Abstract
Immunotherapy using either checkpoint blockade or the adoptive transfer of antitumor lymphocytes has shown effectiveness in treating cancers with high levels of somatic mutations—such as melanoma, smoking-induced lung cancers and bladder cancer—with little effect in other common epithelial cancers that have lower mutation rates, such as those arising in the gastrointestinal tract, breast and ovary1–7. Adoptive transfer of autologous lymphocytes that specifically target proteins encoded by somatically mutated genes has mediated substantial objective clinical regressions in patients with metastatic bile duct, colon and cervical cancers8–11. We present a patient with chemorefractory hormone receptor (HR)-positive metastatic breast cancer who was treated with tumor-infiltrating lymphocytes (TILs) reactive against mutant versions of four proteins—SLC3A2, KIAA0368, CADPS2 and CTSB. Adoptive transfer of these mutant-protein-specific TILs in conjunction with interleukin (IL)-2 and checkpoint blockade mediated the complete durable regression of metastatic breast cancer, which is now ongoing for > 22 months, and it represents a new immunotherapy approach for the treatment of these patients.
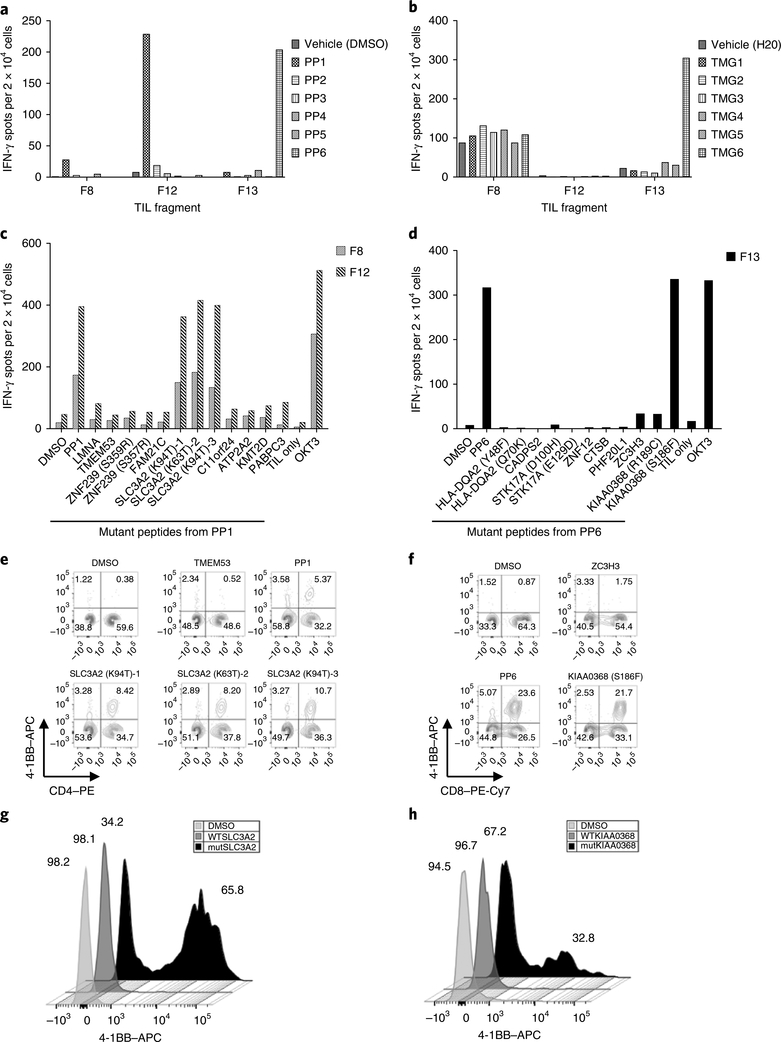
A 49-year-old woman with estrogen receptor (ER)-positive and ERB-B2 receptor tyrosine kinase 2 (ERBB2; also known as HER2)-negative metastatic breast cancer that was refractory to multiple lines of chemotherapy was enrolled in a clinical trial, NCT01174121, designed to determine the ability of autologous tumor-infiltrating lymphocytes (TILs) to mediate tumor regression in patients with metastatic epithelial cancers. Whole-exome sequencing (WES) and RNA sequencing (RNA-seq) of a right breast subcutaneous lesion revealed the presence of 62 nonsynonymous somatic mutations (Supplementary Table 1). This mutation burden fell within the interquartile range (IQR 30–95.5, median 47) of a published cohort of 140 patients with HR+HER2– metastatic breast cancer, whereas primary luminal A breast cancers, generally ER+HER2–, harbor fewer mutations (IQR 26–39.5, median 32)12–14. TIL fragments lymphocyte populations derived from culturing small pieces of the patient’s tumor in high-dose IL-2 (Supplementary Table 2) were initially screened against all of the mutations by using pulsed peptide pools (PPs 1–6) or transfected mRNAs (tandem minigenes (TMGs) 1–6) expressed in autologous antigen-presenting cells (APCs). TIL fragments F8 and F12 showed reactivity against PP1 but not the corresponding TMG1 (Fig. 1a,b). TIL F13 showed reactivity against both peptide pool PP6 and TMG6 (Fig. 1a,b). Individual peptide screens of the TIL fragments showed F8 and F12 recognized the solute carrier family 3 member 2 (SLC3A2) protein (Fig. 1c). Due to alternative splicing, a variant in the SLC3A2 gene produced three separate peptides, in which each contained the same mutation (p.lysine>threonine) but different C terminal regions (Supplementary Table 1). All three SLC3A2 peptides were recognized by F8 and F12 TILs (Fig. 1c). The reactive F13 TIL recognized mutant ECM29 proteasome adaptor and scaffold (ECPAS; also known as KIAA0368) (Fig. 1d). SLC3A2 and KIAA0368 reactivity was mediated by CD4+ and CD8+ T cells, respectively (Fig. 1e,f and data not shown).
Fig. 1 |. TIL populations from patient 4136 recognize autologous mutant SLC3A2 and KIAA0368 antigens.
a, Interferon (IFN)-γ production, as determined by ELISPOT assay, following coculture with autologous B cells pulsed with the respective peptide pools, showing that TIL fragments F8 and F12 recognize a component of peptide pool PP1 and that F13 recognizes a component of peptide pool PP6. b, IFN-γ production following coculture with autologous B cells that were electroporated with TMG RNA, showing that fragment F13 also recognizes one of the TMGs within TMG6. c, IFN-γ production following coculture with autologous B cells that were pulsed with the individual peptides within peptide pool PP1, showing that TIL fragments F8 and F12 recognize the mutSLC3A2 (p.Lys94Thr, p.Lys63Thr) peptides. d, IFN-γ production following coculture with autologous B cells that were pulsed with the individual peptides within peptide pool PP6, showing that F13 specifically recognizes the mutKIAA0368 (p.Ser186Phe). e, Flow cytometry analysis of F12 T cells after coculture with B cells that were pulsed with mutSLC3A2 peptides, showing that 4–1BB expression was upregulated. No upregulation was observed when mutant transmembrane protein 53 (TMEM53) or other mutant peptides from PP1 (not shown) were tested. f, Flow cytometry analysis of F13-reactive T cells after coculture with B cells that were pulsed with mutKIAA0368 (p.Ser186Phe) peptide, showing that 4–1BB expression was upregulated. No upregulation was observed when mutant zinc finger CCCH-type containing 3 (ZC3H3) or other mutant peptides from PP6 (not shown) were tested. In e and f, cells were gated on CD3, and data are representative of at least three independent experiments. g, Flow cytometry analysis of allogeneic T cells that were transduced with the sequences encoding TCR K, which specifically recognized the 18-mer mutSLC3A2 peptide (amino acid sequence LLASSDPPALASTNAEVT), as shown by upregulation of 4–1BB. These results are representative of all seven TCRs that were found to specifically recognize the mutSLC3A2 peptide. h, Flow cytometry analysis of allogeneic T cells that were transduced with sequences encoding TCR R, which specifically recognized the 25-mer mutKIAA0368 (p.Ser186Phe) peptide, as shown by upregulation of 4–1BB. In g and h, cells were gated on CD3, and data are representative of two independent experiments. WT, wild type.
