Abstract
Background: Mutations in splicing factor 3b subunit 1 (SF3B1) have been reported to be associated with a favorable prognosis, while the prognostic impact of tet methylcytosine dioxygenase 2 (TET2) mutations is still controversial. The clinical significance of combined SF3B1 and TET2 mutation is even more uncertain. In this study, the clinical consequences of concurrent double SF3B1/TET2 mutation were compared with isolated SF3B1 or TET2 mutation. Materials and Methods: The demographics, diagnosis, cytogenetic abnormalities, and overall survival time of 130 patients with isolated SF3B1 (n=48) or TET2 mutation (n=54), or double SF3B1/TET2 mutation (n=28) were compared by next-generation sequencing. Results: Patients with double mutation were found to be significantly older than patients with isolated TET2 mutation. Patients with double mutation or isolated SF3B1 mutation were less likely to be diagnosed with acute myeloid leukemia than patients with isolated TET2 mutation. Patients with myelodysplasia had a higher percentage of double or isolated SF3B1 mutation, while patients with myeloproliferative neoplasms had a higher percentage of isolated TET2 mutation. Patients with double mutation more frequently had increased ring sideroblasts similarly to patients with isolated SF3B1 mutation. The percentage of patients with normal cytogenetics or good cytogenetic abnormalities was significantly higher in patients with double mutation than those with isolated mutation. Finally, in patients with myelodysplasia and normal cytogenetics, the median survival time in those with double mutation was significantly longer than in those with isolated SF3B1 mutation, even though the overall survival curve was not statistically significant. Conclusion: TET2 mutation appeared not to have additional effects when combined with SF3B1, and patients with double mutation appeared to have at least as, good as or even better prognosis than patients with isolated mutation.
Keywords: SF3B1, TET2, mutation, myeloid neoplasm
Myeloid neoplasms mostly include myelodysplastic syndrome (MDS), myeloproliferative neoplasm (MPN), and acute myeloid leukemia (AML). MDS and MPN are heterogeneous groups of clonal stem cell disorders with an inherent tendency to progress and transform into AML. Diagnosis of MDS/MPN/AML was traditionally made using morphology and cytogenetic studies on peripheral blood or bone marrow biopsies. With progress of next-generation sequencing, gene mutation profiling is playing a more and more important role in the diagnosis, stratification, and identification of therapeutic targets for patients with MDS/MPN/AML.
Recurrent somatic mutations in genes involved in DNA methylation [DNA methyltransferase 3 alpha (DNMT3A), tet methylcytosine dioxygenase 2 (TET2), isocitrate dehydrogenase 1/2 (IDH1/2)], post-translational chromatin modification [enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), ASXL transcriptional regulator 1 (ASXL1)], RNA splicing [splicing factor 3b subunit 1 (SF3B1), serine and arginine rich splicing factor 2 (SRSF2), U2 small nuclear RNA auxiliary factor 1 (U2AF1), Zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2 (ZRSR2)], DNA damage response [tumor protein P53 (TP53)], transcriptional regulation [runt related transcription factor 1 (RUNX1), BCL6 corepressor (BCOR), ETS variant 6 (ETV6)], signal transduction [Cbl proto-oncogene (CBL), NRAS proto-oncogene, GTPase (NRAS), Janus kinase 2 (JAK2)], and cohesin complex [stromal antigen 2 (STAG2)] have been identified in myeloid neoplasms (1-5). Interpretation of mutation profiles of patients with myeloid neoplasm is becoming a daily practice and challenge for pathologists, as well as for clinicians. It is essential to understand the clinical significance of these gene mutations both when present alone and in combination. SF3B1 and TET2 are two most commonly mutated genes in myeloid neoplasms. It is therefore of great interest to understand the clinical significance of these two genes when they are mutated alone or in combination.
SF3B1, encoding the subunit 1 of the splicing factor 3b complex, is an essential component of the U2 snRNP that recognizes the 3’ splice site at intron–exon junctions. SF3B1 is the most commonly mutated gene in MDS. SF3B1 mutation are seen in 57-75% of patients with refractory anemia with ring sideroblasts and 6-18% of patients with other subtypes of MDS (6,7). Most mutations in SF3B1 are missense mutations that affect invariant positions of this gene, predominantly at residues K700, K666, R625, and H662, but the specific effect of these mutations on protein function remains unknown. The presence of mutational hot spots and the absence of frame shift or nonsense changes suggest that the large majority of mutations are gain-of-function or change-of-function mutations (8). The close association between SF3B1 mutation and the ring sideroblastic phenotype is consistent with a causal link. SF3B1 is the first gene to be strongly associated with a specific feature of MDS. Mutation of SF3B1 confers a favorable prognosis in MDS and MDS/MPN cases and is associated with lower risk of progression to AML compared with patients with wild-type SF3B1 (6,9).
