Abstract
Fibrotic responses involve multiple cellular processes, including epigenetic changes. Epigenetic changes are sensitive to alterations in the tissue microenvironment such as the flux of tricarboxylic acid (TCA) cycle metabolites. TCA metabolites directly regulate epigenetic states, in part by regulating histone modification–related enzymes. Glutaminolysis is a critical metabolic process by which glutamine is converted to glutamate by glutaminase and then to α-ketoglutarate (α-KG), a TCA cycle metabolite. Idiopathic pulmonary fibrosis (IPF) is a disease characterized by aberrant metabolism, including enhanced glutaminolysis. IPF fibroblasts are apoptosis resistant. In this study, we explored the relationship between glutaminolysis and the resistance to apoptosis of IPF fibroblasts. Inhibition of glutaminolysis decreased expression of XIAP and survivin, members of the inhibitor of apoptosis protein (IAP) family. α-KG is a cofactor for JMJD3 histone demethylase, which targets H3K27me3. In the absence of glutamine, JMJD3 activity in fibroblasts is significantly decreased, whereas H3K27me3 levels are increased. Chromatin immunoprecipitation assays confirmed that JMJD3 directly interacts with XIAP and survivin promoter regions in a glutamine-dependent manner. Exogenous α-KG partially restores JMJD3 function and its interaction with the XIAP and survivin promoter regions under glutamine-deficient conditions. Interestingly, α-KG upregulates XIAP, but not survivin, suggesting differential α-KG–dependent and –independent mechanisms by which glutamine regulates these IAPs. Our data demonstrate a novel mechanism of metabolic regulation in which glutaminolysis promotes apoptosis resistance of IPF fibroblasts through epigenetic regulation of XIAP and survivin.
Keywords: glutamine, apoptosis, H3K27me3, JMJD3, IPF fibroblasts
Clinical Relevance
Idiopathic pulmonary fibrosis fibroblasts are apoptosis resistant. This study demonstrates a novel mechanism whereby glutaminolysis epigenetically upregulates the antiapoptotic genes XIAP and survivin in idiopathic pulmonary fibrosis fibroblasts.
Recent studies indicate that fibrotic diseases such as idiopathic pulmonary fibrosis (IPF) are characterized by aberrant metabolism (1, 2), and myofibroblast differentiation is associated with metabolic reprogramming characterized by increased oxidative phosphorylation and aerobic glycolysis (3, 4). Glutaminolysis converts the nonessential amino acid glutamine (Gln) to glutamate through glutaminase (GLS) and then to α-ketoglutarate (α-KG) by glutamate dehydrogenase (5) to enter the tricarboxylic acid (TCA) cycle. This is a key metabolic process that is required for myofibroblast differentiation (6), supporting an important role for glutaminolysis in fibrotic lung disease (2, 6, 7). GLS was reported to be positively correlated with growth rate in normal cells and malignancy in cancers (8). There are two GLS isoforms, GLS1 (the kidney type) and GLS2 (the liver type), and upregulation of GLS1 is associated with increased rates of proliferation (9). GLS1 is significantly increased in lung fibrosis and in fibroblasts treated with the profibrotic mediator transforming growth factor-β (7).
Acquisition of an apoptosis-resistant phenotype contributes to the accumulation of myofibroblasts in the context of aberrant wound repair and fibrosis (10, 11). Studies in cancer cells have shown that extracellular Gln levels modulate apoptosis susceptibility, and Gln deprivation can sensitize cancer cells to apoptosis (12, 13). The precise mechanisms by which cellular metabolism regulates apoptosis are not clearly defined. Recent studies indicate that TCA metabolites can directly affect epigenetic states and alter cell phenotypes by regulating the activities of histone acetyltransferases and histone demethylases (5). Specifically, α-KG is a required cofactor for a subset of histone demethylases, the Jumonji-domain–containing (JMJC) enzymes (14). JMJD3 (jumonji domain–containing protein 3, also called lysine-specific demethylase 6 [KDM6B]) and UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome) belong to the JMJC demethylase family and are H3K27-specific demethylases (15, 16).
Given previous studies demonstrating a role for glutaminolysis in the regulation of cancer cell apoptosis (12) and our studies indicating that glutaminolysis regulates lung myofibroblast differentiation (6), in this work we sought to explore how Gln metabolism may regulate the apoptosis susceptibility of IPF fibroblasts, and to determine whether myofibroblast phenotypes are influenced by metabolically mediated epigenetic changes.
Methods
For details regarding the methods used in this work, see the data supplement.
Cell Culture and Treatments
Human primary IPF lung fibroblasts were derived from deidentified tissues from the University of Alabama at Birmingham Tissue Procurement Facility, which is approved by the University of Alabama at Birmingham Institutional Review Board. The diagnosis of IPF was made by means of a multidisciplinary approach according to American Thoracic Society/European Respiratory Society guidelines (17). All primary cell lines were used before passage 5. When the cells were ∼80% confluent, the full medium (with 2 mM L-Gln) was changed to reduced L-Gln at 0.1 or 0 mM. In some experiments, 2 mM α-KG (AK126628, Ark Pharm Inc.) was added to L-Gln–free medium. The cells were harvested for various assays after 48 hours or as indicated.
siRNA Transfection
siRNA transfections were performed with Opti-MEM (Thermo Fisher Scientific) using lipofectamine RNAi/MAX (Invitrogen). The sequences for GLS1 (NM_0149054) siRNA and nontargeting siRNA are listed in Table 1. IPF lung fibroblasts were kept in full medium (2 mM L-Gln) for 48 hours after transfection and then subjected to various assays.
