Abstract
Extracellular deoxyribonucleases (DNases) contribute to the spread of pathogenic bacteria through the evasion from host innate immunity. Our main objective was to evaluate the production of extracellular DNases by human and bovine Streptococcus agalactiae clinical strains and perform a correlation of genetic lineages and DNase activity with capsular type, genetic determinants, clinical origin (colonization and infection), and host (human or bovine). DNase activity was evaluated by qualitative and quantitative assays for a collection of 406 human (n = 285) and bovine (n = 121) strains. All (121/121) bovine were isolated from mastitis and revealed to be DNase (+), indicating a putative pathogenic role in this clinical scenario. From the human S. agalactiae strains, 86% (245/285) showed DNase activity, among which all strains belonging to capsular types, namely, Ia, Ib, III-2, and IV. All CC17 strains (n = 58) and 56/96 (58.3%) of the CC19 displayed DNase activity. DNase (–) strains belonged to the CC19 group. However, the subcharacterization of CC19 S. agalactiae strains through multiple-locus variable number tandem repeat analysis (MLVA), antibiotic resistance, mobile elements, and surface proteins did not provide any distinction among DNase producers and non-producers.
The production of DNases by all human CC17 strains, about two-fifths of human CC19, and all bovine strains, suggest an important contribution of DNases to hypervirulence.
Keywords: bovine and human strains, CC17, CC19, DNase activity assays, extracellular DNases, Streptococcus agalactiae
Introduction
Genetic lineages of Streptococcus agalactiae, such as genotype III ST17, have been considered highly virulent because of their association with the development of meningitis in neonates. This may be due to their intrinsic invasive characteristics that would turn them more prone to invade the central nervous system than other lineages [1–3]. However, the molecular mechanisms underlying the transition from colonization to infection have yet to be disclosed, and while the exact pathogenic features of S. agalactiae remain unrevealed, many virulence factors have been proposed to explain S. agalactiae-infection-related pathogenesis. These include the serotype-specific polysaccharide capsule, secreted enzymes, such as hyaluronidase and C5a peptidase [2, 4–6], and the activity of extracellular deoxyribonucleases (DNases) [7]. Extracellular DNase production, described for S. pyogenes [8], S. pneumoniae [9], Staphylococcus aureus [10], and S. agalactiae [7, 11], was shown to promote neutrophil-mediated resistance and the spread of infection in vivo [12]. DNases would protect streptococci from being killed by neutrophil extracellular traps (NETs), by degrading this neutrophil-mediated antimicrobial system (most associated to Gram positives). However, bacteria might escape NET entrapment through the production of nucleases. The identification of the major nuclease (nuclease A) coding gene (gbs0661) in S. agalactiae, in 2013 [7], provided the genetic background for the observed DNase activity first reported in 1980 [11].
As there remains a little knowledge regarding DNase activity in human and no data concerning bovine S. agalactiae strains, we aimed to determine DNase activity (qualitatively and quantitatively) in strains of different origin (human and bovine) and different capsular types to evaluate possible associations with genetic features.
Materials and Methods
Strain Collection
S. agalactiae reference strains belonging to different genetic lineages were used in this study: 2603V/R (genotype: V/ST110) (ATCC BAA-611, GenBank accession number NC004116), COH1 (genotype: III/ST17) (GenBank accession number AAJR01000000), NEM316 (genotype: III/ST23) (ATCC 12403, GenBank accession number NC004368), 090R (genotype: Ia/ST25) (ATCC 12386, GenBank accession number NZ_LCXR00000000.1), and A909 (genotype: Ia/ST7) (ATCC BAA-1138, GenBank accession number NC007432.1). Clinical strains of bovine (n = 121) and human (n = 285) origin were also used.
All bovine S. agalactiae strains included in this study were associated with clinical and sub clinical mastitis cases identified in different farms of Portugal in 2002–2003 and in 2011–2013, n = 60 and n = 61, respectively. These strains were previously described elsewhere [13, 14].
For the human S. agalactiae collection (colonization and invasive infection), a stratified sample (n = 285) from an enlarged set involving the collections of the Portuguese National Institute of Health (NIH), Portugal, and the Institute of Medical Microbiology and Hygiene (IMMH), Ulm University, Germany, was used. These strains have been previously described [15–20]. The stratification of the different capsular types to be included in the study was statistically established by using the computer program SPSS (SPSS software, version 16.0, IBM; Fisher Exact test, significance of 5%), based on the distribution of the capsular types (data not shown). The desirable number of genotype II invasive strains in neonatal infections was not achieved due to the low prevalence of this genotype, as described by others [18].