The TET family of enzymes (TET1, TET2, TET3), mapping to chromosome 4q24, modulate hydroxymethylation by catalyzing the conversion of 5-methylcytosine to 5-hydroxymethylcytosine, which is an intermediate of DNA methylation that blocks the building of silencing proteins to methylated DNA. TET2 mutations identified in MDS are thought to result in a loss of catalytic activity. Loss or mutation of TET2 impairs its catalytic activity leading to accumulation of 5-methylcytosine. There are conflicting reports on whether TET2 mutation results in DNA hypermethylation (10). TET2 mutations are found in 20-25% of patients with MDS but occur at 30-60% in those with chronic myelomonocytic leukemia (11,12). The prognostic impact of TET2 mutation on survival in patients with MDS is controversial. Large cohort studies showed that the TET2 mutation did not appear to impact on overall survival (OS) (13-15). However, alterations of the TET2 gene were reported to negatively affect outcome of patients with chronic myelomonocytic leukemia (16). In one study, TET2 mutations were reported as predictors of inferior survival in post-transplant patients (17), while another study reported that TET2 mutation was an independent favorable prognostic factor in MDS (18).
There have been few studies reporting the clinical significance when SF3B1 and TET2 are mutated together, which occurs in some patients. It is still unknown if double SF3B1/TET2 mutation confer better or worse prognosis than these mutations in isolation. In addition, most of the studies to date have investigated the mutation of SF3B1 or TET2 in the background of other mutated genes, which could be confounding factors. Therefore, in this study, we compared patients with isolated SF3B1 mutation, isolated TET2 mutation, and double SF3B1/TET2 mutation in order to better understand the clinical significance of combined SF3B1/TET2 mutation.
Materials and Methods
Selection of patients. The Institutional Review Board (IRB) of the Institute approved this study (MCC17964). All patients with suspected myeloid neoplasms were submitted to next-generation sequencing. All patients with next-generation data from February 2014 to July 2017 (around 3,000 patients) were retrieved per IRB protocols. Patients with isolated SF3B1 mutation, isolated TET2 mutation, or double SF3B1/TET2 mutation only were selected for this study. The pathology reports and pathology slides were reviewed by two Board-certified hematopathologists to confirm the diagnosis. Diagnoses were rendered following the WHO 2008 classification for hematopoietic malignancies (19). The cytogenetic results of all these patients, including karyotyping and fluorescence in situ hybridization (FISH) study results [mostly FISH for MDS panel, including del(5q), del(7q), del(17p), del(20), and trisomy 8] were also retrieved from the electronic database and analyzed.
Next-generation sequencing analysis. Next-generation sequencing for the following most commonly mutated genes were analyzed since February 2014 by commercial or in-house at labs with College of American Pathologists/Clinical Laboratory Improvement Amendments (CAP/CLIA) certification: ASXL1, CBL, DNMT3A, ETV6, EZH2, IDH1, IDH2, JAK2, KIT, MPL proto-oncogene, thrombopoietin receptor (MPL), nucleophosmin 1 (NPM1), NRAS proto-oncogene, GTPase (NRAS), PHD finger protein 6 (PHF6), RUNX1, SET binding protein 1 (SETBP1), SF3B1, SRSF2, TET2, TP53, U2AF1, ZRSR2. The in-house next-generation sequencing platform was validated against the results of the same outside laboratory for quality control at the validation stage, and confirmed that the results were comparable. In-house targeted next-generation sequencing was performed using Illumina MiSeq or NexSeq500 instruments (Illumina, San Diego, CA, USA). DNA was isolated from bone marrow aspirates or peripheral blood with QiaAmp DNA extraction kit and the QiaCube robot (Qiagen, Germantown, MD, USA) and quantitated by Nanodrop spectrophotometry (Agilent, Santa Clara, CA, USA). Library construction was performed with 250 ng of genomic DNA using amplicon-based capture for the targeted genes. FastQ files were aligned using Novalign, and various bioinformatics tools (Samtools, Varscan, and Freebayes) in Clinical Genomicist Workspace (PierianDX™, St Louis, MO, USA) were used to make variant calls. The outside laboratory isolated DNA from bone marrow aspirates or peripheral blood and then the coding regions of 21 genes were amplified by polymerase chain reaction, and interrogated by next-generation sequencing technology.
All tests reported variants with an allelic frequency of 5% or more. For patients with next-generation sequencing testing on more than one instance, a gene mutated in any of the tests was considered to be mutated for that patient. A gene that was found to be mutated in multiple samples from the same patient was counted only once when enumerating the number of patients that were positive for the mutation of that gene. Known benign single nucleotide polymorphisms were excluded. Variants with more than 1% minor allelic frequency in dbSNP or NHLBI Exome Sequencing Project (ESP) database (Seattle, WA, USA) were also considered non-pathogenic mutations and excluded.
