents used singly. Thus in the presence of sapanisertib, pevonedistat's proapoptotic effects were neutralized.
Interestingly, although sapanisertib and pevonedistat were antagonistic in terms of cytotoxicity and apoptosis induction, they both inhibited each other's signal pathway activation. As shown in Figure 2C, after 24-hour or 48-hour treatment either alone or in combination, sapanisertib and pevonedistat decreased the phosphorylation of mTOR on Serine 2448 (a marker for mTORC1 activity)/Serine 2481 (a marker for mTORC2 activity) sites as well as the phosphorylation of mTOR signaling downstream molecules, 4E-BP1 on serine 65 and S6RP on Serine 240/244. As Deptor, a natural mTOR signal endogenous inhibitor, is a substrate of CRL/SCFbTrCP E3 ubiquitin ligase [27], [32], [33], we postulated that inhibition of the Neddylation pathway by pevonedistat might cause Deptor accumulation. However, pevonedistat treatment alone did not cause Deptor accumulation. Deptor expression increased only with sapanisertib exposure or with the combination of the two compounds. These data suggest that inhibition of mTOR by sapanisertib efficiently blocked Deptor destruction, consistent with a previous study which showed that Deptor accumulation was induced by another mTOR inhibitor, PP242 [33]. Pevonedistat failed to stabilize Deptor in AML cells, indicating that Deptor degradation might not be accomplished through the NEDD8 pathway or that CRL1/SCFbTrCP E3 ubiquitin ligase might not have strong activity in these AML cells. In addition, pevonedistat only partially and indirectly inhibited mTOR. Similarly, sapanisertib also inhibited NEDD8-cullin expression, but the inhibition intensity was not as great as with pevonedistat alone or in combination, suggesting that sapanisertib only partially inhibited the neddylation pathway.
Differentiation Effects of Pevonedistat Are Attenuated by the mTOR Inhibitor Sapanisertib
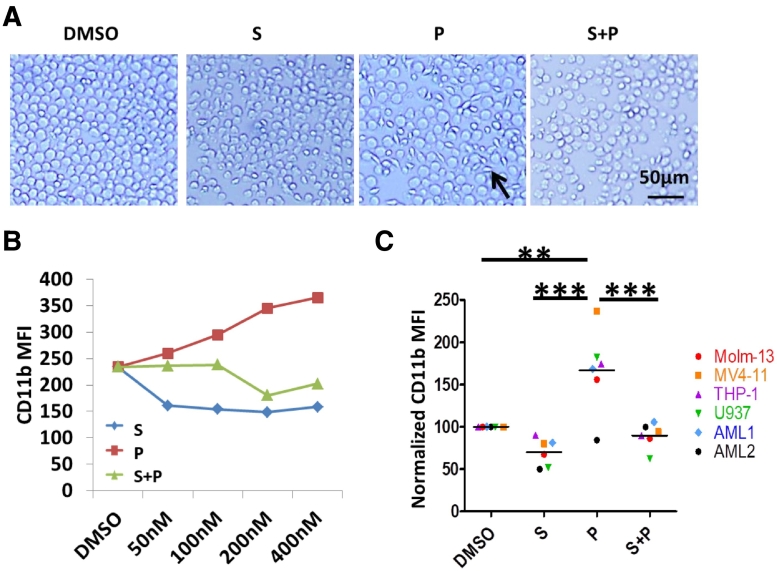
Pevonedistat induced Molm-13 cell morphology changes, but this was not observed in other AML cell lines (Figure 3A). By light microscopy, the normally round Molm-13 cells became elongated and fibroblast-like with variation in shape and size. The cells were still in suspension and did not adhere to the tissue culture plate. Similar to the effect on apoptosis, sapanisertib restored pevonedistat induced Molm-13 cell morphology changes.
Figure 3.
P increased AML cell CD11b expression was attenuated by S. A. Molm-13 cells were treated with 400 nM of S, P or S + P for 24 hours and the cell morphology changes were observed under phase contrast microscopy (bar, 50 μm). B. Molm-13 was incubated with different concentrations of S, P or S + P for 24 hours. Flow cytometry analysis was done to determine CD11b expression. MFI = mean fluorescence intensity. C. CD11b expression in various AML cell lines and primary AML samples (n = 6) treated with 400 nM of P, S or combination for 24 hours was analyzed via flow cytometry and normalized to expression with vehicle DMSO incubation alone. **P < .01; ***P < .001 using one-way ANOVA followed by Newman–Keuls Multiple Comparison Test.
