Abstract
Epigenetics refers to the regulation of gene expression mainly by changes in DNA methylation and modifications of histone proteins without altering the actual DNA sequence. While epigenetic modifications are essential for normal cell differentiation, several driver mutations in leukemic pathogenesis have been identified in genes that affect epigenetic processes, such as DNA methylation and histone acetylation. Several therapeutic options to target epigenetic alterations in acute myeloid leukemia (AML) have been successfully tested in preclinical studies and various drugs have already been approved for use in clinical practice. Among these already approved therapeutics are hypomethylating agents (azacitidine and decitabine) and isocitrate dehydrogenase inhibitors (ivosidenib, enasidenib). Other agents such as bromodomain-containing epigenetic reader proteins and histone methylation (e.g. DOT1L) inhibitors are currently in advanced clinical testing. As several epigenetic therapies have only limited efficacy when used as single agents, combination therapies that target AML pathogenesis at different levels and exploit synergistic mechanisms are also in clinical trials. Combinations of either epigenetic therapies with conventional chemotherapy, different forms of epigenetic therapies, or epigenetic therapies with immunotherapy are showing promising early results. In this review we summarize the underlying pathophysiology and rationale for epigenetically-based combination therapies, review current preclinical and clinical data and discuss the future directions of epigenetic therapy combinations in AML.
Keywords: acute myelogenous leukemia, acute myeloid leukemia, AML, combination therapy, DNA methylation, epigenetic therapy, histones, histone deacetylase inhibitors, hypomethylating agent
Introduction
Acute myeloid leukemia (AML) is the most common form of acute leukemias in adults and, while pathogenetically heterogenous, it is typically caused by genetic events affecting hematopoietic progenitor or stem cells leading to the clonal proliferation of abnormally differentiated or undifferentiated myeloid cells.1 Despite the identification of several genetic abnormalities underlying AML, the mainstay of AML therapy for fit patients for several decades has been intensive induction chemotherapy with anthracyclines and cytarabine, followed by consolidation chemotherapy or allogeneic hematopoietic stem cell transplantation.2,3 However, intensive chemotherapy is generally only an option for younger and medically fit patients. Unfortunately, the majority of AML patients are older than 65 years with multiple comorbidities and are often ineligible for intensive chemotherapy with long-term survival rates of 10% or less.4–6
Histone proteins provide a scaffold for storage of DNA within the nucleus of eukaryotic cells. The macromolecular complex of DNA and histone proteins is referred to as chromatin.7 Chromatin modification altering interactions between DNA and histones is a highly dynamic process in cells affecting transcription, DNA repair and replication.7 Epigenetics are most commonly defined as changes to the chromatin structure which can occur in the form of DNA methylation, histone modifications, and changes to higher-order chromatin structures that ultimately affect gene expression.8,9 Epigenetic regulators can be broadly classified as ‘writers’ (e.g. DNA and histone methyltransferases), ‘erasers’ (e.g. histone deacetylase) or ‘readers’ (e.g. bromodomain-containing proteins).10 The importance of epigenetics in cancer development has been well documented for several decades for both solid and hematologic malignancies.11,12 Especially in AML, several specific mutations affecting epigenetic processes such as histone modification [Enhancer of Zeste Homologue 2 (EZH2) and the additional sex combs-like gene (ASXL1)], regulation of DNA methylation (DNMT3A, TET2) and enzymes regulating metabolism (IDH1/2) with epigenetic consequences have been identified as important players in pathogenesis of the disease and often with prognostic implications.2,10,13–15
DNA methylation and DNA hydroxymethylation are key epigenetic pathways that have been linked to malignant transformation by inactivating tumor suppressor genes.7,16 Mutations in genes affecting DNA methylation (e.g. DNMT3A) and demethylation (e.g. TET2) often cause silencing of target genes and are found in up to 22% and 23% of AML patients, respectively.17,18 DNA methyltransferase (DNMT) inhibitors, which are also known as hypomethylating agents (HMAs), such as 5-azacytidine (5-AZA) and its analogue 5-aza-2’deoxycytidine (decitabine) have been used for over a decade in the treatment of patients with AML who are unfit for intensive induction chemotherapy and those with myelodysplastic syndromes (MDS). Trials have shown a significantly prolonged survival for treatment with HMAs compared with conventional care in MDS, with trends for better survival in AML, but the therapeutic effects of HMAs seen in clinical trials have been difficult to reproduce in the real-world setting and treatment failure is common.19–22
Besides DNA methylation, histone acetylation is a highly dynamic process of modifying gene transcription that is tightly regulated by the competing activity of histone lysine acetyltransferases and histone deacetylases (HDACs) with histone acetylation often leading to a more accessible chromatin structure that promotes gene transcription.7 Mutations in other genes affecting histone modifying enzymes such as EZH2 and ASXL1 are found in up to 30% of AML patients.13,14,23 HDAC inhibitors are a heterogenous group of molecules that increase histone acetylation which promote transcription of various genes mediating cell differentiation, cell cycle regulation and apoptosis.24 Several studies using HDAC inhibitors as monotherapy for AML have yielded disappointing results with response rates less than 20%.24
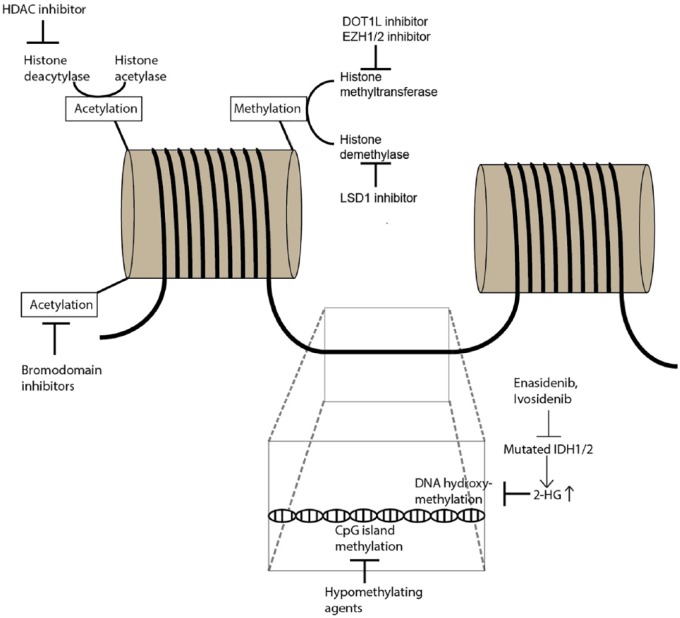
Overall, the therapeutic efficacy of HMAs and HDAC inhibitors are limited when used as single agents. Combination strategies of epigenetic therapy with either conventional chemotherapy, immunotherapy, or other forms of targeted therapies such as fms-related tyrosine kinase 3 (FLT3) inhibitors or the B-cell leukemia/lymphoma-2 (BCL-2) inhibitor venetoclax are currently in different stages of clinical testing and will be the focus of this review. Figure 1 illustrates epigenetic mechanisms and potential therapeutic targets.
Figure 1.
Overview of epigenetic mechanisms and selected therapeutic interventions.
Epigenetics refer to the modification of the chromatin structure without altering the base pair sequence of the DNA itself which is an essential process for regulating gene transcription and cell differentiation in both physiological conditions and malignant cell transformation. Epigenetic modifications can occur in the form of DNA methylation/hydroxymethylation, histone protein modifications (acetylation, methylation) and changes to higher-order chromatin structures. Methylation of CpG islands in the DNA generally suppress gene transcription and is mediated by DNA methyltransferases (DNMT). Alterations in DNA methylation have been linked to AML development and treatment with hypomethylating agents (e.g. azacitidine, decitabine) that inhibit DNMT has been successfully used in AML patients. On the other hand, DNA hydroxymethylation enhances gene transcription and is mediated by α-ketoglutarate-dependent enzymes such as TET2. IDH1/2 mutations lead to the formation of the oncometabolite 2-hydroxyglutarate instead of α-ketoglutarate which blocks DNA hydroxymethylation. The action of mutated IDH1/2 can be blocked by enasidenib and ivosidenib which restores function of enzymes orchestrating DNA hydroxymethylation. The DNA double-strand is stored in cells as a complex with histone proteins. Acetylation of histone proteins reduces the access of transcription factors to the DNA strand and thereby prevents gene transcription. Histone acetylation status is regulated by balancing the activity of histone deacetylases and histone acetylases which can be therapeutically targeted by bromodomain inhibitors and histone deacetylase (HDAC) inhibitors. Methylation and demethylation of histone proteins can occur at different sites of the histone molecule and is mediated by histone methyltransferases and histone demethylases. DOT1L is a histone H3K79 methyltransferase while EZH1/2 methylates histone H3K27 and both have both implicated in leukemogenesis and can be targeted by specific inhibitors. Histone demethylation can be blocked by LSD1 inhibitors.