Table 1.
siRNA Sequences and Primer Sequences for PCR
| Name | Sequence | |
|---|---|---|
| GLS1 (NM_0149054) | siRNA | Sense: 5′-UGCAACGUUUCAGUCUGAAAGAGAA-3′ |
| Antisense: 5′-UUCUCUUUCAGACUGAAACGUUGCA-3′ | ||
| Nontargeting | siRNA | Sense: 5′-UAAGGCUAUGAAGAGAUACUU-3′ |
| Antisense: 5′-GUAUCUCUUCAUAGCCUUAUU-3′ | ||
| XIAP (ENSG000000101966) | RT-PCR | F: 5′-TGTTTCAGCATCAACACTGGCACG -3′ |
| R: 5′- TGCATGACAACTAAAGCACCGCAC -3′ | ||
| ChIP-qPCR | F: 5′- TGCCTGCTTAAATATTACTTTCCTCAAAA-3′ | |
| R: 5′- ACTACACGACCGCTAAGAAACATTCT-3′ | ||
| Survivin (ENSG000000089685) | RT-PCR | F: 5′- AGCCCTTTCTCAAGGACCAC-3′ |
| R: 5′- CAGCTCCTTGAAGCAGAAGAA-3′ | ||
| ChIP-qPCR | F: 5′- ACCACGCCCAGCTAATTT -3′ | |
| R: 5′- CATCACTTGAGTCCTGGAGTTC -3′ | ||
| β-actin | RT-PCR | F: 5′-TGCTATCCAGGCTGTGCTAT-3′ |
| R: 5′- AGTCCATCACGATGCCAG T-3′ |
Definition of abbreviations: ChIP = chromatin immunoprecipitation; F = forward; GLS1 = glutaminase 1; R = reverse.
Apoptosis Assays
After 32 hours of incubation in media with 0, 0.1, or 2 mM of Gln, the cells were treated with Fas ligand (FasL; S8689, Sigma) at 200 ng/ml and cycloheximide (Cell Signaling) at 500 ng/ml or with vehicle control for 16 hours to induce apoptosis. Caspase-3 activity was measured as described previously (18) or subjected to Annexin-V FITC/propidium iodide with a kit (MBL International Corp.) (19).
RNA Extraction and Real-Time RT-PCR
RNA was extracted with an RNeasy Mini Kit (Qiagen) and transcribed into cDNA with a cDNA synthesis kit (Takara Bio). Real-time RT-PCR was performed in triplicate and normalized to β-actin using the ΔΔCt method (20). Primers are listed in Table 1.
Protein/Nuclear Extraction, Immunoblotting, and Demethylase Activity Assays
Whole-cell lysates were collected and quantified with a Micro BCA Protein Assay kit (Thermo Scientific). Nuclear extracts were collected with an EpiQuick Nuclear Extraction kit (Epigentek). Lysates or nuclear extracts were subjected to SDS-PAGE and Western immunoblotting as previously described (21). Anti-XIAP (#2042), anti-survivin (#2808), anti–β-actin (#2128), and anti-H3 (#9715) were obtained from Cell Signaling; anti-GLS1 (cat #12885-1-AP) was obtained from Proteintech; and H3K27me3 (#39155) was obtained from Active Motif. Demethylase JMJD3/UTX activity was measured with nuclear extracts using a kit (#P-3084) from Epigentek.
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) assays (ab500, Abcam) were performed according to the manufacturer’s protocol with minor modifications (22). Antibody against JMJD3 (ab38113) was obtained from Abcam. ChIP-DNA was amplified by real-time PCR with the primers listed in Table 1, using SYBR Green PCR Master Mix (Life Technologies). Results were normalized to input DNA.
Statistical Analysis
Data are presented as the mean ± SD. All data were statistically analyzed using GraphPad Prism 5.0. One-way ANOVA was used to compare multiple groups, and Student’s paired t test was used for comparisons between two groups. A P value of <0.05 was considered to be statistically significant.
Results
Gln Availability Regulates the Apoptosis Susceptibility of IPF Lung Fibroblasts
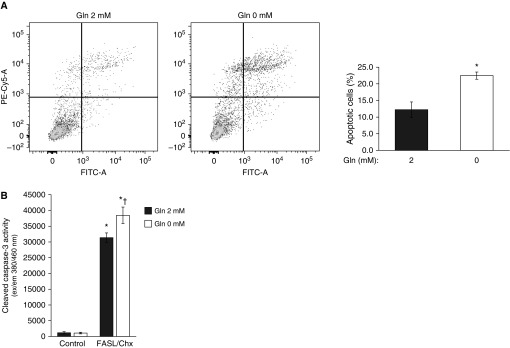
A hallmark of fibrotic diseases, including IPF, is fibroblast acquisition of an apoptosis-resistant phenotype (10). Because Gln may regulate apoptosis pathways (23), we examined the effects of exogenous Gln on susceptibility to apoptosis of IPF fibroblasts. IPF fibroblasts were maintained in media with either 2 mM or 0 mM Gln for 32 hours, and then exposed to a combination of FasL and cycloheximide to induce apoptosis. The lack of an exogenous source of Gln predisposed the IPF fibroblasts to increased rates of apoptosis, as assessed by flow-cytometric analysis of Annexin-V (Figure 1A) and by cleaved caspase-3 activity assays (Figure 1B). This observation suggested that Gln metabolism may regulate apoptosis-/survival-related genes to confer apoptosis resistance to IPF lung fibroblasts.
Figure 1.
The apoptosis assays in idiopathic pulmonary fibrosis (IPF) fibroblasts. Primary IPF fibroblasts were maintained in Dulbecco’s modified Eagle’s medium with or without 2 mM glutamine (Gln) for 32 hours. The cells were then treated with or without a combination of Fas ligand (FasL) and cycloheximide (Chx) for 16 hours before apoptosis was measured. Similar data were obtained in at least three different cell lines; the data shown are from three independent experiments in one representative cell line. (A) Apoptosis was assessed using flow cytometry. Apoptotic cells were labeled with Annexin V-FITC/propidium iodide. Data are given as the percentage (mean ± SD) of apoptotic fibroblasts to total cells. *P < 0.05 compared with the 2 mM Gln group. (B) Cells were collected to measure cleaved caspase-3 activity. *P < 0.05, caspase-3 activity in FasL/Chx-treated cells versus control (non–FasL/Chx-treated) cells; †P < 0.05, 0 mM Gln group compared with the 2 mM Gln group in FasL/Chx-treated cells.