To further investigate the effect, we compared expression of the cell surface marker, CD11b, which is a macrophage cell marker used to indicate differentiation, between vehicle and pevonedistat treated cells using flow cytometry. We observed a graded increase of CD11b protein levels with a corresponding increase in pevonedistat concentration in Molm-13 cells (Figure 3B). At the highest concentration of pevonedistat tested (400 nM), the protein expression increased 1.5 fold in the Molm-13 line compared to DMSO vehicle control. Sapanisertib decreased CD11b expression and also reduced pevonedistat induced CD11b expression. Significantly increased CD11b expression was also found in other pevonedistat treated AML cell lines and primary cells, and mTOR inhibition blocked these effects (Figure 3C). (Fig. S3), Effects of pevonedistat and sapanisertib on expression of CD11c and CD36, both also variably used as differentiation markers in myelomonocytic AML cells lines, differed amongst the cell lines as shown in Fig. S3.
mTOR Inhibition by Sapanisertib Led AML Cells into a Dormant/Quiescent Status
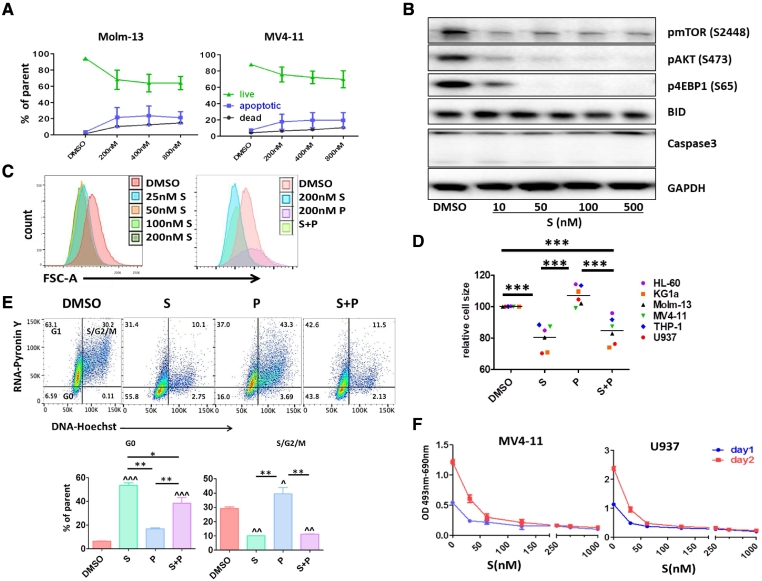
To further understand the effects of sapanisertib, we treated AML cells with varying concentrations of sapanisertib for 1 or 2 days followed by flow cytometry, immunoblotting, apoptosis determination, cell cycle analysis and XTT assay. As shown in Figure 4A, sapanisertib did not significantly induce cell death as measured by Annexin V and 7AAD apoptosis analysis after 2 days exposure at concentrations up to 800 nM in Molm-13 and MV4–11 cells. As expected, sapanisertib exposure for 2 days markedly inhibited mTOR phosphorylation as well as mTORC1 downstream mediators p4E-BP1 and the mTORC2 mediator pAKT expression in both primary cells and in cell lines (shown here with U937) in a concentration dependent manner; however, there was no effect on BID or cleaved caspase 3 expression in the cells as shown in Figure 4B, suggesting that sapanisertib, blocking both mTORC1 and mTORC2 activities, did not induce AML apoptosis.
Figure 4.
S led AML cells into a dormant/ quiescent status. A. Annexin V and 7AAD apoptosis flow cytometry analysis shows that S did not significantly induce cell death. Molm-13 and MV4–11 cells were treated with different concentrations of S for 48 hours and the percentage of live, apoptotic and dead cell was determined by flow cytometry. B. U937 cells were treated with different concentrations of S for 48 hours and western blot analysis was performed. C. Molm-13 cells were treated with different concentrations of S or S and P combination for 1 day followed by flow cytometry cell size analysis. Representative flow cytometry figures show that S sharply shifted FSC to the left at the concentration as low as 25 nM. D. S significantly reduced AML cell size. After 1 day of exposure, AML cell size was determined using FSC-A parameter by flow cytometry (***P < .001) in the presence of S, P, or S + P. E. S remarkably arrested AML cells in G0-phase. After 24 hours 200 nM P or S alone or combination treatment, RNA/DNA levels were analyzed using pyronin Y (RNA) and Hoechst (DNA) staining following by flow cytometry. ^P < .05, ^^P < .01, ^^^P < .001 compared to vehicle DMSO control; *P < .05, **P < .01 using one-way ANOVA followed by Newman–Keuls Multiple Comparison Test. F. MV4–11 and U937 cells were treated with different concentrations of S for 1 or 2 days, cell metabolic activity was measured by XTT assay.
