Abstract
Dronedarone is used to treat patients with cardiac arrhythmias and has been reported to be associated with liver injury. Our previous mechanistic work demonstrated that DNA damage-induced apoptosis contributes to the cytotoxicity of dronedarone. In this study, we examined further the underlying mechanisms and found that after a 24-h treatment of HepG2 cells, drone-darone caused cytotoxicity, G1-phase cell cycle arrest, suppression of topoisomerase II, and DNA damage in a concentration-dependent manner. We also investigated the role of cytochrome P450s (CYPs)-mediated metabolism in the dronedarone-induced toxicity using our previously established HepG2 cell lines expressing individually 14 human CYPs (1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5, and 3A7). We demonstrated that CYP3A4, 3A5, and 2D6 were the major enzymes that metabolize dronedarone, and that CYP3A7, 2E1, 2C19, 2C18, 1A1, and 2B6 also metabolize dronedarone, but to a lesser extent. Our data showed that the cytotoxicity of dronedarone was decreased in CYP3A4-, 3A5-, or 2D6-overexpressing cells compared to the control HepG2 cells, indicating that the parent dronedarone has higher potency than the metabolites to induce cytotoxicity in these cells. In contrast, cytotoxicity was increased in CYP1A1-overexpressing cells, demonstrating that CYP1A1 exerts an opposite effect in dronedarone’s toxicity, comparing to CYP3A4, 3A5, or 2D6. We also studied the involvement of topoisomerase II in dronedarone-induced toxicity, and demonstrated that the overexpression of topoisomerase II caused an increase in cell viability and a decrease in γ-H2A.X induction, suggesting that suppression of topoisomerase II may be one of the mechanisms involved in dronedarone-induced liver toxicity.
Keywords: Dronedarone, Liver toxicity, DNA damage, Topoisomerase II, CYP3A4, CYP3A5, CYP2D6
Introduction
Dronedarone is a first-line antiarrhythmic drug used to treat patients with non-permanent atrial fibrillation or atrial flutter. From its approval in July 2009–October 2011, approximately 1.3 million dronedarone prescriptions were dispensed and around 278,000 patients received dronedarone prescriptions from outpatient retail pharmacies in the United States (https://www.fda.gov/Drugs/DrugSafety/ucm283933.htm). Dronedarone has been generally considered to be safe; however, dronedarone increased mortality in patients suffering from New York Heart Association classes (NYHA) III and IV heart failure (Kober et al. 2008) or permanent atrial fibrillation (Connolly et al. 2011). Thus, the drone-darone is contraindicated in patients with advanced NYHA class or with permanent atrial fibrillation (De Ferrari and Dusi 2012). In addition, unpredictable idiosyncratic drug-induced liver injury (DILI) occurred in a small portion of patients. In particular, two cases of severe acute liver failure requiring liver transplantation were reported by January 2011 (De Ferrari and Dusi 2012). The adverse events have led the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA) to issue warnings about possible severe hepatotoxicity of dronedarone. Since dronedarone is still in use and reports of hepatotoxicity continue (Rizkallah et al. 2016), evaluating the toxicity of dronedarone and studying its underlying mechanisms are an urgent priority. Thus far, only a few studies has investigated the mechanisms of dronedarone-induced toxicity, with mitochondrial dysfunction (Felser et al. 2013, 2014; Serviddio et al. 2011) and DNA damage-induced apoptosis (Chen et al. 2018) being reported.
It is known that DILI is associated with cytochrome P450s (CYP)-mediated bioactivation (Funk and Roth 2017; Thompson et al. 2016). Using primary human hepatocytes and hepatic microsomes, a study demonstrated that CYP3A4, 3A5, and 2D6 were the three major CYP iso-forms responsible for metabolizing dronedarone to its major metabolite N-desbutyl-dronedarone, with metabolic potency in the order of CYP3A4 ≈ 3A5 > 2D6, as calculated by the oxidation rate (Klieber et al. 2014). Other CYP isoforms (CYP2C8, 2C19, and 1A1) have also been shown to be involved in the metabolism of dronedarone and/or its metabolites. The inactivation of CYP3A4 and 3A5 by dronedarone and N-desbutyl-dronedarone has also been reported based upon in vitro enzyme kinetics experiments (Hong et al. 2016). However, it is currently not known whether metabolism is associated with the cytotoxic effects of dronedarone and which metabolizing enzymes are responsible.
Previously, we have established a panel of 14 HepG2-derived cell lines that stably express individual human CYPs, including 1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5, and 3A7. The characterization of these 14 cell lines and their use in studying metabolism-associated toxicity have been demonstrated (Wu et al. 2016; Xuan et al. 2016). In the current study, we used these cell lines to identify systematically the metabolic activity of each CYP toward dronedarone by evaluating the production of metabolites using mass spectrometry analysis. In addition, the mechanisms underlying dronedarone-induced toxicity were investigated after a 24 h exposure of dronedarone to cells at clinically relevant concentrations. Since we previously demonstrated that a 4-h treatment of dronedarone could induce DNA damage and topoisomerase II suppression in HepG2 cells (Chen et al. 2018), we further studied the direct involvement of topoisomerase II in dronedarone-induced DNA damage by over-expressing the topoisomerase II gene. Furthermore, we extensively examined the roles of human CYP isoforms in dronedarone-induced liver toxicity.
Materials and methods
Chemicals and reagents
Dronedarone, dimethyl sulfoxide (DMSO), Williams’ medium E, and propidium iodide were from Sigma–Aldrich (St. Louis, MO, USA). N-Desbutyl-dronedarone, and dronedarone-d6 were purchased from TLC Pharmaceutical Standards Ltd. (Aurora, Ontario, Canada). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Lawrenceville, GA, USA). Antibiotic–antimycotic was from Life Technologies (Grand Island, NY, USA). SmaI and NheI restriction enzymes were purchased from New England Biolabs (Ipswich, MA, USA). For Western blotting assays, primary antibodies against the CDK2, CDK4, CDK6, cyclin D1, cyclin D3, and γ-H2A.X (Ser139) were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibody for CYP3A5 was obtained from Abcam (Cambridge, MA, USA). Antibodies for topoisomerase II, CYP3A4, CYP2D6, and GAPDH were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell culture and treatment with dronedarone
The HepG2 human hepatoma cell line was from the American Type Culture Collection (ATCC; Manassas, VA, USA). HepG2 cells were cultured in Williams’ medium E complete media containing 10% FBS and 1 × antibiotic antimycotic at 37 °C in a humidified atmosphere with 5% CO2. The passage number did not excess 10. Unless otherwise specified, cells were seeded at a density of 2.5 × 105 cells/ml in a volume of 100 μl per well in 96-well plates, or in a volume of 8 ml in 60 mm tissue culture dishes, or 16 ml in 100 mm tissue culture dishes. Cells were cultured for 24 h prior to treatment with dronedarone or the DMSO vehicle control. The final concentration of DMSO was 0.1%.
