Abstract
Background
The objective of this study was to assess an in-house loop-mediated isothermal amplification (LAMP) platform for malaria parasite detection and identification on species level.
Methods
LAMP primers specific for the human Plasmodium spp., namely, P. falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi, as well as genus-specific primers, were tested against a composite gold standard comprising microscopy from thick and thin blood films, commercial genus-specific Meridian illumigene Malaria LAMP, in-house real-time polymerase chain reaction (PCR), and commercial fast-track diagnostics (FTD) Malaria differentiation PCR.
Results
Of the 523 blood samples analyzed, the composite gold standard indicated 243 Plasmodium-species-DNA-containing samples (46.5%). Sensitivity and specificity of the analyzed genus- and species-specific LAMP primers were 71.0%–100.0% and 90.8%–100.0%, respectively. The influence of parasitemia was best documented for P. falciparum-specific LAMP with sensitivity values of 35.5% (22/62) for microscopically negative samples containing P. falciparum DNA, 50% (19/38) for parasitemia ≤50/μL, 84% (21/25) for parasitemia ≤500/μL, and 100% (92/92) for parasitemia >500/μL.
Conclusions
In our hands, performance characteristics of species-specific in-house LAMP for malaria lack reliability required for diagnostic laboratories. The use of the easy-to-apply technique for surveillance purposes may be considered.
Keywords: Malaria, loop-mediated isothermal amplification, screening, species discrimination, non-endemic
Introduction
Close to the beginning of the new millennium, first approaches of loop-mediated isothermal amplification (LAMP)-based identification of malaria have been described [1]. In the meantime, numerous publications on this molecular approach have been published [2–20] and summarized in reviews [21–23]. Many evaluations were performed with limited sample sizes only [24–26], but there were also some studies with sample sizes of several hundred patients [27, 28]. Based on the ongoing progress in the development of molecular diagnostic tools for malaria, organizations like the Société de Pathologie Infectieuse de Langue Française (SPILF) have recommended molecular tools [29] like LAMP as useful approaches even for initial screening of patients with suspected malaria. The aim is the identification of easy-to-implement alternatives to microscopy, which is largely investigator-dependent, so its reliability is considerably affected by individual factors like the level of training and professional skills.
Automated LAMP approaches have the potential of use as easy-to-teach point-of-care testing (POCT) systems, making them interesting alternatives to classic polymerase chain reaction (PCR) [30, 31] for the detection of malaria parasites in human blood samples [21, 32, 33], not only in industrialized countries but also in tropical areas with restricted laboratory infrastructure.
The easy application of LAMP facilitates the further development of malaria-LAMP point-of-care applications, like the combination of LAMP with lateral flow platforms [34–36] or the in situ application on microscopic slides [37], as well as LAMP from blood samples on filter papers [38, 39]. Even from body fluids other than blood like saliva and urine, LAMP-based detection of Plasmodium spp. DNA has been attempted [40, 41]. Although species-specific identification can be addressed by LAMP [15, 21], species-specific malaria LAMP is not commercially available so far, and lacking capability for quantitative assessments remains a disadvantage of the technique. Nevertheless, the LAMP approach can support therapeutic decisions if other alternatives are not available.
Between April and December 2017, the German National Reference Center for Tropical Pathogens, Bernhard Nocht Institute for Tropical Medicine, conducted a study with several hundred blood samples of patients with suspicion of malaria for the evaluation of a commercial malaria LAMP approach [28]. From those samples, more than 20% were positive for Plasmodium spp. DNA. Residual material of nucleic acid extractions were frozen for further studies, thus enabling the here described approach for the evaluation of in-house LAMP primers not only for the identification of Plasmodium spp. DNA but also for its discrimination on species level.
In addition, a small number of frozen nucleic acid extractions of hemolytic samples from patients with suspicion of malaria were used for a proof-of-principle assessment with such even more complex sample matrices.
The aim was to set up a LAMP platform based on a semi-automated, easy-to-apply approach with standardized reagents and automated interpretation of the results. For this platform, primers were evaluated regarding their test characteristics with the above-mentioned, well-characterized set of samples.
Methods
Samples
Residual materials of nucleic acid extractions of 523 fresh ethylenediaminetetraacetic acid (EDTA) blood samples from patients with clinical suspicion of malaria, i.e., a subset of a sample set which was assessed during a previous study at the Bernhard Nocht Institute for Tropical Medicine between April and December 2017 [28], were included. The number was chosen to ensure a nearly equal distribution of positive and negative samples for the case-control-based study approach. Nucleic acid extraction had been performed with the EZ1 DNA Blood 200-μL Kit (Qiagen, Hilden, Germany) as described by the manufacturer and by others [42]. Afterwards, the samples were stored at –80 °C.
The samples were assessed in a blinded manner. This means that the investigators did not know the results of reference testing while performing the in-house LAMP analyses.
Reference Testing
Reference testing for the presence of Plasmodium spp. DNA was based on 3 approaches, i.e., microscopy (as an indirect method of proving the presence of DNA by showing the whole microorganisms), LAMP, and PCR. From all the EDTA blood samples, the results of Giemsa-stained [43] thick and thin blood films for Plasmodium spp. including quantification in line with WHO recommendations, as well as the results of genus-specific Meridian illumigene Malaria LAMP (product reference 280925, Meridian Bioscience Inc., Cincinnati, Ohio, USA), were available from the previous study [28]. Only in the case of contradicting results, i.e., discrepant results of microscopy and illumigene Malaria LAMP, confirmation testing had been done in line with the previous study protocol [28] with the molecular reference method of the Bernhard Nocht Institute, i.e., by genus-specific real-time PCR targeting Plasmodium spp. (RealStar Malaria PCR kit 1.0, altona Diagnostics Ltd., Hamburg, Germany) and by species-specific real-time PCR targeting P. falciparum, P. knowlesi, P. vivax, P. ovale, and P. malariae as described elsewhere [31, 42, 44]. In addition, all nucleic acid extractions were assessed with the FTD Malaria differentiation PCR (product reference 800613, Fast Track Diagnostics, Sliema, Malta) according to the manufacturer-s instructions on a Corbett Rotor-Gene 6000 or a Rotor-Gene Q (Qiagen) from the stored nucleic acid extractions. Of note, this real-time multiplex PCR discriminates P. falciparum, P. vivax, P. malariae, and P. ovale.
In-house-LAMP Testing
LAMP primers were designed by Amplex Diagnostics GmbH (Gars-Bahnhof, Germany) using the software LAMP Designer V1.10 (PREMIER Biosoft Int., Palo Alto, USA). Thereby, 18S rRNA gene-based approaches as applied by others [15, 45] were discouraged by the software due to high risks of cross reactions because of sequence similarities, so other targets were favoured. Newly designed primers targeted the histone deacetylase gene sal-1 of P. falciparum and P. knowlesi, as well as the dihydrofolate reductase gene (DHFR) of P. vivax (Table 1). For genus-specific Plasmodium spp. LAMP, published primers [46] targeting the mitochondrial sequence PgMt19 were used. As repeated approaches failed to identify useful LAMP primers for the merozoite surface protein gene msp-1, the lactate dehydrogenase gene LD, and the DHFR gene of P. malariae and P. ovale, previously published LAMP primers for the 18S rRNA gene of these species were chosen [45] in spite of the above-mentioned risk of cross-reactions.
Table 1.