Statistical analysis. Two-way Fisher’s exact test was used to calculate the p-values when comparing the percentages of patients with AML, >15% ring sideroblasts, different cytogenetic categories, and female/male ratio. Kaplan–Meier survival analysis was used to compare the overall survival levels. Two-way Mann–Whitney U-test was used when comparing the medians of the ages, OS time, hemoglobin levels, white blood cell counts, platelet counts, blast counts, and bone marrow cellularity (i.e. the percentage of cells in the total bone marrow space). A p-value of less than 0.05 was defined as statistically significant.
Results
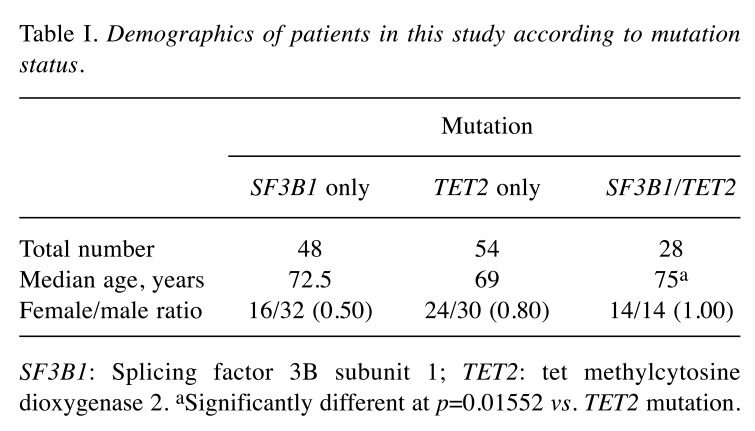
Patient characteristics. A total of 130 patients were identified, including 48 with only SF3B1 mutation, 54 with only TET2 mutation, and 28 patients with only double mutation (Table I). The median age was lowest in patients with isolated TET2 mutation and highest in patients with double mutations (p=0.01552). The female-to-male ratio was higher in patients with double mutations or TET2 mutation than in those with SF3B1 mutation, but not statistically significantly so. Double mutations appeared to be equally distributed in female and male patients, while isolated SF3B1 mutations were more prevalent in male patients. K700E was the most common mutation of SF3B1 in patients with isolated SF3B1 mutation or double mutation (data not shown).
Table I. Demographics of patients in this study according to mutation status.
SF3B1: Splicing factor 3B subunit 1; TET2: tet methylcytosine dioxygenase 2. aSignificantly different at p=0.01552 vs. TET2 mutation.
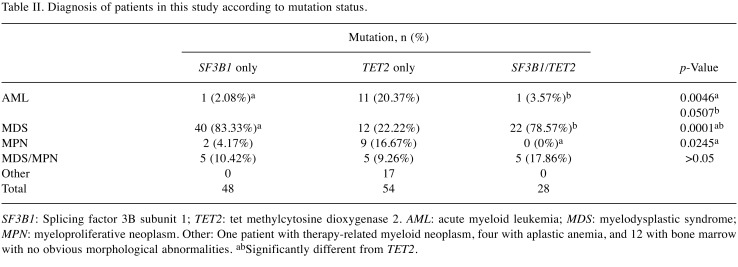
Comparison of diagnostic categories. The diagnostic categories of the patients included in this study are listed in Table II. Most of the patients in this study had MDS (n=74). We calculated the percentage of patients with AML in each mutation group. The percentage of AML in patients with isolated SF3B1 mutation was significantly lower than that of those with isolated TET2 mutation (p=0.0046). The percentage of AML in patients with double mutation was similarly lower than in patients with isolated TET2 mutation, with a borderline p-value of 0.0507, probably due to fewer patient samples. In patients with MDS, SF3B1 and double mutations were significantly more frequent than TET2 mutations (p=0.0001). In patients with MPN, TET2 mutations were significantly more frequent than double SF3B1/TET2 mutations (p=0.0245), and also more frequent than isolated SF3B1 mutations but not statistically significantly so (p>0.5).
Table II. Diagnosis of patients in this study according to mutation status.
SF3B1: Splicing factor 3B subunit 1; TET2: tet methylcytosine dioxygenase 2. AML: acute myeloid leukemia; MDS: myelodysplastic syndrome; MPN: myeloproliferative neoplasm. Other: One patient with therapy-related myeloid neoplasm, four with aplastic anemia, and 12 with bone marrow with no obvious morphological abnormalities. abSignificantly different from TET2.