Capsular Genotyping, Multilocus Sequence Typing (MLST)
Distribution of capsular genotypes is shown in Table 1. Capsular genotyping for most of the bovine strains has been previously described [13, 14]; for 13 of them (from the 2011–2013 group), the capsular genotype could not be identified because polymerase chain reaction (PCR) amplification of cpsD-cpsE-cpsF did not succeed. Capsular types designated as “New” identify strains, evidencing no similarity with alleles included in the cpsD-E-F database [14, 17]. For the human S. agalactiae collection, all the capsular genotypes have been identified for the purposes of prior studies of the Portuguese NIH and the IMMH group [15–19].
Table 1.
Collection of S. agalactiae strains used in the present study
| Capsular serotype/genotype | Number of human colonizing strains (n = 157)a | Number of human invasive strains (n = 128)b | Number of bovine strains from 2002–2003 (n = 60)c | Number of bovine strains from 2011–2013 (n = 61)d |
|---|---|---|---|---|
| Ia | 20 | 20 | ||
| Ib | 10 | 10 | ||
| II | 39 | 17 | ||
| III-I | 29 | 29 | ||
| III-2 | 29 | 29 | ||
| III-3 | 1 | |||
| IV | 10 | 3 | ||
| V | 20 | 20 | 14 | |
| New cpsBOve 1 | 6 | 1 | ||
| New cpsBOV 2 | 11 | |||
| New cpsBOV 3 | 24 | 28 | ||
| New cpsBOV 4 | 1 | |||
| New cpsBOV 5 | 3 | |||
| New cpsBOV 6 | 10 | |||
| New cpsBOV 7 to new cpsBOV 15 | 1 isolate of each type |
MLST allelic profile or sequence type (ST) was known for most of the strains included in prior publications [13–18]. Whenever the MLST was missing, it was determined as previously described [21], where clonal complexes (CCs) were defined for isolates sharing six or seven identical alleles. For seven bovine isolates, the ST could not be determined.
The study collection involved 3 bovine S. agalactiae genetic lineages, CC1, CC23, and CC61, and five human S. agalactiae genetic lineages, CC1, CC12, CC17, CC19, and CC23.
DNase Activity – Qualitative Assays
Qualitative evaluation of DNase production was performed for all S. agalactiae strains; this was done by inoculation of DNA–methyl green agar plates (Oxoid, Basingstoke, England) or DNase test agar with toluidine blue plates (Sigma-Aldrich, USA). S. agalactiae strains, namely, 090R, COH1, or NEM316, were used as positive controls, and 2603V/R as a negative control. The interpretation of the results was done after 24 h of incubation at 37 °C in an atmosphere of 5% CO2. Strains were considered DNase producers when displaying transparent halos around colonies of S. agalactiae in DNA–methyl green agar plates, or a characteristic bright rose-pink zone in DNase test agar with toluidine blue.
DNase Activity - Semi-Quantitative and Quantitative Assays
The semi-quantitative assessment of the activity of DNases was performed according to that described by Sumby et al. [8], with modifications, in order to confirm the results of the qualitative testing. Briefly, S. agalactiae strains were inoculated in 5 mL of Todd Hewitt broth supplemented with 0.5% yeast extract (Oxoid), culture supernatant from stationary phase was centrifuged (10 min, 3000 rpm), syringe-filtrated (0.2 μm), before incubation at 37 °C with1 μg of double-stranded DNA (dsDNA) (atr amplicon [21]) in the presence of 1× M buffer (Roche). Filtrated supernatants (10 μL) were incubated for 1 h, 2 h, 4 h and “overnight” (~ 17 h) in order to evaluate the digestion of DNA in a final volume of 50 μL. Nuclease reaction was stopped by adding ethylenediaminetetraacetic acid EDTA (0.5 M, pH 8.0) at 4 °C. DNA digestion was analyzed by electrophoresis in 1% agarose gel. S. agalactiae strains 090R, COH1, or NEM316 were used as positive controls. A negative control consisting on a reaction mixture without supernatant was used in all experiments.
Qualitative and semi-quantitative assays yielded identical results. An example of semi-quantitive testing is shown in Figure 1. The quantitative evaluation of the activity of DNases was only performed for a selection of S. agalactiae strains, including null or residual producers in qualitative and semi-quantitative assays. The quantitative DNase assays were performed as described above, and the amount of DNA present in each sample was determined by measuring at the same time points of incubation. For this purpose, the fluorescent Pico-Green dye (Invitrogen) was used to quantify the dsDNA according to the manufacturer's instructions (https://www.thermofisher.com/order/catalog/product/P7589). Briefly, 1 mL of 1× Quant-iT PicoGreen was added to an equal volume of each supernatant, previously diluted in 1× Tris–EDTA (TE) buffer. After 5 min of incubation at room temperature, in the dark, we proceeded to the fluorescence measurement (Fluorimeter – Anthos Zenith 3100) in 96-well microtiter plates (Corning 96 Well Clear Flat Bottom Polystyrene Black TC, Costar, USA). To calculate the concentration of remnant DNA in each sample, a standard curve with 4 solutions of phage Lambda DNA of known concentrations (1, 10, 100, and 1000 ng/mL) was used: y = 3675x (x = concentration of dsDNA ng/mL, y = fluorescence) (see supporting information Figure 1). Each obtained fluorescence value was subtracted to the blank solution (Pico-Green 1× + 1× TE buffer at a ratio of 1:1) fluorescence value. S. agalactiae strains, namely 090R, COH1, and NEM316 were used as positive controls, and 2603V/R as a negative control. The results were based on 2 independent experiments.