Glutaminolysis Regulates the Expression of Antiapoptotic Genes in IPF Fibroblasts
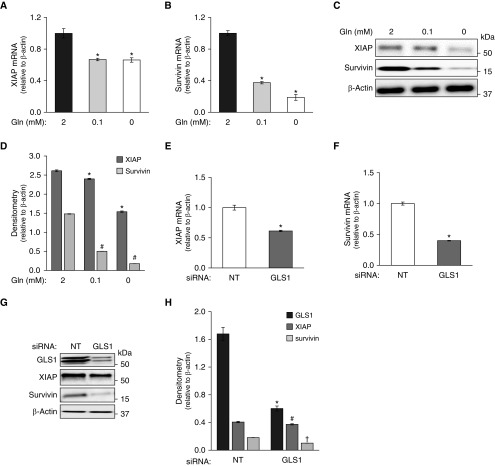
We then examined whether specific genes that contribute to apoptosis resistance may be controlled by Gln metabolism. X-linked inhibitor of apoptosis (XIAP) and survivin belong to the inhibitor of apoptosis protein (IAP) family (24), and have been reported to be upregulated in IPF and to regulate apoptosis susceptibility in lung fibroblasts (25, 26). We found these genes to be significantly downregulated in Gln-deficient media (Gln 0 and 0.1 mM) at both the RNA (Figures 2A and 2B) and protein (Figures 2C and 2D; Figure E1B in the data supplement) levels. This was not an effect on global gene expression, as other apoptosis-related genes, such as BID, were not affected under the same conditions (Figure E1A).
Figure 2.
Expression of the antiapoptotic genes XIAP and survivin. (A–D) IPF fibroblasts were cultured in media with decreasing concentrations of extracellular Gln (2, 0.1, and 0 mM) for 48 hours. * or #, P < 0.05 compared to control of 2 mM Gln of XIAP or survivin expression. (A and B) Expression of XIAP (A) and survivin (B) mRNA was determined by qRT-PCR. (C and D) Expression of XIAP and survivin proteins was assessed by Western blotting, with β-actin as the loading control. (D) Densitometry analysis of XIAP- and survivin-associated signals detected (ratio to β-actin) in C. (E–H) IPF fibroblasts were transfected with nontargeting (NT) or glutaminase 1 (GLS1) siRNA (100 nM) in medium containing 2 mM Gln. (E and F) Expression of XIAP (E) and survivin (F) mRNA was assessed 48 hours after transfection by qRT-PCR. *P < 0.05 compared to siRNA NT control of XIAP or survivin expression. (G and H) Expression of XIAP and survivin proteins was assessed 48 hours after transfection by Western blotting. (H) Densitometry analysis of XIAP- and survivin-associated signals detected (ratio to β-actin) in G. *, #, †, P < 0.05 compared to siRNA NT control group of GLS1, XIAP or survivin, respectively. The results are expressed as mean ± SD from three independent experiments of a representative cell line.
Because GLS1 is the key enzyme regulating glutaminolysis, which converts Gln to glutamate in IPF lung fibroblasts (6), we next examined the effects of silencing this gene with siRNA in the presence of Gln (2 mM)-containing medium. Silencing GLS1 recapitulated the downregulation of XIAP and survivin in Gln-deficient media (Figures 2E–2H), further supporting a role for Gln-glutamate flux in regulating the expression of these antiapoptotic genes.
Gln Metabolism Regulates Histone Modification H3K27me3 in IPF Fibroblasts
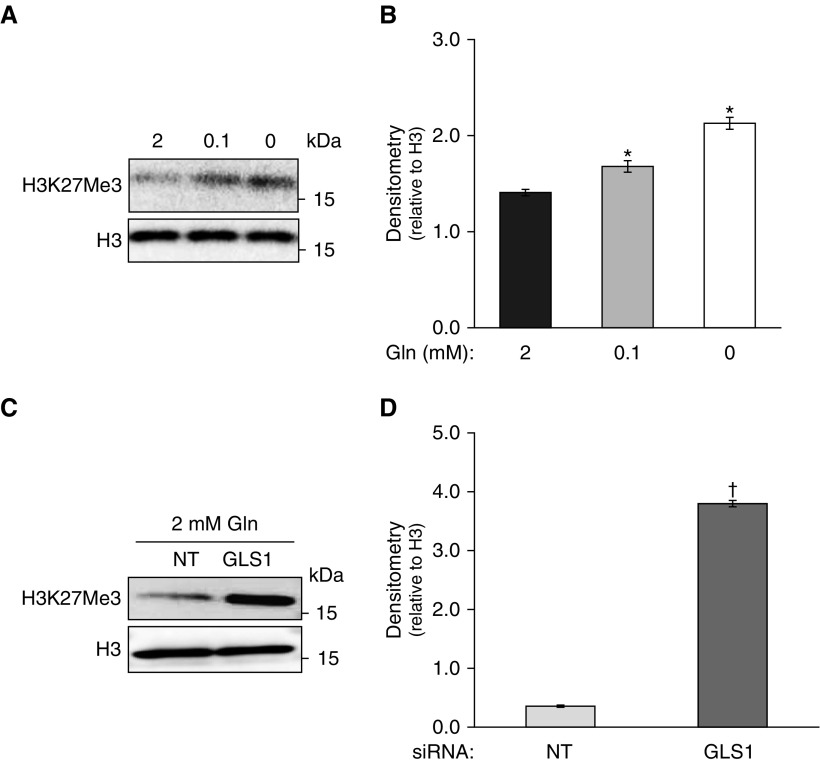
Histone modifications are critical for gene regulation of most biological processes (27), and this may be influenced by the availability of TCA metabolites (28). Gln has been reported to alter histone methylation status (16, 29). We examined whether reduced Gln metabolism alters H3K27me3. With reduced Gln, we observed that levels of H3K27me3 were significantly increased in IPF fibroblasts (Figures 3A, 3B, and E2A). Consistent with that finding, GLS1 silencing similarly increased H3K27me3 levels (Figures 3C and 3D), supporting the concept that glutaminolysis controls the epigenetic reprogramming of IPF fibroblasts. Other histone modifications, such as H3K9Me3 and H3K9Ac, showed no significant changes under the same conditions as in Figure 3A (Figure E2B). Together, these data support a critical role for Gln metabolism in regulating the levels of H3K27me3 in IPF fibroblasts.