Mass spectrometry analyses of dronedarone and N‑desbutyl‑dronedarone
After 24-h exposure to 6 μM dronedarone, cells and super-natant from each individual cell line were harvested and collected. The cell lysates were obtained by adding 100 μl Nanopure water, followed by three cycles of freeze and thaw. The concentrations of the cell lysates were determined using a Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, CA, USA). The supernatants were diluted using four volumes of acetonitrile containing 100 ng/ml of the internal standard dronedarone-d6, vortexed and then sonicated for 10 min. Samples were centrifuged at 12,000 rpm for 5 min and the supernatant was loaded into sample vials for UPLC–MS analysis. Two μl of the supernatant were injected onto a Waters ACQUITY UPLC System coupled with a Waters ACQUITY QDa Mass Detector. Drone-darone, N-desbutyl-dronedarone, and the dronedaroned6 were eluted on an ACQUITY UPLC HSS T3 column (2.1 mm × 50 mm, 1.8 μm) at 40 °C with mobile phases of LC–MS grade water (A) and acetonitrile (B), both containing 0.1% formic acid, at a flow rate of 0.5 ml/min. Elution started with 10% solvent B followed by a linear gradient of 10–90% solvent B in 0.8 min, keeping solvent B at 90% for 0.6 min, returning to 10% B in 0.1 min, and maintained for 1.0 min to re-equilibrate the column. The eluate was detected by mass spectrometry with an electrospray ion source operating in the positive ion mode (ESI+) using single ion recording (SIR). The monitored (M+H)+ ions were m/z 557.26 for dronedarone, m/z 563.29 for dronedarone-d6, and m/z 501.23 for N-desbutyl-dronedarone. Dronedarone and N-desbutyl-dronedarone were quantified using calibration curves ranging from 1.6 to 1000 ng/ml using Waters Empower 3 software. The results were expressed as ng analyte/mg protein or ng analyte/ml media.
MTS cell viability assay
Cell viability was determined using a CellTiter 96® AQueous One Solution Reagent (MTS, Promega Corporation, Madison, WI, USA) as previously described (Ren et al. 2016).
Lactate dehydrogenase assay
The cytotoxicity of dronedarone was assessed using a lac-tate dehydrogenase (LDH) assay as described previously (Li et al. 2012).
Cell cycle analysis
HepG2 cells were seeded in 6-well plates at a density of 1 × 106 cells/well and cultured for 24 h prior to treatment with dronedarone or the DMSO vehicle control. Cell cycle phase distribution was analyzed by flow cytometry. Briefly, treated cells were trypsinized and fixed on ice in 1 ml 70% cold ethanol for 1 h. After washing with cold PBS, cells were incubated in PBS containing 0.2 μg/μl RNase A (Qiagen, Valencia, CA, USA) at 37 °C for 1 h. Propidium iodide (PI; Sigma-Aldrich) was added to the cells at a final concentration of 10 μg/ml and then the cells were stained overnight at 4 °C. The DNA content was measured the following day by FACScanto II flow cytometry (BD Biosciences, San Jose, CA, USA). The cell cycle data were analyzed using FlowJo® software (FlowJo, LLC, Ashland, OR, USA).
Western blot analysis
Cells were plated in 60 or 100 mm tissue culture dishes and treated with dronedarone. After treating for specified times and concentrations, whole-cell lysates were prepared using RIPA buffer containing Halt Protease Inhibitor Cocktail (ThermoFisher Scientific, Waltham, MA, USA). The concentrations of the protein samples were determined using a Bio-Rad Protein Assay (Bio-Rad Laboratories). Standard Western blots were performed. Depending on the proteins of interest, antibodies were selected against CDK-2, CDK-4, CDK-6, cyclinD1, cyclinD3, γ-H2A.X (Ser139), topoisomerase II, CYP3A4, CYP3A5, and CYP2D6 followed by an incubation with secondary antibody conjugated with horseradish peroxidase (HRP) (Santa Cruz Biotechnology). GAPDH was used as the internal control. The protein signals were determined and quantified with a FluroChem E System (ProteinSimple, San Jose, CA, USA).
RNA isolation and real‑time PCR assay
Total RNA was isolated using the RNeasy system (Qiagen, Germantown, MD, USA). The purity and quality of RNA were examined as described previously (Chen et al. 2014). cDNAs were generated by reverse transcription of 2 μg of total RNA using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Quantitative real-time PCR for topoisomerase II was performed as described previously (Chen et al. 2013) to evaluate relative gene expression. Data normalization and analysis were conducted as described previously (Guo et al. 2009).
Transient overexpression of topoisomerase IIα in HepG2 cells
The cDNA of human topoisomerase IIα was amplified by PCR and cloned into SmaI and NheI restriction sites of the lentiviral expression vector pLv-EF1α-MCS-IRES-puro. The generated lentiviral vector or empty vector and lentiviral packaging mix (pMDL-G, pRSV-REV, and pVSV-G) were co-transfected into 293T cells to generate lentiviruses. The titrations of the generated lentiviruses were measured by antibiotics selection according to the manufacturer’s instruction (Biosettia). HepG2 cells were infected with the lentiviruses at a multiplicity of infection of 10. The overexpression level of topoisomerase IIα was determined by real-time PCR at 48 h post-infection, and then the topoisomerase IIα-overexpressing cells and empty vector-transduced cells were exposed to dronedarone for another 24 h.
Statistical analyses
Data are presented as the mean ± standard deviation (SD) of at least three independent experiments. Analyses were performed using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). Statistical significance was determined by one-way analysis of variance (ANOVA) followed by the Dunnett’s tests for pairwise-comparisons or two-way ANOVA followed by the Bonferroni post-test. The difference was considered statistically significant when p was less than 0.05.
Results
Metabolism of dronedarone in 14 individual CYP‑overexpressing HepG2 cells
To explore the metabolism of dronedarone, an analytical method utilizing UPLC mass spectrometry was established to determine the amount of parent dronedarone, and its major metabolite N-desbutyl-dronedarone, after dronedarone treatment. The mass spectrometry analysis was performed based on the SIR monitoring of dronedarone (m/z 557.26), N-desbutyl-dronedarone (m/z 501.23), and the internal standard dronedarone-d6 (m/z 563.29). The retention times of dronedarone and dronedarone-d6 were 0.95 min while N-desbutyl-dronedarone eluted at 0.87 min. The linear quantification ranged from 1.6 to 1000 ng/ml, with excellent linearity of R2 = 0.997 for both dronedarone and N-desbutyl-dronedarone.