Newly designed LAMP primers for this study
| Species | P. falciparum | P. knowlesi | P. vivax |
|---|---|---|---|
| Target gene | sal-1 | sal-1 | DHFR |
| Primer F3 | 5‘-AAG-CAA-CGG-ATT-GTC-CTG-3‘ | 5‘-GCA-TGG-TGT-AAT-CCT-CCA-G-3‘ | 5‘-AGA-ACT-GCA-ATG-CGT-TGA-AC-3‘ |
| Primer B3 | 5‘-GCA-TCA-TCT-GTC-ATA-CCA-TCA-3‘ | 5‘-GAA-GCC-ACA-GAG-AAT-CCG-3‘ | 5‘-CCA-CAC-CTG-TTC-TGT-CTC-C-3‘ |
| Primer forward inner primer (FIP) (F1c + F2) | 5‘-GCA-ATC-CTC-CTG-ACC-AAT-TCA-CGC-TGG-AGC-TTC-GAT-AGA-TG-3‘ | 5‘-AGC-AGT-CGT-GTG-CAG-GTG-TTA-CAC-AGA-TAT-CAG-CAC-AGT-G-3‘ | 5‘-AAG-CTG-AAG-TAC-ACG-AGG-TCG-CGT-GGC-AGA-AGA-ACA-ACG-3‘ |
| Primer backward inner primer (BIP) (B1c + B2) | 5‘-ATG-GAG-ATG-GTG-TTG-AGG-AAG-CAC-ACC-AAC-ATC-CGT-AAT-ATC-TC-3‘ | 5‘-GAG-TCA-ACT-CGT-TCA-CAT-CAC-TCT-TCG-CTA-ATC-GTG-TCG-TAC-AA-3‘ | 5‘-CCG-GAG-CAC-CTG-CAA-GAT-TTG-ATG-ATG-TCG-TAT-ATG-ATG-CCT-AG-3‘ |
| Primer LoopF | 5‘-CAT-ATG-TCA-GCA-CAA-TGG-TGA-T-3‘ | 5‘-TAG-ATG-GAG-CTT-CTA-AGC-TGA-AC-3‘ | 5‘-TCT-TCC-TCT-GCC-TCG-G-3‘ |
| Primer LoopB | 5‘-ACA-CAT-CGT-GTT-ATG-ACA-GT-3‘ | 5‘-GTG-GTC-TGT-ACA-CCT-CCA-TG-3‘ | 5‘-CAA-CAG-CCT-GAA-GAT-TAA-GCA-G-3‘ |
The LAMP reactions were run in a Genie II Mk2 system (Amplex Diagnostics GmbH) using standard conditions, as detailed in Table 2. Run time was set at 40 min. Temperature optimum for the primers for Plasmodium spp. was 66 °C for P. falciparum, P. knowlesi, and P. vivax and 61 °C for P. ovale and P. malariae. A LAMP signal was considered as non-specific if its melting temperature was beyond the range of the minimum and maximum melting temperature as observed for truly positive samples.
Table 2.
LAMP reaction conditions on the Genie II Mk2 system (Amplex Diagnostics GmbH)
| Software on the LAMP devices | eazyReport™-IVD-software (Amplex Diagnostics GmbH) |
| Master mix | Isothermal master mix ISO-001 (OptiGene Ltd., Horsham, UK) |
| Dilution of individual LAMP primers for the mixing of primer mix | 100 pmol/μL |
| Proportions of primer dilutions of the primers | 5 pmol + 5 pmol + 20 pmol + 20 pmol + 10 pmol + 10 pmol |
| F3 + B3 + BIP + FIP + LF + LB in the primer mix | |
| Composition of LAMP reaction mix | 25 μL containing 2 μL primer mix, 8 μL sample, and 15 μL isothermal master mix |
| Reaction time | 40 min |
| Normalization threshold value | 70,000 arbitrary units |
| Amplification threshold value | 10,000 arbitrary units |
| Minimum ratio | 0.025 |
| Time of first measurement | 3 min after initiation of the reaction |
Optimum reaction temperatures and detection limits were defined using serial dilutions of positive control plasmids (pEX-A2 vector backbone, Eurofins Genomics, Brussels) containing sequence inserts as shown in Supplementary material 1. Detection limits as detected with the plasmid dilutions are shown in Table 3.
Supplementary material 1:
Sequence inserts for the positive control plasmids.
| Sequence insert for the positive control plasmid of the Plasmodium genus LAMP |
| 5‘- CAATTGTGTTCATAGCTAGAGTACGTAAGGAAAAGGAAAGGTTAACCGCTATCAAATG GCGAGAAGGGAAGTGTGTTTCCATAGAAACCTTCATATATACTATGCTGACTTGAGTA ATGATAAATTGATAGTATCAGCTATCCATAGTTAATTGATTCCGTTTTGACCGGTCATT TTCTTTGCCTGGAGGTTACGTCCATACAGTTATAAGCAAGTGGAATGTTAGAAGCAAA CACTAGCGGTGGAACACATTGTTTCATTTGATAGTAAACACTATACCTTACCAATCTAT TTGAACTTGAACAAGGTTCCATT-3’ |
| Sequence insert for the positive control plasmid of the P. falciparum LAMP |
| 5‘- GCCCCCAATGTCGAACCGCAAGAAGGTGGCCTACTTCCACGACCCCGACATCGGGA GCTACTACTACGGGGCGGGGCACCCGATGAAGCCGCAGAGAATCCGCATGACGCAC TCGCTGATAGTCTCGTACAACCTGTATAAGTACATGGAGGTGTACCGGCCCCACAAA AGTGACGTAAACGAACTGACCCTCTTCCACGACTACGAATATGTAGATTTCCTCTCCT CCATTTCGATGGAAAATTATCGTGAGTTTACATACCAACTGAAGCGATTCAATGTTGG GGAAGCAACCGACTGTCCTGTGTTCGATGGCCTCTTCCAATTCCAGCAGTCATGTGC AGGTGCTTCTATAGATGGGGCAGCCAAGCTGAATCACCACTGTGCGGATATCTGCGT GAACTGGTCAGGCGGATTGCA-3‘ |
| Sequence insert for the positive control plasmid of the P. vivax LAMP |
| 5‘- CCCACCAGTGAAGGGACAAAGAATGAACCGTTCAGCCCGCGGACCTTTAGGGGTCT GGGCAATAAGGGGACTCTCCCATGGAAATGCAACTCCGTCGATATGAAGTACTTCAG CTCGGTGACGACCTACGTGGATGAGTCAAAGTATGAGAAGCTAAAGTGGAAGAGGGA GAGGTACCTACGAATGGAAGCCTCACAGGGGGGGGGTGACAACACAAGCGGTGGTG ACAACACACACGGTGGTGACAACGCCGACAAGCTGCAAAACGTCGTGGTCATGGGG AGAAGCAGCTGGGAGAGCATCCCCAAGCAGTACAAGCCGCTCCCAAACAGAATCAAC GTCGTGCTTTCCAAGACGCTAACAAAGGAAGACGTGAAGGAAAAGGTCTTCATAATTG ACAGCATAGATGACCTACTGCTGCTCTTAAAGAAGCTGAAGTACTACAAATGCTTCAT CATTGGGGGAGCACAAGTTTATAGGGAATGCCTAAGTAGAAACTTAATCAAGCAGATC TACTTCACGAGGATCAACGGCGCCTACCCGTGTGACGTCTTCTTCCCCGAGTTTGAC GAAAGCCAGTTTCGGGTGACGTCCGTCAGTGAGGTGTACAACAGCAAGGGCACCAC TCTGGACTTTTTGGTTTACAGCAAGGTGGGGGGGGGAGTTGACGGGGGCGCCTCCA ACGGGAGCACTGCGGACAGCGCTTCGGAGAACTGCAATGCGTTGAACTGCAATGCG CCGAAATGTAGCGCCCCGAACTGCCGCTCCCCCAATGGGGGGACCGCACAGCAGG GCGAATGGGGAAAGGGCCCCGCCGCGTGCCCGTGGCAGAAGAACAACGCCGAGGC AGAGGAAGACGACCTCGTGTACTTCAGCTTTAACAACAAAGTGGGGGAGAAAAACCC GGAGCACCTGCAAGATTTTAAAATTTACAACAGCCTGAAGATTAAGCAGCACCCAGAG TACCAATACCTAGGCATCATATACGACATCATCATGAATGGAAACAAACAAGGAGACA GAACAGGTGTGGGAGTGATGAGCAAATTTGGCTACATGATGAAATTTAATTTAAGTGA A-3‘ |
| Sequence insert for the positive control plasmid of the P. malariae LAMP |