Figure 3.
Histone H3K27me3 levels in IPF fibroblasts under different statuses of Gln metabolism. (A and B) IPF fibroblasts were cultured in the presence of decreasing concentrations of extracellular Gln (2, 0.1, and 0 mM) for 48 hours. Histone H3K27me3 levels were assessed by Western blotting, with H3 as the loading control. (B) Densitometry analysis of H3K27me3 signals detected (ratio to H3) in A. The results are expressed as mean ± SD from three independent experiments in a representative cell line. (C and D) IPF fibroblasts were transfected with NT or GLS1 siRNA (100 nM) in media containing 2 mM Gln. (C) Histone H3K27me3 levels were assessed by Western blotting, with H3 as the loading control. (D) Densitometry analysis of H3K27me3 signals detected (ratio to H3) in C. The results are expressed as mean ± SD from three independent experiments in a representative cell line. *P < 0.05 compared with control of 2 mM Gln; †P < 0.05 compared with control of NT siRNA.
The Gln Metabolite α-KG Regulates JMJD3/UTX Activity and H3K27me3 Levels in IPF Fibroblasts
H3K27 methylation is specifically targeted by the JMJD3/UTX demethylases (15). These enzymes contain the JMJC domain and use α-KG as a cofactor for their activity (14). α-KG is a metabolite that is largely derived from Gln metabolism and has been reported to be upregulated in lung myofibroblasts (7). We hypothesized that the observed increase in H3K27me3 levels under conditions of reduced glutaminolysis may be a consequence of decreased α-KG generated from Gln metabolism. To examine this possibility, we measured JMJD3/UTX activity in IPF fibroblasts cultured in the presence and absence of Gln, and observed significantly decreased activity in Gln-free conditions (Figure 4A). Importantly, the addition of cell-permeable α-KG to the culture medium restored JMJD3/UTX activity (Figure 4A). In parallel, the increased levels of H3K27me3 in Gln-free conditions was partially reversed by addition of exogenous α-KG (Figures 4B, 4C, and E2C). Together, these experiments indicate that reduced levels of α-KG, at least partially, account for the decreased activity of JMJD3/UTX demethylases and increased levels of H3K27me3 under conditions of reduced glutaminolysis.
Figure 4.
α-Ketoglutarate (KG) affects the activity of the histone demethylases JMJD3 and UTX, and H3K27me3 levels. (A) Nuclear extract was collected to measure JMJD3/UTX activity from IPF fibroblasts in culture media with 2 mM Gln or 0 mM Gln, or 0 mM Gln with 2 mM α-KG for 48 hours. The results are expressed as mean ± SD from three independent experiments in a representative cell line. *P < 0.05 compared with 2 mM Gln; ‡P < 0.05 compared with 0 mM Gln. (B) JMJD3/UTX substrate H3K27me3 levels changed under the same conditions as in A, assessed by Western blotting, with total H3 as the loading control. (C) Densitometry analysis of H3K27me3 signals detected (ratio to H3) in B. The results are expressed as mean ± SD from three independent experiments in a representative cell line. *P < 0.05 compared with 2 mM Gln; †P < 0.05 compared with 0 mM Gln.
α-KG Regulates the Gene Expression of XIAP, but Not Survivin
Because α-KG rescued the activity of histone demethylases and related H3K27me3 under conditions of Gln depletion, we examined whether α-KG could restore XIAP and survivin expression as well. Interestingly, although XIAP expression was partially restored with the introduction of exogenous α-KG in Gln-free medium, at both the mRNA (Figure 5A) and protein (Figures 5C and 5D) levels, the expression of survivin was not significantly altered (Figures 5B–5D and E3). This indicates that Gln metabolism may regulate the expression of apoptotic-related genes by α-KG–dependent and –independent mechanisms.
Figure 5.
α-KG can partially upregulate XIAP but not survivin. (A and B) IPF fibroblasts were cultured in the presence of extracellular Gln at 2 mM or 0 mM, or 0 mM with added α-KG (2 mM) for 48 hours. (A and B) Expression of XIAP (A) and survivin (B) mRNA was determined by qRT-PCR. (C and D) Expression of XIAP and survivin proteins was assessed by Western blotting, with β-actin as the loading control. (D) Densitometry analysis of XIAP- and survivin-associated signals detected (ratio to β-actin) in C. The results are expressed as mean ± SD from three independent experiments in a representative cell line. * or #, P < 0.05, compared with the expression of XIAP (*) or survivin (#) of 2 mM Gln.
