Abstract
Epidermal growth factor receptor (EGFR)-targeted cancer therapy requires an accurate estimation of EGFR expression in tumors to identify responsive patients, monitor therapeutic effect, and estimate prognosis. The EGFR molecular imaging is an optimal method for evaluating EGFR expression in vivo accurately and noninvasively. In this review, we discuss the recent advances in EGFR-targeted molecular imaging in cancer, with a special focus on the development of imaging agents, including epidermal growth factor (EGF) ligand, monoclonal antibodies, antibody fragments, Affibody, and small molecules. Each substrate or probe, whether it is an endogenous ligand, antibody, peptide, or small molecule labeled with fluorochrome or radionuclide, has unique advantages and limitations. Antibody-based probes have high affinity but a long metabolic cycle and therefore offer poor imaging quality. Affibody molecules promise to surpass antibody-based probes due to their small size, stable chemical properties, and high affinity to the target. Small-molecule probes are safe, have favorable pharmacokinetics, and show high affinity and specificity, in addition to having an ideal size, but are inadequate for delayed imaging after injection due to their fast clearance.
Keywords: EGFR, molecular imaging, targeted molecular probe, Affibody molecule, cancer
Background
Clinical Relevance of the EGFR Signaling Pathway
The epidermal growth factor receptor (EGFR) is 1 of 4 members of the human EGFR (hEGFR) family of receptor tyrosine kinases.1 The EGFR is a single-chain transmembrane glycoprotein comprising an extracellular ligand-binding domain, a transmembrane region, and an intracellular tyrosine kinase domain2 and is known to be involved in several physiological processes, such as cell growth, proliferation, differentiation, and apoptosis. Furthermore, it is well documented that EGFR plays a crucial role in cancer formation, progression, metastasis, and angiogenesis.3-5 Preclinical data show that over 70% of malignant tumors have abnormal expression of EGFR, including bladder, lung, and breast cancers.6-9 In addition, clinical trials have shown that, besides being associated with poor prognosis and short survival time, EGFR overexpression is a reliable predictor of tumor invasiveness and chemotherapy and radiotherapy resistance.10,11 Thus, EGFR has emerged as a potential diagnostic and therapeutic biomarker in cancers.12
A variety of EGFR-targeted drugs approved by the Food and Drug Administration (FDA) are available for clinical applications, including monoclonal antibodies (mAbs) such as cetuximab (IMC-C225, Erbitux) and panitumumab (ABX-EGF, Vectibix), which block the extracellular ligand-binding domain of the receptor and tyrosine kinase inhibitors (TKIs) such as gefitinib, which prevent the activation of the cytoplasmic kinase portion.13 However, a wide range of clinical effects has been reported, from minor or no clinical benefits to nearly complete response. For instance, in a phase III clinical trial on 1125 patients with advanced non-small-cell lung cancer (NSCLC) treated for 15 months with cetuximab and chemotherapy (n = 557) or chemotherapy alone (n = 568), patients receiving cetuximab therapy survived for longer than those in the chemotherapy-alone group.14 However, other studies on unselected (not screened for gene mutation or protein expression) patients failed to achieve similar results. The multicenter study (Iressa Survival Evaluation in Lung Cancer) revealed that unselected patients receiving gefitinib had no clinical benefits compared to the placebo group in NSCLC.15,16 These studies have raised awareness for the need to evaluate EGFR expression before targeted treatment in order to select responsive patients and predict clinical results more accurately.
Methods to detect expression levels of EGFR protein include biopsy and serological detection. Although biopsy and immunohistochemistry are currently the most commonly used methods, they present several limitations. First, biopsy involves an invasive examination and is therefore not suitable for repeated interventions. Second, tumor heterogeneity may reduce the accuracy of the biopsy results.17-19 Indeed, different EGFR expression levels may be obtained within the same tumor or between the primary tumors and the metastatic lesions.20 Furthermore, EGFR expression levels in tumors can change during treatment and may also be influenced by the detection method.21-23 Serological detection, such as enzyme-linked immunosorbent assay, is simple, fast, and reproducible but has low sensitivity, poor specificity, a high rate of false positives,24,25 and a relatively long time lag for positive results.26 Finally, the results between serological examinations and biopsy are not always consistent,27 and serology cannot confirm lesion location or provide quantitative information. It has therefore become necessary to develop an accurate noninvasive method for detecting EGFR expression in vivo, to select patients potentially responsive to EGFR-targeted treatment, to monitor changes in EGFR expression levels during treatment, and to guide the selection of adequate clinical treatments. In recent years, the development of molecular imaging has enabled the visualization of cells, molecules, and metabolic processes in real time in vivo. Thus, EGFR-targeted imaging in vivo could be a valuable tool for noninvasive identification of EGFR expression.
Relationship Between EGFR Signaling Pathway and Cancer
The EGFR binding to its natural ligand, epidermal growth factor (EGF), activates the Ras/Raf/MEK/ERK-MAPK, PI3K-Akt(PKB)/mTOR, and JAK/STAT signaling pathways, thereby promoting proliferation, differentiation, migration, and apoptosis inhibition.3-5 Numerous studies have shown that EGFR is upregulated in most malignancies and that it plays a crucial role in phenotypic transformation and maintenance. Indeed, EGFR activation is closely associated with tumor angiogenesis, metastasis, and treatment resistance.11,28 In addition to directing affecting cellular proliferation and survival, EGFR is a key mediator in biochemical and molecular events underpinning carcinogenesis.29 The signaling pathways downstream of EGFR have multiple crossing sites with oncogenes, such as Ras, PI3K, PLC-γ, and STAT, which in turn can regulate each other, ultimately leading to oncogenesis.30,31 Furthermore, EGFR downstream signaling pathways can also interact with the focal adhesion kinase pathways to regulate cell proliferation and invasion.32,33 The EGFR activation leads to upregulation of signal transducer and activator of transcription 3 (STAT3), specificity protein 1, and hypoxia-inducible factors, which induce expression of vascular endothelial growth factor (VEGF), a central mediator of angiogenesis.33 The EGFR is also closely related to programmed cell death ligand 1 (PD-L1).34-36 Indeed, there is a correlation between EGFR pathway activation and a signature of immunosuppression manifested by upregulation of programmed cell death 1 (PD-1), PD-L1, cytotoxic T-lymphocyte antigen-4, and multiple tumor-promoting inflammatory cytokines.
The studies discussed earlier are merely a small selection from a vast amount of research demonstrating the essential role of EGFR in malignant transformation. Thus, EGFR-targeted imaging has strong potential as an invaluable tool for tumor diagnostics and therapy efficacy monitoring.
Review
Molecular Imaging Agents Based on EGF Ligand
The EGF (∼6.4 kDa) is the natural ligand of EGFR and has therefore high affinity to the receptor. Velikyan et al37 labeled hEGF with gallium-68 ([68Ga]Ga-DOTA-hEGF) to image EGFR using positron emission tomography (PET) in a preclinical model. Although EGFR-positive A431 tumors (squamous cell carcinoma [SCC]) could be visualized with micro-PET, the imaging quality was not ideal due to high tracer uptake in the abdominal region. Li et al38 used the positron emitter fluorine-18 to label EGF ([18F]F-FBEM-cEGF) for EGFR-targeted imaging in vivo. Micro-PET images of UM-SCC1 tumors (human head and neck squamous cell carcinoma [HNSCC]) were clearly visible in comparison to the contralateral background. The tumor uptake (%ID/g, percentage of the injected dose per gram of organ or tissue) of [18F]F-FBEM-cEGF was 2.60 ± 0.59, 1.87 ± 0.44, and 0.98 ± 0.33%ID/g at 30, 60, and 120 minutes postinjection (pi), respectively. Both liver and kidneys showed high uptake at 30 minutes pi at 7.23 ± 0.51 and 15.5 ± 1.21%ID/g, respectively; however, the tracer was rapidly washed out of these tissues (Figure 1). In another study, EGF and anti-EGFR mAb (mAb 528, ∼150 kDa) were labeled with indium-111 ([111In]In-DTPA-hEGF and [111In]In-DTPA-mAb 528).39 Biodistribution and tumor imaging assays were performed after injection of the probes into mice bearing xenografts from cells with low (MCF-7), medium (MDA-MB-231), and high (MDA-MB-468) EGFR expression. The maximum uptake of [111In]In-DTPA-mAb 528 in MDA-MB-468 breast cancer cells (21.6%ID/g vs 2.2%ID/g) was higher than that of [111In]In-DTPA-hEGF, and similar results were obtained with tumor-to-normal tissue ratios. Thus, radiolabeled anti-EGFR mAbs show more promise than peptide-based radiopharmaceuticals such as hEGF for future studies in patients with cancer, as they have higher tumor uptake and provide better visualization of tumors at lower tumor-to-blood ratios.
Figure 1.
A, PET/CT overlay of UM-SCC1 tumor-bearing mice imaged with [18F]F-FBEM-cEGF at 30 minutes after injection. B, Decay-corrected whole-body coronal microPET images of UM-SCC1 tumor-bearing mice at 30, 60, and 120 minutes after injection of 3.7 MBq (100 μCi) of [18F]F-FBEM-cEGF. Tumors are indicated by arrows. Adapted and reproduced with permission from Li et al.38 PET indicates positron emission tomography; CT, computed tomography; SCC, squamous cell carcinoma; EGF, epidermal growth factor.