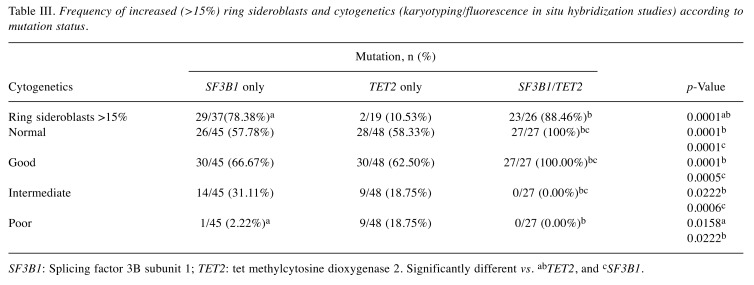
Comparison of the cytogenetic abnormalities. To better understand the impact of double SF3B1/TET2 mutations on prognosis, we also analyzed the results of cytogenetic studies, either by FISH studies or karyotyping. Most of the patients in this study had cytogenetic test results by karyotyping, FISH or both, except for a few patients in each group. Given the lack of cytogenetics data for these few patients, the percentages of patients in each category were calculated by dividing the number of patients in that category by the total number of patients tested cytogenetically (Table III). Three cytogenetic risk categories (good, intermediate, or poor), as defined by the International Prognostic Scoring System (20), were used in this study due to limited number of the patients. These were defined as follows: Good: Normal cytogenetics, -Y, del(5q) alone, or del(20q) alone; Poor: chromosome 7 anomalies or three or more abnormalities; Intermediate: other abnormalities. We also performed subgroup comparison focusing on patients with normal cytogenetics (which belong to the good cytogenetics category). Patients with good-risk cytogenetics usually have better prognosis than patients with poor- or intermediate-risk cytogenetic abnormalities. All 27 patients (100%) with double mutations had normal cytogenetics.
Table III. Frequency of increased (>15%) ring sideroblasts and cytogenetics (karyotyping/fluorescence in situ hybridization studies) according to mutation status.
SF3B1: Splicing factor 3B subunit 1; TET2: tet methylcytosine dioxygenase 2. Significantly different vs. abTET2, and cSF3B1.
Of patients with good cytogenetics, the percentage with double mutations (100%) was significantly higher than that of patients with isolated SF3B1 (p=0.0005) or isolated TET2 (p=0.0001) mutations. Accordingly, patients with intermediate cytogenetics had isolated SF3B1 or TET2 mutations more frequently than double SF3B1/TET2 mutations. Finally, patients with poor cytogenetics had TET2 mutations more frequently than isolated SF3B1 (p=0.0158) or double mutations (p=0.0222).
Comparison of frequency of patients with ring sideroblasts. SF3B1 mutations are associated with the presence of increased ring sideroblasts. Therefore, we compared the percentages of patients with ring sideroblasts in these three groups of patients (Table III). Not all the patients were evaluated for ring sideroblasts at the time of bone marrow biopsies; thus, the percentages of patients with more than 15% ring sideroblasts were calculated only for those tested. As expected, patients with isolated SF3B1 mutations more frequently had an increased level of ring sideroblasts than patients with isolated TET2 mutation (p=0.0001). Patients with double mutations also more frequently had more than 15% ring sideroblasts, at a rate significantly higher than those with isolated TET2 mutations (p=0.0001).
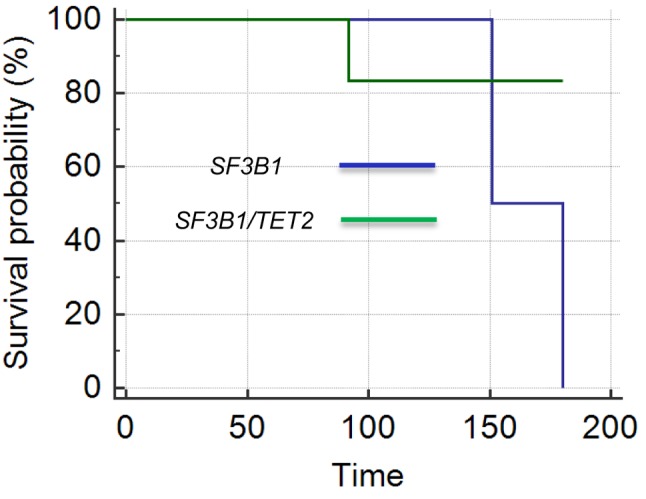
Comparison of OS. In this study, we tried to compare the clinical outcomes of patients with double mutation versus single SF3B1 or TET2 mutations. Most of the patients in this study were diagnosed with MDS, which was therefore selected for this comparison. Cytogenetic abnormalities are known to affect prognosis in myeloid neoplasms. Therefore, to avoid the confounding effects of cytogenetic abnormalities, we only compared OS of patients with MDS with normal cytogenetics. Of these patients, there were 21 with isolated SF3B1 mutation, 22 with double mutation, and six with isolated TET2 mutation. The patients with TET2 mutations were too few for a meaningful analysis. Therefore, OS for patients with double mutation was compared with that of those with isolated SF3B1 mutation. The median OS of patients with double mutations (73 months) was significantly longer than that of patients with isolated SF3B1 mutation (20 months) (p=0.02144). However, Kaplan–Meier survival analysis as shown in Figure 1 was not statistically significant (p=0.6076), which might be due to inadequate patients collected for this study or no actual difference in their OS.