JMJD3 Associates with the Gene Promoter Regions of both XIAP and Survivin
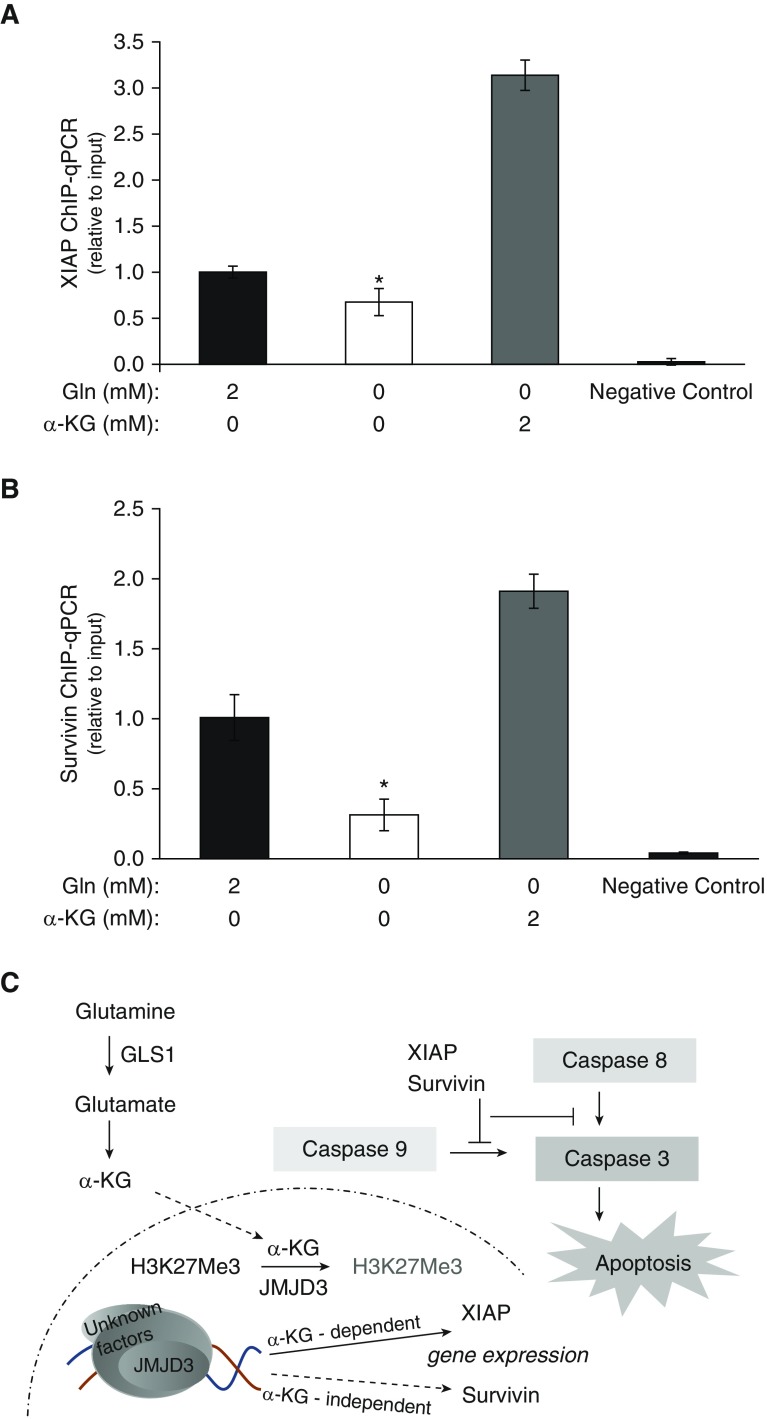
Based on our observation that, in contrast to XIAP levels, survivin levels failed to recover with α-KG reconstitution, we explored whether this TCA metabolite, α-KG, mediates differential association of the JMJD3 demethylase with the promoter regions of these genes. ChIP assays showed that the association of both genes with JMJD3 was significantly reduced by Gln depletion, but was enhanced upon addition of α-KG (Figures 6A and 6B). These data suggest that despite the regulation of JMJD3 binding and activity by Gln metabolism, XIAP and survivin transcriptions are differentially controlled.
Figure 6.
Association of JMJD3 with XIAP and survivin, as assessed by chromatin immunoprecipitation (ChIP) assays. (A and B) Association of JMJD3 with (A) XIAP and (B) survivin in IPF fibroblasts cultured in media with 2 or 0 mM Gln, or 0 mM Gln with 2 mM α-KG for 48 hours. Quantitative ChIP assays were performed to analyze the association of XIAP and survivin with the histone demethylase JMJD3. DNA was immunoprecipitated with the specific antibody JMJD3. Bars represent the relative levels of the PCR product of the XIAP or survivin promoter region’s association with JMJD3. qPCR data were analyzed using the 2−ΔΔCt method, and results normalized to input DNA were expressed as fold changes relative to cells in 2 mM Gln. Bar graphs represent mean ± SD from the average of three independent experiments in a representative cell line. *P < 0.05 compared with 2 mM Gln. (C) Schematic demonstrating that the glutaminolysis metabolite α-KG affects JMJD3, which further affects H3K27me3 levels and the expression of XIAP and survivin in an α-KG–dependent or –independent pathway, which alters the IPF fibroblasts’ susceptibility to apoptosis.
Discussion
Gln is the most abundant amino acid in the body and serves as a critical precursor of many important biological processes (30). In recent years, metabolic reprogramming has been recognized as one of the hallmarks of cancer (31), and restriction of Gln metabolism has been shown to inhibit tumor growth through various mechanisms, including apoptosis induction (23). In line with reports of mechanistic similarities between cancer and IPF biology (32, 33), enhanced glutaminolysis has also been shown in IPF lungs (2, 7). In this report, we show that Gln contributes to the apoptosis resistance of IPF lung fibroblasts, and that reduced Gln metabolism sensitizes IPF fibroblasts to FasL-induced apoptosis, suppresses antiapoptotic gene expression, and alters the epigenetic status of the cells (Figure 6C). These results provide novel insights into the mechanisms by which epigenetic reprogramming mediated by Gln metabolism regulates IPF fibroblast susceptibility to apoptosis. To our knowledge, this is the first study to demonstrate that the antiapoptotic genes XIAP and survivin are regulated by Gln metabolism through α-KG–dependent and –independent mechanisms.