| 5‘- AATAACAATGCAAGGCCAAATTTTGGTTTTGCAATTGGAATGATGGGAATTTAAAACCT TCCCAGAAGGCAATTGGAGGGCAAGTCTGGTGCCAGCAGCCGCGGTAATTCCAGCT CCAATAGCGTATATTAAAATTGTTGCAGTTAAAACGCTCGTAGTTGAATTTCAAGGAAT CAATATTTTAAGTAATGCTTTGTATATTTATAACATAGTTGTACGTTAAGAATAACCGCC AAGGCTTATATTTTTTCTGTTACATTTTGTTTTATTAATATATATATGCGTTCTTATTATA AAAATGATTCTTTTTAAAATTCTTTTGTATAATTTTTTATGCATGGGAATTTTGTTACTTT GAGTAAATTAGAGTGTTCAAAGCAAACAGTTAAAACAGTTTCTGTGTTTGAATACTACA GCATGGAATAACAAAATTGAACAAGTCAGAAGTTTGTTCTTTTTTCTTATTTTGGCTTA GTTACGATTAATAGGAGTAGCTTGGGGGCATTTGTATTCAGATGTCAGAGGTGAAATT CTTAGATTTTCTGGAGACAAGCAACTGCGAAAGCATATGCCTAAAATACTTCCATTAAT CAAGAACGAAAGTTAAGGGAGTGAAGACGATCAGATACCGTCGTAATCTTAACCATAA ACTATGCCGACTAGGTGTTGGATGATAGTGTAAAAAATAAAAG-3‘ |
| Sequence insert for the positive control plasmid of the P. ovale LAMP |
| 5‘- CCATTTCATGGTTTTGTAATTGGAATGATGGGAATTTAAAACCTTCCCAAAATTCAATT GGAGGGCAAGTCTGGTGCCAGCAGCCGCGGTAATTCCAGCTCCAATAGCGTATATTA AAATTGTTGCAGTTAAAACGCTCGTAGTTGAATTTCAAAGAATCAATATTTTAAGTAAT GCTTTTGGTTTAAGGTGCTTAGAACAATACAACGTATCTGCTCTTTGCATTCCTTATCC AAAATGCGTTCTTATTAATAAAAAGGATTCTTTTTAAAATCTCCTATACTTTCTTGTACT GGAGATTTTGTTACTTTGAGTAAATTAGAGTGTTCAAAGCAAACAGTTAATGCATTTTA CTGCGTTTGAATACTACAGCATGGAATAACAAAATTGAACAAGTCAAAACTCTGTTCCT TTTTCTTATTTTGGCTTAGTTACGATTAATAGGAGTAGCTTGGGGGCATTTGTATTCAG ATGTCAGAGGTGAAATTCTTAGATTTTCTGGAGACAAACAACTGCGAAAGCATTTGCC TAAAATACTTCCATTAATCAAGAACGAAAGTTAAGGGAGTGAAGACGATCAGATACCG TCGTAATCTTAACCATAAACTATGCCGACTAGGTTTTGGATGAAAGATTTTTAAATAAG AAAATTCCTTTTGGAAATTTCTTAGATTGCTTCCTTCAGTACCTTATGAGAAATCAAAGT CTTTGGGTTCTGGGGCGAGTATTCGCGCAAGCGAGAAAGTTAAAAGAATTGACGGAA GGGCACCACCAGG-3’ |
| Sequence insert for the positive control plasmid of the P. knowlesi LAMP |
| 5‘- TTCTGACATCTTTGCATGGTGTAATCCTCCAGACCAATTTACACAGATATCAGCACAGT GGTGGTTCAGCTTAGAAGCTCCATCTATGGATGCACCTGCACACGACTGCTGAAATT GGAATAACCCATCAAAGACAGGACAGTCTGTCGCTTCTCCTACATTGAAACGTTTCAA TTGGCATGTGAAATCACGATAATTTTCCATGGAAATGGAAGATAAGAAATCTACATATT CGTAATCATGGAAGAGAGTCAACTCGTTCACATCACTCTTGTGTGGTCTGTACACCTC CATGTATTTGTATAAATTGTACGACACGATTAGCGAGTGTGTCATTCGGATTCTCTGTG GCTTCATCGGATGGCCTGCT-3‘ |
Table 3.
Detection limits of LAMP approaches as defined with positive control plasmids
| LAMP approach | Detection limits (calculated copy numbers per reaction) |
|---|---|
| Plasmodium genus LAMP | 104 |
| P. falciparum LAMP | 1 |
| P. vivax LAMP | 8 |
| P. malariae LAMP | 29 |
| P. ovale LAMP | 1 |
| P. knowlesi LAMP | 1 |
Case Definitions
Based on the reference diagnostics as described above, a sample was considered as truly positive for the presence of DNA of the Plasmodium sp. which was identified by the assessed in-house-LAMP approach if:
either the in-house-LAMP result matched the diagnostic results from the previous study [28] based on microscopy, Meridian illumigene Malaria LAMP or reference PCR of the Bernhard Nocht Institute (the latter only performed in the case of contradicting results between microscopy and illumigene LAMP as detailed elsewhere [28])
or the in-house-LAMP result matched the results of the FTD Malaria differentiation PCR irrespective of the microscopic results.
If none of these prerequisites was matched, an in-house-LAMP signal in the expected melting temperature range of truly positive samples was considered as false positive.
In the case of discrepancy between species identification by microscopy and species identification by PCR, the result of less investigator-dependent PCR was accepted as more likely to be true.
A negative in-house-LAMP reaction was considered as truly negative, if no other approach from the reference diagnostics panel indicated a mismatching positive result. If in-house-LAMP was negative, while microscopy was concordantly negative and only one other molecular approach showed a positive result, the result of the other molecular approach was considered as potentially false positive.
The comparator for the LAMP-assessment was defined by these case definitions. Figure 1 shows the diagnostic approaches as detailed above. Only illumigene Malaria LAMP was performed without prior nucleic acid extraction.
Figure 1.
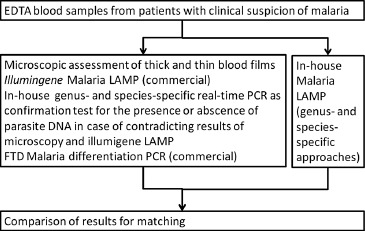
Flow chart of the performed diagnostic algorithm with fresh EDTA blood samples
Statistics
Sensitivity and specificity were descriptively demonstrated for this case-control-based assessment using the software Microsoft Excel version 2007 (Microsoft Corporation, Redmond, USA).
Proof-of-Principle Assessment with Hemolytic EDTA Blood
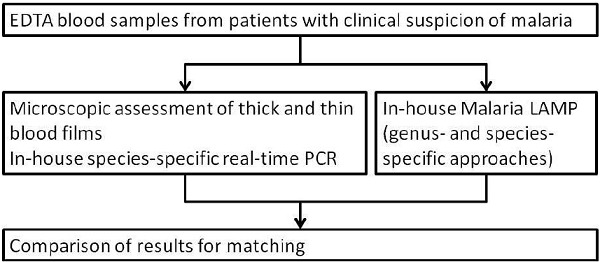
Residual materials from a total of 247 hemolytic EDTA blood samples of patients with suspicion of malaria, which had already been used in previous evaluation studies [31, 42], were included as a proof-of-principle assessment of the LAMP approach with such complex sample materials. All samples had been assessed by microscopy as described above and by SYBR-green-based, species-specific in-house real-time PCR as described elsewhere [30, 31, 42]. In the case of discrepancy between microscopy-based and PCR-based species differentiation, the result of less-investigator-dependent PCR was accepted as more likely to be true. As the available quantities of residual materials of nucleic acid extractions varied, not all assessments could be performed for all samples. Figure 2 shows a flow diagram of the assessments.