To identify neoantigen-reactive T cell clonotypes, mutant-peptide-reactive cells were sorted on the basis of the T cell receptor (TCR)-β variable (TRBV) region or high expression levels of 4–1BB (CD137, a marker of T cell activation) and then subjected to single-cell sequencing. TILs from reactive fragments (F12 and F13) were FACS-sorted by individual TRBV family-member-specific antibodies and stimulated with APCs loaded with mutant (mut)-SLC3A2 or mutKIAA0368. TCR-focused deep sequencing of the sorted cells identified an oligoclonal population of TCR-encoding sequences, with two dominant TRBV (TRBV19–01 and TRBV05–01) and three dominant TCR-α variable (TRAV) sequences (TRAV08–06, TRAV38–01 and TRAV02–01) (Supplementary Fig. 1a). Of the six tested TCR pairs, two TCRs (TRBV19–01–TRAV02–01 (TCR K) and TRBV05–01–TRAV38–01 (TCR L)) recognized mutSLC3A2 as demonstrated by ELISPOT assays for detection of IFN-γ-producing cells (Supplementary Fig. 1b) and upregulation of 4–1BB (Fig. 1g and data not shown). In addition, a single KIAA0368-reactive TCR clonotype was identified (TRAV21–01–TRBV06–05) (Fig. 1h and Supplementary Fig. 1b,c). Alternatively, 4–1BBhi cells were sorted for single-cell sequencing. Five additional mutSLC3A2-specific TCR pairs were identified (Supplementary Table 3). No new mutKIAA0368-reactive TCRs were identified. In total, seven mutSLC3A2-reactive TCRs demonstrated specific and highly avid recognition of mutSLC3A2 to concentrations as low as 10 ng/ml (Supplementary Fig. 2a). The response was human leukocyte antigen (HLA)-DRB1*07:01-restricted, and the minimal epitope was located within the amino acid sequence, DPPALASTNAEVT (Supplementary Fig. 2b–f). The mutKIAA0368-reactive TCR, TRBV06–05–TRAV21–01 (TCR R), recognized the mut-KIAA0368 peptide at concentrations as low as 100 ng/ml (Fig. 1h and Supplementary Fig. 3a). In addition, Vβ13.1+ (TRBV06–05, TRBV06–06 and TRBV06–09)-sorted F13 TILs specifically recognized TMG6-electroporated APCs, demonstrating that the mutKIAA0368 protein was efficiently processed and presented (Supplementary Fig. 3b). The CD8+ mutKIAA0368-specific TCR recognized the predicted epitope (10-mer, MPYGYVLNEF, rank 0.06, 9.5 nM), which was restricted by HLA-B*35:01 (Supplementary Fig. 3c,d).
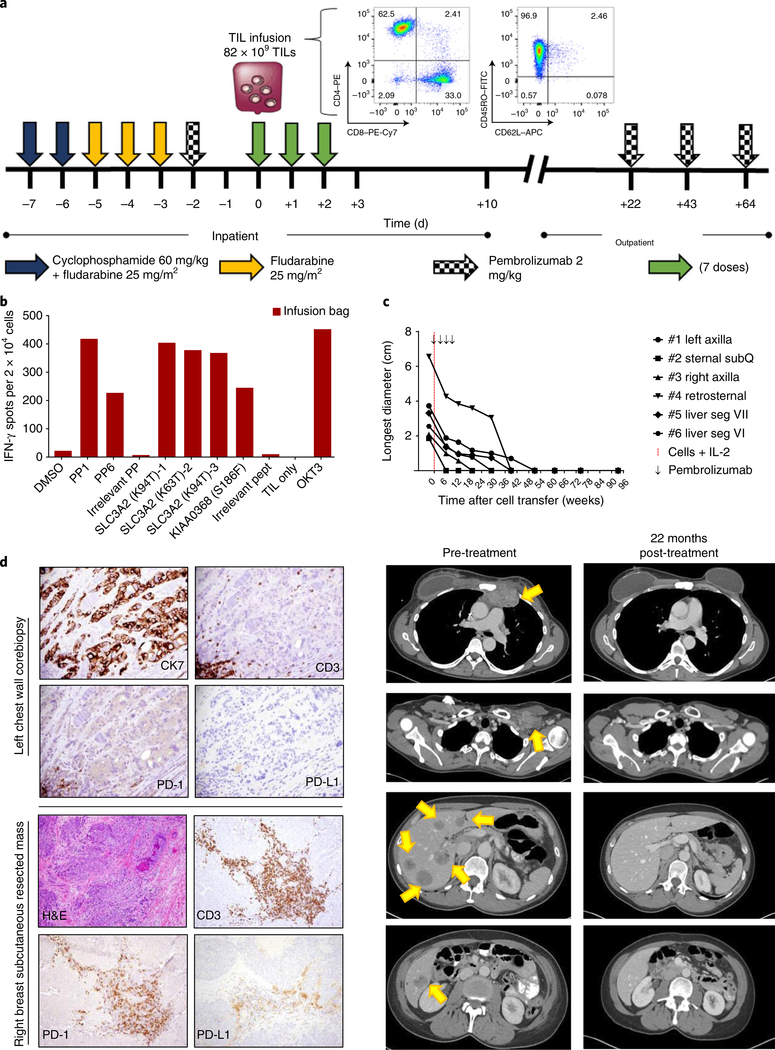
The patient was treated with 8.2 × 1010 TILs as described in Fig. 2a. The infused cells were predominantly CD4+ (62.5%), with an effector memory (CD62L–CD45RO+) phenotype. The infusion product maintained its reactivity to mutSLC3A2 and mutKIAA0368 after rapid expansion of the T cells from the fragments (Fig. 2b), and approximately 21% of the infused CD3+ cells expressed programmed cell death 1 (PD-1). At the first evaluable time point, 6 weeks after cell transfer, we found that the target tumor burden had reduced 51%; at the most recent evaluation (22 months after cell transfer), all target and nontarget lesions have radiographically resolved (Fig. 2c).
Fig. 2 |. Adoptive transfer of autologous TILs targeting immunogenic tumor mutations mediated tumor regression.
a, Treatment schema, with characteristics of the infusion product. Initial gating for flow cytometry analysis was done on live CD3+ cells. b, Interferon (IFN)-γ production, as determined by ELISPOT assay, showing that the infusion product, consisting of TILs expanded from fragments 8, 12 and 13 maintained their reactivity to mutSLC3A2 and mutKIAA0368. c, Top, response curves of target lesions (tumor size measurements). All lesions resolved 1 year after TIL transfer, and the patient continued to demonstrate complete response 22 months after cell infusion and 20 months after the last dose of pembrolizumab. Bottom, cross-sectional imaging was obtained 1 week before cell infusion (pre-treatment) and 22 months after infusion (22 months post-treatment). Arrows indicate target lesions (from top to bottom: retrosternal mediastinum, left axilla with clinical brachial plexopathy and compressed axillary vein, and multiple liver segments). SubQ, subcutaneous. d, Top, images showing pre-treatment tumor biopsy of a left chest wall mass (present at time of treatment), demonstrating ductal breast adenocarcinoma with scattered peripheral PD-1+ lymphocytes and few intratumoral lymphocytes (magnification: 40× ). Bottom, images showing the subcutaneous tumor that served as the source of TILs, with intratumoral PD-1+ lymphocytes and PD-L1+ stroma. Tumor cells were negative for PD-L1 expression (magnification: H&E-stained images, 10×; CD3-, PD-1- or PD-L1-stained images, 20× ).