Molecular Imaging Agents Based on Anti-EGFR Antibodies
Cetuximab (C225, Erbitux) was the first FDA-approved mAb that blocks the ligand-binding domain of EGFR with an affinity comparable to the natural ligand. With a dissociation constant (KD) of 1.0 nmol/L, cetuximab competes for binding to EGFR, thereby inhibiting receptor activation and subsequent autophosphorylation and ultimately inducing its internalization and degradation.40 The overall pharmacokinetics of cetuximab are slow (elimination rate ranging from 65 to 95 hours), partly because of the large size (∼152 kDa) of the antibody.41 Consequently, cetuximab has become a promising new therapy agent in oncology and is increasingly used in clinical trials, mainly in combination with chemotherapy or radiotherapy.42,43 Cetuximab is a human/mouse chimeric immunoglobulin G1 (IgG1) mAb, whereas panitumumab is a human IgG2 mAb that binds to a different epitope of the EGFR antigen than cetuximab, and is used in the treatment of patients with chemotherapy-refractory metastatic colorectal carcinoma (CRC).44 The EGFR is internalized upon binding to cetuximab or panitumumab, and the ligand–receptor complexes are targeted for lysosomal degradation, resulting in signal attenuation.45
As the biological half-life of cetuximab in the blood is 65 to 95 hours,41 a radioactive tracer with a long half-life is needed to visualize its uptake. Cetuximab labeled with indium-111 (half-life = 2.83 days) and iodine-125 (half-life = 60.14 days) was assessed with single-photon emission computed tomography (SPECT) imaging in nude mice bearing HNSCC FaDu cell.46 [111In]In-cetuximab showed significantly higher uptake in tumors than [125I]I-cetuximab (P = .002) at all time points, and similar results were obtained with tumor-to-blood ratios (6.03 ± 1.69 vs 1.91 ± 0.72). [125I]I-IBPA-cetuximab is a new bifunctional linker for radiohalogenation of antibodies (IBPA, N-(4-isothiocyanatobenzyl)-2-(3-(tributylstannyl)phenyl)acetamide [patent no. 10-1550399KR]). Kim et al47 showed that the tumor uptake value of [125I]I-IBPA-cetuximab was higher than that of [125I]I-cetuximab for up to 168 hours in athymic mice bearing human colorectal adenocarcinoma LS174T tumor xenografts (12.42 ± 1.63%ID/g vs 7.10 ± 1.54%ID/g at 48 hours after injection). The thyroidal uptake value of [125I]I-IBPA-cetuximab (0.09 ± 0.05%ID/g) after injection was ∼8-fold lower than that of [125I]I-cetuximab (0.69 ± 0.36%ID/g), with a statistically significant difference (P < .005). Given that [125I]I-IBPA-cetuximab is stable and resistant to deiodination in vivo, IBPA shows great potential as a bifunctional linker for radioiodination of internalizing mAbs for in vivo applications, including radioimmunotherapy. Another study48 revealed that [111In]In-DTPA-cetuximab accumulated in colorectal HCT-15 xenograft tumors (50 and 250 mm3), whereas the tumor-to-muscle ratio in the large tumor was 7.5-fold, further suggesting that [111In]In-DTPA-cetuximab may prove valuable for early diagnosis of EGFR-positive tumors in the clinical practice. The PET images with [111In]In-DTPA-cetuximab show high spatial resolution, good signal-to-noise ratio, and the tumor-to-muscle and tumor-to-blood ratios are comparable to those of [89Zr]Zr-DFO-cetuximab (half-life of approximately 78 hours)49 and [64Cu]Cu-DOTA-cetuximab (half-life of approximately 12.7 hours; 2.96 ± 0.40 vs 12.4 ± 0.50 at 4 hours, respectively).50 However, [64Cu]Cu-labeled cetuximab was observed to have a better biodistribution profile than [111In]In-DTPA-cetuximab at 48 hours pi.51 Cai et al52 uncovered a positive correlation between EGFR expression and uptake of [64Cu]Cu-DOTA-cetuximab in tumor-bearing mouse models. The conjugate was cleared mainly through the hepatobiliary system, with little to no renal uptake or renal clearance being observed.
Over recent years, cancer immunotherapy has attracted significant research interest within the scientific and medical communities. Immuno-PET provides comprehensive information about tumor location, phenotype, susceptibility to therapy, and treatment response, particularly to radioimmunotherapy. Immuno-PET, micro-SPECT/computed tomography (CT), and biodistribution assays showed that specific uptake of radiolabeled cetuximab in esophageal squamous cell carcinoma (ESCC) tumors correlated to EGFR expression levels.53 Tumor uptake of [64Cu]Cu-cetuximab and [177Lu]Lu-cetuximab in mice bearing TE-8 (ESCC cell line) xenografts peaked at 48 and 120 hours (17.5 ± 4.4%ID/g vs 55.7 ± 6.5%ID/g, respectively). Radioimmunotherapy with [177Lu]Lu-cetuximab (half-life = 6.7 days) showed significant inhibition of tumor growth (P < .01) and marked reduction in [18F]F-fluorodeoxyglucose (FDG) standard uptake value (SUV), when compared to the control on day 14 after treatment (0.66 ± 0.12 vs 0.94 ± 0.12, P < .05). These results suggest that radiopharmaceutical [64Cu]Cu-PCTA-cetuximab/[177Lu]Lu-PCTA-cetuximab may be useful as a diagnostic tool for patient selection and as a potent radioimmunotherapy agent in EGFR-positive ESCC tumors.
Fluorescence imaging is among the most widely utilized molecular imaging methods. Cetuximab labeled with IRDye800CW, a near-infrared fluorescent dye, was assessed by optical imaging in nude mice bearing HNSCC cell lines (SCC5 and SCC1).54 Cetuximab-IRDye800CW showed specific and high-affinity binding to EGFR (KD = 0.31 nmol/L). Both PET and fluorescence imaging have complementary features, particularly in the clinical setting. Indeed, PET is especially well suited for whole-body evaluation, whereas fluorescence imaging is more adequate for the analysis of superficial tissue layers at the cellular level. To facilitate translational imaging research, ideally these complementary imaging modalities would seamlessly be applied both preclinically in animal models and clinically in particular patient groups. To achieve this, Cohen et al55 developed a cetuximab probe dual-labeled with IRDye800CW and zirconium-89, without impairment of immunoreactivity and pharmacokinetics of the mAb. Biodistribution assays of [89Zr]Zr-cetuximab-IRDye800CW in A431 xenograft-bearing nude mice revealed that uptake in tumors (∼20%ID/g) was higher than in normal tissues and organs (no more than 10%ID/g), except in the liver (above 20%ID/g).56 Bevacizumab, recombinant humanized mAb, is the first approved drug by FDA to inhibit tumor angiogenesis. It can bind to human VEGF and block its biological activity. Dual-mode imaging of [89Zr]Zr-bevacizumab-IRDye800CW in a FaDu xenograft-bearing nude mouse showed that tumors could be clearly visualized both by optical and PET imaging. However, the liver could only be visualized by PET imaging, probably because it is a deep-seated organ.56
In summary, mAb probes can not only provide a clear imaging of EGFR expression in tumors but may also detect EGFR expression levels based on probe uptake quantifications. However, the results from studies addressing these questions are not always in agreement. For instance, no correlation was found between the evaluation of EGFR expression using flow cytometry and the accumulation of radiopharmaceutical in mice bearing human SCC xenografts. Niu et al showed that EGFR expression was higher in SCC1 than in UM-SCC-22B HNSCC cells.57 In contrast, SCC1 tumors had much lower [64Cu]Cu-DOTA-cetuximab accumulation than UM-SCC-22B tumors. Furthermore, [64Cu]Cu-DOTA-panitumumab accumulation in tumor-bearing mice revealed that probe uptake was higher in UM-SCC-22B low EGFR expression EGFR xenografts than in SQB20 high EGFR expression xenografts at 30 hours pi (31.42 ± 10.77%ID/g vs 31.42 ± 10.77%ID/g).58 Aerts et al59 found a similar disparity between [89Zr]Zr-labeled cetuximab tumor uptake and in vivo EGFR expression levels, but uptake in normal tissue was consistent across different tumor models, suggesting that additional pharmacokinetic or pharmacodynamic mechanisms affect antibody-targeted therapy. These mechanisms may also explain why receptor expression levels alone are not sufficient to predict patient response.