Figure 1.
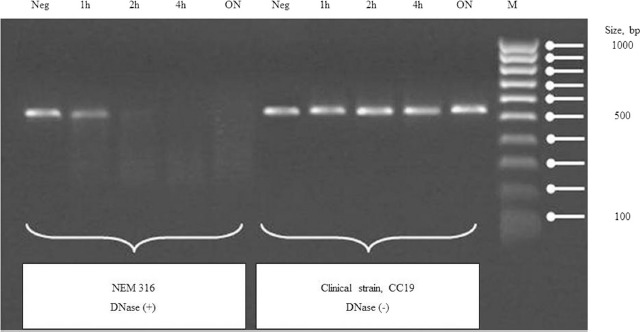
Semi-quantitative assay in gel electrophoresis for the evaluation of DNase activity. 1 μg of DNA (amplicon atr) incubated with 10 μl of S. agalactiae culture supernatant (NEM316 vs CC19 clinical strain) for 1h, 2h, 4h, overnight at 37°C. Neg, Negative control: without culture supernatant; ON, Overnight; M, Molecular weight Ladder.
Supporting information Figure 1.
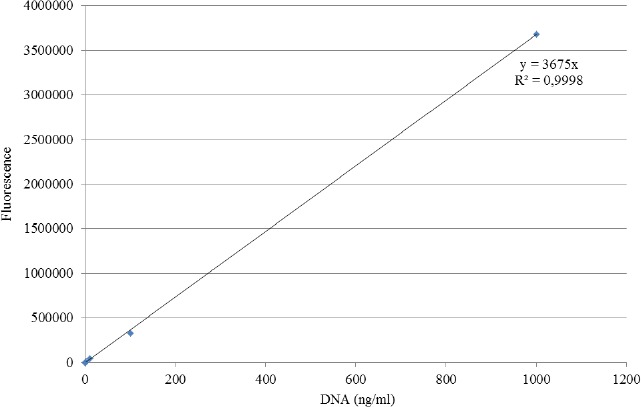
Standard curve for quantitative DNase assays A standard curve with four solutions of phage Lambda DNA of known concentrations (1, 10, 100 and 1000 ng/ml) was used: y = 3675x (x = concentration of dsDNA ng/ml, y = fluorescence).
Multiple-Locus Variable Number Tandem Repeat Analysis (MLVA)
For determining the MLVA genotype, the variable number of tandem repeats present in SAG2, SAG3, SAG4, SAG7, SAG21, and SAG22 genes according to nucleotide sequences of the reference strains, namely, 2603V/R, NEM316, and A909, was evaluated; this was done as described elsewhere [22].
S. agalactiae Antimicrobial Resistance Profile
Antimicrobial resistance testing (penicillin G, erythromycin, clindamycin, and vancomycin) was performed by Epsilometer test (E-test) according to Clinical and Laboratory Standards Institute guidelines [23] and by the detection of macrolide-resistance-associated genes (ermTR, ermB, and mefA) through PCR amplification, as described elsewhere [24].
Alpha-like Protein (Alp) Family and Mobile Genetic Elements
The major surface-localized protein antigens of S. agalactiae belong to a family of surface proteins named Alp family, which contains large internal tandem repeats and is a potential virulence factor. A multiplex PCR was used for direct identification of the Alp family (alpha-C, rib, epsilon, and alp2–alp4 genes) [25, 26].
Genes encoding the surface proteins ScpB and Lmb are located on a composite transposon that appears to be a hot spot for integration of two MGEs, GBSi1, or IS1548. Several MGEs are genetic markers for particular genetic lineages. Indeed, CC17 is usually marked by the group II intron GBSi1, whereas CC19 is marked by the insertion sequence IS1548.
The presence of two mobile genetic elements, namely, IS1548 and GBSi1, within the scpB–lmb intergenic region was evaluated by PCR, as previously described [27]. In the absence of mobile genetic elements, the presence of the flanking genes (scpB and lmb) was assessed.
Statistical Analysis
Associations between categorical variables (DNase producers/non-producers and colonization/invasive infection) were calculated using Chi-square with a significance level of 5%. All presented statistical results were obtained using SPSS, version 16.0 (SPSS Inc.).