Several prior studies have shown that deficient Gln metabolism can enhance apoptosis in both normal cells and cancer cells through a variety of mechanisms. For example, studies in rat intestinal epithelial cells showed that Gln deprivation induced apoptosis (34), which involves the activation of caspase-3 (35). Similarly, in human colon cancer cells, Gln was shown to inhibit TNF-α–related apoptosis-inducing ligand apoptosis, although the mechanism involved in Gln-mediated protection from apoptosis was not identified (36). Consistent with a role for Gln metabolism in apoptosis resistance, other studies reported that Gln deprivation–induced apoptosis activates different pathways and apoptosis-related genes (37, 38). Due to the heterogeneous nature of IPF fibroblasts, different fibroblast populations would be expected to show some differences in glutaminolysis and/or responses to metabolites. Our data demonstrate that some IPF fibroblasts are more sensitive than others to changes in Gln metabolism. Nonetheless, we observed that Gln deprivation consistently led to decreased expression of the IAP family members XIAP and survivin, and increased activated caspase-3 in IPF lung fibroblasts. The increased apoptosis associated with decreased levels of XIAP and survivin in IPF fibroblasts is consistent with prior studies demonstrating that these IAPs are expressed at increased levels in IPF fibroblasts (25, 26, 39–41), further supporting a role for IAP-mediated fibroblast resistance to apoptosis in IPF.
Emerging evidence demonstrates that cellular metabolism can regulate epigenetic states (42–44). Jumonji-domain (Jmjc-KDM)–containing histone demethylases require the metabolite of glutaminolysis, α-KG, as their cofactor (45). Consistently, we found that deficient glutaminolysis impaired JMJD3 demethylase activity, which was restored by exogenous addition of cell-permeable α-KG. The methylation levels of histone are controlled by a balance of the activities of its respective demethylases and methyltransferases, which could be affected by various metabolic factors. For example, the H3K27me3 methyltransferase EZH2 (29) is regulated by hypoxia-inducible factor 1 (46), which has been reported to be affected by Gln metabolism (47). Currently, the specific role of EZH2 in regulating H3K27me3 in IPF fibroblasts is not known, although it is unlikely that EZH2 expression or activity is directly regulated by Gln. Studies in other cell types have demonstrated that Gln deprivation may affect a different set of histone methylation modifications (16, 29), which could explain the variability of responses in different tissues.
Histone demethylases are known to regulate gene transcription (48–50). JMJD3 was reported to regulate gene expression independently of its demethylase activity (51–53). Our data show that in Gln-deficient medium, exogenous α-KG promotes binding of JMJD3 to the promoter regions of XIAP and survivin. Surprisingly, despite similar binding of JMJD3 to the XIAP and survivin promoters, only XIAP expression levels were dependent on α-KG, suggesting that additional mechanisms are involved in survivin gene transcription (54). Epigenetically, survivin can be regulated by DNA methylation (55) and histone modification (56), both of which can be affected by Gln metabolism (57). A study in cancer cells indicated that p53 recruits DNA methyltransferase 1 to the survivin gene promoter to downregulate its expression (58). PI3K/Akt and NF-κB are implicated in survivin transcriptional regulation (40, 59, 60), and both of these pathways are modulated by Gln metabolism (61). Other factors, such as hypoxia-inducible factor 1α, which is altered by Gln metabolism in IPF fibroblasts (6), can also regulate survivin expression (54, 62). Studies are underway to explore the mechanism by which Gln metabolism regulates survivin expression in our experimental model.
The role of Gln metabolism in the regulation of cancer cell survival and proliferation has generated substantial interest in targeting Gln metabolism as a therapeutic strategy in cancer (12, 63). Emerging evidence suggests that glutaminolysis is critical for the pathogenesis of pulmonary fibrosis (6, 7). Our current study demonstrates that interfering with Gln metabolism sensitizes IPF fibroblasts to undergo apoptosis, a mechanism that is essential for the resolution of fibrosis in the lungs and other organs. This novel mechanism by which Gln metabolism regulates the fibroblast phenotype through epigenetic mechanisms supports the potential for interventions to restrict Gln metabolism in fibrotic diseases such as IPF.
Supplementary Material
Footnotes
Supported by National Institutes of Health grants R01AG050567 (Y.Y.S.), R01HL141195 (J.C.H.), P01HL114470, and R01AG046210, and VA Merit Award I01BX003056 (V.J.T.).