To assess the potential influence of pembrolizumab, an anti-PD-1 monoclonal antibody, on the patient’s tumors, retrospective analysis for the expression of PD-1 and one of its ligands, PD-L1, was performed on two tumor samples obtained before adoptive cell transfer. Biopsy of a chest wall lesion that was refractory to multiple treatments demonstrated one small focal area of CD3+PD-1+ cells with no expression of PD-L1 on the tumor or stroma (Fig. 2d, top). This tumor regressed following adoptive cell transfer. The tumor resected as the source of TILs was a rapidly growing right breast lesion and found to have abundant CD3+PD-1+ intratumoral T cells with PD-L1+ stroma; however, there was no evidence of PD-L1 expression on the tumor cells (Fig. 2d, bottom). PD-1 expression in peripheral blood was 11.4% of CD3+ cells at 6 weeks after transfer, and it remained relatively constant at later time points—9% of CD3+ cells (at 9 months after transfer) and 7.9% of CD3+ cells (at 2 years after transfer).
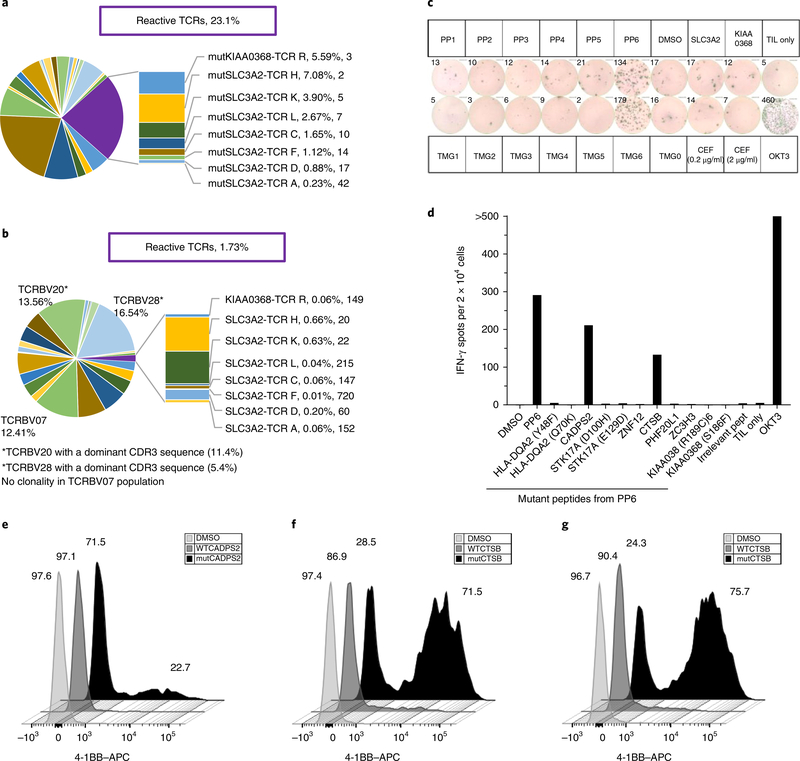
TCR-focused deep sequencing of the infused cell population showed that the unique TRBV complementarity determining region 3 (CDR3)-encoding sequences of the eight mutation-reactive clonotypes (seven SLC3A2-reactive and one KIAA0368-reactive TCRs) accounted for 23.1% of the productive sequences at the time of the cell transfer (Fig. 3a). Evaluation of the peripheral blood 6 weeks after cell transfer revealed that these known mutant-protein-reactive TCRs persisted at a level of 1.73% of all productive TRBV sequences. Additional analysis showed three TRBV families (TRBV07, 12.41%; TRBV20, 13.56%; TRBV28, 16.54%) had significant expansion in the blood at 6 weeks after transfer, with TRBV20 and TRBV28 containing at least one dominant TCR clonotype (β-chain CDR3: CSASDGSTTGSTYEQYF, 11.4%; and CDR3: CASRPPGYEQYF, 5.4%, respectively) of unknown reactivity (Fig. 3b). TRBV20+-sorted cells were nonreactive against the peptide pools, TMGs and peptides from cytomegalovirus (CMV), Epstein–Barr virus (EBV) and influenza virus (CEF peptides; data not shown). FACS-sorted TRBV28+ cells showed specific recognition of PP6 and TMG6 (Fig. 3c). The TRBV28+ cells specifically recognized two mutant proteins: calcium-dependent secretion activator 2 (CADPS2; p.Arg1266His) and cathepsin B (CTSB; p.Asp159His) (Fig. 3d). Single-cell sequencing revealed a dominant TCR pair, TRBV28–01–TRAV26–01 (TCR J, 82% of all TCR sequences), that specifically recognized mutCADPS2 and was HLA-C*04–01-restricted, and the minimal epitope recognized was the predicted 9-mer, TYDTVHRHL (Fig. 3e, Supplementary Fig. 4a–c and Supplementary Table 4). Two TCR pairs were identified to be reactive specifically to mutCTSB (TCR M and TCR O, both TRBV28–01–TRAV12–01) with unique CDR3 regions (Fig. 3f, Supplementary Fig. 4d and Supplementary Table 4). Both CTSB-specific TCRs were HLA class II, DRB1*07:01 restricted, and the minimal epitope was located within the amino acid sequence GYNSYSVSNSEKHIM (Supplementary Fig. 4e,f). Repeat analysis of the T cell repertoire in the infusion bag, using ultra-deep sequencing, revealed the presence of the CADPS2-specific and CTSB-specific TCRs (TCR J and TCR M, respectively), at frequencies <0.001%, whereas the second CTSB-specific TCR (TCR O) was undetectable.
Fig. 3 |. Persistence of known mutant-reactive TCR clonotypes at time of infusion and identification of new dominant clonotypes of unknown reactivity present in an apheresis product obtained 6 weeks after treatment.
a, TRBV diversity of the infused TILs with eight unique CDR3s (antigen, TCR, %, rank) associated with mutant-protein-specific reactive TCRs that account for 23.1% of the infused product. Unlabeled sections represent different TCRBV families (TCRBV01–30) without known reactivity. b, TRBV diversity of peripheral blood sampled at 6 weeks after infusion (post treatment). Known reactive TCR clonotypes accounted for 1.73% of circulating lymphocytes. The TCRBV20 and TCRVB28 populations were FACS-sorted from post-treatment peripheral blood for additional screening of mutant-peptide reactivity. c, The TRBV28 population was screened against peptide pools and TMGs as previously described, and it demonstrated concordant reactivity to PP6 and TMG6 by an IFN-γ ELISPOT assay. Data are representative of a single experiment and were confirmed by individual peptide experiments. d, The TCRBV28 population demonstrated reactivity against peptides encoded by two nonsynonymous mutations within PP6—CADPS2 (p.Arg1266His) and CTSB (p.Asp159His). TCRs were isolated from CD3+4–1BB+ peripheral blood lymphocytes (PBLs) following coculture with autologous B cells pulsed with either mutCADPS2 or mutCTSB. e, Flow cytometry analysis showing that allogeneic peripheral blood T cells that were transduced with constructs encoding TCR J (TRBV28–01–TRAV26–02) specifically recognized the 25-mer mutCADPS2 peptide and not the WT peptide, as shown by upregulation of 4–1BB. f, Flow cytometry analysis showing that allogeneic peripheral blood T cells that were transduced with constructs encoding TCR M (left) or TCR O (right), both of which are TRBV28–01–TRAV12–01, specifically recognized the 25-mer mutCTSB peptide, and minimal recognition was observed for the WT CTSB peptide, as shown by upregulation of 4–1BB. Data in e and f are representative of two independent experiments.