Ethics
No ethical approval was obtained because our work involves only in vitro assays involving prototype strains and anonymized strains from laboratory collections. No relation to specific individuals is performed. No personal data are involved in the present study.
Results
DNase Assays
All bovine S. agalactiae strains (121/121) and 86% (245/285) of the human S. agalactiae strains displayed DNase activity.
All human strains belonging to capsular types Ia, Ib, III-2, and IV displayed DNase activity; the percentage of DNase producing strains among types II, III-1, and V varied between 82% and 88%, 48% and 59%, and 90%, respectively, depending on the clinical origin (colonization or invasive infection), and sequence type/clonal complex (Tables 2 and 3).
Table 2.
Evaluation of the DNase activity by qualitative and semi-quantitative assays among S. agalactiae of human origin (ST = sequence type; CC = clonal complex)
| Strains with DNase activity (frequency) | ||
|---|---|---|
| [STs (frequency)] | ||
| CCs (frequency) | ||
| Capsular type | Colonization | Invasive infection |
| Ia | 100% (20/20) | 100% (20/20) |
| [ST23 (17/20); ST144 (2/20); ST24 (1/20)] | [ST23 (19/20); ST24 (1/20)] | |
| CC23 (20/20) | CC23 (20/20) | |
| Ib | 100% (10/10) | 100% (10/10) |
| [ST8 (5/10); ST12 (2/10); ST1 (1/10); ST10 (1/10); ST563 (1/10)] | [ST10 (5/10); ST8 (4/10); ST12 (1/10)] | |
| CC1 (2/10); CC12 (8/10) | CC12 (10/10) | |
| II | 82% (32/39) | 88% (15/17) |
| [ST 28 (10/32); ST12 (8/32); ST44 (4/32); ST10 (3/32); ST2 (2/32); ST43 (1/32); ST154 (1/32); ST249 (1/32); ST347 (1/32); ST 472 (1/32)] | [ST 12 (5/15); ST28 (4/15); ST19 (3/15); ST10 (1/15); ST22a (1/15); ST42 (1/15)] | |
| CC19 (17/32); CC12 (12/32); CC1 (2/32); CC23 (1/32) | CC19 (8/15); CC12 (6/15) | |
| III-1 | 48% (14/29) | 59% (17/29) |
| [ST19 (8/14); ST27 (2/14); ST106 (2/14); ST286 (1/14); ST369 (1/14)] | [ST19 (17/29)] | |
| CC19 (14/14) | CC19 (17/17) | |
| III-2 | 100% (29/29) | 100% (29/29) |
| [ST17 (28/29); ST287 (1/29)] | [ST17 (29/29)] | |
| CC17 (29/29) | CC17 (29/29) | |
| IV | 100% (10/10) | 100% (3/3) |
| [ST196 (3/10); ST1 (1/10); ST2 (2/10); ST3 (1/10); ST162 (1/10); ST23 (1/10); ST10 (1/10)] | [ST66 (1/3); ST8 (1/10); ST23 (1/10)] | |
| CC1 (7/10); CC23 (2/10); CC12 (1/10) | CC1 (1/3); CC23 (1/3); CC12 (1/3) | |
| V | 90% (18/20) | 90% (18/20) |
| [ST1 (5/18); ST2 (13/18)] | [ST1 (15/18); ST2 (1/18); ST23 2/18)] | |
| CC1 (18/18) | CC1 (16/18); CC23 (2/18) | |
aSingleton ST.
Table 3.
Evaluation of the DNase activity by qualitative and semi-quantitative assays among S. agalactiae of bovine origin
| Strains with DNase activity (frequency); | ||
|---|---|---|
| [STs (frequency)] CCs (frequency) | ||
| Capsular type | Collection of 2002–2003 | Collection of 2011–2013 |
| III-3 | 100% (1/1) [ST23 (1/1)] CC23 (1/1) |
|
| V | 100% (14/14) [ST2 (14/14)] CC1 (14/14) |
|
| New cpsBOV 1 | 100% (6/6) [ST2 (6/6)] CC1 (6/6) 100% (11/11) |
100% (1/1) [ST554 (1/1)] CC61 (1/1) |
| New cpsBOV 2 | [ST61 (11/11)] CC61 (11/11) 100% (24/24) |
100% (28/28) |
| New cpsBOV 3 | [ST61 (4/24); ST554 (20/24)] CC61 (24/24) |
[ST61 (3/28); ST554 (23/28); ST2 (1/28); ST N.D. (1/28)] CC1 (1/28)] CC61 (26/28) |
| New cpsBOV 4 | 100% (1/1) [ST61 (1/1)] CC61 (1/1) |
|
| New cpsBOV 5 | 100% (3/3) [ST2 (3/3)] CC1 (3/3) |
|
| New cpsBOV 6 | 100% (10/10) [ST61 (7/10); ST554 (2/10); ST N.D. (1/10)] CC61 (9/10) |
|
| New cpsBOV 7 | 100% (1/1) [ST554 (1/1)] CC61 (1/1) |
|
| New cpsBOV 8 | 100% (1/1) [ST554 (1/1)] CC61 (1/1) |
|
| New cpsBOV 9 | 100% (1/1) [ST554 (1/1)] CC61 (1/1) |
|
| New cpsBOV 10 | 100% (1/1) [ST554 (1/1)] CC61 (1/1) |
|
| New cpsBOV 11 | 100% (1/1) [ST554 (1/1)] CC61 (1/1) |
|
| New cpsBOV 12 | 100% (1/1) [ST554 (1/1)] CC61 (1/1) |
|
| New cpsBOV 13 | 100% (1/1) [ST554 (1/1)] CC61 (1/1) |
|
| New cpsBOV 14 | 100% (1/1) [ST61 (1/1)] CC61 (1/1) |
|
| New cpsBOV 15 | 100% (1/1) [ST N.D. (1/1)] |
|
ST = Sequence type; CC = Clonal Complex; New cpsBOV = new bovine capsular type; N.D. = not determined.