Combination of HMAs with other epigenetic therapy
Combinations of HMAs and HDAC inhibitors
In vitro studies showed a synergistic effect of HDAC inhibitors and HMAs25 leading to several clinical trials that combined HMAs and HDAC inhibitors in both AML and MDS (Table 1).26–33 While most studies that showed synergistic effects have been single-arm studies, subsequent multi-arm studies comparing a combination of HMAs and HDAC inhibitors with HMA monotherapy have yielded disappointing results. Two large phase II trials combining 5-AZA with HDAC inhibitors (entinostat or vorinostat) failed to provide any survival benefit compared with 5-AZA monotherapy.28,30,31 This might be due to higher rates of hematologic side effects in the combination therapy groups that led to earlier discontinuation of the treatment. As a molecular correlate of the lower response rate for the combination therapy, the reversal of promoter methylation was lower compared with 5-AZA monotherapy.30 Additionally, the HDAC inhibitors used in these studies are a very heterogenous group in terms of their cellular targets and these pleotropic effects may have contributed to the excess toxicity seen in clinical trials leading to shortened treatment duration and insufficient drug exposure as potential explanations for the lack of clinical efficacy. Furthermore, reversal of histone acetylation may only be one of their mechanisms of action and additional biomarkers to predict response are needed.24,34 Future challenges for this combination approach of HMAs and HDAC inhibitors that need to be addressed are optimization of the sequence and dose of drug administration as pharmacodynamic antagonism might have been an issue in these initial trials as well as the choice of the HDAC inhibitor itself with a need for more selective HDAC inhibitors. However, both entinostat which specifically targets histone deacetylases and the less selective drug vorinostat which is also acting on other protein deacetylases have yielded comparable results at least for MDS but this might not necessarily be true for AML as well.30,31 It remains to be seen if the newer HDAC inhibitors such as belinostat, pracinostat, or panobinostat provide any additional benefit.34,35 So far, data from a phase II study in elderly patients with AML (ClinicalTrials.gov identifier: NCT01912274) testing the pan-HDAC inhibitor pracinostat in combination with 5-AZA showed a median overall survival (OS) of 19.1 months and a composite complete remission (CRc) rate of 52% which exceeds historical data for 5-AZA alone36 and has led to a phase III trial of 5-AZA ± pracinostat that is currently recruiting patients (ClinicalTrials.gov identifier: NCT03151408). In high-risk MDS patients, however, the combination of pracinostat and 5-AZA failed to improve outcomes which is potentially related to a higher rate of adverse events in the combination group that led to an earlier discontinuation of treatment.37 Finally, guadecitabine (SGI-110), a novel decitabine analogue with a better bioavailability due to greater resistance to deamination, has yielded promising response rates in both elderly newly diagnosed AML patients and relapsed/refractory (RR)-AML making it an interesting target for combination therapy with HDAC inhibitors38,39
Table 1.
Overview of selected clinical trials for combination of HMAs and HDAC inhibitors in AML treatment.
| Drug combination | Phase | n | Inclusion criteria | Outcomes | Trial registration | Reference |
|---|---|---|---|---|---|---|
| 5-AZA + Vorinostat + gemtuzumab ozogamicin | I/II | 52 | RR-AML | CR: 23%, CRi: 19% | NCT00895934 | Walter and colleagues26 |
| 5-AZA + valproic acid + ATRA | I/II | 53 | AML or HR-MDS | ORR: 42% (pretreated), 52% (untreated); median response duration: 26 weeks |
NCT00326170 | Soriano and colleagues 27 |
| 5-AZA ± entinostat | II | 47 | Therapy-related AML or MDS | Median OS: 13 months (5-AZA alone) versus 6 months (combination) HN: 46% (5-AZA alone) versus 17% (combination) |
NCT00313586 | Prebet and colleagues28 |
| 5-AZA ± entinostat | II | 149 | HR-MDS, AML | HN: 32% (5-AZA alone) versus 27% (combination); median OS: 18 months (5-AZA alone) versus 13 months (combination) |
NCT00313586 | Prebet and colleagues30 |
| 5-AZA + phenylbutyrate | II | 29 | HR-MDS, ND-AML, RR-AML | ORR: 38% (4 CR) | Gore and colleagues33 | |
| Decitabine + vorinostat | I | 71 | RR or ND-AML, IR- or HR-MDS | Response rate: concurrent versus sequential schedule in ND-AML (46% versus 14%), RR-AML (15% versus 0%) and MDS (60% versus 0%) | NCT00479232 | Kirschbaum and colleagues29 |
| Decitabine + valproic acid | I/II | 54 | ND-AML, RR-AML, HR-MDS | ORR: 22% (10 CR) median OS: 15.3 months in responders versus 4.9 months in nonresponders; untreated: ORR: 50% |
NCT00075010 | Garcia-Manero and colleagues40 |
| Decitabine + valproic acid | II | 149 | HR-MDS, ND-AML | ORR: 51% (decitabine alone) versus 35% (combination); median OS: 9.6 months (decitabine alone) versus 7.9 months (combination) |
NCT00414310 | Issa and colleagues 32 |
| Pracinostat + 5-AZA | II | 50 | Age > 65 years, ND-AML, secondary AML | CRc: 52%, median OS: 19.1 months | NCT01912274 | Garcia-Manero and colleagues36 |
5-AZA, 5-azacytidine; AML, acute myeloid leukemia; CR, complete remission; CRc, composite complete remission; Cri, complete remission with incomplete cell count recovery; HN, hematologic normalization; HR-MDS, high-risk myelodysplastic syndrome; IR-MDS, intermediate-risk myelodysplastic syndrome; NCT, ClinicalTrials.gov identifier; ND, new diagnosis; ORR, overall response rate; OS, overall survival; RR, relapsed/refractory.
Combination of HMAs and isocitrate dehydrogenase inhibitors
Methylation of cytosine-guanine dinucleotides (CpG) within the DNA is one of the key epigenetic mechanisms that regulate gene transcription and is mediated mainly by a tightly-controlled enzyme called DNA methyltransferase.11,41 On the other hand, 5-hydroxymethylation of these cytosine residues by α-ketoglutarate-dependent enzymes such as ten eleven translocation (TET) DNA methylases leads to DNA demethylation.42,43 Under physiological conditions, isocitrate dehydrogenase (IDH) catalyzes the conversion of isocitrate to α-ketoglutarate in the Krebs cycle. However, mutations in IDH1 and IDH2, which are observed in 8% and 12% of AML patients, respectively, lead to the generation of the neometabolite 2-hydroxyglutarate (2-HG).44 By competitively inhibiting α-ketoglutarate-dependent enzymes such as egg-laying-defective nine (EGL-9) prolyl hydroxylases, TET DNA methylases, and Jumonji C (JmjC) domain-containing histone demethylases, 2-HG causes DNA and histone hypermethylation leading to an impaired differentiation of myeloid precursor cells.45,46 IDH inhibitors have been shown to lower levels of 2-HG and to restore differentiation of leukemic cells which ultimately led to overall response rates (ORRs) of around 40% in RR-AML patients and provided proof of principle that epigenetic therapies are a promising target in AML treatment.44,47,48 However, potential nonepigenetic, leukemogenic effects of IDH mutations include impaired DNA repair by homologous recombination and impaired regulation of the transcription factor HIF1α which has been shown to inhibit hematopoietic stem cell and leukemic cell proliferation.49–51 The effect of IDH inhibitors on these processes is still incompletely understood and the subject of ongoing research.