Figure 1. Comparison of the overall survival times (months) for patients with myelodysplastic syndrome with normal cytogenetics according to the presence of double vs. isolated mutations of splicing factor 3B subunit 1 (SF3B1) and tet methylcytosine dioxygenase 2 (TET2). The overall survival time for patients with double mutations was not significantly different from that of those with isolated SF3B1 mutations (p=0.6076).
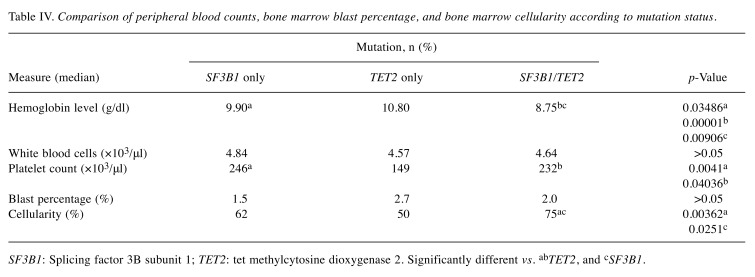
Comparison of hematological characteristics. Finally, we compared the median peripheral blood counts, hemoglobin levels, as well as the bone marrow blast percentages and the bone marrow cellularity in patients (Table IV). The median hemoglobin level of patients with double mutation was significantly lower than that of patients with isolated SF3B1 mutations, which was significantly lower than that of the patients with isolated TET2 mutations. The platelet count in patients with isolated SF3B1 mutation or double mutation was found to be significantly higher than that of patients with isolated TET2 mutations (p=0.0041 and p=0.04036 respectively). Finally, the median bone marrow cellularity of patients with double mutation was significantly higher than that of patients with isolated SF3B1 (p=0.0251) and those with isolated TET2 (p=0.00362) mutations. However, our Institute is a tertiary referral hospital and most patients had been treated with transfusion etc. before next-generation sequencing testing. Therefore, these values may not be representative and the comparisons may not be meaningful.
Table IV. Comparison of peripheral blood counts, bone marrow blast percentage, and bone marrow cellularity according to mutation status.
SF3B1: Splicing factor 3B subunit 1; TET2: tet methylcytosine dioxygenase 2. Significantly different vs. abTET2, and cSF3B1.
Discussion
The progression of myeloid neoplasms and their transformation to AML involves genetic and epigenetic aberrations (4). Risk stratification of myeloid neoplasms based on a prognostic scoring system determines the therapeutic approach. The treatment of low-risk patients is conservative, while the treatment of high-risk MDS involves the use of hypomethylating agents and allogeneic hematopoietic stem cell transplantation in younger patients (21). However, gene mutations are playing increasingly more important roles in risk stratification and therapeutic decision making in myeloid neoplasms.
SF3B1 and TET2 are two of the most commonly mutated genes in myeloid neoplasms. SF3B1 mutations have been reported to be prevalent in MDS, especially in the presence of ring sideroblasts, and were associated with better overall and leukemia-free survival (6,7,22). In one study, unsupervised statistical analysis of somatic mutations and morphological diagnosis showed that MDS associated with SF3B1 mutations was a distinct entity from MDS with ring sideroblasts with non-mutated SF3B1 gene (23). Based on data from these studies, patients with MDS and SF3B1 mutation can be classified as having either refractory anemia with ring-sideroblasts or refractory cytopenia with multilineage dysplasia and ring sideroblasts based on the mere presence of ring sideroblasts regardless of their percentage (24). SF3B1 mutations in patients with MDS were associated with thrombocytosis, increased ring sideroblasts, and lower blasts percentage, and a favorable prognosis and prolonged survival (24).
Approximately 20% of patients with MDS have TET2 mutation, and this gene is also frequently mutated in chronic myelomonocytic leukemia and AML (15,25). In addition, TET2 mutations are relatively frequent in patients with myeloproliferative neoplasms, in whom dysplastic features are very infrequently present (26). The clinical significance of TET2 mutations in myeloid neoplasms is still unclear, with most literature reporting no impact (27-28), some reporting poor prognosis (29-33), and a few reporting good prognosis (18,34,35). However, TET2 mutations have been shown to predict response to DNMT inhibitor in MDS (36,37).
So far, very few studies have studied the consequence of combined mutations of SF3B1 and TET2. Somatic recurrent TET2 mutations have been found in normal elderly individuals with clonal hematopoiesis (38). SF3B1 mutations have also been found in normal blood samples from 2728 patients with cancer but without hematopoietic neoplasm, suggesting that these mutations may represent pre-malignant events that cause clonal hematopoietic expansion (39). The presence of SF3B1 or TET2 mutations in normal elderly individuals indicates that accumulation of isolated somatic mutations may not cause direct disease development. However, cooperation with other genes that are frequently mutated in myeloid and other hematopoietic cancer might result in clonal expansion (40). In one study, TET2 loss acts with SF3B1(K700E) to cause a more severe erythroid and long-term hematopoietic stem cell phenotype (41).