It has been reported that mTOR inhibition could reduce cell size [34], [35], [36], so we compared the Molm-13 cell size with or without sapanisertib treatment using a flow cytometry geometric mean forward scatter area (FSC-A) parameter. After one day of sapanisertib exposure, there was a sharp leftward shift in the FSC-A histogram compared with DMSO vehicle control, indicating that cell size decreased (Figure 4C). We found that pevonedistat exposure for 24 hours did not have significant influence on cell size changes, whereas treatment with sapanisertib and pevonedistat together followed the same trend as sapanisertib alone in reducing cell size, indicating that pevonedistat presence was not able to overcome the effect induced by sapanisertib (Figure 4D). Fig. S4 also demonstrates that spanisertib was able to reduce protein content in the MV4–11 and Molm-13 cell lines in keeping with the noted effects on cell size.
We also analyzed cell cycle profiles generated by the two compounds alone or combination. We found that pevonedistat and sapanisertib resulted in different profiles. Using pyronin Y to label RNA and Hoechst 33342 to label DNA, we found that only 1 day of exposure to sapanisertib induced an accumulation of cells in the G0 phase of the cell cycle (Figure 4E) whereas cell numbers in S phase and G2/M phase were significantly decreased. Pevonedistat alone increased cell population in S/G2/M cell cycle phases; however, the RNA levels detected by pyronin Y staining were much lower than that of DMSO vehicle control, suggesting that DNA replication occurred with very low levels of RNA transcription. When cells were treated with the two compounds together, the cell cycle profiles were similar to the one with sapanisertib alone, indicating that sapanisertib presence prevented pevonedistat induced S/G2/M accumulation.
Since mTOR signaling regulates cell proliferation, we also performed an XTT cytotoxicity assay to detect the cellular metabolic activities. XTT measures cell viability based on the activity of mitochondrial enzymes in live cells that reduce XTT and are inactivated shortly after cell death. After one day exposure, sapanisertib significantly decreased cell proliferation rate (Figure 4F), suggesting that sapanisertib induced growth inhibition is not mainly through apoptosis induction, but rather through suppression of metabolic activity.
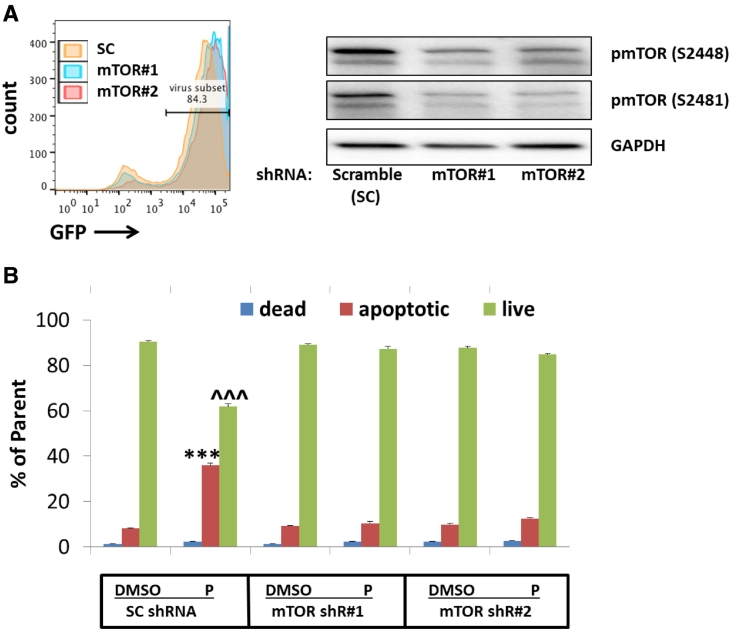
Pevonedistat Induced AML Cytotoxicity Was Attenuated by mTOR shRNA
To confirm the role of mTOR inhibition on pevonedistat-induced AML apoptosis, we targeted mTORC1/2 by knocking down mTOR expression with small hairpin RNAs (shRNA). Down-regulation of mTOR expression mirrored the effects of sapanisertib (Figure 5). mTOR inhibition by shRNAs resulted in decreased AML cell size (Fig. S5) and arrest of cells in G0-G1 phase. In addition, pevonedistat-induced cell death was significantly impeded in the cells infected with mTOR shRNA lentivirus compared to scrambled shRNA (P < .01). Therefore, we validated that mTOR signal inactivation protects AML cells from NAE inhibitor pevonedistat-induced apoptosis using both pharmacological and genetic approaches. To further study the mechanism which might explain an mTOR inhibitor's ability to induce resistance to pevonedistat, we tested rapamycin, an mTORC1 inhibitor without mTORC2 inhibitory effects. Concurrent exposure to rapamycin did not significantly alter the effects of pevonedistat and did not change cell size and cell cycle status. In addition, rapamycin did not inhibit the phosphorylation of 4E-BP1, suggesting that mTORC1 inhibition alone may be insufficient to inhibit the effects of pevonedistat, thereby suggesting a role for mTORC2 (Fig. S6).