Since one of the mechanisms by which IDH mutations contribute to leukemogenesis is DNA hypermethylation, combination therapy of IDH inhibitors and HMAs seems to be a promising therapeutic strategy. Preclinical data have suggested a synergistic effect of enasidenib and 5-AZA leading to the initiation of a clinical phase I/II trial of enasidenib and 5-AZA (ClinicalTrials.gov identifier: NCT02677922) and a phase III trial of ivosidenib and 5-AZA for newly diagnosed AML patients ineligible for standard intensive chemotherapy (ClinicalTrials.gov identifier: NCT03173248).45,52,53 While neither of these trials has been published in a peer-reviewed journal yet, data available in abstract form seem promising with an ORR of 73% [n = 11; complete remission (CR): 50%] in treatment-naïve, IDH1-mutated AML patients treated with ivosidenib and 5-AZA.52 Preliminary data for the enasidenib + 5-AZA trial showed a 66% response rate (n = 6 patients; CR: 33%) in patients with newly diagnosed AML with nausea and hyperbilirubinemia being the most common adverse events.54
Combination of HMAs and novel epigenetic therapies
So-called ‘epigenetic readers’ comprise a class of proteins that specifically bind to distinct DNA or histone modifications and constitute the third class of epigenetic regulators besides ‘epigenetic writers’ and ‘epigenetic erasers’.10 Rearrangements of the MLL/KMT2A gene are one of the first abnormalities in epigenetic regulators that has been linked to leukemogenesis and are associated with a poor prognosis.55,56 Additionally, the MLL gene can fuse with several different other genes leading to the interaction with the histone H3K79 methyltransferase DOT1L causing the downstream transcriptional activation of various genes such as HOXA9 and MEIS that have been linked to leukemogenesis.10,57–59 Preclinical studies of small molecule inhibitors of DOT1L (EPZ004777, EPZ-5676) showed a high level of suppression of HOX expression leading to induction of cell differentiation and antileukemic activity.60–62 A phase I human study of a DOT1L inhibitor (EPZ-5676; pinometostat) in 51 AML patients with MLL gene rearrangements showed a decreased level of H3K79 methylation and inhibition of HOXA9 in some patients. However, the clinical benefit was very modest with formal clinical responses in only 2 out of 51 patients.63 Potential explanations for these results include the heterogeneity of MLL fusion proteins with different levels of sensitivity to DOT1L inhibition and uncertainty about the optimal dose of pinometostat. As aberrant HOX expression is present in all AML cases with nucleophosmin (NPM1) gene mutations and DOT1L inhibition has also been shown to be effective in DNMT3A-mutated patients these patient subgroups might be particularly responsive to this approach.60,61 The combination of pinometostat with HMAs such as 5-AZA has also been successfully tested in animal models and might be a feasible approach in humans as well.64
MLL fusion proteins also interact with BET bromodomains which are epigenetic readers involved in the regulation of RNA polymerase II promoting the transcription of important regulatory genes such as BCL2 and c-MYC.65 Various BET inhibitors have shown strong in vitro antileukemic effects especially in MLL-mutated AML65–68 leading to several early-phase clinical trials testing BET inhibitors [FT-1101 (ClinicalTrials.gov identifier: NCT02543879), MK-8628 (ClinicalTrials.gov identifier: NCT02698189), RO6870810 (ClinicalTrials.gov identifier: NCT02308761), GSK525762 (ClinicalTrials.gov identifier: NCT01943851), and INCB054329 (ClinicalTrials.gov identifier: NCT02431260)] for use in human AML patients.69 In a phase I trial using the BET inhibitor OTX015 (ClinicalTrials.gov identifier: NCT01713582), only 3 out of 41 pretreated patients (36 with AML) achieved a CR or CR with incomplete platelet count recovery (CRi)70 suggesting that BET inhibitors alone might not be an adequate treatment strategy. Therefore, further studies are warranted to identify predictive biomarkers and to develop strategies for combination therapy of BET inhibitors with FLT3-inhibitors, HMAs, or HDAC inhibitors which have intriguing in vitro data.71 Careful patient selection for future clinical trials is also warranted as preclinical studies have shown higher efficacy in FLT3-ITD-mutated and NPM1-mutated leukemia cells.72,73
Similar to DNA methylation, histone methylation and demethylation are highly dynamic processes that can lead to either activation or repression of transcription depending on the specific location of the modified lysine residue.74 Histone methylation is catalyzed by specific methyltransferases such as MLL1/2, DOT1L, and EZH1/2, while histone demethylation is mainly regulated by the activity of lysine-specific histone demethylase (LSD) 1 and LSD2.74,75 Mutations in the involved genes and resulting overexpression of the encoded enzymes have been documented in both primary and secondary AML.74 Pharmacological inhibition of LSD1 (ORY-1001, GSK2879552, IMG-7289) has been shown in animal models to induce differentiation of AML blasts and has been tested in phase I/II clinical trials (ClinicalTrials.gov identifiers: NCT02177812, NCT02842827, EUDRACT no. 2013-002447-29).76–80 While a phase I trial of the LSD1 inhibitor GSK2879552 has been stopped recently due to a negative risk-benefit assessment, preclinical data suggest synergistic effects of the combination of LSD1 inhibitors with the HDAC inhibitor panobinostat and all-trans-retinoic acid (ATRA).81 LSD1 inhibition has been shown to increase HDAC inhibition and to block activity of the transcription factor c-Myc which can be further enhanced by addition of HDAC inhibitors.77,81
EZH1 and EZH2 are epigenetic regulators that methylate histones (H3K27) and repress transcription of target genes. Dysfunction of EZH1/2 has been associated with unregulated self-renewal of leukemic stem cells and a poor prognosis of AML and MDS patients harboring these mutations.82,83 Recent preclinical studies showed synergistic effects of EZH2 inhibition with the HMA decitabine and the HDAC inhibitor panobinostat.83–85 The underlying mechanism is the enhanced effect on gene silencing by simultaneously preventing DNA methylation by HMAs or histone acetylation with HDAC inhibitors in combination with increasing the pro-transcriptional effect of histone methylation by EZH2 inhibition.83 Additionally, trimethylation of histone H3K27 by EZH2 has been shown to mark genes for silencing by DNA methylation and is one of the mechanisms that has been linked with decitabine resistance.86–88 However, neither of these combinations has yet been tested in clinical trials.
Combination of epigenetic therapy with conventional chemotherapy
Combination of HMAs and conventional chemotherapy
Several in vitro studies have shown synergistic antileukemic effects of HMAs and anthracycline or cytarabine, which might be explained by the ability of 5-AZA to induce deoxycytidine kinase which phosphorylates cytarabine to its active phosphorylated form.89–91 HMAs could also act as chemosensitizers that restore expression of tumor suppressor genes and thereby susceptibility to chemotherapy.92 The combination of HMAs and cytarabine is especially interesting for older patients with AML who are ineligible for intensive chemotherapy as both agents when used alone have only shown moderate and transient response rates.93,94 Initial phase I studies in AML patients tested the sequential application of HMAs followed by cytarabine and daunorubicin chemotherapy and showed CR rates of up to 83% in the absence of increased toxicity.92,95,96 However, a subsequent larger phase II study in elderly AML patients comparing 5-AZA + cytarabine/daunorubicin (‘7 + 3’) induction chemotherapy with induction chemotherapy alone not only failed to show any survival benefit but rather led to increased adverse events.97 Identification of molecular biomarkers as response predictors, fine-tuning of the treatment schedules (e.g. greater delay between 5-AZA and chemotherapy to decrease toxicity) and testing this combination in patients with lower cytogenetic risk might be valid approaches for future trials. There are several clinical trials currently active and enrolling patients (ClinicalTrials.gov identifiers: NCT03417427, NCT01839240, NCT02275663) with the largest of such being a phase II study aiming to recruit 200 patients for epigenetic priming with 5-AZA or decitabine as a single agent prior to standard chemotherapy (ClinicalTrials.gov identifier: NCT03164057). Table 2 provides an overview of selected published clinical trials combining epigenetic therapies with conventional chemotherapy.
Table 2.
Overview of selected clinical trials for combination of epigenetic therapies with conventional chemotherapy in AML treatment.
| Drug combination | Phase | n | Inclusion criteria | Outcomes | Trial registration | Reference |
|---|---|---|---|---|---|---|
| Decitabine + (7 + 3) versus 7 + 3 alone | I | 30 | ND-AML<60 years with less-than-favorable karyotype | CR: 83% | NCT00538876 | Scandura and colleagues92 |
| 5-AZA + (7+3) | I | 6 | ND-AML >60 years | Median OS: 266 days, no dose-limiting toxicity | NCT00915252 | Krug and colleagues95 |
| Decitabine + low-dose idarubicin/cytarabine | I/II | 30 | RR-AML, HR-MDS | CR: 67% | ChiCTR-OPC-15005771. | Ye and colleagues96 |
| 5-AZA + (7+3) versus 7 + 3 alone | II | 214 | ND-AML >60 years | Median OS: 15 months (combination) versus 21 months 5-AZA + (7 + 3) | NCT00915252 | Muller-Tidow and colleagues97 |
| Decitabine + clofarabine + idarubicin + cytarabine; followed by consolidation of decitabine + clofarabine + idarubicin + cytarabine | II | 54 | RR-AML (⩽ salvage 2) <65 years | CR: 48% CR/CRi; 46% proceed to allo-HSCT | NCT01794702 | Jain and colleagues98 |
| vorinostat + idarubicin + cytarabine | II | 75 | HR-MDS, ND-AML <65 years | ORR: 85% (76% CR) | NCT00656617 | Garcia-Manero and colleagues99 |
| Vorinostat + etoposide + cytarabine | I | 21 | RR-AML, RR-ALL, secondary AML, CML in blast crisis | Response rate: concurrent versus sequential schedule in ND-AML (46% versus 14%), RR-AML (15% versus 0%) and MDS (60% versus 0%) | NCT00357305 | Gojo and colleagues100 |
| panobinostat + idarubicin + cytarabine | I/II | 38 | ND-AML >65 years | CR: 64%; median OS: 17 months |
NCT00840346 | Ocio and colleagues101 |
| 7 + 3 versus idarubicin + high-dose cytarabine versus idarubicin + vorinostat | III | 738 | ND-AML <60 years | CR: 75–79% for all groups; better outcomes for 7 + 3 for favorable cytogenetics |
NCT0180233 | Garcia-Manero and colleagues102 |
5-AZA, 5-azacitidine; AML, acute myeloid leukemia; CML, chronic myelogenous leukemia; CR, complete remission; CRi, complete remission with incomplete cell count recovery; HR-MDS, high-risk myelodysplastic syndrome; HSCT, hematopoietic stem cell transplant; NCT, ClinicalTrials.gov identifier; ND, new diagnosis; ORR, overall response rate; OS, overall survival; Ref, reference; RR, relapsed/refractory.