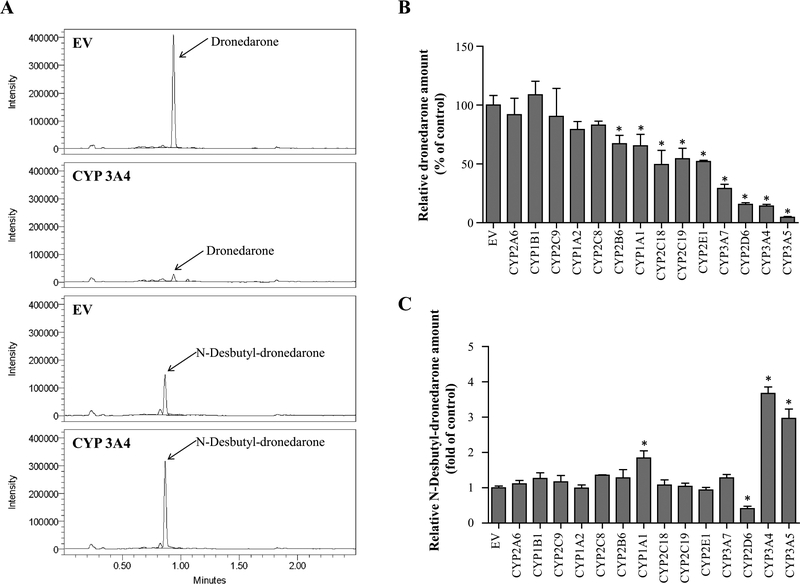
A battery of CYP-overexpressing HepG2 cell lines (Xuan et al. 2016) was used to investigate which human CYP iso-forms are the major enzymes contributing to the metabolism of dronedarone. Fourteen cell lines, each of which overex-presses a different human CYP isoform, including CYP1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, 2E1, 3A4, 3A5, and 3A7, were examined independently. The overexpression of each CYP was previously evaluated and verified by real-time PCR and Western blots (Xuan et al. 2016). Figure 1a shows representative HPLC–MS chromatograms of the dronedarone and N-desbutyl-dronedarone detected in CYP3A4-overexpressing HepG2 cells and empty vector (EV) transduced control cells after incubation with 6 μM dronedarone for 24 h. Significantly decreased levels of the parent dronedarone were observed in cell lysates and their corresponding supernatant for nine CYP-overexpression HepG2 cell lines (CYP3A4, 3A5, 2D6, 3A7, 2E1, 2C19, 2C18, 1A1, and 2B6), indicating these CYPs were capable of metabolizing dronedarone (Fig. 1b). CYP2D6, 3A4, and 3A5 were the three major enzymes responsible for dronedarone’s metabolism under our experimental conditions in CYP-overexpressing cell lines. Compared with EV control, increased levels of N-desbutyl-dronedarone were found in CYP1A1-, 3A4-, and 3A5-overexpressing cells, while decreased levels were observed in CYP2D6-overex-pressing cell lysates and their corresponding supernatants (Fig. 1c).
Fig. 1.
Metabolism of dronedarone in individual CYP-overexpressing HepG2 cells. UPLC-QDa mass spectrometry chromatograms for dronedarone and its metabolite N-desbutyl-dronedarone in extract from HepG2-empty vector control (EV) and HepG2-CYP3A4 over-expressing cells treated with 6 μM dronedarone for 24 h. b, c Fourteen CYP-overexpressing HepG2 cell lines were exposed to 6 μM dronedarone for 24 h. The total amount of dronedarone (b) and its metabolite N-desbutyl-dronedarone (c) in cell lysate and culture media were quantified with LC–MS. The results shown are relative values normalized to EV control. Data represent mean ± SD from three independent experiments. *p < 0.05 compared with EV control
The roles of CYP‑mediated metabolism in dronedarone‑induced toxicity
Our previous study investigated dronedarone-induced toxicity after 6 h treatment and demonstrated that there was significant cytotoxicity when cells were treated with drone-darone at 10 μM for 6 h (Chen et al. 2018). It has been reported that dronedarone can be detected within 24 h incubation in metabolically competent human primary hepatocytes (Klieber et al. 2014). Therefore, in the current study, the cytotoxicity of dronedarone was initially investigated at various concentrations below 10 μM for a longer time (24 h). Human plasma concentration of dronedarone has been reported to reach 0.28 μM in 7 days following oral administration of 400 mg twice daily (Dorian 2010). However, under certain circumstances, the plasma concentration can be significantly higher. For instance, the plasma concentration of dronedarone was reported to be increased by 25-fold when ketoconazole, a potent CYP3A4 inhibitor, was co-administered (Patel et al. 2009). Thus, the concentrations used in the current investigation (≤ 10 μM) are in the range of reported plasma concentrations and are clinically relevant.
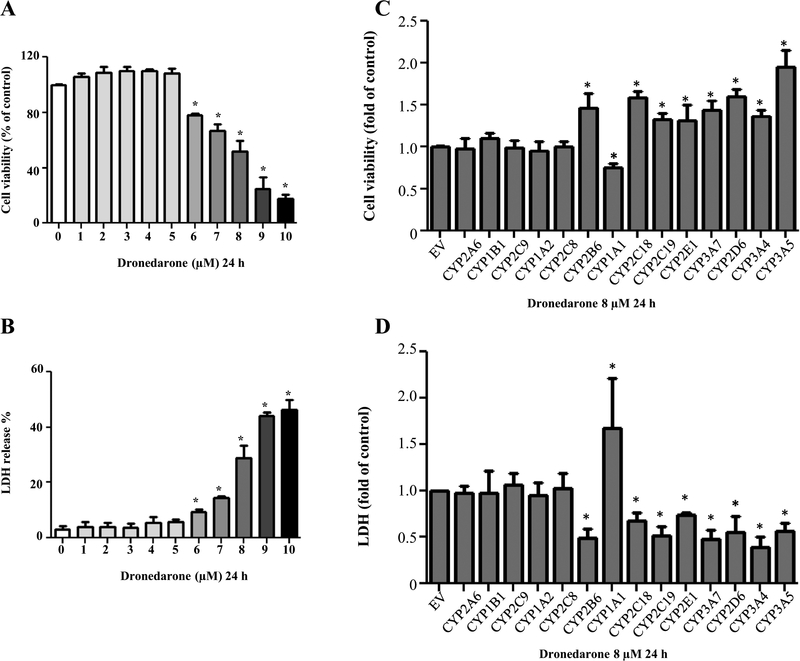
Dronedarone treatment of HepG2 cells above 6 μM showed a concentration-dependent decrease in cell viability using MTS assays (Fig. 2a). At 6 μM, dronedarone decreased the cell viability to about 78% of that of the DMSO control. Moreover, cell viability was decreased to about 17% in the cells treated with 10 μM of dronedarone, indicating significant cellular growth inhibition and injury. The severity of cell death caused by dronedarone was assessed by LDH release. The elevated LDH release was first shown at 6 μM dronedarone treatment, and the effect was more profound at higher concentrations. A 46% release of LDH occurred at 10 μM dronedarone treatment, implicating significant cell death and cytotoxicity at this high concentration of dronedarone treatment (Fig. 2b). Based on the results described above, further toxicity and mechanistic studies were conducted at the time point of 24 h and the concentrations between 6 and 10 μM.
Fig. 2.
Dronedarone induces cellular damage in HepG2 cells and HepG2 cells overexpressing various CYP isoforms. HepG2 cells were exposed to dronedarone at 1–10 μM for 24 h, with DMSO as the vehicle control and cytotoxicity was measured using MTS assay (a) and LDH assay (b). c Empty vector (EV)-transduced HepG2 cells or HepG2 overexpressing human CYP cells were treated with 8 μM dronedarone for 24 h and cytotoxicity was determined using MTS assay (c) and LDH assay (d). The results shown are mean ± SD from three independent experiments. *p < 0.05 compared with t DMSO control or EV control
The effect of CYP-mediated metabolism on cytotoxicity of dronedarone was investigated by comparing the cytotoxicity of dronedarone at a concentration of 8 μM in empty vector control cells and 14 CYP-overexpressing cells. The cytotoxicity was assessed using two toxicity detection methods: MTS assays and LDH release assays. The cytotoxicity was altered in all nine CYP-overexpressing cell lines that were found capable of metabolizing dronedarone (Fig. 1). As shown in Fig. 2c, d, the metabolism catalyzed by CYP3A5, 3A4, 2D6, 3A7, 2E1, 2C19, 2C18, and 2B6 significantly attenuated dronedarone-induced cytotoxicity; whereas the metabolism catalyzed by CYP1A1 exacerbated dronedarone-caused cell death, implying that the metabolism is associated with the cytotoxicity of dronedarone, either enhancing or decreasing cytotoxicity of dronedarone.