Clinical research using [89Zr]Zr-cetuximab (half-life of approximately 78 hours) PET imaging demonstrated a strong correlation between uptake and treatment response, and further clinical validation suggested that [89Zr]Zr-cetuximab PET may be an innovative patient selection method for cetuximab treatment in patients with advanced CRC60 and head and neck cancer.61 In 2016, a phase I clinical trial was performed by Van et al62 to determine the safety of using [89Zr]Zr-cetuximab for assessing tumor uptake, with the aim of identifying patients with cancer most likely to benefit from targeted treatment. [89Zr]Zr-cetuximab administration schedules of 2 consecutive doses of 60 MBq, or a single dose of 120 MBq, were shown to be safe for patients (toxicity according to the CTCAE 3.0 scoring system). The recommended dose for future trials was 60 MBq, with a minimum time interval for scanning of 6 days. [89Zr]Zr-cetuximab dosimetry has recently been studied by Makris et al in patients with CRC.63 The liver received the highest absorbed dose of 2.60 ± 0.78 mGy/MBq, followed by the kidneys, spleen, and lungs, whereas the effective whole-body dose was 0.61 ± 0.09 mSv/MBq. The ultimate aim of these studies is to determine the feasibility and the optimal conditions for using these tracers in the treatment, monitoring, and therapy response prediction of cancer. In addition, anti-EGFR tracers could potentially aid in determining nodal metastatic disease.
Molecular Imaging Agents Based on Antibody Fragments
Despite the encouraging results in preclinical and clinical studies, molecular imaging with mAbs presents many limitations. As they have large molecular weight (∼150 kDa), mAbs produce high imaging background, low tumor-to-background ratio, and poor imaging quality, particularly in the first hours after injection. Furthermore, their large size also limits penetration into the tumor area, which reduces probe access to the molecular target, thereby affecting imaging quality. To overcome these technical challenges, in recent years significant research efforts have been made to develop antibody fragments, instead of whole antibodies, for molecular imaging.
Fab is an antigen-binding fragment of any antibody. However, in this review, Fab specifically refers to a human anti-EGFR antibody fragment (∼55 kDa) and can recognize and bind to the extracellular domain of EGFR in its native conformation.64 Fab retains the targeting specificity of whole IgG but can link to therapeutic payloads (such as radionuclides). Furthermore, Fab can be engineered into diagnostic and therapeutic tracers, as it is nonimmunogenic in vivo and its superior biodistribution and blood clearance properties compare to IgG. Xu et al64 showed that the affinity of [125I]I-Fab bound to A431 (high EGFR expression) and U118 (moderate EGFR expression) was 3- and 2-fold greater, respectively, than that of NIH 3T3 (EGFR-negative mouse fibroblast). The monovalent F(ab) fragment has only 1 antigen-binding region, whereas the polyvalent F(ab′)2 fragment has 2 antigen-binding regions, which are bound together by disulfide bonds. The F(ab′)2 fragment produces 2 univalent Fab′ fragments and a free sulfur group that can be used to bind other molecules. Cetuximab-F(ab′)2 (∼110 kDa) was labeled with [111In]In by Van et al.65 Micro-SPECT imaging showed high uptake in EGFR-expressing HNSCC xenografts. Specifically, the probe uptake was 5.7 ± 1.1%ID/g (SCCNij202, high-), 7.5 ± 2.2%ID/g (SCCNij153, moderate-), 2.7 ± 0.3%ID/g (SCCNij185, moderate-), 2.2 ± 0.7%ID/g (SCCNij167, negative-), and 1.7 ± 0.6%ID/g (mice receiving a blocking dose by cetuximab). Tumor uptake of [111In]In-cetuximab-F(ab′)2 was proportionally associated with cetuximab treatment response in 3 out of 4 xenograft models. The same team conducted another study comparing [111In]In-cetuximab-F(ab′)2 and [111In]In-cetuximab.66 Tumor uptake of [111In]In-cetuximab-F(ab′)2 was significantly lower than that of [111In]In-cetuximab (10.3 ± 5.2%ID/g vs 26.9 ± 3.3%ID/g) at 24 hours pi, and tumor-to-blood ratios and tumor-to-muscle ratios at 24 hours pi were significantly higher for [111In]In-cetuximab-F(ab′)2 (31.4 ± 3.8 vs 1.7 ± 0.2, 107.0 ± 17.0 vs 69.7 ± 3.9, respectively; Figure 2). These studies suggest that cetuximab-F(ab′)2 is more suitable than whole antibody for EGFR visualization, and potentially for selecting responsive patients for treatment with EGFR inhibitors.
Figure 2.
Serial micro-SPECT images of [111In]In-cetuximab-F(ab′)2 obtained at 4, 24, and 48 hours after injection and of [111In]In-cetuximab at 4, 24, 48, and 168 hours after injection, including mice that received a blocking dose of unlabeled cetuximab. Tumor in the right flank. Adapted and reproduced with permission from Van et al.66 SPECT indicates single-photon emission computed tomography; pi, postinjection.
[18F]F-FDG is a glucose analog widely used in oncology as a PET radioligand. This probe was designed based on the observation that tumors generally have increased glycolysis rates; however, inflammatory cells are also glycolytically active.67 Thus, detection of high glycolytic activity with [18F]F-FDG in CRC when there is also increased glycolysis due to inflammatory bowel disease may result in poor target-to-background ratio. In contrast, since EGFR overexpression occurs in approximately 80% of cases with CRC 68 and inflammatory cell lineages do not overexpress EGFR, imaging based on EGFR expression in colitis can provide high target-to-background ratios.69 Turker et al70 compared PET imaging with [18F]F-FDG and [64Cu]Cu-DOTA-cetuximab-F(ab′)2 in CT26 (colonic adenocarcinomas) tumor-bearing mice with and without DSS-induced colitis. The tumor-to-background ratio for CT26 tumors was high for both [18F]F-FDG (3.95 ± 0.13) and [64Cu]Cu-DOTA-cetuximab-F(ab′)2 (4.42 ± 0.11) in control mice. However, while the tumor-to-background of the EGFR-targeted probe remained high (3.78 ± 0.06) in colitis, with [18F]F-FDG it was markedly reduced (1.54 ± 0.08). Furthermore, there was a correlation between radiotracer accumulation in spontaneous colonic lesions and EGFR staining level ex vivo, further suggesting that [64Cu]Cu-DOTA-cetuximab-F(ab′)2 is a clinically translatable PET imaging probe for assessing EGFR levels.
Molecular Imaging Agents Based on Affibody Molecules
Affibody molecules (Affibody) are 3-helix bundle nonimmunoglobulins based on a scaffold protein of small size (∼7 kDa, VENKFNKEMRNAYWEIALLPNLNNQQKRAFIRSLYDDPSQSANLLAEAKKLNDAQAPK) derived from the IgG-binding B-domain of staphylococcal protein A analog Z.71 Affibody phage display libraries were created by Smith in 1985 to provide a link between genotype and phenotype and thereby allow the selection of peptides or proteins with desired functions.71 Mutational studies performed on the 58 residues of Affibody have shown that the 3-helix bundle structure is highly tolerant to single, as well as multiple, random amino acid substitutions on the molecular surface involved in the binding to the fragment crystallizable (Fc) region of IgG. Indeed, these residue substitutions affected only the Fc-binding characteristics (the affinity of Affibody), but not its stability or overall structure.71,72 The randomization was targeted to residues Q9, Q10, N11, F13, Y14, L17, and H18 in the first helix and E24, E25, R27, N28, Q32, and K35 positioned either close to (E24) or at the helix 2 surface (Figure 3).73 The isoleucine at position 31 in the Z domain (Figure 3B) was not included in the randomization because this residue was earlier proposed to be important for the helix–helix packing.74 Affibody has high affinity and specificity and is therefore considered a molecular imaging probe of great potential.75 Table 1 summarizes the characteristics of Affibody molecules discussed in this review.
Figure 3.

Graphic representation of the wild-type Z domain (residues 5-58) based on the NMR structure. A Main-chain trace ribbon diagram showing the 3-helix bundle structure. B Space-filling representation of the domain showing the positions of the 13 amino acids located in helices 1 and 2 subjected to the randomization (red). The position of Ile31 (blue) is also shown. Adapted and reproduced with permission from Nord et al.73
Table 1.
Characteristics of the Affibody Molecules Discussed in the Review.
| Affibody | Time | Affinity, nmol/L | Fluorescence Intensity | Tracer | Molecular Structure | Affinity, nmol/L | Specificity | Imaging Mode | Half-Life | Imaging Time, hours | Image Quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZEGFR:955 | First-generation | Monomer | 130-185 | 277 | 125I- | Dimer | / | √ | SPECT | 60.14 d | 4-8 | Poor image resolution |
| Dimer | 50 | 111In- | / | SPECT | 2.83 d | 4 | ||||||
| ZEGFR:1907 | Second-generation | 5-10 (∼5.4) | 6458 | Alexa680- | Monomer | 28.3 | OI | / | 4 | Poor image resolution; low T/B | ||
| Cy5.5- | 43.6 | OI | / | 4 | ||||||||
| FAPdL5**- | 37 | OI | / | / | ||||||||
| 64Cu | 20 | PET | 12.7 h | 4 | High image resolution; high T/B | |||||||
| 18F | 37 | PET | 1.83 h | 1-3 | ||||||||
| 64Cu-NOTA-Au-IONP- | / | PET/MRI/OI | 12.7 h | 4 | ||||||||
| ZEGFR:2377 | / | / | / | 99mTc- | 0.274 | SPECT | 6.01 h | 3 | Poor image resolution | |||
| 111In | 0.9 | SPECT | 2.83 d | 4 | ||||||||
| 89Zr- | 0.16 | PET | 78.4 h | 3 | High T/B | |||||||
Abbreviations: /, no or not mentioned; SPECT, single-photon emission computed tomography; OI, optical imaging; PET, positron emission tomography; MRI, magnetic resonance imaging; T/B, tumor-to-background ratio.