Combination of HDAC inhibitors and conventional chemotherapy
While having only moderate antileukemic effects as single agents, HDAC inhibitors have shown synergistic activity in combination with various forms of conventional chemotherapy such as nucleoside analogues (cytarabine, fludarabine), anthracyclines, and topoisomerase inhibitors (etoposide).40,103–105 The underlying mechanism for this synergy is not completely understood, but it is hypothesized that HDAC inhibitors promote a more open chromatin structure that might allow for better access of topoisomerase inhibitors to the DNA and therefore higher efficacy of chemotherapeutics.105 Other studies investigating the synergy between doxorubicin and panobinostat implicated DNA double-strand breaks and activation of caspase-dependent apoptosis pathways in the antileukemic efficacy.104
This concept has also been tested in various clinical trials. In a phase II trial of vorinostat in combination with cytarabine and idarubicin of 75 newly diagnosed AML or high-risk MDS patients ORRs were 85% and addition of vorinostat did not lead to an increase in toxicity.99 Interestingly, 100% of patients with FLT3-ITD mutations responded and mutations in NRF2 and CYBB were identified as biomarkers associated with a prolonged survival.99 The SWOG 1203 study (ClinicalTrials.gov identifier: NCT0180233) was a phase III clinical trial comparing 7 + 3 induction chemotherapy, idarubicin + high-dose cytarabine, and idarubicin + vorinostat in 738 newly diagnosed AML patients younger than 60 years of age. CR rates were comparable for all treatment arms (75–79%) with significantly better outcomes for patients with favorable cytogenetics in the 7 + 3 arm.102 However, in a cohort of RR-AML or secondary AML patients, the combination of vorinostat with etoposide and cytarabine yielded a response rate of 33%.100 Another trial tested a combination of panobinostat with cytarabine and idarubicin in AML patients >65 years of age.101 Despite a reported longer relapse-free survival, addition of panobinostat in a subset of patients led to dose-limiting degrees of toxicity.101 Future challenges in this field include defining the optimal timing and combination partners as concomitant application of vorinostat and cytarabine had antagonistic effects while sequential administration led to synergy.100
Combination of epigenetic therapy with targeted therapy
Combination of epigenetic therapy and venetoclax
B-cell leukemia/lymphoma-2 (BCL-2) is an anti-apoptotic protein that inhibits cell death by blocking permeability of the mitochondrial outer membrane.106 Though first identified in follicular lymphoma, BCL-2 is also overexpressed in other hematologic malignancies including AML and has been implicated in leukemia stem cell survival.107,108 BCL-2 activity is regulated by a group of small molecules known as BH3-mimetics that bind to and inhibit the BH3 domain of BCL-2 proteins which releases proapoptotic factors from their BCL-2 binding site and triggers apoptosis.109
Venetoclax (ABT-199) is an oral, highly-specific BCL-2 inhibitor which is United States Food and Drug Administration (US FDA)-approved for the treatment of chronic lymphocytic leukemia.110,111 Venetoclax has recently been shown to be modestly effective as a single agent in the treatment of RR-AML or newly diagnosed AML patients who are ineligible for intensive chemotherapy.112 However, early-phase clinical data have shown both an impressive synergistic effect and acceptable safety profile of venetoclax in combination with HMAs (ClinicalTrials.gov identifier: NCT02203773; composite CR: 61%, median OS: 17.5 months) or low-dose cytarabine (ClinicalTrials.gov identifier: NCT02287233; CR/CRi: 62%; median OS: 11.4 months) in the frontline setting in elderly AML patients ineligible for intensive chemotherapy.107,113,114 Acknowledging the remarkable response rates that exceed historical outcomes with 5-AZA alone (composite CR: 28%, median OS: 10.4 months),115 the combination of 5-AZA and venetoclax received breakthrough designation by the US FDA and is currently tested in a phase III clinical trial against 5-AZA monotherapy.106 A similar trial of cytarabine in combination with venetoclax is also currently recruiting (ClinicalTrials.gov identifier: NCT03069352).
The combination of venetoclax and either HMAs or low-dose cytarabine appears to have activity in the AML salvage therapy setting based on data which reported objective response rates of 21% and a median OS of 3.0 months.107 While these numbers seem modest at a first glance, it must be kept in mind that 94% of these patients had been pretreated and 41% of the patients had failed ⩾3 previous lines of therapy.112
On a molecular basis this synergy can be explained by the fact that resistance to venetoclax is mediated by the antiapoptotic proteins BCL-XL and MCL1 which can be overcome by combination therapy with HMAs, daunorubicin or cytarabine.116,117 On the other hand, resistance of leukemic blasts to chemotherapy has been linked to overexpression of BCL-2.118 Therefore, combining venetoclax with HMAs targets resistance mechanisms that have hampered monotherapy with either of these agents and seems to make this combination highly effective.107 Of note, subgroup analysis has shown that patients with IDH1/2 mutations have higher response rates to venetoclax monotherapy which is due to the fact that IDH-mutant AML cells depend on BCL-2 for survival.112,119,120
Combination of epigenetic therapy and FLT3 inhibitors
Addition of the multikinase inhibitor midostaurin to intensive induction chemotherapy was recently shown to yield a higher response rate than chemotherapy alone leading to its US FDA-approval for the treatment of FLT3-mutated AML in conjunction with chemotherapy.121 One potential explanation for treatment failure in FLT3-mutated AML with FLT3 inhibitors is the persistence of leukemic stem cells in a protective bone marrow compartment; animal models have demonstrated that this barrier can be overcome by the combination of FLT3 inhibitors with HMAs.122,123 Another study suggested that one mechanism of resistance to FLT3 inhibitors is higher expression of FLT3 ligand following chemotherapy which could block the action of tyrosine kinase inhibitors on FLT3 kinase.124 Given that the expression of FLT3 ligand is lower in patients treated with HMAs, early-phase clinical trials combining HMAs with the FLT3 inhibitors sorafenib125 and midostaurin,126–128 especially in elderly and RR-AML patients, have been conducted. Despite one study reporting that the addition of midostaurin to HMAs did not provide any additional benefit,128 other studies have reported ORRs of about 25% and a median OS of 22 weeks, which is longer compared with historical data from 5-AZA or midostaurin as single agents.126,127,129,130
In addition to midostaurin, several more specific FLT3 inhibitors such as quizartinib and gilteritinib have been developed and are currently being tested in combination with HMAs (ClinicalTrials.gov identifiers: NCT03661307, NCT01892371, NCT02752035). To date, only abstract data from the combination of 5-AZA and quizartinib have been published. In a phase I/II study of 37 patients with both newly diagnosed and previously treated AML, MDS or chronic myelomonocytic leukemia with FLT3-ITD mutation the combination of 5-AZA and quizartinib showed an overall response (CR, CRi, CRp, partial remission (PR)) rate of 76% with a median OS of 13.4 months.131 An overview of selected completed studies of HMAs in combination with several forms of targeted therapies is provided in Table 3.
Table 3.
Overview of selected clinical trials for combination of hypomethylating agents and targeted therapies in AML treatment.
| Drug combination | Phase | n | Inclusion criteria | Outcomes | Trial registration | Reference |
|---|---|---|---|---|---|---|
| Decitabine ± posaconazole or 5-AZA + venetoclax | I | 47 | ND-AML<65 | CR/CRi: 61%; median OS: 17.5 months |
NCT02203773 | DiNardo and colleagues107 |
| 5-AZA + sorafenib | II | 37 | ND-AML >60 years, RR-AML | CR/CRi: 43%; median OS: 6.2 months ND-AML: CR/CRi: 67% RR-AML: CR/CRi: 33% |
NCT01254890 | Ravandi and colleagues125 |
| 5-AZA + midostaurin | I/II | 54 | RR-AML, ND-AML, HR-MDS, s-AML | ORR: 26% Median OS: 22 weeks |
NCT01202877 | Strati and colleagues127 |
| Decitabine + midostaurin | I | 16 | ND-AML >60 years, RR-AML | CR/CRi: 25%, median response duration: 107 days | NCT01130662 | Williams and colleagues126 |
| 5-AZA + Midostaurin | I/II | 17 | ND-AML >70 years, RR-AML, s-AML | ORR: 18% Median OS: 6 months No difference between ND-AML and RR-AML |
NCT01093573 | Cooper and colleagues128 |
| Quizartinib + 5-AZA or LDAC | I/II | 59 | ND and RR-AML, MDS, CMML | (1) Untreated: ORR: 92%, median OS: 18.6 months (2) Pretreated: ORR: 73%; median OS: 11.25 months |
NCT01892371 | Swaminathan and colleagues131 |
| 5-AZA + nivolumab | IB/II | 51 | RR-AML | CR/CRi: 18% Median OS: 9.3 months |
NCT02397720 | Daver and colleagues132 |
5-AZA, azacitidine; AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia; CR, complete remission; Cri, complete remission with incomplete cell count recovery; HR-MDS, high-risk myelodysplastic syndrome; LDAC, low-dose cytarabine; NCT, ClinicalTrials.gov identifier; ND, new diagnosis; ORR, overall response rate; OS, overall survival; Ref, reference; RR, relapsed/refractory; s-AML, secondary AML.