All DNase (–) strains (40/406; 9.9%; all from human origin) belonged to CC19; however, 56 CC19 strains (involving capsular types II/III-1) within the present study collection displayed DNase activity (Table 2). The quantitative DNase activity over time within the population of CC19 strains is shown in Figure 2. In accordance with these results, prototype strain 2603V/R showed a disability to degrade DNA, whose amount remained nearly constant after 17 h. In contrast, S. agalactiae NEM316, COH1, 090R, and various clinical strains, belonging to CC17, CC23, and CC61 genetic lineages, revealed a substantial digestion of the DNA, reflecting a high production of extracellular DNases (Figure 2). Except ST61 strains (all of bovine origin), the profiles of DNA digestion were quite similar within strains of the same ST (Figure 2).
Figure 2.
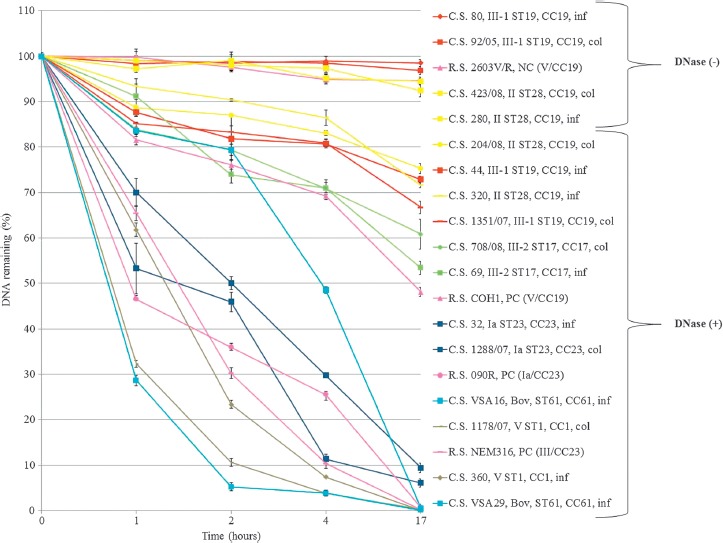
Quantitative DNase assays displaying differential DNase activity between S. agalactiae strains over time. 1 μg of DNA (amplicon atr) incubated with 10 μl of S. agalactiae culture supernatant for 1h, 2h, 4h, overnight at 37°C. Fluorescent PicoGreen dye (Invitrogen) was used to quantify the dsDNA. The graphic displays the mean values of the experiment performed in duplicate. Error bars show ± standard deviation (see Supporting information Table 1). Col, Colonization; Inf, Infection; Bov, Bovine; C.S., Clinical Strain; R.S., Reference Strain; PC, Positive Control; NC, Negative Control
No statistical association between DNase production and invasive infection could be established (P > 0.05) for human S. agalactiae isolates. For bovine, strains from colonization or systemic infections were not available, precluding any statistical evaluation of clinical findings for DNase production.
Sub-Characterization of CC19 Strains
For further phenotypic characterization and to identify putative genetic determinants associated with the production/non production of DNases, CC19 was selected (all strains isolated from humans) (n = 96, 55 colonizing strains, and 41 invasive strains). Based on MLVA, two profiles/genotypes were identified: profile 33 (3,3,3,5,0,2) exhibited by all ST19 strains, and profile 32 (3,3,1,5,0,2) exhibited by ST28 strains.