CYP3A4‑, 3A5‑, and 2D6‑mediated metabolism prevents dronedarone‑induced cell cycle disturbance
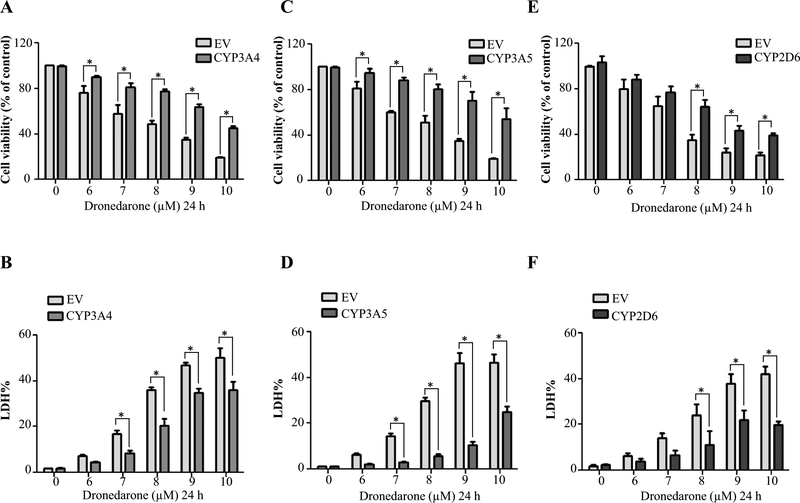
CYP3A4, 3A5, and 2D6 are the enzymes reported to be involved in dronedarone metabolism (Klieber et al. 2014); interestingly, they also showed prominent detoxification effects in our study (Fig. 2c, d). Thus, we explored the mechanisms underlying the detoxification of dronedarone by these particular CYPs. Significant detoxification of dronedarone was confirmed in CYP3A4, 3A5, and 2D6-overexpressing cells when measured with MTS and LDH assays (Fig. 3). A concentration-dependent decrease in cytotoxicity was observed in CYP-overexpressing cells.
Fig. 3.
Effect of metabolism mediated by CYP3A4, 3A5, and 2D6 on dronedarone-induced cytotoxicity. Empty vector (EV)-transduced or CYP3A4, 3A5, and 2D6-overexpressing HepG2 cells were treated with the indicated concentrations of dronedarone for 24 h. Cytotoxicity was measured using MTS assay (a, c, e) and LDH assay (b, d, f). The results shown are mean ± SD from three independent experiments. *p < 0.05 compared with EV control
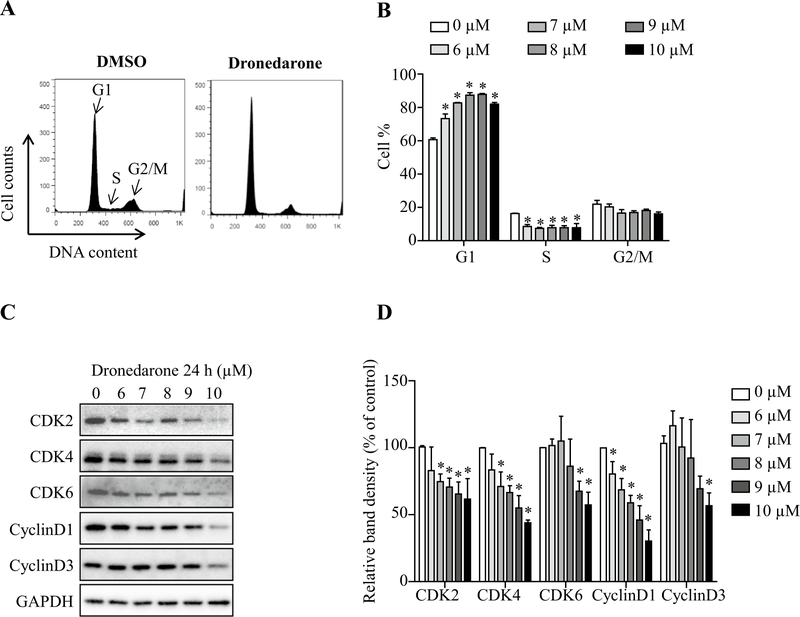
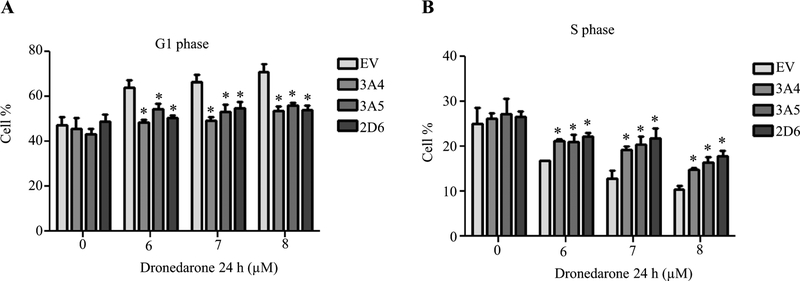
Cell cycle analysis was then used to investigate the possible mechanisms of cellular growth inhibition caused by dronedarone after 24 h exposure. Cell cycle arrest in G1 phase and a concurrent reduction of cells in S phase were found with increased concentrations of dronedarone, while the proportion of cells in G2/M phase remained approximately the same (Fig. 4a, b). Cyclin-dependent kinase (CDK) 2-cyclin D and CDK4/6-cyclin D1/D3 complexes are among the most important proteins involved in controlling cell cycle G1 to S phase progression. As shown in Fig. 4c, d, treatment with dronedarone markedly decreased CDK2, CDK4, CDK6, cyclin D1, and cyclin D3 levels, suggesting dronedarone disturbed cell cycle checkpoint controls and inhibited cellular DNA synthesis. After metabolizing by CYP3A4, 3A5, and 2D6, cell cycle arrest in G1 phase and S phase reduction were both significantly reversed (Fig. 5a, b), indicating that cell cycle perturbation is detoxication-sensitive and the cell cycle perturbation is more likely due to the parental form of dronedarone, rather than its metabolites.
Fig. 4.
Effect of metabolism mediated by CYP3A4, 3A5, and 2D6 on dronedarone-induced cell cycle perturbation. a Flow cytometric analysis for cell cycle distribution. Histograms shown were DNA content analyses for HepG2 cells treated with the indicated concentrations of dronedarone for 24 h. b bar graph depicts the mean percentage of each cell cycle phase ± SD from three independent experiments. *p < 0.05 compared with DMSO control. c, d Expression of cell cycle regulators were determined by Western blotting. GAPDH was used as a loading control. Similar results were obtained from three independent experiments. d Intensities of bands were normalized to the amount of GAPDH. *p < 0.05 versus treatment of DMSO vehicle control
Fig. 5.