Combination of epigenetic therapy and immunotherapy
Evasion of immunosurveillance by epigenetic silencing of genes involved in immune recognition and effector T-cell function is an essential feature of cancer cells and contributes to the immunosuppressive tumor microenvironment.133 HMAs have been shown to increase the expression of leukemia-associated antigens such as NY-ESO-1 and MAGE-A that can potentially trigger an antileukemia immune response.134–138 Additionally, they contribute to immune system activation by increasing the expression of major histocompatibility complex (MHC)-I and co-stimulatory molecules (ICAM1, CD80, CD86).135,138,139 However, at the same time HMAs are also hampering antitumor immune response by upregulating the expression of immune checkpoint molecules such as programmed cell death (PD)-1/programmed death ligand (PD-L)1 and cytotoxic T-lymphocyte-associated protein (CTLA)-4 which may contribute to treatment failure with HMAs.140–142 However, the increased expression of PD-1/PD-L1 with HMA treatment may lead to a greater susceptibility of cancer cells to treatment with PD-1/PD-L1 inhibitors. Preclinical experiments have shown that pretreatment with 5-AZA led to an enhanced response to treatment with immunotherapeutics such as CTLA-4 inhibitors.143 Studies in non-small cell lung cancer patients have provided additional proof of principle that combing HMAs with PD-1/PD-L1 inhibitors sensitized patients to treatment with PD-1 inhibitors.
These findings have inspired various clinical trials that combine HMAs with various PD-1 inhibitors [nivolumab (ClinicalTrials.gov identifier: NCT02397720), and pembrolizumab (ClinicalTrials.gov identifier: NCT02845297)], PD-L1 inhibitors [durvalumab (ClinicalTrials.gov identifier: NCT02775903), and atezolizumab (ClinicalTrials.gov identifier: NCT02508870)], or the CTLA-4 inhibitor ipilimumab (ClinicalTrials.gov identifiers: NCT02890329, NCT02397720). One study demonstrated a 33% ORR (22% CR/CRi) and median OS of 6.3 months in RR-AML patients treated with nivolumab and 5-AZA combination therapy (n = 35), which appears to be better compared with historic controls of 5-AZA alone and the effect seemed to be long lasting.132 However, 11% of patients developed grade 3/4 immune-mediated adverse events that were controlled with corticosteroids except for one death from grade 4 pneumonitis.132 Of note, a clinical trial of 5-AZA with atezolizumab (ClinicalTrials.gov identifier: NCT02508870) has been discontinued due to safety concerns. Future studies to address the safety profile of checkpoint inhibitors are therefore warranted prior to their broader clinical application. There are some early data that suggest a different efficacy of PD-1 versus CTLA-4 inhibition in myeloid neoplasms making studies investigating the combination of different immune checkpoint inhibitors with HMAs an interesting area of research and such trials are currently ongoing [combination of nivolumab and ipilimumab with 5-AZA (ClinicalTrials.gov identifier: NCT02397720)].140 Immune checkpoint inhibitors could also be applied to control minimal residual disease as preclinical data suggest that immune checkpoint pathways contribute to immune system evasion and that these dormant leukemia cells are susceptible to T-cell-mediated cytolysis.144
Future directions
Our understanding of the epigenetic processes underlying AML is still incomplete. With further improvement in and wider availability of diagnostic techniques such as next-generation sequencing, an increasing number of mutations affecting epigenetic regulators is being discovered, but their precise impact on AML development and their suitability as therapeutic targets remains to be elucidated in preclinical experiments. However, identification of these mutations has already led to the development and approval of drugs that specifically target these mutations which has paved the way to a more individualized treatment approach for AML patients. Nevertheless, several important questions remain to be answered.
One of the major challenges remaining is the appropriate selection of patients who are most likely to benefit from an intervention. While targeted therapies such as IDH inhibition with ivosidenib and enasidenib only work in patients with IDH mutations, predicting treatment response to HMAs or HDAC inhibitors is difficult. Methylation of CpG islands has been suggested to be a biomarker predicting response to HMAs.145 However, subsequent studies showed that methylation status is dynamic while patients are undergoing treatment and that it depends rather on the methylation status of certain individual genes than on the overall CpG methylation status.146,147 Additionally, epigenetic therapies such as DOT1L and BET inhibition can be successful even if the target gene is not mutated at all which questions the utility of simple gene mutation testing and underlines the need for broader drug sensitivity testing.13 The combination of different mutations can predict treatment response as well which is seen in the higher response rate of patients with concomitant IDH mutations who are treated with venetoclax. Further studies are warranted to guide appropriate drug selection.112
Various studies have shown that targeting a single genetic mutation such as FLT3 is not sufficient to control or eradicate cancer. Basic research has shown the synergy of different therapeutics in combination can target leukemia cell escape mechanisms which have hampered therapeutic success with single agent therapies. The most promising approaches so far appear to be the use of HMAs in combination with immune checkpoint inhibitors and venetoclax. Additionally, it remains to be seen if more specific epigenetic therapies such as EZH2 or DOT1L inhibitors will increase the response rates compared with ‘older’ HMAs or HDAC inhibitors.
Finally, preleukemic hematopoietic stem cells already harbor some mutations in genes that are involved in epigenetic changes such as DNA methylation and histone modification.148 Interestingly, these progenitor cells can survive induction chemotherapy and have been linked to disease relapse.149 As some of the earliest mutations occurring during the transformation of normal HSCs to leukemia clones are seen in IDH1/2 and DNMT3A148 targeting these mutations either early in the disease course or to maintain minimal residual disease following chemotherapy might be an interesting target that is already being explored in clinical trials (e.g. ClinicalTrials.gov identifier: NCT01757535).150
Conclusion
Epigenetic therapy in AML is still in its infancy but is a rapidly evolving and highly promising field. While early forms of epigenetic therapies like the HMAs 5-AZA and decitabine have shown modest effects, more targeted therapies such as IDH inhibitors have shown a better response and survival rates in appropriately selected patients. Combination therapies of either epigenetic therapies with conventional chemotherapy, different forms of epigenetic therapies, or epigenetic therapies with immunotherapy promise even higher success rates even in elderly and RR-AML patients as they target cell proliferation at different levels. However, further research is needed to identify biomarkers that predict response to therapy and guide drug selection for individual patients.
Acknowledgments
Amer Zeidan is a Leukemia and Lymphoma Society Scholar in Clinical Research and is also supported by a NCI’s Cancer Clinical Investigator Team Leadership Award (CCITLA).
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: A.M.Z. had a consultancy with and received honoraria from AbbVie, Otsuka, Pfizer, Celgene, Ariad, Agios, Novartis, Acceleron, Astellas, Daiichi Sankyo and Takeda; and received honoraria from and was a speaker for Takeda. The remaining authors declare no competing financial interests.
ORCID iD: Jan Philipp Bewersdorf  https://orcid.org/0000-0003-3352-0902
https://orcid.org/0000-0003-3352-0902
Contributor Information
Jan Philipp Bewersdorf, Department of Internal Medicine, Section of Hematology, Yale University School of Medicine, New Haven, CT, USA.
Rory Shallis, Department of Internal Medicine, Section of Hematology, Yale University School of Medicine, New Haven, CT, USA.