ZEGFR:955 is a typical representative of the first-generation anti-EGFR Affibody molecules, which form dimers that bind EGFR on cultured cells with low nanomolar affinity.76 A comparative study on the imaging performance of [125I]I-(ZEGFR:955)2, EGF, and cetuximab in A431 cells showed that all probes have specificity for EGFR.77 The cellular uptake of [125I]I-(ZEGFR:955)2 and cetuximab peaked at ∼4 to 8 hours, retaining 20% and 25% after 48 hours, while the uptake of EGF peaked at 2 hours and disappeared almost completely after 24 hours. Competitive inhibition experiments showed that EGF and cetuximab can block binding of [125I]I-(ZEGFR:955)2 to EGFR with an efficiency of up to ∼97% to 98%, indicating that these 3 probes bind to the same, or nearly the same, binding site of EGFR. In addition, Nordberg et al78 demonstrated that [111In]In-(ZEGFR:955)2 accumulated specifically in A431 cells (38.0% ± 1.2%) and retained for 72 hours, while in vivo A431 tumor uptake was 3.8 ± 1.4%ID/g at 4 hours after injection, and the tumor-to-blood ratio was about 9.1.
ZEGFR:1907 is an outstanding representative of second-generation anti-EGFR Affibody molecules. Compared to ZEGFR:955, the affinity of ZEGFR:1907 is nearly 30-fold higher76 and its fluorescence intensity approximately 20-fold stronger.79 Furthermore, ZEGFR:1907 shows no unspecific cross-reaction to any serum protein or to other EGFR family members.79 First proposed by Nord et al71, Affibody connected to free cysteine80,81 could be easily labeled site specifically via the cysteine thiol groups (–SH).82 Ac-Cys-ZEGFR:1907 showed higher affinity to A431 cells when labeled with Alexa680 than with Cy5.5 (KD values of 28.3 and 43.6 nmol/L, respectively).83,84 However, the image quality was poor due to high background.83 To obtain a clear image, Wang et al85 labeled ZEGFR:1907 with fluorogen-activating protein dL5** (FAPdL5**). FAP-Affibody retained the ability of FAP to increase fluorescence intensity while showing the specificity and sensitivity of Affibody targeted to EGFR in A431 cells (KD value is 37 nmol/L). As this fluorophore can be linked to a variety of substances and retain low nanomolar KD values, it may be possible to directly target radionuclides and nanophase materials for PET/SPECT imaging or magnetic resonance imaging (MRI)/optical imaging.
Zhao et al assessed the feasibility of using Affibody Ac-Cys-ZEGFR:1907 labeled with copper-64 or Alexa680 to evaluate EGFR expression in hepatocellular carcinoma (HCC) tumor models with different EGFR expression levels.86 Hep3B (high expression) and PLC/PRF/5 (moderate expression) xenografts could be distinguished clearly from the background from 1 hour pi by PET imaging, with the signal reaching a peak at 24 hours and tumor uptakes of 8.52 ± 1.10 and 6.46 ± 0.59%ID/g, respectively. PLC/PRF/5 xenografts were visible by optical imaging at 4 hours pi with a low tumor-to-normal tissue ratio (maximum 1.6 at 1 hour pi). However, the fluorescent signal was weak in Hep3B xenografts, probably because of the abundance of tumor blood vessels, which may hinder or absorb the fluorescence signal (a well-known limitation of optical imaging). Finally, no signal was detected in HepG2 (no expression) xenografts.
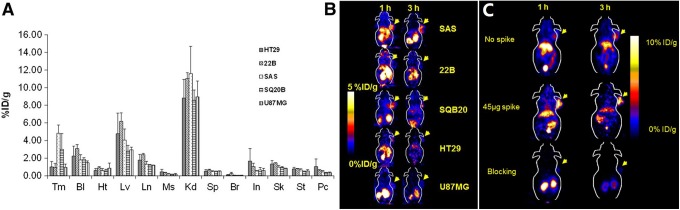
Fluorine-18 is a more clinically suitable radionuclide due to its shorter physical half-life. Miao et al developed [18F]F-FBEM-Cys-ZEGFR:1907 and assessed tumor uptake using different EGFR-expressing lines (A431, SQ20B and SAS, high EGFR; U87MG and 22B, low EGFR; HT29, no EGFR).87 Biodistribution assays revealed moderate-to-high tumor uptake (1.0%-4.8%ID/g) in HT29, U87MG, 22B, SQ20B, and SAS xenograft models (HT29, colon cancer; U87MG, glioma; SQ20B, 22B, and SAS, head and neck cancer; Figure 4A). Radioactive accumulation in all other organs was low, except in the liver and kidneys in A431 xenografts (9.28 and 3.78%ID/g at 3 hours pi, respectively). However, accumulation of ZEGFR:1907 labeled with copper-64 in the liver and kidneys was higher (18.99 and 88.45%ID/g at 4 hours pi, respectively).88 Small animal PET also revealed rapid tumor targeting and accumulation of [18F]F-FBEM-Cys-ZEGFR:1907 in several EGFR-positive tumors (Figure 4B). Interestingly, radioactive uptake in A431 xenograft tumors was clearly increased upon co-injection with 45 µg of Ac-Cys-ZEGFR:1907 (3.9%ID/g vs 8.1%ID/g at 3 hours pi), whereas 500 µg of Ac-Cys-ZEGFR:1907 blocked uptake (8.1%ID/g vs 1.0%ID/g at 3 hours pi, 88% inhibition; Figure 4C).
Figure 4.
A, Biodistribution of [18F]F-FBEM-Cys-ZEGFR:1907 at 3 hours after injection in 6 xenograft models with different EGFR expression levels (n = 3). B, Representative [18F]F-FBEM-Cys-ZEGFR:1907 PET of 5 tumor xenograft models with EGFR overexpression: SQ20B, SAS, and 22B (head and neck cancer); HT29 (colon cancer); U87MG (glioma cancer). C, Small-animal PET of [18F]F-FBEM-Cys-ZEGFR:1907 in A431 (epithelial cancer) xenograft model co-injected with 0, 45, and 500 μg (blocking dose) of Ac-Cys-ZEGFR:1907. Adapted and reproduced with permission from Miao et al.87 EGFR indicates epidermal growth factor receptor; PET: positron emission tomography; Tm, tumor; Bl, blood; Ht, heart; Lv, liver; Ln, lungs; Ms, muscle; Kd, kidney; Sp, spleen; Br, brain; In, intestine; Sk, skin; St, stomach; Pc, pancreas.
A multimodal molecular imaging probe based on iron oxide nanoparticles (IONPs), [64Cu]Cu-NOTA-Au-IONP-Ac-Cys-ZEGFR:1907, was recently developed by Yang et al for PET, optical, and MRI in EGFR-positive tumors.89 In vitro and in vivo studies showed that the nanoprobe has high specificity and sensitivity, and excellent tumor contrast in PET and MRI of A431 cells and xenografts. Tumor accumulation was 3.5%ID/g at 4 hours and peaked to 4.6%ID/g at 24 hours after injection in small animal PET imaging, and the maximum tumor-to-muscle ratio was about 6 at 24 hours pi. Comparing with the specific targeted imaging, the blocking group clearly showed lower tumor uptake (with unlabeled Ac-Cys-ZEGFR:1907; 1.9%ID/g vs 4.6%ID/g at 24 hours pi, P < .05). Furthermore, the overlap between the reflection signal of the gold component DAPI (4′,6-diamidino-2-phenylindole), immunofluorescence staining in optical imaging showed that the nanoprobe was targeted to the cell surface of EGFR-positive cells. In MRI, the signal intensity dropped 44% in tumor sites at 8 hours after injection, suggesting that Au-IONP could serve as an efficient MR contrast agent. Finally, the PET and MRI results were highly correlated, confirming the feasibility of performing multichannel multifunctional molecular imaging with these probes.
Notably, ZEGFR:2377 appears to have similar affinity for hEGFR and mouse EGFR.90–92 The cross-reactivity between hEGFR and mouse EGFR is useful for assessing specificity and biodistribution in preclinical models, which is particularly relevant in EGFR-expressing normal tissues in humans. [99mTc]Tc-ZEGFR:2377 showed specificity to 3 different EGFR expression cell lines (A431, epithelial carcinoma; MDA468, breast cancer; and PC3, prostatic cancer) and was successfully used for EGFR quantification.93,94 The KD value of [99mTc]Tc-ZEGFR:2377 was in the picomolar range (274 pM),93 similarly to [89Zr]Zr-DFO-ZEGFR:2377 (160 pM).95 Studies in mice bearing A431 xenografts demonstrated that the tumor uptake of [99mTc]Tc-ZEGFR:2377 was 3.6 ± 1.0 and 2.5 ± 0.4%ID/g at 3 and 24 hours after injection, respectively. At 3 hours, tumor-to-blood and tumor-to-muscle ratios were 1.8 and 7.8, respectively, and increased to 8.0 and 17.5 at 24 hours. Nevertheless, radioactivity uptake in the liver, but mostly in the kidneys, was high (5.6 ± 0.6 and 146 ± 19%ID/g, respectively), indicating that the probe has high hydrophilicity and is excreted mostly via the urinary system.