Empty vector (EV)-transduced or CYP3A4, 3A5, and 2D6-overexpressing HepG2 cells were treated with the indicated concentrations of dronedarone for 24 h. a, b Cell cycle distribution was measured by flow cytometer. Bar graph represents the mean percentage of cell cycle G1 phase (a) and S phase (b) ± SD from three independent experiments. *p < 0.05 compared with EV control
Dronedarone causes DNA damage via topoisomerase IIα suppression
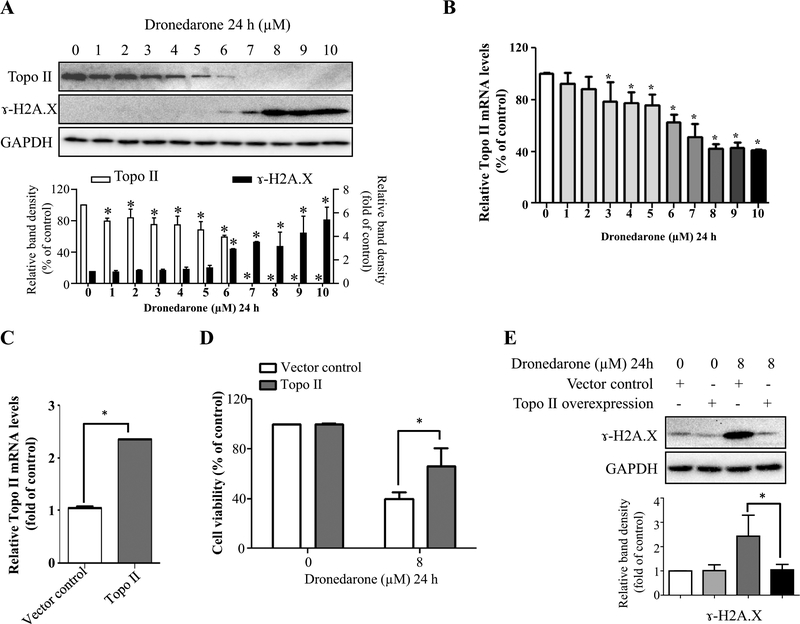
Generally, cell cycle G1 arrest appears in response to DNA damage or incomplete replication. Our previous study revealed that with high concentrations and a 4-h exposure, dronedarone caused severe DNA damage and topoisomerase IIα suppression. However, whether the topoisomerase IIα suppression is an upstream regulator of DNA damage or is associated with dronedarone-induced toxicity has not been investigated. Therefore, in this study, we determined the role of DNA damage and topoisomerase IIα in the cells treated with dronedarone for 24 h at concentrations from 1 to 10 μM. As shown in Fig. 6a, starting at 6 μM, the treatment of HepG2 cells with dronedarone initiated DNA damage, as demonstrated by a concentration-dependent increase of histone H2A.X phosphorylation at serine 139 (γ-H2A.X), a hallmark of double-strand DNA breakage in cells (Rogakou et al. 1998). In contrast to the significant induction of γ-H2A.X, the expression of topoisomerase IIα decreased in a concentration-dependent manner. Moreover, starting from 7 μM, topoisomerase IIα protein became undetectable, indicating that the dronedarone treatment caused a complete depletion of topoisomerase IIα (Fig. 6a).
Fig. 6.
Suppressed topoisomerase II levels contribute to dronedarone-induced DNA damage in HepG2 cells. a Total cellular proteins were extracted after dronedarone treatment at indicated concentrations for 24 h. The levels of topoisomerase II and γ-H2A.X were measured by Western blotting. GAPDH was used as a loading control. Similar results were obtained from three independent experiments. Intensities of bands were normalized to the amount of GAPDH. *p < 0.05 versus treatment of DMSO vehicle control. b Total RNA were extracted after dronedarone treatment at indicated concentrations for 24 h. c HepG2 cells were infected with lentivirus carrying topoisomerase II. The gene expression level of topoisomerase II was measured by real-time PCR. Human GAPDH was used as an internal control to normalize the amount of cDNA template. The results shown are mean ± SD from three independent experiments. *p < 0.05 compared with DMSO control (b) or vector control (c). d, e Empty vector or topoisomerase II transduced HepG2 cells were treated with 8 μM dronedarone for 24 h. d Cytotoxicity was measured using MTS assay. *p < 0.05 compared with vector control. e The expression level of γ-H2A.X was detected by Western blotting. GAPDH was used as a loading control. Similar results were obtained from three independent experiments. Intensities of bands were normalized to the amount of GAPDH. *p < 0.05 compared with vector control
We also examined the gene expression of topoisomerase IIα by real-time PCR. Starting from as low as 3 μM, topoisomerase IIα mRNA decreased significantly in a concentration-dependent manner. It is worth noting that the decrease of topoisomerase IIα occurred at both transcriptional and translational levels; however, the decrease level of topoisomerase IIα protein was more severe than changes in its mRNA transcript (Fig. 6a, b). Although the mechanism needs to be studied, we speculate that dronedarone modulates topoisomerase IIα at the post-transcriptional level more profoundly than the transcriptional level.
To investigate further the role of topoisomerase IIα in dronedarone-induced cytotoxicity, we transiently overex-pressed topoisomerase IIα in HepG2 cells using a lentivirus system. Real-time PCR showed a 2.4-fold overexpression of topoisomerase IIα (Fig. 6c) in comparison to the vector control. Dronedarone-induced cytotoxicity was markedly reduced by the increased expression of topoisomerase IIα compared with the vector control (Fig. 6d). Furthermore, we explored the effect of topoisomerase IIα on dronedarone-induced DNA damage. As shown in Fig. 6e, the overexpression of topoisomerase IIα reduced the extent of DNA damage caused by 8 μM dronedarone, as indicated by the decreased induction of γ-H2A.X in comparison with the same treatment of vector control cells. These results show that both dronedarone-induced DNA damage and cytotoxicity are ameliorated by the overexpression of topoisomerase IIα.
CYP3A4‑, 3A5‑, and 2D6‑mediated metabolism attenuates dronedarone‑induced DNA damage and topoisomerase IIα‑suppression
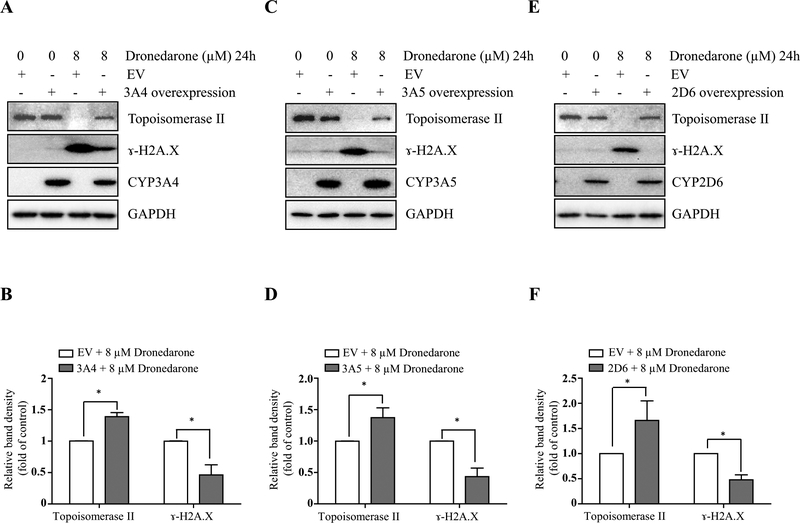
We then investigated the contributions of the metabolism in dronedarone-induced DNA damage and topoisomerase IIα suppression. DNA damage and topoisomerase IIα suppression were remarkably attenuated in the cells over-expressing CYP3A4, 3A5, or 2D6. As shown in Fig. 7, a decreased induction of γ-H2A.X and an increased expression of topoisomerase IIα in CYP3A4-, 3A5-, and 2D6-overexpressing cells were observed when compared with empty vector control cells under the same treatment of 8 μM for 24 h. The overexpression of CYP3A4, 3A5, and 2D6 was confirmed by Western blotting (Fig. 7).
Fig. 7.