Maximilian Stahl, Leukemia Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Amer M. Zeidan, Department of Internal Medicine, Section of Hematology, Yale University School of Medicine, 333 Cedar Street, PO Box 208028, New Haven, CT 06520-8055, USA.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017; 129: 424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood 2016; 127: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018; 559: 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Podoltsev NA, Stahl M, Zeidan AM, et al. Selecting initial treatment of acute myeloid leukaemia in older adults. Blood Rev 2017; 31: 43–62. [DOI] [PubMed] [Google Scholar]
- 6. Wang R, Zeidan AM, Halene S, et al. Health care use by older adults with acute myeloid leukemia at the end of life. J Clin Oncol 2017; 35: 3417–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012; 150: 12–27. [DOI] [PubMed] [Google Scholar]
- 8. Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med 2018; 378: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med 2012; 367: 647–657. [DOI] [PubMed] [Google Scholar]
- 10. Sun Y, Chen BR, Deshpande A. Epigenetic regulators in the development, maintenance, and therapeutic targeting of acute myeloid leukemia. Front Oncol 2018; 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358: 1148–1159. [DOI] [PubMed] [Google Scholar]
- 12. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 13. Stahl M, Kohrman N, Gore SD, et al. Epigenetics in cancer: a hematological perspective. PLoS Genet 2016; 12: e1006193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and prognosis in acute myeloid leukemia. N Engl J Med 2016; 374: 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cancer Genome Atlas Research N; Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011; 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marcucci G, Metzeler KH, Schwind S, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol 2012; 30: 742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metzeler KH, Maharry K, Radmacher MD, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 2011; 29: 1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol 2009; 10: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cashen AF, Schiller GJ, O’Donnell MR, et al. Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. J Clin Oncol 2010; 28: 556–561. [DOI] [PubMed] [Google Scholar]
- 21. Stahl M, DeVeaux M, Montesinos P, et al. Hypomethylating agents in relapsed and refractory AML: outcomes and their predictors in a large international patient cohort. Blood Adv 2018; 2: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeidan AM, Stahl M, DeVeaux M, et al. Counseling patients with higher-risk MDS regarding survival with azacitidine therapy: are we using realistic estimates? Blood Cancer J 2018; 8: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim TK, Gore SD, Zeidan AM. Epigenetic therapy in acute myeloid leukemia: current and future directions. Semin Hematol 2015; 52: 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ball B, Zeidan A, Gore SD, et al. Hypomethylating agent combination strategies in myelodysplastic syndromes: hopes and shortcomings. Leuk Lymphoma 2017; 58: 1022–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cameron EE, Bachman KE, Myohanen S, et al. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 1999; 21: 103–107. [DOI] [PubMed] [Google Scholar]
- 26. Walter RB, Medeiros BC, Gardner KM, et al. Gemtuzumab ozogamicin in combination with vorinostat and azacitidine in older patients with relapsed or refractory acute myeloid leukemia:a phase I/II study. Haematologica 2014; 99: 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soriano AO, Yang H, Faderl S, et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood 2007; 110: 2302–2308. [DOI] [PubMed] [Google Scholar]
- 28. Prebet T, Sun Z, Ketterling RP, et al. Azacitidine with or without Entinostat for the treatment of therapy-related myeloid neoplasm: further results of the E1905 North American leukemia intergroup study. Br J Haematol 2016; 172: 384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirschbaum M, Gojo I, Goldberg SL, et al. A phase 1 clinical trial of vorinostat in combination with decitabine in patients with acute myeloid leukaemia or myelodysplastic syndrome. Br J Haematol 2014; 167: 185–193. [DOI] [PubMed] [Google Scholar]
- 30. Prebet T, Sun Z, Figueroa ME, et al. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: results of the US Leukemia Intergroup trial E1905. J Clin Oncol 2014; 32: 1242–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sekeres MA, Othus M, List AF, et al. Randomized phase II study of azacitidine alone or in combination with lenalidomide or with vorinostat in higher-risk myelodysplastic syndromes and chronic myelomonocytic leukemia: North American intergroup study SWOG S1117. J Clin Oncol 2017; 35: 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Issa JP, Garcia-Manero G, Huang X, et al. Results of phase 2 randomized study of low-dose decitabine with or without valproic acid in patients with myelodysplastic syndrome and acute myelogenous leukemia. Cancer 2015; 121: 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gore SD, Baylin S, Sugar E, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res 2006; 66: 6361–6369. [DOI] [PubMed] [Google Scholar]
- 34. Stahl M, Gore SD, Vey N, et al. Lost in translation? Ten years of development of histone deacetylase inhibitors in acute myeloid leukemia and myelodysplastic syndromes. Expert Opin Investig Drugs 2016; 25: 307–317. [DOI] [PubMed] [Google Scholar]
- 35. Gimsing P. Belinostat: a new broad acting antineoplastic histone deacetylase inhibitor. Expert Opin Investig Drugs 2009; 18: 501–508. [DOI] [PubMed] [Google Scholar]
- 36. Garcia Manero G, Atallah E, Khaled SK, et al. A phase 2 study of pracinostat and azacitidine in elderly patients with acute myeloid leukemia (AML) not eligible for induction chemotherapy: response and long-term survival benefit. Blood 2016; 128: 100. [Google Scholar]
- 37. Garcia-Manero G, Montalban-Bravo G, Berdeja JG, et al. Phase 2, randomized, double-blind study of pracinostat in combination with azacitidine in patients with untreated, higher-risk myelodysplastic syndromes. Cancer 2017; 123: 994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roboz GJ, Kantarjian HM, Yee KWL, et al. Dose, schedule, safety, and efficacy of guadecitabine in relapsed or refractory acute myeloid leukemia. Cancer 2018; 124: 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kantarjian HM, Roboz GJ, Kropf PL, et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol 2017; 18: 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, et al. Phase 1/2 study of the combination of 5-aza-2’-deoxycytidine with valproic acid in patients with leukemia. Blood 2006; 108: 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003; 349: 2042–2054. [DOI] [PubMed] [Google Scholar]
- 42. Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ito S, D’Alessio AC, Taranova OV, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Amatangelo MD, Quek L, Shih A, et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 2017; 130: 732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abou Dalle I, DiNardo CD. The role of enasidenib in the treatment of mutant IDH2 acute myeloid leukemia. Ther Adv Hematol 2018; 9: 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012; 483: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. DiNardo CD, Stein EM, de Botton S, et al. Durable remissions with ivosidenib in IDH1-Mutated relapsed or refractory AML. N Engl J Med 2018; 378: 2386–2398. [DOI] [PubMed] [Google Scholar]
- 48. Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017; 130: 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nassereddine S, Lap CJ, Haroun F, et al. The role of mutant IDH1 and IDH2 inhibitors in the treatment of acute myeloid leukemia. Ann Hematol 2017; 96: 1983–1991. [DOI] [PubMed] [Google Scholar]
- 50. Sulkowski PL, Corso CD, Robinson ND, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med 2017; 9: pii: eaal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Losman JA, Looper RE, Koivunen P, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013; 339: 1621–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stein E, DiNardo CD, Jang JH, et al. AGILE: a phase 3, multicenter, randomized, placebo-controlled study of ivosidenib in combination with azacitidine in adult patients with previously untreated acute myeloid leukemia with an IDH1 mutation. J Clin Oncol 2018; 36(Suppl. 15): TPS7074. [Google Scholar]
- 53. DiNardo CD, Stein AS, Fathi AT, et al. Mutant isocitrate dehydrogenase (mIDH) inhibitors, enasidenib or ivosidenib, in combination with azacitidine (AZA): preliminary results of a phase 1b/2 study in patients with newly diagnosed acute myeloid leukemia (AML). Blood 2017; 130: 639. [Google Scholar]
- 54. DiNardo CD, Stein AS, Stein EM, et al. Mutant IDH (MIDH) inhibitors, ivosidenib or enasidenib, with azacitidine (AZA) in patients with acute myeloid leukemia (AML). Abstract #S1562 Presented at the EHA 23rd Congress, 17 June 2018, Stockholm, Sweden. [Google Scholar]
- 55. Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012; 366: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Caligiuri MA, Strout MP, Lawrence D, et al. Rearrangement of ALL1 (MLL) in acute myeloid leukemia with normal cytogenetics. Cancer Res 1998; 58: 55–59. [PubMed] [Google Scholar]
- 57. Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005; 121: 167–178. [DOI] [PubMed] [Google Scholar]
- 58. Stahl M, Lu BY, Kim TK, et al. Novel therapies for acute myeloid leukemia: are we finally breaking the deadlock? Target Oncol 2017; 12: 413–447. [DOI] [PubMed] [Google Scholar]