Figure 5.
mTOR knockdown decreased AML cell S phase percentage and attenuated P induced AML cell apoptosis. A. primary AML blast cells were infected with 10moi mTOR or scramble shRNA lentiviruses. Three days after infection, mTOR expression was tested by immunoblotting. B. Seven days after infection, Molm-13 was treated with 200 nM P for 24 hours. The cells were stained with Annexin V and 7AAD and cell apoptosis was detected using flow cytometry. **P < .01 compared to SC shRNA DMSO apoptotic control; ^P < .05 compared to SC shRNA DMSO live cell control using two-way ANOVA followed by Bonferroni post-tests.
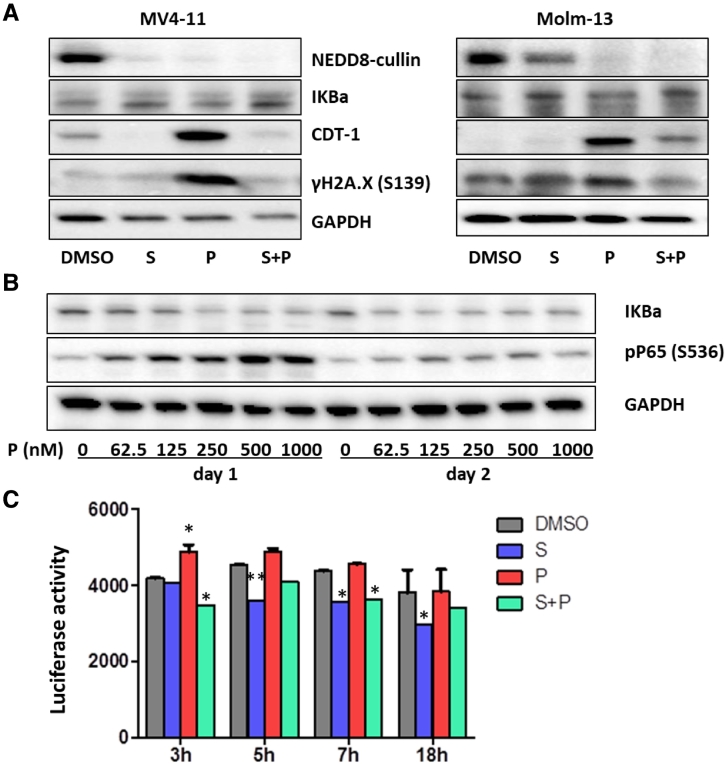
Pevonedistat did Not Initiate Cytotoxic Effects Through Inhibition of NFκB Signaling, But Rather Through DNA Re-Replication/Damage in AML Cells
According to previous studies, there are at least two mechanisms for the cytotoxic effects of pevonedistat in B-cell lymphoma [37]. One is through NFκB signaling inhibition by blocking IκBα ubiquitination and the other is via DNA re-replication by blocking CDT1 degradation. To determine the possible causes of the apoptotic effects induced by pevonedistat in AML cell lines, we analyzed both NFκB signaling and DNA damage response. By using western blotting in MV4–11 and Molm-13 cells, we could not detect phosphorylated IκBα (pIκBα, Ser32) using the well cited antibody from Cell Signaling or the accumulation of IκBα protein after pevonedistat exposure compared to control (Figures 2C and 6A). In U937 cells treated with pevonedistat, we detected a dose response increase in phosphorylated P65 (Ser536) and a slight decrease of IκBα at day1, but by day 2 the levels of pP65 were back to normal (Figure 6B). By using an NFκB reporter assay in U937 and KG1a cells, we detected a slight increase of NFκB signaling at the beginning (3–6 hours) of pevonedistat exposure (Figure 6C). These data indicate that the cytotoxic effects induced by pevonedistat on AML cells might not be initiated through NFκB inhibition.
Figure 6.
Pevonedistat did not initiate cytotoxic effects through inhibition of NFκB signaling, but through DNA re-replication/damage in AML cells. A. AML cell lines MV4–11 and Molm-13 cells were treated with S, P or S + P for 16 hours and western blot analysis was performed to detect the levels of NEDD8-cullin, IKBα, CDT-1 and γH2A.X (S139). GAPDH was probed as loading control. B. U937 cells were treated with different concentrations of P for 24 or 48 hours. Western blot analysis was performed to detect NFκB pathway markers. C. KG1a NFκB luciferase reporter cells were treated with 400 nM of S, P or S + P for 3, 5, 7, or 18 h. At the end of time point, the luciferase activity was detected. *P < .05, **P < .01 compared to DMSO vehicle control using two-way ANOVA followed by Bonferroni post-tests.