Figure 2.
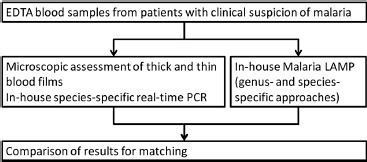
Flow chart of the performed diagnostic algorithm with hemolytic EDTA blood samples
Ethics
The blinded use of residual materials from the diagnostic routine for test comparison and evaluation purposes was allowed by the Ethics Committee of the Medical Association of Hamburg, Germany (registration number WF-046/16) in line with national laws.
Results
Proportion of Positive Samples for the Case-Control-Based Approach
In line with the case definitions above, 243 out of 523 EDTA blood samples (46.5%) were classified as positive for the presence of Plasmodium spp. DNA. The identification on species level showed 217 out of 243 samples containing P. falciparum (89.3%), 17 out of 243 samples containing P. vivax (7.0%), 6 out of 243 samples containing P. malariae (2.5%), and 7 out of 243 samples containing P. ovale (2.9%). P. knowlesi was not detected. Further, there were 3 samples (1.2%) with a mixed infection of P. falciparum and P. vivax, as well as 1 sample (0.4%) with a mixed infection of P. ovale and P. falciparum.
Test Characteristics of In-House Malaria LAMP
Genus-specific in-house malaria LAMP identified 234 out of 243 samples, resulting in a sensitivity of 96.3% (Table 4), comprising 66 samples without parasites in microscopy, 45 at the microscopic detection limit of ≤50 parasites/μL, 6 between 50 and 99/μL, 30 between 100 and 999/μL, 43 between 1,000 and 9,999/μL, 20 between 10,000 and 99,999/μL, and 33 with ≥1% infected erythrocytes. The mean time-to-positivity was 15.1 min (±5.3 min) (standard deviation [SD]), with a median of 14 min. The 9 false negative samples comprised 5 samples with parasitemia below the microscopic detection limit, but also 1 sample with P. vivax and 1 with P. ovale with parasitemia of ≤50/μL for each sample, and 2 P. falciparum-positive samples with parasitemia of 112/μL and 1%, respectively. Of note, the latter 2 false negative results were not reproducible, when the assessments were exemplarily repeated after unblinding at the end of the study. In addition, 3 false positive signals were detected, resulting in specificity of 98.9% (277/280) (Table 4). The mean time-to-positivity was 20.8 min (±18.2 min) with a median of 23 min. In addition, there were 26 non-specific LAMP-signals outside the temperature range of positive samples, which was between 82.95 °C and 83.93 °C. The mean time-to-positivity was 3.9 min (±6.2 min), with a median of 1.5 min.
Table 4.
Sensitivity and specificity of malaria LAMP in truly positive and negative malaria samples according to the case definitions as presented in the Materials and Methods section
| Sensitivity | Specificity | |
|---|---|---|
| Plasmodium genus LAMP | 96.3% (234/243) | 98.9% (277/280) |
| P. falciparum LAMP | 71.0% (154/217) | 90.8% (278/306) |
| P. vivax LAMP | 82.4% (14/17) | 95.3% (482/506) |
| P. malariae LAMP | 100% (6/6) | 100% (517/517) |
| P. ovale LAMP | 100% (7/7) | 98.8% (511/517) |
| P. knowlesi LAMP | n.a. | 92.9% (486/523) |
n.a. = not assessable.
P. falciparum-specific LAMP showed sensitivity of 71.0%, correctly identifying 154 out of 217 P. falciparum positive samples (Table 4). There was a marked influence of parasitemia. While only 35.5% (22/62) samples were positive by LAMP in the case of submicroscopic positive samples, the sensitivity was 50% (19/38) for samples with parasitemia ≤50/μL and 84% (21/25) for samples with parasitemia between 51/μL and 500/μL. All samples with higher parasitemia (92/92) were positive in LAMP. Time-to-positivity in truly positive samples was 27.3 (±6.1) min with a median of 26.0 min. The specificity was 90.8% (278/306) with 2 false positive results in samples with P. ovale, 1 false positive result in a sample with P. vivax, and 25 false positive results in negative samples. Time-to-positivity in false positive samples was 32.0 (±13.2) min with a median of 38.5 min. There was a wide melting temperature range between 80.57 °C and 86.11 °C for truly positive samples, which was, however, associated with only 3 non-specific reactions with negative samples outside this range. Time-to-positivity in those samples with non-specific signals was 1.6 (±0.1) min with a median of 1.5 min.
P. vivax-specific LAMP identified 82.4% (14/17) P. vivax-positive samples (Table 4). The failed reactions comprised 1 submicroscopically positive sample and 2 out of 5 (40%) samples with parasitemia ≤50/μL. All positive samples with higher parasitemia were correctly identified. Time-to-positivity of truly positive samples was 17.2 (±10.3) min with a median of 12.4 min. The melting temperature of positive samples ranged between 86.04 °C and 87.01 °C. Within this temperature range, 24 false positive results were detected, comprising 15 samples with P. falciparum and 9 negative samples. Time-to-positivity in false positive samples was 36.4 (±4.2) min with a median of 38.1 min. The specificity was 95.3% (482/506) (Table 4). The observed 31 non-specific LAMP signals outside this temperature range comprised 18 samples with P. falciparum and 13 negative samples. Time-to-positivity in the case of non-specific signals was 31.1 (±13.1) min with a median of 37.0 min.
P. malariae-specific LAMP showed a sensitivity of 100% (6/6) and specificity of 100% (517/517). Parasitemia of positive samples was low, ranging from submicroscopic parasite densities to 360/μL. The melting temperature range was 85.12 °C to 85.82 °C. Time-to-positivity in truly positive samples was 19.7 (±2.1) min with a median of 19.9 min. A total of 7 non-specific signals were observed, comprising 4 samples with P. falciparum and 3 negative samples. Time-to-positivity in the case of non-specific signals was 28.0 (±15.9) min with a median of 39.3 min.
P. ovale-specific LAMP was characterized by 100% sensitivity (7/7) with 98.8% specificity (511/517). Parasitemia in the positive samples ranged from <50/μL to 20,900/μL. Time-to-positivity in truly positive samples was 16.9 (±9.9) min with a median of 13.3 min. False positive results within the melting temperature range of truly positive samples between 84.13 °C and 85.22 °C were observed for 5 samples with P. falciparum and 1 negative sample. Time-to-positivity in false positive samples was 29.5 (±8.1) min with a median of 28.5 min. In addition, 13 samples comprising 7 negative samples and 4 samples with P. falciparum, as well as 1 sample with P. vivax and 1 sample with P. malariae, showed non-specific signals. Time-to-positivity in samples with non-specific signals was 23.4 (±17.3) min with a median of 32.8 min.
As P. knowlesi was not observed within the assessed samples, a sensitivity value cannot be defined. Assuming a melting temperature range of 84 °C to 87 °C based on the melting temperature of the positive control plasmids between 85 °C and 86 °C ± 1 °C, a total of 37 false positive signals were observed. These comprised 20 samples with P. falciparum, 1 sample with P. vivax, and 16 negative samples, resulting in a specificity of 92.9% (486/523). Time-to-positivity in false positive samples was 36.1 (±8.3) min with a median of 38.8 min. Outside this hypothetical melting temperature range, 17 non-specific signals were observed, comprising 12 samples with P. falciparum, 3 samples with P. vivax, and 2 negative samples. Time-to-positivity in the case of non-specific signals was 11.2 (±13.8) min with a median of 4.0 min.