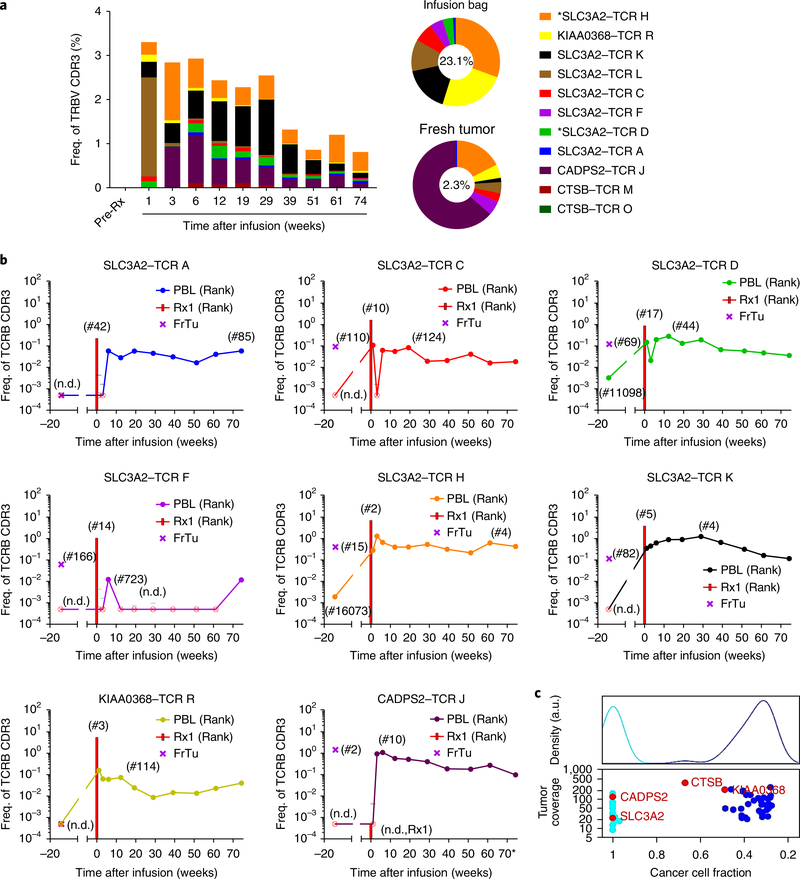
In total, 11 TCR clonotypes recognized the four neoantigens (SLC3A2, KIAA0368, CADPS2 and CTSB) for patient 4136. Only two TCRs, both of which recognized mutSLC3A2 (TRBV7–9–TRAV26–2 (TCR D) and TRBV19–TRAV9–2 (TCR H)) were detectable in peripheral blood prior to treatment (0.003% and 0.002% of all TRBV sequences, respectively). Following the cell infusion, we evaluated the in vivo persistence of known neoantigen-reactive TCRs in the peripheral blood. The median cumulative frequency during weeks 3–12 was 2.83% of the peripheral repertoire, and the known reactive clonotypes comprised 0.81% of the post-infusion repertoire at 17 months after infusion (Fig. 4a). Eight of the 11 neoantigen-reactive TCRs persisted in the patient’s peripheral blood at least 17 months after treatment (Fig. 4b). In addition, the dominant clonotypes that were identified 6 weeks after transfer persisted.
Fig. 4 |. Persistence of the 11 mutant-reactive TCR clonotypes from cell infusion to 17 months after cell transfer.
a, Of the 11 clonotypes, only 2 were detected in pre-treatment peripheral blood (denoted by the asterisk) and comprised 0.005% of all CDR3 sequences. The total percentage of reactive clonotypes identified in the peripheral blood was highest at the earliest time point (3.29%), but eight of these clonotypes persisted 17 months after treatment, comprising 0.81% of all CD3+ cells. Clonotypes (of the TRBV20 and TRBV28 families; see Fig. 3b) of unknown reactivity that dominated the peripheral blood at 6 weeks after transfer persisted as 3.6% of productive CDR3 sequences at 17 months after transfer (data not shown). Sequences corresponding to TCR J were detected only with ultra-deep sequencing of the infusion bag, but TCR J was a dominant intratumoral clonotype that expanded after treatment. b, The pattern of persistence in peripheral blood for each of the eight mutant-reactive clonotypes that persisted at 17 months after transfer. Bars indicate the percentage of infusion product. Rank (in parentheses) indicates relative position of the unique CDR3 sequence when all productive CDR3 sequences are sorted in decreasing frequency. Crossed circles indicate ‘not detected’ (n.d.) with a cut-off frequency of 0.0005. For the +1-week and +3-week samples, the lower limits of detection were 0.0044 and 0.0016, respectively. Rx1, treatment 1. FrTu, fresh tumor. c, Clonal architecture of the resected tumor, with recognized neoantigens identified. The top panel provides a histogram of the density of the nonsynonymous mutations depicted in the bottom panel. a.u., arbitrary units.
To better understand the heterogeneity of the nonsynonymous mutations within the patient’s tumor, a clonality analysis was performed on the resected tumor DNA. This analysis revealed that 20/62 (32%) mutations were clonal, 37/62 (60%) were subclonal, and 5 (8%) mutations could not be determined. MutSLC3A2 and mutCADPS2 were clonal (cancer cell fraction (CCF): 0.999, each), whereas, mutCTSB and mutKIAA0368 seemed to be subclonal (CCF: 0.669 and 0.489, respectively) (Fig. 4c).
This highly personalized therapy identified reactive T cells against multiple HLA-class-I- and HLA-class II-restricted neoantigens that were presented by the autologous tumor, enriching a potentially narrower repertoire of mutant-reactive T cells than bulk TILs alone. Targeting multiple tumor antigens may overcome potential tumor escape mechanisms, such as downregulation of class I molecules or tumor heterogeneity. By screening for reactivity on the basis of nonsynonymous mutations, T cells that recognized other classes of potential neoantigens were deliberately not selected but may have been a component of the infusion product. The post-treatment blood contained an oligoclonal T cell population of unknown specificity, which may or may not have been tumor reactive. It is likely that the expansion of T cells with unknown reactivity resulted from nonspecific homeostatic reconstitution after lymphodepletion, or it may represent epitope spreading with tumor-stimulated clonal expansion to neoantigens not yet identified, potentially triggered or enhanced by checkpoint inhibitor blockade. It has been shown previously that preparative lymphodepletion can create an immune milieu that is primed for lymphocyte expansion, with high circulating levels of IL-7 and IL-1515. While the increased homeostatic cytokines can aid the proliferation of infused autologous TILs, it is likely that the entire reconstituting lymphocyte population may compete for these growth factors.