- 59. Stein EM, Tallman MS. Mixed lineage rearranged leukaemia: pathogenesis and targeting DOT1L. Curr Opin Hematol 2015; 22: 92–96. [DOI] [PubMed] [Google Scholar]
- 60. Rau RE, Rodriguez BA, Luo M, et al. DOT1L as a therapeutic target for the treatment of DNMT3A-mutant acute myeloid leukemia. Blood 2016; 128: 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kuhn MW, Hadler MJ, Daigle SR, et al. MLL partial tandem duplication leukemia cells are sensitive to small molecule DOT1L inhibition. Haematologica 2015; 100: e190–e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kuhn MW, Song E, Feng Z, et al. Targeting chromatin regulators inhibits leukemogenic gene expression in NPM1 mutant leukemia. Cancer Discov 2016; 6: 1166–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stein EM, Garcia-Manero G, Rizzieri DA, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 2018; 131: 2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Klaus CR, Iwanowicz D, Johnston D, et al. DOT1L inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs and hypomethylating agents in MLL-rearranged leukemia cells. J Pharmacol Exp Ther 2014; 350: 646–656. [DOI] [PubMed] [Google Scholar]
- 65. Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011; 478: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011; 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Saenz DT, Fiskus W, Qian Y, et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia 2017; 31: 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Coude MM, Braun T, Berrou J, et al. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget 2015; 6: 17698–17712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brinda B, Khan I, Parkin B, et al. The rocky road to personalized medicine in acute myeloid leukaemia. J Cell Mol Med 2018; 22: 1411–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Berthon C, Raffoux E, Thomas X, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol 2016; 3: e186–e195. [DOI] [PubMed] [Google Scholar]
- 71. Astorgues-Xerri L, Canet-Jourdan C, Bekradda M, et al. OTX015, a BET-bromodomain (BET-BRD) inhibitor, potentiates the in vitro effects of chemotherapy drugs and targeted agents in human leukemic cell lines. Eur J Cancer 2014; 50(Suppl. 6): 183. [Google Scholar]
- 72. Dawson MA, Gudgin EJ, Horton SJ, et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia 2014; 28: 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fiskus W, Sharma S, Qi J, et al. BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD. Mol Cancer Ther 2014; 13: 2315–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Castelli G, Pelosi E, Testa U. Targeting histone methyltransferase and demethylase in acute myeloid leukemia therapy. Onco Targets Ther 2018; 11: 131–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Magliulo D, Bernardi R, Messina S. Lysine-specific demethylase 1A as a promising target in acute myeloid leukemia. Front Oncol 2018; 8: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cusan M, Cai SF, Mohammad HP, et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU. 1- and C/EBPalpha-dependent enhancers in AML. Blood 2018; 131: 1730–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Harris WJ, Huang X, Lynch JT, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012; 21: 473–487. [DOI] [PubMed] [Google Scholar]
- 78. Fang J, Ying H, Mao T, et al. Upregulation of CD11b and CD86 through LSD1 inhibition promotes myeloid differentiation and suppresses cell proliferation in human monocytic leukemia cells. Oncotarget 2017; 8: 85085–85101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Maes T, Mascaro C, Tirapu I, et al. ORY-1001, a potent and selective covalent KDM1A Inhibitor, for the treatment of acute leukemia. Cancer Cell 2018; 33: 495–511. e12. [DOI] [PubMed] [Google Scholar]
- 80. Somervaille T, Salamero O, Montesinos P, et al. Safety, Phamacokinetics (PK), Pharmacodynamics (PD) and preliminary activity in acute leukemia of Ory-1001, a first-in-class inhibitor of lysine-specific histone demethylase 1A (LSD1/KDM1A): initial results from a first-in-human phase 1 study. Blood 2016; 128: 4060. [Google Scholar]
- 81. Fiskus W, Sharma S, Shah B, et al. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia 2014; 28: 2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fujita S, Honma D, Adachi N, et al. Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia 2018; 32: 855–864. [DOI] [PubMed] [Google Scholar]
- 83. Momparler RL, Cote S, Momparler LF, et al. Inhibition of DNA and histone methylation by 5-Aza-2’-Deoxycytidine (Decitabine) and 3-Deazaneplanocin-A on antineoplastic action and gene expression in myeloid leukemic cells. Front Oncol 2017; 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fiskus W, Wang Y, Sreekumar A, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood 2009; 114: 2733–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fiskus W, Buckley K, Rao R, et al. Panobinostat treatment depletes EZH2 and DNMT1 levels and enhances decitabine mediated de-repression of JunB and loss of survival of human acute leukemia cells. Cancer Biol Ther 2009; 8: 939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vire E, Brenner C, Deplus R, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006; 439: 871–874. [DOI] [PubMed] [Google Scholar]
- 87. Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007; 39: 232–236. [DOI] [PubMed] [Google Scholar]
- 88. Si J, Boumber YA, Shu J, et al. Chromatin remodeling is required for gene reactivation after decitabine-mediated DNA hypomethylation. Cancer Res 2010; 70: 6968–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Neil GL, Berger AE, Bhuyan BK, et al. Combination chemotherapy of L1210 leukemia with 1-beta-D-arabinofuranosylcytosine and 5-azacytidine. Cancer Res 1976; 36: 1114–1120. [PubMed] [Google Scholar]
- 90. Avramis VI, Mecum RA, Nyce J, et al. Pharmacodynamic and DNA methylation studies of high-dose 1-beta-D-arabinofuranosyl cytosine before and after in vivo 5-azacytidine treatment in pediatric patients with refractory acute lymphocytic leukemia. Cancer Chemother Pharmacol 1989; 24: 203–210. [DOI] [PubMed] [Google Scholar]
- 91. Li K, Hu C, Mei C, et al. Sequential combination of decitabine and idarubicin synergistically enhances anti-leukemia effect followed by demethylating Wnt pathway inhibitor promoters and downregulating Wnt pathway nuclear target. J Transl Med 2014; 12: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Scandura JM, Roboz GJ, Moh M, et al. Phase 1 study of epigenetic priming with decitabine prior to standard induction chemotherapy for patients with AML. Blood 2011; 118: 1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kantarjian HM, Thomas XG, Dmoszynska A, et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J Clin Oncol 2012; 30: 2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J Clin Oncol 2010; 28: 562–569. [DOI] [PubMed] [Google Scholar]
- 95. Krug U, Koschmieder A, Schwammbach D, et al. Feasibility of azacitidine added to standard chemotherapy in older patients with acute myeloid leukemia–a randomised SAL pilot study. PLoS One 2012; 7: e52695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ye XN, Zhou XP, Wei JY, et al. Epigenetic priming with decitabine followed by low-dose idarubicin/cytarabine has an increased anti-leukemic effect compared to traditional chemotherapy in high-risk myeloid neoplasms. Leuk Lymphoma 2016; 57: 1311–1318. [DOI] [PubMed] [Google Scholar]
- 97. Muller-Tidow C, Tschanter P, Rollig C, et al. Azacitidine in combination with intensive induction chemotherapy in older patients with acute myeloid leukemia: the AML-AZA trial of the study alliance leukemia. Leukemia 2016; 30: 555–561. [DOI] [PubMed] [Google Scholar]
- 98. Jain N, Ravandi F, Garcia-Manero G, et al. Decitabine followed by clofarabine, idarubicin, and cytarabine (DAC-CIA) in relapsed/refractory acute myeloid leukemia (AML). Blood 2016; 128: 2817. [Google Scholar]
- 99. Garcia-Manero G, Tambaro FP, Bekele NB, et al. Phase II trial of vorinostat with idarubicin and cytarabine for patients with newly diagnosed acute myelogenous leukemia or myelodysplastic syndrome. J Clin Oncol 2012; 30: 2204–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gojo I, Tan M, Fang HB, et al. Translational phase I trial of vorinostat (suberoylanilide hydroxamic acid) combined with cytarabine and etoposide in patients with relapsed, refractory, or high-risk acute myeloid leukemia. Clin Cancer Res 2013; 19: 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ocio EM, Herrera P, Olave MT, et al. Panobinostat as part of induction and maintenance for elderly patients with newly diagnosed acute myeloid leukemia: phase Ib/II panobidara study. Haematologica 2015; 100: 1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Garcia-Manero G, Othus M, Pagel JM, et al. SWOG S1203: a randomized phase III study of standard cytarabine plus daunorubicin (7+3) therapy versus idarubicin with high dose cytarabine (IA) with or without vorinostat (IA+V) in younger patients with previously untreated acute myeloid leukemia (AML). Blood 2016; 128: 901. [Google Scholar]
- 103. Kim MS, Blake M, Baek JH, et al. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res 2003; 63: 7291–7300. [PubMed] [Google Scholar]
- 104. Maiso P, Colado E, Ocio EM, et al. The synergy of panobinostat plus doxorubicin in acute myeloid leukemia suggests a role for HDAC inhibitors in the control of DNA repair. Leukemia 2009; 23: 2265–2274. [DOI] [PubMed] [Google Scholar]