Regarding the antibiotic resistance testing, no CC19 strain was resistant to penicillin (minimum inhibitory concentration [MIC] between 0.032 and 0.125 μg/mL) and vancomycin (MIC between 0.25 and 1 μg/mL). In addition, no invasive
CC19 strain displayed resistance to macrolides, contrasting to colonizing CC19 strains, which have shown a macrolide resistance rate of 31% (17/55; III-1, 16 strains; II, one strain). Nine of these 17 strains showed simultaneous resistance to erythromycin and clindamycin, MIC ≥ 256 mg/mL, attributable to the presence of ermB gene. The remaining CC19 strains were only resistant to erythromycin, 2 ≤ MICs ≤ 6 μg/mL, presenting ermA (n = 8) and mefA (n = 1). There was no statistical correlation between antibiotic resistance profile and DNase activity among CC19 strains.
The screening of mobile genetic elements within the scpB-lmb intergenic region and the study of Alp genes also failed to distinguish among human CC19 DNase producers and non-producers. In fact, 79.2% (76/96) of the CC19 strains carried the IS1548, whereas 100% (96/96) displayed the rib gene.
Discussion
Our study aimed to evaluate DNase production among a human strain collection statistically stratified to represent the different S. agalactiae capsular types (except genotype II because of the lack of invasive strains of this capsular type in the collection available for the study) and to correlate DNase production with clinical and genotypic features.
In bovine S. agalactiae, to our best knowledge, this is the first study reporting DNase activity, and a high DNase activity was determined, in particular, among strains belonging to the ST61 lineage. Considering that neither colonization strains, nor bovine DNase non-producer strains were available, no further association could be established.
Qualitative and semi-quantitative assays are easy to execute and have been proposed to determine DNase activity [8]. However, they are inaccurate to fully characterize the production of DNases, and for this reason, a quantitative methodology was applied in the present study. Using this approach, 100% and 86% of the bovine and human strains, respectively, displayed DNase activity. For human strains, these results corroborate the earlier findings [11] describing exactly the same 86% of DNase producers among strains from human origin.
ST17 strains belonging to the capsular type III have long been associated with meningitis and late onset disease of the newborn. These strains display secreted and surface-exposed factors, among which DNases that have been related to virulence [2, 4]; accordingly, in our study, all ST17 strains were DNase producers. Thus, although less often related to meningitis (in contrast to ST17 strains), CC19 strains attracted our attention, as this less studied group involves both DNase (+) and DNase (–) strains, a phenotype that was only found among the CC19 genetic lineage. These strains belonged to capsular types II, III-1, and V, contrasting with CC17 strains, which were all capsular type III-2 and DNase producers. The production of DNases by all CC17 strains, in contrast to the absence of DNase production in 41.7% (40/96) of the CC19 strains, could suggest a contribution of DNases to hypervirulence. However, in the present study, CC19 strains were involved in both colonization and invasive infection; thus, CC19 strains also could not be associated with colonization, not corroborating the hypothesis proposed by others [3].
In our attempt to deeper characterize the subpopulation of CC19 producer and non-producer strains, we used MLVA, as some authors defined MLVA as an epidemiological tool displaying higher discriminatory power than MLST [22]. However, the polymorphism of tandem repeats obtained by this technique only reinforced the clonality of CC19 strains previously obtained by MLST. Thus, MLVA did not provide the separation of CC19 strains in clones associated with the production/non production of extracellular DNases. Also, the study of Alp genes and the search for mobile genetic elements in the intergenic region scpB-lmb (located on a composite transposon and a hot spot for integration of IS1548 or group II intron GBSi1) did not provide new evidence for distinguishing CC19 subpopulations according to their DNase activity. In addition, DNase coding genes and antibiotic resistance genes do not seem to be genetically related, as no association between antibiotic resistance and DNase production could be established.
All bovine strains (all DNase (+)) were isolated from mastitis, highlighting the need for extensive studies in cattle. In fact, udder infections by S. agalactiae DNase (+) strains might induce serious economic repercussions over milk production, as DNase production might be advantageous to overcome bovine host mammary tissue NETs and thus facilitating infection establishment; if this hypothesis is confirmed in future studies, DNases could be useful for the development of a vaccine for preventing S. agalactiae mastitis.
In conclusion, this study contributes for the perception of DNase production among S. agalactiae from human and, for the first time, bovine origin, characterizing 406 strains regarding the capsular type, sequence type, and antimicrobial resistance factors. The high percentage of strains evidencing DNase activity (100% of bovine origin and 86% of human origin), including all CC17 human strains, seems to corroborate the hypothesis of a major role of this enzyme in the evasion processes from the host defense mechanisms, namely NETs, and a putative role in hypervirulence. The circumstance that all bovine strains were DNase (+) and all were isolated from mastitis highlights the need for an extensive evaluation of S. agalactiae infections in bovine, to fully understand whether DNase production is required for the development of mastitis. Comparative whole-genome sequencing studies together with a large-scale transcriptomic analysis involving strains of diverse origin should be crucial to deeply scalp the molecular features associated to DNase release and activity.