Most previous reports studied the significance of SF3B1 or TET2 mutations in the background of other mutated genes, which could be confounding factors and biased the results or conclusions. In this study, we only included patients with isolated SF3B1, isolated TET2, or isolated SF3B1/TET2 mutations, therefore reducing the influences of other co-mutated genes, even though coexisting mutations in other not-tested genes cannot be ruled out.
In our study and consistent with previous knowledge, SF3B1 was mutated most often in patients with MDS and TET2 in patients with MPN. Double SF3B1/TET2 mutations were also found to be most common in patients with MDS. Patients with isolated TET2 mutations were more likely to be diagnosed with AML than patients with isolated SF3B1 mutations, indicating better prognosis for those with isolated SF3B1 mutations. Interestingly, the rate of AML in patients with double mutations was similarly low as for those with isolated SF3B1 mutation, suggesting that the effect of additional TET2 mutations on top of SF3B1 mutations is weak or non-existent.
SF3B1 mutations are associated with increased ring sideroblasts, while TET2 mutations are not. In our study, similarly high percentages of patients with double SF3B1/TET2 mutation had increased ring sideroblasts as patients with isolated SF3B1 mutations, also indicating minimal effect of additional TET2 mutations.
Cytogenetic abnormalities play an important role in the stratification of patients with myeloid neoplasms, and poor cytogenetic abnormalities are associated with poor prognosis. In our study, patients with double SF3B1/TET2 mutations all had normal cytogenetics, suggesting better prognosis for patients with the double mutation. The median age of patients with double SF3B1/TET2 mutations was significantly higher than that of patients with isolated TET2 mutations and slightly higher than those with isolated SF3B1 mutations, also appearing to suggest that double SF3B1/TET2 mutation carries similar or even better prognosis than isolated mutations. However, this may have been due to the fact that older patients might be more likely to have two genes mutated than younger patients. The fact that patients with double SF3B1/TET2 mutation had significantly longer median OS time than those with isolated SF3B1 mutations, even though not statistically significant by Kaplan–Meier analysis, also suggests better or similar prognosis for patients with double mutations when compared with patients with isolated SF3B1 mutations. This may not be surprising since SF3B1 mutations are known to be associated with favorable prognosis, while TET2 mutations were also associated with favorable prognosis in a few studies (18,34,35).
In summary, our study indicated that double SF3B1/TET2 mutations might confer similar or potentially better prognosis than isolated SF3B1 or TET2 mutations. This information will be helpful in understanding the roles gene mutations play in myeloid neoplasms, and in interpretation of gene mutations for clinical practice. However, due to the small number of patients included in this study and limited genes tested in our panels, larger cohorts studies are necessary to confirm the findings and the interpretations.
Conflicts of Interest
The Authors declare that they have no conflict of interest in regard to this study.
References
- 1.Gangat N, Patnaik MM, Tefferi A. Myelodysplastic syndromes: Contemporary review and how we treat. Am J Hematol. 2016;91:76–89. doi: 10.1002/ajh.24253. [DOI] [PubMed] [Google Scholar]
- 2.Martínez-Avilés L, Besses C, Álvarez-Larrán A, Torres E, Serrano S, Bellosillo B. TET2, ASXL1, IDH1, IDH2, and c-CBL genes in JAK2- and MPL-negative myeloproliferative neoplasms. Ann Hematol. 2012;91:533–541. doi: 10.1007/s00277-011-1330-0. [DOI] [PubMed] [Google Scholar]
- 3.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ades L, Itzykson R, Fenaux P. Myelodysplastic syndromes. Lancet. 2014;383:2239–2252. doi: 10.1016/S0140-6736(13)61901-7. [DOI] [PubMed] [Google Scholar]