CYP3A4-, CYP3A5-, and CYP2D6-mediated metabolism attenuates dronedarone-induced DNA damage and topoisomerase IIα suppression. Empty vector (EV)-transduced or CYP3A4-, 3A5-, and 2D6-overexpressing HepG2 cells were treated with 8 μM dronedarone for 24 h. Total cellular proteins were extracted after dronedarone treatment. The expression levels of γ-H2A.X, topoisomerase II, CYP3A4 (a, b), CYP 3A5 (c, d) and CYP 2D6 (e, f) were determined by Western blotting. GAPDH was used as a loading control. Similar results were obtained from three independent experiments. Intensities of bands were normalized to the amount of GAPDH (b, d, f). *p < 0.05 compared with EV control
Taken together, these data suggest that the toxicity of dronedarone is mainly caused by the parent compound.
Discussion
Previously, we studied the toxic effects of dronedarone in HepG2 cells, a cell line that has low metabolic activity, and also in hepatic progenitor cell line HepaRG, a terminally differentiated human primary hepatic cell line that expresses key metabolic enzymes (Guillouzo et al. 2007; Guo et al. 2011). Significant cytotoxicity was observed in both cell lines, but HepG2 cells displayed a higher sensitivity to dronedarone than the metabolically competent HepaRG cells (Chen et al. 2018). These results stimulated the current investigation to determine which drug-metabolizing enzymes are most important in the toxicity or detoxification of dronedarone, i.e., whether they enhance or reduce its toxicity. Among 14 CYP isoforms examined using a panel of HepG2 derivatives that individually overexpress a single CYP, 9 CYP isoforms (CYP3A4, 3A5, 2D6, 3A7, 2E1, 2C19, 2C18, 1A1, and 2B6) showed the capability to metabolize dronedarone (Fig. 1). CYP-catalyzed metabolism impacted the toxicity of dronedarone; in particular, metabolism by CYP1A1 enhanced the cytotoxicity, whereas the other 8 CYPs attenuated the cytotoxicity induced by drone-darone (Fig. 2).
In our study, three CYPs (2D6, 3A4, and 3A5) were identified to be the major enzymes responsible for dronedarone metabolism. These results are in a good agreement with a study that analyzed the metabolic pathways of dronedarone using recombinant human enzymes, human liver micro-somes, and primary human hepatocytes (Klieber et al. 2014). CYP2D6, 3A4, and 3A5 were the three main CYP isoforms shown to metabolize dronedarone to N-desbutyldronedarone by 94, 90, and 50%. CYP2D6 and CYP3A4 were able to further oxidize N-desbutyl-dronedarone to additional metabolites by 55 and 21% (Klieber et al. 2014). In agreement with these findings, our current study showed a significant increase in N-desbutyl-dronedarone in CYP3A4-and CYP3A5-overexpressing cells. However, we observed a decreased production of N-desbutyl-dronedarone in CYP2D6-overexpression cells (Fig. 1c). This decrease in N-desbutyl-dronedarone may be due to its further metabolism (Klieber et al. 2014) in CYP2D6-overexpressing cells.
Dronedarone was developed as an alternative drug to amiodarone, which causes a rare but severe liver injury when used to treat cardiac arrhythmias patients. Although their chemical structures are quite similar, dronedarone is a substitute which lacks the 3,5-diiodophenyl moiety of amiodarone. These structural differences decrease dronedarone’s lipophilicity and result in reduced liver toxicity. A previous study demonstrated that the metabolism of amiodarone by CYP 3A4 significantly increased the cytotoxicity of amiodarone, and that its main metabolite, desethylamiodarone, was the major contributor of this cytotoxicity (Wu et al. 2016). CYP3A4 constitutes approximately 28% of the total CYP enzymes present in human liver and plays a critical role in drug metabolism (Burkina et al. 2017). Although dronedarone and amiodarone have similar structures and both are metabolized by CYP3A4, our results indicate that the impact of CYP3A4-mediated metabolism is distinctly different between the two drugs. The toxicity of dronedarone was decreased, while the toxicity of amiodarone was aggravated by CYP3A4 metabolism.
Our previous study on dronedarone-induced cytotoxicity revealed that the suppression of topoisomerase IIα might be a potential mechanism for DNA damage (Chen et al. 2018). In this study, we further studied the role of topoisomerase IIα in the toxicity of dronedarone by overex-pressing topoisomerase IIα in HepG2 cells. We attempted to establish a HepG2-derived cell line that expresses topoisomerase IIα stably; however, the increased topoisomerase IIα expression led to cell death and the topoisomerase IIα expression declined and was eventually lost after a certain culture period. Thus, transient transduction of topoisomerase IIα was used in this study. Our data demonstrated that topoisomerase IIα is involved in the toxicity of dronedarone because the overexpression of topoisomerase IIα partially but significantly attenuated DNA damage and increased cell viability (Fig. 6d, e). In CYP3A4-, 3A5-, and 2D6-overex-pressing cells, the inhibitory effects on topoisomerase IIα were attenuated and the DNA damage and cytotoxicity were also attenuated (Fig. 7), suggesting the parent dronedarone is more likely than its metabolites to interfere with topoisomerase IIα expression. It has been reported that some drugs and naturally occurring compounds inhibit topoisomerases II, leading to DNA damage and liver toxicity (Chen et al. 2013; Poulsen et al. 2014; Zhang et al. 2015). In agreement with these studies, our current study highlights the role of topoisomerase IIα in drug-induced liver toxicity.
In this study, we found that overexpression of CYP1A1 aggravated the cytotoxicity of dronedarone. CYP1A1 can be induced by environmental compounds, pharmaceuticals, and herbal dietary supplements (Brown et al. 2017; Denison and Nagy 2003; Guo et al. 2010; Hu et al. 2007; Li et al. 2011). CYP1A1 is known to be responsible for the metabolism of pro-carcinogens such as polycyclic aromatic hydrocarbons (Moorthy et al. 2015; Shimada 2006). The toxicological and clinical importance of CYP1A1 in drug metabolism warrants further investigation (Howard et al. 2017; Lin et al. 2017; Ma and Lu 2007) and our results suggest that precaution should be taken in patients with elevated CYP1A1 activity.
A key finding of our study is that CYP isoforms (CYP3A4, 3A5, and 2D6) were identified to be involved in the metabolism of dronedarone and that metabolism by these CYPs counteracted the cytotoxicity of dronedarone (Fig. 7). Our results suggest that increased hepatic exposure to the parent form of dronedarone may occur in individuals with low levels of these enzymes, and as a consequence, may increase their susceptibility to dronedarone-induced liver toxicity. Many factors impact the expression and activity of CYPs, and inter-individual variability in the expression of these enzymes among humans has been well documented (Yang et al. 2010, 2013; Zhou et al. 2009). A substantial variation in the expression of these CYP isoforms among 427 human liver samples was reported with 641-fold differences in CYP3A4, 112-fold differences in CYP3A5, and 49-fold differences in CYP2D6 (Yang et al. 2013). Multiple factors, including genetic (Yang et al. 2010; Zhou et al. 2009), epigenetic (Koturbash et al. 2015), environmental factors (Rendic and Guengerich 2010), and disease/health status of the individuals (Rendic and Guengerich 2010), attribute to the inter-individual variability. As to genetic polymorphisms, CYP3A4*20 (Westlind-Johnsson et al. 2006) and *26 (Werk et al. 2014), CYP3A5*3B and *3C (Hustert et al. 2001), CYP3A5*8 and *9 (Lee et al. 2003), CYP2D6*3A, *3B, and CYP2D6*4B (Kagimoto et al. 1990) were reported to produce non-functional or dramatically decreased enzyme activities compared to their wild-type enzymes. For patients carrying these genetic variants, special precautions should be taken when dronedarone is prescribed, since impaired CYP3A4, 3A5 or 2D6 may increase the risk of dronedarone-induced toxicity.