We then checked the expression of the DNA replication-licensing factor CDT1. CDT1 is one of the substrates of both CRL1-SCFskp2 [38] and CRL4CDT2 [39]. We found that blocking the neddylation pathway with pevonedistat significantly increased CDT1 protein levels in MV4–11 and Molm-13 cells after 16 hours of exposure, suggesting that CRL1 and CRL4 Ub ligase activity was inhibited. Not surprisingly, sapanisertib decreased CDT1 expression and the combination treatment showed a slight increase of CDT1 (Figure 6A). As pevonedistat induces DNA damage in chronic lymphocytic leukemia B cells [40], we tested DNA damage marker γH2A.X (the Serine 139 site phosphorylation of the minor histone H2A variant H2A.X) expression upon pevonedistat exposure as well. We found that treatment of AML cell lines MV4–11 and Molm-13 with pevonedistat resulted in an increase of γH2A.X expression (Figure 6A) and sapanisertib completely abolished pevonedistat-induced γH2A.X expression. These results together with a rise in cleaved PARP (Figures 1C and 2C) and cell cycle changes (Figure 4E) indicate that pevonedistat initiated AML cell apoptosis most likely through DNA re-replication and DNA damage and that sapanisertib attenuation of pevonedistat induced apoptosis might occur in part through inhibition of pevonedistat induced DNA re-replication and DNA damage.
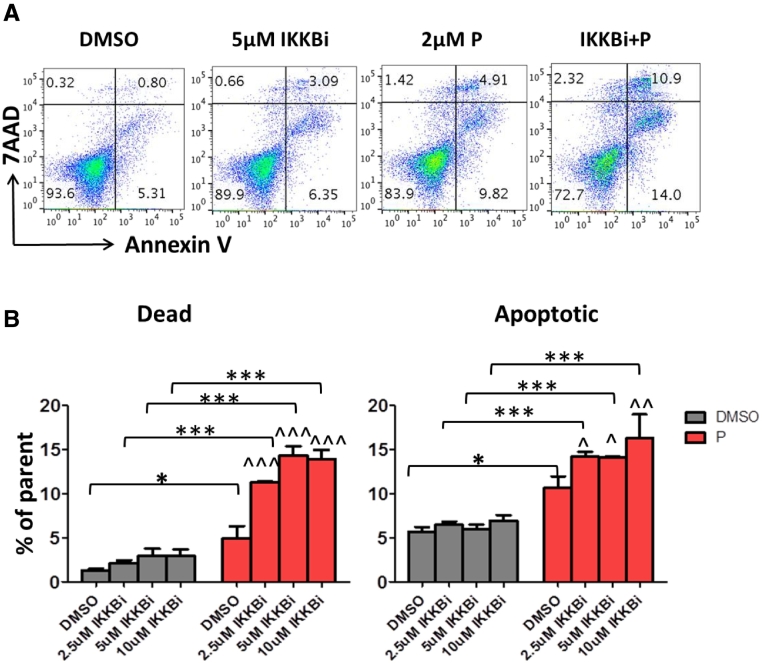
NFκB Signal Pathway Inhibitor (IKKB Inhibitor IV) Enhanced Pevonedistat Induced Cytotoxicity
Since the NFκB pathway has anti-apoptosis activity and we detected a slight increase in NFκB activity in KG1a and U937 cells, we postulated that inhibition of NFκB signaling could have synergetic effects of inducing cytotoxicity with pevonedistat. To test this hypothesis, we pretreated the pevonedistat resistant AML cell line KG1a with different concentrations of IKKB inhibitor IV (IKKBi) for 1 hour and then added pevonedistat to the cell culture for another 48 hours followed by flow cytometry apoptosis/ cell death analysis. Treatment with IKKBi alone for 48 hours had minimal effect on cytotoxicity; at a concentration of 5 μM, the cell death/apoptosis effects reached the maximum (4%/6%, respectively). Treatment with high concentration pevonedistat (2 μM) alone was able to induce cell apoptosis to a greater extent than did 5 μM IKKBi alone (10% vs 5%), while the combination of these 2 compounds enhanced cell death and apoptosis, indicating that NFκB inhibition could interact with neddylation inhibition to induce apoptosis (Figure 7).
Figure 7.
NFκB signal IKKB inhibitor enhanced P induced cytotoxicity. KG1a cells were pretreated with different concentrations of IKKB inhibitor (IKKBi) for 1 hour, followed by 2 μM of P treatment for 48 hours. The cells were stained with Annexin V and 7AAD and cell apoptosis was detected using flow cytometry. A. Representative flow figures are shown. B. Data are means ± SD (duplicate). *P < .05; ***P < .001; ^P < .05; ^^P < .01; ^^^P < .001 compared to pevonedistat treatment by one-way ANOVA followed by Newman–Keuls Multiple Comparison Test.
Discussion
In the present study, we investigated the effect of the neddylation inhibitor pevonedistat, the mTOR pathway inhibitor sapanisertib and the combination of these two compounds on AML cells. We were interested in targeting both neddylation and mTOR pathways to see if there were any synergistic effects of pathway inhibition in AML. We found that as a single agent treatment, both of these two compounds had significant antitumor activities in AML. However, there were antagonistic effects observed when the two compounds were administrated simultaneously.