Molecular Imaging Agents Based on Other Peptides
Vosjan et al96 and Huang et al97 developed novel tracers based on Nanobody (minimal antigen-binding fragment of the heavy chain in antibody, ∼15 kDa). [99mTc]Tc-8B6 could specifically bind to EGFR-positive cells (A431 and DU145), but not to EGFR-negative cells (NIH3T3). Tumor uptake revealed by SPECT imaging was 5.2 ± 0.5 and 1.8 ± 0.3%IA/cm3 at 3 hours after injection for A431 and DU145 xenografts, and the corresponding tumor-to-background ratios were 7.4 ± 1.0 and 3.2 ± 0.9, respectively. Thus, EGFR-binding nanobodies have high specificity and selectivity toward EGFR-overexpressing cells.
Another probe, [64Cu]Cu-fibronectin, developed by Hackel et al,98 exhibited EGFR-dependent binding to multiple cell lines in culture. The tracer displayed good tumor accumulation (3.4 ± 1.0%ID/g at 1 hour), retention (2.7 ± 0.6%ID/g at 24 hours), and specificity (8.6 ± 3.0 tumor-to-muscle ratio, 8.9 ± 4.7 tumor-to-blood ratio at 1 hour). Specific targeting was revealed by the low accumulation of the probe in low EGFR-expressing MDA-MB-435 (mammary carcinoma) tumors (0.7 ± 0.8%ID/g at 1 hour). Furthermore, the fibronectin nonbinding control showed poor localization to EGFR-overexpressing xenografts (0.8 ± 0.2%ID/g at 1 hour), indicating the probe has high specificity to EGFR. With its high stability, low background, and high target specificity and retention, [64Cu]Cu-fibronectin is a promising EGFR-targeting molecular imaging agent.
Nanofitins are cysteine-free protein scaffolds derived from the hyperstable DNA-binding protein Sac7d (∼7 kDa, 66 amino acids) of Sulfolobus acidocaldarius.99 The in vivo tumor-targeting and imaging profile of radiolabeled anti-EGFR Nanofitin Cys-B10, [18F]F-FBEM-Cys-B10, was investigated by Goux et al in xenograft models.100 The EGFR-positive tumors (A431) were clearly delineated when compared to EGFR-negative tumors (H520), and with a significant tumor-to-background contrast. [18F]F-FBEM-Cys-B10 showed significantly higher retention in A431 tumors than in H520 tumors at 2.5 hours pi, with an uptake A431-to-H520 ratio of 2.53 ± 0.18 and a tumor-to-blood ratio of 4.55 ± 0.63. This was the first study reporting that Nanofitin scaffold is a viable EGFR-targeting PET radiotracer, thereby promoting the development of valuable alternative PET-based approaches for in vivo imaging of EGFR-expressing tumors.
Molecular Imaging Agents Based on Small Molecule
Small molecule-based probes (molecular weight <500 Da) display high affinity and selectivity and adequate lipophilicity or hydrophily and are therefore currently the most promising molecular tracers for in vivo imaging. Song et al developed [18F]F-IRS ([18F]F-N-(3-chloro-4-fluorophenyl)-7-(2(2-(2-(2-(4-fluorine)ethoxy)ethoxy)-ethoxy)-6-(3-morpholinopropoxy)quinazolin-4-amine) and assessed its feasibility for detecting mutant EGFR in NSCLC, particularly the EGFR 19 exon-deleted mutation.101 Figure 5 shows the chemical structure of [18F]F-IRS. Cellular uptake assays showed a rapid and significant uptake of [18F]F-IRS in HCC827 (EGFR 19 exon-deleted mutation) cells at 7.51% after 15 minutes of incubation, and reaching a peak at 14.07% after 120 minutes. The PET/CT images revealed a high accumulation of [18F]F-IRS in HCC827 tumors at 120 minutes; however, probe uptake was difficult to detect in H1975 (EGFR L858R/T790M mutation), H520 (negative EGFR), and H358 (EGFR wild-type) tumors at all time points. Although probe uptake in HCC827 tumors reached 4.27%ID/g at 120 minutes pi in biodistribution assays, in H1975, H520, and H358 tumors [18F]F-IRS uptake was significantly lower at 1.71, 1.62, and 1.68%ID/g at 120 minutes pi, respectively. HCC827 tumor uptake was blocked by gefitinib (1.21%ID/g at 120 minutes), suggesting that [18F]F-IRS binds specifically to EGFR 19 exon-deleted mutation.
Figure 5.
Chemical structure of F-IRS.
Another molecular tracer, [18F]HO-J (a series of HO compounds with a 4-(anilino)pyrido[3,4-d]pyrimidine scaffold),102 was designed and synthesized for discriminating between L858R and L858R/T790M mutant EGFR in NSCLC in PET imaging. Biodistribution assays revealed that [18F]HO-J displayed a high tumor-to-muscle, tumor-to-blood, and tumor-to-lung ratios in H3255 (L858R mutation) tumor-bearing mice (13, 3, and 3 at 3 hours pi, respectively). Co-injection of excess AZD9291 (highly selective EGFR mutant inhibitors, IC50 of exon 19 deletion type, and L858R/T790M EGFR and wild-type EGFR were 12.92, 11.44, and 493.8 nM, respectively) resulted in a significant decrease in 54.3% in tumor uptake in H3255 xenografts at 3 hours pi, without any significant effect in other tissues, which demonstrates that tracer uptake is specific for EGFR-TK. In addition, H3255 tumors could be visualized more clearly than H1975 (L858R/T790M mutation) tumors at 3 hours pi by PET imaging.
PD153035 is the most studied small molecule quinazoline imaging agent. Dai et al assessed the efficacy of [11C]C-PD153035 (PD153035, 4-N-(3-bromoanilino)-6,7-dimethoxyquinazoline) PET imaging in detecting EGFR-TKI sensitivity in NSCLC models.103 They found that cells highly sensitive to EGFR-TKIs exhibited higher [11C]C-PD153035 uptake (HCC827 and PC9 cells), whereas A549 cells, which are moderately sensitive to EGFR-TKIs, showed higher uptake than EGFR-TKI-resistant H1975 cells. Radioactive accumulations were positively correlated with phosphorylated EGFR expression in all cells. The PET/CT showed that radioactivity was highest in HCC827 xenografts and the radioactivity in PC9 xenografts was higher than that in A549 and H1975 xenografts. Patients with advanced chemotherapy-refractory NSCLC were prospectively enrolled in a trial of erlotinib and imaged by [11C]C-PD153035 PET/CT to predict survival.104 Patients with higher SUVmax lived for over twice as long as patients with lower SUVmax (median overall survival = 11.4 months vs 4.6 months, P = .002; progression-free survival [PFS] = 4.4 months vs 1.8 months, P < .001). These preliminary results suggest that [11C]C-PD153035 PET/CT may be a noninvasive and rapid method for identifying patients with refractory advanced NSCLC likely to respond to EGFR-TKIs.
Zhang et al105 explored the feasibility of using [99mTc]Tc-3PRGD2 ([[99mTc]Tc(HYNIC-3P-RGD2)(tricine)(TPPTS)]: where HYNIC is 6-hydrazi nonicotinyl; 3P-RGD2 is PEG4-E[PEG4-c(RGDfK)]2; PEG4 is 15-amino-4,7,10,13-tetrao-xapentadecanoic acid; and TPPTS is trisodium triphenylphosphine-3,3′,3″-trisulfonate) SPECT/CT imaging to monitor the efficacy of EGFR-TKI therapy in patients with advanced-stage lung adenocarcinoma. The tumor-to-nontumor ratio in the partial response and stable disease groups was 35.8% and 8.9%, respectively, while in the progressive disease group was 76.1%. For receiver operator characteristic analysis, using a cutoff value of 23.8% decreased the tumor-to-nontumor ratio. The sensitivity and specificity in identifying responders were 80.0% and 87.5%, respectively. The median PFS with responders was 18 months, and with nonresponders was 7 months (P = .006). Thus, [99mTc]Tc-3PRGD2 imaging can evaluate the early response to EGFR-targeted therapy and predict PFS in patients with lung adenocarcinoma.
In 2018, Sun et al106 developed [18F]F-MPG (N-(3-chloro-4-fluorophenyl)-7-(2-(2-(2-(2-18F-fluoroethoxy) ethoxy) ethoxy) ethoxy)-6-methoxyquinazolin-4-amine) and evaluated its suitability for noninvasive PET imaging and for the quantification of EGFR-activating mutation status in preclinical models of NSCLC, as well as in patients with primary and metastatic NSCLC undergoing EGFR-TKI treatment. In NSCLC animal models, there was a significant correlation (R 2 = .9050) between [18F]F-MPG PET uptake and activating EGFR mutation status. In patients with NSCLC (n = 75), an overlap of 84.29% was found between the evaluation of EGFR activation by [18F]F-MPG uptake and tissue biopsy. In addition, the patients who showed greater response to EGFR-TKIs (81.58% vs 6.06%) and had a longer median PFS (348 days vs 183 days) also had a high [18F]F-MPG PET SUVmax (≥2.23 vs <2.23). Thus, [18F]F-MPG promises to be a powerful method for precisely quantifying EGFR-activating mutation status in patients with NSCLC, and ultimately for noninvasively identifying patients sensitive to EGFR-TKIs and for monitoring the efficacy of EGFR-TKI therapy.