Supporting information Table 1.
Standard deviations obtained in quantitative DNase assays
| Strains | Time (hours) | Standard deviation (%) |
|---|---|---|
| 1351/07 | 1 | 0,09 |
| 2 | 3,9 | |
| 4 | 0,85 | |
| 17 | 1,4 | |
| 92/05 | 1 | 1,69 |
| 2 | 0,76 | |
| 4 | 0,56 | |
| 17 | 0,78 | |
| 44 | 1 | 0,86 |
| 2 | 2,77 | |
| 4 | 0,94 | |
| 17 | 0,76 | |
| 80 | 1 | 0,38 |
| 2 | 1,89 | |
| 4 | 1,03 | |
| 17 | 0,8 | |
| 204/08 | 1 | 1,82 |
| 2 | 0,35 | |
| 4 | 0,58 | |
| 17 | 1,1 | |
| 423/08 | 1 | 0,38 |
| 2 | 1,89 | |
| 4 | 1,03 | |
| 17 | 0,92 | |
| 320 | 1 | 1,61 |
| 2 | 0,23 | |
| 4 | 1,71 | |
| 17 | 0,55 | |
| 280 | 1 | 2,57 |
| 2 | 1,23 | |
| 4 | 0,9 | |
| 17 | 1,3 | |
| 1288/07 | 1 | 5,57 |
| 2 | 2,16 | |
| 4 | 1,12 | |
| 17 | 0,9 | |
| 32 | 1 | 3,1 |
| 2 | 1,43 | |
| 4 | 0,36 | |
| 17 | 1,01 | |
| 708/08 | 1 | 1,6 |
| 2 | 2,1 | |
| 4 | 1,95 | |
| 17 | 3,3 | |
| 69 | 1 | 2,7 |
| 2 | 1,89 | |
| 4 | 1,3 | |
| 17 | 1,46 | |
| 1178/07 | 1 | 0,72 |
| 2 | 0,9 | |
| 4 | 0,65 | |
| 17 | 1,1 | |
| 360 | 1 | 1,47 |
| 2 | 0,89 | |
| 4 | 0,26 | |
| 17 | 0,67 | |
| VSA16 | 1 | 0,8 |
| 2 | 1,2 | |
| 4 | 0,71 | |
| 17 | 0,3 | |
| VSA29 | 1 | 1,2 |
| 2 | 0,85 | |
| 4 | 0,63 | |
| 17 | 1,32 | |
| NEM316 | 1 | 1,6 |
| 2 | 1,2 | |
| 4 | 1,08 | |
| 17 | 0,95 | |
| COH1 | 1 | 1,11 |
| 2 | 0,98 | |
| 4 | 0,74 | |
| 17 | 0,95 | |
| 090R | 1 | 0,35 |
| 2 | 0,68 | |
| 4 | 1,25 | |
| 17 | 0,8 | |
| 2603V/R | 1 | 1,23 |
| 2 | 0,64 | |
| 4 | 1,09 | |
| 17 | 0,55 |
Footnotes
Funding Sources
This work was supported by Fundação para a Ciência e Tecnologia, Portugal [grant numbers PTDC/SAU-MII/105114/2008, PTDC/CVT-EPI/6685/2014, SFRH/BD/48231/2008, and SFRH/BD/118350/2016].
Authors' Contributions
All authors had full access to all data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. MJB, ISS, JPG, and BS conceptualized, designed, and supervised the study and performed the analysis and interpretation of data. ISS, CF, and CAB obtained funding. CF, CAB, IS, and VD performed the wet lab experiments, performed research, and analyzed data. IS performed statistical analysis. CF, CAB, IS, and MJB wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Héry-Arnaud G, Guillaume B, Lanotte P, et al. Acquisition of Insertion Sequences and the GBSi1 Intron by Streptococcus agalactiae Isolates Correlates with the Evolution of the Species. J Bacteriol. 2005;187:6248–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manning SD, Springman AC, Lehotzky E, Lewis MA, Whittam TS, Davies HD. Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J Clin Microbiol. 2009;47:1143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martins ER, Pessanha MA, Ramirez M, et al. Analysis of group B streptococcal isolates from infants and pregnant women in Portugal revealing two lineages with enhanced invasiveness. J Clin Microbiol. 2007;45:3224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brochet M, Couvé E, Zouine M, et al. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 2006;8:1227–43. [DOI] [PubMed] [Google Scholar]
- 5.Florindo C, Gomes JP, Rato MG, et al. Molecular epidemiology of group B streptococcal meningites in children beyond the neonatal period from Angola. J Med Microbiol. 2011;60:1276–80. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopal L. Understanding the regulation of Group B Streptococcal virulence factors. Fut Microbiol. 2009;4:201–21. doi: 10.2217/17460913.4.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derré-Bobillot A, Cortes-Perez NG, Yamamoto Y, et al. Nuclease A (Gbs0661), an extracellular nuclease of Streptococcus agalactiae, attacks the neutrophil extracellular traps and is needed for full virulence. Mol Microbiol. 2013;89:518–31. [DOI] [PubMed] [Google Scholar]
- 8.Sumby P, Barbian KD, Gardner DJ, et al. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. PNAS. 2005;102:1679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Bio. 2006;16:401–7. [DOI] [PubMed] [Google Scholar]