Even though there was variability, most AML cell lines and primary cells were sensitive to pevonedistat treatment. In addition, pevonedistat increased expression of myeloid differentiation markers in some cell lines, arrested AML cells in S/G2/M cell cycle phases, and induced DNA re-replication and DNA damage, consistent with previous reports [41], [42]. Surprisingly, with pre- or co-treatment with sapanisertib, the dual mTORC1/2inhibitor, almost all the effects of pevonedistat were attenuated. Sapanisertib not only decreased pevonedistat induced apoptosis and reduced apoptosis markers such as cleaved caspase3/8 and cleaved PARP levels, it also restored anti-apoptosis protein XIAP levels. The differentiation of the AML cells and S/G2/M arrest induced by pevonedistat were also inhibited by simultaneous sapanisertib exposure.
We found that sapanisertib alone induced limited cell death in most AML cell lines and primary AML blasts, but it was able to inhibit cell cycle progression, reduce cell size, and protect from apoptosis as has been described in other systems [43]. Pevonedistat and sapanisertib therefore exhibit distinct anti-tumor effects on AML cells, which are cytotoxic and cytostatic effects, respectively.
It has been reported that pevonedistat induced cytotoxicity involves several mechanisms, such as NFκB inactivation [6], [37], reactive oxygen species induction [41], DNA re-replication and DNA damage [6], [44], [45], [46], [47], proteotoxic/ER stress, and unfolded protein response [48]. We observed that pevonedistat elicited S/G2/M accumulation in AML cells, consistent with the induction of the DNA re-replication process through up-regulation of the DNA replication licensing factor CDT1, a substrate of both CRL1/SCFskp2 and CRL4CDT2. Consequently, DNA damage increased as evidenced by PARP cleavage and γH2AX expression, and this was followed by cell apoptosis and death. These results suggest that pevonedistat induced AML cytotoxicity is most likely through DNA re-replication and DNA damage response. Sapanisertib as an antagonist of pevonedistat further confirmed this possibility, for sapanisertib diminished pevonedistat-mediated DNA damage exhibited by decreased CDT1 accumulation, reduced PARP cleavage and γH2AX expression, and arrest of cells in G0 phase.
Since the NFκB inhibitory protein IκBα is a substrate of CRL1/ SCFβTrCP [49], through neddylation inhibition, pevonedistat blocks IκBα degradation, and as a consequence, the NFκB pathway is inactivated. However, in our experimental system, we could not detect IκBα accumulation after pevonedistat treatment in the most sensitive cell lines MV4–11 or Molm-13. Moreover, our data also showed a slight elevation of P65 phosphorylation in U937. Recently, Zhou et al. showed that pevonedistat treatment alone induced P65 phosphorylation, but pevonedistat was able to inhibit belinostat induced NFκB activation [45]. Our results suggest that pevonedistat induced cytotoxicity was most likely due to DNA re-replication and DNA damage, but not NFκB inhibition in AML cells.
We found that pevonedistat alone slightly increased NFκB activity in KG1a and U937 NFκB reporter cells in early phase of treatment. Based on this finding, we speculated that inhibition of the NFκB pathway was likely able to sensitize the effects of pevonedistat in KG1a cells, the most resistant AML cell line to pevonedistat. We found that pretreatment of IKKB inhibitor IV modestly but significantly increased KG1a cell death and apoptosis induced by pevonedistat, suggesting that inhibition of NFκB activity can improve the effect of neddylation inhibition in AML cell lines largely insensitive to pevonedistat.
Although both the mTOR pathway endogenous inhibitor Deptor [32], [33] and NFκB pathway suppressor IκBα [49] are bound and ubiquitylated by CRL1/ SCFβTrCP with consequent proteasomal degradation in HEK293 or Hela cells, the two CRL1 substrates had different sensitivities to neddylation inhibition in AML cells. Pevonedistat failed to stabilize Deptor and IκBα in AML cells, indicating that their degradation might not be completely accomplished through the NEDD8 pathway or that CRL1/SCFβTrCP E3 ubiquitin ligase might not have strong activity in these AML cells. Milhollen et al. [37] found that there might be differential effects of NAE inhibition on individual CRL complexes or their substrates, and different cell types have different CRL activities and levels of different substrates. Therefore, understanding different CRL activities and different mechanisms of response to pevonedistat in different tumor types may allow for appropriate tailoring of pevonedistat therapeutic development.