Author Contributions: L.B. and X.T. performed the experiments. L.B., K.B., M.H., J.C.H., V.J.T., and Y.Y.S. analyzed the data. J.C.H. and V.J.T. edited the manuscript. Y.Y.S. designed and coordinated the study, and drafted the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0180OC on August 21, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Kang YP, Lee SB, Lee JM, Kim HM, Hong JY, Lee WJ, et al. Metabolic profiling regarding pathogenesis of idiopathic pulmonary fibrosis. J Proteome Res. 2016;15:1717–1724. doi: 10.1021/acs.jproteome.6b00156. [DOI] [PubMed] [Google Scholar]
- 2.Zhao YD, Yin L, Archer S, Lu C, Zhao G, Yao Y, et al. Metabolic heterogeneity of idiopathic pulmonary fibrosis: a metabolomic study. BMJ Open Respir Res. 2017;4:e000183. doi: 10.1136/bmjresp-2017-000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie N, Tan Z, Banerjee S, Cui H, Ge J, Liu RM, et al. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. Am J Respir Crit Care Med. 2015;192:1462–1474. doi: 10.1164/rccm.201504-0780OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard K, Logsdon NJ, Ravi S, Xie N, Persons BP, Rangarajan S, et al. Metabolic reprogramming is required for myofibroblast contractility and differentiation. J Biol Chem. 2015;290:25427–25438. doi: 10.1074/jbc.M115.646984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong CC, Qian Y, Yu J. Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene. 2017;36:3359–3374. doi: 10.1038/onc.2016.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard K, Logsdon NJ, Benavides GA, Sanders Y, Zhang J, Darley-Usmar VM, et al. Glutaminolysis is required for transforming growth factor-β1-induced myofibroblast differentiation and activation. J Biol Chem. 2018;293:1218–1228. doi: 10.1074/jbc.RA117.000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge J, Cui H, Xie N, Banerjee S, Guo S, Dubey S, et al. Glutaminolysis promotes collagen translation and stability via alpha-ketoglutarate mediated mTOR activation and proline hydroxylation. Am J Respir Cell Mol Biol. 2018;58:378–390. doi: 10.1165/rcmb.2017-0238OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lora J, Alonso FJ, Segura JA, Lobo C, Márquez J, Matés JM. Antisense glutaminase inhibition decreases glutathione antioxidant capacity and increases apoptosis in Ehrlich ascitic tumour cells. Eur J Biochem. 2004;271:4298–4306. doi: 10.1111/j.1432-1033.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Gómez C, Campos-Sandoval JA, Alonso FJ, Segura JA, Manzanares E, Ruiz-Sánchez P, et al. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem J. 2005;386:535–542. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thannickal VJ, Horowitz JC. Evolving concepts of apoptosis in idiopathic pulmonary fibrosis. Proc Am Thorac Soc. 2006;3:350–356. doi: 10.1513/pats.200601-001TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drakopanagiotakis F, Xifteri A, Polychronopoulos V, Bouros D. Apoptosis in lung injury and fibrosis. Eur Respir J. 2008;32:1631–1638. doi: 10.1183/09031936.00176807. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Cui H. Targeting glutamine induces apoptosis: a cancer therapy approach. Int J Mol Sci. 2015;16:22830–22855. doi: 10.3390/ijms160922830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko YG, Kim EY, Kim T, Park H, Park HS, Choi EJ, et al. Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J Biol Chem. 2001;276:6030–6036. doi: 10.1074/jbc.M006189200. [DOI] [PubMed] [Google Scholar]
- 14.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 15.Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JMJC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders YY, Liu H, Zhang X, Hecker L, Bernard K, Desai L, et al. Histone modifications in senescence-associated resistance to apoptosis by oxidative stress. Redox Biol. 2013;1:8–16. doi: 10.1016/j.redox.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders YY, Hagood JS, Liu H, Zhang W, Ambalavanan N, Thannickal VJ. Histone deacetylase inhibition promotes fibroblast apoptosis and ameliorates pulmonary fibrosis in mice. Eur Respir J. 2014;43:1448–1458. doi: 10.1183/09031936.00095113. [DOI] [PubMed] [Google Scholar]
- 20.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39:610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders YY, Kumbla P, Hagood JS. Enhanced myofibroblastic differentiation and survival in Thy-1(-) lung fibroblasts. Am J Respir Cell Mol Biol. 2007;36:226–235. doi: 10.1165/rcmb.2006-0178OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders YY, Liu H, Liu G, Thannickal VJ. Epigenetic mechanisms regulate NADPH oxidase-4 expression in cellular senescence. Free Radic Biol Med. 2015;79:197–205. doi: 10.1016/j.freeradbiomed.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs BC, Bode BP. Stressing out over survival: glutamine as an apoptotic modulator. J Surg Res. 2006;131:26–40. doi: 10.1016/j.jss.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 25.Ajayi IO, Sisson TH, Higgins PD, Booth AJ, Sagana RL, Huang SK, et al. X-linked inhibitor of apoptosis regulates lung fibroblast resistance to Fas-mediated apoptosis. Am J Respir Cell Mol Biol. 2013;49:86–95. doi: 10.1165/rcmb.2012-0224OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sisson TH, Maher TM, Ajayi IO, King JE, Higgins PD, Booth AJ, et al. Increased survivin expression contributes to apoptosis-resistance in IPF fibroblasts. Adv Biosci Biotechnol. 2012;3:657–664. doi: 10.4236/abb.2012.326085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Pastor B, Cosentino C, Mostoslavsky R. A tale of metabolites: the cross-talk between chromatin and energy metabolism. Cancer Discov. 2013;3:497–501. doi: 10.1158/2159-8290.CD-13-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan M, Reid MA, Lowman XH, Kulkarni RP, Tran TQ, Liu X, et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat Cell Biol. 2016;18:1090–1101. doi: 10.1038/ncb3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Vancheri C. Idiopathic pulmonary fibrosis and cancer: do they really look similar? BMC Med. 2015;13:220. doi: 10.1186/s12916-015-0478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horowitz JC, Osterholzer JJ, Marazioti A, Stathopoulos GT. “Scar-cinoma”: viewing the fibrotic lung mesenchymal cell in the context of cancer biology. Eur Respir J. 2016;47:1842–1854. doi: 10.1183/13993003.01201-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papaconstantinou HT, Hwang KO, Rajaraman S, Hellmich MR, Townsend CM, Jr, Ko TC. Glutamine deprivation induces apoptosis in intestinal epithelial cells. Surgery. 1998;124:152–159, discussion 159–160. [PubMed] [Google Scholar]