Although limited responses to pembrolizumab treatment have been reported in patients with triple-negative breast cancer (TNBC), there are no published data to support pembrolizumab activity in HR+ breast cancer. The biopsies of the chest wall and subcutaneous masses shown in Fig. 2 revealed no expression of PD-L1, suggesting that pembrolizumab treatment likely had minimal to no impact in this patient. In a related clinical trial, 2 of 12 patients, one with bile duct cancer10 and the other with colon cancer9, received mutant-protein-reactive TILs without the administration of pembrolizumab, and both patients underwent objective cancer regressions. On the basis of prior studies, transferred lymphocytes expressed little to no PD-1 at the time of infusion but re-expressed PD-1 1 month after the infusion16. Although a portion of this patient’s infusion product did express PD-1, the level of PD-1 expression in the peripheral blood remained relatively constant during and after pembrolizumab administration. The combination strategy of pembrolizumab treatment with cell transfer was an attempt to prevent PD-1/PD-L1-mediated T cell inhibition in the tumor microenvironment at the time of cell infusion. The patient’s complete tumor regression does not seem consistent with a response to a short course of single-agent pembrolizumab.
The rapid identification of mutant-protein-specific TCR sequences by single-cell sequencing may provide a means of engineering neo-antigen reactivity into autologous PBLs with a high proliferative potential, which may be more effective than the direct use of TILs. Metastatic breast cancer can be a heterogeneous disease, and this patient demonstrated that a personalized therapy tailored to target the unique somatic mutations presented by the autologous tumor of the affected individual can mediate complete durable cancer regression.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41591–018-0040–8.
Methods
Study design.
This is an ongoing phase 2 study designed to evaluate the safety and efficacy of adoptive cell transfer of selected, mutant-protein-reactive TILs plus pembrolizumab treatment following lymphocyte depletion in patients with metastatic epithelial cancer (NCT01174121). Specifically, patients with metastatic breast cancer must have been refractory to at least two systemic regimens. This study was approved by the Institutional Review Board (IRB) of the NCI, and all relevant ethical regulations were followed. Informed consent was obtained from all patients.
Clinical history.
Patient 4136—a 49-year-old woman with metastatic ER+HER2– breast cancer—was initially diagnosed with ductal carcinoma in situ (marker status unknown) with no invasive component identified after a left modified radical mastectomy. After a disease-free interval of 10 years, recurrence was identified in multiple lymph nodes (axillary, supraclavicular, mediastinal and paratracheal) and confirmed by biopsy (100% ER+, 40% PR+, HER2–). After a short response to nanoparticle-albumin-bound (nab)-paclitaxel treatment, persistent disease was found to be refractory to multiple endocrine therapies and chemotherapies (capecitabine, vinorelbine, docetaxel, doxorubicin and cyclophosphamide), and biopsy (Fig. 2d, top) of a chest wall mass was > 50% ER+, PR– and HER2–. She developed progressive disease during lucitanib therapy and was enrolled in a clinical trial (NCT01174121), as described above. A metastatic right breast subcutaneous lesion was resected (ER–PR–HER2–) and processed for identification of nonsynonymous somatic mutations in the tumor and for the generation of TILs (Fig. 2d, bottom). In the 4 months between resection and treatment, the patient received everolimus and demonstrated clinical and radiologic progression before receiving the clinical treatment described below. The patient never received radiation therapy and did not harbor known germline mutations in the BRCA1 and BRCA2 oncogenes, or other known deleterious somatic mutations in high-penetrance genes involved in breast cancer susceptibility.
Treatment and analysis of response.
Administration of autologous TILs was defined as day 0. Preparative nonmyeloablative lymphodepleting chemotherapy consisted of cyclophosphamide (60 mg per kg body weight (mg/kg)) and fludarabine (25 mg/m2), which was administered on days –7 and –6, followed by additional doses of fludarabine alone on days –5 to –3. A single dose of pembrolizumab (2 mg/kg) was administered on day –2. On day 0, the patient received an intravenous infusion of 8.2 × 1010 TILs supported by administration of aldesleukin (720,000 IU/kg) every 8 h to tolerance (seven doses) and pembrolizumab (days +21, +42 and +63). The patient developed transient grade 3 hypophosphatemia and expected grade 4 hematologic cytopenias during preparative chemotherapy. After cell infusion and during the course of IL-2 administration, the patient developed grade 3 febrile neutropenia, which resolved with antibiotic therapy. There were no grade 3 or 4 adverse events during outpatient pembrolizumab administration. There was no evidence of off-target effects on normal tissue. Cross-sectional imaging was performed before treatment and at regular intervals after treatment. Response was monitored using Response Evaluation Criteria in Solid Tumors (RECIST) 1.0 parameters.
Whole-exome sequencing (WES), RNA sequencing (RNA-seq) and determination of mutation clustering.
WES and RNA-seq were performed at the Surgery Branch (SB), NCI. Genomic DNA (gDNA) and total RNA was purified from fresh tumor (FrTu) or ten 10-μm OCT sections and matched normal apheresis samples using the Qiagen AllPrep DNA/RNA kit (Qiagen), as per the manufacturer’s suggestions. Whole-exome library construction and exon capture of approximately 20,000 coding genes were done using Agilent Technologies SureSelectXT Target Enrichment System for paired-end libraries coupled with Human All Exon V6 RNA bait (Agilent Technologies). Exome libraries were prepped using 3 μg gDNA from fresh tumor tissue samples or 200 ng gDNA from the OCT sections, as per the manufacturer’s protocol. WES libraries were subsequently sequenced on a NextSeq 500 desktop sequencer (Illumina). Paired-end sequencing was done with an Illumina High-output flow cell kit (300 cycles). The mean sequencing depth for the following samples were: FrTu (118× ), OCT (120× ) and normal blood (116× ). The percentage of tumor in each sample (tumor purity) was 26.6% for both the FrTu and OCT samples, as estimated with the bioinformatics program ‘allele-specific copy-number analysis of tumors’ (ASCAT). An RNA-seq library was prepared using 2 μg of total RNA from fresh tumor with the Illumina TruSeq Stranded Total RNA library prep kit, as per the manufacturer’s protocol. The RNA-seq library was paired-end-sequenced on a NextSeq 500 desktop sequencer (Illumina). Exome-sequencing alignments were performed using novoalign MPI from novocraft (http://www.novocraft.com/) to human genome build hg19. Duplicates were marked using Picard’s MarkDuplicates tool. Insertion and deletion (indel) realignment and base recalibration were performed according to the GATK best-practices workflow (https://www.broadinstitute.org/gatk/). Once data were realigned and recalibrated, samtools (http://samtools.sourceforge.net) was used to create tumor and normal pileup files, and Varscan2 (http://varscan.sourceforge.net) was used to call somatic variants using the following criteria: tumor and normal read counts of 6 or greater, variant-allele frequency (VAF) ≥ 5% and tumor-variant reads ≥ 3. These variants were then annotated using Annovar (http://annovar.openbioinformatics.org).
For RNA-seq, alignments were performed by using the STAR (https://github.com/alexdobin/STAR) two-pass method to human genome build hg19. Duplicates were marked using Picard’s MarkDuplicates tool. Reads were split and trimmed using the GATK SplitNTrim tool, after which indel realignment and base recalibration were performed using the GATK toolbox. A pileup file was created using the final recalibrated bam file and samtools mpileup. Finally, variants were called using Varscan2.
Variants were further selected to increase the confidence of neoantigen identification. Mutations for immunological screening were selected based on their presence in both the tumor exome and the transcriptome, and the RNA variants must have had VAF > 10%, reads > 1. The mutations in patient 4,136 that were used for neoantigen identification are listed in Supplementary Table 1.