- 5.Delic S, Rose D, Kern W, Nadarajah N, Haferlach C, Haferlach T, Meggendorfer M. Application of an NGS-based 28-gene panel in myeloproliferative neoplasms reveals distinct mutation patterns in essential thrombocythaemia, primary myelofibrosis and Polycythaemia vera. Br J Haematol. 2016;175(3):419–426. doi: 10.1111/bjh.14269. [DOI] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, Mudie LJ, Stephens PJ, McLaren S, Massie CE, Tarpey PS, Varela I, Nik-Zainal S, Davies HR, Shlien A, Jones D, Raine K, Hinton J, Butler AP, Teague JW, Baxter EJ, Score J, Galli A, Della Porta MG, Travaglino E, Groves M, Tauro S, Munshi NC, Anderson KC, El-Naggar A, Fischer A, Mustonen V, Warren AJ, Cross NC, Green AR, Futreal PA, Stratton MR, Campbell PJ. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patnaik MM1, Lasho TL, Hodnefield JM, Knudson RA, Ketterling RP, Garcia-Manero G, Steensma DP, Pardanani A, Hanson CA, Tefferi A. SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood. 2012;119:569–572. doi: 10.1182/blood-2011-09-377994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellagatti A, Boultwood J. The molecular pathogenesis of the myelodysplastic syndromes. Eur J Haematol. 2015;95:3–15. doi: 10.1111/ejh.12515. [DOI] [PubMed] [Google Scholar]
- 9.Visconte V, Makishima H, Maciejewski JP, Tiu RV. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia. 2012;26:2447–2454. doi: 10.1038/leu.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, Berthon C, Adès L, Fenaux P, Beyne-Rauzy O, Vey N, Braun T, Haferlach T, Dreyfus F, Cross NC, Preudhomme C, Bernard OA, Fontenay M, Vainchenker W, Schnittger S, Birnbaum D, Droin N, Solary E. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31:2428–2436. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 13.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine R. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, Raza A, Kantarjian H, Levine RL, Neuberg D, Garcia-Manero G, Ebert BL. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gäken J, Lea NC, Przychodzen B, Mian SA, Nasser EE, Shooter C, Westwood NB, Strupp C, Gattermann N, Maciejewski JP, Germing U, Mufti GJ. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 16.Kosmider O, Gelsi-Boyer V, Ciudad M, Racoeur C, Jooste V, Vey N, Quesnel B, Fenaux P, Bastie JN, Beyne-Rauzy O, Stamatoulas A, Dreyfus F, Ifrah N, de Botton S, Vainchenker W, Bernard OA, Birnbaum D, Fontenay M, Solary E, Groupe Francophone des Myélodysplasies TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94:1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bejar R, Stevenson KE, Caughey B, Lindsley RC, Mar BG, Stojanov P, Getz G, Steensma DP, Ritz J, Soiffer R, Antin JH, Alyea E, Armand P, Ho V, Koreth J, Neuberg D, Cutler CS, Ebert BL. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32:2691–2698. doi: 10.1200/JCO.2013.52.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosmider O, Gelsi-Boyer V, Cheok M, Grabar S, Della-Valle V, Picard F, Viguié F, Quesnel B, Beyne-Rauzy O, Solary E, Vey N, Hunault-Berger M, Fenaux P, Mansat-De Mas V, Delabesse E, Guardiola P, Lacombe C, Vainchenker W, Preudhomme C, Dreyfus F, Bernard OA, Birnbaum D, Fontenay M, Groupe Francophone des Myélodysplasies TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 19.Swerdlow H, Campo Elias, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Wardiman JW. WHO Classification of Tumors of Hematopoietic and Lymphoid Tissue. WHO Press, Geneva, 2008 [Google Scholar]
- 20.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, Ohyashiki K, Toyama K, Aul C, Mufti G, Bennett J. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079–2088. [PubMed] [Google Scholar]
- 21.Gill H, Leung AY, Kwong YL. Molecular and cellular mechanisms of myelodysplastic syndrome: Implications on targeted therapy. Int J Mol Sci. 2016;17:440. doi: 10.3390/ijms17040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visconte V, Rogers HJ, Singh J, Barnard J, Bupathi M, Traina F, McMahon J, Makishima H, Szpurka H, Jankowska A, Jerez A, Sekeres MA, Saunthararajah Y, Advani AS, Copelan E, Koseki H, Isono K, Padgett RA, Osman S, O’Keefe C, Maciejewski JP, Tiu RV. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 2012;120:3173–3186. doi: 10.1182/blood-2012-05-430876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malcovati L, Papaemmanuil E, Ambaglio I, Elena C, Gallì A, Della Porta MG, Travaglino E, Pietra D, Pascutto C, Ubezio M, Bono E, Da Vià MC, Brisci A, Bruno F, Cremonesi L, Ferrari M, Boveri E, Invernizzi R, Campbell PJ, Cazzola M. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood. 2014;124:1513–1521. doi: 10.1182/blood-2014-03-560227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patnaik MM, Hanson CA, Sulai NH, Hodnefield JM, Knudson RA, Ketterling RP, Lasho TL, Tefferi A. Prognostic irrelevance of ring sideroblast percentage in World Health Organization-defined myelodysplastic syndromes without excess blasts. Blood. 2012;119:5674–5677. doi: 10.1182/blood-2012-03-415356. [DOI] [PubMed] [Google Scholar]