Taken together, our data clearly demonstrate that both the neddylation pathway inhibitor pevonedistat and the mTOR pathway inhibitor sapanisertib have anti-tumor effects on AML. However, their anti-tumor mechanisms are distinct, and mTOR inhibition protected AML cells from apoptosis induced by pevonedistat through reducing cell metabolic activities and decreasing DNA damage responses (DDR). Further investigation of how these agents might be best utilized in therapy of AML is warranted, but our data would suggest that concurrent use of these agents would not be effective. Each has been successfully combined with other classes of agents. For example, pevonedistat combinations with histone deaceytlase inhibitors [45], hypomethylating agents [50], cytarabine [51], and bcl-2 inhibitors [52] are being examined. Our data also suggest that its use in combination with other NF-ΚB inhibitors might also have additive or synergistic effects. Given the cytostatic effects of sapanisertib, its addition after an apoptosis—inducing agent might also have efficacy in AML suppression.
Acknowledgments
Acknowledgements
The authors would like to thank Allison Berger for proof reading. This work was funded in part by a grant from the When Everyone Survives (WES) Foundation.
Disclosure of Potential Conflicts of Interest
The authors declare no competing financial interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.01.001.
Appendix A. Supplementary data
Supplementary material
References
- 1.Martelli AM, Evangelisti C, Chiarini F, Grimaldi C, McCubrey JA. The emerging role of the phosphatidylinositol 3-kinase/ akt/mammalian target of rapamycin signaling network in cancer stem cell biology. Cancers (Basel) 2010;2(3):1576–1596. doi: 10.3390/cancers2031576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Drakos E, Grammatikakis I, Schlette EJ, Li J, Leventaki V, Staikou-Drakopoulou E, Patsouris E, Panayiotidis P, Medeiros LJ. mTOR signaling is activated by FLT3 kinase and promotes survival of FLT3-mutated acute myeloid leukemia cells. Mol Cancer. 2010;9:292. doi: 10.1186/1476-4598-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow S, Minden MD, Hedley DW. Constitutive phosphorylation of the S6 ribosomal protein via mTOR and ERK signaling in the peripheral blasts of acute leukemia patients. Exp Hematol. 2006;34(9):1183–1191. doi: 10.1016/j.exphem.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Furqan M, Mukhi N, Lee B, Liu D. Dysregulation of JAK-STAT pathway in hematological malignancies and JAK inhibitors for clinical application. Biomark Res. 2013;1(1):5. doi: 10.1186/2050-7771-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griessinger E, Frelin C, Cuburu N, Imbert V, Dageville C, Hummelsberger M, Sirvent N, Dreano M, Peyron JF. Preclinical targeting of NF-kappaB and FLT3 pathways in AML cells. Leukemia. 2008;22(7):1466–1469. doi: 10.1038/sj.leu.2405102. [DOI] [PubMed] [Google Scholar]
- 6.Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 conjugation Pathway and its relevance in cancer biology and therapy. Genes Cancer. 2010;1(7):708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14(12):1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 9.Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell. 2011;19(2):168–176. doi: 10.1016/j.ccr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6(10):1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 11.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 12.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ, Xiong Y. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 2008;22(7):866–871. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118(1):83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 15.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23(11):1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 16.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 17.Swords RT, Erba HP, DeAngelo DJ, Bixby DL, Altman JK, Maris M, Hua Z, Blakemore SJ, Faessel H, Sedarati F. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. Br J Haematol. 2015;169(4):534–543. doi: 10.1111/bjh.13323. [DOI] [PubMed] [Google Scholar]
- 18.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15(12):3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 19.Pevonedistat plus azacitidine versus single-agent azacitidine as first-line treatment for participants with higher-risk myelodysplastic syndromes (HR MDS), chronic myelomonocytic leukemia (CMML), or low-blast acute myelogenous leukemia (AML) (PANTHER) https://clinicaltrials.gov/ct2/show/NCT03268954