WES data for patient 4136 were used in the copy-number analysis. The data analysis was performed in the R statistical environment, version 3.3.0. The segmented copy number, cellularity and ploidy were determined using Sequenza v2.1.2, with normal sample as references and hg19 coordinates. The CCF of each mutation was estimated by integrating the local copy number, tumor purity (obtained from Sequenza) and VAF. All mutations with a read depth > 3 and VAF > 5% were clustered using PyClone v1.3.0 Dirichlet process clustering17. We ran PyClone with 50,000 iterations and a burn-in of 1,000.
TIL generation.
TILs were generated as previously described18. Cells from each TIL culture were phenotypically characterized by flow cytometry before cryopreservation, by staining for the CD3, CD4, CD8 and CD56 surface markers. Following the identification of neoantigen-reactive TILs, TIL cultures derived from fragments 8, 12 and 13 were selected for treatment and were expanded into high numbers using a rapid expansion protocol (REP) using IL-2, anti-CD3 antibody (clone OKT3, Miltenyi Biotec) and irradiated feeders18. All cells were cultured at 37 °C with 5% CO2 for 2 weeks before infusion.
Preparation of antigen-presenting cells (APCs) and EBV-B cells for TIL screening.
Autologous CD19+ B cells were sorted from the patient’s apheresis sample using anti-CD19 microbeads (Miltenyi Biotec) and cultured with irradiated (6,000 rad) NIH-3T3 cells that stably expressed CD40L (3T3-CD40L, authenticated by FACS for CD40L) at a 1:1 ratio in B cell medium comprised of Iscove’s modified Dulbecco’s medium (IMDM) (ThermoFisher Scientific) supplemented with 10% human serum and 200 U/ml IL-4 (Peprotech). CD14+ dendritic cells were sorted and transferred to a T150 flask, incubated with dendritic cell medium (RPMI 1640 medium supplemented with 5% human serum, 2 mM ʟ-glutamine, 800 IU/ml granulocyte–macrophage–colony-stimulating factor (GM-CSF; Peprotech) and 200 U/ml IL-4 (Peprotech)). Partially HLA-DR-matched allogeneic EBV-B cells (authenticated in the Surgery Branch by HLA typing) were grown in R10 medium (RPMI 1640 medium supplemented with 10% fetal bovine serum and 2 mM ʟ-glutamine).
Generation of tandem minigene (TMG) constructs and in-vitro-transcribed (IVT) RNA.
TMGs were constructed as previously described10. Twelve minigenes were included in each TMG construct for this study. Plasmids encoding the minigenes were linearized with the restriction enzyme NsiI, and each linearized plasmid was used as a template for in vitro transcription using the mMESSAGE mMACHINE T7 Transcription Kit (ThermoFisher Scientific), as per the manufacturer’s instructions.
RNA transfection and peptide-pulsing of autologous antigen-presenting cells.
Dendritic or B cells (APCs) were transfected with TMG RNA using the Neon Transfection System (ThermoFisher Scientific), as per the manufacturer’s instructions. Briefly, APCs were resuspended in the kit’s Resuspension Buffer at a concentration 1 × 107 cells/ml. The APCs were mixed with TMG RNA (10 μg per 1 × 106 APCs) just before the electroporation, aspirated into the Neon pipette and electroporated with either ‘1,500 V, 30 ms and one pulse’ for dendritic cells or ‘1,500 V, 10 ms and three pulses’ for B cells in the Neon device (ThermoFisher Scientific). Following electroporation, cells were immediately transferred into 24- or 48-well plates with dendritic cell (D) or B cell medium, respectively, supplemented with the appropriate cytokines and incubated overnight at 37 °C and 5% CO2.
All peptides were synthesized by Genscript, reconstituted in DMSO at a stock concentration of 50 mg/ml and were used to pulse APCs either individually or as a mixture of peptides (peptide pools). The peptide pools consisted of 12 long peptides that corresponded exactly to the peptides encoded by the respective TMG (Supplementary Table 1). Autologous dendritic or B cells (APCs) were harvested, washed, resuspended at 1.0 × 106 cells/ml in D or B cell medium, respectively, and incubated with 10 μg/ml of the individual peptide or the peptide pools (10 μg/ml of each peptide), unless otherwise specified, either for 2 h or overnight at 37 °C at 5% CO2. Following pulsing, the APCs were centrifuged, resuspended in fresh D or B cell medium without cytokines in concentration 1 × 106 cells/ml and used immediately in coculture assays.
Coculture assays: IFN-γ enzyme-linked immunospot (ELISPOT) assay, enzyme-linked immunosorbent assay (ELISA) and flow cytometry for activation marker 4–1BB.
Coculture conditions
T cells were incubated with peptide-pulsed or TMG-electroporated APCs at a ratio of 1:2.5 or 1:5 for dendritic or B cells, respectively. In addition, vehicle (DMSO)-pulsed APCs or T cell alone, with or without plate-bound OKT3 (1–10 μg/ml), were used as controls. TILs were rested in complete medium (CM; RPMI 1640 medium supplemented 10% human serum, 25 mM HEPES, 2 mM ʟ-glutamine, with or without 55 μM β-mercaptoethanol (ThermoFisher Scientific)) with or without IL-2 (300–6,000 IU/ml) for several hours to several days before coculture. PBLs were cultured in LCM medium (AIM-V medium CTS (Gibco) complemented with 5% human serum, 25 mM HEPES buffer, 1× MEM non-essential amino acids (Corning), ʟ-glutamine and 55 μM β-mercaptoethanol) supplemented with 300 IU/ml IL-2. Before each coculture, the cytokine-containing medium was removed, and the TILs or PBLs were resuspended in cytokine-free medium. Typically, an equal volume (100 μl) of T cells and APCs were mixed together for total volume of 200 μl/well in 96-well plates. All cocultures were performed in exogenous cytokine-free medium at 37 °C and 5% CO2 for 18–24 h. For cocultures intended to be used for sorting, the ratio of T cells to APCs was typically 1:2 to 1.0:2.5 regardless of the type of APCs. Cell sorting was carried out using the BD FACSAria IIu instrument (BD Biosciences) at the FACS facility, Surgery Branch, NCI.
IFN-γ enzyme-linked immunospot (ELISPOT) assay.
IFN-γ ELISPOT assays were performed in MultiScreen-IP filter plates (EMD Millipore). Each plate was pretreated with 50 μl 35% or 70% ethanol/well for < 2 min, washed 4× with Ultrapure water (Quality Biological) and then coated with 10 μg/ml of an IFN-γ capture antibody (100 μl/well, clone: 1-D1K, Mabtech, diluted in PBS) overnight at 4 °C. Anti-CD3 antibody (clone: OKT3, 1–10 μg/ml) was added to the positive-control wells. At the day of coculture, each plate was washed 5× with PBS and blocked with complete medium without IL-2 for at least 30 min at room temperature. After overnight coculture (18–24 h), the cells were harvested and transferred into a round-bottom 96-well plate for flow cytometry staining and analysis. Each ELISPOT plate was washed 5× with PBS containing 0.05% Tween 20 (MP Biomedicals) and then incubated for 2 h with 1 μg/ml, 0.22-μm filtered anti-human-IFN-γ detection antibody (clone: 7-B6–1, Mabtech, 100 μl/well, diluted in PBS + 0.5% FBS). Next, each plate was washed 5× with PBS and incubated for 1 h with streptavidin–ALP (Mabtech, 100 μl/well, 1:3,000 diluted with PBS + 0.5% FBS), followed by three washes with ddH2O and development with 0.45-μm-filtered BCIP–NBT substrate solution (KPL, 100 μl/well) for 5–10 min. The reaction was stopped by rinsing thoroughly with cold tap water. After they completely dried, each ELISPOT plate was scanned and counted using an ImmunoSpot plate reader and associated software (Cellular Technologies).