Likewise, epigenetic factors can modulate the expression or activity of these CYP isoforms. CYP3A4 is down-regulated by microRNA hsa-miR-224–5p (Yu et al. 2018), and CYP2D6 is suppressed by microRNA hsa-miR-370–3p (Zeng et al. 2017). Therefore, drugs or other factors that increase the expression of hsa-miR-224–5p and/or hsa-miR-370–3p, in turn, downregulate CYP3A4 or 2D6, may contribute to an adverse reaction to dronedarone.
Some physiological factors influence the expression and activity of CYPs. For instance, men have a lower CYP3A4 activity; while women have lower activity of other CYPs including CYP2E1 and 1A2 (Cotreau et al. 2005; Scandlyn et al. 2008; Waxman and Holloway 2009). Autoimmune hepatitis, chronic renal failure, and diabetes have been reported to affect the metabolism of drugs by inhibiting key enzymes in the liver, such as CYP2D6, 3A4/5, and 1A2 (He et al. 2015). It is particularly important to note that CYP3A4, 3A5 and 2D6 can be inhibited by a number of drugs, foods, and dietary supplements. For instance, quinidine (an antiar-rhythmic agent) is an inhibitor of CYP2D6 and ketoconazole (an antifungal agent) is an inhibitor of CYP3A4 (https://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm093664.htm#table1–2). Goldenseal inhibited the activity of CYP2D6 and 3A4 and black cohosh inhibited CYP2D6 in healthy subjects (Gurley et al. 2005, 2008). Grapefruit juice can produce irreversible inhibition of CYP3A4 activity (Bailey et al. 2003; Harris et al. 2003). It should be emphasized that the inhibition of these CYP isoforms is clinically significant, since inhibition-mediated drug/food–drug interactions may contribute to increased intracellular dronedarone levels within liver cells, which consequently increases the likelihood of dronedarone-induced hepatotoxicity.
In summary, our study suggests that CYP-mediated metabolism is associated with dronedarone-induced toxicity. CYP3A4, 3A5, and 2D6 were found to be the main metabolizing enzymes and overexpression of these CYPs decreased the cytotoxicity of dronedarone. On the other hand, overexpression of CYP1A1 increased the cytotoxicity of drone-darone. Our study further revealed that the mechanism for dronedarone-induced DNA damage and cell death involves topoisomerase IIα, and these processes are also metabolism-dependent. It is important to note that this work was conducted in hepatic cells that overexpress CYP450s, and that this may not necessarily reflect the situation for human populations. Nonetheless, individuals with low CYP3A4, 3A5, or 2D6 activity may have higher risk of dronedarone-induced liver toxicity.
Acknowledgements
This work was supported by U.S. FDA’s intramural grant program.
Footnotes
Conflict of interest The authors declare that no conflict of interest.
Publisher's Disclaimer: Disclaimer This article is not an official guidance or policy statement of the U.S. FDA. No official support or endorsement by the U.S. FDA is intended or should be inferred.
References
- Bailey DG, Dresser GK, Bend JR (2003) Bergamottin, lime juice, and red wine as inhibitors of cytochrome P450 3A4 activity: comparison with grapefruit juice. Clin Pharmacol Ther 73:529–537 [DOI] [PubMed] [Google Scholar]
- Brown MR, Garside H, Thompson E et al. (2017) From the cover: development and application of a dual rat and human AHR activation. Assay Toxicol Sci 160:408–419 [DOI] [PubMed] [Google Scholar]
- Burkina V, Rasmussen MK, Pilipenko N, Zamaratskaia G (2017) Comparison of xenobiotic-metabolising human, porcine, rodent, and piscine cytochrome P450. Toxicology 375:10–27 [DOI] [PubMed] [Google Scholar]
- Chen S, Wan L, Couch L et al. (2013) Mechanism study of goldenseal-associated DNA damage. Toxicol Lett 221:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Xuan J, Couch L et al. (2014) Sertraline induces endoplasmic reticulum stress in hepatic cells. Toxicology 322C:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ren Z, Yu D, Ning B, Guo L (2018) DNA damage-induced apoptosis and mitogen-activated protein kinase pathway contribute to the toxicity of dronedarone in hepatic cells. Environ Mol Mutagen. 10.1002/em.22173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SJ, Camm AJ, Halperin JL et al. (2011) Dronedarone in high-risk permanent atrial fibrillation. N Engl J Med 365:2268–2276 [DOI] [PubMed] [Google Scholar]
- Cotreau MM, von Moltke LL, Greenblatt DJ (2005) The influence of age and sex on the clearance of cytochrome P450 3A substrates. Clin Pharmacokinet 44:33–60 [DOI] [PubMed] [Google Scholar]
- De Ferrari GM, Dusi V (2012) Drug safety evaluation of dronedarone in atrial fibrillation. Expert Opin Drug Saf 11:1023–1045 [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43:309–334 [DOI] [PubMed] [Google Scholar]
- Dorian P (2010) Clinical pharmacology of dronedarone: implications for the therapy of atrial fibrillation. J Cardiovasc Pharmacol Ther 15:15S–8S [DOI] [PubMed] [Google Scholar]
- Felser A, Blum K, Lindinger PW, Bouitbir J, Krahenbuhl S (2013) Mechanisms of hepatocellular toxicity associated with dronedarone—a comparison to amiodarone. Toxicol Sci 131:480–490. (2012/11/09 edn) [DOI] [PubMed] [Google Scholar]
- Felser A, Stoller A, Morand R et al. (2014) Hepatic toxicity of drone-darone in mice: role of mitochondrial beta-oxidation Toxicology. Vol 323, pp 1–9 [DOI] [PubMed] [Google Scholar]
- Funk C, Roth A (2017) Current limitations and future opportunities for prediction of DILI from in vitro. Arch Toxicol 91:131–142 [DOI] [PubMed] [Google Scholar]
- Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C (2007) The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact 168:66–73 [DOI] [PubMed] [Google Scholar]
- Guo L, Li Q, Xia Q, Dial S, Chan PC, Fu P (2009) Analysis of gene expression changes of drug metabolizing enzymes in the livers of F344 rats following oral treatment with kava extract. Food Chem Toxicol 47:433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Mei N, Xia Q, Chen T, Chan PC, Fu PP (2010) Gene expression profiling as an initial approach for mechanistic studies of toxicity and tumorigenicity of herbal plants and herbal dietary supplements. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 28:60–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Dial S, Shi L et al. (2011) Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos 39:528–538. (2010/12/15 edn) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley BJ, Gardner SF, Hubbard MA et al. (2005) In vivo effects of goldenseal, kava kava, black cohosh, and valerian on human cytochrome P450 1A2, 2D6, 2E1, and 3A4/5 phenotypes. Clin Pharmacol Ther 77:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley BJ, Swain A, Hubbard MA et al. (2008) Supplementation with goldenseal (Hydrastis canadensis), but not kava kava (Piper methysticum), inhibits human CYP3A activity in vivo. Clin Pharmacol Ther 83:61–69 [DOI] [PubMed] [Google Scholar]
- Harris RZ, Jang GR, Tsunoda S (2003) Dietary effects on drug metabolism and transport. Clin Pharmacokinet 42:1071–1088 [DOI] [PubMed] [Google Scholar]
- He ZX, Chen XW, Zhou ZW, Zhou SF (2015) Impact of physiological, pathological and environmental factors on the expression and activity of human cytochrome P450 2D6 and implications in precision medicine. Drug Metab Rev 47:470–519 [DOI] [PubMed] [Google Scholar]