- 20.Brenner AK, Andersson Tvedt TH, Bruserud O. The complexity of targeting PI3K-Akt-mTOR signalling in human acute myeloid leukaemia: the importance of leukemic cell heterogeneity, neighbouring mesenchymal stem cells and immunocompetent cells. Molecules. 2016;21(11) doi: 10.3390/molecules21111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17(8):666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 22.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altman JK, Sassano A, Platanias LC. Targeting mTOR for the treatment of AML. New agents and new directions. Oncotarget. 2011;2(6):510–517. doi: 10.18632/oncotarget.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altman JK, Sassano A, Kaur S, Glaser H, Kroczynska B, Redig AJ, Russo S, Barr S, Platanias LC. Dual mTORC2/mTORC1 targeting results in potent suppressive effects on acute myeloid leukemia (AML) progenitors. Clin Cancer Res. 2011;17(13):4378–4388. doi: 10.1158/1078-0432.CCR-10-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghobrial IM, Siegel DS, Vij R, Berdeja JG, Richardson PG, Neuwirth R, Patel CG, Zohren F, Wolf JL. TAK-228 (formerly MLN0128), an investigational oral dual TORC1/2 inhibitor: A phase I dose escalation study in patients with relapsed or refractory multiple myeloma, non-Hodgkin lymphoma, or Waldenstrom's macroglobulinemia. Am J Hematol. 2016;91(4):400–405. doi: 10.1002/ajh.24300. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol Cell. 2011;44(2):317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui D, Xiong X, Zhao Y. Cullin-RING ligases in regulation of autophagy. Cell Div. 2016;11:8. doi: 10.1186/s13008-016-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashton JM, Balys M, Neering SJ, Hassane DC, Cowley G, Root DE, Miller PG, Ebert BL, McMurray HR, Land H. Gene sets identified with oncogene cooperativity analysis regulate in vivo growth and survival of leukemia stem cells. Cell Stem Cell. 2012;11(3):359–372. doi: 10.1016/j.stem.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Y, Kaufman JL, Bernal L, Torre C, Matulis SM, Harvey RD, Chen J, Sun SY, Boise LH, Lonial S. MLN4924, an NAE inhibitor, suppresses AKT and mTOR signaling via upregulation of REDD1 in human myeloma cells. Blood. 2014;123(21):3269–3276. doi: 10.1182/blood-2013-08-521914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44(2):304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44(2):290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16(12):1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24(15):6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Readinger JA, DuBois W, Janka-Junttila M, Robinson R, Pruitt M, Bliskovsky V, Wu JZ, Sakakibara K, Patel J. Constitutive reductions in mTOR alter cell size, immune cell development, and antibody production. Blood. 2011;117(4):1228–1238. doi: 10.1182/blood-2010-05-287821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116(9):1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278(33):30854–30858. doi: 10.1074/jbc.C300251200. [DOI] [PubMed] [Google Scholar]
- 39.Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature. 2003;423(6942):885–889. doi: 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- 40.Paiva C, Godbersen JC, Berger A, Brown JR, Danilov AV. Targeting neddylation induces DNA damage and checkpoint activation and sensitizes chronic lymphocytic leukemia B cells to alkylating agents. Cell Death Dis. 2015;6:e1807. doi: 10.1038/cddis.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115(18):3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 42.Khalife J, Radomska HS, Santhanam R, Huang X, Neviani P, Saultz J, Wang H, Wu YZ, Alachkar H, Anghelina M. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia. 2015;29(10):1981–1992. doi: 10.1038/leu.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fumarola C, La Monica S, Alfieri RR, Borra E, Guidotti GG. Cell size reduction induced by inhibition of the mTOR/S6K-signaling pathway protects Jurkat cells from apoptosis. Cell Death Differ. 2005;12(10):1344–1357. doi: 10.1038/sj.cdd.4401660. [DOI] [PubMed] [Google Scholar]
- 44.Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71(8):3042–3051. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- 45.Zhou L, Chen S, Zhang Y, Kmieciak M, Leng Y, Li L, Lin H, Rizzo KA, Dumur CI, Ferreira-Gonzalez A. The NAE inhibitor pevonedistat interacts with the HDAC inhibitor belinostat to target AML cells by disrupting the DDR. Blood. 2016;127(18):2219–2230. doi: 10.1182/blood-2015-06-653717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70(24):10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blank JL, Liu XJ, Cosmopoulos K, Bouck DC, Garcia K, Bernard H, Tayber O, Hather G, Liu R, Narayanan U. Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer Res. 2013;73(1):225–234. doi: 10.1158/0008-5472.CAN-12-1729. [DOI] [PubMed] [Google Scholar]
- 48.Leclerc GM, Zheng S, Leclerc GJ, DeSalvo J, Swords RT, Barredo JC. The NEDD8-activating enzyme inhibitor pevonedistat activates the eIF2alpha and mTOR pathways inducing UPR-mediated cell death in acute lymphoblastic leukemia. Leuk Res. 2016;50:1–10. doi: 10.1016/j.leukres.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999;13(3):284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith PG, Traore T, Grossman S, Narayanan U, Carew JS, Lublinksky A, Kuranda M, Milhollen M. Azacitidine/decitabine synergism with the NEDD8-activating enzyme inhibitor MLN4924 in pre-clinical AML models. Blood. 2011;118(21):267. [Google Scholar]
- 51.Nawrocki ST, Kelly KR, Smith PG, Keaton M, Carraway H, Sekeres MA, Maciejewski JP, Carew JS. The NEDD8-activating enzyme inhibitor MLN4924 disrupts nucleotide metabolism and augments the efficacy of cytarabine. Clin Cancer Res. 2015;21(2):439–447. doi: 10.1158/1078-0432.CCR-14-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knorr KL, Schneider PA, Meng XW, Dai H, Smith BD, Hess AD, Karp JE, Kaufmann SH. MLN4924 induces Noxa upregulation in acute myelogenous leukemia and synergizes with Bcl-2 inhibitors. Cell Death Differ. 2015;22(12):2133–2142. doi: 10.1038/cdd.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material