Flow cytometry for activation markers.
T cell activation was assessed by staining for upregulation of 4–1BB (CD137) and OX40 (CD134); however, upregulation of OX40 seemed inconsistent with T cell activation, as compared to 4–1BB. Therefore, 4–1BB was used as the T cell activation marker throughout the manuscript. Specifically, following coculture, harvested cells were pelleted and resuspended in FACS buffer (PBS containing 0.5% BSA (Sigma-Aldrich) and 2 mM EDTA (Crystalgen)). The cells were surface-stained with the appropriate antibodies for 30 min, at 4 °C in the dark, followed by 2× washes with cold FACS buffer and resuspension in FACS buffer containing propidium iodide (PI; 1 μg/ml, Sigma Aldrich). All data acquisition was performed using a BD FACSCanto I or FACSCanto II flow cytometer (BD Biosciences), and data analysis was performed using the FlowJo software (FlowJo, LLC). All of the data were gated on single (forward scatter (FSC)-H versus FSC-W, and side scatter (SSC)-H versus SSC-W), live (PI–) cells.
Enzyme-linked immunosorbent assay (ELISA).
In cocultures in which adherent cells (COS7 and HEK-CIITA) were used, the assessment of IFN-γ secretion was performed by ELISA. Briefly, flat-bottom 96-well plates were pretreated with 1 μg/ml of a human IFN-γ capture antibody (clone: 2G1, ThermoFisher Scientific), at least 24 h before the assay, at 4 °C. At the day of the assay, the plates were blocked with 150 μl of 4% BSA in PBS for at least 30 min at room temperature. Meanwhile, coculture supernatant was harvested or thawed, and 50 μl of each sample was added per well, after the plate-blocking step. Each plate was incubated for 60 min, followed by three washes with ELISA buffer (PBS containing 0.05% Tween 20) and incubation for 1 h with 0.5 μg/ml biotinylated anti-human-IFN-γ detection antibody (clone: B133.5, ThermoFisher Scientific, 50 μl/well, diluted in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS). Each plate was then washed 3× with ELISA buffer and incubated for 30–45 min with 125 ng/ml streptavidin–horseradish peroxidase (ThermoFisher Scientific, 150 μl/well, diluted in PBS with 4% BSA), followed by three washes with ELISA buffer. Finally, the plate was developed with TMB substrate (100 μl/well, ThermoFisher Scientific) in the dark, and the reaction was stopped with 100 μl/well of 0.1 M H2SO4 (Sigma Aldrich). Each ELISA plate was measured using a SpectraMax 190 microplate reader (Molecular Devices) and the associate software SoftMax Pro 6.2.2 (Molecular Devices). Data analysis was performed with Microsoft Excel software.
Flow cytometry antibodies and procedures.
The following human-protein-specific flow cytometry antibodies were purchased from BD Biosciences: CD3– APC-H7 (clone: SK7), CD3–APC-Cy7 (clone: SK7), CD3–APC (clone: SK7), CD137(4–1BB)–APC (clone: 4B4–1), CD4–PE (clone: RPA-T4), CD4–PE-Cy7 (clone: SK3), CD8–PE-Cy7 (clone: RPA-T8), CD8–APC-H7 (clone: SK1), CD62L–APC (clone: DREG-56), CD45RO–FITC (clone: UCHL1), CD279(PD-1)–APC (clone MIH4), mouse IgG1κ (isotype control)-APC. The following antibodies from Beckman Coulter were also used here: TCR-Vβ3–FITC (clone: CH92), TCR-Vβ13.1–PE (clone IMMU 222) and TCR-Vβ17–PE (clone: E17.5F3.15.13). For the assessment of the TCR repertoire, the Beta Mark TCR Vβ Repertoire KIT (Beckman Coulter) was used. Also, the following mouse-protein-specific flow cytometry antibodies were used: TCR-β-chain–PE (clone: H57–597, BD Biosciences) and TCR-β-chain–FITC (clone: H57–597, BD Biosciences). Standard procedures, as described above, were used for all cell-surface staining, data acquisition and analysis. For the investigation of PD-1 expression by flow cytometry, TILs (infusion product) or post-infusion peripheral blood mononuclear cells (PBMCs) were thawed and rested overnight in CM medium without cytokines before staining.
TCR identification and construction.
TCR high-throughput sequencing.
TRAV- and TRBV- targeted high-throughput sequencing, following DNA extraction, was performed on pelleted, snap-frozen TILs, PBLs, sorted T cells or fresh tumor samples by Adaptive Biotechnologies, using the ImmunoSEQ assay platform. All TCR frequencies were calculated based only on the productive TCR rearrangements.
Single-cell RT–PCR.
The single-cell RT–PCR protocol for the identification of neoantigen-reactive TCR pairs has been previously described19. Briefly, 4–1BB+ cells were sorted into RT–PCR buffer, following coculture with neoantigen-pulsed APCs. The reverse-transcription and first-amplification reactions were performed with the One-Step RT-PCR kit (Qiagen) and multiplex PCR with multiple Vα- and Vβ-region primers. For the second amplification reaction, the first RT–PCR products were used as templates and multiple internally nested Vα- and Vβ-region primers, with one internally nested primer for Cα- and Cβ-region each, and HotStarTaq DNA polymerase (Qiagen) were used. The PCR products were purified and sequenced by the Sanger method with an internally nested Cα- or Cβ-region primer (Beckmann Coulter).
Single-cell sequencing.
Activated (4–1BB+) T cells were sorted following stimulation with a neoantigen and subjected to the Fluidigm C1 system (Fluidigm) to prepare single-cell RNA-seq samples, according to the manufacturer’s protocol. Single-cell RNA-seq samples were then sequenced by the Illumina MiSeq system (Illumina), and the data were analyzed by an in-house bioinformatics program. Paired TRAV and TRBV sequences were extracted and synthesized as follows.
Full-length TCR construction.
Full-length TCR-encoding sequences were constructed by fusing the sequence encoding the CDR3 region of a putative reactive TCR (provided by sequencing analysis) with the partly missing conserved V- and J-region-encoding sequences, which were obtained from the IMGT online database (http://www.imgt.org). The sequence encoding the resulting human V-(D)-J region was fused to the modified mouse TRAC or TRBC region. The modifications in the mouse TCR constant regions were previously described20,21. Use of the mouse TCR constant regions promoted pairing of the introduced TCR-encoding genes by excluding pairing with the genes encoding the endogenous TCRs and facilitated identification of the transduced T cells by flow cytometry using anti-mouse-TCR-β-chain antibodies. Sequences encoding the full-length TCRs were codon-optimized, synthesized and cloned into the MSGV1 retroviral vector (Genscript). When sequences for the TCR α-chains and β-chains were cloned into the same vector, a furin SGSG P2A linker was added between the two chains.
TCR transduction of peripheral blood T cells.