- 10.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, Kockritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun. 2010;2:576–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrieri P, Gray ED, Wannamaker LW. Biochemical and immunological characterization of the extracellular nucleases of Group B streptococci. J Exp Med. 1980;151:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracelular traps kill bacteria. Science. 2004;303:1532–5. [DOI] [PubMed] [Google Scholar]
- 13.Almeida A, Alves-Barroco C, Sauvage E, et al. Persistence of a dominant bovine lineage of group B Streptococcus reveals genomic signatures of host adaptation. Environ Microbiol. 2016;18:4216–29. https://doi.org/10.1111/1462-2920.13550. [DOI] [PubMed] [Google Scholar]
- 14.Rato MG, Bexiga R, Nunes SF, Cavaco LM, Vilela CL, Santos-Sanches I. Antimicrobial resistance and molecular epidemiology of streptococci from bovine mastitis. Vet Microbiol. 2013;161:286–94. [DOI] [PubMed] [Google Scholar]
- 15.Florindo C, Damião V, Lima J, et al. Accuracy of prenatal culture in predicting intrapartum group B Streptococcus colonization status. J. Mat.-Fetal and Neon. Med. 2014;27:640–2. [DOI] [PubMed] [Google Scholar]
- 16.Florindo C, Damião V, Silvestre I, et al. Epidemiological surveillance of 11 colonizing group B Streptococcus epidemiology in the Lisbon and Tagus Valley regions, Portugal (2005 to 2012): emergence of a new epidemic type IV/clonal complex 17 clone. Euro Surveill. 2014;19(23):pii20825. [DOI] [PubMed] [Google Scholar]
- 17.Florindo C, Viegas S, Paulino A, Rodrigues E, Gomes JP, Borrego MJ. Molecular characterization and antimicrobial susceptibility profiles in Streptococcus agalactiae colonizing strains: association of erythromycin resistance with subtype III-I genetic clone family. Clin. Microbiol. Infect. 2010;16:1458–63. [DOI] [PubMed] [Google Scholar]
- 18.Fluegge K, Wons J, Spellerberg B, et al. Genetic differences between invasive and noninvasive neonatal group B streptococcal isolates. Pediat. Infect. Dis. J. 2011;30:1027–31. [DOI] [PubMed] [Google Scholar]
- 19.Fluegge K, Supper S, Siedler A, Berner R. Serotype distribution of invasive Group B Streptococcal isolates in infants: results from a nationwide active laboratory surveillance study over 2 years in Germany. Clin. Infect. Dis. 2005;40:760–3. [DOI] [PubMed] [Google Scholar]
- 20.Fluegge K, Supper S, Siedler A, Berner R. Antibiotic susceptibility in neonatal invasive isolates of Streptococcus agalactiae in a 2-year nationwide surveillance study in Germany. Antimicrob. Agents and Chemoter. 2004;48(11):4444–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones N, Bohnsack JF, Takahashi S, et al. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 2003;41:2530–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haguenoer E, Baty G, Pourcel C, et al. A multi locus variable number of tandem repeat analysis (MLVA) scheme for Streptococcus agalactiae genotyping. BMC Microbiol. 2011;11:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standard Institute – CLSI. Performance Standards for Antimicrobial Susceptibility Testing M100-S19. Wayne. 2009; 19th Informational Supplement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gygax SE, Schuyler JA, Kimmel LE, Trama JP, Mordechai E, Adelson ME. Erythromycin and clindamycin resistance in group B Streptococcal clinical isolates. Antimicrob Agents Chemother. 2006;50:1875–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creti R, Fabretti F, Orefici G, Hunolstein C. Multiplex PCR Assay for direct Identification of Group B Streptococcal Alpha-Protein-Like Protein Genes. J Clin Microbiol. 2004;42:1326–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gherardi G, Imperi M, Baldassarri L, et al. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B Streptococci in Italy. J Clin Microbiol. 2007;45:2909–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Safadi R, Amor S, Héry-Arnaud G, et al. Enhanced expression of lmb gene encoding laminin-binding protein in Streptococcus agalactiae strains harboring IS1548 in scpB-lmb intergenic region. PLoS One. 2010;5:e10794. [DOI] [PMC free article] [PubMed] [Google Scholar]