Conclusions
Molecular-targeted therapy is the future of cancer treatment. Clinical research on EGFR-targeting therapy shows that modulating EGFR expression has remarkable therapeutic effects. The assessment of curative effect resulting from EGFR-targeted molecular imaging overcomes the limitations of traditional monitoring methods based on tumor volume or pathological and serological examinations using tumor markers. The EGFR molecular imaging may provide more accurate information in the quantification of EGFR expression in vivo, thus providing a reliable basis for screening patients, evaluating treatment efficacy, and estimating prognosis. The development of more high stability, high affinity, and high specific and also suitable hydrophilic and lipophilic probes would open the way for the clinical translation of EGFR molecular imaging. In the future, if imaging radionuclide could be replaced by the therapeutic radionuclide used in tumor-targeting drugs, imaging would harmonize with local treatment. Such an integration of diagnosis and treatment could bring revolutionary progress in tumor treatment with added clinical benefits that would significantly improve the lives of patients and their families.
Footnotes
Authors’ Note: XS initiated and oversaw the project. All authors were responsible for the article. The manuscript was drafted by WC and XS and was discussed and critically revised by XS and BS. All authors approved the final draft of the manuscript. Ethics Approval and Consent to Participate: No studies with human participants or animals performed by any of the authors were included in the article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Research was supported, in part, by the National Basic Research Program of China (2015CB931800), National Natural Science Foundation of China (81627901, 81471724, 31210103913, 81101088, 81130028), Heilongjiang Province Foundation for Returned Overseas Chinese Scholars, and the Key Laboratory of Molecular Imaging Foundation (College of Heilongjiang Province).
References
- 1. Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat Rev Mol Cell Biol. 2001;2(2):127. [DOI] [PubMed] [Google Scholar]
- 2. Kondo I, Shimizu N. Mapping of the human gene for epidermal growth factor receptor (EGFR) on the p13→q22 region of chromosome 7. Cytogenet Cell Genet. 1983;35(1):9–14. [DOI] [PubMed] [Google Scholar]
- 3. Suzuki S, Dobashi Y, Sakurai H, Nishikawa K, Hanawa M, Ooi A. Protein overexpression and gene amplification of epidermal growth factor receptor in nonsmall cell lung carcinomas. An immunohistochemical and fluorescence in situ hybridization study. Cancer. 2005;103(6):1265–1273. [DOI] [PubMed] [Google Scholar]
- 4. Eberhard A, Kahlert S, Goede V, Hemmerlein B, Plate KH, Augustin HG. Heterogeneity of angiogenesis and blood vessel maturation in human tumors: implications for antiangiogenic tumor therapies. Cancer Res. 2000;60(5):1388–1393. [PubMed] [Google Scholar]
- 5. Cataldo VD, Gibbons DL, Pérez-Soler R, Quintás-Cardama A. Treatment of non-small-cell lung cancer with erlotinib or gefitinib. N Engl J Med. 2011;364(10):947–955. [DOI] [PubMed] [Google Scholar]
- 6. Libermann TA, Nusbaum HR, Razon N, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313(5998):144–147. [DOI] [PubMed] [Google Scholar]
- 7. Rikimaru K, Tadokoro K, Yamamoto T, Enomoto S, Tsuchida N. Gene amplification and overexpression of epidermal growth factor receptor in squamous cell carcinoma of the head and neck. Head Neck. 1992;14(1):8–13. [DOI] [PubMed] [Google Scholar]
- 8. Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. CRC Crit Rev Oncol/Hematol. 1995;19(3):183–232. [DOI] [PubMed] [Google Scholar]
- 9. Gardmark T, Wester K, DeLa Torre M, Carlsson J, Malmstrom PU. Analysis of HER2 expression in primary urinary bladder carcinoma and corresponding metastases. BJU Int. 2005;96(7):440; author reply-1. [DOI] [PubMed] [Google Scholar]
- 10. Douillard JY, Cunningham D, Roth AD. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 1936;351(351):337–345. [DOI] [PubMed] [Google Scholar]
- 11. Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(suppl 4):S9–S15. [DOI] [PubMed] [Google Scholar]
- 12. Sato JD, Kawamoto T, Le AD, Mendelsohn J, Polikoff J, Sato GH. Biological effects in vitro of monoclonal antibodies to human epidermal growth factor receptors. Mol Biol Med. 1983;1(5):511. [PubMed] [Google Scholar]
- 13. Sharafinski ME, Ferris RL, Ferrone S, Grandis JR. Epidermal growth factor receptor targeted therapy of squamous cell carcinoma of the head and neck. Head Neck. 2010;32(10):1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pirker R, Pereira JA. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009;373(9674):1525. [DOI] [PubMed] [Google Scholar]
- 15. Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer). Lancet. 2005;366(9496):1527. [DOI] [PubMed] [Google Scholar]
- 16. Xu C, Zhou Q, Wu YL. Can EGFR-TKIs be used in first line treatment for advanced non-small cell lung cancer based on selection according to clinical factors? – a literature-based meta-analysis. J Hematol Oncol. 2012;5(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bambury RM, Power DG, O’Reilly S. Intratumor heterogeneity and branched evolution. N Engl J Med. 2012;366(22):2132–2133; discussion 3. [DOI] [PubMed] [Google Scholar]
- 18. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72(19):4875–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vignot S, Besse B, André F, Spano JP, Soria JC. Discrepancies between primary tumor and metastasis: a literature review on clinically established biomarkers. CRC Crit Rev Oncol/Hematol. 2012;84(3):301–313. [DOI] [PubMed] [Google Scholar]
- 21. Jens SR, Dan S, Mattias SM, et al. First-in-human molecular imaging of HER2 expression in breast cancer metastases using the 111In-ABY-025 affibody molecule. J Nucl Med. 2014;55(5):730–735. [DOI] [PubMed] [Google Scholar]
- 22. Sandström M, Lindskog K, Velikyan I, et al. Biodistribution and radiation dosimetry of the anti-HER2 affibody molecule 68Ga-ABY-025 in breast cancer patients. J Nucl Med. 2016;57(6):867–871. [DOI] [PubMed] [Google Scholar]
- 23. Sörensen J, Velikyan I, Sandberg D, et al. Measuring HER2-receptor expression in metastatic breast cancer using [68Ga]ABY-025 affibody PET/CT. Theranostics. 2016;6(2):262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schiller JH. Noninvasive monitoring of tumors. N Engl J Med. 2008;359(359):418–420. [DOI] [PubMed] [Google Scholar]
- 25. Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet. 2011;378(9804):1727–1740. [DOI] [PubMed] [Google Scholar]
- 26. Onn A, Herbst RS. Molecular targeted therapy for lung cancer: the lancet. Lancet. 2005;366(9496):1507–1508. [DOI] [PubMed] [Google Scholar]
- 27. Molina R, Filella X, Augé JM, et al. Tumor markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Tumor Biol. 2003;24(4):209–218. [DOI] [PubMed] [Google Scholar]
- 28. Lo HW, Hsu SC, Aliseyed M, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7(6):575. [DOI] [PubMed] [Google Scholar]
- 29. Hynes NE, Macdonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21(2):177–184. [DOI] [PubMed] [Google Scholar]
- 30. Sebastian S, Settleman J, Reshkin SJ, Azzariti A, Bellizzi A, Paradiso A. The complexity of targeting EGFR signalling in cancer: from expression to turnover. Biochim Biophys Acta. 2006;1766(1):120–139. [DOI] [PubMed] [Google Scholar]
- 31. Luwor R, Taylor L, Wang B, Zhu H. Tumor-associated EGFR over-expression specifically activates Stat3 and Smad7 resulting in desensitization of TGF-β signaling. Nat Publ Group. 2008;26(673):574. [Google Scholar]
- 32. Kim SH, Kim SH. Antagonistic effect of EGF on FAK phosphorylation/dephosphorylation in a cell. Cell Biochem Funct. 2008;26(5):539–547. [DOI] [PubMed] [Google Scholar]
- 33. Larsen AK, Ouaret D, Ouadrani KE, Petitprez A. Targeting EGFR and VEGF(R) pathway cross-talk in tumor survival and angiogenesis. Pharmacol Therapeut. 2011;131(1):80–90. [DOI] [PubMed] [Google Scholar]
- 34. Akbay EA, Koyama S, Carretero J, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3(12):1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Azuma K, Ota K, Kawahara A, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol. 2014;25(10):1935. [DOI] [PubMed] [Google Scholar]