Comparison of LAMP and Microscopy. If only microscopically positive samples were counted, sensitivity would change to 97.7% (173/177) for Plasmodium genus, 86.6% (131/153) for P. falciparum, 87.5% (14/16) for P. vivax, 100% (5/5) for P. malariae, and 100% (6/6) for P. ovale.
Evaluation of LAMP with Hemolytic Blood Samples. To assess malaria LAMP with the more complex sample matrices of hemolytic blood, a small proof-of-principle assessment was performed. Among a total of 168 malaria-positive hemolytic EDTA blood samples with parasitemia from submicroscopic levels up to 10%, all showed a positive signal in the Plasmodium genus-specific malaria LAMP (Table 5). For 1 sample, positivity was confirmed neither by microscopy nor by species-specific SYBR-green PCR but by commercial genus-specific PCR (altona Diagnostics) only. The mean time to detect a positive result was 11.1 (±3.3) min, and the median time was 10.5 min. The melting temperature range for positive samples was between 82.48 °C and 85.15 °C. Among 79 negative samples, 7 false positive results were detected, resulting in specificity of 91.1% (72/79) (Table 5). The time-to-positivity for the false positive signals was 16.5 (±16.1) min with a median of 13.3 min. In addition, 1 non-specific signal was observed 1.8 min after the onset of measuring.
Table 5.
Sensitivity and specificity of malaria LAMP in truly positive and negative hemolytic malaria samples according to the case definitions as presented in the Materials and Methods section
| Sensitivity | Specificity | |
|---|---|---|
| Plasmodium genus-specific LAMP | 100% (168/168) | 91.1% (72/79) |
| P. falciparum LAMP | 92.0% (126/137) | 99.1% (107/108) |
| P. vivax LAMP | 100% (16/16) | 100% (230/230) |
| P. malariae LAMP | 100% (3/3) | 99.2% (243/245) |
| P. ovale LAMP | 100% (8/8) | 95.0% (228/240) |
| P. knowlesi LAMP | n.a. | 98.4% (242/246) |
n.a. = not assessable. Different total numbers of assessments are due to varying volumes of available residual sample material, not allowing for performing all assays with all samples.
Focusing on P. falciparum-specific LAMP, 126 out of 137 (92.0%) P. falciparum-positive hemolytic samples were correctly identified (Table 5). Parasitemia in the 11 false negative samples comprised 7 microscopically negative samples, 3 samples with parasitemia ≤50/μL, and 1 sample with parasitemia of 64/μL. The time-to-positivity for truly positive samples was 25.3 (±4.0) min with a median of 25.0 min. In contrast, specificity was 99.1% with 1 false positive signal in a sample without plasmodia DNA as detected by the reference methods that became positive 29.5 min after the onset of measuring (Table 5). In another negative sample, a non-specific signal outside the observed melting temperature range between 81.58 °C and 82.88 °C for positive hemolytic samples was observed after 2.8 min.
P. vivax-specific LAMP-primers correctly identified 100% of P. vivax-positive hemolytic blood samples (16/16) (Table 5) with parasitemia ranging from submicroscopic parasite density up to 19,000/μL. The observed melting temperature range for the hemolytic samples was 85.72 °C to 86.41 °C. The time-to-positivity for the truly positive samples was 12.5 (±1.7) min with a median of 12.5 min. No false positive signals within the melting temperature range were observed, resulting in 100% specificity (230/230) (Table 5). However, 18 non-specific signals were detected outside this range, comprising 12 samples with P. falciparum, 5 negative samples, and 1 sample with P. ovale. The time-to-positivity for the non-specific signals was 38.5 (±1.6) min with a median of 39.4 min.
Focusing on P. ovale-specific LAMP, 100% of hemolytic blood samples (8/8) with P. ovale were correctly identified (Table 5). Parasite density ranged from submicroscopic parasitemia to 714/μL, the melting temperature of positive samples was between 84.65 °C and 85.02 °C. The time-to-positivity for the truly positive samples was 18.0 (±2.9) min with a median of 17.9 min. A total of 12 false positive signals were observed, which occurred in samples with P. falciparum without exemption, resulting in specificity of 95% (228/240) (Table 5). The time-to-positivity for the false positive samples was 29.2 (±6.0) min with a median of 29.0 min. Further, 17 non-specific signals were observed outside the melting temperature range of truly positive samples. Again, this phenomenon was only observed in samples with P. falciparum. The time-to-positivity for the samples with non-specific signals was 32.8 (±6.6) min with a median of 35.8 min.
For P. malariae-specific LAMP-primers, a sensitivity of 100% (3/3) was observed for the assessed hemolytic samples (Table 5). Parasitemia ranged from 1400/μL to 2640/μL. The melting temperature of the truly positive samples ranged from 84.82 °C to 86.02 °C. The time-to-positivity for the truly positive samples was 21.0 (±0.9) min with a median of 21.3 min. Two false positive samples were observed, comprising 1 negative sample, which became positive 3.8 min after the onset of measuring, and 1 sample with P. falciparum, which became positive after 24 min. The resulting specificity was 99.2% (243/245) (Table 5). In addition, 26 non-specific signals were observed in 14 negative samples and 9 samples with P. falciparum, as well as in 1 sample with P. vivax, 1 sample with P. ovale, and 1 sample containing unidentified Plasmodium spp. The time-to-positivity for the non-specific signals was 5.2 (±4.5) min with a median of 4.5 min.
P. knowlesi-positive hemolytic samples were not available, not allowing for calculations of sensitivity of P. knowlesi-specific LAMP-primers. Assuming a likely melting temperature range between 84 °C and 87 °C based on the values measured with the positive control plasmid ±1 °C, 4 false positive samples were observed, comprising 2 samples with P. falciparum, 1 sample with P. vivax, and 1 negative sample. Accordingly, the observed specificity for hemolytic samples was 98.4% (242/246) (Table 5). The time-to-positivity for the false positive samples was 39.5 (±0.4) min with a median of 39.5 min.
Discussion
Other than for genus-specific LAMP for malaria [28], commercial products for species-specific malaria LAMP are still missing so far. The present study was performed to assess the diagnostic characteristics of genus- and species-specific in-house LAMP for the detection of malaria after automated nucleic acid extraction from fresh EDTA blood, as well as from hemolytic blood samples. As suggested by previous assessments [24–28, 46], genus-specific LAMP targeting mitochondrial DNA shows very good sensitivity and specificity of more than 98% each [28] in preparations from fresh EDTA blood samples compared with other microscopic and molecular diagnostic approaches. However, the observed failed genus-specific malaria LAMP reaction in a sample with high parasitemia of 1% P. falciparum is bothersome. Although this finding was not reproducible after unblinding of the study, this fact does not solve the problem that the diagnosis might have been missed in a real-life diagnostic setting. Next to this, the specificity of the assessed genus-specific LAMP approach considerably drops if appropriate pre-analytic conditions cannot be maintained, and hemolytic samples have to be assessed. Anyway, even in fresh EDTA samples, a recently assessed commercial genus-specific malaria LAMP approach scored better [28] than the here assessed approach. Considering the fact that this commercial approach [24–28] did not even require sophisticated nucleic acid extraction procedures and could be operated as a real point-of-care system in this way, the genus-specific malaria LAMP approach which was used in this study did not challenge the present state of the art. Other study groups have introduced malaria LAMP approaches without nucleic acid extraction as well [1, 47].