- 25.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 26.McPherson S, McMullin MF, Mills K. Epigenetics in myeloproliferative neoplasms. J Cell Mol Med. 2017;21(9):1660–1667. doi: 10.1111/jcmm.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaidzik VI, Paschka P, Späth D, Habdank M, Köhne CH, Germing U, von Lilienfeld-Toal M, Held G, Horst HA, Haase D, Bentz M, Götze K, Döhner H, Schlenk RF, Bullinger L, Döhner K. TET2 mutations in acute myeloid leukemia (AML): Results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol. 2012;30:1350–1357. doi: 10.1200/JCO.2011.39.2886. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, Lin Z, Cheng K, Fang Z, Li Z, Luo Y, Xu B. Prognostic role of TET2 deficiency in myelodysplastic syndromes: A meta-analysis. Oncotarget. 2017;8(26):43295–43305. doi: 10.18632/oncotarget.17177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohgami RS, Ma L, Merker JD, Gotlib JR, Schrijver I, Zehnder JL, Arber DA. Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod Pathol. 2015;28:706–714. doi: 10.1038/modpathol.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metzeler KH, Maharry K, Radmacher MD, Mrózek K, Margeson D, Becker H, Curfman J, Holland KB, Schwind S, Whitman SP, Wu YZ, Blum W, Powell BL, Carter TH, Wetzler M, Moore JO, Kolitz JE, Baer MR, Carroll AJ, Larson RA, Caligiuri MA, Marcucci G, Bloomfield CD. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2011;29(10):1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, Huberman K, Thomas S, Dolgalev I, Heguy A, Paietta E, Le Beau MM, Beran M, Tallman MS, Ebert BL, Kantarjian HM, Stone RM, Gilliland DG, Crispino JD, Levine RL. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohgami RS, Ma L, Merker JD, Gotlib JR, Schrijver I, Zehnder JL, Arber DA. Next-generation sequencing of acute myeloid leukemia identifies the significance of TP53, U2AF1, ASXL1, and TET2 mutations. Mod Pathol. 2015;28(5):706–714. doi: 10.1038/modpathol.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cher CY, Leung GM, Au CH, Chan TL, Ma ES, Sim JP, Gill H, Lie AK, Liang R, Wong KF, Siu LL, Tsui CS, So CC, Wong HW, Yip SF, Lee HK, Liu HS, Lau JS, Luk TH, Lau CK, Lin SY, Kwong YL, Leung AY. Next-generation sequencing with a myeloid gene panel in core-binding factor AML showed KIT activation loop and TET2 mutations predictive of outcome. Blood Cancer J. 2016;6(7):e442. doi: 10.1038/bcj.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamazaki J, Taby R, Jelinek J, Raynal NJ, Cesaroni M, Pierce SA, Kornblau SM, Bueso-Ramos CE, Ravandi F, Kantarjian HM, Issa JP. Hypomethylation of TET2 target genes identifies a curable subset of acute myeloid leukemia. J Natl Cancer Inst 108(2): 2015;pii:djv323. doi: 10.1093/jnci/djv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohamed AM, Balsat M, Koering C, Maucort-Boulch D, Boissel N, Payen-Gay L, Cheok M, Mortada H, Auboeuf D, Pinatel C, El-Hamri M, Tigaud I, Hayette S, Dumontet C, Cros E, Flandrin-Gresta P, Nibourel O, Preudhomme C, Thomas X, Nicolini FE, Solly F, Guyotat D, Campos L, Michallet M, Ceraulo A, Mortreux F, Wattel E. TET2 exon 2 skipping is an independent favorable prognostic factor for cytogenetically normal acute myelogenous leukemia (AML): TET2 exon 2 skipping in AML. Leuk Res. 2017;56:21–28. doi: 10.1016/j.leukres.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O, Quesnel B, Vey N, Gelsi-Boyer V, Raynaud S, Preudhomme C, Adès L, Fenaux P, Fontenay M, Groupe Francophone des Myelodysplasies (GFM) Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25:1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 37.Traina F, Visconte V, Elson P, Tabarroki A, Jankowska AM, Hasrouni E, Sugimoto Y, Szpurka H, Makishima H, O’Keefe CL, Sekeres MA, Advani AS, Kalaycio M, Copelan EA, Saunthararajah Y, Olalla Saad ST, Maciejewski JP, Tiu RV. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28:78–87. doi: 10.1038/leu.2013.269. [DOI] [PubMed] [Google Scholar]
- 38.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, Hassimi M, Socci N, Bhatt PK, Gonen M, Mason CE, Melnick A, Godley LA, Brennan CW, Abdel-Wahab O, Levine RL. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, Ozenberger BA, Welch JS, Link DC, Walter MJ, Mardis ER, Dipersio JF, Chen F, Wilson RK, Ley TJ, Ding L. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganguly BB, Kadam NN. Mutations of myelodysplastic syndromes (MDS): An update. Mutat Res Rev Mutat Res. 2016;769:47–62. doi: 10.1016/j.mrrev.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 41.Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, Schneider RK, Lord AM, Wang L, Gambe RG, McConkey ME, Ali AM, Raza A, Yu L, Buonamici S, Smith PG, Mullally A, Wu CJ, Fleming MD, Ebert BL. Physiologic expression of SF3B1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30:404–417. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]