- 36. Tang Y, Fang W, Zhang Y, et al. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs. Oncotarget. 2015;6(16):14209–14219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Velikyan I, Sundberg AL, Lindhe O, et al. Preparation and evaluation of (68)Ga-DOTA-hEGF for visualization of EGFR expression in malignant tumors. J Nucl Med. 2005;46(11):1881–1888. [PubMed] [Google Scholar]
- 38. Li W, Gang N, Lang L, et al. PET imaging of EGF receptors using [18F]FBEM-EGF in a head and neck squamous cell carcinoma model. Eur J Nucl Med Mol Imaging. 2012;39(2):300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reilly RM, Kiarash R, Sandhu J, et al. A comparison of EGF and MAb 528 labeled with 111In for imaging human breast cancer. J Nucl Med. 2000;41(5):903–911. [PubMed] [Google Scholar]
- 40. Mendelsohn J. Epidermal growth factor receptor inhibition by a monoclonal antibody as anticancer therapy. Clin Cancer Res. 1997;3(2):2703–2707. [PubMed] [Google Scholar]
- 41. Fracasso PM, Rd BH, Arquette MA, et al. A phase 1 escalating single-dose and weekly fixed-dose study of cetuximab: pharmacokinetic and pharmacodynamic rationale for dosing. Clin Cancer Res. 2007;13(3):986–993. [DOI] [PubMed] [Google Scholar]
- 42. Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354(6):567–578. [DOI] [PubMed] [Google Scholar]
- 43. Lammering G. Molecular predictor and promising target: will EGFR now become a star in radiotherapy? Radiother Oncol. 2005;74(2):89–91. [DOI] [PubMed] [Google Scholar]
- 44. Nayak TK, Garmestani K, Milenic DE, Baidoo KE, Brechbiel MW. HER1-targeted 86Y-panitumumab possesses superior targeting characteristics than 86Y-cetuximab for PET imaging of human malignant mesothelioma tumors xenografts. PLoS One. 2011;6(3):e18198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lammerts van Bueren JJ, Bleeker WK, Bøgh HO, et al. Effect of target dynamics on pharmacokinetics of a novel therapeutic antibody against the epidermal growth factor receptor: implications for the mechanisms of action. Cancer Res. 2006;66(15):7630. [DOI] [PubMed] [Google Scholar]
- 46. Hoeben BA, Molkenboerkuenen JD, Oyen WJ, et al. Radiolabeled cetuximab: dose optimization for epidermal growth factor receptor imaging in a head-and-neck squamous cell carcinoma model. Int J Cancer. 2011;129(4):870. [DOI] [PubMed] [Google Scholar]
- 47. Kim EJ, Kim BS, Choi DB, Chi SG, Choi TH. Enhanced tumor retention of radioiodinated anti-epidermal growth factor receptor antibody using novel bifunctional iodination linker for radioimmunotherapy. Oncol Rep. 2016;35(6):3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shih BB, Chang YF, Cheng CC, et al. SPECT imaging evaluation of (111)indium-chelated cetuximab for diagnosing EGFR-positive tumor in an HCT-15-induced colorectal xenograft. J Chin Med Assoc. 2017;80(12):766–773. [DOI] [PubMed] [Google Scholar]
- 49. Perk LR, Visser GW, Vosjan MJ, et al. (89)Zr as a PET surrogate radioisotope for scouting biodistribution of the therapeutic radiometals (90)Y and (177)Lu in tumor-bearing nude mice after coupling to the internalizing antibody cetuximab. J Nucl Med. 2005;46(11):1898–1906. [PubMed] [Google Scholar]
- 50. Ping LW, Meyer LA, Capretto DA, Sherman CD, Anderson CJ. Receptor-binding, biodistribution, and metabolism studies of 64Cu-DOTA-cetuximab, a PET-imaging agent for epidermal growth-factor receptor-positive tumors. Cancer Biother Radiol. 2008;23(2):158. [DOI] [PubMed] [Google Scholar]
- 51. Wen X, Wu QP, Ke S, et al. Conjugation with (111)In-DTPA-poly(ethylene glycol) improves imaging of anti-EGF receptor antibody C225. J Nucl Med. 2001;42(10):1530–1537. [PubMed] [Google Scholar]
- 52. Cai W. Quantitative PET of EGFR expression in xenograft-bearing mice using 64Cu-labeled cetuximab, a chimeric anti-EGFR monoclonal antibody. Eur J Nucl Med Mol I. 2007;34(6):850. [DOI] [PubMed] [Google Scholar]
- 53. Song IH, Lee TS, Park YS, et al. Immuno-PET imaging and radioimmunotherapy of 64Cu-/177Lu-labeled anti-EGFR antibody in esophageal squamous cell carcinoma model. J Nucl Med. 2016;57(7):1105–1111. [DOI] [PubMed] [Google Scholar]
- 54. Day KE, Sweeny L, Kulbersh B, Zinn KR, Rosenthal EL. Preclinical comparison of near-infrared-labeled cetuximab and panitumumab for optical imaging of head and neck squamous cell carcinoma. Mol Imaging Biol. 2013;15(6):722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cohen R, Stammes MA, de Roos IH, Stigter-van Walsum M, Visser GW, van Dongen GA. Inert coupling of IRDye800CW to monoclonal antibodies for clinical optical imaging of tumor targets. EJNMMI Res. 2011;1(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cohen R, Vugts DJ, Stigter-Van WM, Visser GW, van Dongen GA. Inert coupling of IRDye800CW and zirconium-89 to monoclonal antibodies for single- or dual-mode fluorescence and PET imaging. Nat Protoc. 2013;8(5):1010. [DOI] [PubMed] [Google Scholar]
- 57. Milenic DE, Wong KJ, Baidoo KE, et al. Cetuximab: preclinical evaluation of a monoclonal antibody targeting EGFR for radioimmunodiagnostic and radioimmunotherapeutic applications. Cancer Biother Radiol. 2008;23(5):619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Niu G, Li Z, Xie J, Le QT, Chen X. PET of EGFR antibody distribution in head and neck squamous cell carcinoma models. J Nucl Med. 2009;50(7):1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aerts HJ, Dubois L, Perk L, et al. Disparity between in vivo EGFR expression and 89Zr-labeled cetuximab uptake assessed with PET. J Nucl Med. 2009;50(1):123–131. [DOI] [PubMed] [Google Scholar]
- 60. Menke-van der Houven van Oordt CW, Gootjes EC, Huisman MC, et al. 89Zr-cetuximab PET imaging in patients with advanced colorectal cancer. Oncotarget. 2015;6(30):30384–30393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Even AJ, Hammingvrieze O, Van EW, et al. Quantitative assessment of zirconium-89 labeled cetuximab using PET/CT imaging in patients with advanced head and neck cancer: a theragnostic approach. Oncotarget. 2016;8(3):3870–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Van LJ, Even AJ, Aerts HJ, et al. PET imaging of zirconium-89 labelled cetuximab: a phase I trial in patients with head and neck and lung cancer. Radiother Oncol. 2016;122(2):267–273. [DOI] [PubMed] [Google Scholar]
- 63. Makris NE, Boellaard R, Van LA, et al. PET/CT-derived whole-body and bone marrow dosimetry of 89Zr-cetuximab. J Nucl Med. 2015;56(2):249–254. [DOI] [PubMed] [Google Scholar]
- 64. Xu N, Cai GW. Molecular imaging application of radioiodinated anti-EGFR human Fab to EGFR-overexpressing tumor xenografts. Anticancer Res. 2009;29(10):4005. [PubMed] [Google Scholar]
- 65. Dijk LKV, Hoeben BAW, Stegeman H, et al. 111 In-cetuximab-F(ab′) 2 SPECT imaging for quantification of accessible epidermal growth factor receptors (EGFR) in HNSCC xenografts. Radiother Oncol. 2013;108(3):484. [DOI] [PubMed] [Google Scholar]
- 66. van Dijk LK, Hoeben BA, Kaanders JH, Franssen GM, Boerman OC, Bussink J. Imaging of epidermal growth factor receptor expression in head and neck cancer with SPECT/CT and 111In-labeled cetuximab-F(ab′)2. J Nucl Med. 2013;54(12):2118. [DOI] [PubMed] [Google Scholar]
- 67. Bettenworth D, Reuter S, Hermann S, et al. Translational 18F-FDG PET/CT imaging to monitor lesion activity in intestinal inflammation. J Nucl Med. 2013;54(5):748. [DOI] [PubMed] [Google Scholar]
- 68. Goetz M, Ziebart A, Foersch S, et al. In vivo molecular imaging of colorectal cancer with confocal endomicroscopy by targeting epidermal growth factor receptor. Gastroenterology. 2010;138(2):435–446. [DOI] [PubMed] [Google Scholar]
- 69. Dubé PE, Yan F, Punit S, et al. Epidermal growth factor receptor inhibits colitis-associated cancer in mice. J Clin Invest. 2012;122(8):2780–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Turker NS, Heidari P, Kucherlapati R, Kucherlapati M, Mahmood U. An EGFR targeted PET imaging probe for the detection of colonic adenocarcinomas in the setting of colitis. Theranostics. 2014;4(9):893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nord K, Nilsson J, Nilsson B, Uhlén M, Nygren PÅ. A combinatorial library of an α-helical bacterial receptor domain. Protein Eng. 1995;8(6):601–608. [DOI] [PubMed] [Google Scholar]