Compared with recent reviews of the literature [21–23], both the sensitivity and the specificity of the newly developed and evaluated LAMP primers for P. falciparum and P. vivax targeting chromosomal genes fell short of our expectations. The relatively high proportion of samples with submicroscopic and very low parasitemia in this study is a likely reason for the observed low sensitivity. For both species-specific LAMP approaches, a clear association between parasitemia and likelihood of observing a positive LAMP signal was detected. Due to the relatively high numbers of samples with low parasite density of P. falciparum, the phenomenon is especially well documented for P. falciparum LAMP. The reduced reliability of P. falciparum LAMP up to a parasite density of 500/μL is bothersome, as the LAMP approach does not score better than traditional immunochromatographic rapid diagnostic test (RDT) devices in this way. The observed specificity was dissatisfying as well, as it leads to interpretation problems in the case of positive test results in low-endemicity settings. Interestingly, both sensitivity and specificity were better in hemolytic blood. The latter phenomenon could be explained by increased inhibition in the hemolytic blood [48], making the detection of late non-specific LAMP signals less likely.
For P. malariae and P. ovale, we had to establish LAMP approaches targeting the 18S ribosomal RNA gene as described [15, 45], although this choice was discouraged by the primer design software due to the likelihood of cross-reactions (data not shown). We did so because attempts with other genes had failed as mentioned above. In initial test series (data not shown), the designs by Han et al. [45] showed less cross-binding and non-specific reactions than the designs by Lau et al. [15] and were therefore chosen. Both with fresh EDTA blood and hemolytic blood, the LAMP primers targeting the 18S rRNA gene of P. malariae scored very well with sensitivity and specificity of >99% each. For the P. ovale LAMP approach, however, the expected specificity problems were observed with increased abundance in the hemolytic samples. Cross-reactions with P. falciparum were the most frequent reasons for false positive signals with the P. ovale LAMP primers targeting the 18S rRNA gene. Due to such problems regarding the identification of reliable species-specific LAMP primers, other authors suggested on-site-sequencing of LAMP-products by MINION™ nanopore sequencing [49]. Due to the costs of such an approach, this is, however, an approach of mere academic value at present.
Due to the complete absence of P. knowlesi in the assessed samples, no sensitivity values for the P. knowlesi-specific primers could be calculated. However, based on a likely spread around the melting temperature of the positive control plasmid, a considerably number of false positive results could be identified. As already observed for P. falciparum- and P. vivax-specific LAMP, the frequency of false positive results was lower in hemolytic materials than in nucleic acid extractions of fresh EDTA blood samples. Studies in endemic areas of P. knowlesi are necessary to thoroughly evaluate the performance of LAMP with blood samples of patients with P. knowlesi [50–53].
In summary, the assessed LAMP approaches both for genus- and species-specific identification of Plasmodium spp. did not achieve the better performance characteristics as described by others [15, 27, 32, 54, 55] with sensitivity ranging from 94.8% to 100% and specificity ranging from 96.7% to 100% in previous studies [15, 27, 54]. Newly developed LAMP protocols targeting chromosomal genes were associated with sensitivity problems in samples with submicroscopic and very low parasitemia. However, incidental failures of the LAMP reaction as observed for the genus-specific approach with an individual highly parasitemic sample limit the diagnostic value of the introduced LAMP approach as well.
Considering the limitations as observed in this study, the easy-to-apply diagnostic approach may theoretically serve as a tool for epidemiologic screenings at population level in areas where sophisticated diagnostic strategies cannot be applied [56–62]. In such settings, the known performance characteristics of the described LAMP schemes can be used to calculate the true prevalence in the assessed populations based on diagnostic accuracy-adjusted methods [63]. This is well in line with previous recommendations for the use of malaria-specific LAMP for surveillance purposes [64, 65].
For diagnostic purposes, however, the observed performance characteristics especially of species-specific malaria LAMP targeting chromosomal genetic elements were dissatisfying. Our approach clearly failed to overcome the obstacles which prevented the commercial availability of species-specific malaria LAMP so far. Our experience may be of value for other groups working on the establishing of reliable species-specific malaria LAMP approaches by clearly demonstrating the limitations that we observed.
Conclusions
The observed species-specific in-house malaria LAMP protocols showed limitations regarding sensitivity and specificity, which make them unsuitable for use in the diagnostic routine setting. The genus-specific approach showed higher reliability but did not score better than an available commercial platform, which works as a real point-of-care approach without previous nucleic acid extraction [28]. If any, one might consider the use of our described easy-to-teach and easy-to-apply procedure as an option for surveillance screenings on population level in resource-limited settings, if diagnostic accuracy-adjusted methods are applied to calculate the true prevalence based on the known test characteristics.
Figure 1.
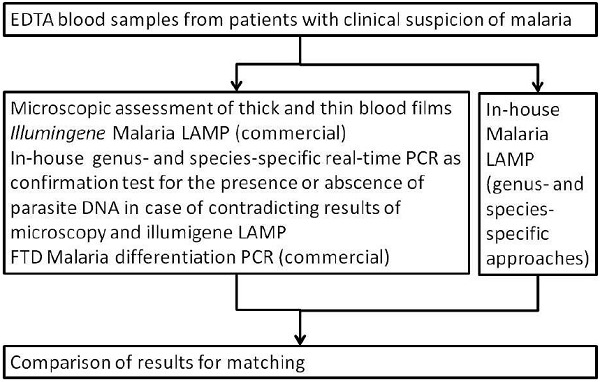
Figure 2.
Acknowledgements
Simone Priesnitz, Annett Michel, and Danny Pergande are gratefully acknowledged for excellent technical assistance.
Footnotes
Funding Sources
The nucleic acid extraction, the FTD Malaria differentiation PCRs and the LAMP assessments were funded by grant 28K2-S-45 1618 “Evaluation of automated loop-mediated isothermal amplification (LAMP) for easy and rapid proof of malaria for tropical deployments” of the German Ministry of Defense (MoD) awarded to Hagen Frickmann and Ralf Matthias Hagen. The sponsor did not have any role in the collection, analysis, and interpretation of data, in the writing of the report, and in the decision to submit the article for publication.
Authors' Contribution
RMH, SP, ET and HF planned the study. LW designed the in-house LAMP primers. HK, MH, SR and RH performed the experiments. HK and HF analyzed the result and wrote the paper. All authors have read and corrected the manuscript.
Conflict of Interest
Nothing to report.