Transduction of autologous or allogeneic peripheral blood T cells was conducted as has been previously described8. Briefly, autologous or allogeneic apheresis samples were stimulated with 50 ng/ml soluble OKT3 antibody and 300 IU/ml IL-2 for 2 d before retroviral transduction. Supernatants from transient retroviral transductions were generated by co-transfecting the retroviral packaging cell line 293GP with the TCRencoding MSGV1 plasmid(s) (1.5 μg/well) and the envelope-encoding plasmid RD114 (0.75 μg/well). Retroviral supernatants were collected at 24 or 48 h after transfection and used to transduce the activated T cells into retronectin-coated plates (Takara). Transduced T cells were kept in LCM medium for at least 7 d, when the transduction efficiency was examined by flow cytometry using CD3 and mouse TCR β-chain (mTCR) staining.
Minimal epitope prediction.
HLA-class-I-bound epitopes were predicted using the netMHCpan 3.0 algorithm, and candidate epitopes were selected for testing based on the highest percentile ranking.
Identification of HLA restriction.
Identification of HLA restriction by HLA-blocking antibodies.
In some cases, the HLA restriction was determined by blocking antibodies against HLA class I (pan-HLA-class-I, clone: w6/32, eBioscience) or HLA class II (pan-HLA-class-II, clone Tu39, BD Biosciences) molecules or against a specific HLA class II locus, DR (clone L243, BioLegend), DP (clone B7/21, Leinco Technologies), DQ (clone SPVL3, Beckman Coulter) or isotype control IgG2a, κ (BD Biosciences). At the day of coculture, autologous pulsed B cells were pelleted and resuspended (1 × 106 cells/ml) in cytokine-free B cell medium with the appropriate HLA-blocking antibody (50 μg/ml). Following a 3.5-h incubation, B and T cells were cocultured overnight for assessment of antigen-recognition blockade.
Identification of HLA restriction by HLA transfection of COS-7 (class I) or HEK-CIITA (class II) cell lines.
COS-7 cells were transfected with plasmids encoding HLA-class I molecules that were matched to those expressed by patient 4136. COS-7 cells were plated a day before the transfection in concentration 2.5 × 104 or 1 × 105/well. On the day of the transfection, medium was replaced with fresh medium, and the cells were transfected with a HLA-expressing plasmid (100 ng/well) or no plasmid (mock), using Lipofectamine 2000 transfection reagent (ThermoFisher Scientific), as per the manufacturer’s protocol. For the determination of the HLA restriction for mutKIAA0368, some of the COS-7 wells were co-transfected with both the HLA-encoding plasmid and the TMG6-expressing (200 ng/well) plasmid. Six hours after transfection, some wells were pulsed with peptides, and all cells were rested overnight at 37 °C and 5% CO2. The next day, medium was replaced with fresh medium, and transfected COS-7 cells were cocultured with 1 × 104 or 1 × 105/well T cells, in medium without cytokines, for 18–20 h, for assessment of recognition.
Similarly, HEK-CIITA cells (1 × 105/well) were transfected with a plasmid encoding HLA-DRB1*07:01 or no plasmid (mock), using Lipofectamine 2000. HEK-CIITA cells are HEK cells that stably express the CIITA regulator to allow expression of HLA-class II molecules. A group of HLA-transfected HEK-CIITA cells were pulsed with 10 μg/ml peptide overnight, washed and cocultured with 2 × 104 to 1 × 105 T cells, in medium without cytokines, for 16–18 h. In all of the experiments, untransduced cells were also used as a negative control.
Identification of HLA restriction by assessing TCR recognition of allogeneic B cells.
Allogeneic APCs (B cells) that were partially matched or mismatched at HLA-DR alleles with those of patient 4136 (Supplementary Table 5) were pulsed with the mutSLC3A2 or mutCTSB (10 μg/ml) and were cocultured with transduced T cells expressing the specific TCR molecules, as described above.
Immunohistochemical staining.
Surgical specimens were fixed in 10% neutral-buffered formalin, routinely processed, embedded in paraffin tissue blocks and sectioned at 5 μm for routine histology. For immunohistochemistry, 5-μm tissue sections were prepared and placed on poly-ʟ-lysine-coated glass slides. Following antigen retrieval, tissue sections were incubated with the following primary antibodies: anti-CD3 (clone 2GV6, prediluted, 790–4341, Ventana Medical Systems), anti-CK7 (clone SP52, prediluted, 790–4462, Roche), anti-PD-1 (clone NAT105, prediluted, 760–4895, Ventana Medical Systems) and anti-PD-L1 (clone SP142, 1:3 dilution, MA4421, Spring Bioscience). The staining for anti-CD3, anti-CK7 and anti-PD1 was performed on the Roche Ventana Medical Systems BenchMark ULTRA automated IHC platform using the ultraView Universal DAB Detection Kit (Ventana Medical Systems); the staining for anti-PDL-1 was performed on a Leica Autostainer Bond Polymer Refine Detection platform, following standard laboratory protocols established by the histology section of the Laboratory of Pathology at the NIH.
Statistical analysis.
We used maximum-likelihood estimation22 to calculate the expected mutation copy number. Determination of whether the observed VAF was significantly different from the expected VAF at P < 0.01 used a proportion test (prop.test in R). Data are reported as the mean ± s.e.m., as specified, and the values were calculated using GraphPad Prism 7.0. The number of experiments in each case is provided in the figure legends.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability.
Exome and RNA-seq fastq files for sample 4136 can be found on NCBI’s Sequence Read Archive (SRA). The patient 4136 Biosample ID is SAMN07333238; sequencing data can be found under BioProject PRJNA342632 for exome data and PRJNA243084 for RNA-seq data.
Supplementary Material
Acknowledgements
The authors would like to thank A. Mixon and S. Farid of the Surgery Branch FACS Core for assistance with data acquisition and cell sorting, and J. Yang and E. Tran for their valuable discussions. This work was supported by the Center for Cancer Research at the National Cancer Institute (NCI) at the US National Institutes of Health (NIH).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41591–018-0040–8.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reck M et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 375, 1823–1833 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Robert C et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomized dose-comparison cohort of a phase 1 trial. Lancet 384, 1109–1117 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Powles T et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 515, 558–562 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawood S et al. Trends in survival over the past two decades among white and black patients with newly diagnosed stage IV breast cancer. J. Clin. Oncol. 26, 4891–4898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le DT et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamanishi J et al. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J. Clin. Oncol. 33, 4015–4022 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Tran E et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran E et al. T cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran E et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevanović S et al. Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 356, 200–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefebvre C et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 13, e1002201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudley ME et al. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J. Clin. Oncol. 26, 5233–5239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abate-Daga D et al. Expression profiling of TCR-engineered T cells demonstrates overexpression of multiple inhibitory receptors in persisting lymphocytes. Blood 122, 1399–1410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assadipour Y et al. Characterization of an immunogenic mutation in a patient with metastatic triple-negative breast cancer. Clin. Cancer Res. 23, 4347–4353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J et al. Simplified method of the growth of human tumor-infiltrating lymphocytes (TIL) in gas-permeable flasks to numbers needed for patient treatment. J. Immunother. 35, 283–292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasetto A et al. Tumor- and neoantigen-reactive T cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol. S 4, 734–743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA & Morgan RA Enhanced antitumor activity of murine–human hybrid T cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR–CD3 stability. Cancer Res. 66, 8878–8886 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haga-Friedman A, Horovitz-Fried M & Cohen CJ Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J. Immunol. 188, 5538–5546 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Aldrich JRA Fisher and the making of maximum likelihood. 1912–1922. Stat. Sci 12, 162–176 (1997). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Exome and RNA-seq fastq files for sample 4136 can be found on NCBI’s Sequence Read Archive (SRA). The patient 4136 Biosample ID is SAMN07333238; sequencing data can be found under BioProject PRJNA342632 for exome data and PRJNA243084 for RNA-seq data.