- 72. Nilsson B, Moks T, Jansson B, et al. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1(2):107–113. [DOI] [PubMed] [Google Scholar]
- 73. Nord K, Gunneriusson E, Ringdahl J, Ståhl S, Uhlén M, Nygren P. Binding proteins selected from combinatorial libraries of an |[alpha]|-helical bacterial receptor domain. Nat Biotechnol. 1997;15(8):772. [DOI] [PubMed] [Google Scholar]
- 74. Cedergren L, Andersson R, Jansson B, Uhlén M, Nilsson B. Mutational analysis of the interaction between staphylococcal protein A and human IgG1. Protein Eng. 1993;6(4):441. [DOI] [PubMed] [Google Scholar]
- 75. Orlova A, Feldwisch J, Abrahmsén L, Tolmachev V. Update: affibody molecules for molecular imaging and therapy for cancer. Cancer Biother Radiol. 2007;22(5):573–584. [DOI] [PubMed] [Google Scholar]
- 76. Friedman M, Nordberg E, Höidén-Guthenberg I, et al. Phage display selection of Affibody molecules with specific binding to the extracellular domain of the epidermal growth factor receptor. Protein Eng Des Sel. 2007;20(4):189–199. [DOI] [PubMed] [Google Scholar]
- 77. Nordberg E, Friedman M, Göstring L, et al. Cellular studies of binding, internalization and retention of a radiolabeled EGFR-binding affibody molecule. Nucl Med Biol. 2007;34(6):609–618. [DOI] [PubMed] [Google Scholar]
- 78. Nordberg E, Orlova A, Friedman M, et al. In vivo and in vitro uptake of 111In, delivered with the affibody molecule (ZEGFR:955)2, in EGFR expressing tumour cells. Oncol Rep. 2008;19(4):853–857. [DOI] [PubMed] [Google Scholar]
- 79. Friedman M, Orlova A, Johansson E, et al. Directed evolution to low nanomolar affinity of a tumor-targeting epidermal growth factor receptor-binding Affibody molecule. J Mol Biol. 2008;376(5):1388–1402. [DOI] [PubMed] [Google Scholar]
- 80. Uhlén M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984;259(3):1695–1702. [PubMed] [Google Scholar]
- 81. Myers JK, Oas TG. Preorganized secondary structure as an important determinant of fast protein folding. Nat Struct Biol. 2001;8(6):552–558. [DOI] [PubMed] [Google Scholar]
- 82. Tolmachev V, Orlova A, Nilsson FY, Feldwisch J, Wennborg A, Abrahmsén L. Affibody molecules: potential for in vivo imaging of molecular targets for cancer therapy. Expert Opin Biol Ther. 2007;7(4):555–568. [DOI] [PubMed] [Google Scholar]
- 83. Miao Z, Ren G, Liu H, Jiang L, Cheng Z. Cy5.5-labeled affibody molecule for near-infrared fluorescent optical imaging of epidermal growth factor receptor positive tumors. J Biomed Opt. 2010;15(3):218–226. [DOI] [PubMed] [Google Scholar]
- 84. Qi S, Zheng M, Liu H, Xu Y, Feng Y, Cheng Z. Evaluation of four affibody-based near-infrared fluorescent probes for optical imaging of epidermal growth factor receptor positive tumors. Bioconjugate Chem. 2012;23(6):1149–1156. [DOI] [PubMed] [Google Scholar]
- 85. Wang Y, Telmer CA, Schmidt BF, et al. Fluorogen activating protein–affibody probes: modular, no-wash measurement of epidermal growth factor receptors. Bioconjugate Chem. 2015;26(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhao P, Yang X, Qi S, et al. Molecular imaging of hepatocellular carcinoma xenografts with epidermal growth factor receptor targeted affibody probes. Biomed Res Int. 2013;2013(1):759057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Miao Z, Ren G, Liu H, Qi S, Wu S, Cheng Z. PET of EGFR expression with an 18F-labeled affibody molecule. J Nucl Med. 2012;53(7):1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Miao Z, Ren G, Liu H, Jiang L, Cheng Z. Small-animal PET imaging of human epidermal growth factor receptor positive tumor with a 64Cu labeled affibody protein. Bioconjugate Chem. 2010;21(5):947–954. [DOI] [PubMed] [Google Scholar]
- 89. Yang M, Cheng K, Qi S, et al. Affibody modified and radiolabeled gold-iron oxide hetero-nanostructures for tumor PET, optical and MR imaging. Biomaterials. 2013;34(11):2796–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tolmachev V, Orlova A. Influence of labelling methods on biodistribution and imaging properties of radiolabelled peptides for visualisation of molecular therapeutic targets. Curr Med Chem. 2010;17(24):2636–2655. [DOI] [PubMed] [Google Scholar]
- 91. Tolmachev V, Rosik D, Wållberg H, et al. Imaging of EGFR expression in murine xenografts using site-specifically labelled anti-EGFR 111In-DOTA-Z EGFR:2377 affibody molecule: aspect of the injected tracer amount. Eur J Nucl Med Mol Imaging. 2010;37(3):613–622. [DOI] [PubMed] [Google Scholar]
- 92. Malmberg J, Tolmachev V, Orlova A. Imaging agents for in vivo molecular profiling of disseminated prostate cancer – targeting EGFR receptors in prostate cancer: comparison of cellular processing of [111In]-labeled affibody molecule Z(EGFR:2377) and cetuximab. Int J Oncol. 2011;38(4):1137–1143. [DOI] [PubMed] [Google Scholar]
- 93. Andersson KG, Oroujeni M, Garousi J, et al. Feasibility of imaging of epidermal growth factor receptor expression with ZEGFR:2377 affibody molecule labeled with 99mTc using a peptide-based cysteine-containing chelator. Int J Oncol. 2016;49(6):2285–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cheng Q, Wållberg H, Grafström J, Li L, Thorell JO. Preclinical PET imaging of EGFR levels: pairing a targeting with a non-targeting Sel-tagged affibody-based tracer to estimate the specific uptake. EJNMMI Res. 2016;6(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Garousi J, Andersson KG, Mitran B, et al. PET imaging of epidermal growth factor receptor expression in tumours using 89Zr-labelled ZEGFR:2377 affibody molecules. Int J Oncol. 2016;48(4):1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Vosjan MJ, Perk LR, Roovers RC, et al. Facile labelling of an anti-epidermal growth factor receptor nanobody with 68Ga via a novel bifunctional desferal chelate for immuno-PET. Eur J Nucl Med Mol Imaging. 2011;38(4):753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Huang L, Gainkam LV, Vanhove C, et al. SPECT imaging with 99mTc-labeled EGFR-specific nanobody for in vivo monitoring of EGFR expression. Mol Imaging Biol. 2008;10(3):167. [DOI] [PubMed] [Google Scholar]
- 98. Hackel BJ, Kimura RH, Gambhir SS. Use of (64)Cu-labeled fibronectin domain with EGFR-overexpressing tumor xenograft: molecular imaging. Int J Med Radiol. 2012;263(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mouratou B, Schaeffer F, Guilvout I, et al. Remodeling a DNA-binding protein as a specific in vivo inhibitor of bacterial secretin PulD. Proc Natl Acad Sci U S A. 2007;104(46):17983–17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Goux M, Becker G, Gorré H, et al. Nanofitin as a new molecular imaging agent for diagnosis of EGFR overexpressing tumors. Bioconjugate Chem. 2017;28(9):2361–2371. [DOI] [PubMed] [Google Scholar]
- 101. Song Y, Xiao Z, Wang K, et al. Development and evaluation of18F-IRS for molecular imaging mutant EGF receptors in NSCLC. Scientific Rep. 2017;7(1):3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kimura H, Okuda H, Arimitsu K, et al. Development of a PET imaging probe for discrimination of secondary mutation in the epidermal growth factor receptor. J Nucl Med. 2016;57(Suppl 2):1060. [Google Scholar]
- 103. Dai D, Li XF, Wang J, et al. Predictive efficacy of (11) C-PD153035 PET imaging for EGFR-tyrosine kinase inhibitor sensitivity in non-small cell lung cancer patients. Int J Cancer. 2016;138(4):1003. [DOI] [PubMed] [Google Scholar]
- 104. Meng X, Loo BW, Jr, Ma L, Murphy JD, Sun X, Yu J. Molecular imaging with 11C-PD153035 PET/CT predicts survival in non-small cell lung cancer treated with EGFR-TKI: a pilot study. J Nucl Med. 2011;52(10):1573–1579. [DOI] [PubMed] [Google Scholar]
- 105. Zhang Z, Zhao X, Ding C, Wang J, Zhang J, Wang F. (99m)Tc-3PRGD2 SPECT/CT imaging for monitoring early response of EGFR-TKIs therapy in patients with advanced-stage lung adenocarcinoma. Cancer Biother Radiol. 2016;31(7):238. [DOI] [PubMed] [Google Scholar]
- 106. Sun X, Xiao Z, Chen G, et al. A PET imaging approach for determining EGFR mutation status for improved lung cancer patient management. Sci Transl Med. 2018;10(431):eaan8840. [DOI] [PubMed] [Google Scholar]