References
- 1.Poon LL, Wong BW, Ma EH, Chan KH, Chow LM, Abeyewickreme W, et al. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem. 2006;52:303–6. [DOI] [PubMed] [Google Scholar]
- 2.Chen JH, Lu F, Lim CS, Kim JY, Ahn HJ, Suh IB, et al. Detection of Plasmodium vivax infection in the Republic of Korea by loop-mediated isothermal amplification (LAMP). Acta Trop. 2010;113:61–5. [DOI] [PubMed] [Google Scholar]
- 3.Buates S, Bantuchai S, Sattabongkot J, Han ET, Tsuboi T, Udomsangpetch R, et al. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) for clinical detection of Plasmodium falciparum gametocytes. Parasitol Int. 2010;59:414–20. [DOI] [PubMed] [Google Scholar]
- 4.Lucchi NW, Demas A, Narayanan J, Sumari D, Kabanywanyi A, Kachur SP, et al. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS One. 2010;5:e13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdul-Ghani R, Al-Mekhlafi AM, Karanis P. Loop-mediated isothermal amplification (LAMP) for malarial parasites of humans: would it come to clinical reality as a point-of-care test? Acta Trop. 2012;122:233–40. [DOI] [PubMed] [Google Scholar]
- 6.Lu F, Gao Q, Zhou H, Cao J, Wang W, Lim CS, et al. Molecular test for vivax malaria with loop-mediated isothermal amplification method in central China. Parasitol Res. 2012;110:2439–44. [DOI] [PubMed] [Google Scholar]
- 7.Han ET. Loop-mediated isothermal amplification test for the molecular diagnosis of malaria. Expert Rev Mol Diagn. 2013;13:205–18. [DOI] [PubMed] [Google Scholar]
- 8.Patel JC, Oberstaller J, Xayavong M, Narayanan J, DeBarry JD, Srinivasamoorthy G, et al. Real-time loop-mediated isothermal amplification (RealAmp) for the species-specific identification of Plasmodium vivax. PLoS One. 2013;8:e54986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiang MS, Greenhouse B, Rosenthal PJ. Point of care testing for malaria using LAMP, loop mediated isothermal amplification. J Infect Dis. 2014;210:1167–9. [DOI] [PubMed] [Google Scholar]
- 10.Mohon AN, Elahi R, Khan WA, Haque R, Sullivan DJ, Jr, Alam MS. A new visually improved and sensitive loop mediated isothermal amplification (LAMP) for diagnosis ofsymptomatic falciparum malaria. Acta Trop. 2014;134:52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oriero EC, Okebe J, Jacobs J, Van Geertruyden JP, Nwakanma D, D-Alessandro U. Diagnostic performance of a novel loop-mediated isothermal amplification (LAMP) assay targeting the apicoplast genome for malaria diagnosis in a field setting in sub-Saharan Africa. Malar J. 2015;14:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oriero CE, Van Geertruyden JP, Jacobs J, D-Alessandro U, Nwakanma D. Validation of an apicoplast genome target for the detection of Plasmodium species using polymerase chain reaction and loop mediated isothermal amplification. Clin Microbiol Infect. 2015;21:686.e1–7. [DOI] [PubMed] [Google Scholar]
- 13.Port JR, Nguetse C, Adukpo S, Velavan TP. A reliable and rapid method for molecular detection of malarial parasites using microwave irradiation and loop mediated isothermal amplification. Malar J. 2014;13:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britton S, Cheng Q, Grigg MJ, Poole CB, Pasay C, William T, et al. Sensitive Detection of Plasmodium vivax Using a High-Throughput, Colourimetric Loop Mediated Isothermal Amplification (HtLAMP) Platform: A Potential Novel Tool for Malaria Elimination. PLoS Negl Trop Dis. 2016;10:e0004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau YL, Lai MY, Fong MY, Jelip J, Mahmud R. Loop-Mediated Isothermal Amplification Assay for Identification of Five Human Plasmodium Species in Malaysia. Am J Trop Med Hyg. 2016;94:336–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohon AN, Lee LD, Bayih AG, Folefoc A, Guelig D, Burton RA, et al. NINA-LAMP compared to microscopy, RDT, and nested PCR for the detection of imported malaria. Diagn Microbiol Infect Dis. 2016;85:149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ocker R, Prompunjai Y, Chutipongvivate S, Karanis P. MALARIA DIAGNOSIS BY LOOP-MEDIATED ISOTHERMAL AMPLIFICATION (LAMP) IN THAILAND. Rev Inst Med Trop Sao Paulo. 2016;58:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perera RS, Ding XC, Tully F, Oliver J, Bright N, Bell D, et al. Development and clinical performance of high throughput loop-mediated isothermal amplification for detection of malaria. PLoS One. 2017;12:e0171126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh R, Singh DP, Savargaonkar D, Singh OP, Bhatt RM, Valecha N. Evaluation of SYBR green I based visual loop-mediated isothermal amplification (LAMP) assay for genus and species-specific diagnosis of malaria in P. vivax and P. falciparum endemic regions. J Vector Borne Dis. 2017;54:54–60. [PubMed] [Google Scholar]
- 20.Zhang Y, Yao Y, Du W, Wu K, Xu W, Lin M, et al. Development of loop-mediated isothermal amplification with Plasmodium falciparum unique genes for molecular diagnosis of human malaria. Pathog Glob Health. 2017;111:247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oriero EC, Jacobs J, Van Geertruyden JP, Nwakanma D, D-Alessandro U. Molecular-based isothermal tests for field diagnosis of malaria and their potential contribution to malaria elimination. J Antimicrob Chemother. 2015;70:2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth JM, Korevaar DA, Leeflang MM, Mens PF. Molecular malaria diagnostics: A systematic review and meta-analysis. Crit Rev Clin Lab Sci. 2016;53:87–105. [DOI] [PubMed] [Google Scholar]
- 23.Mukry SN, Saud M, Sufaida G, Shaikh K, Naz A, Shamsi TS. Laboratory Diagnosis of Malaria: Comparison of Manual and Automated Diagnostic Tests. Can J Infect Dis Med Microbiol. 2017;2017:9286392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucchi NW, Gaye M, Diallo MA, Goldman IF, Ljolje D, Deme AB, et al. Evaluation of the Illumigene Malaria LAMP: A Robust Molecular Diagnostic Tool for Malaria Parasites. Sci Rep. 2016;6:36808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Koninck AS, Cnops L, Hofmans M, Jacobs J, Van den Bossche D, Philippé J. Diagnostic performance of the loop-mediated isothermal amplification (LAMP) based illumigene(®) malaria assay in a non-endemic region. Malar J. 2017;16:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rypien C, Chow B, Chan WW, Church DL, Pillai DR. Detection of Plasmodium Infection by the illumigene Malaria Assay Compared to Reference Microscopy and Real-Time PCR. J Clin Microbiol. 2017;55:3037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponce C, Kaczorowski F, Perpoint T, Miailhes P, Sigal A, Javouhey E, et al. Diagnostic accuracy of loop-mediated isothermal amplification (LAMP) for screening patients with imported malaria in a non-endemic setting. Parasite. 2017;24:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frickmann H, Hinz R, Rojak S, Bonow I, Ruben S, Wegner C, et al. Evaluation of automated loop-mediated amplification (LAMP) for routine malaria detection in blood samples of German travelers—A cross-sectional study. Travel Med Infect Dis. 2018;24:25–30. [DOI] [PubMed] [Google Scholar]
- 29.No authors listed Prise en charge et prevention du paludisme d’importation. Mise à jour 2017 des RPC 2007. Available from: http://www.infectiologie.com/UserFiles/File/spilf/recos/2017-palu-texte-final-flash.pdf (last accessed 31st July 2018).