- 35.Papaconstantinou HT, Chung DH, Zhang W, Ansari NH, Hellmich MR, Townsend CM, Jr, et al. Prevention of mucosal atrophy: role of glutamine and caspases in apoptosis in intestinal epithelial cells. J Gastrointest Surg. 2000;4:416–423. doi: 10.1016/s1091-255x(00)80022-0. [DOI] [PubMed] [Google Scholar]
- 36.Evans ME, Jones DP, Ziegler TR. Glutamine prevents cytokine-induced apoptosis in human colonic epithelial cells. J Nutr. 2003;133:3065–3071. doi: 10.1093/jn/133.10.3065. [DOI] [PubMed] [Google Scholar]
- 37.Paquette JC, Guérin PJ, Gauthier ER. Rapid induction of the intrinsic apoptotic pathway by L-glutamine starvation. J Cell Physiol. 2005;202:912–921. doi: 10.1002/jcp.20194. [DOI] [PubMed] [Google Scholar]
- 38.Fuchs BC, Perez JC, Suetterlin JE, Chaudhry SB, Bode BP. Inducible antisense RNA targeting amino acid transporter ATB0/ASCT2 elicits apoptosis in human hepatoma cells. Am J Physiol Gastrointest Liver Physiol. 2004;286:G467–G478. doi: 10.1152/ajpgi.00344.2003. [DOI] [PubMed] [Google Scholar]
- 39.Sisson TH, Ajayi IO, Subbotina N, Dodi AE, Rodansky ES, Chibucos LN, et al. Inhibition of myocardin-related transcription factor/serum response factor signaling decreases lung fibrosis and promotes mesenchymal cell apoptosis. Am J Pathol. 2015;185:969–986. doi: 10.1016/j.ajpath.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horowitz JC, Ajayi IO, Kulasekaran P, Rogers DS, White JB, Townsend SK, et al. Survivin expression induced by endothelin-1 promotes myofibroblast resistance to apoptosis. Int J Biochem Cell Biol. 2012;44:158–169. doi: 10.1016/j.biocel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashley SL, Sisson TH, Wheaton AK, Kim KK, Wilke CA, Ajayi IO, et al. Targeting inhibitor of apoptosis proteins protects from bleomycin-induced lung fibrosis. Am J Respir Cell Mol Biol. 2016;54:482–492. doi: 10.1165/rcmb.2015-0148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaelin WG., Jr Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335–345. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinnaird A, Zhao S, Wellen KE, Michelakis ED. Metabolic control of epigenetics in cancer. Nat Rev Cancer. 2016;16:694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 44.Lempradl A, Pospisilik JA, Penninger JM. Exploring the emerging complexity in transcriptional regulation of energy homeostasis. Nat Rev Genet. 2015;16:665–681. doi: 10.1038/nrg3941. [DOI] [PubMed] [Google Scholar]
- 45.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 46.Pang B, Zheng XR, Tian JX, Gao TH, Gu GY, Zhang R, et al. EZH2 promotes metabolic reprogramming in glioblastomas through epigenetic repression of EAF2-HIF1α signaling. Oncotarget. 2016;7:45134–45143. doi: 10.18632/oncotarget.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michalak KP, Mackowska-Kedziora A, Sobolewski B, Wozniak P. Key roles of glutamine pathways in reprogramming the cancer metabolism. Oxid Med Cell Longev. 2015;2015:964321. doi: 10.1155/2015/964321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 49.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wissmann M, Yin N, Müller JM, Greschik H, Fodor BD, Jenuwein T, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W, et al. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 2012;26:1364–1375. doi: 10.1101/gad.186056.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q, et al. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell. 2013;152:1037–1050. doi: 10.1016/j.cell.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boidot R, Végran F, Lizard-Nacol S. Transcriptional regulation of the survivin gene. Mol Biol Rep. 2014;41:233–240. doi: 10.1007/s11033-013-2856-0. [DOI] [PubMed] [Google Scholar]
- 55.Hattori M, Sakamoto H, Satoh K, Yamamoto T. DNA demethylase is expressed in ovarian cancers and the expression correlates with demethylation of CpG sites in the promoter region of c-erbB-2 and survivin genes. Cancer Lett. 2001;169:155–164. doi: 10.1016/s0304-3835(01)00499-2. [DOI] [PubMed] [Google Scholar]
- 56.Acquati S, Greco A, Licastro D, Bhagat H, Ceric D, Rossini Z, et al. Epigenetic regulation of survivin by Bmi1 is cell type specific during corticogenesis and in gliomas. Stem Cells. 2013;31:190–202. doi: 10.1002/stem.1274. [DOI] [PubMed] [Google Scholar]
- 57.van der Knaap JA, Verrijzer CP. Undercover: gene control by metabolites and metabolic enzymes. Genes Dev. 2016;30:2345–2369. doi: 10.1101/gad.289140.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Estève PO, Chin HG, Pradhan S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. J Biol Chem. 2007;282:2615–2625. doi: 10.1074/jbc.M606203200. [DOI] [PubMed] [Google Scholar]
- 59.Dan HC, Jiang K, Coppola D, Hamilton A, Nicosia SV, Sebti SM, et al. Phosphatidylinositol-3-OH kinase/AKT and survivin pathways as critical targets for geranylgeranyltransferase I inhibitor-induced apoptosis. Oncogene. 2004;23:706–715. doi: 10.1038/sj.onc.1207171. [DOI] [PubMed] [Google Scholar]
- 60.Sethi G, Ahn KS, Xia D, Kurie JM, Aggarwal BB. Targeted deletion of MKK4 gene potentiates TNF-induced apoptosis through the down-regulation of NF-kappa B activation and NF-kappa B-regulated antiapoptotic gene products. J Immunol. 2007;179:1926–1933. doi: 10.4049/jimmunol.179.3.1926. [DOI] [PubMed] [Google Scholar]
- 61.Hou YC, Chiu WC, Yeh CL, Yeh SL. Glutamine modulates lipopolysaccharide-induced activation of NF-κB via the Akt/mTOR pathway in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2012;302:L174–L183. doi: 10.1152/ajplung.00066.2011. [DOI] [PubMed] [Google Scholar]
- 62.Chen YQ, Zhao CL, Li W. Effect of hypoxia-inducible factor-1alpha on transcription of survivin in non-small cell lung cancer. J Exp Clin Cancer Res. 2009;28:29. doi: 10.1186/1756-9966-28-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol. 2012;23:362–369. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.