- Hong Y, Chia YM, Yeo RH et al. (2016) Inactivation of human cytochrome P450 3A4 and 3A5 by dronedarone and N-desbutyl dronedarone. Mol Pharmacol 89:1–13 [DOI] [PubMed] [Google Scholar]
- Howard JT, Ashwell MS, Baynes RE, Brooks JD, Yeatts JL, Maltecca C (2017) Gene co-expression network analysis identifies porcine genes associated with variation in metabolizing fenbendazole and flunixin meglumine in the liver. Sci Rep 7:1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sorrentino C, Denison MS, Kolaja K, Fielden MR (2007) Induction of cyp1a1 is a nonspecific biomarker of aryl hydrocarbon receptor activation: results of large scale screening of pharmaceuticals and toxicants in vivo and in vitro. Mol Pharmacol 71:1475–1486 [DOI] [PubMed] [Google Scholar]
- Hustert E, Haberl M, Burk O et al. (2001) The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11:773–779 [DOI] [PubMed] [Google Scholar]
- Kagimoto M, Heim M, Kagimoto K, Zeugin T, Meyer UA (1990) Multiple mutations of the human cytochrome P450IID6 gene (CYP2D6) in poor metabolizers of debrisoquine. Study of the functional significance of individual mutations by expression of chimeric genes. J Biol Chem 265:17209–17214 [PubMed] [Google Scholar]
- Klieber S, Arabeyre-Fabre C, Moliner P et al. (2014) Identification of metabolic pathways and enzyme systems involved in the in vitro human hepatic metabolism of dronedarone, a potent new oral anti-arrhythmic drug. Pharmacol Res Perspect 2:e00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober L, Torp-Pedersen C, McMurray JJ et al. (2008) Increased mortality after dronedarone therapy for severe heart failure. N Engl J Med 358:2678–2687 [DOI] [PubMed] [Google Scholar]
- Koturbash I, Tolleson WH, Guo L et al. (2015) microRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark Med 9:1153–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Usmani KA, Chanas B et al. (2003) Genetic findings and functional studies of human CYP3A5 single nucleotide polymorphisms in different ethnic groups. Pharmacogenetics 13:461–472 [DOI] [PubMed] [Google Scholar]
- Li Y, Mei H, Wu Q et al. (2011) Methysticin and 7,8-dihydromethysticin are two major kavalactones in kava extract to induce CYP1A1. Toxicol Sci 124:388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Couch L, Higuchi M, Fang JL, Guo L (2012) Mitochondrial dys-function induced by sertraline, an antidepressant agent. Toxicol Sci 127:582–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Kostov R, Huang JT, Henderson CJ, Wolf CR (2017) Novel pathways of ponatinib disposition catalyzed by CYP1A1 involving generation of potentially toxic metabolites. J Pharmacol Exp Ther 363:12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Lu AY (2007) CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab Dispos 35:1009–1016 [DOI] [PubMed] [Google Scholar]
- Moorthy B, Chu C, Carlin DJ (2015) Polycyclic aromatic hydrocarbons: from metabolism to lung cancer. Toxicol Sci 145:5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel C, Yan GX, Kowey PR (2009) Dronedarone. Circulation 120:636–644 [DOI] [PubMed] [Google Scholar]
- Poulsen KL, Olivero-Verbel J, Beggs KM, Ganey PE, Roth RA (2014) Trovafloxacin enhances lipopolysaccharide-stimulated production of tumor necrosis factor-alpha by macrophages: role of the DNA damage response. J Pharmacol Exp Ther 350:164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Chen S, Zhang J, Doshi U, Li AP, Guo L (2016) Endoplasmic reticulum stress induction and ERK1/2 activation contribute to nefazodone-induced toxicity in hepatic cells. Toxicol Sci 154:368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic S, Guengerich FP (2010) Update information on drug metabolism systems–2009, part II: summary of information on the effects of diseases and environmental factors on human cytochrome P450 (CYP) enzymes and transporters. Curr Drug Metab 11:4–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizkallah J, Kuriachan V, Brent Mitchell L (2016) The use of dronedarone for recurrent ventricular tachycardia: a case report and review of the literature. BMC Res Notes 9:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868 [DOI] [PubMed] [Google Scholar]
- Scandlyn MJ, Stuart EC, Rosengren RJ (2008) Sex-specific differences in CYP450 isoforms in humans. Expert Opin Drug Metab Toxicol 4:413–424 [DOI] [PubMed] [Google Scholar]
- Serviddio G, Bellanti F, Giudetti AM et al. (2011) Mitochondrial oxidative stress and respiratory chain dysfunction account for liver toxicity during amiodarone but not dronedarone administration. Free Radic Biol Med 51:2234–2242 [DOI] [PubMed] [Google Scholar]
- Shimada T (2006) Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab Pharmacokinet 21:257–276 [DOI] [PubMed] [Google Scholar]
- Thompson RA, Isin EM, Ogese MO, Mettetal JT, Williams DP (2016) Reactive metabolites: current and emerging risk and hazard assessments. Chem Res Toxicol 29:505–533 [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Holloway MG (2009) Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol 76:215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werk AN, Lefeldt S, Bruckmueller H et al. (2014) Identification and characterization of a defective CYP3A4 genotype in a kidney transplant patient with severely diminished tacrolimus clearance. Clin Pharmacol Ther 95:416–422 [DOI] [PubMed] [Google Scholar]
- Westlind-Johnsson A, Hermann R, Huennemeyer A et al. (2006) Identification and characterization of CYP3A4*20, a novel rare CYP3A4 allele without functional activity. Clin Pharmacol Ther 79:339–349 [DOI] [PubMed] [Google Scholar]
- Wu Q, Ning B, Xuan J, Ren Z, Guo L, Bryant MS (2016) The role of CYP 3A4 and 1A1 in amiodarone-induced hepatocellular toxicity. Toxicol Lett 253:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan J, Chen S, Ning B, Tolleson WH, Guo L (2016) Development of HepG2-derived cells expressing cytochrome P450s for assessing metabolism-associated drug-induced liver toxicity. Chem Biol Interact 255:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhang B, Molony C et al. (2010) Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res 20:1020–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Price ET, Chang CW et al. (2013) Gene expression variability in human hepatic drug metabolizing enzymes and transporters. PLoS One 8:e60368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Wu L, Gill P et al. (2018) Multiple microRNAs function as self-protective modules in acetaminophen-induced hepatotoxicity in humans. Arch Toxicol 92:845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Chen Y, Wang Y et al. (2017) MicroRNA hsa-miR-370–3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol 140:139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen S, Mei H et al. (2015) Ginkgo biloba leaf extract induces DNA damage by inhibiting topoisomerase II activity in human hepatic cells. Sci Rep 5:14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF, Liu JP, Chowbay B (2009) Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev 41:89–295 [DOI] [PubMed] [Google Scholar]