- 105. Rosato R, Hock S, Dent P, et al. LBH-589 (panobinostat) potentiates fludarabine anti-leukemic activity through a JNK- and XIAP-dependent mechanism. Leuk Res 2012; 36: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Konopleva M, Letai A. BCL-2 inhibition in AML - an unexpected bonus? Blood 2018; 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. DiNardo CD, Rausch CR, Benton C, et al. Clinical experience with the BCL2-inhibitor venetoclax in combination therapy for relapsed and refractory acute myeloid leukemia and related myeloid malignancies. Am J Hematol 2018; 93: 401–407. [DOI] [PubMed] [Google Scholar]
- 108. Lagadinou ED, Sach A, Callahan K, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013; 12: 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Green DR. A BH3 mimetic for killing cancer cells. Cell 2016; 165: 1560. [DOI] [PubMed] [Google Scholar]
- 110. Roberts AW, Davids MS, Pagel JM, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med 2016; 374: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pullarkat VA, Newman EM. BCL2 Inhibition by venetoclax: targeting the Achilles’ heel of the acute myeloid leukemia stem cell? Cancer Discov 2016; 6: 1082–1083. [DOI] [PubMed] [Google Scholar]
- 112. Konopleva M, Pollyea DA, Potluri J, et al. Efficacy and biological correlates of response in a phase ii study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov 2016; 6: 1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wei A, Strickland SA, Roboz GJ, et al. Phase 1/2 study of venetoclax with low-dose cytarabine in treatment-naive, elderly patients with acute myeloid leukemia unfit for intensive chemotherapy: 1-year outcomes. Blood 2017; 130: 890. [Google Scholar]
- 114. DiNardo CD, Pollyea DA, Jonas BA, et al. Updated safety and efficacy of venetoclax with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 2017; 130: 2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Dombret H, Seymour JF, Butrym A, et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood 2015; 126: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bogenberger JM, Kornblau SM, Pierceall WE, et al. BCL-2 family proteins as 5-Azacytidine-sensitizing targets and determinants of response in myeloid malignancies. Leukemia 2014; 28: 1657–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Niu X, Zhao J, Ma J, et al. Binding of released Bim to Mcl-1 is a mechanism of intrinsic resistance to ABT-199 which can be overcome by combination with daunorubicin or cytarabine in AML cells. Clin Cancer Res 2016; 22: 4440–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pan R, Ruvolo VR, Wei J, et al. Inhibition of Mcl-1 with the pan-Bcl-2 family inhibitor (-)BI97D6 overcomes ABT-737 resistance in acute myeloid leukemia. Blood 2015; 126: 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chyla B, Daver N, Doyle K, et al. Genetic biomarkers of sensitivity and resistance to venetoclax monotherapy in patients with relapsed acute myeloid leukemia. Am J Hematol 2018; 93: E202–E205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Chan SM, Thomas D, Corces-Zimmerman MR, et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat Med 2015; 21: 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med 2017; 377: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Garz AK, Wolf S, Grath S, et al. Azacitidine combined with the selective FLT3 kinase inhibitor crenolanib disrupts stromal protection and inhibits expansion of residual leukemia-initiating cells in FLT3-ITD AML with concurrent epigenetic mutations. Oncotarget 2017; 8: 108738–108759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Parmar A, Marz S, Rushton S, et al. Stromal niche cells protect early leukemic FLT3-ITD+ progenitor cells against first-generation FLT3 tyrosine kinase inhibitors. Cancer Res 2011; 71: 4696–4706. [DOI] [PubMed] [Google Scholar]
- 124. Sato T, Yang X, Knapper S, et al. FLT3 ligand impedes the efficacy of FLT3 inhibitors in vitro and in vivo. Blood 2011; 117: 3286–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ravandi F, Alattar ML, Grunwald MR, et al. Phase 2 study of azacytidine plus sorafenib in patients with acute myeloid leukemia and FLT-3 internal tandem duplication mutation. Blood 2013; 121: 4655–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Williams CB, Kambhampati S, Fiskus W, et al. Preclinical and phase I results of decitabine in combination with midostaurin (PKC412) for newly diagnosed elderly or relapsed/refractory adult patients with acute myeloid leukemia. Pharmacotherapy 2013; 33: 1341–1352. [DOI] [PubMed] [Google Scholar]
- 127. Strati P, Kantarjian H, Ravandi F, et al. Phase I/II trial of the combination of midostaurin (PKC412) and 5-azacytidine for patients with acute myeloid leukemia and myelodysplastic syndrome. Am J Hematol 2015; 90: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Cooper BW, Kindwall-Keller TL, Craig MD, et al. A phase I study of midostaurin and azacitidine in relapsed and elderly AML patients. Clin Lymphoma Myeloma Leuk 2015; 15: 428–432.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Fischer T, Stone RM, Deangelo DJ, et al. Phase IIB trial of oral Midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J Clin Oncol 2010; 28: 4339–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tawfik B, Sliesoraitis S, Lyerly S, et al. Efficacy of the hypomethylating agents as frontline, salvage, or consolidation therapy in adults with acute myeloid leukemia (AML). Ann Hematol 2014; 93: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Swaminathan M, Kantarjian HM, Daver N, et al. The combination of quizartinib with azacitidine or low dose cytarabine is highly active in patients (Pts) with FLT3-ITD mutated myeloid leukemias: interim report of a phase I/II trial. Blood 2017; 130: 723. [Google Scholar]
- 132. Daver N, Garcia-Manero G, Basu S, et al. Efficacy, safety, and biomarkers of response to azacitidine and nivolumab in relapsed/refractory acute myeloid leukemia: a non-randomized, open-label, phase 2 study. Cancer Discov 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Heninger E, Krueger TE, Lang JM. Augmenting antitumor immune responses with epigenetic modifying agents. Front Immunol 2015; 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Almstedt M, Blagitko-Dorfs N, Duque-Afonso J, et al. The DNA demethylating agent 5-aza-2’-deoxycytidine induces expression of NY-ESO-1 and other cancer/testis antigens in myeloid leukemia cells. Leuk Res 2010; 34: 899–905. [DOI] [PubMed] [Google Scholar]
- 135. Srivastava P, Paluch BE, Matsuzaki J, et al. Immunomodulatory action of the DNA methyltransferase inhibitor SGI-110 in epithelial ovarian cancer cells and xenografts. Epigenetics 2015; 10: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Srivastava P, Paluch BE, Matsuzaki J, et al. Induction of cancer testis antigen expression in circulating acute myeloid leukemia blasts following hypomethylating agent monotherapy. Oncotarget 2016; 7: 12840–12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Goodyear O, Agathanggelou A, Novitzky-Basso I, et al. Induction of a CD8+ T-cell response to the MAGE cancer testis antigen by combined treatment with azacitidine and sodium valproate in patients with acute myeloid leukemia and myelodysplasia. Blood 2010; 116: 1908–1918. [DOI] [PubMed] [Google Scholar]
- 138. Gbolahan OB, Zeidan AM, Stahl M, et al. Immunotherapeutic concepts to target acute myeloid leukemia: focusing on the role of monoclonal antibodies, hypomethylating agents and the leukemic microenvironment. Int J Mol Sci 2017; 18: pii: E1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wang LX, Mei ZY, Zhou JH, et al. Low dose decitabine treatment induces CD80 expression in cancer cells and stimulates tumor specific cytotoxic T lymphocyte responses. PLoS One 2013; 8: e62924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Daver N, Boddu P, Garcia–Manero G, et al. Hypomethylating agents in combination with immune checkpoint inhibitors in acute myeloid leukemia and myelodysplastic syndromes. Leukemia 2018; 32: 1094–10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Yang H, Bueso-Ramos C, DiNardo C, et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia 2014; 28: 1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Orskov AD, Treppendahl MB, Skovbo A, et al. Hypomethylation and up-regulation of PD-1 in T cells by azacytidine in MDS/AML patients: a rationale for combined targeting of PD-1 and DNA methylation. Oncotarget 2015; 6: 9612–9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2017; 169: 361. [DOI] [PubMed] [Google Scholar]
- 144. Saudemont A, Quesnel B. In a model of tumor dormancy, long-term persistent leukemic cells have increased B7-H1 and B7.1 expression and resist CTL-mediated lysis. Blood 2004; 104: 2124–2133. [DOI] [PubMed] [Google Scholar]
- 145. Shen L, Kantarjian H, Guo Y, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol 2010; 28: 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Hiller JK, Schmoor C, Gaidzik VI, et al. Evaluating the impact of genetic and epigenetic aberrations on survival and response in acute myeloid leukemia patients receiving epigenetic therapy. Ann Hematol 2017; 96: 559–565. [DOI] [PubMed] [Google Scholar]
- 147. Zhang LY, Yuan YQ, Zhou DM, et al. Impact of global and gene-specific DNA methylation in de novo or relapsed acute myeloid leukemia patients treated with decitabine. Asian Pac J Cancer Prev 2016; 17: 431–437. [DOI] [PubMed] [Google Scholar]
- 148. Corces-Zimmerman MR, Hong WJ, Weissman IL, et al. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A 2014; 111: 2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Jan M, Majeti R. Clonal evolution of acute leukemia genomes. Oncogene 2013; 32: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Roboz GJ, Montesinos P, Selleslag D, et al. Design of the randomized, Phase III, QUAZAR AML Maintenance trial of CC-486 (oral azacitidine) maintenance therapy in acute myeloid leukemia. Future Oncol 2016; 12: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]