- 30.Mangold KA, Manson RU, Koay ES, Stephens L, Regner M, Thomson RB, Jr, et al. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol. 2005; 43: 2435–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagen RM, Hinz R, Tannich E, Frickmann H. Comparison of two real-time PCR assays for the detection of malaria parasites from hemolytic blood samples - Short communication. Eur J Microbiol Immunol (Bp). 2015;5:159–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marti H, Stalder C, González IJ. Diagnostic accuracy of a LAMP kit for diagnosis of imported malaria in Switzerland. Travel Med Infect Dis. 2015;13:167–71. [DOI] [PubMed] [Google Scholar]
- 33.Chiodini PL. LAMP in the context of travellers- malaria: a shining light? Travel Med Infect Dis. 2015;13:126–7. [DOI] [PubMed] [Google Scholar]
- 34.Yongkiettrakul S, Jaroenram W, Arunrut N, Chareanchim W, Pannengpetch S, Suebsing R, et al. Application of loop-mediated isothermal amplification assay combined with lateral flow dipstick for detection of Plasmodium falciparum and Plasmodium vivax. Parasitol Int. 2014;63:777–84. [DOI] [PubMed] [Google Scholar]
- 35.Kongkasuriyachai D, Yongkiettrakul S, Kiatpathomchai W, Arunrut N. Loop-Mediated Isothermal Amplification and LFD Combination for Detection of Plasmodium falciparum and Plasmodium vivax. Methods Mol Biol. 2017;1572: 431–43. [DOI] [PubMed] [Google Scholar]
- 36.Mallepaddi PC, Lai MY, Podha S, Ooi CH, Liew JW, Polavarapu R, et al. Development of Loop-Mediated Isothermal Amplification-Based Lateral Flow Device Method for the Detection of Malaria. Am J Trop Med Hyg. 2018. doi: 10.4269/ajtmh.18–0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hashimoto M, Sakamoto H, Ido Y, Tanaka M, Yatsushiro S, Kajimoto K, et al. In situ loop-mediated isothermal amplification (LAMP) for identification of Plasmodium species in wide-range thin blood smears. Malar J. 2018;17:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aydin-Schmidt B, Xu W, González IJ, Polley SD, Bell D, Shakely D, et al. Loop mediated isothermal amplification (LAMP) accurately detects malaria DNA from filter paper blood samples of low density parasitaemias. PLoS One. 2014;9:e103905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamura M, Makimura K, Ota Y. Evaluation of a new rapid molecular diagnostic system for Plasmodium falciparum combined with DNA filter paper, loop-mediated isothermal amplification, and melting curve analysis. Jpn J Infect Dis. 2009;62:20–5. [PubMed] [Google Scholar]
- 40.Singh R, Savargaonkar D, Bhatt R, Valecha N. Rapid detection of Plasmodium vivax in saliva and blood using loop mediated isothermal amplification (LAMP) assay. J Infect. 2013;67:245–7. [DOI] [PubMed] [Google Scholar]
- 41.Ghayour Najafabadi Z, Oormazdi H, Akhlaghi L, Meamar AR, Nateghpour M, Farivar L, et al. Detection of Plasmodium vivax and Plasmodium falciparum DNA in human saliva and urine: loop-mediated isothermal amplification for malaria diagnosis. Acta Trop. 2014;136:44–9. [DOI] [PubMed] [Google Scholar]
- 42.Frickmann H, Hinz R, Hagen RM. Comparison of an automated nucleic acid extraction system with the column-based procedure. Eur J Microbiol Immunol (Bp). 2015;5:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giemsa G. Eine Vereinfachung und Vervollkommnung meiner Methylenblau-Eosin-Färbemethode zur Erzielung der Romanowsky-Nocht’schen Chromatinfärbung. Centralblatt für Bakteriologie. 1904;32:307–13. [Google Scholar]
- 44.Hecht LS, Wegner C, Bunte N, Tannich E, Hess M. A real-time PCR based Malaria-typing assay for the detection and differentiation of all five human pathogenic Plasmodium species. Poster P80. 12. Kongress für Infektionskrankheiten und Tropenmedizin. 2014; Cologne, Germany. [Google Scholar]
- 45.Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, et al. Detection of 4 Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol. 2007;45:2521–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polley SD, Mori Y, Watson J, Perkins MD, González IJ, Notomi T, Chiodini PL, Sutherland CJ. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol. 2010;48:2866–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Modak SS, Barber CA, Geva E, Abrams WR, Malamud D, Ongagna YS. Rapid Point-of-Care Isothermal Amplification Assay for the Detection of Malaria without Nucleic Acid Purification. Infect Dis (Auckl). 2016;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Färnert A, Arez AP, Correia AT, Björkman A, Snounou G, do Rosário V. Sampling and storage of blood and the detection of malaria parasites by polymerase chain reaction. Trans R Soc Trop Med Hyg. 1999;93:50–3. [DOI] [PubMed] [Google Scholar]
- 49.Imai K, Tarumoto N, Misawa K, Runtuwene LR, Sakai J, Hayashida K, et al. A novel diagnostic method for malaria using loop-mediated isothermal amplification (LAMP) and MinION™ nanopore sequencer. BMC Infect Dis. 2017;17:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iseki H, Kawai S, Takahashi N, Hirai M, Tanabe K, Yokoyama N, et al. Evaluation of a loop-mediated isothermal amplification method as a tool for diagnosis of infection by the zoonotic simian malaria parasite Plasmodium knowlesi. J Clin Microbiol. 2010;48:2509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau YL, Fong MY, Mahmud R, Chang PY, Palaeya V, Cheong FW, et al. Specific, sensitive and rapid detection of human Plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J. 2011;10:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Britton S, Cheng Q, Grigg MJ, William T, Anstey NM, McCarthy JS. A Sensitive, Colorimetric, High-Throughput Loop-Mediated Isothermal Amplification Assay for the Detection of Plasmodium knowlesi. Am J Trop Med Hyg. 2016;95:120–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piera KA, Aziz A, William T, Bell D, González IJ, Barber BE, et al. Detection of Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax using loop-mediated isothermal amplification (LAMP) in a co-endemic area in Malaysia. Malar J. 2017;16:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel JC, Lucchi NW, Srivastava P, Lin JT, Sug-Aram R, Aruncharus S, et al. Field evaluation of a real-time fluorescence loop-mediated isothermal amplification assay, RealAmp, for the diagnosis of malaria in Thailand and India. J Infect Dis. 2014;210:1180–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goyal K, Kaur H, Sehgal A, Sehgal R. RealAmp Loop-Mediated Isothermal Amplification as a Point-of-Care Test for Diagnosis of Malaria: Neither Too Close nor Too Far. J Infect Dis. 2015;211:1686. [DOI] [PubMed] [Google Scholar]
- 56.Labarre P, Gerlach J, Wilmoth J, Beddoe A, Singleton J, Weigl B. Non-instrumented nucleic acid amplification (NINA): instrument-free molecular malaria diagnostics for low-resource settings. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:1097–9. [DOI] [PubMed] [Google Scholar]
- 57.Sirichaisinthop J, Buates S, Watanabe R, Han ET, Suktawonjaroenpon W, Krasaesub S, et al. Evaluation ofloop-mediated isothermal amplification (LAMP) for malaria diagnosis in a field setting. Am J Trop Med Hyg. 2011;85:594–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao ZY, Zhou HY, Xia H, Xu S, Zhu HW, Culleton RL, et al. Adaptation of a visualized loop-mediated isothermal amplification technique for field detection of Plasmodium vivax infection. Parasit Vectors. 2011;4: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Britton S, Cheng Q, Sutherland CJ, McCarthy JS. A simple, high-throughput, colourimetric, field applicable loop-mediated isothermal amplification (HtLAMP) assay for malaria elimination. Malar J. 2015;14:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vallejo AF, Martínez NL, González IJ, Arévalo-Herrera M, Herrera S. Evaluation of the loop mediated isothermal DNA amplification (LAMP) kit for malaria diagnosis in P. vivax endemic settings of Colombia. PLoS Negl Trop Dis. 2015;9:e3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kemleu S, Guelig D, Eboumbou Moukoko C, Essangui E, Diesburg S, Mouliom A, et al. A Field-Tailored Reverse Transcription Loop-Mediated Isothermal Assay for High Sensitivity Detection of Plasmodium falciparum Infections. PLoS One. 2016;11:e0165506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Serra-Casas E, Manrique P, Ding XC, Carrasco-Escobar G, Alava F, Gave A, et al. Loop-mediated isothermal DNA amplification for asymptomatic malaria detection in challenging field settings: Technical performance and pilot implementation in the Peruvian Amazon. PLoS One. 2017;12:e0185742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogan WJ, Gladen B. Estimating prevalence from the results of a screening test. AmJ Epidemiol. 1978;107:71–6. [DOI] [PubMed] [Google Scholar]
- 64.Lucchi NW, Ndiaye D, Britton S, Udhayakumar V. Expanding the malaria molecular diagnostic options: opportunities and challenges for loop-mediated isothermal amplification tests for malaria control and elimination. Expert Rev Mol Diagn. 2018;18:195–203. [DOI] [PubMed] [Google Scholar]
- 65.Tambo M, Auala JR, Sturrock HJ, Kleinschmidt I, Bock R, Smith JL, et al. Evaluation of loop-mediated isothermal amplification as a surveillance tool for malaria in reactive case detection moving towards elimination. Malar J. 2018;17:255. [DOI] [PMC free article] [PubMed] [Google Scholar]


